Significance
Success with immune checkpoint therapy has led to a proliferation of antibody therapeutics with distinct biophysical and pharmacological properties. Antibodies experience poor penetration in solid tumors because of their size, and blood-based measurements fail to reflect their activity at the tumor. While of fundamental importance to establish dose–exposure–response relationships, pharmacologic activity of antibodies within tumors is technically challenging to study in real time because of the lack of quantitative tools. Here, we show that target pharmacodynamics can yield insights into pharmacologic activity of antibodies in tumors and elucidate therapeutic response. Our approach of using small-molecule probes for rapid quantification of target pharmacodynamics to interrogate the pharmacologic activity of antibodies has broad implications for drug development and guiding therapy.
Keywords: immune checkpoint therapy, pharmacodynamics, pharmacokinetics, drug disposition, PET
Abstract
Macromolecules such as monoclonal antibodies (mAbs) are likely to experience poor tumor penetration because of their large size, and thus low drug exposure of target cells within a tumor could contribute to suboptimal responses. Given the challenge of inadequate quantitative tools to assess mAb activity within tumors, we hypothesized that measurement of accessible target levels in tumors could elucidate the pharmacologic activity of a mAb and could be used to compare the activity of different mAbs. Using positron emission tomography (PET), we measured the pharmacodynamics of immune checkpoint protein programmed-death ligand 1 (PD-L1) to evaluate pharmacologic effects of mAbs targeting PD-L1 and its receptor programmed cell death protein 1 (PD-1). For PD-L1 quantification, we first developed a small peptide-based fluorine-18–labeled PET imaging agent, [18F]DK222, which provided high-contrast images in preclinical models. We then quantified accessible PD-L1 levels in the tumor bed during treatment with anti–PD-1 and anti–PD-L1 mAbs. Applying mixed-effects models to these data, we found subtle differences in the pharmacodynamic effects of two anti–PD-1 mAbs (nivolumab and pembrolizumab). In contrast, we observed starkly divergent target engagement with anti–PD-L1 mAbs (atezolizumab, avelumab, and durvalumab) that were administered at equivalent doses, correlating with differential effects on tumor growth. Thus, we show that measuring PD-L1 pharmacodynamics informs mechanistic understanding of therapeutic mAbs targeting PD-L1 and PD-1. These findings demonstrate the value of quantifying target pharmacodynamics to elucidate the pharmacologic activity of mAbs, independent of mAb biophysical properties and inclusive of all physiological variables, which are highly heterogeneous within and across tumors and patients.
Solid tumors present passive and active barriers to drug penetration that lead to drug resistance, poor low response rates, and poor overall survival. In particular, little is known about the pharmacokinetics (PK) and pharmacodynamics (PD) of monoclonal antibodies (mAbs) in the tumor bed, a class of drugs which has emerged as a highly successful class of anticancer therapeutics (1). Studying the PK and PD parameters of mAbs in situ and in real time is essential for optimizing dosing, and for designing and developing more effective therapeutics. Macromolecular drugs such as mAbs show especially poor solid tumor penetration, perhaps explaining why a significant percentage of patients receiving mAb therapeutics do not respond (2). The lack of accurate tumor drug penetration measurements of mAbs limits our ability to optimize therapies (3).
Unfortunately, tissue- and cell-based methods used for quantifying drug concentrations, such as mass spectrometry, microscopy, and thermal shift assays, cannot be implemented directly in patients (4–6). Notably, mAb distribution in the tumor following systemic administration is heterogeneous and influenced by biophysical properties of the mAb, tumor type, and location (7). Additionally, the extent and duration of target engagement of mAbs in the tumor can be significantly shorter than those measured by in vitro PD assays that fail to capture the impact of the local tumor microenvironment (TME) on mAb target engagement and PK (8). While mAbs labeled with radionuclides are routinely used to gain insights into mAb PK and disease site concentrations and thus to infer biologic activity (9), development of radiolabeled analogs for every therapeutic mAb created against every target presents a formidable challenge in the rapidly growing market of mAbs and their biosimilars. More importantly, this approach is costly, clinical implementation is unrealistic, and the approach cannot be personalized. Consequently, there is an ongoing need for a methodology that 1) provides a quantitative measure of mAb activity at the tumor in real time; 2) is agnostic to the biophysical properties of the mAb; and 3) accounts for both intrinsic and extrinsic peculiarities of tumor type and location. Measuring drug concentrations and their effects at the site of action in vivo and in real time is a challenge and a missing piece in delivering the promise of personalized medicine (10). Addressing this challenge by conventional means would require repetitive sampling at the disease site, which poses significant practical and technical limitations for widespread diseases like cancer.
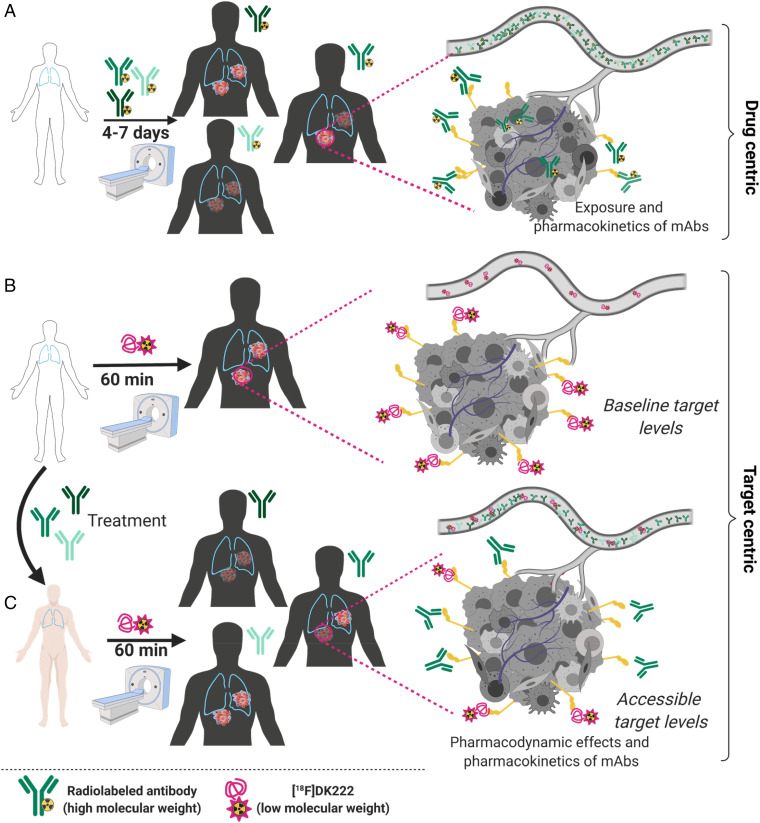
Here we propose a noninvasive approach using positron emission tomography (PET) that is based on quantifying the PD changes (accessible target levels) in the tumor bed to gain insights into in situ PK and PD of mAb therapeutics (Fig. 1).
Fig. 1.
PD measures as a means to understand the pharmacologic behavior of antibodies within the tumor bed. (A) Distinct radiolabeled mAbs binding the same antigen could exhibit differential drug exposure in different lesions, as they are subjected to heterogeneity in tumor intrinsic and extrinsic factors that influence mAb PK. Also, it is not practical to assess the effects of drug penetration following a therapeutic dose of a mAb due to the long half-life of mAbs. (B) Low–molecular-mass imaging agents provide an assessment of baseline target expression in all lesions and often demonstrate better permeability than mAbs. These agents can be selected for high affinity and tractable PK to fit within the constraints of clinical practice. (C) Low–molecular-mass imaging agents can be used to gain insights into inter- and intratumoral levels of an accessible target following a therapeutic dose of mAb. Those measurements account for the effects of tumor intrinsic and extrinsic factors on drug penetration and target occupancy without any interruption to therapy. Created with BioRender.com.
PET is a powerful noninvasive tool that can be used to quantify target levels anywhere in the body using molecularly targeted imaging agents administered at tracer doses (10). Although PET is routinely used to quantify PD effects of central nervous system–targeting therapeutics (11), its quantitative power has not been similarly applied to define the activity of mAb therapeutics. Thus, we hypothesized that accessible target levels in tumors, measured using PET, could elucidate the pharmacologic activity of mAb treatment, and could function as a common denominator to compare the activity of different mAbs. In developing this approach, we rationalized that custom-designed peptide (or small molecule or protein) -derived PET radiotracers, which bind a target of interest with a weaker affinity than the parent mAb and are designed to be rapidly eliminated from the body, could be used to quantify the accessible target levels at the disease site. Such an approach would provide a repetitive, noninvasive, real-time, personalized measure of mAb exposure and activity in situ and within a time frame that fits the standard clinical workflow. To test the hypothesis, we selected mAbs targeting immune checkpoint proteins that have been transformative in treating multiple cancer types.
We also recognized that durable responses observed with immune checkpoint therapeutics have led to the development of multiple mAb conjugates targeting the same antigen and various therapy combinations to further improve efficacy (12). To date, there has been no common denominator or proximal biomarkers to assess the PD effects resulting from therapeutic activity of those mAbs in the tumor bed (13). Patient selection for these clinical trials is often guided by analysis of tumor mutation burden, microsatellite instability status, or determination of programmed death-ligand 1 (PD-L1) levels by immunohistochemistry (IHC) (14). PD-L1 IHC has emerged as an important predictive and prognostic biomarker to guide PD-(L)1 therapies (14). However, understanding the dynamics of PD-L1 during therapy is key to elucidating the PK and PD of therapeutics targeting programmed cell death protein 1 (PD-1) and PD-L1 and improving their efficacy. To this end, we developed a peptide-based fluorine-18–labeled PET imaging agent that provides high-contrast PD-L1–specific images and quantifies PD-L1 levels within 60 min of injection. Because PD-L1 expression in the TME is indicative of adaptive immune responses unleashed by therapeutics targeting PD-1, we investigated the effect of different anti–PD-1 (aPD-1) therapeutics (nivolumab and pembrolizumab) on PD-L1 levels in the TME and their relevance to PET measures. Also, the manner in which different PD-L1 therapeutics engage the target at the tumor remains poorly understood. To address this, we evaluated the effect of mAb dose and time on PD-L1 dynamics in the tumor for three anti–PD-L1 (aPD-L1) mAbs (atezolizumab, avelumab, and durvalumab). Our studies uncover insights into the PK and PD of different aPD-1 and aPD-L1 therapeutics at the tumor and reveal how such measurements can inform tumor response to therapy. Moreover, our data support the use of noninvasive PD measurements for optimizing immune checkpoint therapy and have important implications for therapeutic mAb dose finding, therapy response assessment, and drug development.
Results
Synthesis and In Vitro Evaluation of a Hydrophilic PD-L1–specific PET Imaging Agent.
PD-L1 detection using IHC is a guiding tool for PD-1/PD-L1 therapy (15); however, tools to quantify total PD-L1 levels in all lesions noninvasively have emerged only recently and are in early clinical evaluation (16, 17). Quantifying PD-L1 dynamics presents a different challenge, however, due to the need for PET imaging agents that provide high-contrast images within the standard clinical workflow. To address this need, we previously developed a PD-L1–specific peptide-based imaging agent, [64Cu]WL12, that detects tumor PD-L1 levels (18). However, [64Cu]WL12 shows high nonspecific accumulation in several tissues (18, 19). To improve the imaging properties, we identified a hydrophilic peptide and generated a radiofluorinated analog using the aluminum fluoride (AlF) method to facilitate clinical translation.
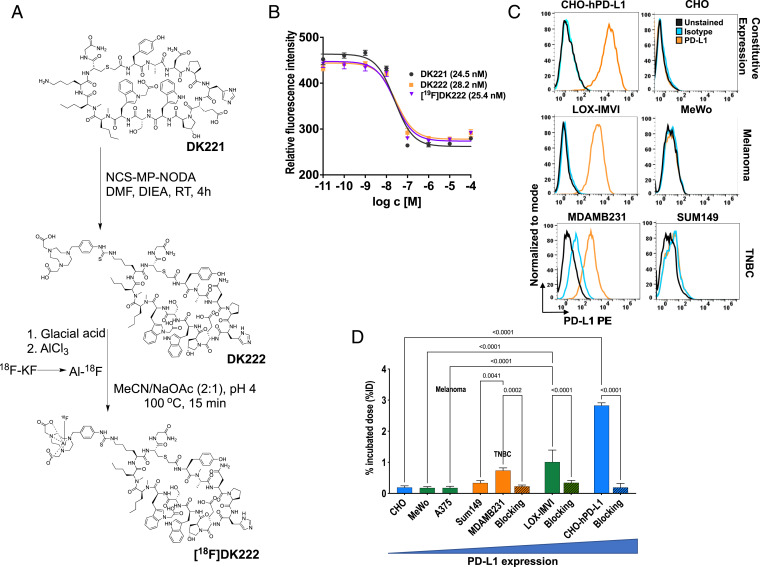
DK221 is a 14–amino acid human PD-L1–specific cyclic peptide with three carboxylate groups and a free lysine amine (20). To modify DK221 for radiolabeling, a bifunctional chelator, NCS-MP-NODA (2,2′-(7-(4-isothiocyanatobenzyl)-1,4,7-triazonane-1,4-diyl)diacetic acid), was conjugated to the free lysine amine to generate DK222. The NODA chelator was used for radiofluorination to produce [18F]DK222 as well as a nonradioactive analog, [19F]DK222 (Fig. 2A and SI Appendix, Fig. S1). To characterize the binding affinity of the peptide analogs to PD-L1, we performed a competitive PD-1:PD-L1 inhibition assay. We observed that peptide analogs inhibit PD-L1 binding to PD-1 in a dose-dependent fashion with half-maximal inhibitory concentration (IC50) values of 24.5, 28.2, and 25.4 nM for DK221, DK222, and [19F]DK222, respectively (Fig. 2B). [18F]Fluoride radiolabeling of peptides using the AlF method retains the hydrophilicity of the binding moiety (21). Thus, we synthesized the radiolabeled analog [18F]DK222 by the AlF method and observed robust radiochemical yields (34.85 ± 1.7%, n = 62) and moderate specific activity of 284 ± 56 mCi/μmol (10.51 ± 2.07 GBq/μmol, n = 25), and found it to be stable in vitro for 4 h (SI Appendix, Fig. S2).
Fig. 2.
Synthesis and in vitro characterization of [18F]DK222. (A) Structure and schema for the preparation of [18F]DK222. (B) Ability of DK221, DK222, and the nonradioactive [19F]DK222 to inhibit PD-1:PD-L1 interaction was assessed in a protein-based fluorescence resonance energy transfer assay. The IC50 values are shown in parentheses. Error bars indicate SD (n = 3). (C) Flow cytometry analysis of PD-L1 levels in various cell lines. CHO-hPD-L1 refers to CHO cells stably transduced with human PD-L1. (D) [18F]DK222 binding to cells with variable PD-L1 expression. Cells were incubated with 1 μCi [18F]DK222 for 30 min at 4 °C in the presence or absence of 1 µM unmodified peptide (blocking dose) and washed thoroughly, and cell-bound activity was measured. Error bars indicate SD (n = 3). Significance was determined by unpaired t test.
To assess the specificity of [18F]DK222 to PD-L1, we performed cell-binding assays. We selected CHO cells with constitutive human PD-L1 (hPD-L1) expression (denoted CHO-hPD-L1) and multiple cancer cell lines of triple-negative breast cancer (TNBC) (MDAMB231 and SUM149) and melanoma (LOX-IMVI, MeWo, and A375) origin. Radioactivity uptake in cells incubated with [18F]DK222 reflected the variable levels of surface PD-L1 expression observed by flow cytometry (Fig. 2 C and D) in the order CHO-hPD-L1 > LOX-IMVI > MDAMB231 > SUM149. A375, CHO, and MeWo cells, which expressed low PD-L1 levels, exhibited the least [18F]DK222 binding. Also, binding studies performed in the presence of a 1 μM excess of the nonradioactive parent DK221 peptide showed >90% reduction in radioactivity uptake in PD-L1–positive cells, validating specificity (P < 0.0001; Fig. 2D). Taken together, these in vitro results provided evidence that [18F]DK222 specifically binds PD-L1 and competes with PD-1:PD-L1 interaction.
Evaluation of [18F]DK222 Biodistribution in Mouse Models of TNBC.
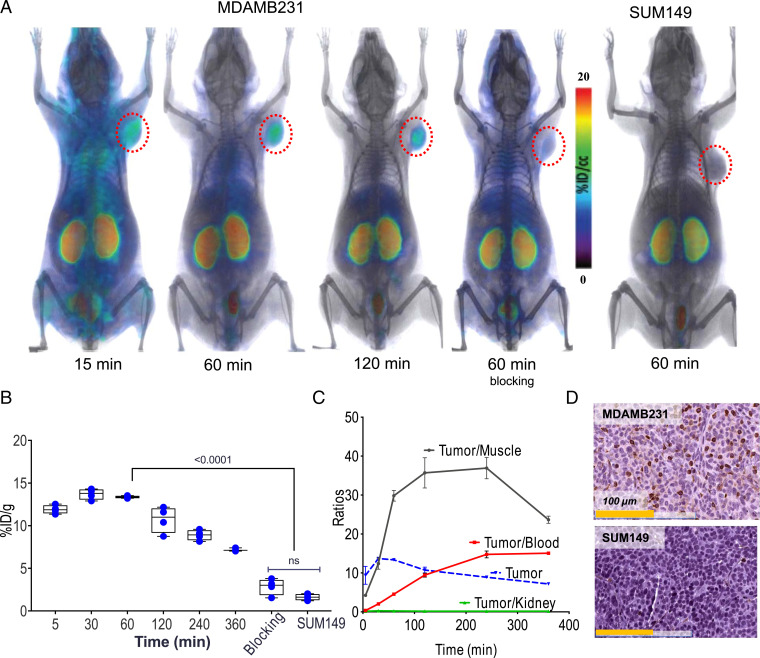
To gain insight into the PK and biodistribution of [18F]DK222, we performed PET imaging studies in immunocompromised NSG (nonobese, diabetic, severe-combined immunodeficient gamma) mice harboring PD-L1–positive MDAMB231 xenografts. PET images acquired at 15, 60, and 120 min after [18F]DK222 injection showed high radiotracer accumulation in tumors as early as 15 min and provided high-contrast images at 60 to 120 min (Fig. 3A). Kidneys also showed a high uptake of radioactivity at all time points investigated. Dramatically reduced uptake of radiotracer was observed in MDAMB231 tumors of mice that received a blocking dose of DK221, and in SUM149 tumors, which express low PD-L1 levels.
Fig. 3.
In vivo kinetics of [18F]DK222 in mice bearing TNBC xenografts. NSG mice with human tumor xenografts were injected with 200 μCi (7.4 MBq) [18F]DK222, and PET-CT images were acquired at different time points. Blocking-dose mice received 50 mg/kg of DK221 30 min prior to radiotracer injection. (A) PET-CT images of mice acquired at 15, 60, and 120 min. Significant uptake of [18F]DK222 can be seen in high–PD-L1–expressing MDAMB231 tumors but not in mice receiving a blocking dose or low–PD-L1–expressing SUM149 tumors (n = 3 or 4). (B) Uptake of [18F]DK222 in MDAMB231 and SUM149 tumors from biodistribution studies (n = 4 or 5). ns, not significant. (C) Time–activity curves derived from biodistribution data (SI Appendix, Table S1). Error bars indicate SD (n = 4 or 5). (D) IHC staining for PD-L1 in MDAMB231 and SUM149 tumors. Significance was determined by unpaired t test.
To validate imaging studies and to quantify [18F]DK222 biodistribution in normal tissues, we conducted ex vivo measurements at 5, 30, 60, 120, 240, and 360 min after radiotracer injection (SI Appendix, Table S1). [18F]DK222 uptake remained consistently high in tumors for 4 h after injection (Fig. 3B). Time–activity curves plotted from the biodistribution data (expressed as the percentage of injected dose per gram of tissue; %ID/g) showed high accumulation and retention of [18F]DK222 in MDAMB231 tumors (Fig. 3C). A steady decrease in radioactivity with time was observed in blood, muscle, and all other tissues that contributed to high image contrast (SI Appendix, Table S1). Consistent with PET, uptake of [18F]DK222 was consistently higher in tumors and kidneys. The tumor-to-blood and tumor-to-muscle ratios at 60 min were 4.5 ± 0.2 and 30.0 ± 1.3, respectively. [18F]DK222 uptake in SUM149 tumors was 88% (P < 0.0001) less than that seen with MDAMB231 tumors at 60 min. Also, mice receiving a blocking dose of DK221 showed a 79% reduction in [18F]DK222 uptake (P < 0.0001; SI Appendix, Table S1). Furthermore, administration of a range of competing nonradioactive doses of DK221 reduced uptake in MDAMB231 tumors in a dose-dependent manner, but not in SUM149 tumors or other tissues (SI Appendix, Fig. S3), further illustrating the specificity of the interaction. These observations were confirmed by IHC, which showed strong and weak PD-L1 expression in MDAMB231 and SUM149 tumors, respectively (Fig. 3D). Based on the high tumor uptake and contrast observed, we conducted all imaging and biodistribution studies discussed hereafter at 60 min.
In Vivo Validation of [18F]DK222 Specificity in Melanoma Xenograft Models.
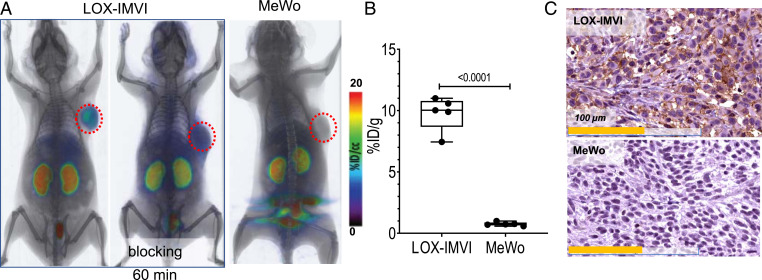
We next sought to validate the PD-L1 specificity of [18F]DK222 in melanoma models. NSG mice with high–PD-L1–expressing LOX-IMVI or low–PD-L1–expressing MeWo melanoma xenografts were injected with [18F]DK222. High accumulation of radioactivity was observed in LOX-IMVI tumors in imaging and biodistribution studies, but not in MeWo tumors or in mice that received a blocking dose of DK221 (P < 0.0001) (Fig. 4 A and B and SI Appendix, Fig. S4). Histological analysis further corroborated the PET findings (Fig. 4C).
Fig. 4.
[18F]DK222 PET in mice with human melanoma xenografts shows high-contrast images at 60 min. NSG mice with human melanoma xenografts were injected with ∼200 μCi (7.4 MBq) [18F]DK222 for PET imaging studies or 50 μCi (1.85 MBq) [18F]DK222 for biodistribution studies. Analyses were conducted at 60 min after injection. (A) Whole-body volume rendered PET-CT images show high and specific uptake of [18F]DK222 in LOX-IMVI tumors that express high PD-L1 but not in mice receiving a blocking dose or bearing low–PD-L1–expressing MeWo tumors (n = 3 or 4). (B) Tumor uptake of [18F]DK222 at 60 min, as measured by ex vivo biodistribution studies (n = 5). Error bars indicate SD (n = 4 or 5). Significance was determined by unpaired t test. (C) IHC staining for PD-L1 in LOX-IMVI and MeWo tumors. P values were determined by unpaired t test.
Overall, in vivo and ex vivo measurements in four different tumor models validated the specificity of [18F]DK222 for PD-L1, and demonstrated its potential to quantify variable PD-L1 levels across different tumor types.
Human Radiation Dosimetry Estimates.
Biodistribution data collected from MDAMB231 tumor-bearing mice were decay-corrected and used to calculate residence times of human organs. The source organ time-integrated activity coefficients (residence times), human organ absorbed doses, human organ absorbed dose per unit accumulated activity (mGy/MBq), and PK parameters for the urinary bladder are listed in SI Appendix, Fig. S4 B–D. These residence times were used as input in OLINDA/EXM to obtain the absorbed organ dose for 18F and adult reference human female phantom. The predicted human organ absorbed doses calculated by this approach are listed in SI Appendix, Table S2. The organs receiving the highest dose are the kidneys (3.74E-01 rem/mCi), followed by the liver (4.91E-02 rem/mCi), adrenals (3.83E-02 rem/mCi), and lungs (3.80E-02 rem/mCi). Based on a single-dose guideline of 5 rem, 13.4 mCi [18F]DK222 may be safely administered to obtain PET images.
Quantifying PD Effects of aPD-1 mAbs at the Tumor with [18F]DK222.
The PD-1/PD-L1 pathway represents a cornerstone for combination immune checkpoint blockade regimens (22). Many such combination therapies converge on the production of interferon γ (IFNγ), which is inextricably linked to PD-L1 levels (23). PD-L1 in the TME is indicative of robust cytolytic activity that is unleashed by the therapeutics targeting PD-1 (23). Reinvigoration of exhausted T cells can be detected in blood as early as 3 wk in patients receiving aPD-1 therapeutics. In contrast, the activity of immune checkpoint blockade therapies at the tumor remains poorly understood (24), and involves cytolytic activity of immune cells, IFNγ secretion, and induction of PD-L1 levels in the tumor bed (25). Thus, we surmised that tumor PD-L1 levels would serve as a proximal biomarker to measure the PD effects of aPD-1 therapeutics. We therefore sought to quantify the changes in tumor PD-L1 levels induced by different aPD-1 therapeutics using PD-L1 PET, and to correlate those measures with immunological responses.
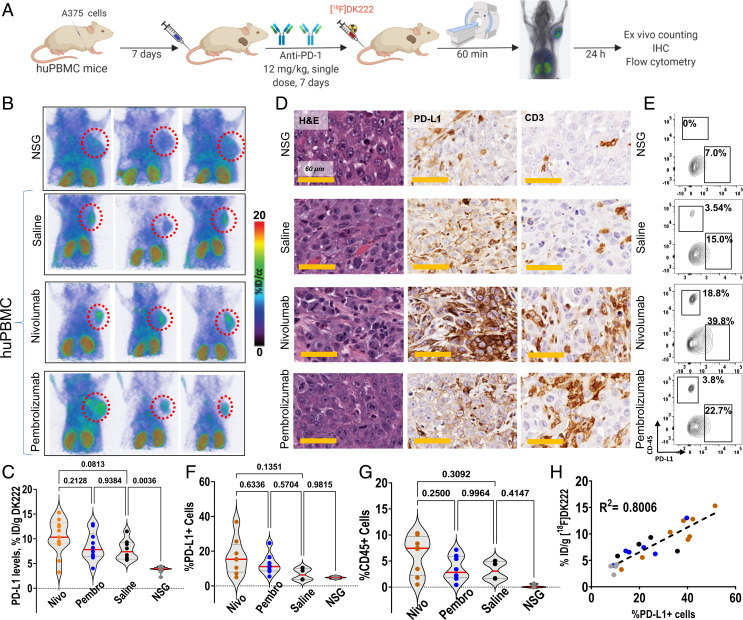
First, we assessed the effect of the adaptive immune response in melanoma cells by assessing changes in PD-L1 levels induced by IFNγ treatment. LOX-IMVI, A375, and MeWo melanoma cells treated with IFNγ showed a two- and fourfold increase in cell-surface PD-L1 levels in LOX-IMVI and A375 cells, respectively, whereas no differences were observed in MeWo cells (SI Appendix, Fig. S5A). To quantify the differences in tumor PD-L1 levels as a measure of adaptive immune response to treatment with different aPD-1 mAbs, we employed a humanized mouse model known as huPBMC, wherein NSG mice are humanized with peripheral blood mononuclear cells (PBMCs). huPBMC mice bearing A375 melanoma xenografts were treated with a single dose of aPD-1 mAbs (12 mg/kg). One week later, tumor PD-L1 levels were measured by [18F]DK222 PET and ex vivo counting (SI Appendix, Fig. S5A). Tumor-bearing huPBMC mice treated with saline and NSG mice treated with both pembrolizumab and nivolumab served as controls. When assessed for possible differences in PD-L1 levels, A375 tumors in huPBMC mice showed elevated [18F]DK222 uptake compared with NSG mice, indicating immune cell–mediated PD-L1 up-regulation (%ID/g 8.5 vs. 3.9; P = 0.0002; SI Appendix, Fig. S5B). We also observed increased [18F]DK222 uptake in the kidneys and spleen of huPBMC mice compared with those of NSG mice (SI Appendix, Fig. S5C). In contrast, no significant differences in [18F]DK222 uptake were observed in other tissues. These results established the potential for [18F]DK222 to distinguish tumors with high vs. low PD-L1 levels, and perhaps also identify tumors that exclude immune cells.
Next, we sought to assess the PD effects of different aPD-1 therapeutics at the tumor site. First, we assessed differences in tumor PD-L1 levels between treatment groups in huPBMC mice. Notably, three of six huPBMC mice treated with saline showed high [18F]DK222 uptake. In contrast, a substantial number of mice treated with aPD-1 mAbs showed high [18F]DK222 uptake, with some variability in the tumors (Fig. 5B). Biodistribution studies showed a 148, 85, and 76% increase in median [18F]DK222 uptake in mice treated with nivolumab, pembrolizumab, or saline, respectively, compared with NSG mice (Fig. 5C), validating PET data and revealing differences in therapy-induced PD-L1 levels in the TME. Analysis of tumor sections from aPD-1 mAb-treated mice showed increased presence of PD-L1 and CD3, consistent with the observed increase in [18F]DK222 uptake (Fig. 5D and SI Appendix, Fig. S5D). Taken together, these results indicate that different PD-1 therapeutics exert differing PD effects at the tumor, which can be measured based on changes in tumor PD-L1 levels.
Fig. 5.
[18F]DK222 uptake correlates with total PD-L1 levels in tumors induced by different aPD-1 therapeutics. (A) Experimental schematic. huPBMC mice with A375 melanoma tumors were treated with a single dose of 12 mg/kg of nivolumab, pembrolizumab, or saline for 7 d. On day 8, mice underwent imaging studies at 60 min after [18F]DK222 injection. The next day, biodistribution studies were carried out and the collected tumors were used to characterize immune responses. Data are pooled from two experiments. (B) PET-CT images of nivolumab- or pembrolizumab-treated mice show increased [18F]DK222 uptake in the tumors (n = 4). (C) [18F]DK222 uptake in tumors quantified by biodistribution studies (n = 8 to 13). Significance was determined by one-way ANOVA using Tukey’s multiple-comparisons test. (D) IHC analysis of tumor sections from imaging mice shows increased immunoreactivity for PD-L1 and CD3 in nivolumab- and pembrolizumab-treated mice compared with saline-treated controls and NSG mice. H&E, hematoxylin and eosin. (E) Flow cytometry plots showing CD45+ cells and total PD-L1 levels in representative mice (n = 4 to 8). (F and G) Flow cytometry analysis of the percentage of PD-L1+ cells (tumor and immune cells) (F) and percentage of CD45+ cells (G) (n = 4 to 7). (H) Correlation between [18F]DK222 uptake from bioactivity assays and PD-L1+ cell percentage from flow cytometry analysis. Simple linear regression and Pearson coefficient are used. Additional statistical analyses are provided in SI Appendix, Fig. S5J.
To validate that the observed [18F]DK222 uptake is indeed PD-L1–specific, we extracted tumors from mice and performed flow cytometry analysis to quantify PD-L1 levels. We observed increased [18F]DK222 uptake and total PD-L1 levels in aPD-1 treatment groups, which is supported by increased accumulation of CD45+CD8+ immune cells in tumors (Fig. 5 E–G and SI Appendix, Fig. S5 E and F). We noted a strong correlation between [18F]DK222 uptake and both total PD-L1 levels in the tumors (R2 = 0.80; P < 0.0001; Fig. 5H) and tumor cell–specific PD-L1 levels (R2 = 0.71; P < 0.0001; SI Appendix, Fig. S5G). In contrast, correlation between [18F]DK222 uptake and immune cell–specific PD-L1 levels was lower, perhaps due to the small contribution from immune cell PD-L1 levels to the total PD-L1 levels in the TME in this model (R2 = 0.57; P < 0.0001; SI Appendix, Fig. S5H). [18F]DK222 uptake in the spleen did not correlate with PD-L1 levels (SI Appendix, Fig. S5I). These data establish that [18F]DK222 PET can be used to quantify PD-L1 dynamics induced by aPD-1 treatments.
Next, to test the hypothesis that [18F]DK222 uptake can be used to quantify differential effects of aPD-1 mAbs in the tumor bed, we used the fixed-effects statistical analysis model to quantify heterogeneity in induced PD-L1 levels in the tumor bed. In the fixed-effects model, both fixed aPD-1 mAbs are treated as specific choices to be compared against one another. Thus, we set out to answer the following question: Does nivolumab or pembrolizumab treatment induce PD-L1 more effectively over time at a given dose? Each aPD-1 mAb was compared against saline, and the results are tabulated in SI Appendix, Fig. S5J. We observed subtle but not statistically significant differences in induced PD-L1 levels between nivolumab and pembrolizumab when these agents were administered at same dose, with nivolumab treatment resulting in greater PD-L1 expression in this model system. Taken together, our data demonstrate that [18F]DK222 measurements in the tumor bed can be used to compare the PD effects of different aPD-1 mAbs early on in the course of treatment.
Quantifying Accessible Tumor PD-L1 Levels During aPD-L1 Treatment.
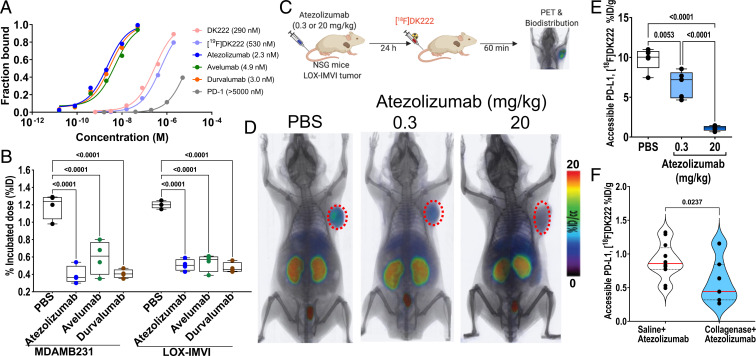
To quantify accessible PD-L1 levels using [18F]DK222, first, we studied the interaction of the peptide analogs and aPD-L1 mAbs with recombinant PD-L1 using biolayer interferometry. We found that the PD-L1–peptide affinity, while stronger than the PD-1:PD-L1 affinity, is at least 100-fold weaker than PD-L1–aPD-L1 mAb affinity for all three anti–PD-L1 mAbs (Fig. 6A and SI Appendix, Fig. S6 A and B). This suggests that [18F]DK222 will not interfere with anti–PD-L1 therapy at the tracer concentrations used (low nanomolar). Indeed, MDAMB231 and LOX-IMVI cells incubated with 1 µCi [18F]DK222 in the presence or absence of 60 nM PD-L1 mAb exhibited a >65% reduction in uptake (P < 0.0001), indicating that cell-membrane PD-L1 levels were occupied by mAbs (Fig. 6B). These data demonstrate that [18F]DK222 has the potential to quantify accessible PD-L1 levels in vivo, enabling real-time measurement of PD-L1 occupancy during treatment.
Fig. 6.
Anti–PD-L1 therapy-induced changes in accessible PD-L1 levels quantified using [18F]DK222. (A) Biolayer interferometry analysis of the affinity of [19F]DK222, anti–PD-L1 mAbs, and PD-1 for immobilized PD-L1. [18F]DK222 affinity for PD-L1 is 100-fold weaker than that of anti–PD-L1 mAbs. (B) Binding of [18F]DK222 to MDAMB231 or LOX-IMVI cells in the absence or presence of aPD-L1 mAbs. Cells were incubated with [18F]DK222 (1 μCi/mL) at 4 °C for 30 min in the presence or absence of 60 nM aPD-L1 mAbs. [18F]DK222 binding to PD-L1 is significantly inhibited in the presence of mAbs. Error bars indicate SD (n = 3). Significance was determined by one-way ANOVA using Tukey’s multiple-comparisons test. (C) Experimental schematic for atezolizumab PD study. Mice with LOX-IMVI tumors were treated with a single dose of atezolizumab (0.3 or 20 mg/kg) for 24 h prior to [18F]DK222 injection. Imaging and biodistribution studies were carried out at 60 min after [18F]DK222 injection. (D and E) Whole-body volume rendered PET-CT images (D) (n = 3) and ex vivo biodistribution studies (E) (n = 5) show reduced [18F]DK222 uptake in LOX-IMVI tumors with increased atezolizumab dose. Error bars indicate SD. Significance was determined by one-way ANOVA using Tukey’s multiple-comparisons test. (F) NSG mice with LOX-IMVI tumors were treated with saline or collagenase (0.1%, 100 μL, intravenously) for 100 min, followed by a single dose of atezolizumab for 24 h (3 mg/kg). Ex vivo biodistribution studies were carried out at 60 min after [18F]DK222 injection (n = 11 or 12), revealing that collagenase treatment induced a significant reduction in [18F]DK222 uptake. Significance was determined by unpaired t test.
To confirm these in vitro observations in vivo, NSG mice with LOX-IMVI tumors were treated for 24 h with a single dose of 0.3 or 20 mg/kg of atezolizumab or saline as control. [18F]DK222 PET images showed a high accumulation of radioactivity in the tumors of saline-treated control mice. In contrast, signal intensity in tumors was drastically reduced in mice receiving 20 mg/kg of mAb (Fig. 6 C and D). Importantly, there was also a modest reduction in signal intensity in the 0.3 mg/kg dose group. Ex vivo analyses showed 89 (P < 0.0001) and 32% (P < 0.01) reduction in [18F]DK222 uptake in tumors in mice treated with 20 and 0.3 mg/kg of atezolizumab, respectively, compared with saline-treated mice, suggestive of differences in accessible PD-L1 levels in the tumors across treatment groups (Fig. 6E and SI Appendix, Fig. S6C).
To further establish the sensitivity of [18F]DK222 PET to quantify therapy-induced changes in accessible PD-L1 levels in the TME, we studied the effects of collagenase treatment on PD-L1 target engagement by atezolizumab. Intratumoral or intravenous preadministration of collagenase has been shown to increase the distribution of macromolecules and viral vectors within tumors (26, 27). Mice bearing LOX-IMVI tumors that received collagenase (0.1%, 100 μL) + atezolizumab (3 mg/kg) showed significantly decreased tumor uptake of [18F]DK222 (P = 0.0237) compared with mice that received saline + atezolizumab (Fig. 6D). Minimal differences were observed in other tissues for mice treated with atezolizumab in the absence or presence of collagenase (SI Appendix, Fig. S6 E and F). These data indicate an enhanced PD-L1 engagement by atezolizumab when given in combination with collagenase.
Taken together, these in vitro and in vivo results demonstrate the potential of [18F]DK222 PET to measure accessible tumor PD-L1 levels in real time and determine the extent to which lesions are saturated by drug treatment under various administration conditions.
Accessible Tumor PD-L1 Levels Provide Insights into PK and PD Effects of aPD-L1 mAbs within the Tumor Bed.
The effectiveness of different mAbs targeting PD-L1 in the TME may be heterogeneous because of differing PK and PD properties, which remain uncharacterized. [18F]DK222 can bind accessible PD-L1, and thus [18F]DK222 PET signal shows the extent to which PD-L1 remains unoccupied; the lower the signal, the better the PD-L1 mAb targeting efficiency. Insights gained into PK and PD of mAbs during a trial round of immunotherapy could further guide the choice of mAb for treatment.
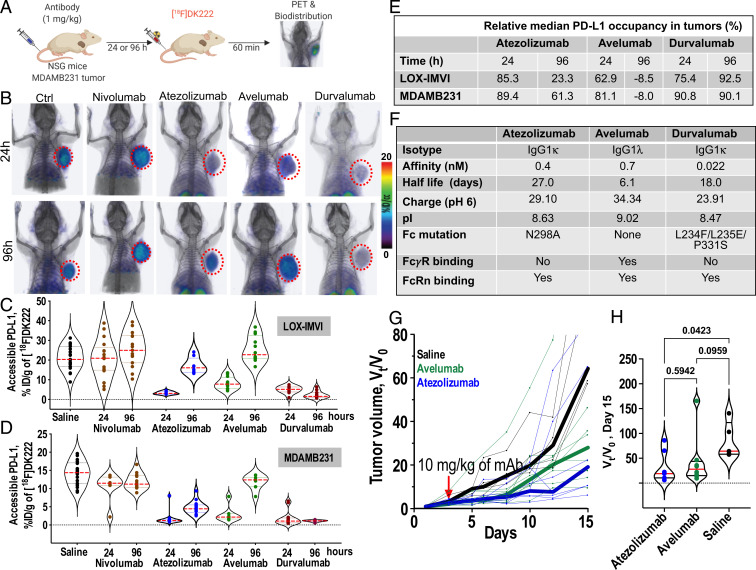
We therefore sought to evaluate the potential of [18F]DK222 to detect the heterogeneity in binding of different mAbs, and thus establish whether it can be used to guide the choice between the multiple mAbs available for treatment. For this experiment, we compared three aPD-L1 mAbs, atezolizumab, avelumab, and durvalumab (12), and nivolumab was used as an a priori negative control. Separate groups of animals were injected with a single 1 mg/kg dose of mAb and, after either 24 or 96 h, each group was injected with [18F]DK222 for PET or ex vivo analysis of accessible PD-L1 (Fig. 7A). The experiment was performed in two different tumor models, LOX-IMVI and MDAMB231.
Fig. 7.
Pharmacologic activity of different aPD-L1 therapeutics quantified at the tumor using [18F]DK222 PET. (A) Experimental schematic. NSG mice were treated with atezolizumab, avelumab, or durvalumab (1 mg/kg) for 24 and 96 h prior to [18F]DK222 injection. Nivolumab (1 mg/kg) and saline were used as controls. Imaging and biodistribution studies were carried out at 60 min after [18F]DK222 injection. Data are pooled from two independent experiments. (B) PET-CT images of [18F]DK222 uptake in LOX-IMVI tumors in mice treated with 1 mg/kg of mAbs for 24 and 96 h capturing differing PD-L1 occupancy and PK at the tumor site (n = 3). (C and D) [18F]DK222 uptake in tumors quantified by biodistribution in LOX-IMVI (C; n = 8 to 19) and MDAMB231 (D; n = 7 to 18) tumor-bearing mice. (E) Median PD-L1 occupancy in LOX-IMVI and MDAMB231 tumors treated with aPD-L1 mAbs relative to mice treated with nivolumab for 24 h. (F) Biophysical, molecular, and PK properties of aPD-L1 mAbs. pI and charge values are from ref. 44. (G and H) Tumor growth curves of MC38-hPD-L1 tumors from mice administered a single dose of atezolizumab or avelumab treatment (10 mg/kg, n = 5 to 8) 5 d after tumor inoculation. Tumor volumes are normalized to pretreatment volume (V0) and median growth curves are shown as thick lines. Vt/V0 measurements on day 15 are provided (H). Statistical significance was determined by one-way ANOVA using uncorrected Fisher’s least significant difference test.
PET images of LOX-IMVI tumor-bearing mice at the 24-h time point showed a significant reduction in tumor uptake of [18F]DK222 in all groups treated with aPD-L1 mAbs, indicative of successful PD-L1 blockade. In contrast, [18F]DK222 uptake in nivolumab-treated control animals was similar to that of saline-treated mice. A significant rebound in tumor uptake of [18F]DK222 was observed at 96 h in mice treated with avelumab and to a lesser extent atezolizumab, but not in mice treated with durvalumab (Fig. 7B). No differences in tumor uptake of [18F]DK222 at 24 vs. 96 h were observed for nivolumab-treated mice, suggesting that [18F]DK222 uptake is dependent on changes in aPD-L1 mAb occupancy.
To test the hypothesis that [18F]DK222 uptake can be used to quantify the PD and PK properties of aPD-L1 mAbs in the tumor bed, we used two different statistical analysis strategies to quantify the heterogeneity of therapeutic mAb binding. First, in the random-effects model, the three mAbs were considered a random sampling of the various mAbs available. This analysis was designed to answer the following question: How much of the variance in the [18F]DK222 signal can be explained by 1) the fact that different mAbs are being used, or 2) the possibility that these mAbs may each have different kinetics between 24 and 96 h? To answer this question, the three aPD-L1 mAbs were selected as random effects, and aPD-L1 mAbs vs. inactive mAb (nivolumab), time points, and the overall difference of active treatments between time points were selected as the fixed effects. The random and fixed effects are jointly estimated in a mixed linear regression model and the measure of heterogeneity is defined by the intraclass correlation coefficient (ICC). The ICC can range between 0 and 1, wherein 1 represents the greatest possible variation of PK and PD among the aPD-L1 mAbs. The ICC of the random PD-only effect model in LOX-IMVI tumors is 0.23 (vs. no random effects, P = 8.9 × 10−5), indicating that 23% of variance in [18F]DK222 signal (%ID/g) comes from differences in PD-L1 occupancy. The ICC of the random PK–PD effect model is 0.36 (vs. no random effects, P = 3 × 10−6; vs. random PD model, P = 0.0014), indicating that 36% of the variance in the [18F]DK222 %ID/g comes from the divergent PK and PD properties of different mAbs in the tumor bed. Similarly, in the MDAMB231 tumor model, the ICC of the random PD-only effect model is 0.54 (vs. no random effects, P = 0). The ICC of the random PK–PD effect model is 0.77 (vs. no random effects, P = 0; vs. random PD model, P = 0), indicating that 77% of the variance in the MDAMB231 %ID/g comes from the divergent PK and PD properties of different mAbs in the tumor bed.
The second model employed was the fixed-effects model, wherein each of the three fixed aPD-L1 mAbs was thought of as a specific choice to be compared against each other. We set out to answer the following question: Which of atezolizumab, avelumab, and durvalumab specifically engages PD-L1 more effectively over time? To answer this question, each aPD-L1 mAb (specific saturation PD), each time point (overall PK, 24 and 96 h), and each mAb*time combination (mAb-specific PK) was considered as a fixed effect, and were estimated together in an ordinary linear regression model. The results of this analysis are given as 1) the difference in accessible PD-L1 levels (%ID/g) for each of the PD-L1 mAbs at 24 h vs. nivolumab, and 2) the difference in accessible PD-L1 levels at 96 vs. 24 h for mice treated with an aPD-L1 mAb as compared with 96- vs. 24-h accessible PD-L1–level differences in mice treated with nivolumab.
[18F]DK222 uptake in LOX-IMVI tumors and tissues of mice treated with different mAbs and at 24 and 96 h is shown in Fig. 7C, and mean tumor %ID/g of different mAbs at both time points is tabulated in SI Appendix, Fig. S7A. The mean LOX-IMVI tumor [18F]DK222 %ID/g for nivolumab-treated control mice was similar to that of saline control–treated mice, and uptake changed very little between 24 and 96 h. In contrast, all PD-L1 mAbs showed significantly lower mean tumor %ID/g and accessible PD-L1 levels than nivolumab at the 24-h time point. The accessible PD-L1 levels at 96-h posttreatment were 60 and 80% higher in the atezolizumab and avelumab groups, respectively, compared with those observed at 24 h posttreatment (P < 0.001). However, accessible PD-L1 levels for durvalumab were reduced by 70% or more, and were similar at 96 vs. 24 h, suggesting a longer-term blockade of PD-L1 by durvalumab. These observations were corroborated in an MDAMB231 xenograft model (Fig. 7D). Tables comparing median PD-L1 occupancy levels and mean tumor %ID/g of different mAbs at the 24- and 96-h time points are shown in Fig. 7E and SI Appendix, Fig. S7B, respectively. These results motivate the use of our approach to elucidate nuances of the kinetics and temporal behavior of mAbs in the tumor bed.
In sum, the random- and fixed-effects statistical analyses reveal that aPD-L1 mAbs have differential PK and PD in the tumor bed, and that these differences influence accessible PD-L1 levels over time. Furthermore, these results clearly demonstrate the potential of PET to quantify accessible target levels in order to gain real-time insights into the pharmacological activity of mAbs at the tumor site.
Having elucidated the differential in PK and PD properties of various aPD-L1 mAbs within the TME, we sought to define the factors contributing to these observed differences. Many factors contribute to the complex plasma and intratumoral PK/PD of mAbs, including nonspecific neonatal Fc receptor (FcRn)–dependent clearance mechanisms, target-mediated clearance mechanisms (target-mediated drug disposition; TMDD), mAb affinity and PK, and high tumor interstitial pressure (2, 7). Biophysical and structural features of aPD-L1 mAbs (summarized in Fig. 7F) indicate that, despite both having ablated Fcγ receptor (FcγR) binding and possessing similar half-lives, atezolizumab and durvalumab exhibit different PK and PD properties in the tumor bed (Fig. 7 B–D). This observation suggests that differences in FcγR binding (and consequent differences in antibody-dependent cellular cytotoxicity and antibody-dependent cellular phagocytosis) do not account for discrepancies in PK and PD parameters at the tumor site. mAb interaction with FcRn is known to prolong the serum half-life of mAbs (28). All three mAbs bind FcRn (29). However, whereas atezolizumab and durvalumab exhibit long half-lives (27 and 18 d, respectively) (30), avelumab exhibits a short half-life (∼6 d) (30), indicating that differential FcRn interactions are not responsible for differences in serum persistence. Instead, the short half-life of avelumab has been attributed to its high isoelectric point (pI) (31). Taken together, this existing evidence suggests that FcRn interactions are not the primary cause for the observed differences in PK and PD.
Interestingly, mAbs with high pI are anticipated to have increased clearance, as well as increased tissue and tumor distribution (32). Although its high pI explains the short half-life of avelumab, high levels of accessible PD-L1 (and with high variance) in the tumors of avelumab-treated mice indicate that either avelumab penetrates poorly or clears from the tumor rapidly compared with atezolizumab and durvalumab (Fig. 7 C–E). In fact, treatment with the mAb that has the lowest pI, durvalumab, resulted in the lowest levels of accessible PD-L1 levels by 96 h and with the least variance, demonstrating stronger target engagement. These observations suggest that differences in charge and pI of mAbs impact intratumoral target engagement of mAbs; however, these differences do not by themselves fully explain the divergent PK and PD properties observed for the three aPD-L1 mAbs.
TMDD, a phenomenon mediated by antigen–mAb interaction, is known to play an important role in mAb clearance. All three aPD-L1 mAbs studied are known to exhibit TMDD (29). Multiple factors including binding affinity, mAb dosing, PD-L1 density, and PD-L1 turnover and internalization rates affect the extent and depth of TMDD (7). Among these factors, binding affinities of the mAbs are distinctive, whereas mAb dosing, PD-L1 density, and PD-L1 turnover and internalization rates are fixed, suggesting that affinity properties partially govern the intratumoral PK and PD behavior of mAbs (SI Appendix, Fig. S7 C and D).
Collectively, these data indicate that charge, pI, affinity, and PK properties are major determinants of the observed intratumoral PK and PD differences for the three aPD-L1 mAbs investigated in this study.
To test whether differences in PK and PD within the tumor bed correlate with the efficacy of treatment, C57BL/6 mice bearing syngeneic MC38 colon tumors stably expressing human PD-L1 (MC38-hPD-L1) were treated with saline control or with a single dose of either atezolizumab or avelumab and tumor growth was monitored (SI Appendix, Fig. S7F). Cross-reactivity of atezolizumab and avelumab with human and murine PD-L1 allowed for their use in this model (33, 34). Note that [18F]DK222 selectively binds human PD-L1. [18F]DK222 PET images of those mice acquired at 120 h following mAb treatment showed higher accessible hPD-L1 levels in avelumab-treated mice (2/2) compared with atezolizumab-treated (1/2) mice (SI Appendix, Fig. S7G). Both the atezolizumab and avelumab treatment groups exhibited delayed tumor growth and a reduction in tumor volume compared with the saline control group. Although not statistically significant at the tested dose, median tumor growth curves showed greater tumor inhibition in the atezolizumab compared with the avelumab treatment group (Fig. 7 G and H), consistent with the superior PD-L1 blockade in the tumor bed for atezolizumab vs. avelumab. Taken together, these data reveal that differential mAb PK–PD properties within tumors have implications for efficacy of the treatment.
Discussion
Here, using PET, we demonstrate that measurements of accessible target levels in tumors can provide insights into PK and PD of mAbs directly within the tumor bed. To make those measurements, we first developed a highly specific 18F-labeled PET imaging agent for PD-L1, [18F]DK222, and evaluated its specificity, biodistribution, and human radiation dose estimates. [18F]DK222 enabled quantification of the dynamic changes in PD-L1 levels in the tumor bed, and also allowed in vivo assessment and comparison of the PD effects of different aPD-1 mAbs at the tumor. In addition, our approach allowed us to delineate the differential effects of various aPD-L1 mAbs on PD-L1 occupancy within tumors, and to relate target engagement to efficacy of treatment. With a growing number of immunooncology clinical trials and a shrinking number of patients per clinical trial, such noninvasive PD measures could form a bridge between preclinical experiments and clinical studies, and assist in drug development, effective dose determination, and therapy optimization. Our approach captured and quantified the contributions of mAb properties, peculiarities of tissue and tumor characteristics, and other individual variables into a single noninvasive quantitative parameter measured as the accessible target level. Since these PD measures are agnostic to disease type, location, and treatment, they enable the application of this approach in a broad range of cancers. Moreover, our versatile strategy is generalizable to any aPD-1/aPD-L1 mAb, and our work provides a roadmap for developing similar reagents to characterize other targeted molecular therapeutics.
Parameters that are often used to define drug behavior, such as volume of distribution, half-life, and clearance, provide important information on the in vivo drug disposition and for devising logical therapeutic regimens (30); however, these parameters fail to inform drug accessibility for targeted tumor cells. Tumor penetration of large therapeutics such as mAbs is complex, slow, and passive, and it hinges on their ability to navigate the tumor interstitium (35). mAbs conjugated with radionuclides are routinely used to gain insights into their biodistribution and target expression, and nearly 26 such agents are currently in clinical trials (9). For example, recent studies with Zr-89–labeled atezolizumab have highlighted the potential of PET to quantify intra- and intertumoral heterogeneity in PD-L1 expression (16).
Despite these advances, there remains a need for imaging agents that provide high-contrast images and also are compatible with a standard clinical workflow. Here we show that the [18F]DK222 peptide exhibits PK and biodistribution features that are distinct from those of reported PD-L1 imaging agents (16, 33, 34, 36–39). Moreover, [18F]DK222 possesses all the salient features required for routine clinical use, including high affinity and specificity for PD-L1 levels, and high image contrast at 60 min.
Radiolabeled mAb accumulation in tumors could be indicative of tumor response to therapy. In a previously reported study, [89Zr]atezolizumab signal in the tumors acquired after multiple days of radiotracer injection was found to be a better predictor of tumor response to atezolizumab therapy than IHC and RNA sequencing–based predictive biomarkers (16). While these results provide important evidence to support the use of a radiolabeled mAb to monitor mAb distribution, [89Zr]mAb imaging is impractical for head-to-head comparisons between mAb therapeutics, or to gain deeper insights into their distribution and activity at the tumor (12). As we show here, radiopharmaceuticals with high affinity and faster PK, such as [18F]DK222, can be useful beyond baseline PD-L1–level quantification and for therapy guidance. Such measurements have the potential to evaluate in situ pharmacologic activity of different aPD-L1 mAbs, as shown by the prolonged target engagement that we observed for durvalumab compared with other aPD-L1 mAbs in our preclinical models.
Importantly, these PD measures, as captured by mixed-effects statistical analyses, encapsulate multiple factors that influence mAb concentrations, including PD-L1 surface expression levels and turnover, complex serum and tumor kinetics (or fate) of mAbs at the tumor, and tumor-intrinsic parameters such as high interstitial pressure and poor vascularity, which impede mAb penetration and accumulation. Our method proved capable of measuring distinct PK characteristics of three mAbs (atezolizumab, avelumab, and durvalumab) and the tumor residence kinetics of these mAbs do not mirror circulating half-life profiles but reflect a combination of factors including mAb charge, pI, and affinity for PD-L1 (40). Moreover, implementation of our approach in the absence or presence of collagenase illustrated the sensitivity of our technique, and revealed its capacity to report on changes in drug penetration and distribution.
The approach and findings of the current study have significant implications for accelerating drug development and evaluation and improving treatments. Predictive computational models are routinely used in clinical development for optimizing mAb dosing and selecting appropriate therapies (41), but personalized cancer treatment based on preclinical mechanistic models can often be misleading due to lack of translatability to humans. The noninvasive measurements shown here, if applied to patients, could help bridge the gap between predictive models and human therapy. Additionally, the rapid emergence of next-generation mAb therapeutics, such as probodies that are specifically activated in the TME (42) and multispecific mAb conjugates that enable higher-avidity binding by allowing simultaneous binding to multiple targets (43), introduces a multitude of compounds that are likely to exhibit PK properties that diverge from traditional in silico models. These novel therapeutics will thus require new approaches such as in situ measurement of PD effects, which take into account their pharmacological activity at the site of the tumor.
Our “target-centric” rather than “drug-centric” approach is particularly effective for evaluating cumulative effects of drug penetration and target-specific retention of mAbs in tumors, independent of their biophysical characteristics. We have shown that quantitative PD of the target can be used as a common denominator for the evaluation of pharmacologic activity of various mAbs targeting different proteins (PD-L1 and PD-1), which is a key step toward integrating experimental PD data into mathematical models. One could enhance the predictive power of in silico models by integrating tumor PD data, prior drug information, and data from patients to optimize the mAb dose needed to maximize therapeutic response. As development and use of mAbs and biosimilars grow, applying our target-centric approach could facilitate selection of drugs with the desired tumor penetration and targeting properties, and their optimized dosing to reduce therapeutic resistance. Furthermore, use of our tools for quantification of PK and PD features at the tumor site could advance the design of personalized therapies tailored to the unique properties of each patient. Overall, our approach empowers the molecular understanding of drug activity at the site of disease, which can transform cancer therapeutic development.
Materials and Methods
Chemicals.
DK221 was custom-synthesized by CPC Scientific with >95% purity. NCS-MP-NODA was purchased from CheMatech Macrocycle Design Technologies (C110). All other chemicals were purchased from Sigma-Aldrich or Fisher Scientific.
Cell-Culture Reagents and Antibodies.
All cell-culture reagents were purchased from Invitrogen. The aPD-L1 mAbs (atezolizumab, avelumab, and durvalumab) and aPD-1 mAbs (nivolumab and pembrolizumab) were purchased from the Johns Hopkins School of Medicine Pharmacy.
Synthesis of DK222.
DK221 is a 14–amino acid cyclic peptide with the sequence cyclo-(-Ac-Tyr-NMeAla-Asn-Pro-His-Glu-Hyp-Trp-Ser-Trp(carboxymethyl)-NMeNle-NMeNle-Lys-Cys-)-Gly-NH2. It was previously reported as peptide 6297 (20). The NODA-conjugated analog of DK221 (cyclo-(-Ac-Tyr-NMeAla-Asn-Pro-His-Glu-Hyp-Trp-Ser-Trp(carboxymethyl)-NMeNle-NMeNle-Lys(NODA_NCS[18F]AlF)-Cys-)-Gly-NH2) was prepared as described in SI Appendix, Methods and referred to as DK222. The purified DK222 was characterized by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Calculated [M+H]+: 2349.68; observed: 2349.06 (SI Appendix, Figs. S1 and S2).
[18F]DK222 Radiopharmaceutical Preparation.
The [18F]fluoride (noncarrier added) received from the Johns Hopkins University PET Center cyclotron was trapped on a preconditioned Chromafix 30-PS-HCO3 cartridge. The cartridge was subsequently washed with metal-free water (5 mL). 18F was eluted from the cartridge with 100 µL of 0.4 M KHCO3. The pH of the solution was adjusted to ∼4.0 with 10 µL of metal-free glacial acetic acid, followed by the addition of 20 µL of 2 mM AlCl3.6H2O in 0.1 M sodium acetate buffer (pH 4). The resulting solution was incubated at room temperature for 2 to 4 min to form an Al18F complex. The precursor DK222 (∼100 μg, 42 nmol) was dissolved in 300 µL of a 2:1 solution of acetonitrile and NaOAc (0.1 M, pH 4.0) and then added to the vial containing Al18F. The resulting reaction mixture was heated at 110 °C for 15 min. Then, the reaction vial was cooled to room temperature and diluted with 400 µL deionized water. The obtained aqueous solution containing the radiolabeled product was purified on a reversed-phase high-performance liquid chromatography (RP-HPLC) system (Varian ProStar) with a 1260 Infinity photodiode array detector (Agilent Technologies). A semipreparative C-18 Luna column (5 mm, 10 × 250 mm; Phenomenex) was used with a gradient elution starting with 50% methanol (0.1% trifluoroacetic acid; TFA) and reaching 90% methanol in 30 min at a flow rate of 5 mL/min with water (0.1% TFA) as cosolvent. The radiolabeled product, [18F]DK222, eluting at a retention time of ∼16.2 min, was collected, evaporated under high vacuum, formulated with saline containing 10% EtOH, sterile-filtered, and used for in vitro and in vivo evaluation. The radiochemical purity, chemical identity, and in vitro stability HPLC chromatograms are shown in SI Appendix, Fig. S2.
Cell Culture.
Seven cell lines were used for in vitro and in vivo evaluation: MDAMB231 and SUM149 (TNBC), LOX-IMVI, MeWo, and A375 (melanoma), CHO, and CHO cells constitutively expressing PD-L1 (hPD-L1).
Cell lines and cell-culture conditions, PD-L1 protein expression, binding studies using biolayer interferometry, detection of PD-L1 expression by flow cytometry, and in vitro binding assays with [18F]DK222 are described in SI Appendix, Methods.
In Vivo Studies.
All mouse studies were conducted under Johns Hopkins University Animal Care and Use Committee–approved protocols. Xenografts were established in 5- to 6-wk-old, male or female, NSG mice obtained from the Johns Hopkins University Immune Compromised Animal Core. huPBMC mice were purchased from The Jackson Laboratory and used for experiments as is.
Xenograft Models.
Mice were implanted in the rostral end with MDAMB231 (2 × 106, orthotopic), SUM149 (5 × 106, orthotopic), LOX-IMVI (5 × 106, intradermal), MeWo (5 × 106, intradermal), or A375 (2 × 106, intradermal) cells. Cells were inoculated in opposite flanks if two cell lines were used, with the cell line expressing high PD-L1 on the right side of the mouse. Mice with tumor volumes of 200 to 400 mm3 were used for treatment, imaging, or biodistribution experiments.
PET–Computed Tomography Imaging of Mouse Xenografts.
To determine the in vivo distribution and pharmacokinetics of [18F]DK222, we acquired PET images at multiple time points. Mice with MDAMB231 tumors were injected with ∼200 μCi (7.4 MBq) of [18F]DK222 in 200 μL of saline intravenously (n = 3) and anesthetized under 3% isoflurane prior to being placed on the scanner. PET images were acquired at 15, 60, and 120 min or at 60 min only after radiotracer injection in two bed positions at 5 min per bed in an ARGUS small-animal PET-CT (computed tomography) scanner (Sedecal) as described. A CT scan (512 projections) was performed at the end of each PET scan for anatomical coregistration.
PET imaging, ex vivo biodistribution, and flow cytometry analysis of tumor component details are described in SI Appendix, Methods.
aPD-1 and aPD-L1 mAb Dosing Studies.
Data Analysis.
Statistical analyses were performed using Prism 8 software (GraphPad Software). Unpaired Student’s t test and one- or two-way ANOVA were utilized for column, multiple-column, and grouped analyses, respectively. P values < 0.05 were considered statistically significant. Statistical methods for analyzing mAb activity in the tumor bed are described in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Dr. Martin Pomper for support and the Johns Hopkins University PET Center for 18F production. We thank Addgene for providing plasmids. Grant funding for this study was provided by the Allegheny Health Network–Johns Hopkins Cancer Research Fund (to S.N.), NIH 1R01CA236616 (to S.N.), and NIH P41EB024495. Core resources (flow cytometry, histology, and imaging) were supported by NIH P30CA006973. This work was also supported by the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program, under Award W81XWH-16-1-0323. Opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense.
Footnotes
Competing interest statement: D.K. and S.N. are coinventors on a pending US patent covering [18F]DK222 and as such are entitled to a portion of any licensing fees and royalties generated by this technology. This arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict-of-interest policies. S.N. is a consultant for and receives funding from Precision Molecular, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107982118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Bartelink I. H., et al., Tumor drug penetration measurements could be the neglected piece of the personalized cancer treatment puzzle. Clin. Pharmacol. Ther. 106, 148–163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain R. K., Physiological barriers to delivery of monoclonal antibodies and other macromolecules in tumors. Cancer Res. 50 (suppl. 3), 814s–819s (1990). [PubMed] [Google Scholar]
- 3.Ribas A., Wolchok J. D., Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller M. A., Weissleder R., Imaging of anticancer drug action in single cells. Nat. Rev. Cancer 17, 399–414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon G. M., Niphakis M. J., Cravatt B. F., Determining target engagement in living systems. Nat. Chem. Biol. 9, 200–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez Molina D., et al., Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Glassman P. M., Balthasar J. P., Physiologically-based pharmacokinetic modeling to predict the clinical pharmacokinetics of monoclonal antibodies. J. Pharmacokinet. Pharmacodyn. 43, 427–446 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Arlauckas S. P., et al., In vivo imaging reveals a tumor-associated macrophage-mediated resistance pathway in anti-PD-1 therapy. Sci. Transl. Med. 9, eaal3604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries E. G. E., et al., Integrating molecular nuclear imaging in clinical research to improve anticancer therapy. Nat. Rev. Clin. Oncol. 16, 241–255 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Weissleder R., Schwaiger M. C., Gambhir S. S., Hricak H., Imaging approaches to optimize molecular therapies. Sci. Transl. Med. 8, 355ps16 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Hargreaves R. J., Rabiner E. A., Translational PET imaging research. Neurobiol. Dis. 61, 32–38 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Yu J. X., et al., Trends in clinical development for PD-1/PD-L1 inhibitors. Nat. Rev. Drug Discov. 19, 163–164 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Anagnostou V., et al., Immuno-oncology trial endpoints: Capturing clinically meaningful activity. Clin. Cancer Res. 23, 4959–4969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu S., et al., Comparison of biomarker modalities for predicting response to PD-1/PD-L1 checkpoint blockade: A systematic review and meta-analysis. JAMA Oncol. 5, 1195–1204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaughlin J., et al., Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol. 2, 46–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bensch F., et al., 89Zr-atezolizumab imaging as a non-invasive approach to assess clinical response to PD-L1 blockade in cancer. Nat. Med. 24, 1852–1858 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Niemeijer A. N., et al., Whole body PD-1 and PD-L1 positron emission tomography in patients with non-small-cell lung cancer. Nat. Commun. 9, 4664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar D., et al., Peptide-based PET quantifies target engagement of PD-L1 therapeutics. J. Clin. Invest. 129, 616–630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterjee S., et al., Rapid PD-L1 detection in tumors with PET using a highly specific peptide. Biochem. Biophys. Res. Commun. 483, 258–263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller M. M., et al., Macrocyclic Inhibitors of the PD1/PDL1 and CD80 (B7-1)/PD-L1 Protein/Protein Interactions (Bristol-Myers Squibb, 2016). [Google Scholar]
- 21.McBride W. J., et al., Improved 18F labeling of peptides with a fluoride-aluminum-chelate complex. Bioconjug. Chem. 21, 1331–1340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Topalian S. L., Drake C. G., Pardoll D. M., Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minn A. J., Wherry E. J., Combination cancer therapies with immune checkpoint blockade: Convergence on interferon signaling. Cell 165, 272–275 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Huang A. C., et al., A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat. Med. 25, 454–461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taube J. M., et al., Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci. Transl. Med. 4, 127ra37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolor A., Szoka F. C. Jr, Digesting a path forward: The utility of collagenase tumor treatment for improved drug delivery. Mol. Pharm. 15, 2069–2083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKee T. D., et al., Degradation of fibrillar collagen in a human melanoma xenograft improves the efficacy of an oncolytic herpes simplex virus vector. Cancer Res. 66, 2509–2513 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Roopenian D. C., Akilesh S., FcRn: The neonatal Fc receptor comes of age. Nat. Rev. Immunol. 7, 715–725 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Centanni M., Moes D. J. A. R., Trocóniz I. F., Ciccolini J., van Hasselt J. G. C., Clinical pharmacokinetics and pharmacodynamics of immune checkpoint inhibitors. Clin. Pharmacokinet. 58, 835–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheng J., et al., Clinical pharmacology considerations for the development of immune checkpoint inhibitors. J. Clin. Pharmacol. 57 (suppl. 10), S26–S42 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Wilkins J. J., et al., Time-varying clearance and impact of disease state on the pharmacokinetics of avelumab in Merkel cell carcinoma and urothelial carcinoma. CPT Pharmacometrics Syst. Pharmacol. 8, 415–427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boswell C. A., et al., Effects of charge on antibody tissue distribution and pharmacokinetics. Bioconjug. Chem. 21, 2153–2163 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Chatterjee S., et al., A humanized antibody for imaging immune checkpoint ligand PD-L1 expression in tumors. Oncotarget 7, 10215–10227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jagoda E. M., et al., Immuno-PET imaging of the programmed cell death-1 ligand (PD-L1) using a zirconium-89 labeled therapeutic antibody, avelumab. Mol. Imaging 18, 1536012119829986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain R. K., Barriers to drug delivery in solid tumors. Sci. Am. 271, 58–65 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Donnelly D. J., et al., Synthesis and biologic evaluation of a novel 18F-labeled adnectin as a PET radioligand for imaging PD-L1 expression. J. Nucl. Med. 59, 529–535 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Lesniak W. G., et al., Development of [18F]FPy-WL12 as a PD-L1 specific PET imaging peptide. Mol. Imaging 18, 1536012119852189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maute R. L., et al., Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc. Natl. Acad. Sci. U.S.A. 112, E6506–E6514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heskamp S., et al., Noninvasive imaging of tumor PD-L1 expression using radiolabeled anti-PD-L1 antibodies. Cancer Res. 75, 2928–2936 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Tan S., et al., Distinct PD-L1 binding characteristics of therapeutic monoclonal antibody durvalumab. Protein Cell 9, 135–139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agoram B. M., Martin S. W., van der Graaf P. H., The role of mechanism-based pharmacokinetic-pharmacodynamic (PK-PD) modelling in translational research of biologics. Drug Discov. Today 12, 1018–1024 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Giesen D., et al., 89Zr-labeled anti-PD-L1 CX-072 PET imaging in human xenograft and syngeneic tumors. Ann. Oncol. 30 (suppl. 1), I4 (2019). [Google Scholar]
- 43.Ravi R., et al., Bifunctional immune checkpoint-targeted antibody-ligand traps that simultaneously disable TGFβ enhance the efficacy of cancer immunotherapy. Nat. Commun. 9, 741 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kingsbury J. S., et al., A single molecular descriptor to predict solution behavior of therapeutic antibodies. Sci. Adv. 6, eabb0372 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.