Significance
The human placenta contains progenitors that give rise to highly specialized trophoblast cell types, and failures in their differentiation are associated with placental pathologies. Importantly, transcription factors controlling these cell fate decisions in humans are poorly understood. Here, we uncovered MSX2 as a human-specific regulator of trophoblast cell identity and implicate its role in placental development and disease. We found that MSX2 interacts and cobinds many target genes with components of the SWI/SNF chromatin remodeling complex, suggesting a mechanistic link. Given the critical function of SWI/SNF in gene expression, organogenesis, and disease, we provided characterization of its composition and possible role in the placental context.
Keywords: placenta, MSX2, human trophoblast stem cells, syncytiotrophoblast, SWI/SNF complex
Abstract
Multiple placental pathologies are associated with failures in trophoblast differentiation, yet the underlying transcriptional regulation is poorly understood. Here, we discovered msh homeobox 2 (MSX2) as a key transcriptional regulator of trophoblast identity using the human trophoblast stem cell model. Depletion of MSX2 resulted in activation of the syncytiotrophoblast transcriptional program, while forced expression of MSX2 blocked it. We demonstrated that a large proportion of the affected genes were directly bound and regulated by MSX2 and identified components of the SWItch/Sucrose nonfermentable (SWI/SNF) complex as strong MSX2 interactors and target gene cobinders. MSX2 cooperated specifically with the SWI/SNF canonical BAF (cBAF) subcomplex and cooccupied, together with H3K27ac, a number of differentiation genes. Increased H3K27ac and cBAF occupancy upon MSX2 depletion imply that MSX2 prevents premature syncytiotrophoblast differentiation. Our findings established MSX2 as a repressor of the syncytiotrophoblast lineage and demonstrated its pivotal role in cell fate decisions that govern human placental development and disease.
The placenta is a vital organ that sustains mammalian development in utero. It provides an interface for gas, nutrients, and metabolite exchange and serves hormonal as well as immunological functions. The human placenta comprises three major trophoblast cell lineages: 1) the extravillous trophoblast (EVT) that invades the maternal decidua and remodels its spiral arteries, 2) the multinuclear syncytiotrophoblast (ST) that provides the site of exchange and produces placental hormones, and 3) the cytotrophoblast (CT), a multipotent progenitor population that, depending on the location, can give rise to EVT and ST (SI Appendix, Fig. S1A) (1). Precise and coordinated differentiation of these cell types is a prerequisite for a successful pregnancy. Failures in cell fate commitment, as well as impaired trophoblast development and function, may lead to placental disorders, including fetal growth restriction, miscarriage, and preeclampsia (2). However, the underlying molecular causes of these pathologies remain largely unknown.
The recent establishment of human trophoblast stem cells (hTSCs) from CT has the potential to revolutionize trophoblast research. hTSCs self-renew and are multipotent, as they can differentiate into both ST and EVT. Overall, global gene expression patterns, DNA methylation profiling, and functional analysis have demonstrated that hTSCs and their in vitro ST and EVT derivatives faithfully recapitulate the in vivo counterparts and provide an excellent model to study molecular mechanisms driving human placental development and disease (3).
Placental growth and development are orchestrated by the spatially and temporally coordinated actions of various transcription factors (TFs). Even though murine and human placentas exert equivalent functions, they are morphologically different, and their governing TF networks overlap only partially. While TEA domain transcription factor 4 (TEAD4), GATA binding protein 3 (GATA3), caudal type homeobox 2 (CDX2), E74-like ETS transcription factor 5 (ELF5), and transcription factor AP-2 gamma (TFAP2C) are expressed in both mouse TSCs (mTSCs) and hTSCs, and their respective in vivo counterparts, eomesodermin (Eomes) and estrogen-related receptor beta (Esrrb), are expressed exclusively in mouse (4, 5). Both mouse and human placentas comprise STs resulting from the fusion of progenitor cells and constitute the direct mother–embryo interface. However, the molecular mechanisms, particularly the TF networks, driving ST development and function are species specific and remain elusive (4, 5).
To identify the human-specific TF operating in the CT progenitor layer of the placenta, we screened the available gene expression datasets for TFs present in CT and absent from ST and EVT lineages. We used the hTSC in vitro model combined with functional genetics, chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq), and immunoprecipitation followed by mass spectrometry to functionally test a candidate and illuminate its mode of action. As a result, we uncovered MSX2 as a central regulator of cell fate decisions in the human trophoblast and provided molecular insights into the underlying mechanisms.
Results
Depletion of MSX2 Results in Loss of Silencing of Syncytiotrophoblast Genes.
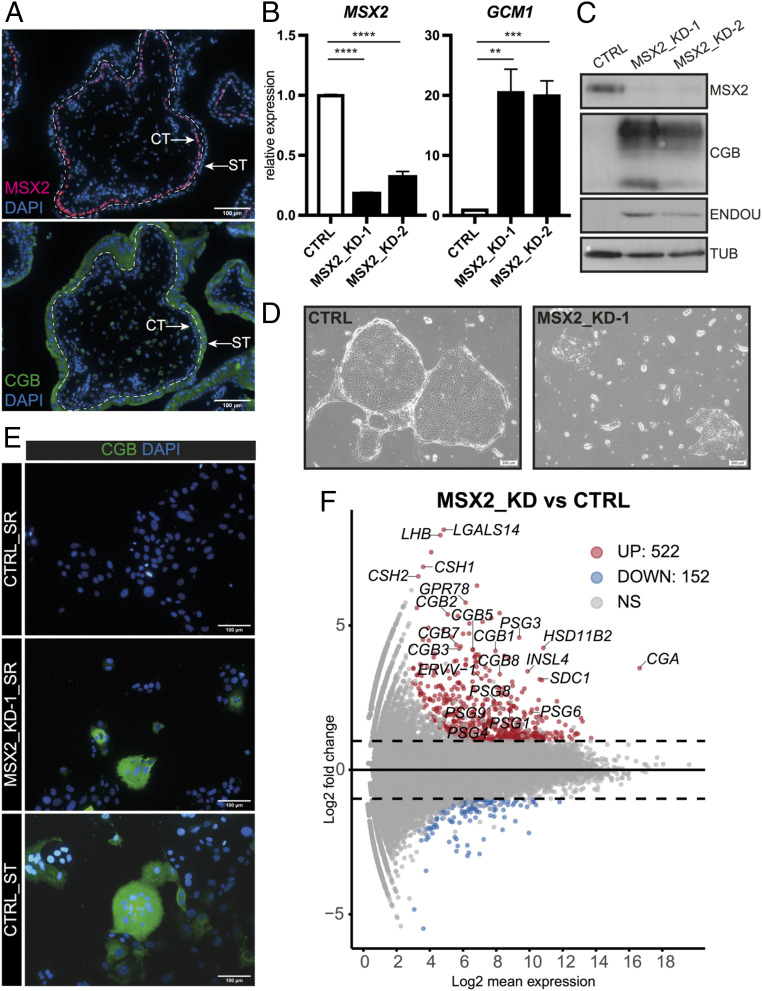
Our previous RNA profiling revealed that the TF MSX2 is highly expressed in CT and down-regulated upon in vitro ST differentiation, prompting us to address its role in the human trophoblast (6). First, we demonstrated that MSX2 is specific to the human trophoblast, compared to the mouse (SI Appendix, Fig. S1B). Next, we examined the expression of MSX2 in the first-trimester human placenta and showed that it was expressed in CT but not in the ST compartment, as indicated by costaining with the ST marker chorionic gonadotropin beta (CGB) (Fig. 1A). Similarly, MSX2 was highly expressed in hTSCs and down-regulated upon ST differentiation induced by 2 µM forskolin (SI Appendix, Fig. S1C), demonstrating hTSCs as a reliable model to address the role of MSX2 in the human placenta. To determine its function, we attempted to knock out MSX2 in hTSCs but could not retrieve clones, possibly due to the essential role of MSX2. Therefore, we depleted MSX2 using two different short hairpin (sh) RNAs (MSX2_KD-1 and MSX2_KD-2) along with a control shRNA (CTRL) by lentiviral transduction. MSX2 transcript levels were reduced in the MSX2_KD-1 and MSX2_KD-2 lines by up to 80%, and these results were confirmed on the protein level (Fig. 1 B and C). Strikingly, while the control cells formed tight, epithelial colonies, the MSX2_KDs gradually lost the hTSC morphology and proliferative capacity, indicating spontaneous differentiation (Fig. 1D). Expression analysis revealed a substantial up-regulation of ST markers including CGB, glial cells missing transcription factor 1 (GCM1), and endonuclease, poly(U) specific (ENDOU) despite culture conditions supporting hTSC self-renewal (SR) (Fig. 1 B, C, and E). We observed pockets of multinucleated structures that secreted CGB and had reduced levels of zonula occludens-1 (ZO-1), features typical for ST (Fig. 1E and SI Appendix, Fig. S1D). These effects were specific to MSX2 depletion, as the doxycycline (dox)-inducible expression of the mouse Msx2 coding sequence, which is resistant to the shRNAs targeting human MSX2, fully rescued the MSX2_KD phenotype (SI Appendix, Fig. S1 E and F).
Fig. 1.
Depletion of MSX2 results in loss of silencing of ST genes. (A) Immunofluorescence staining of a first-trimester human placenta for MSX2 and CGB, counterstained with DAPI. A dashed line separates ST from CT. (B and C) RT-qPCR (B) Western blot (C) expression and analysis of CTRL and MSX2-depleted (MSX2_KD-1 and MSX2_KD-2) hTSC lines cultured in stem cell conditions for MSX2 and ST markers GCM1, CGB, and ENDOU. (B) The bars represent a mean of four (n = 4) biological replicates with SEM ****P < 0.0001, ***P < 0.001, **P < 0.01. (C) Tubulin (TUB) serves as a loading control. (D) Phase contrast microscope images of hTSCs 10 d after viral transduction with MSX2_KD1 or CTRL constructs. (E) Immunofluorescence staining for CGB of control (CTRL_SR) and MSX2-depleted (MSX2_KD-1_SR) hTSC lines cultured in SR conditions and CTRL after 4 d of ST differentiation (CTRL_ST); DAPI indicates nuclei. (F) MA plot from differentially expressed genes between MSX2-depleted (MSX2_KD1 [n = 3] and MSX2_KD2 [n = 1]) and CTRL (n = 3). Analysis (cutoff: |log2FC| > 1, p adj < 0.05) revealed 522 up-regulated and 152 down-regulated genes in MSX2_KD lines compared to CTRL. Significantly up- (UP) and down-regulated (DOWN) genes are indicated in red and blue, respectively. Dashed black line highlights |log2FC| > 1. NS: not significant.
To gain a global overview of gene expression changes caused by depletion of MSX2, we performed an RNA sequencing (RNA-seq) analysis on MSX2_KD-1, MSX2_KD-2, and CTRL cells. We identified 152 down-regulated and 522 up-regulated genes (cutoff: |log2FC| > 1, adjusted P value [p adj] < 0.05), in line with the reported function of MSX2 as a transcriptional repressor (7) (Fig. 1F and SI Appendix, Fig. S1G). Among the up-regulated genes, we noted numerous ST markers including glycoprotein hormones, alpha polypeptide (CGA), chorionic gonadotropin subunit beta 1 (2, 3, 5, 7, 8) [CGB1 (2, 3, 5, 7, 8)], luteinizing hormone subunit beta (LHB), pregnancy specific beta-1-glycoprotein 1 (3, 4, 6, 8, 9) [PSG1 (3, 4, 6, 8, 9)], chorionic somatomammotropin hormone 1 (CSH1), chorionic somatomammotropin hormone 2 (CSH2), endogenous retrovirus group V member 1, envelope (ERVV-1), endogenous retrovirus group V member 2, envelope (ERVV-2), insulin like 4 (INSL4), syndecan 1 (SDC1), hydroxysteroid 11-beta dehydrogenase 2 (HSD11B2), G protein-coupled receptor 78 (GPR78), galectin 14 (LGALS14), solute carrier family 6 member 2 (SLC6A2), and cytochrome P450 family 19 subfamily A member 1 (CYP19A1) (Fig. 1F and SI Appendix, Fig. S1H). Importantly, their expression levels were comparable to the in vitro differentiated ST (SI Appendix, Fig. S1H). Gene ontology (GO) term enrichment analysis revealed enrichment of biological processes related to female pregnancy, hormone and steroid metabolism, and regulation of glycoprotein biosynthesis among the up-regulated genes and in terms pertaining to epithelium development and monocarboxylic acid metabolic process among the down-regulated genes (SI Appendix, Fig. S1I). Taken together, MSX2 is required for hTSC self-renewal and its depletion results in the massive loss of silencing of ST genes, suggesting that MSX2 prevents access to ST fate.
Ectopic Expression of MSX2 Blocks Syncytiotrophoblast Cell Fate.
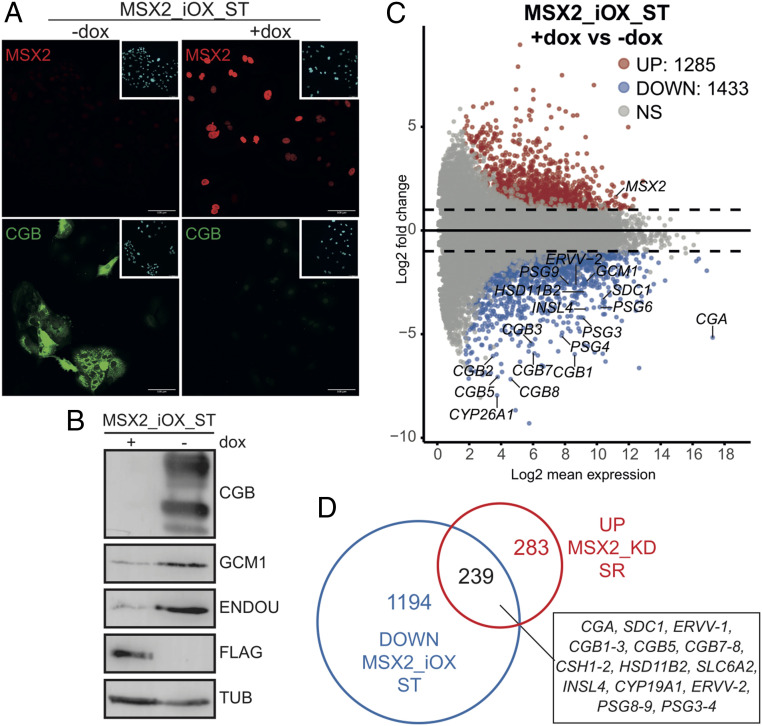
As depletion of MSX2 leads to derepression of ST genes, supporting the role of MSX2 as a transcriptional repressor, we reasoned that forced expression of MSX2 would block ST differentiation. To test this hypothesis, we cloned a Flag-tagged human MSX2 protein-coding sequence under the control of a dox-inducible promoter, generated a stable hTSC line (MSX2_iOX) (SI Appendix, Fig. S2A) and concomitantly induced ST differentiation and ectopic expression of MSX2 using forskolin and dox, respectively. After a 72-h treatment, we observed a robust formation of syncytia and CGB expression in nontreated cells and lack thereof in dox-treated, MSX2-expressing cells (Fig. 2A). Instead, they formed large, flattened colonies with clearly visible cell borders (SI Appendix, Fig. S2C). Gene expression analysis revealed induction of ST markers including GCM1, SDC1, ERVW-1, ENDOU, and CGB in control but not in the dox-treated MSX2_iOX cells (Fig. 2B and SI Appendix, Fig. S2B). To examine the global gene expression changes, we performed RNA-seq on untreated and dox-treated MSX2_iOX cells cultured in SR and for 6 d in ST conditions. Our analysis confirmed the down-regulation of self-renewal markers (TEAD4, TP63, and ELF5) upon ST induction in both −dox and +dox. Interestingly, the expected differentiation-induced up-regulation of a variety of ST markers including CGA, CGB1 (2, 3, 5, 7, 8), PSG1 (3, 4, 6, 8, 9), ERVV-1, ERVV-2, GCM1, SDC1, CYP26A1, HSD11B2, and others was only observed in −dox but not in +dox treatment (Fig. 2C and SI Appendix, Fig. S2 D and E). We identified 1,285 up-regulated and 1,433 down-regulated genes in +dox versus −dox MSX2_iOX ST cells (cut off: |log2FC| > 1, p adj < 0.05) (Fig. 2C and SI Appendix, Fig. S2D). The GO term enrichment analysis of the down-regulated genes confirmed enrichment for terms related to female pregnancy, response to peptide hormone, amino acid transport, and embryonic placenta development, whereas of the up-regulated genes, in terms related to the regulation of cell division and epithelial development (SI Appendix, Fig. S2F). Since we demonstrated that depletion of MSX2 led to spontaneous ST differentiation in SR conditions and, conversely, forced expression of MSX2 prevented ST differentiation, we sought to identify a shared set of misregulated genes. Based on the RNA-seq datasets, we compared genes that were up-regulated upon MSX2 depletion in SR conditions to those down-regulated upon MSX2 forced expression in ST conditions and revealed that 239 genes were shared (i.e., commonly misregulated) (Fig. 2D). These results demonstrate that MSX2 acts as a strong transcriptional repressor that presides over a network of ST-specific genes and regulates key cell fate decisions during trophoblast differentiation.
Fig. 2.
Ectopic expression of MSX2 blocks ST cell fate. (A) Immunofluorescence staining for MSX2 and CGB in hTSC line carrying a dox-inducible MSX2 transgene (MSX2_iOX) and differentiated to ST in the presence (+dox) or absence (−dox) of dox. (B) Western blot analysis for ST markers CGB, GCM1, and ENDOU in hTSC line carrying a dox-inducible MSX2 transgene and differentiated to ST in the presence (+dox) or absence (−dox) of dox. The MSX2 transgene carries a 3xFlag-tag; tubulin was used as a loading control. (C) MA plot from differential expressed genes between MSX2_iOX ST +dox (n = 2) and MSX2_iOX ST −dox (n = 3). Analysis (cutoff: |log2FC| > 1, p adj < 0.05) revealed 1,285 up-regulated and 1,433 down-regulated genes in MSX2_iOX ST +dox compared to MSX2_iOX ST−dox. Significantly up- (UP) and down-regulated (DOWN) genes are indicated in red and blue, respectively. Dashed black line highlights log fold changes of −2 and 2. NS: not significant. (D) Venn diagram showing overlap of genes that were up-regulated upon MSX2 depletion (MSX2_KD) in SR conditions (compared to KD control) and genes that were down-regulated (i.e., did not get up-regulated) upon induced expression of MSX2 transgene (MSX2_iOX) during ST differentiation (compared to −dox CTRL). The Venn diagram is based on the RNA-seq analysis detailed in Figs.1F and 2C.
MSX2 Binds and Silences Syncytiotrophoblast Genes in hTSCs.
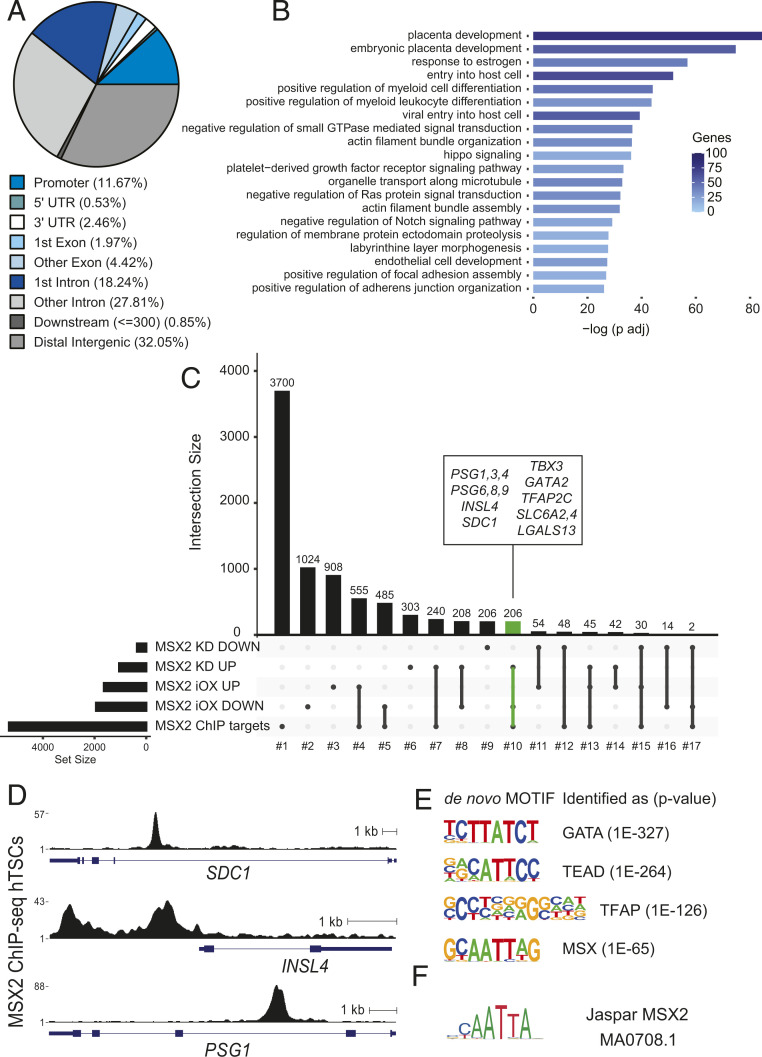
To explore which of the genes that are misregulated upon genetic manipulations of MSX2 are directly bound and regulated by MSX2, we performed ChIP-seq in hTSCs. We uncovered 11,494 MSX2 binding sites, of which nearly 12% were located in promoters, 46% in intronic regions, and 32% in distal intergenic areas, suggesting enrichment of enhancers among MSX2 peaks (Fig. 3A). The Genomic Regions Enrichment of Annotations Tool (GREAT) analysis of the MSX2-bound regions revealed enrichment for terms related to placental development, response to estrogen, HIPPO signaling, and negative regulation of both NOTCH and MAPK signaling, among others (Fig. 3B). To further narrow down the MSX2 functional targets, we overlaid genes misregulated upon MSX2 depletion or overexpression during ST differentiation with those bound by MSX2. The analysis provided us with 17 gene sets illustrating the various links between genes bound by MSX2 and misregulated upon MSX2 perturbations (Fig. 3C and SI Appendix, Fig. S3 A and B). The functional enrichment analysis of the resulting gene sets showed, for instance, an enrichment of the Neurotrophin and EGFR pathways among MSX2 targets that were only down-regulated (set No. 5) in MSX2iOX STs, suggesting important regulation of those pathways during the ST differentiation. Among MSX2 targets that were only up-regulated in MSX2_KD (set No. 7), hTSC enrichment for vitamin C metabolism may suggest its relevance during the exit from hTSC multipotency (SI Appendix, Fig. S3C). Interestingly, the 206 MSX2 targets that were both up-regulated in MSX2_KD cells and down-regulated upon MSX2 overexpression during ST differentiation (set No. 10) were enriched for the terms female pregnancy and PI3K/Akt pathway components (SI Appendix, Fig. S3C) and contained many genes associated with ST, including the PSG family, INSL4, SDC1, T-box transcription factor 3 (TBX3), solute carrier family 6 member 4 (SLC6A4), SLC6A2, (SLC40A1), aldo-keto reductase family 1 member B (AKR1B1), NUCB2 (nucleobindin 2), galectin 13 (LGALS13), protein kinase C zeta (PRKCZ) and interleukin 1 receptor type 1 (IL1R1) (Fig. 3 C and D). These genes are direct targets of MSX2 and are associated with the ST lineage, but their precise functional role has yet to be determined.
Fig. 3.
MSX2 binds and silences ST genes in hTSC. (A) Proportion of MSX2 ChIP-seq peaks overlapping genomic features in hTSC. (B) GO term enrichment of MSX2 peaks. (C) Upset plot depicting the overlap between genes bound by MSX2 (identified by ChIP-seq) and genes that were deregulated upon MSX2 depletion or upon forced MSX2 expression during ST differentiation (based on RNA-seq analysis). Genes that are MSX2 bound and affected by either perturbation are highlighted in green. Names are indicated in the inset. (D) RPKM normalized MSX2 binding profiles at selected loci identified in C. (E) De novo motifs identified by HOMER. (F) Matching of the de novo motif similar to MSX2 with the deposited MSX2 motif from Jaspar.
To gain better molecular insights into the MSX2-driven gene regulation, we performed de novo motif analysis using HOMER. The top four identified motifs shared high similarity with the GATA, TEAD, TFAP, and MSX motifs (Fig. 3E). Comparison of the de novo motif for MSX with published MSX2 motifs (8) confirmed its identity (Fig. 3F). Moreover, our data suggest extending the currently described human MSX2 motif (8) by a flanking G/C nucleotide on each side (Fig. 3 E and F). A specific search identified MSX2 de novo motif instances (Fig. 3E) in 69% of the significantly reproducible MSX2 peaks (P value 1E-9, SI Appendix, Fig. S3D), further validating the motif search and supporting the specificity of the ChIP-seq data. Furthermore, known motifs for GATA3, TEAD4, and TFAP2C were strongly enriched in these MSX2 peaks (SI Appendix, Fig. S3E), suggesting potential cooperation between those factors and MSX2. Several GATA, TFAP2, and TEAD TF family members are key trophoblast regulators in mice and humans (5). TFAP2C, TFAP2A, GATA3, and GATA2 were reported to cooperatively regulate early trophoblast specification in vitro (9). Similarly, TEAD4 is a crucial controller of the CT identity, as its depletion in hTSCs results in loss of self-renewal and differentiation (9). Overall, our findings highlight MSX2 as a key regulator within the TF network of human trophoblast development.
MSX2 Cooperates with Components of the SWI/SNF Complex.
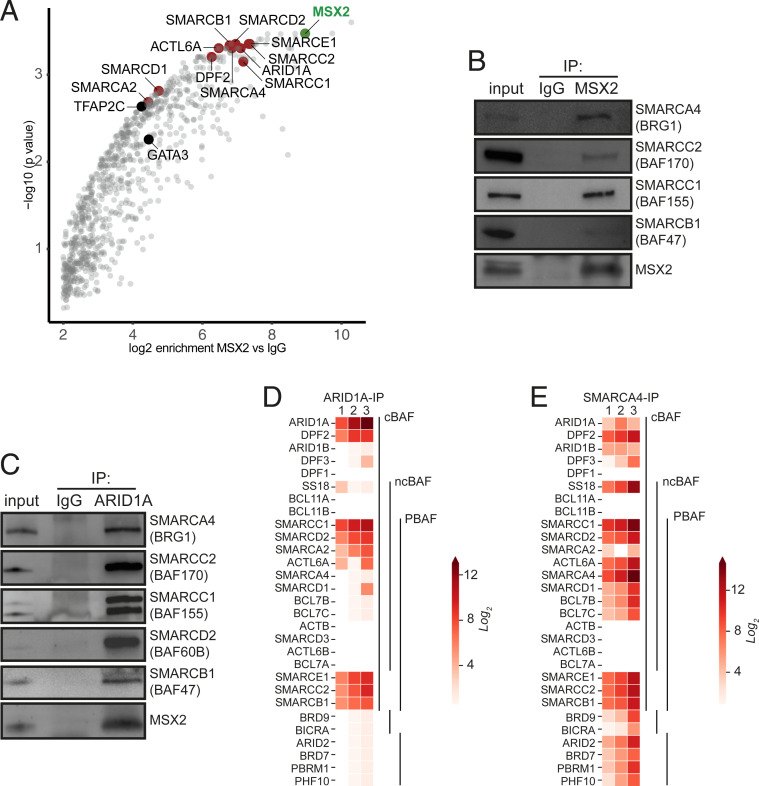
To control gene expression, TFs usually cooperate with chromatin-modifying and remodeling complexes as well as with other TFs. To gain new molecular insights into the MSX2 mode of action, we set out to determine its interactome by rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) (10). Using this unbiased protein identification approach, we uncovered MSX2 in addition to many high-confidence interaction partners (Fig. 4A and SI Appendix, Fig. S4A). Among them were the key trophoblast regulators GATA3 and TFAP2C. These interactions, together with our previous findings that both GATA3 and TFAP2C DNA binding motifs are overrepresented in the MSX2 ChIP-Seq peaks, raise the exciting possibility that these three factors closely cooperate to regulate transcriptional outputs in hTSCs. Strikingly, we also identified numerous components of the mammalian SWI/SNF complex as robust MSX2 interactors: SMARCA4, SMARCA2, SMARCC2, SMARCC1, SMARCB1, SMARCE1, ARID1A, SMARCD1, SMARCD2, DPF2, and ACTL6A but no other protein complexes (Fig. 4A and SI Appendix, Fig. S4A). SWI/SNF is a chromatin remodeling complex that, upon recruitment by TFs, shifts nucleosomes along the DNA and modifies chromatin accessibility, resulting in context-dependent transcriptional activation or repression (11–13). We confirmed several of these interactions by coimmunoprecipitation followed by Western blot analysis (Fig. 4 B and C). The mammalian SWI/SNF encompasses 29 subunits that assemble into three distinct complexes: canonical BAF (cBAF), polybromo-associated BAF (PBAF), and noncanonical BAF (ncBAF), each of which comprises common as well as specific subunits. The combinatorial assemblies of specific subunits, including numerous paralogs, result in unique subcomplexes with cell-type–specific functions (11–13). Based on the detected subunits, our results indicate that MSX2 cooperates specifically with the cBAF complex. To get an overview of all SWI/SNF complexes operating in hTSCs and to shed light on their paralog composition, we performed RIME for the SMARCA4 (BRG1) and ARID1A subunits. SMARCA4 is the ATPase subunit shared by all three complexes, while ARID1A is specific to cBAF. Our results confirmed ARID1A as part of the cBAF complex and provided insights into its specific composition in hTSCs (SMARCC1, DPF2, SMARCE1, SMARCD2, SMARCC2, SMARCB1, SMARCA2, and ACTL6A) (Fig. 4D and SI Appendix, Fig. S4B). Interestingly, we observed that the ARID1A interaction with SMARCA2 (BRM) was much stronger compared to SMARCA4 (BRG1) (Fig. 4D and SI Appendix, Fig. S4B). The SMARCA4 interactome analysis demonstrated that besides cBAF, SMARCA4 interacts with specific components of the PBAF complex (ARID2, PBRM1, PHF10, and BRD7) as well as the ncBAF complex (BRD9 and BICRA) (Fig. 4E and SI Appendix, Fig. S4 C and D). In addition to subcomplexes, we also identified different subunit paralogs that are incorporated in a mutually exclusive manner, such as SMARCA4 (BRG1) and SMARCA2 (BRM), ARID1A and ARID1B, and DPF2 and DPF3, as well as SMARCD1 and SMARCD2.
Fig. 4.
MSX2 interacts with components of the cBAF complex. (A) MSX2 interactome identified in hTSC. Red and black mark selected MSX2 binding partners: components of the cBAF complex and prominent TFs, respectively. MSX2 is marked in green. The analysis is based on three biological replicates; IgG was used as CTRL. (B) MSX2 immunoprecipitates analyzed by Western blot probed with anti-MSX2, anti-SMARCA4 (BRG1), anti-SMARCC2 (BAF170), anti-SMARCC1 (BAF155), and anti-SMARCB1 (BAF47/INI1), confirming the prominent interaction between MSX2 and cBAF complex components. (C) ARID1A immunoprecipitates analyzed by Western blot probed with anti-SMARCA4 (BRG1), anti-SMARCC2 (BAF170), anti-SMARCC1 (BAF155), anti-SMARCD2 (BAF60B), anti-SMARCB1 (BAF47), and anti-MSX2. (D) Heat map representing the log2 fold change ratio of ARID1A and IgG normalized areas of SWI/SNF components identified by mass spectrometry in ARID1A immunoprecipitates. (E) Heat map representing the log2 fold change ratio of SMARCA4 and IgG normalized areas of SWI/SNF components identified by mass spectrometry in SMARCA4 immunoprecipitates.
Taken together, we identified the cBAF complex as a strong MSX2 interactor, showed that cBAF, PBAF, and ncBAF complexes operate in hTSCs, and illuminated another layer of complexity in their paralog composition.
MSX2-cBAF Cobound Regions Display Active but Not Repressive Histone Marks.
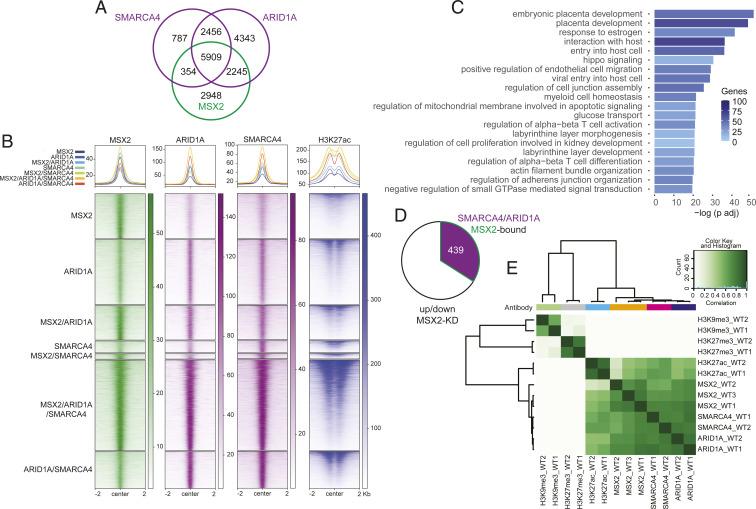
The interaction between MSX2 and the cBAF complex suggests that they may cobind and coregulate the expression of shared target genes. To test this, we performed ChIP-seq using antibodies against SMARCA4 and ARID1A. Feature distribution showed that over 40% of their binding sites were located +/−10 kb from the transcriptional start site, in agreement with cBAF occupying distal regulatory elements in other systems (14, 15) (SI Appendix, Fig. S5A). We then intersected the SMARCA4-, ARID1A-, and MSX2-bound regions and demonstrated that 5,909 were cooccupied by all three factors (Fig. 5 A and B). The GREAT analysis of these triple-bound regions revealed enrichment for terms related to placental development, response to estrogen, HIPPO signaling, and glucose transport (Fig. 5C), implying a functional link. We also identified 354 and 2,245 regions cooccupied only by SMARCA4/MSX2 and ARID1A/MSX2, respectively (Fig. 5 A and B). The latter likely represent regions cobound by MSX2 and the SMARCA2 (BRM)-specific cBAF subcomplex. In addition, 2,948 regions were bound exclusively by MSX2, indicating a distinct, cBAF-independent mode of regulation. Finally, 2,456 regions were cooccupied by SMARCA4 and ARID1A but not by MSX2 (Fig. 5 A and B). Next, we assigned regions to genes and asked how many of the genes cooccupied by SMARCA4/ARID1A/MSX2 were misregulated upon MSX2 depletion. By intersecting the corresponding ChIP-seq and RNA-seq datasets, we identified 439 genes that likely represent the direct targets of MSX2/cBAF (Fig. 5D). Importantly, this group contained ST-related genes, including PSG family members, INSL4, SDC1, TBX3, PGF, CYP19A1, SLC6A2, and SLC6A4 among others, implying coregulation by MSX2 and cBAF. In summary, MSX2 interacts with the cBAF complex and cobinds a large proportion of genomic regions and MSX2-dependent genes.
Fig. 5.
MSX2 and cBAF complex cobind trophoblast genes marked by H3K27ac. (A) Venn diagram depicting the overlap between MSX2-, ARID1A-, and SMARCA4-bound regions in hTSCs based on irreproducible discovery rate (IDR) analysis. (B) Heat map of the RPKM normalized ChIP-seq signal of MSX2, ARID1A, SMARCA4, and H3K27ac in regions defined by the peak overlaps of A and S5C. The H3K27ac signal is additionally normalized to drosophila spike-in chromatin. (C) GO term enrichment of the MSX2, ARID1A, and SMARCA4 cooccupied regions. (D) Pie chart of genes deregulated upon MSX2 depletion that are also bound by MSX2, ARID1A, and SMARCA4. (E) Correlation heat map of ChIP-seq counts of MSX2, ARID1A, SMARCA4, H3K27ac, and the repressive histone marks H3K27me3 and H3K9me3.
Control of gene expression by TFs often involves regulation of chromatin accessibility and structure. Active chromatin regulatory regions typically display the H3K27ac and H3K4me3 histone modifications, while repressive chromatin correlates with H3K27me3 and H3K9me3. To determine the chromatin status of regions bound by MSX2 and cBAF in hTSCs and gain insights into the molecular mechanism, we performed ChIP-seq using anti-H3K27ac, anti-H3K27me3, and anti-H3K9me3 antibodies. The analysis revealed virtually no overlap between MSX2-, SMARCA4-, and ARID1-bound regions with either H3K27me3 or H3K9me3 (Fig. 5E and SI Appendix, Fig. S5B). The lack of correlation between H3K27me3 and MSX2-cBAF confirms the overall antagonistic relationship between the mammalian SWI/SNF complexes and the polycomb repressive complex 2 (PRC2), which deposits the H3K27me3 mark (16–18). Similarly, in agreement with previous reports in other systems (19), the general overlap between H3K27me3 and H3K9me3 repressive modifications was limited in hTSCs. In stark contrast, the active H3K27ac mark displayed a strong association with single, double, and triple MSX2, SMARCA4, and ARID1 peaks (Fig. 5 B and E and SI Appendix, Fig. S5C). The SWI/SNF complexes are known to bind to enhancers, regulate H3K27ac deposition therein, and control the expression of target genes (14, 18, 20). Whether and to what extent the H3K27ac regions we identified here act as enhancers will require further characterization. Taken together, consistent with the mass spectrometry data, MSX2 and cBAF components show a high overlap of genome occupancy that correlates with H3K27ac but not with silent chromatin histone marks.
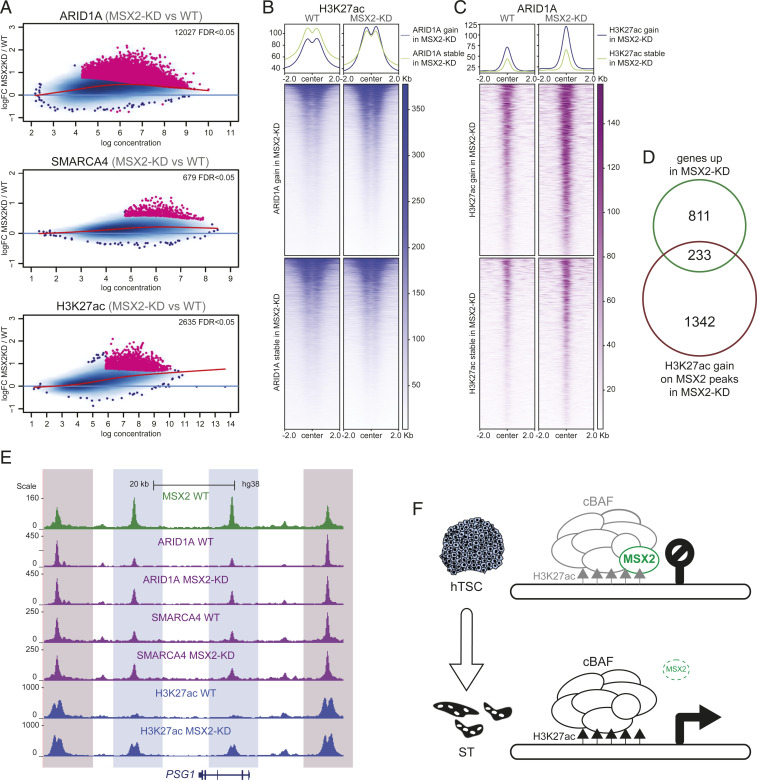
Depletion of MSX2 Leads to an Increase in cBAF Occupancy and H3K27ac Levels.
We demonstrated that MSX2 controls transcription of a subset of genes and cooperatively binds target regions with the cBAF complex. These observations raised the possibility that MSX2 could recruit the cBAF complex to shared target genes. To test this, we depleted MSX2 and profiled the genome-wide binding sites of SMARCA4 and ARID1 in MSX2_KD-1 and wild-type (WT) hTSCs by ChIP-seq. Surprisingly, the differential binding analysis of all MSX2 target regions revealed that binding of ARID1A and SMARCA4 did not diminish but considerably increased upon depletion of MSX2 (Fig. 6A). We next asked whether the increase in occupancy by the cBAF components was accompanied by changes in levels of histone modifications. The differential binding analysis demonstrated that levels of H3K27ac dramatically increased at the MSX2 target regions upon depletion of MSX2 (Fig. 6 A–C and SI Appendix, Fig. S6 A–D). MSX2-bound regions that gained ARID1A binding upon MSX2 depletion, but not those that did not, simultaneously exhibited increased H3K27ac in MSX2-KD hTSCs (Fig. 6B). Similarly, MSX2-bound regions that gained SMARCA4 binding upon MSX2 depletion also showed an increase in H3K27ac (SI Appendix, Fig. S6A). In agreement with this, MSX2 peaks that gained H3K27ac upon MSX2 depletion simultaneously showed increased ARID1A and SMARCA4 occupancy in MSX2-KD hTSCs (Fig. 6C and SI Appendix, Fig. S6B). A significant proportion of the MSX2 peaks that gain H3K27ac upon MSX2 depletion correspond to genes that were up-regulated upon MSX2 depletion (Fig. 6D). This group contained syncytiotrophoblast-related genes including PSG family members, TBX3, SDC1, and SLC6A4. For instance, in the PSG gene cluster, while depletion of MSX2 resulted in a gain of cBAF/H3K27ac occupancy in some regions, binding in others remained stable, highlighting their functional link (Fig. 6E and SI Appendix, Fig. S6E). Taken together, these results suggest that removal of MSX2 unleashes cBAF-mediated activation of syncytiotrophoblast genes. In summary, all the presented evidence is consistent with the model that MSX2 reinforces the hTSC state by attenuating cBAF-driven trophoblast differentiation (Fig. 6F).
Fig. 6.
MSX2 depletion leads to an increase in both cBAF occupation and H3K27ac signal. (A) MA plots of MSX2 binding sites, differentially enriched for ARID1A, SMARCA4, and H3K27ac in MSX2-KD versus WT. Log2 fold change is plotted as a function of the log normalized ChIP-seq reads. (Top) ARID1A, (Middle) SMARCA4, (Bottom) H3K27ac. (B) Heat map and profile plot of the H3K27ac ChIP-seq signal at MSX2-bound regions differentially enriched for ARID1A in MSX2-KD versus WT hTSCs and compared to an equally sized set of random MSX2-bound regions. (C) Heat map and profile plot of the ARID1A ChIP-seq signal at MSX2-bound regions differentially enriched for H3K27ac in MSX2-KD versus WT hTSCs compared to an equally sized set of random MSX2-bound regions. (D) Venn diagram depicting the overlap between genes up-regulated upon MSX2 depletion (MSX2-KD) and those that gain H3K27ac on MSX2 peaks in MSX2-KD. (E) Genome browser tracks of MSX2, ARID1A, SMARCA4, and H3K27ac signal in WT and MSX2-KD hTSCs at the PSG1 locus. MSX2: green, ARID1A and SMARCA4: purple, H3K27ac: blue. Differentially enriched regions are highlighted in light blue, and regions without change are highlighted in light red. (F) Model: in self-renewing hTSCs, MSX2 attenuates the cBAF complex to prevent premature activation of differentiation genes. MSX2 depletion unleashes the cBAF complex and H3K27Ac at differentiation genes leading to ST specification.
Discussion
During the first trimester of pregnancy, successful placentation relies on a delicate balance between self-renewal of multipotent CT progenitors and their timely and coordinated differentiation to ST and EVT lineages. Importantly, studies on transcriptional mechanisms controlling this precarious balance are scarce and focus mainly on transcriptional activators. Here, we identified MSX2 as the key human-specific determinant of CT versus ST cell identity. Our studies demonstrate that the primary function of MSX2 is that of a transcriptional repressor promoting stemness and self-renewal indirectly by preventing premature expression of inappropriate ST lineage markers in CT cells. The combined RNA-seq and ChIP-seq results did not indicate that MSX2 positively regulates the expression of known stemness/self-renewal factors. Therefore, we propose MSX2 as a repressor of the ST lineage.
Previous reports demonstrated that MSX2/Msx2 deficiency results in impaired osteogenesis, chondrogenesis, and defective tooth, hair follicle, and mammary gland development (21–23). A common theme of these diverse phenotypes was faulty progenitor proliferation and their imbalanced differentiation, consistent with our observations that depletion of MSX2 in hTSCs resulted in precocious ST differentiation. These findings have important implications, as the underlying causes of various placental pathologies, including severe forms of fetal growth restriction and preeclampsia, may have their roots in defective ST development and function.
Our observations that forced expression of MSX2 blocks ST differentiation align with previous reports showing that ectopic expression of mouse Msx1 and Msx2 negatively regulates differentiation in multiple mesenchymal and epithelial cell types including muscle, adipocytes, cartilage, bone, and mammary gland epithelium (24). Consistent with its function as an inhibitor of differentiation, increased expression of MSX2 was detected in several cancers and correlated with tumor invasiveness (25). The role of MSX2 as a strong determinant of cell identity was also demonstrated in human embryonic stem cells (hESCs), in which forced expression of MSX2 resulted in precocious mesendoderm differentiation. Conversely, MSX2-deficient hESCs were severely impaired in mesendodermal differentiation (26). Likewise, MSX2 was reported to initiate and accelerate a molecular program driving mesenchymal stem/stromal cell specification (27). Taken together, these observations suggest that, independently of the context, expression of MSX2/Msx2 marks a transient population of multipotent, proliferative progenitors that, as development progresses and MSX2/Msx2 expression vanishes, will further differentiate into specialized cell types.
We found that regions bound by MSX2 contain the DNA binding motifs for GATA3, TFAP2C, and TEAD4 TFs and that both GATA3 and TFAP2C are MSX2 interactors, suggesting close cooperation between these CT-expressed factors (28, 29). In the mouse, Tfap2c, Tead4, and Gata3 are critical transcriptional regulators of trophoblast identity that preside over and partially coregulate networks of trophoblast genes in mouse TSCs, and their depletion leads to loss of self-renewal and differentiation (9, 28, 30–33). Interestingly, TEAD4 and its cofactor YAP1 were recently demonstrated to reinforce stemness and prevent ST differentiation in hTSCs (9, 34). Thus, while TEAD4 and YAP1 have a dual role as transcriptional activators of proliferation genes and repressors of differentiation genes (9, 34), MSX2 appears to primarily prevent ST gene up-regulation.
Our mass spectrometry analysis identified numerous components of the cBAF chromatin remodeling complex as the most prominent interactors of MSX2. The mammalian SWI/SNF complexes cBAF, PBAF, and ncBAF are characterized by different subunit compositions. The resulting heterogeneity ensures functional and context-dependent diversity in driving lineage-specific gene expression programs as exemplified by the unique, stage-specific SWI/SNF complexes found in ESCs, neural progenitors, postmitotic neurons, cardiac lineage, and cancer (12, 13, 35–37). Our RIME analysis for MSX2, ARID1A, and SMARCA4 revealed that cBAF, PBAF (specific subunits: ARID2, PBRM1, PH10, and BRD7), and ncBAF (specific subunits: BRD9 and BICRA) complexes exist in hTSCs, but only cBAF cooperates with MSX2. Interestingly, both PBAF and ncBAF complexes operate in mESCs, but their specific subunits were not detected in hESCs (38–40). The subunit composition of the hTSC cBAF complex is similar to the one operating in hESCs (except for the BRM subunit) and distinct from the one in mESCs (35, 40). Notably, alternative variants (paralogs) exist for several of the subunits, and only one of them can be incorporated into the complex at a given polymorphic position. For instance, we detected both SMARCA4 and SMARCA2, ARID1A and ARID1B, and DPF2 and DPF3, as well as SMARCD1 and SMARCD2. However, each pair is incorporated into the complex mutually exclusively, indicating that several different subcomplexes operate in hTSCs. Further thorough biochemical and functional analyses are required to dissect their specific functions.
We found that MSX2 and the cBAF complex interact but also cobind a large proportion of target genes. Among the regulated gene targets, we identified established regulators of ST fate GATA2 and TBX3 TF (41, 42), as well as pregnancy hormones including PSG family, PGF, INSL4, and LGALS13, highlighting the importance of MSX2-cBAF for the development of the human placenta. MSX2/SMARCA4/ARID1A cobound peaks are largely devoid of H3K27me3 and H3K9me3 in hTSCs. These observations are in agreement with findings in mESCs, in which cBAF and the H3K27me3-associated PRC2 complex play primarily opposing roles but also cooperate to silence a small group of genes (16, 17, 37). Similarly, the considerable overlap between cBAF binding and H3K27ac is commonly observed, in particular at enhancer regions, and depletion of cBAF subunits results in a reduction of H3K27ac (14, 15, 20, 43). In contrast, in hESCs, cBAF was reported to preferentially bind poised enhancers devoid of H3K27ac, and depletion of SMARCA4 resulted in increased levels of H3K27ac, suggesting a role of cBAF in the repression of lineage-specific genes (40). While in hTSCs, the function of cBAF still needs to be determined by depletion experiments, our results are consistent with an activating role of cBAF during trophoblast differentiation.
Since the depletion of MSX2 resulted in an up-regulation of many genes cobound by MSX2 and cBAF, we tested whether MSX2 recruits cBAF to bring about the silencing of these target genes. However, loss of MSX2 instead increased both SMARCA4/ARID1A binding and levels of H3K27ac, indicating that MSX2 does not recruit cBAF to repress the targets. Our results, therefore, favor an alternative mechanism, by which cBAF binds genes that are poised for activation and MSX2 keeps their transcriptional induction in check (Fig. 6F). In vivo, in the floating villi, this mechanism could ensure a seamless conversion from the MSX2+ CT to the adjacent MSX2− ST lineage without a transition state. As disruption of individual cBAF subunits leads to diverse phenotypes, including mouse early embryonic lethality, it will be paramount to carefully tease apart their roles during human trophoblast development to test this model.
Taken together, our findings place the repressor MSX2 at the center of the human trophoblast specification process, link it to the cBAF chromatin remodeling complex, and uncover its vital role in the context of placental development and function.
Materials and Methods
Cell Culture.
hTSC lines CT30 and CT27 were a generous gift from Hiroaki Okae (Department of Informative Genetics Environment and Genome Research Center Tohoku University Graduate School of Medicine, Sendai, 980-8575, Japan). The cells were cultured, passaged, and differentiated as described (5) with minor modifications. For details, see SI Appendix.
Genetic Modifications.
Lentiviral knock-down was carried out as described (44); for details, see SI Appendix. To generate the inducible MSX2 expression construct, we cloned the coding sequence of MSX2, including the 3xFlag tag, into PiggyBac-Tre-Dest-rTA-HSV-neo (a kind gift from Joerg Betschinger, Friedrich Miescher Institute for Biomedical Research, Maulbeerstrasse 66, 4058 Basel, Switzerland). After transfection, the hTSCs were selected with 300 μg/mL G418 and expression was induced with 1 μg/mL dox.
Protein isolation and Western blot analyses.
Whole cell lysates were prepared with lysis buffer (20 mM Tris HCl pH 7.5, 137 mM NaCl, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 1% Triton X-100, 10% glycerol and 1.5mM MgCl2). The following primary antibodies were used: anti-MSX2 (HPA005652, Sigma-Aldrich, 1:800), anti-human Chorionic Gonadotropin (hCG) (A0231, Dako, 1:1,000), anti-ENDOU (HAP067448, Sigma-Aldrich, 1:1,000), anti-tubulin (ab6160, Abcam, 1:2,000), anti-FlagM2 (F1804, Sigma-Aldrich, 1:2,000), anti-GCM1 (HPA011343, Sigma-Aldrich, 1:1,000).
RT-qPCR.
RNA was extracted using the RNeasy Mini Kit (Qiagen) and treated with DNaseI (Qiagen) according to the manufacturer’s protocol. Complementary DNA was synthesized using 1.5 to 3 µg RNA primed with random hexamers according to the RevertAid Reverse Transcriptase protocol (Thermo Fisher Scientific, EP0442). DNA was diluted and qPCR performed using GoTaq qPCR Master Mix (A6002, Promega). Results are shown as means of the indicated number of biological replicates (n) ± SEM. Statistical significance was determined using a two-tailed, unpaired t test. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, ns: not significant. Primer sequences are provided in SI Appendix.
RNA-Seq.
RNA was extracted using the RNeasy mini kit (Qiagen) and treated with DNaseI (Qiagen). Indexed libraries were prepared with 500 ng RNA using QuantSEQ 3′ messenger RNA-Seq Library Prep Kit FWD for Illumina (015.96, Lexogen) according to the manufacturer’s recommendations. The libraries were pooled and sequenced with a 100-bp single-end (MSX2_iOX) and 50-bp single-end protocol on Illumina HiSEq 2500 sequencer. For details of the bioinformatic analysis, see SI Appendix.
Immunofluorescence.
Placental tissue (seventh week of gestation) was embedded in paraffin and processed as described (45). Utilization of tissues and all experimental procedures were approved by the Medical University of Vienna ethics boards (No. 084/2009) and required written informed consent. Cells were fixed in 4% paraformaldehyde/phosphate-buffered saline (PBS) for 20 min at 4 °C, permeabilized, and blocked for 30 min in 4% donkey serum and 0.1% Triton X-100 in PBS. The following primary antibodies with given dilutions were used: anti-MSX2 (HPA005652, Sigma-Aldrich, 1:250), hCG (A0231, Dako, 1:300), anti-ENDOU (HAP067448, Sigma-Aldrich, 1:250), and anti-ZO-1 (ab216880, Abcam, 1:1,000). Alexa Fluor–conjugated secondary antibodies (A-21206, A-21207, Invitrogen) were applied at 1:1,000 in 4% donkey serum and 0.1% Tween-20 in PBS blocking solution. Cells were counterstained with DAPI and imaged using Zeiss Imager A2 microscope with Zen 2012 software.
RIME.
RIME was carried out as described (10), using anti-MSX2 (HPA005652, Sigma-Aldrich), anti-ARID1A (12354, Cell Signaling) and anti-SMARCA4 (ab110641, Abcam) antibodies.
Coimmunoprecipitation.
Cells were harvested and resuspended in Hunt Buffer (20mM Tris HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40), followed by three freeze–thaw cycles for whole protein extraction. Protein G magnetic Dynabeads (10004D, Invitrogen) were preblocked with 1 mg/mL bovine serum albumin and 5 µg rabbit anti-MSX2 (HPA005652, Sigma-Aldrich), 10 µg anti-ARID1A (12354, Cell Signaling), anti-SMARCA4 (ab110641, Abcam) or rabbit normal Immunoglobulin G (IgG) (NI01, Sigma-Aldrich, and ab172730, Abcam) were conjugated to the beads for 1 h at room temperature. Affinity purification by prebound beads was performed with 1,000 µg proteins overnight at 4 °C with rotation. The next morning, beads were washed in Hunt Buffer and eluted. The following antibodies were used: anti-MSX2 (HPA005652, Sigma-Aldrich, 1:800), anti-SMARCA4 (sc-17796, Santa-Cruz, 1:1,000), anti-SMARCA4 (ab110641, Abcam, 1:1,000), anti-BAF170 (sc-17838, Santa-Cruz, 1:500), anti-BAF155 (sc-48350, Santa-Cruz, 1:500), anti-BAF47 (sc-166165, Santa-Cruz, 1:500), and anti-SMARCD2/BAF60B (sc-101162, Santa-Cruz, 1:500).
ChIP-Seq.
Immunoprecipitations were carried out as described (46) using anti-MSX2 (HPA005652, Sigma-Aldrich) anti-ARID1A (12354, Cell Signaling), anti-SMARCA4 (ab110641, Abcam), anti-trimethyl histone H3 (Lys27) (07-442, Merck Millipore), anti-trimethyl histone H3 (Lys9) (07-449, Merck Millipore), and anti-histone H3K27 (ACM-39685, Active Motif/THP) antibody; for details, including the bioinformatic analysis, see SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the Austrian Science Fund (Grant P-31738-B26 awarded to P.A.L. and M.K.). Sequencing was performed at the Vienna Biocenter Core Facilities Next-Generation Sequencing Unit. We are grateful to Sasha Mendjan and Malte Mederacke for valuable comments.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2105130118/-/DCSupplemental.
Data Availability
Raw and processed next-generation sequencing datasets were deposited at the National Center for Biotechnology Information Gene Expression Omnibus repository under accession number GSE165970. Materials, data, and associated protocols, including code and scripts, are available upon request.
References
- 1.Turco M. Y., Moffett A., Development of the human placenta. Development 146, dev163428 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Brosens I., Pijnenborg R., Vercruysse L., Romero R., The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 204, 193–201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okae H., et al., Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Latos P. A., Hemberger M., From the stem of the placental tree: Trophoblast stem cells and their progeny. Development 143, 3650–3660 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Hemberger M., Hanna C. W., Dean W., Mechanisms of early placental development in mouse and humans. Nat. Rev. Genet. 21, 27–43 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Haider S., et al., Self-renewing trophoblast organoids recapitulate the developmental program of the early human placenta. Stem Cell Reports 11, 537–551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catron K. M., et al., Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. Mol. Cell. Biol. 15, 861–871 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma L., Golden S., Wu L., Maxson R., The molecular basis of Boston-type craniosynostosis: The Pro148–>His mutation in the N-terminal arm of the MSX2 homeodomain stabilizes DNA binding without altering nucleotide sequence preferences. Hum. Mol. Genet. 5, 1915–1920 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Saha B., et al., TEAD4 ensures postimplantation development by promoting trophoblast self-renewal: An implication in early human pregnancy loss. Proc. Natl. Acad. Sci. U.S.A. 117, 17864–17875 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohammed H., et al., Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc. 11, 316–326 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Centore R. C., Sandoval G. J., Soares L. M. M., Kadoch C., Chan H. M., Mammalian SWI/SNF chromatin remodeling complexes: Emerging mechanisms and therapeutic strategies. Trends Genet. 36, 936–950 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Kadoch C., Crabtree G. R., Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci. Adv. 1, e1500447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Y., Chen X., Zhang W., Mammalian SWI/SNF chromatin remodeling complexes in embryonic stem cells: Regulating the balance between pluripotency and differentiation. Front. Cell Dev. Biol. 8, 626383 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alver B. H., et al., The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat. Commun. 8, 14648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., et al., SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 49, 289–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho L., et al., esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 13, 903–913 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadoch C., Copeland R. A., Keilhack H., PRC2 and SWI/SNF chromatin remodeling complexes in health and disease. Biochemistry 55, 1600–1614 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Zhang W., et al., The BAF and PRC2 complex subunits Dpf2 and Eed antagonistically converge on Tbx3 to control ESC differentiation. Cell Stem Cell 24, 138–152.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawkins R. D., et al., Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6, 479–491 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander J. M., et al., Brg1 modulates enhancer activation in mesoderm lineage commitment. Development 142, 1418–1430 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satokata I., et al., Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat. Genet. 24, 391–395 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Babajko S., de La Dure-Molla M., Jedeon K., Berdal A., MSX2 in ameloblast cell fate and activity. Front. Physiol. 5, 510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkie A. O., et al., Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nat. Genet. 24, 387–390 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Hu G., Lee H., Price S. M., Shen M. M., Abate-Shen C., Msx homeobox genes inhibit differentiation through upregulation of cyclin D1. Development 128, 2373–2384 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Satoh K., Hamada S., Shimosegawa T., MSX2 in pancreatic tumor development and its clinical application for the diagnosis of pancreatic ductal adenocarcinoma. Front. Physiol. 3, 430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q., et al., MSX2 mediates entry of human pluripotent stem cells into mesendoderm by simultaneously suppressing SOX2 and activating NODAL signaling. Cell Res. 25, 1314–1332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L., et al., MSX2 initiates and accelerates mesenchymal stem/stromal cell specification of hPSCs by regulating TWIST1 and PRAME. Stem Cell Reports 11, 497–513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Home P., et al., GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J. Biol. Chem. 284, 28729–28737 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee C. Q. E., et al., What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Reports 6, 257–272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auman H. J., et al., Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development 129, 2733–2747 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Kidder B. L., Palmer S., Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res. 20, 458–472 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee B.-K., et al., Super-enhancer-guided mapping of regulatory networks controlling mouse trophoblast stem cells. Nat. Commun. 10, 4749 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ralston A., et al., Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395–403 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Meinhardt G., et al., Pivotal role of the transcriptional co-activator YAP in trophoblast stemness of the developing human placenta. Proc. Natl. Acad. Sci. U.S.A. 117, 13562–13570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho L., et al., An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U.S.A. 106, 5181–5186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hota S. K., et al., Dynamic BAF chromatin remodeling complex subunit inclusion promotes temporally distinct gene expression programs in cardiogenesis. Development 146, dev174086 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Son E. Y., Crabtree G. R., The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am. J. Med. Genet. C. Semin. Med. Genet. 166C, 333–349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaeser M. D., Aslanian A., Dong M.-Q., Yates J. R. III, Emerson B. M., BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 283, 32254–32263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gatchalian J., et al., A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat. Commun. 9, 5139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X., et al., Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports 3, 460–474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milano-Foster J., et al., Regulation of human trophoblast syncytialization by histone demethylase LSD1. J. Biol. Chem. 294, 17301–17313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lv B., et al., Single-cell RNA sequencing reveals regulatory mechanism for trophoblast cell-fate divergence in human peri-implantation conceptuses. PLoS Biol. 17, e3000187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Y., et al., Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152, 248–261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Latos P. A., et al., Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev. 29, 2435–2448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haider S., et al., Notch1 controls development of the extravillous trophoblast lineage in the human placenta. Proc. Natl. Acad. Sci. U.S.A. 113, E7710–E7719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Latos P. A., et al., Fgf and Esrrb integrate epigenetic and transcriptional networks that regulate self-renewal of trophoblast stem cells. Nat. Commun. 6, 7776 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed next-generation sequencing datasets were deposited at the National Center for Biotechnology Information Gene Expression Omnibus repository under accession number GSE165970. Materials, data, and associated protocols, including code and scripts, are available upon request.