Significance
Hyperpolarization-activated cyclic nucleotide-gated (HCN) channels are essential for the rhythmic activity of pacemaker cells in the heart and brain. How the movement of the voltage sensor, which is triggered by hyperpolarization of the membrane voltage, causes HCN channels to open is not clear. Here, we found that in HCN channels, the voltage sensor moves in two steps in response to hyperpolarizations and that the second voltage sensor step correlates with gate opening. A better understanding of HCN channel gating will aid in the development of better antiarrhythmic drugs.
Keywords: molecular models, S4-to-gate coupling, spHCN, S4 movement, voltage clamp fluorometry
Abstract
Rhythmic activity in pacemaker cells, as in the sino-atrial node in the heart, depends on the activation of hyperpolarization-activated cyclic nucleotide-gated (HCN) channels. As in depolarization-activated K+ channels, the fourth transmembrane segment S4 functions as the voltage sensor in hyperpolarization-activated HCN channels. But how the inward movement of S4 in HCN channels at hyperpolarized voltages couples to channel opening is not understood. Using voltage clamp fluorometry, we found here that S4 in HCN channels moves in two steps in response to hyperpolarizations and that the second S4 step correlates with gate opening. We found a mutation in sea urchin HCN channels that separate the two S4 steps in voltage dependence. The E356A mutation in S4 shifts the main S4 movement to positive voltages, but channel opening remains at negative voltages. In addition, E356A reveals a second S4 movement at negative voltages that correlates with gate opening. Cysteine accessibility and molecular models suggest that the second S4 movement opens up an intracellular crevice between S4 and S5 that would allow radial movement of the intracellular ends of S5 and S6 to open HCN channels.
Hyperpolarization-activated cyclic nucleotide-gated (HCN) channels are important for generating pacemaker activity in cardiomyocytes and neurons (1, 2). The rhythmic and autonomous activity of these cells is triggered by the activation of a depolarizing Na+ current through HCN channels at hyperpolarized voltages after an action potential that drives the membrane potential back to the threshold for firing another action potential. Studies in patients and animal models have found that impaired function of HCN channels is associated with diseases like neuropathic pain (3, 4), epilepsies (5, 6), and cardiac arrhythmias (7–9). HCN channels belong to the same superfamily as voltage-gated K+ (Kv) channels. But, in contrast to most Kv channels, which are opened by depolarization, HCN channels are opened by hyperpolarization of the cell membrane (10–12). In addition to voltage, HCN channels are further opened by cyclic nucleotides that bind to a cyclic nucleotide-binding domain in the cytoplasmic C-terminal region (13–15). Both HCN and Kv channels are tetramers and each of their subunits has six transmembrane segments, where segments S1 through S4 form the voltage-sensing domain (VSD) and segments S5 through S6 form the pore domain (PD) (14, 16). Similar to Kv channels, the gate of HCN channels is formed by the most C-terminal region of the four S6 helices (14, 17–19) and their pore selectivity filter contains the typical GYG of the potassium signature sequence, but not the complete TVGYG signature sequence, which might explain why HCN channels are only slightly more selective for K+ than for Na+ (1, 14, 20). In addition, both HCN and Kv channels sense changes in the membrane potential through the positively charged S4 segment that moves across the membrane in response to changes in the membrane potential (21–24). However, in HCN channels the gate opens when S4 moves inward at hyperpolarized voltages, whereas Kv channels open when S4 moves outwards at depolarized voltages (22). Despite many studies into the voltage-gating mechanism of HCN channels (22, 23, 25–28), it is still not completely understood how inward S4 movement in response to a hyperpolarization opens HCN channels. We here study S4 movement in channels carrying mutations at the interface between the VSD and the PD to understand how hyperpolarization opens HCN channels.
In contrast to most Kv channels, HCN channels have a nondomain swapped architecture, which means that the VSD interacts with the PD from the same subunit (14, 16). In domain-swapped Kv channels (Kv1 to -7), in which the VSD is next to the PD from the neighboring subunit, it is assumed that the long S4-S5 linker transduces the S4 movement to the opening of the gate by pulling on the lower S6 (16, 29). In rEAG (Kv10.1), a nondomain swapped depolarization-activated Kv channel, and spHCN (Strongylocentrotus purpuratus HCN, sea urchin HCN), a nondomain swapped hyperpolarization-activated HCN channel, cutting the short S4-S5 linker does not prevent gating (25, 30), which shows that the voltage-gating mechanism in these nondomain-swapped channels does not require a covalent link between the S4 and S5 segments. Instead, several studies have suggested that noncovalent interactions between S4 and S5 are required for the opening of HCN channels by hyperpolarization (25, 26, 28, 31, 32). In addition, the S4 segment of HCN channels is two helical turns longer than the S4 of depolarization-activated Kv channels. Recently, three different groups showed that in HCN channels S4 moves inward by around 10 Å in response to hyperpolarization, and that there is a kink in the most C-terminal part of S4 when S4 moves inwards (27, 33, 34). But, how the inward movement of voltage sensor S4 causes channel opening is not clear from these studies.
At the S4-S5 interface of HCN channels, there are several residues that are absolutely conserved in HCN channels but that are different in depolarization-activated Kv channels. We mutated these conserved residues, one at a time, in the sea urchin HCN channel (spHCN) and used voltage clamp fluorometry (VCF) (21) to test their role in HCN gating. The VCF technique allows us to simultaneously detect gate movement, by recording the currents through the channels, and S4 movement, by recording the fluorescence emitted by fluorophores attached to the extracellular end of S4. We found that S4 moves in two steps and that the second S4 movement correlates with channel opening. We also show that the second S4 movement changes the intracellular accessibility of residues L340C in S4 and N370C in S5, as if an internal crevice opens between the voltage sensors and the pore, thereby allowing the intracellular pore to expand and open the channel.
Results
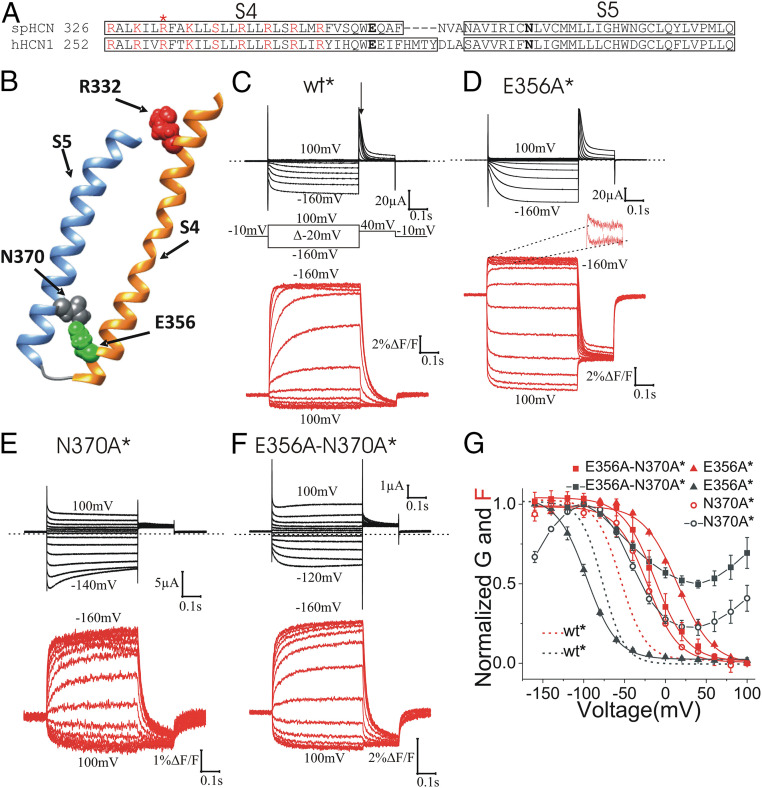
E356A and N370A Mutations Destabilize the Outward State of S4.
Among the conserved residues at the interface between S4 and S5 of the HCN channel family, a glutamate in the C-terminal S4 (E356 in spHCN) and an asparagine in the N-terminal S5 (N370 in spHCN) (Fig. 1A) seem to interact in the HCN1 cryoelectron microscopy (cryo-EM) structure with the S4 outward and closed gate (14) (Fig. 1B). To test the role of those residues in gating, we mutated E356 and N370 to alanine, respectively, to determine whether or not their interaction is important for S4 movement or S4-to-gate coupling. Since glutamate and asparagine are polar residues able to form hydrogen bonds, and considering the distance between these residues in the S4 outward cryo-EM structure (14), we hypothesized that the interaction between E356 and N370 in the S4 outward state is a hydrogen bond. We used VCF to measure S4 movement by the fluorescence emitted from fluorophores attached to S4 and gate opening by the current through the channel (21). We first introduced the mutation R332C in S4 in the E356A and N370A single mutants and in the E356A-N370A double mutant, and then labeled these channels with the fluorophore Alexa-488 (we indicate channels with Alexa-488 attached to R332C with an *). We have previously shown that the fluorescence from Alexa-488–labeled R332C (wt*) channels is a good reporter for S4 movement in spHCN channels (21) (Fig. 1C). The E356A, N370A, and E356A-N370A mutations shifted the fluorescence versus voltage, F(V), relationship in a positive direction along the voltage axis (Fig. 1 D–G and SI Appendix, Table S1) compared with wt*, as if a hydrogen bond between E356 and N370 stabilizes S4 in its outward position in wt* channels. The effects of E356A and N370A on the voltage dependence of the fluorescence were not additive (SI Appendix, Table S1). A thermodynamic mutant cycle analysis showed that the coupling energy between E356 and N370 is ∼2.06 kcal/mol [a coupling energy > 0.89 kcal/mol has been suggested to be a significant interaction (35)], suggesting a significant interaction between E356 and N370 that stabilize S4 in the outward position.
Fig. 1.
E356 and N370 stabilize S4 in the outward state. (A) Amino acid sequence alignment of S4 and S5 segments of spHCN and hHCN1 channels. Box indicates the extend of the α-helical regions of S4 and S5. The asterisk (*) indicates R332, which was mutated to cysteine for fluorescence labeling. Positively charged residues in S4 are marked in red. E356 and N370 are marked in bold. (B) spHCN homology model showing the interaction of E356 and N370 in closed channels with S4 outward. (C–F) Representative current (black) and fluorescence (red) from (C) wt*, (D) E356A* (second component zoomed out in Inset), (E) N370A*, and (F) E356A-N370A* channels. Arrow in C marks where tail currents were measured for G(V)s. (G) Normalized G(V) and F(V) curves of wt*, E356A*, N370A*, and E356A-N370A* channels. Mean ± SEM. In a previous publication we showed that a polar residue is needed at position 370 to close completely HCN channels at positive voltages (28).
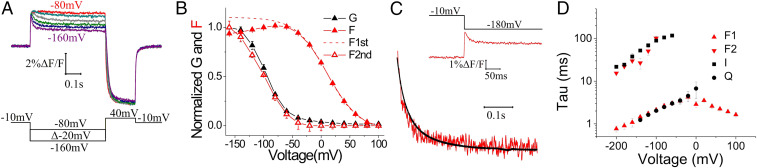
A Second S4 Movement Is Revealed by E356A Mutation.
Surprisingly, in contrast to the F(V) of E356A*, the conductance versus voltage, G(V), relationship of E356A* was shifted slightly to more negative voltages (Fig. 1G). At +80 mV, all the E356A* channels are closed and the fluorescence signal is at a minimum, suggesting that all S4s are in their resting outward state. Around −40 mV, the F(V) of the E356A* channels reaches its maximum value, suggesting that all S4s are in their activated inward state. However, at this voltage, the conductance of E356A* channels is still close to zero, suggesting that the initial S4 movement is not sufficient to open the gate. We noticed, however, that a second fluorescence component occurs in a similar voltage range as gate opening (Figs. 1 D, Inset, and 2 A and B, and SI Appendix, Table S1). In addition, the time courses of the second fluorescence component and the current overlapped (Fig. 2 C and D), as if channel opening is favored by the second S4 movement. At extreme voltages, the fluorescence from wt* and E356A* looks similar (SI Appendix, Fig. S1), as if similar S4 movements occur in both of these channels. We also measured gating currents from E356A* channels by introducing the P435Y mutation that makes spHCN channels nonconductive (SI Appendix, Fig. S2). The kinetics of the gating currents overlayed with the kinetics of the first fluorescence component (Fig. 2D), as if this fluorescence component tracks the gating charge movement. The second S4 movement is most likely too slow, as suggested by the slow kinetics of the second fluorescence component (Fig. 2D), to be measured as gating currents.
Fig. 2.
E356A channels display a second fluorescence component that correlates with opening. (A) Representative traces of the second component of the E356A* fluorescence stepping to voltages between −60 mV and −160 mV from a holding potential of −10 mV. (B) Normalized G(V), F(V), and the second component of the F(V) curves of E356A* channels. The F(V) of the first component was estimated by fitting a single Boltzmann curve to the F(V) at voltages for which only the first component was observed and then extrapolating this curve to more negative voltages (dashed line). Then, the F(V) of the second component (open triangles) was calculated as the difference between the estimated first component of the F(V) (dashed line) and the measured F(V) (solid triangles). (C) Overlay of current (black) and second component (i.e., the decaying component) of E356A* fluorescence (red) in response to a voltage step to −180 mV (Inset shows complete fluorescence trace). (D) Exponential time constant of the ionic currents (I) and the first (F1) and second (F2) fluorescence component for E356A* and the gating currents (Q) for E356A-P435Y*. Mean ± SEM (n = 4).
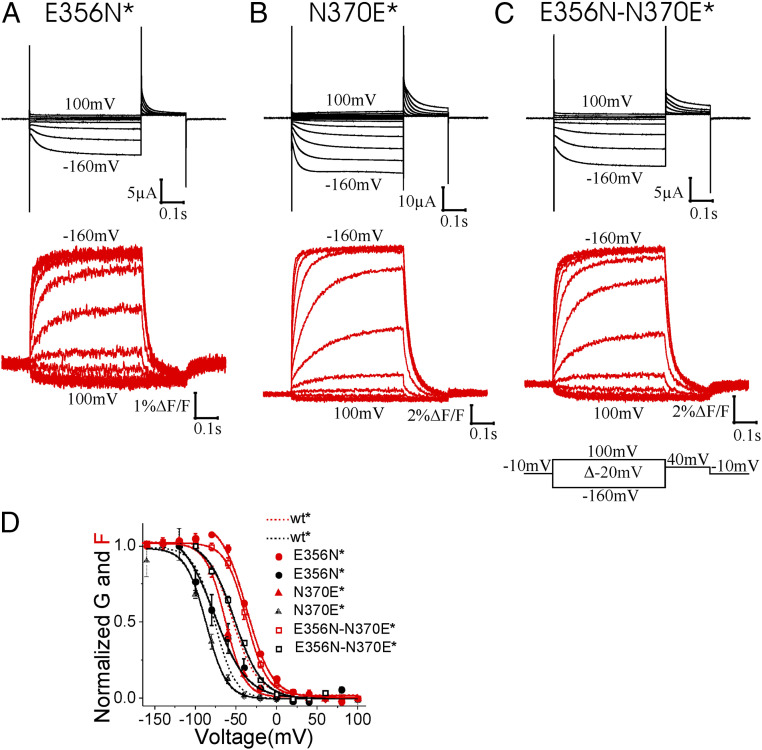
A Polar Interaction Between 356 and 370 Is Required for Normal Gating.
To further study the role of the E356-N370 interaction in stabilizing S4 in the outward state, we mutated E356 and N370 while retaining the hydrogen bond-forming capabilities at those positions. Mutations E356N, E356Q, N370E, N370D, and E356N-N370E—all of which retain the hydrogen bond-forming capabilities between residues 356 and 370—induced only small shifts in the F(V) relationship compared with wt* channels (Fig. 3 and SI Appendix, Fig. S3 and Table S1), supporting our hypothesis that a hydrogen bond between these two residues stabilizes S4 in its outward position.
Fig. 3.
Channels with hydrogen-bonding residues at E356 and N370 gate normally. Representative traces of the current (black) and fluorescence (red) from (A) E356N*, (B) N370E*, and (C) E356N-N370E* channels. (D) Normalized G(V) and F(V) curves of wt*, E356N*, N370E*, and E356N-N370E* channels. Mean ± SEM.
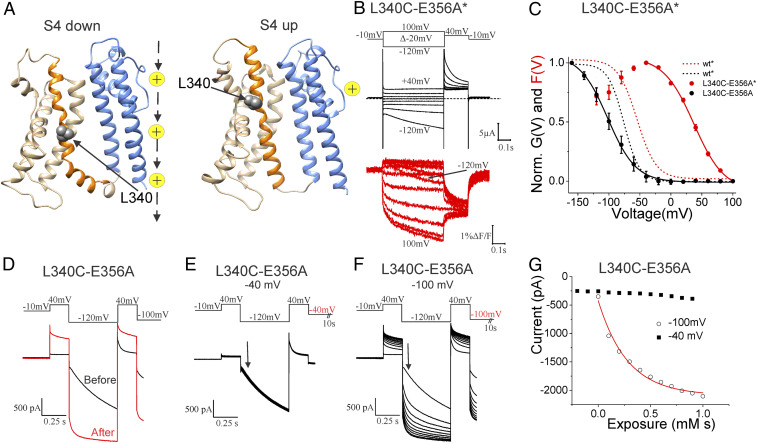
L340C-E356A Intracellular Accessibility Changes in the Voltage Range of the Second S4 Movement.
To determine what type of conformational changes the two fluorescence components are reporting on in HCN channels, we measured the intracellular accessibility of a cysteine introduced in S4 at different voltages. We and others have previously shown that the accessibility of cysteines introduced in S4 of HCN channels changes at both the intracellular and extracellular ends of S4 between the closed and open states of HCN channels (22, 23, 36). To test S4 conformational changes at different voltages, we selected residue L340 after analyzing molecular models of the spHCN channel in the S4 outward/closed state and in the S4 inward/open state (28) (Fig. 4A).
Fig. 4.
Intracellular accessibility of S4 increases in voltage range of the second S4 movement. (A) spHCN molecular models in the S4 inward/open gate (Left) and S4 outward/closed gate (Right) states (28). Only the transmembrane region of one channel subunit is shown for clarity. S1–S3 (light gray), S4 (orange), L340 (gray space-filled), and S5-S6 (blue). Yellow ions to indicate location of ion conducting pore. (B) Representative traces of the current (black) and fluorescence (red) from L340C-E356A* channels. (C) Normalized G(V) and F(V) curves of wt* and L340C-E356A* channels. Mean ± SEM. G(V) and the first component of F(V) were fitted by a single Boltzmann function. (D) Currents before (black) and after (red) application of 10 μM MTSET for 100 s to the cytosolic side of L340C-E356A channels while voltage clamped at −100 mV. (Upper) Voltage protocol. (E and F) Currents during the application of MTSET to L340C-E356A channels held mainly (10 s of 11.5 s) at (E) −40 mV or (F) −100 mV. Currents from E and F were measured from the same patch. (Upper) Voltage protocol. The step at the end of the protocol lasted 10 s. (G) Currents measured at the arrow in E and F from L340C-E356A channels during MTSET application at −40 mV (closed squares, n = 3) and −100 mV (open circles, n = 3: fitted with an exponential function).
We tested the intracellular accessibility of L340C to the membrane impermeable thiol reactive 2-(trimethylammounium)ethyl methanethiosulfonate (MTSET) reagent and determined whether a change in the accessibility of the introduced cysteine occurs in the voltage range for the first fluorescence component or in the voltage range for the second fluorescence component and the G(V) in the background of the E356A mutation (Fig. 4 B and C). We have previously shown that L340C is intracellularly accessible when L340C channels are open at hyperpolarized voltages and that the intracellular accessibility decreases drastically when L340C channels are closed at depolarized voltages (22). As for L340C channels (22), modification by MTSET turns L340C-E356A channels into almost completely voltage-independent open channels (Fig. 4D). We here found that L340C is intracellularly accessible at −100 mV to MTSET applied on the cytosolic side of L340C-E356A channels and that the intracellular accessibility decreases drastically already at −40 mV (Fig. 4 E–G).
The modification rates of L340C-E356A was 4,082 ± 1,347 M−1s−1 (n = 3) at −100 mV and decreased to <136 ± 38 M−1s−1 (n = 3) at −40 mV (Fig. 4G). These rates are similar to the rates we found earlier for MTSET modification of L340C in open (at −100 mV: 2,400 ± 1,100 M−1s−1) and closed (at +50mV: <100 ± 50 M−1s−1) L340C channels, respectively (22). That the MTSET-modification rate changes drastically between −100 mV and −40 mV, the voltage range for the second fluorescence component (Fig. 4C), shows that the conformational change during the first fluorescence component is not enough to expose L340C. An additional conformational change during the second fluorescence component is necessary to expose L340C to the intracellular solution. Similarly, we found that the modification rate of N370C in S5 in E356A-N370C channels was much faster at −100 mV than at −40 mV (SI Appendix, Fig. S4). The intracellular accessibility changes of L340C and N370C in the background of the E356A mutant support a model with three positions of S4: 1) at +80 mV an outward position in which L340C and N370C are intracellularly inaccessible; 2) at −40 mV an intermediate position, at which L340C and N370C are still mainly inaccessible to the intracellular solution; and 3) at −100 mV an inward position in which L340C and N370C are accessible to the intracellular solution.
Discussion
Despite the progress that has been made on elucidating the voltage-gating mechanism of HCN channels (22, 25, 26, 28, 37), it is still not clear how inward S4 movement opens HCN channels in response to hyperpolarizations. It has been suggested that the S4-S5 linker is not required for gating in HCN channels (25) and that instead, noncovalent interactions at the interface between S4 and S5 are critical for gating in HCN channels (25, 26, 28). We here tested whether two conserved residues in the HCN family, E356 and N370, that are at the interface between S4 and S5 in the cryo-EM hHCN1 structure, play an important part in S4 movement and the coupling between S4 movement and the gate. We first showed that an interaction between the E356 in lower S4 and N370 in lower S5 stabilizes S4 in its outward position. Mutations of either E356 or N370 shift the voltage dependence of S4 movement to more positive voltages, as if E356 and N370 stabilize the S4 outward state by forming a hydrogen bond. Surprisingly, even if S4 movement in E356A* channels is shifted by around +70 mV compared with wt* channels, channel opening is shifted to more negative voltages than wt* channels. This shows that in the E356A* channel the gate is still closed even when all the S4s have moved inward, suggesting that the main S4 charge movement—which we interpret as the first component of the fluorescence signal—is not sufficient for gate opening. A previous study showed that modification by extracellular MTSET of two cysteines inserted in the most extracellular part of S4 in hHCN1, homologous to V331C and T333C in spHCN, inhibited channel opening (36), as if S4 immobilization in the outward state by extracellular MTSET impedes gate opening. Therefore, considering the data we present here and previous experimental results, we suggest that the main S4 movement is required but not sufficient to open the gate. Instead, our data here, from VCF and MTSET accessibility studies, show that there is a second conformational change in and around S4 that correlates with channel opening.
We, therefore, propose that S4 moves in two steps and that channel opening is favored by the second S4 movement (Fig. 5). This second S4 movement is most likely also present in wt* HCN channels, but the overlapping voltage dependence of the first and second S4 movements in wt* HCN channels makes it hard to quantify the second S4 movement. The overlapping voltage dependence of the two S4 movements makes the second fluorescence component to show up as an increase in fluorescence at less negative voltages, as no increase at intermediate negative voltages, and as a decrease in fluorescence at extreme negative voltages (SI Appendix, Figs. S1, S5, and S6). The second component is evident in wild-type and mutant channel recordings, but it is more clearly quantified in the E356A mutation that separates the two S4 movements to a larger degree. A simple kinetic model of wt* and E356A* HCN channels with two S4 movements is consistent with our data (SI Appendix, Figs. S5 and S6). The E356A* channel is reminiscent of the ILT mutation in Shaker K+ channels (38) and wild-type cardiac KCNQ1/KCNE1 channels (39), with an initial S4 movement that does not directly open the gate and a second S4 movement that opens the gate.
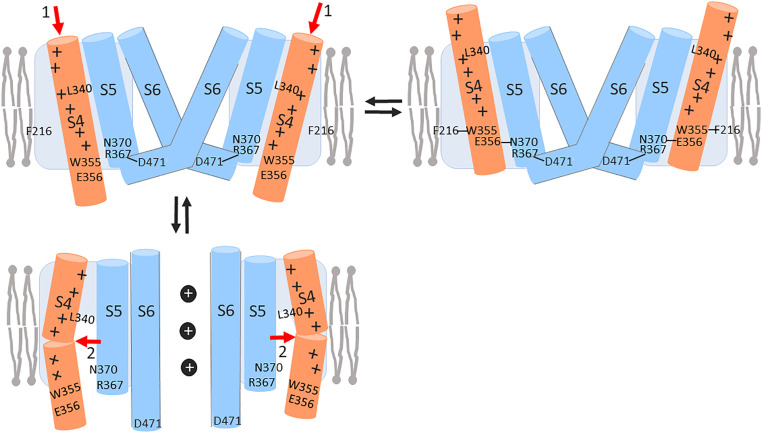
Fig. 5.
Two-step S4 movement opens HCN channels. (Upper Right) In the closed state with S4 outward, there is a E356-N370 hydrogen bond, which stabilizes S4 outward. We and others have previously shown that F216-W355 and R367-D471 interactions stabilize a closed gate (28). (Upper Left) Closed state with S4 intermediate. Upon hyperpolarization S4 moves inward to the intermediate state, here shown as an initial inward S4 movement (red 1 arrows). The inward S4 movement breaks the E356-N370 hydrogen bond. (Lower Left) Open state with S4 inward. The initial inward S4 movement is followed by a subsequent tilting S4 movement (red 2 arrows), which separates S4 from S5, opening a hydrophilic crevice around both segments and allowing the gate to open at hyperpolarized voltages. The opening of the crevice during the second S4 movement exposes L340 to the intracellular solution. We have previously shown that a hydrophilic residue at N370 stabilizes an open gate (28).
So, what are the molecular rearrangements during these two conformational changes that lead to channel opening in HCN channels? We created molecular models of the spHCN channel in the S4 outward/closed state, the S4 intermediate/closed state, and the S4 inward/open state by using the existing hHCN1 cryo-EM structures and the open hERG cryo-EM structure (14, 27, 40) (SI Appendix, Fig. S7). EAG and ERG channels are the closest relatives in sequence and structure to HCN channels and the hERG channel was solved in an open state. We therefore use the hERG channel pore as a model for the open HCN channels. The models of the three states were stable in long molecular dynamics (MD) simulations, similar to other cryo-EM structures of related channels in MD simulations (SI Appendix, Fig. S8). In our models, the first S4 movement is like a classic helical screw motion that vertically translates charges across the membrane and across the electrical field (Movie S1). The second S4 movement does not significantly translate charges vertically, but an opening of the aqueous crevice alters the electrical field across the protein and alters the voltage sensed at the individual S4 charges (Movie S1). Therefore, both S4 movements will generate movement of gating charge and voltage is the main driver for both conformational changes. Gating charge estimates from the three molecular models are consistent with the amount of gating charge in our kinetic model to explain the S4 movement and channel gating (SI Appendix, Fig. S9). The breaking of the hydrogen bond between E356 and N370 is part of the energetics that allows S4 to move inward to the intermediate state and thereby contributes to determining at what voltage range the first S4 movement occurs. The opening of the intracellular aqueous crevice around S4 and S5 and the hydration of residues like N370 is part of the energetics of the second S4 movement, and therefore contributes to determining at what voltage range the second S4 movement and subsequent channel opening occurs.
This second S4 movement is consistent with recent publications that propose a model in which the C-terminal part of S4 tilts after the main inward S4 movement has occurred in HCN channels (27, 33, 34). In the S4 inward state, the second S4 movement, possibly produced by tilting of the C-terminal S4, spatially separates S4 and S5, opening a hydrophilic crevice between the S4 and S5 segments. Based on a gating model that we recently proposed (28), the formation of this crevice allows S5 to radially translate and S6 to slightly rotate, thereby opening the pore. Previous cysteine accessibility studies of HCN channels have shown that in response to changes in membrane potential there are more residues that change their accessibility on the intracellular end of S4 compared with the extracellular end of S4 (36). Bell et al. (36) suggested that this was due to the opening of a hydrophilic crevice around the intracellular end of S4 in response to hyperpolarizations. The Chanda laboratory found that the intracellular access of F186C in S2 increased in response to hyperpolarizations in support of a crevice opening up around the intracellular end of S4 (33). The accessibility data of L340C in S4 and N370C in S5 presented here is consistent with that the second S4 movement in HCN channels creates a hydrophilic crevice between S4 and S5 that leads to channel opening. MTSET accessibility in our three molecular model states is consistent with the cysteine accessibility data from this and other studies (22, 33, 36).
In summary, our experiments reveal a second S4 movement and the opening of an intracellular crevice around S4 that correlates with channel opening, as if HCN channel opening at hyperpolarized voltages are favored by these conformational changes.
Methods
Molecular Biology.
Point mutations were performed in the gene encoding the spHCN channel using site-directed mutagenesis (QuikChange Stratagene). The expression plasmid for the spHCN channel gene was pGEM-HE. All the mutant DNAs were checked for correct sequences (Genewiz). The DNA was linearized with NheI and cRNA was obtained using the T7 mMessage mMachine transcription kit (Ambion). RNA concentrations were measured using UV spectroscopy and were between 1 µg/µL and 5 µg/µL.
Expression System.
The R332C mutation was introduced in all the mutant channels, unless otherwise noted, to label the channels with Alexa-488 maleimide for fluorescence measurements. For simplicity, we identified the mutant channels only by the mutation under study (omitting R332C) and an asterisk that represents S4 labeling at R332C. cRNA at 1 to 5 µg/µL was injected into the cytoplasm of stage IV and V Xenopus laevis oocytes. The oocytes were incubated for 2 to 3 d at 10 °C for membrane expression of the channels.
VCF.
The cells expressing the channels were labeled in a bath solution with 100 µM Alexa-488 C5-maleimide (Molecular Probes) and 2.5 mM Na-pyruvate for 30 min at 8 °C. After washing, oocytes were placed with the animal pole “up” in a bath housed on a Leica DMLFS upright fluorescence microscope. Light was focused on the animal pole of the oocyte through a 20× objective (NA 1.0) and was passed through a filter cube from Chroma 41026 (HQ495/×30, Q515LP, HQ545/50 m). Whole-cell ionic currents were measured with the two-electrode voltage clamp technique using an Axon Geneclamp 500B amplifier (Axon Instruments). Microelectrodes were pulled using borosilicate glass, filled with a 3 M KCl solution, and had resistances between 0.5 and 0.9 MΩ. All experiments were performed at room temperature, but after labeling the oocytes were kept on ice to prevent internalization of labeled channels. Data were filtered at 1 kHz, digitized at 5 kHz (Axon Digidata 1322 A), and monitored and collected using pClamp software (Axon Instruments). Fluorescence signals were low-pass Bessell-filtered (Frequency Devices) at 200 Hz and digitized at 1 kHz. The bath solution (pH 7.5) contained in: 96 mM NaCl, 2 mM KCl, 1.8 mM MgCl2, and 5 mM Hepes. In addition, 100 µM LaCl3 was added to block oocytes endogenous currents. HCN currents were isolated by subtracting the currents in the presence of 1 mM of the HCN channel blocker ZD7288 (Tocris Bioscience).
Thermodynamic Mutant Cycle Analysis.
The F(V) curve of the spHCN channel was used to evaluate the change in the free-energy difference between the closed state and the open state caused by the mutation. Free energy was calculated as , where z and V1/2 were obtained from the G(V) and F is the Faraday’s number. The perturbation in free energy by a mutation relative to the wild-type was calculated as . The coupling energy between each pair of residues was calculated as
| [1] |
If the two residues are independent of each other, the sum of the free-energy changes of the two single mutations will be equal to the free-energy change of its corresponding double mutation. Conversely, if the two residues are interacting with each other, the sum of the free-energy changes of the two single mutations will be different from the free-energy change of the double mutation. A coupling energy greater than 0.89 kcal/mol (35, 41) is considered to be significant.
Cysteine Accessibility.
For intracellular accessibility, currents were measured in the inside-out configuration using an Axon 200B amplifier and pClamp 9.0 (Axon Instruments). Patches were recorded with 1 to 2 MΩ pipettes containing: 89 mM KCl, 15 mM Hepes, 0.4 mM CaCl2, 0.8 mM MgCl2 (pH = 7.4 adjusted with KOH). The intracellular solution contained in: 100 mM KCl, 10 mM Hepes, 1 mM EGTA, 0.1 mM CaCl2, and 0.5 mM MgCl2, and 100 µM cAMP (pH 7.1 adjusted with KOH). Next, 10 µM MTSET diluted in the internal solution at the moment of the experiment was continuously perfused to the patches using a rapid perfusion system (Biologic Science Instruments RSC-160). MTSET was stored as aliquots of 5-mM stock solution at −20 °C and an aliquot was thawed and kept on ice during each day of experiments.
Gating Currents.
Gating currents were measured with the cut-open oocyte technique. Currents were filtered at 5 kHz and digitized at 20 kHz. External solution: 96 mM NMDG-Mes, 2.3 mM MgCl2, 5 mM Hepes, pH = 7.4. Internal solution: 96 mM NMDG-Mes, 2.3 mM MgCl2, 5 mM Hepes, 1 mM EGTA, pH = 7.1. Capacitance and leak currents were subtracted off-line from voltage steps between +80 and +100 mV.
Kinetic Modeling.
The fluorescence and currents for the 12-state model of the spHCN channel were simulated using the Berkeley Madonna program. We made a global fitting of both current and fluorescence for multiple voltage steps (−40 mV to +160 mV) using the Madonna Berkeley fitting module. Since the two components are not easily distinguishable in wild-type channels, we kept the z values and ΔF values obtained for E356A as fixed parameters in fitting the wild-type data.
Molecular Modeling.
The homology model of the closed spHCN channel with S4 outward was created using the Swiss-model server (https://swissmodel.expasy.org/) with the closed hHCN1 with S4 outward cryo-EM structure (14) as template. The model of the open spHCN channel with S4 inward was created using the closed hHCN1 with S4 inward cryo-EM structure as template for the VSD and the open hERG with S4 outward cryo-EM structure (40) as template for the pore. The homology model of the closed spHCN channel with S4 in the intermediate state was created using the closed hHCN1 with S4 inward cryo-EM structure as template but straighten the S4 and replacing S1 to S3 with those of the closed hHCN1 with S4 outward cryo-EM structure (14). Morph videos from S4 outward/closed to S4 intermediate/closed, and from S4 intermediate/closed to S4 inward/open states were created in University of California San Francisco Chimera 1.13rc software. Those videos were merged using Adobe Premiere Pro-2020 (Adobe) to obtain the molecular model shown in Movie S1 that shows the predicted closed-to-open transition in the spHCN channel.
All-Atom MD Simulations.
Refined homology models of HCN channels in three different conformational states were selected for all-atom MD simulations. The protein–membrane systems were built and solvated in 150 mM of KCl using CHARMM-GUI membrane builder protocol (steps 1 to 7) with POPC bilayer (42). The solvated proteins were simulated then for 150 ns in the presence of weak harmonic restraints acting on heavy protein atoms with the NAMD2.13 software package (43, 44). The CHARMM36M and updated CHARMM36 force-fields were used for protein and lipids, respectively (45, 46). The water was modeled with TIP3P potential (47) and ion parameters included NBFIX corrections. Production runs were performed for 1.9 µs for all three systems. The production runs were executing in a semi-isotropic (NPaT) ensemble at temperature 315 K maintained by the Nosé–Hoover thermostat (48). The time step for the production runs was set to 2 fs and the trajectories were saved every 240 ps. Nonbonded and long-range electrostatic interactions were evaluated every 2 and 6 fs, respectively. Long-range electrostatics were calculated using the k-Gaussian Ewald method implemented to enhance performance on ANTON2 platform (49) with a 64 × 64 × 64 grid. SHAKE was used to constrain all bonds involving hydrogen atoms.
Gating Charge Calculations.
The gating charge was calculated from an ensemble of structures (1,700 for each state). The gating charges are computed using the Poisson–Boltzmann voltage equation, as previously described (50–52). Briefly, each of the frames contained only a protein/lipid system with lipid charges set to zero. The optimized protein/lipid radius was used to solve Poisson–Boltzmann equation with an orthorhombic grid covering an entire system. The grid resolution was set to 0.4 Å. Both protein and lipid atoms were assigned a dielectric constant of 2 and water was assigned a dielectric value of 80. The ionic concentration was set to 150 mM and a focusing method was used to obtain converged electrostatic potential for each of the frame with a coarse-grain grid resolution of 1.5 Å. Poisson–Boltzmann calculations were performed with the PBEQ module of CHARMM42b1 program.
Statistics and Data Analysis.
Average values are shown as mean ± SEM. The G(V) and F(V) curves were fitted to a Boltzmann equation using the Origin program (Origin 9.0.0, OriginLab Corporation). The Boltzmann equation: G(V) = Amin + (Amax − Amin)/(1 + exp((V − V1/2)/K)), where Amax and Amin are the maximum and minimum, respectively, V1/2 is the voltage where 50% of the maximal conductance level is reached, and K is the slope factor. Parameters of the fits and n are shown in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
This project was supported by NIH Grant R01GM139164 (to H.P.L.). Partial support was provided by Natural Sciences and Engineering Research Council of Canada Grant RGPIN-2021-02439 (to S.Y.N.). Molecular dynamics simulations and analysis were performed on the Canada Foundation for Innovation supported by the GlaDOS cluster at the University of Calgary, and on the West-Grid/Compute Canada clusters under a Research Allocation Award (to S.Y.N.). The production molecular dynamics simulations were performed on the Anton 2 computer with access provided by the Pittsburgh Supercomputing Center and D. E. Shaw Research through NIH Grant R01GM116961.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102036118/-/DCSupplemental.
Data Availability
The data that support the findings of this study are available in Dataset S1. All other study data are included in the main text and supporting information.
References
- 1.Biel M., Wahl-Schott C., Michalakis S., Zong X., Hyperpolarization-activated cation channels: From genes to function. Physiol. Rev. 89, 847–885 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Sartiani L., Mannaioni G., Masi A., Novella Romanelli M., Cerbai E., The hyperpolarization-activated cyclic nucleotide-gated channels: From biophysics to pharmacology of a unique family of ion channels. Pharmacol. Rev. 69, 354–395. (2017). [DOI] [PubMed] [Google Scholar]
- 3.Chaplan S. R., et al., Neuronal hyperpolarization-activated pacemaker channels drive neuropathic pain. J. Neurosci. 23, 1169–1178 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Q., Xing G. G., Tu H. Y., Han J. S., Wan Y., Inhibition of hyperpolarization-activated current by ZD7288 suppresses ectopic discharges of injured dorsal root ganglion neurons in a rat model of neuropathic pain. Brain Res. 1032, 63–69 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Strauss U., et al., An impaired neocortical Ih is associated with enhanced excitability and absence epilepsy. Eur. J. Neurosci. 19, 3048–3058 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Marini C., et al., HCN1 mutation spectrum: From neonatal epileptic encephalopathy to benign generalized epilepsy and beyond. Brain 141, 3160–3178 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Nof E., et al., Point mutation in the HCN4 cardiac ion channel pore affecting synthesis, trafficking, and functional expression is associated with familial asymptomatic sinus bradycardia. Circulation 116, 463–470 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Schulze-Bahr E., et al., Pacemaker channel dysfunction in a patient with sinus node disease. J. Clin. Invest. 111, 1537–1545 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stieber J., Hofmann F., Ludwig A., Pacemaker channels and sinus node arrhythmia. Trends Cardiovasc. Med. 14, 23–28 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Brown H. F., DiFrancesco D., Noble S. J., How does adrenaline accelerate the heart? Nature 280, 235–236 (1979). [DOI] [PubMed] [Google Scholar]
- 11.Gauss R., Seifert R., Kaupp U. B., Molecular identification of a hyperpolarization-activated channel in sea urchin sperm. Nature 393, 583–587 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Mayer M. L., Westbrook G. L., A voltage-clamp analysis of inward (anomalous) rectification in mouse spinal sensory ganglion neurones. J. Physiol. 340, 19–45 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiFrancesco D., Tortora P., Direct activation of cardiac pacemaker channels by intracellular cyclic AMP. Nature 351, 145–147 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Lee C. H., MacKinnon R., Structures of the human HCN1 hyperpolarization-activated channel. Cell 168, 111–120.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wainger B. J., DeGennaro M., Santoro B., Siegelbaum S. A., Tibbs G. R., Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature 411, 805–810 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Long S. B., Campbell E. B., Mackinnon R., Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Rothberg B. S., Shin K. S., Phale P. S., Yellen G., Voltage-controlled gating at the intracellular entrance to a hyperpolarization-activated cation channel. J. Gen. Physiol. 119, 83–91 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin K. S., Rothberg B. S., Yellen G., Blocker state dependence and trapping in hyperpolarization-activated cation channels: Evidence for an intracellular activation gate. J. Gen. Physiol. 117, 91–101 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Holmgren M., Jurman M. E., Yellen G., Gated access to the pore of a voltage-dependent K+ channel. Neuron 19, 175–184 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Ludwig A., Zong X., Jeglitsch M., Hofmann F., Biel M., A family of hyperpolarization-activated mammalian cation channels. Nature 393, 587–591 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Bruening-Wright A., Elinder F., Larsson H. P., Kinetic relationship between the voltage sensor and the activation gate in spHCN channels. J. Gen. Physiol. 130, 71–81 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mannikko R., Elinder F., Larsson H. P., Voltage-sensing mechanism is conserved among ion channels gated by opposite voltages. Nature 419, 837–841 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Vemana S., Pandey S., Larsson H. P., S4 movement in a mammalian HCN channel. J. Gen. Physiol. 123, 21–32 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson H. P., Baker O. S., Dhillon D. S., Isacoff E. Y., Transmembrane movement of the shaker K+ channel S4. Neuron 16, 387–397 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Flynn G. E., Zagotta W. N., Insights into the molecular mechanism for hyperpolarization-dependent activation of HCN channels. Proc. Natl. Acad. Sci. U.S.A. 115, E8086–E8095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowgill J., et al., Bipolar switching by HCN voltage sensor underlies hyperpolarization activation. Proc. Natl. Acad. Sci. U.S.A. 116, 670–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee C. H., MacKinnon R., Voltage sensor movements during hyperpolarization in the HCN channel. Cell 179, 1582–1589.e1587 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramentol R., Perez M. E., Larsson H. P., Gating mechanism of hyperpolarization-activated HCN pacemaker channels. Nat. Commun. 11, 1419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long S. B., Campbell E. B., Mackinnon R., Voltage sensor of Kv1.2: Structural basis of electromechanical coupling. Science 309, 903–908 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Lörinczi É., et al., Voltage-dependent gating of KCNH potassium channels lacking a covalent link between voltage-sensing and pore domains. Nat. Commun. 6, 6672 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai H. C., Grabe M., Jan Y. N., Jan L. Y., The S4 voltage sensor packs against the pore domain in the KAT1 voltage-gated potassium channel. Neuron 47, 395–406 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Grabe M., Lai H. C., Jain M., Jan Y. N., Jan L. Y., Structure prediction for the down state of a potassium channel voltage sensor. Nature 445, 550–553 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Kasimova M. A., et al., Helix breaking transition in the S4 of HCN channel is critical for hyperpolarization-dependent gating. eLife 8, e53400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai G., Aman T. K., DiMaio F., Zagotta W. N., The HCN channel voltage sensor undergoes a large downward motion during hyperpolarization. Nat. Struct. Mol. Biol. 26, 686–694 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranganathan R., Lewis J. H., MacKinnon R., Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron 16, 131–139 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Bell D. C., Yao H., Saenger R. C., Riley J. H., Siegelbaum S. A., Changes in local S4 environment provide a voltage-sensing mechanism for mammalian hyperpolarization-activated HCN channels. J. Gen. Physiol. 123, 5–19 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryu S., Yellen G., Charge movement in gating-locked HCN channels reveals weak coupling of voltage sensors and gate. J. Gen. Physiol. 140, 469–479 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ledwell J. L., Aldrich R. W., Mutations in the S4 region isolate the final voltage-dependent cooperative step in potassium channel activation. J. Gen. Physiol. 113, 389–414 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barro-Soria R., et al., KCNE1 divides the voltage sensor movement in KCNQ1/KCNE1 channels into two steps. Nat. Commun. 5, 3750. 10.1038/ncomms4750. (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W., MacKinnon R., Cryo-EM structure of the open human ether-a-go-go-related K+ channel hERG. Cell 169, 422–430.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barro-Soria R., et al., KCNE1 and KCNE3 modulate KCNQ1 channels by affecting different gating transitions. Proc. Natl. Acad. Sci. U.S.A. 114, E7367–E7376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jo S., Lim J. B., Klauda J. B., Im W., CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 97, 50–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips J. C., et al., Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips J. C., et al., Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 153, 044130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Best R. B., et al., Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klauda J. B., et al., Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 48.Tuckerman M., Berne B. J., Martyna G. J., Reversible multiple time scale molecular dynamics. J. Chem. Phys. 97, 1990–2001 (1992). [Google Scholar]
- 49.Shaw D. E., et al., “Anton 2: Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer” in SC ’14: Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis (IEEE Press, 2014), pp. 41–53. [Google Scholar]
- 50.Khalili-Araghi F., et al., Calculation of the gating charge for the Kv1.2 voltage-activated potassium channel. Biophys. J. 98, 2189–2198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roux B., Influence of the membrane potential on the free energy of an intrinsic protein. Biophys. J. 73, 2980–2989 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roux B., The membrane potential and its representation by a constant electric field in computer simulations. Biophys. J. 95, 4205–4216 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in Dataset S1. All other study data are included in the main text and supporting information.