Significance
Soil is a complex and competitive environment, forcing its inhabitants to develop strategies against competitors, predators, and pathogens. Identifying and understanding the molecular mechanisms has translational value for medicine, ecology, and agriculture. In this study, we show that a member of important soil-dwelling fungi (Mortierella) forms a tight alliance with toxin-producing bacteria (Mycoavidus) that live within the fungal hyphae and protect their host from nematode attack. This discovery is relevant since Mortierella species correlate with healthy soils and are used as plant growth–promoting fungi in agriculture. Unraveling an ecological role for fungal endosymbionts in Mortierella, our results contribute to the understanding of a mainspring in fungal–endobacterial symbioses and open the possibility for the development of new biocontrol agents.
Keywords: natural products, symbiosis, microbial interactions
Abstract
Fungi of the genus Mortierella occur ubiquitously in soils where they play pivotal roles in carbon cycling, xenobiont degradation, and promoting plant growth. These important fungi are, however, threatened by micropredators such as fungivorous nematodes, and yet little is known about their protective tactics. We report that Mortierella verticillata NRRL 6337 harbors a bacterial endosymbiont that efficiently shields its host from nematode attacks with anthelmintic metabolites. Microscopic investigation and 16S ribosomal DNA analysis revealed that a previously overlooked bacterial symbiont belonging to the genus Mycoavidus dwells in M. verticillata hyphae. Metabolic profiling of the wild-type fungus and a symbiont-free strain obtained by antibiotic treatment as well as genome analyses revealed that highly cytotoxic macrolactones (CJ-12,950 and CJ-13,357, syn. necroxime C and D), initially thought to be metabolites of the soil-inhabiting fungus, are actually biosynthesized by the endosymbiont. According to comparative genomics, the symbiont belongs to a new species (Candidatus Mycoavidus necroximicus) with 12% of its 2.2 Mb genome dedicated to natural product biosynthesis, including the modular polyketide-nonribosomal peptide synthetase for necroxime assembly. Using Caenorhabditis elegans and the fungivorous nematode Aphelenchus avenae as test strains, we show that necroximes exert highly potent anthelmintic activities. Effective host protection was demonstrated in cocultures of nematodes with symbiotic and chemically complemented aposymbiotic fungal strains. Image analysis and mathematical quantification of nematode movement enabled evaluation of the potency. Our work describes a relevant role for endofungal bacteria in protecting fungi against mycophagous nematodes.
A healthy soil nourishes plants and animals, purifies water and air, and promotes sustainable agriculture. Characteristic for highly complex and competitive soil ecosystems are the frequent and direct interactions between all soil-dwelling microorganisms, animals, and plants (1, 2), all of which need to be provided with minerals and carbon sources. Thus, carbon cycling, mainly promoted by fungal saprophytes and decomposers that release nutrients from decaying matter, plays a pivotal role for soil health (3, 4). Fungi belonging to the genus Mortierella are the most common soil-dwelling fungi, ubiquitously distributed in all parts of the world, inhabiting highly diverse niches including the rhizosphere and plant tissues (5–9). Owing to their ability to degrade biopolymers as well as xenobiotics, they not only deliver energy-rich carbon sources but also clear the environment from pollutants (10, 11). Typically associated with healthy soils, Mortierella species are recognized as valuable plant growth–promoting fungi in agriculture (9, 12).
Even so, all fungi, including Mortierella species, are threatened by micropredators such as nematodes (13–15). In order to oppose these predators, fungi have developed a diverse set of defense strategies. These include the production of toxic proteins and nematocidal natural products, hyphal piercing, trapping, egg parasitism, and endoparasitism (13, 16). Information on defense strategies employed by Mortierella species against nematodes is, however, scarce. It is known that Mortierella globalpina traps nematodes by means of its hyphae and penetrates the nematode’s cuticula. In this way, M. globalpina may protect its host plants from plant-parasitic nematodes (e.g., Meloidogyne chitwoodi) (17). Antinematode activities have been implicated for some Mortierella species (18, 19), including Mortierella alpina [against Meloidogyne javanica or Heterodera sp. (20, 21)], but it is not a general trait of Mortierella (21, 22). Apart from the hyphal trapping strategy, insight into the molecular basis of the antinematode activities of Mortierella is missing. Furthermore, on a more general note, it is remarkable that thus far no Mortierella secondary metabolites have been associated with potential protective roles against nematodes.
Here, we report a so far unknown strategy of a Mortierella species to protect itself from nematode attack. We provide evidence that cytotoxic benzolactones initially isolated from fungal cultures are in fact produced by bacterial endosymbionts that have been overlooked thus far. We also show that the bacteria dwelling in the fungal hyphae protect their host from predatory nematodes.
Results and Discussion
Mortierella Fungus Harbors Bacterial Endosymbionts Producing Toxic Macrolactones.
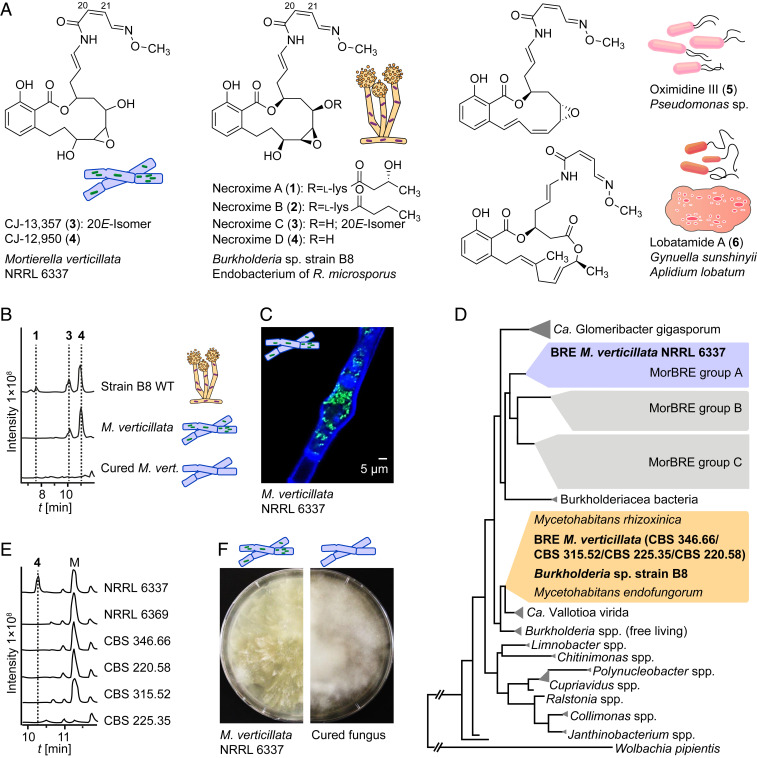
We reasoned that benzolactones CJ-12,950 and CJ-13,357 (Fig. 1A) (23) from cultures of Mortierella verticillata [synonym Podila verticillata (24)] could play a role as nematode defense metabolites. Although the initial report on CJ-12,950 and CJ-13,357 only stated that these compounds enhance the expression of the low-density lipoprotein receptor in human hepatocytes (23), they share the benzolactone enamide architecture with structurally related vATPase inhibitors (25, 26). Moreover, the architectures of CJ-12,950 and CJ-13,357 specifically resemble those of Burkholderia sp. strain B8 produced necroximes A to D (1 to 4), which proved to be cytotoxic (27). Since only the two-dimensional structures of CJ-12,950 and CJ-13,357 had been reported (23), we assigned their absolute configurations by examining the structural relationships with necroximes C and D. Optical rotation comparison, high-performance liquid chromatography (HPLC)–based coelution experiments and comparison of tandem mass spectrometry (MS/MS) fragmentation indicated that necroxime D (4) is identical to CJ-12,950, and necroxime C (3) is identical to CJ-13,357 (Fig. 1B and SI Appendix, Table S4). These assignments were corroborated by comparison of the NMR spectra of purified metabolites (SI Appendix, Table S9).
Fig. 1.
Bacterial origin of cytotoxic benzolactones from M. verticillata cultures. (A) Cytotoxic lactone compounds assigned to endofungal symbionts from the fungus R. microsporus (1–4), M. verticillata (3–4), Pseudomonas sp. (5), and a tunicate and the bacterium Gynuella sunshinyii (6). (B) Metabolic profiles of extracts from Burkholderia sp. strain B8 and M. verticillata NRRL 6337 as symbiont or cured strain as total ion chromatograms in the negative mode. (C) Fluorescence micrograph depicting endosymbionts living in the fungal hyphae; staining with Calcofluor White and Syto9 Green. (D) Phylogenetic relationships of Mortierella symbionts, Burkholderia sp. strain B8, and other bacteria based on 16S rDNA. BRE, Burkholderia-related endosymbiont of Mortierella spp. (E) Metabolic profiles of extracts from M. verticillata NRRL 6337 and other necroxime-negative M. verticillata strains analyzed for endosymbionts in this study as total ion chromatograms in the negative mode. M, medium component. (F) Growth of symbiotic M. verticillata NRRL 6337 in comparison to the cured strain.
Given the bacterial origin of the necroximes (27) and related benzolactones (25, 28–30), we questioned the biosynthetic capability of M. verticillata and sought to identify the true producer. Since several Mortierella spp. have been reported to live in symbiosis with bacteria (31, 32), we suspected an endosymbiont to be the true source of 3 and 4. Yet, a 2018 report investigating the prevalence of Burkholderiaceae-related bacteria within Mortierella spp. stated that strain NRRL 6337 was devoid of endosymbionts (32). Nonetheless, we re-examined the same strain for endosymbionts by staining fungal hyphae with the chitin-binding Calcofluor White dye, and tentative endobacteria with the nucleic acid dye Syto9 Green (Fig. 1C). Fluorescence microscopy revealed the presence of endosymbiotic organisms in M. verticillata NRRL 6337 (SI Appendix, Fig. S1).
To identify the observed bacterial endosymbionts, we cut a small piece of fungal mycelium and extracted holobiont DNA, followed by PCR amplification of the 16S ribosomal DNA (rDNA) region using universal primers. Sequencing of the 16S rDNA region (SI Appendix, Table S1) and BLAST analysis indicated that the symbiont of M. verticillata NRRL 6337 is a Mycoavidus species. Notably, members of this genus have been reported as symbionts of soil-dwelling fungi (32). So far, the full genomes of only three Mycoavidus cysteinexigens strains from Mortierella elongata and Mortierella parvispora have been sequenced (31, 33–35). PCR-amplified bacterial 16S rDNA sequences from other Mortierella fungi, however, revealed further Mycoavidus endosymbionts with three phylogenetically distant clades (Mortierella-associated Burkholderia-related endosymbiont [MorBRE] groups A to C) (32). Through phylogenetic analysis, we found that the Mycoavidus symbiont of M. verticillata NRRL 6337 falls into MorBRE group A (Fig. 1D and SI Appendix, Fig. S3) comprising symbionts of Mortierella humilis, Mortierella gamsii, Mortierella basiparvispora, and M. elongata (M. cysteinexigens). To better understand the occurrence of Mycoavidus endosymbionts in M. verticillata strains, we investigated five additional M. verticillata strains for the presence of endosymbionts. Amplification of the 16S rDNA regions from gDNA of symbionts of these strains revealed a conserved occurrence of Mycoavidus endosymbionts in M. verticillata strains. Interestingly, these additional endosymbionts all fall into another phylogenetic group together with Burkholderia sp. strain B8. Furthermore, analysis of the metabolic profiles of the respective fungi did not show any production of necroximes (Fig. 1 D and E). This finding shows that endosymbionts may frequently occur in Mortierella and other species of the order Mucorales, but they can be phylogenetically different.
To clarify whether bacterial endosymbionts are the true producers of 3 and 4, we aimed at curing M. verticillata NRRL 6337 of its symbiont through the addition of antibiotics (36). Over the course of several months, we subcultivated the fungal strain on agar plates containing kanamycin, ciprofloxacin, or chloramphenicol. During treatment, changes of the fungal growth were noticeable (Fig. 1F). Finally, we confirmed the absence of the symbionts by fluorescence staining, microscopic inspection, and PCR analysis (SI Appendix, Figs. S2 and S4). The metabolic profiling of the symbiont-free fungal strain by liquid chromatography (LC) combined with high-resolution electrospray ionization revealed the complete absence of 3 and 4 (Fig. 1B). These findings indicate that Candidatus Mycoavidus necroximicus is the true producer of the benzolactones.
Ca. M. necroximicus Dedicates 12% of Its Genome to Secondary Metabolism.
To gain insight into the symbiont’s biosynthetic potential, with particular focus on the molecular basis of necroxime biosynthesis, we aimed at sequencing the genome of the endosymbiont. Attempts to isolate and cultivate the endosymbiont in the absence of the fungal host, however, proved to be futile. Methods previously used to axenically cultivate similar fungal endobacteria did not enable growth of the endosymbionts (33, 37), indicating a strong dependence of the bacterial symbiont on the host environment. Thus, we sought to enrich the symbiotic bacteria for DNA isolation. Initially, physical disruption of the host’s mycelium resulted in high levels of contamination with fungal DNA, which complicated the assembly of the endosymbiont's genome. Eventually, we succeeded in retrieving a bacterial cell pellet by filtration and centrifugation of the turbid supernatant of shaking cultures in baffled flasks and isolated the genomic DNA from resuspended bacteria.
The genome of the bacterial endosymbiont was sequenced using a combination of Oxford Nanopore MinION and Illumina NextSeq sequencing, and both data sets were used to generate a hybrid genome assembly. Of the 118 contigs, a single 2.4 Mb contig of putative bacterial origin was identified through homology searches using the Mycoavidus-like 16S rDNA sequence previously amplified from M. verticillata NRRL 6337. Following trimming of overlapping ends (suggesting a circular chromosome) the final 2.2 Mb contig was found to contain 1,768 CDS, 6 rRNAs, 42 tRNAs, and a GC content of 50.6% (genome accession number: PRJNA733818). The 16S rDNA sequence of the new strain has 98.82% nucleotide identity to M. cysteinexigens B1-EBT (33). Even so, genomic comparisons showed an average nucleotide identity of only 81.85% across the two genomes. By current standards for molecular species discrimination, the newly identified Mortierella endosymbiont should be considered a new species (Ca. M. necroximicus) (38, 39).
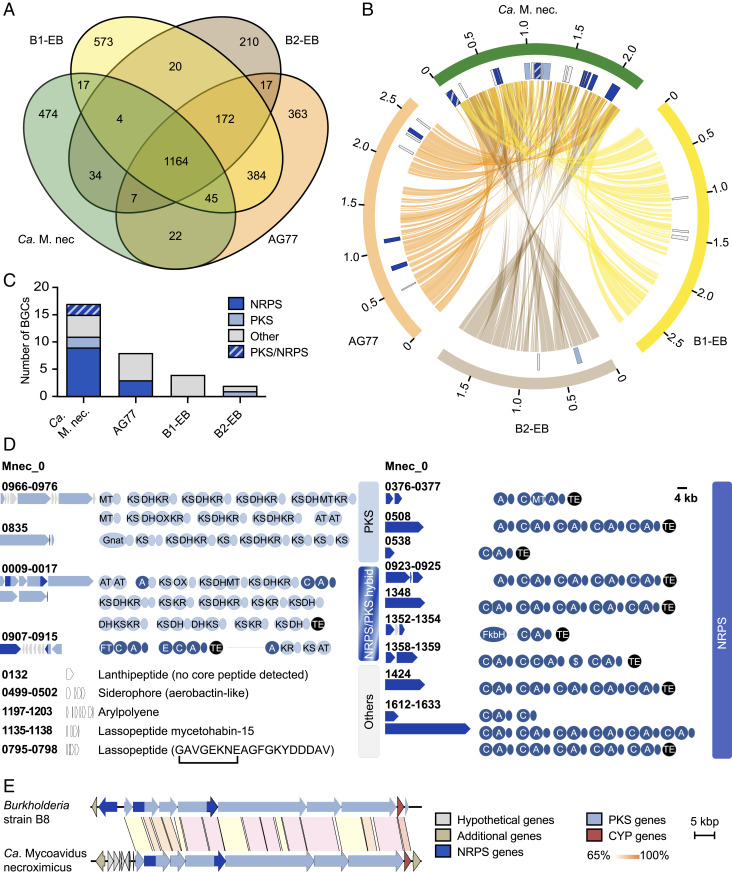
By comparative genomic analyses, we noted that the genomes of the two endofungal strains AG77 and B1-EBT isolated from M. elongata (33, 35) are 400 to 500 kb larger than the genome of Ca. M. necroximicus. Only the genome of strain B2-EB isolated from M. parvispora (34) is smaller (∼500 kb) than the genome of Ca. M. necroximicus (2.2 Mb). When investigating shared protein orthologs, we noted that a core genome encoding 1,164 proteins exists among the four genomes at the 70% identity level (Fig. 2A). However, a further all-versus-all comparison showed B1-EBT and AG77 to be the most closely related as they share ∼75% of their deduced proteome. The B2-EB and Ca. M. necroximicus strains are more distantly related to B1-EBT and AG77, as well as each other, with only a small number of proteins shared exclusively with either B1-EBT (20 and 17 proteins, respectively) or AG77 (17 and 22 proteins, respectively) (Fig. 2B).
Fig. 2.
Comparative genomic analyses of Mycoavidus spp. (A) Number of orthologous proteins among the four Mycoavidus strains at 70% identity. (B) Circos plot of shared protein orthologs, and secondary metabolite loci (detected by antiSMASH v5) in Mycoavidus genomes. Outer blocks (orange, brown, yellow, green) represent genome sizes, while the inner blocks represent genomic positions of secondary metabolite loci. Lines linking the three genomes show position of genes whose proteins are orthologous at 70% identity. Depicted are the genome sequences of M. cysteinexigens strains AG77, B1-EB, B2-EB, and Ca. M. necroximicus (Ca. M. nec.). (C) Number of gene clusters putatively coding for natural products in Mycoavidus spp. detected by antiSMASH and by manual assignment. (D) BGCs and their encoded assembly lines identified from the endofungal Ca. M. necroximicus are displayed. A, adenylation; AT, acyltransferase; C, condensation; DH, dehydratase; E, epimerization; Gnat, GCN5-related N-acetyltransferase; KR, ketoreductase; KS, ketosynthase; MT, methyltransferase; OX, oxygenase; TE, thioesterase domains. Acyl carrier (light blue) and peptidyl carrier proteins (dark blue) are shown as circles without designators. (E) Homologous benzolactone BGCs in the genome of Burkholderia strain B8 and Ca. M. necroximicus.
Whereas biosynthetic gene clusters (BGCs) are present in the genomes of all four studied Mortierella symbionts, antiSMASH analysis (40) revealed that the biosynthetic potential for secondary metabolites is by far the greatest in Ca. M. necroximicus (Fig. 2C). Despite the relatively small genome for Mycoavidus standards, ∼12% of its protein-encoding capacity is dedicated to natural product biosynthesis. We identified nine nonribosomal peptide synthetase (NRPS) gene clusters, two polyketide synthase (PKS) gene clusters, two hybrid PKS/NRPS gene clusters, and five other BGCs (Fig. 2D). Notably, several large PKS and NRPS gene loci present in the Ca. M. necroximicus genome are absent in the genomes of strains B1-EBT, B2-EB, and AG77 (Fig. 2B). This BGC list includes a cryptic BGC (Mcyst_0009–0017) encoding a PKS/NRPS hybrid that shows high similarity to the necroxime assembly line from Burkholderia sp. strain B8 (97% coverage, ∼70% amino acid identity), which has been unequivocally linked to necroxime biosynthesis by targeted gene knockouts (Fig. 2E) (27). The only major difference between the two BGCs is the NRPS gene necA, which is missing in the genome of Ca. M. necroximicus. This finding is in full agreement with the current biosynthetic model, since NecA is responsible for the attachment of the peptide side chain in 1 (Fig. 1A) (27), which is absent in 3 and 4. Furthermore, the architecture of the encoded PKS/NRPS modules is perfectly in line with the biosynthesis of the benzolactone enamide backbone of 3 and 4. Based on these in silico predictions, we inferred that this PKS/NRPS hybrid gene cluster codes for the biosynthesis of 3 and 4 (SI Appendix, Figs. S6 and S8). Together with the metabolic profiling of the cured fungal strain, these data indicate that the bacterial endosymbionts, not the fungus, are the true producers of the benzolactones 3 and 4. CJ-12,950 and CJ-13,357 are thus important additions to the small group of natural products that were believed to be fungal metabolites but are actually produced by bacterial endosymbionts; rhizoxins (41) and rhizonins (42) from symbionts of Rhizopus microsporus (43), and endolides from Stachylidium bicolor (44). From an ecological viewpoint, it is remarkabe that endosymbiotic bacteria were identified as the true producers of the virulence factor of the rice-seedling blight fungus R. microsporus (36, 37, 45). Given the different ecological context of Mortierella, however, we assumed that the necroximes may have another function in microbial interactions.
Necroximes Protect the Fungal Host from Nematode Attacks.
To learn more about the potential role of necroximes (3 and 4) in the ecological context of the Mortierella–Mycoavidus symbiosis, we investigated whether these toxins could impair the growth of, or even kill, competitors. Therefore, we considered that the common natural habitat of Mortierella species, including M. verticillata NRRL 6337, is soil, and that microbial survival in the soil environment is not only determined by the capacity to grow under harsh conditions but also by the ability to defend oneself from (micro)predators (46). Among the most abundant fungal predators are nematodes, which share the same soil habitat as Mortierella (47).
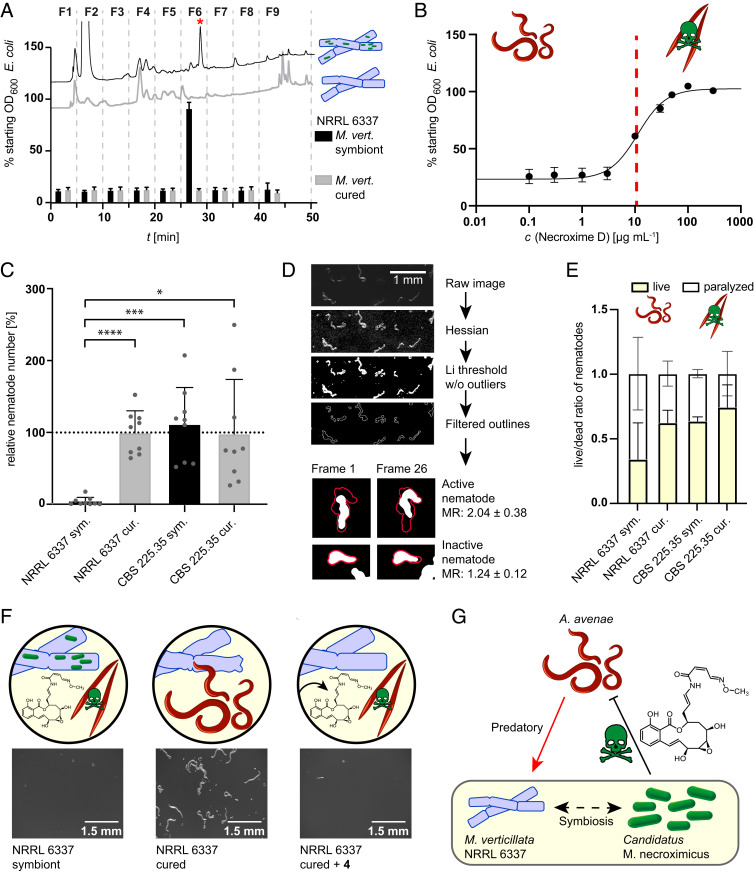
To determine if 3 and 4 or any other endobacteria-derived substance have anthelmintic activity, we first performed a viability assay against the model organism, Caenorhabditis elegans (48). We cultivated both cured (Mycoavidus-free) and symbiotic M. verticillata NRRL 6337 on potato dextrose agar (PDA agar). Cultures were extracted, and each extract was fractionated by preparative HPLC. The individual fractions (F1 to F9) were subsequently tested against C. elegans. Anthelmintic activity in this assay was determined by the ability of C. elegans to feed on a supplied Escherichia coli food source in the presence of the different fractions. Consumption of bacteria indicates unimpeded nematodes, whereas growth of E. coli indicates that the nematodes are negatively affected by the added substances (Fig. 3A). Notably, all fractions of the extract obtained from the cured strain culture were found to be inactive in the C. elegans assay. In contrast, we observed a marked nematocidal activity of fraction 6 from the extract of the symbiotic fungus. By LC/MS measurements we confirmed the presence of 3 and 4 in the active fraction. In order to determine the anthelmintic potency of the major metabolite 4, we performed the viability assay against C. elegans using increasing concentrations of the pure substance and determined an inhibitory concentration at 50% (IC50) value of 11.3 µg ⋅ mL−1 (24.66 µM) (Fig. 3B). Interestingly, the amount of isolated necroximes from fungal cultures grown on agar plates is ∼11 µg ⋅ mL−1. Assuming that the actual concentration in fungal hyphae is slightly higher due to an uneven diffusion into the agar and some loss during the purification steps, we conclude that the concentrations inside and around the fungal mycelium are sufficiently high to fully protect it from mycophagous nematodes.
Fig. 3.
Nematocidal activity of symbiont-derived toxins. (A) Viability assay of C. elegans in presence of extract fractions of symbiotic and cured M. verticillata NRRL 6337. HPLC profiles of extracts are shown with corresponding effect on nematodes, measured as effect on the E. coli optical density (OD). When nematode growth is impaired by the fraction, E. coli cells are not consumed, and thus the OD600 is not altered (error bars represent mean of three biological replicates). The red asterisk represents 4. (B) Toxicity screening of 4 against C. elegans. The red line marks IC50 at 11.3 µg ⋅ mL−1 (24.66 µM; 95% CI, 21.45 to 28.37 µM; error bars as mean of five biological replicates). (C) Nematode counts from propagation assay of M. verticillata and A. avenae cocultures. Bars represent relative nematode numbers compared to the mean of the nematode count from cured M. verticillata NRRL 6337 cultures. cur., cured; sym., symbiotic. *P < 0.02; ***P < 0.001; ****P < 0.0001. Data represent three biological replicates with three technical replicates each. (D) Workflow of image analysis and mathematical evaluation of A. avenae mobility in fungal–nematodal coincubations. Processing of time series is demonstrated by one time frame. Exemplary images of nematodes from two time frames (frame 1 and 26) are shown to illustrate differences in motility. Results of calculated mobility ratios (MR) were used for live or paralyzed/dead categorization. (E) Results of image analysis and mathematical quantification of nematode movement. Bars show ratio between moving/living nematodes and paralyzed/dead nematodes, which were harvested from cocultures of A. avenae with symbiotic M. verticillata NRRL 6337 cultures, cured NRRL 6337 cultures, or CSB 225.35 cultures. Numbers and error bars were calculated from minimal 176 worms from three biological replicates. (F) Stereomicroscopic images and schematic picture of chemical complementation assay with a magnitude of 25×. Sample of nematodes harvested from plates containing symbiotic, cured, or cured and with 4 chemically complemented M. verticillata NRRL 6337 cultures. (G) Schematic summary of tripartite interaction between fungal host, bacterial endosymbiont, and mycophagous nematodes.
In order to corroborate a potential host-protective role of the symbiont-derived toxin, we next focused on a fungivorous nematode. Therefore, we selected Aphelenchus avenae, a predator using a stylet to feed on fungi, which pierces the fungal cell wall and allows the fungivore to ingest the fungal cytoplasm (49, 50). Sharing the same soil habitat, A. avenae represents a realistic predator of Mortierella spp. (51). To investigate the effects of symbiotic and cured M. verticillata strains on the feeding behavior and survival of A. avenae, we determined the number of animals that were harvested from fungal–nematodal cocultures. In addition, we compared the mobility ratios of the nematodes in correlation to the presence or absence of the bacterial symbiont. As a control, we employed the symbiont-bearing, but necroxime-negative, M. verticillata strain CSB 225.35 (Fig. 1E), thus ruling out an influence solely based on the presence of bacterial symbionts.
To determine nematodal propagation rates, we inoculated plate cultures of symbiotic and cured fungi with A. avenae and cocultivated both organisms for 17 to 24 d (three biological triplicates). Subsequently, nematodes were isolated from the cocultivation plates by Baermann funneling (52), transferred onto water-agar plates, and counted by stereomicroscopic visualization. We found that significantly fewer nematodes are able to grow in the presence of the necroxime-producing endosymbionts (Fig. 3C and SI Appendix, Fig. S9 and Tables S5 and S6).
To scrutinize the effect of the toxin on the fitness of the fungivorous nematodes, we harvested the animals from cocultures and determined their movement—and thus the mobility ratios—by image analysis and mathematical quantification (Fig. 3D). Using stereoscopic time series to track their movement, we compared the area covered by each moving nematode during the time series to the area covered solely by its body without movement, allowing us to differentiate active (living) from inactive (dead or paralyzed) animals. Analyzing a minimum of 176 nematodes from three independent experiments, we observed a significant decrease in the mobility ratio of nematodes grown on symbiotic M. verticillata NRRL 6337 compared to cured NRRL 6337 cultures and to necroxime-negative CSB 225.35 cultures (Fig. 3E and SI Appendix, Fig. S10 and Tables S7 and S8).
HPLC analyses of the plate extracts detected necroximes only in symbiotic cultures of NRRL 6337 but not in cured strains or CBS 225.35, correlating once again the toxins with reduced numbers and lower fitness of the nematodes. To unambiguously assign the nematocidal activity in the propagation assay to the necroximes, we repeated the A. avenae assay with the cured Mortierella strain and chemically complemented the major toxin. Specifically, we overlaid the cured strain NRRL 6337 with solutions of 4 in increasing concentrations (25 µM [IC50], 50 µM, 109 µM, and 219 µM). We then compared A. avenae propagation in necroxime-complemented cultures to untreated cured as well as symbiotic fungi by microscopic examination after two weeks of coincubation. For cultures supplemented with 25 µM or 50 µM of 4, we noted a moderate reduction of nematode propagation, whereas in cultures supplemented with 109 µM or 219 µM of 4 the presence of nematodes in the fungus was abolished (Fig. 3F and SI Appendix, Figs. S11 and S12). The elevated concentrations compared to the IC50 value can be explained due to an uneven distribution of 4 into deeper layers of the hydrophobic fungal colony and the ongoing growth of fungal hyphae, which were not wetted with toxin solution. Nonetheless, these experiments unambiguously verified that the chemical complementation restores the anthelmintic effect. Thus, we uncovered an important role of a natural product in the complex tripartite interplay of symbiont, host, and (micro)predator (Fig. 3G).
Conclusions
In this study, we uncovered a previously overlooked bacterial endosymbiont that protects the important soil-dwelling fungus M. verticillata from a fungivorous nematode. Comparative genomics indicate that the yet unculturable bacterial symbionts belong to a new species that is endowed with a high biosynthetic potential. Through metabolic profiling of the symbiotic wild type and cured aposymbiotic fungi, we provide evidence that the endofungal bacteria are the true producers of highly toxic macrolides that were previously believed to be fungal metabolites. Importantly, these compounds (necroximes) efficiently protect the host from nematode attack, as demonstrated by coculture experiments, chemical complementation, and image analyses. Thus, this work not only reveals an ecological role of endofungal bacteria but also introduces a strategy to ward off micropredators. Consequently, the bacterial biosynthesis of necroximes provides an advantage of the fungal–bacterial alliance over other aposymbiotic or necroxime-negative symbiotic M. verticillata strains in the soil niche. Beyond inspiring the discovery of related tactics in symbioses, our findings may set the basis for new biocontrol agents, with the prospect of shielding plant hosts from plant-pathogenic nematodes.
Materials and Methods
Isolation of Natural Products.
For 3 and 4 isolation, M. verticillata NRRL 6337 was cultivated on PDA plates (Bacto, BD) at 26 °C. The culture was extracted twice with 1:1 volume of ethyl acetate overnight. The organic phase was concentrated under reduced pressure and the residue was dissolved in methanol. The extracts were prefractionated on an open Sephadex LH-20-column with methanol as eluent. Necroxime-containing fractions were further purified with a preparative HPLC under following conditions: A, H2O + 0.01% TFA; B, methanol; and 15 to 100% B in 35 min, 15 mL ⋅ min−1 [Phenomenex, Luna, 10 µm, C18(2), 100 Å, 250 × 21.2 mm]. NMR analysis was carried out on a 600 MHz Avance III Ultra Shield (Bruker), and signals were referenced to the residual solvent signal (DMSO-d6).
Identification of Endosymbionts in M. verticillata.
For the preparation of cured fungal strains, fungi were continuously subcultivated at 24 °C on PDA plates containing 40 µg ⋅ mL−1 ciprofloxacin or 50 µg ⋅ mL−1 kanamycin for several months. After phenotypic changes were observed by eye, an agar plate of each fungal culture was extracted with 20 mL ethyl acetate and controlled for the absence of 3 and 4 by LC/MS. Final verification of the cured fungal strains was performed by fluorescence staining (Calcofluor White Stain [Sigma] and SYTO 9 Green [Invitrogen]).
Genome Assembly for Ca. M. necroximicus.
M. verticillata NRRL 6337 was grown in MM9 medium (53) and orbitally shaken at 160 rpm and 26 °C. The turbid supernatant, containing bacteria from disrupted hyphae, was twice filtered through a membrane (pore diameter, 40 µm) and centrifuged (12,000 × g, 25 °C, 10 min) until a stabile pellet occurred. The genomic DNA was extracted with the MasterPure DNA Purification Kit (Epicentre). For long-read sequencing on the MinION platform, DNA quality was evaluated by pulsed-field gel electrophoresis and prepared for sequencing according to the protocol of the Ligation Sequencing kit (Oxford Nanopore). DNA was loaded onto a single MinION flow cell, and data were collected over a 72-h period. DNA was prepared for sequencing on the Illumina NextSeq platform using the Nextera XT DNA preparation kit (Illumina) with ×150 bp paired end chemistry and with a targeted sequencing depth of >50×. Combined MINion and Illumina sequencing data were assembled using the Unicycler hybrid assembler (54) to form a single contig 2.2 Mb containing a 98.82% match to the M. cysteinexigens rDNA gene. The evaluation of secondary metabolite loci was performed with antiSMASH version 5 (55).
Nematode Assays.
Liquid assays for active-fraction determination and potency assessment against C. elegans were conducted as previously described (48). For A. avenae coincubation assay, an aliquot of hyphae of each tested Mortierella strain was transferred to a PDA plate and incubated at 24 °C overnight. Nematodes were sterilized and starved. After one washing step with K-medium, nematodes were resuspended in 300 µL K-medium and aliquots of 50 µL were distributed onto the fresh fungal cultures. Plates were dried and controlled for living nematodes before they were incubated for 17 to 24 d at 20 °C. For the evaluation, nematodes were harvested via Baermann funneling (56). Funneled A. avenae were transferred on 1.5% water-agar plates containing 200 mM geneticin and 50 µg ⋅ mL−1 kanamycin overnight and subsequently monitored with a Zeiss Axio Zoom.V16 Stereomicroscope for worm count and bioinformatics (https://www.jipipe.org/). Remaining plates were extracted with ethyl acetate to control the metabolite production and processed as described before. For the A. avenae chemical complementation assay, an aliquot of hyphae of the respective fungus was transferred into 12-well plates filled with 1 mL PDA and incubated overnight. The 4 dissolved in 200 µL 50% MeOH was applied and evaporated at room temperature. Nematode suspensions of 50 µL were distributed onto the fungi, dried, and coincubated for 14 d at 20 °C. For evaluation, the coculture was removed from the well and washed in 5 mL K-medium overnight. The mixture was filtered through miracloth (Merck) to avoid agar carryover and left at 4 °C for 1 h. The remaining worms were transferred onto 6-well plates containing 5 mL 1.5% water-agar with 200 mM geneticin and 50 µg ⋅ mL−1 kanamycin. After the plates were dried, the worm count from each plate was assessed with a Zeiss Axio Zoom.V16 Stereomicroscope.
Supplementary Material
Acknowledgments
Mortierella strains were supplied by ARS Culture Collection (NRRL) and the Jena Microbial Resource Collection. C. elegans was provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). A. avenae was received as a kind gift from Prof. Dr. M. Künzler (ETH Zürich). We thank E. Bratovanov (HKI) for helpful discussions. Assistance by K. Martin and S. Linde (HKI) is gratefully acknowledged. H.B. and S.P.N. were funded by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) Project No. 239748522 SFB 1127, the Cluster of Excellence “Balance of the Microverse,” and also the Leibniz Award (to C.H.). Z.C. and M.T.F. were funded by the DFG Project No. 316213987 SFB 1278 (Z01). I.R. acknowledges financial support from the European Union Horizon 2020 Research and Innovation Program under the Marie Sklodowska-Curie grant agreement No. 794343. R.G. was funded by the International Leibniz Research School for Microbial and Biomolecular Interactions Jena.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110669118/-/DCSupplemental.
Data Availability
Genome sequence data have been deposited in GenBank (PRJNA733818). The 16S rDNA sequences of the Mortierella endosymbionts were deposited at the NCBI database (BRE_MvertCBS_346.66: MZ330684; BRE_MvertCBS_220.58: MZ330685; BRE_MvertCBS_225.35: MZ330686; BRE_MvertCBS_315.52: MZ330687; BRE_MvertCBS_100561: MZ330688).
References
- 1.Fierer N., Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Bardgett R. D., van der Putten W. H., Belowground biodiversity and ecosystem functioning. Nature 515, 505–511 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Tedersoo L., et al., Fungal biogeography. Global diversity and geography of soil fungi. Science 346, 1256688 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Ozimek E., et al., Synthesis of indoleacetic acid, gibberellic acid and ACC-deaminase by Mortierella strains promote winter wheat seedlings growth under different conditions. Int. J. Mol. Sci. 19, 3218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou D., et al., Deciphering microbial diversity associated with Fusarium wilt-diseased and disease-free banana rhizosphere soil. BMC Microbiol. 19, 161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J., et al., Predicting disease occurrence with high accuracy based on soil macroecological patterns of Fusarium wilt. ISME J. 14, 2936–2950 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D., Sun H., Ma H., Deciphering microbiome related to rusty roots of Panax ginseng and evaluation of antagonists against pathogenic Ilyonectria. Front. Microbiol. 10, 1350 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgington S., Thompson E., Moore D., Hughes K. A., Bridge P., Investigating the insecticidal potential of Geomyces (Myxotrichaceae: Helotiales) and Mortierella (Mortierellacea: Mortierellales) isolated from Antarctica. Springerplus 3, 289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozimek E., Hanaka A., Mortierella species as the plant growth-promoting fungi present in the agricultural soils. Agriculture 11, 7 (2021). [Google Scholar]
- 10.Ellegaard-Jensen L., Aamand J., Kragelund B. B., Johnsen A. H., Rosendahl S., Strains of the soil fungus Mortierella show different degradation potentials for the phenylurea herbicide diuron. Biodegradation 24, 765–774 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Zeng J., et al., Lignocellulosic biomass as a carbohydrate source for lipid production by Mortierella isabellina. Bioresour. Technol. 128, 385–391 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Li F., et al., Mortierella elongata's roles in organic agriculture and crop growth promotion in a mineral soil. Land Degrad. Dev. 29, 1642–1651 (2018). [Google Scholar]
- 13.Künzler M., How fungi defend themselves against microbial competitors and animal predators. PLoS Pathog. 14, e1007184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leveau J. H. J., Preston G. M., Bacterial mycophagy: Definition and diagnosis of a unique bacterial-fungal interaction. New Phytol. 177, 859–876 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Zhang S., Mukherji R., Chowdhury S., Reimer L., Stallforth P., Lipopeptide-mediated bacterial interaction enables cooperative predator defense. Proc. Natl. Acad. Sci. U.S.A. 118, e2013759118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Degenkolb T., Vilcinskas A., Metabolites from nematophagous fungi and nematicidal natural products from fungi as an alternative for biological control. Part I: Metabolites from nematophagous ascomycetes. Appl. Microbiol. Biotechnol. 100, 3799–3812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiLegge M. J., Manter D. K., Vivanco J. M., A novel approach to determine generalist nematophagous microbes reveals Mortierella globalpina as a new biocontrol agent against Meloidogyne spp. nematodes. Sci. Rep. 9, 7521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Topalović O., Hussain M., Heuer H., Plants and associated soil microbiota cooperatively suppress plant-parasitic nematodes. Front. Microbiol. 11, 313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu W., et al., Organic fertilization assembles fungal communities of wheat rhizosphere soil and suppresses the population growth of Heterodera avenae in the field. Front. Plant Sci. 11, 1225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Shammari T. A., Bahkali A. H., Elgorban A. M., El-Kahky M. T., Al-Sum B. A., The use of Trichoderma longibrachiatum and Mortierella alpina against root-knot nematode, Meloidogyne javanica on tomato. J. Pure Appl. Microbiol. 7, 199–207 (2013). [Google Scholar]
- 21.Meyer S., et al., Activity of fungal culture filtrates against soybean cyst nematode and root-knot nematode egg hatch and juvenile motility. Nematology 6, 23–32 (2004). [Google Scholar]
- 22.Hasna M. K., Insunza V., Lagerlöf J., Rämert B., Food attraction and population growth of fungivorous nematodes with different fungi. Ann. Appl. Biol. 151, 175–182 (2007). [Google Scholar]
- 23.Dekker K. A., et al., Novel lactone compounds from Mortierella verticillata that induce the human low density lipoprotein receptor gene: Fermentation, isolation, structural elucidation and biological activities. J. Antibiot. (Tokyo) 51, 14–20 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Vandepol N., et al., Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Divers. 104, 267–289 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyd M. R., et al., Discovery of a novel antitumor benzolactone enamide class that selectively inhibits mammalian vacuolar-type (H+)-atpases. J. Pharmacol. Exp. Ther. 297, 114–120 (2001). [PubMed] [Google Scholar]
- 26.Pérez-Sayáns M., Somoza-Martín J. M., Barros-Angueira F., Rey J. M., García-García A., V-ATPase inhibitors and implication in cancer treatment. Cancer Treat. Rev. 35, 707–713 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Niehs S. P., et al., Mining symbionts of a spider-transmitted fungus illuminates uncharted biosynthetic pathways to cytotoxic benzolactones. Angew. Chem. Int. Ed. Engl. 59, 7766–7771 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayakawa Y., et al., Oximidine III, a new antitumor antibiotic against transformed cells from Pseudomonas sp. II. Structure elucidation. J. Antibiot. (Tokyo) 56, 905–908 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Galinis D. L., McKee T. C., Pannell L. K., Cardellina J. H., Boyd M. R., Lobatamides A and B, novel cytotoxic macrolides from the tunicate Aplidium lobatum. J. Org. Chem. 62, 8968–8969 (1997). [Google Scholar]
- 30.Ueoka R., et al., Genome mining of oxidation modules in trans-acyltransferase polyketide synthases reveals a culturable source for lobatamides. Angew. Chem. Int. Ed. Engl. 59, 7761–7765 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato Y., et al., Detection of betaproteobacteria inside the mycelium of the fungus Mortierella elongata. Microbes Environ. 25, 321–324 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Takashima Y., et al., Prevalence and intra-family phylogenetic divergence of Burkholderiaceae-related endobacteria associated with species of Mortierella. Microbes Environ. 33, 417–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohshima S., et al., Mycoavidus cysteinexigens gen. nov., sp. nov., an endohyphal bacterium isolated from a soil isolate of the fungus Mortierella elongata. Int. J. Syst. Evol. Microbiol. 66, 2052–2057 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Guo Y., et al., Mycoavidus sp. Strain B2-EB: Comparative genomics reveals minimal genomic features required by a cultivable Burkholderiaceae-related endofungal bacterium. Appl. Environ. Microbiol. 86, e01018-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uehling J., et al., Comparative genomics of Mortierella elongata and its bacterial endosymbiont Mycoavidus cysteinexigens. Environ. Microbiol. 19, 2964–2983 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Partida-Martinez L. P., Hertweck C., Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437, 884–888 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Lackner G., Moebius N., Hertweck C., Endofungal bacterium controls its host by an hrp type III secretion system. ISME J. 5, 252–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain C., Rodriguez-R L. M., Phillippy A. M., Konstantinidis K. T., Aluru S., High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yarza P., et al., Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Blin K., et al., antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47 (W1), W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scherlach K., Partida-Martinez L. P., Dahse H. M., Hertweck C., Antimitotic rhizoxin derivatives from a cultured bacterial endosymbiont of the rice pathogenic fungus Rhizopus microsporus. J. Am. Chem. Soc. 128, 11529–11536 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Partida-Martinez L. P., et al., Rhizonin, the first mycotoxin isolated from the zygomycota, is not a fungal metabolite but is produced by bacterial endosymbionts. Appl. Environ. Microbiol. 73, 793–797 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lackner G., Partida-Martinez L. P., Hertweck C., Endofungal bacteria as producers of mycotoxins. Trends Microbiol. 17, 570–576 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Almeida C., et al., Unveiling concealed functions of endosymbiotic bacteria harbored in the ascomycete stachylidium bicolor. Appl. Environ. Microbiol. 84, e00660-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scherlach K., Busch B., Lackner G., Paszkowski U., Hertweck C., Symbiotic cooperation in the biosynthesis of a phytotoxin. Angew. Chem. Int. Ed. Engl. 51, 9615–9618 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Geisen S., et al., The soil food web revisited: Diverse and widespread mycophagous soil protists. Soil Biol. Biochem. 94, 10–18 (2016). [Google Scholar]
- 47.van den Hoogen J., et al., A global database of soil nematode abundance and functional group composition. Sci. Data 7, 103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith M. P., et al., A liquid-based method for the assessment of bacterial pathogenicity using the nematode Caenorhabditis elegans. FEMS Microbiol. Lett. 210, 181–185 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Ragsdale E. J., Crum J., Ellisman M. H., Baldwin J. G., Three-dimensional reconstruction of the stomatostylet and anterior epidermis in the nematode Aphelenchus avenae (Nematoda: Aphelenchidae) with implications for the evolution of plant parasitism. J. Morphol. 269, 1181–1196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmieder S. S., et al., Bidirectional propagation of signals and nutrients in fungal networks via specialized hyphae. Curr. Biol. 29, 217–228.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Yeates G. W., Bongers T., De Goede R. G., Freckman D. W., Georgieva S. S., Feeding habits in soil nematode families and genera-an outline for soil ecologists. J. Nematol. 25, 315–331 (1993). [PMC free article] [PubMed] [Google Scholar]
- 52.Tayyrov A., Schmieder S. S., Bleuler-Martinez S., Plaza D. F., Künzler M., Toxicity of potential fungal defense proteins towards the fungivorous nematodes Aphelenchus avenae and Bursaphelenchus okinawaensis. Appl. Environ. Microbiol. 84, e02051-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hermenau R., et al., Gramibactin is a bacterial siderophore with a diazeniumdiolate ligand system. Nat. Chem. Biol. 14, 841–843 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Wick R. R., Judd L. M., Gorrie C. L., Holt K. E., Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 3, e000132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blanton S., et al., A web-based carepartner-integrated rehabilitation program for persons with stroke: Study protocol for a pilot randomized controlled trial. Pilot Feasibility Stud. 5, 58 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bleuler-Martínez S., et al., A lectin-mediated resistance of higher fungi against predators and parasites. Mol. Ecol. 20, 3056–3070 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequence data have been deposited in GenBank (PRJNA733818). The 16S rDNA sequences of the Mortierella endosymbionts were deposited at the NCBI database (BRE_MvertCBS_346.66: MZ330684; BRE_MvertCBS_220.58: MZ330685; BRE_MvertCBS_225.35: MZ330686; BRE_MvertCBS_315.52: MZ330687; BRE_MvertCBS_100561: MZ330688).