Significance
Successful biologic drug discovery and development involves finding functional as well as developable candidates. Once a candidate has been demonstrated to be functional, the next step is to determine whether it can be translated into a drug product. This requires that the candidate can withstand stresses encountered during manufacturing, shipping, and storage. Additionally, it must be safe, efficacious, and possess good pharmacology. In silico analyses of the variable regions of 77 marketed antibody-based biotherapeutics have revealed five nonredundant physicochemical descriptors. Distributions of these descriptors, observed for marketed biotherapeutics, can help prioritize a drug candidate for experimental testing at early discovery stages, guide engineering efforts to further optimize it, and help increase the productivity of biologic drug discovery and development.
Keywords: biologics, antibody, developability, computation, drug discovery
Abstract
Feeding biopharma pipelines with biotherapeutic candidates that possess desirable developability profiles can help improve the productivity of biologic drug discovery and development. Here, we have derived an in silico profile by analyzing computed physicochemical descriptors for the variable regions (Fv) found in 77 marketed antibody-based biotherapeutics. Fv regions of these biotherapeutics demonstrate significant diversities in their germlines, complementarity determining region loop lengths, hydrophobicity, and charge distributions. Furthermore, an analysis of 24 physicochemical descriptors, calculated using homology-based molecular models, has yielded five nonredundant descriptors whose distributions represent stability, isoelectric point, and molecular surface characteristics of their Fv regions. Fv regions of candidates from our internal discovery campaigns, human next-generation sequencing repertoires, and those in clinical-stages (CST) were assessed for similarity with the physicochemical profile derived here. The Fv regions in 33% of CST antibodies show physicochemical properties that are dissimilar to currently marketed biotherapeutics. In comparison, physicochemical characteristics of ∼29% of the Fv regions in human antibodies and ∼27% of our internal hits deviated significantly from those of marketed biotherapeutics. The early availability of this information can help guide hit selection, lead identification, and optimization of biotherapeutic candidates. Insights from this work can also help support portfolio risk assessment, in-licensing, and biopharma collaborations.
Antibody-based drug products have emerged as the best-selling class of biopharmaceuticals in recent years. However, only 18% of biotherapeutic drug candidates entering Phase I clinical trials today will be available in the market after several years (1). This is because discovery of functional biotherapeutic candidates is only an initial step. Newly discovered drug candidates must translate into drug products via a series of product development processes and clinical trials. Requirements of chemistry, manufacturing, and control (CMC), pharmacology, safety, and efficacy as well as business decisions influence chances of successful translation of discoveries into drug products (2–4). Therefore, drug discovery scientists need to include considerations of developability (5) along with function while nominating a biotherapeutic candidate for development. In recent years, the concept of developability has gained acceptance in the biopharmaceutical industry, and several approaches are being developed as manifest in the book edited by Kumar and Singh (5). However, developability is often interpreted as being limited to CMC and biophysical aspects of drug product development (6–13). Concurrently, the concept of “drug likeness” is also being developed for biopharmaceuticals by analyzing sequences and structural models of antibody-based biotherapeutic candidates in the clinic (14, 15). Both trends can help improve the productivity of biologic discovery and development projects by enabling a greater number of biotherapeutic candidates to reach clinical development (1). There is, however, significant attrition during all three stages of clinical trials (1), and it is therefore important to make a distinction between antibody-based biotherapeutics already in the market and drug candidates in Phases I to III of the clinical trials.
Numerous factors, both intrinsic and extrinsic to the primary sequence, contribute significantly toward successful translation of a biologic drug candidate as it progresses through a complex series of processes. The generation of antibodies against a target is usually the first stage in biotherapeutic drug discovery and development projects. At this stage, only amino acid sequences of potential hits are available. Is it feasible to estimate whether a newly discovered hit has a potential to become a biotherapeutic product? We hypothesize that a truly developable drug candidate should possess an intrinsic physicochemical profile that would embody manufacturability, safety, efficacy, and pharmacology in a holistic manner. Many of these aspects are interrelated because physicochemical characteristics of a biotherapeutic are inherent to its in vivo performance. For example, most aggregation-prone regions found in therapeutic antibody sequences and major histocompatibility complex II T cell immune epitopes overlap with the complementarity determining regions (CDRs) (16–18). Charge and isoelectric point (pI) of therapeutic antibodies play a role in their pharmacokinetics and pharmacodynamics (PK/PD), and clearance (4, 19, 20). Some undesirable events, other than those linked to the specific biology of a target, may also be inherent to a biotherapeutic drug candidate. For example, the generation of the antidrug antibodies (ADAs) against biotherapeutics can affect their immunogenicity, safety, efficacy, stability in human serum, PK/PD, and clearance (21–26). In addition to intrinsic factors, many external elements associated with manufacturing, formulation and product development, safety, toxicology, pharmacology, clinical trial designs and outcomes, and risk versus benefit profile of a biotherapeutic drug candidate in relation to patient indication(s) can also significantly affect its translation.
We have derived an intrinsic physicochemical profile by analyzing sequence and structural attributes of 79 Fv regions from 77 approved antibody-based biotherapeutics via computational means. In this first step, we chose Fv regions of marketed biotherapeutics as the most fundamental aspect to be studied objectively via standardized computational methods because they are amenable to high-throughput modeling, descriptor calculations, and analyses. The intrinsic physicochemical characteristics that can be studied using computational means include a) amino acid sequence patterns such as aggregation-prone regions, chemical liability motifs, and immune epitopes; b) molecular surface features such as three-dimensional locations of charged and hydrophobic residues; and c) physicochemical descriptors, such as pI, charge, and hydrophobicity, computed using homology-based molecular models. Expectedly, computed physicochemical descriptors are related to biophysical aspects of antibody solution behavior such as stability, viscosity, and aggregation (4, 27). However, physicochemical descriptors were also found to be correlated with other aspects such as PK/PD of monoclonal antibodies (4, 28, 29). Recently, Tessier and coworkers have derived amino acid composition–based physicochemical rules to distinguish antibody sequences that drive nonspecific interactions from those that do not (30, 31). Additionally, Finlay et al. have shown that re-engineering the paratope for an anti–PD-1 antibody, camrelizumab, mitigated its poly-specificity issues (32). Therefore, physicochemical attributes encoded by sequence and structural properties of biotherapeutic candidates can influence their behaviors both in vitro and in vivo.
An intrinsic physicochemical profile derived by analyzing molecular sequence and structural characteristics of marketed biotherapeutics, all of which have been proven to withstand different physicochemical stresses during commercial scale manufacturing, shipping, storage, and administration as well as possess acceptable safety, efficacy, and pharmacology as tested in clinical trials and the market, can be valuable toward the following use cases: 1) help prioritize thousands of hits generated from antibody discovery campaigns for initial small scale production and experimental testing; 2) contribute toward lead identification from a pool of hits shown to be functional and of reasonable biophysical quality from initial experiments; 3) guide optimization of the lead molecules for developability by identifying physicochemical properties with suboptimal values; 4) analyze biotherapeutic candidates currently in development from a perspective of portfolio risk assessment; and finally, 5) contribute toward molecular assessments during in-licensing collaborations among academic/industrial partners.
We hypothesize that the holistic physicochemical profile derived in this work shall contribute toward improved productivity of drug discovery and development projects. It should also be mentioned that there is little self-consistent experimental data on marketed biotherapeutics in the public domain yet, although a few noteworthy attempts have been made (14, 33). Analogous to similar rules proposed to guide small molecule drug discovery (34–36), we anticipate that this work will prove useful for biotherapeutics as well.
Results
A General Survey of Antibody-Based Biotherapeutics Currently Available in Market.
Dataset S1 provides names, sequences, and molecular formats of all 78 antibody-based biotherapeutics that were available in the market as of early 2020. Of them, 77 have been approved for human use, and one (lokivetmab) is for the treatment of atopic dermatitis in canines. Of these 78 biotherapeutics, 76 (97%) are monospecific antibodies, and two are bispecific antibodies (emicizumab and blinatumomab). Full-length monoclonal antibody (mAb, 72 out of 78, 92%) is the most common molecular format among these 78 biotherapeutics. Among the full-length mAbs, 51 (71%) are IgG1s, 9 (12%) are IgG2s, and 12 (17%) are IgG4s. Four of the remaining six biotherapeutics are Fabs (namely, abciximab, certolizumab, idarucizumab, and ranibizumab), one is scFv (BiTE, bispecific T cell engager, blinatumomab), and the last one is Fv (moxetumomab). The light chains for 73 (92%) of the 78 biotherapeutics are of kappa (κ) isotype, and the remaining five (6%) are lambda (λ) (Dataset S2). Interestingly, all λ light chains belong to full-length mAbs; heavy chain isotypes in three (avelumab, belimumab, and guselkumab) of them are immunoglobulin G1 (IgG1), and the remaining two (erenumab and evolocumab) are IgG2.
The marketed biotherapeutics serve several therapeutic areas including oncology, inflammation, autoimmune disorders, and chronic diseases, among others (Dataset S2). They also come from diverse sources. Of them, 28 (36%) are fully human, 34 (44%) are humanized, 9 (12%) are chimeric, 6 (8%) are murine, and the last one (lokivetmab) is of canine origin. We focus on 79 Fvs from 77 biotherapeutics for human use in this work and exclude lokivetmab from further analyses.
There Is No Germline Pair Preference among Marketed Biotherapeutics.
Currently marketed antibody-based biotherapeutics utilize diverse germlines. Heavy chains in 15 (19%) of them belong to a single germline, namely, IGHV1-46*01, while the remaining 62 heavy chains are distributed across 32 different germlines as shown in SI Appendix, Fig. S1A. Similarly, 31 light chains belong to one of the following three germlines, IGKV1-39*01, IGKV3-11*01, and IGKV1-33*01 (SI Appendix, Fig. S1B). The remaining 46 light chains belong to 25 different germlines as shown in SI Appendix, Fig. S1B. Table 1 presents the five most frequent variable heavy (VH) and variable light (VL) germline pairs found in 77 marketed biotherapeutics. Interestingly, the most common heavy chain germline IGHV1-46*01 pairs with the most common light chain germline IGKV1-39*01 only five times (Table 1). Additionally, only three Fv regions pair IGHV1-46*01 with IGKV3-11*01, the second most common light chain germline. These observations show that there are no germline pairing preferences among the marketed antibody-based biotherapeutics.
Table 1.
Five most frequent VH and VL germlines in 77 marketed antibody-based biotherapeutics
| VH germline | No. | VL germline | No. | No. of germline pairs |
| IGHV1-46*01 | 15 | IGKV1-39*01 | 11 | 5 |
| IGHV1-46*01 | 15 | IGKV3-11*01 | 11 | 3 |
| IGHV3-23*04 | 5 | IGKV1-NL1*01 | 5 | 3 |
| IGHV3-7*01 | 3 | IGKV3-20*01 | 6 | 2 |
| IGHV1-3*01 | 4 | IGKV1-33*01 | 9 | 2 |
Diversity in CDR Lengths.
Lengths of four out of the six CDR loops remain constant in 77 marketed antibody-based biotherapeutics, while those of the remaining two CDRs vary significantly (SI Appendix, Table S1). The first CDRs in light chains (LCDR1s) range from 10 to 17 (average = 12 ± 2) residues in length, while lengths of LCDR2 (seven residues) and LCDR3 (9 ± 1) loops remain constant. Among the heavy chains, the first and the second CDRs (HCDR1 and HCDR2) are also constant in length (HCDR1, 10 ± 1; HCDR2, 17 ± 1), while the HCDR3 loops vary in length from merely four residues in dinutuximab to 21 residues in erenumab (average length =11 ± 3, SI Appendix, Table S1). These findings agree with an earlier study by Raybould et al. that involved 242 clinical-stage antibodies (15). HCDR3 length diversity in biotherapeutics reflects the diversity of antigens these antibodies bind to since HCDR3 loops are often the major contributors toward antigen binding (16).
CDR Charge Diversity.
CDRs in VL and VH domains of biotherapeutics show significant diversity in charge. The total charge on CDRs in VL and VH domains of 77 marketed biotherapeutics is shown in SI Appendix, Fig. S2. This calculation was performed using amino acid sequences of CDRs and does not consider their conformational attributes. There are 79 Fvs within the 77 marketed biotherapeutics because one of them is a bispecific antibody, and another is a BiTE. Salient observations are described below:
-
1)
Of VL CDRs, 53 (67.1%) are positively charged, 17 (21.25%) are neutral, and 9 (11.25%) are negatively charged. On the other hand, 39 (49.4%) VH CDRs are negatively charged, 27 (34%) are neutral, and only 13 (16%) are positively charged (SI Appendix, Fig. S2A). This leads to CDR charge asymmetry in 56 (∼71%) of the 79 Fv regions. In 32 of these 56 Fvs, the CDRs in VH and VL domains are oppositely charged, while the remaining 24 Fvs contain either the VH or VL domain with the total charge on their CDRs being 0. Similar observations have been previously made for clinical-stage antibodies (15).
-
2)
Out of the 32 Fvs, 30 (∼94%) with oppositely charged CDRs are comprised of the negatively charged CDRs in VH domains and the positively charged CDRs in VL domains. The remaining two oppositely charged Fvs are comprised of the positively charged CDRs in the VH and the negatively charged CDRs in the VL domains (SI Appendix, Fig. S2 B and C).
-
3)
Among the 24 Fvs that contain CDRs with a total charge of 0 in either VH or VL domains, 14 (58%) contain CDRs with 0 total charge in their VH domains and positively charged CDRs in their VL domains. Three Fvs comprise CDRs with zero total charge in VH and negatively charged CDRs in VL domains. Negatively charged CDRs in VH domains and CDRs with zero total charge in VL domains were observed for five Fvs. The remaining two of the 24 Fvs contain positively charged CDRs in VH, and those in VL domains have zero total charge.
-
4)
Of the 79 Fvs, 23 (∼29%) do not show CDR charge asymmetry. CDRs in both the domains have 0 total charge in 10 of these 23 Fvs. Another nine of these 23 Fvs contain positively charged CDRs, and the remaining four contain negatively charged CDRs in both the domains (SI Appendix, Fig. S2 B and C).
CDR Hydrophobicity.
Hydrophobic residues also show differences in their incidences within the CDR loops. The average number of hydrophobic residues (A, I, L, F, W, Y, V, M, G, and P) in LCDR1, LCDR2, and LCDR3 loops of VL are 5 ± 1, 3 ± 1, and 4 ± 1, respectively (SI Appendix, Table S1). They account for 43 ± 7, 38 ± 12, and 42 ± 12% of all the residues in these three light chain CDRs, respectively. In the case of VH domains, the average number of hydrophobic residues in HCDR1, HCDR2, and HCDR3 are 6 ± 1, 9 ± 1, and 7 ± 2, respectively (SI Appendix, Table S1). They account for 58 ± 8, 51 ± 8, and 66 ± 13% of all residues in these three heavy chain CDRs, respectively. In summary, a majority of the residues in light chain CDRs are hydrophilic, while those in heavy chain CDRs are hydrophobic in the 79 Fvs.
The diversities in CDR lengths, charges, and hydrophobicity are likely driven by diversities in the electrostatic and hydrophobic characteristics of the antigens as well as of the epitope regions the marketed biotherapeutics recognize. However, these diversities can also have important implications for their in vitro and in vivo behaviors.
Physicochemical Attributes of the Fv Regions of Marketed Biotherapeutics.
Homology-based molecular models of the 79 Fv regions of the 77 marketed biotherapeutics were used to compute different protein descriptors available in MOE2018. These descriptors cover stability, electrostatic, and hydrophobic features of the Fv regions and their molecular surfaces.
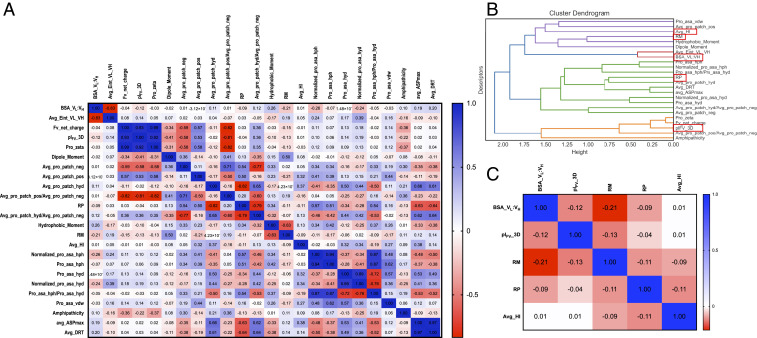
Several computed descriptors such as protein mass, Debye length, and so on remain nearly constant among all the 79 Fvs. These were discarded, and a set of 24 descriptors that show larger variations were selected for further analyses. Pairwise Pearson’s linear correlation coefficients show statistically significant correlations among them (Fig. 1A). A clustering of descriptors based on Pearson correlations among them allowed us to select five nonredundant descriptors with different physicochemical meanings. These descriptors show low values of pairwise correlations with one another (Materials and Methods and Fig. 1 B and C). To test the robustness of the method used in our work, this process was repeated four more times by setting aside 10 randomly selected biotherapeutics. There were minor differences in the clusters due to the smaller number of data points (69 Fvs instead of 79 Fvs). However, this did not affect selection of the five nonredundant descriptors (SI Appendix, Fig. S3 and Dataset S2). The distributions of these five descriptors inform us about stability, electrostatics, and molecular surface properties of Fv regions.
Fig. 1.
(A) Linear correlations among physicochemical descriptors computed from homology-based models of the 79 Fv regions of 77 marketed antibody-based biotherapeutics. Correlations among these descriptors range from −1 (red) to +1 (blue). (B) Cluster analysis of 24 descriptors that show significant variations among different Fv regions. These 24 descriptors were grouped into five clusters shown in assorted colors. The five noncorrelated descriptors selected from these clusters are highlighted in the red boxes. (C) Selected five noncorrelated descriptors. These descriptors demonstrate low statistical correlation coefficients (r < 0.3) among themselves and have different physicochemical meaning. These properties are surface area buried between VL and VH domains (BSA_VL: VH), structure-based isoelectric point (pIFv_3D), ratio of dipole and hydrophobic moments (RM), ratio of charged to hydrophobic surface patches (RP), and hydrophobic anisotropy (Avg_HI).
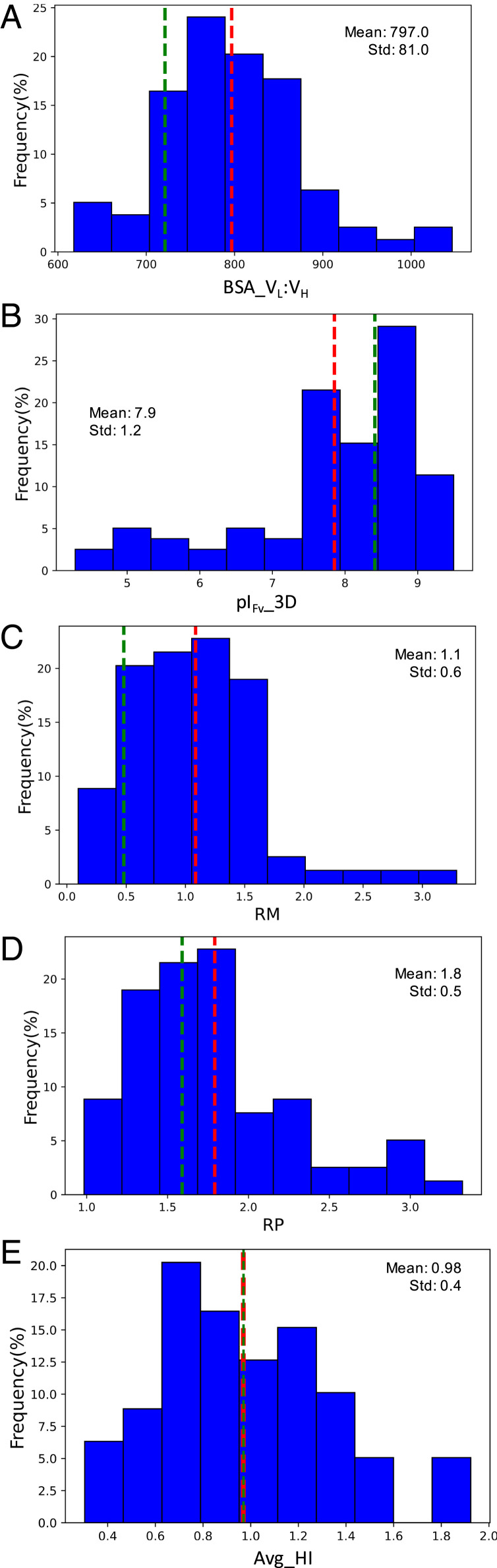
Table 2 and Fig. 2 summarize distributions of the five nonredundant physicochemical descriptors for the 79 Fvs. These distributions were further divided into different classes based on the year of approval for biotherapeutics (older versus newer biotherapeutics, with 2015 arbitrarily chosen as the transition year), route of administration (intravenous versus subcutaneous injections), formulation buffer pH (above or below pH 6), and concentration (low versus high concentration liquid formulations). Intrinsic physicochemical profiles were rederived for each of these classes by reclustering the descriptors (SI Appendix, Fig. S4 and Dataset S2) and computing the values of average, SD, and range for each of the five nonredundant descriptors in each class. These results are also shown in Table 2. No statistically significant differences among the average values of five nonredundant descriptors in these categories were observed (P value = 1.0, Table 2). This suggests that the physicochemical profile derived in this work is truly intrinsic to sequence and structural characteristics of Fv portions of the marketed biotherapeutics. The following text discusses insights gained from these distributions of five nonredundant descriptors for all 79 Fvs.
Table 2.
Average, SD, and range values for the five nonredundant descriptors for 79 Fvs in 77 marketed antibody-based biotherapeutics and different subsets
| Descriptors | BSA_VL: VH (Å2) | pIFv_3D | RM(µD/µH) (D) | RP | Avg_HI |
| All 79 Fvs from 77 biotherapeutics approved for human use | 797 ± 81 | 7.85 ± 1.24 | 1.09 ± 0.58 | 1.79 ± 0.51 | 0.97 ± 0.37 |
| (618 to 1,046) | (4.28 to 9.5) | (0.09 to 3.29) | (0.98 to 3.32) | (0.3 to 1.92) | |
| 43 Fvs from 42 biotherapeutics approved through 2014 | 787 ± 73 | 7.8 ± 1.4 | 1 ± 0.5 | 1.8 ± 0.5 | 1 ± 0.3 |
| (618 to 964) | (4.3 to 9.5) | (0.1 to 2.6) | (1.1 to 3.1) | (0.4 to 1.9) | |
| 36 Fvs from 35 biotherapeutics approved in 2015 and onwards | 808 ± 88 | 8 ± 0.9 | 1.1 ± 0.7 | 1.7 ± 0.5 | 1 ± 0.4 |
| (630 to 1,046) | (5.3 to 9.5) | (0.2 to 3.3) | (1 to 3.3) | (0.3 to 1.9) | |
| 28 Fvs from 27 biotherapeutics approved for subcutaneous route of administration | 798 ± 68 | 8.1 ± 0.9 | 1.1 ± 0.7 | 1.8 ± 0.5 | 1.1 ± 0.4 |
| (666 to 955) | 5.3 to 9.5 | 0.1 to 3.3 | (1.2 to 3.3) | 0.3 to 1.9 | |
| 51 Fvs from 50 biotherapeutics approved for intravenous route of administration | 792 ± 86 | 7.8 ± 1.3 | 1.1 ± 0.6 | 1.7 ± 0.5 | 0.9 ± 0.3 |
| (618 to 1,046) | (4.3 to 9.5) | (0.3 to 2.7) | (1 to 3.1) | (0.3 to 1.8) | |
| 28 Fvs from 28 approved biotherapeutics with a formulation pH < 6 | 797 ± 68 | 8 ± 0.9 | 1.1 ± 0.7 | 1.6 ± 0.5 | 0.9 ± 0.4 |
| (644 to 943) | (5.1 to 9.5) | (0.3 to 3.3) | (1 to 3) | (0.3 to 1.9) | |
| 45 Fvs from 43 approved biotherapeutics with a formulation pH ≥ 6 | 794 ± 88 | 7.8 ± 1.3 | 1.1 ± 0.6 | 1.9 ± 0.5 | 1 ± 0.4 |
| (618 to 1,046) | (4.3 to 9.5) | (0.1 to 2.6) | (1 to 3.3) | (0.3 to 1.9) | |
| 26 Fvs from 25 approved biotherapeutics with Low concentration (≤10 mg/mL) formulation | 794 ± 106 | 7.6 ± 1.7 | 1.1 ± 0.6 | 1.9 ± 0.5 | 1 ± 0.4 |
| (618 to 1,046) | (4.3 to 9.5) | (0.4 to 2.7) | (1.2 to 3.1) | (0.4 to 1.8) | |
| 27 Fvs from 27 approved biotherapeutics with high concentration (≥100 mg/mL) formulation | 797 ± 65 | 8.1 ± 0.8 | 1.1 ± 0.6 | 1.8 ± 0.4 | 1 ± 0.4 |
| (666 to 943) | (5.5 to 9.3) | (0.1 to 3.3) | (1.1 to 3) | (0.3 to 1.9) |
The routes of administration for three biotherapeutics are intradermal, intramuscular, or intravitreal. Seven biotherapeutics did not have documented pH values. When a range for pH values was provided, the midpoint of this range was used. All the datasets in this table yield similar values for the five nonredundant descriptors, and the difference among them are statistically insignificant (χ2 = 0.091 and a P value = 1.0).
Fig. 2.
Distributions of the five nonredundant descriptors for the 79 Fv regions from 77 marketed antibody-based biotherapeutics. (A) Buried surface area between the BSA_VL: VH, (B) pIFv_3D, (C) RM, (D) RP and, (E) Avg_HI. The red dotted lines in these plots show the mean values, and the green dotted lines show values for trastuzumab.
Variable Domain Interface Stability: Surface Area Buried between VL and VH Domains (BSA_VL: VH).
Interaction between the VL and VH domains contributes toward the stability of an Fv region and indicates compatibility between them. Lower compatibility between VL and VH domains make Fv regions more flexible, which can potentially initiate misfolding or domain interface rearrangements and lead to reduced antigen-binding affinity (37, 38). Furthermore, smaller surface areas buried between VL and VH domains can also potentially lower an Fv region’s conformational stability at a given temperature (38). In this dataset, BSA_VL: VH in the 79 Fvs of antibody-based biotherapeutics ranges from 618 to 1,046 Å2 (average = 797 ± 81Å2, Fig. 2A). The red dotted line in Fig. 2A indicates average value. The distribution plot shows that the majority lies near the mean, while a few Fv regions exhibit high and low BSA_VL: VH values. For example, the variable domains of inotuzumab and moxetumomab are highly compatible (BSA_VL: VH, 1,046 and 1,026 Å2, respectively), whereas those in dinutuximab and nivolumab show lower than average compatibilities (BSA_VL: VH, 629 and 618 Å2, respectively). Fig. 2A also shows the value of BSA_VL: VH for trastuzumab (736 Å2) as a green dotted line. Throughout the plots of physicochemical descriptors, we have used trastuzumab as a reference because this antibody has been known to possess good physicochemical attributes (14).
Structure-Based pI of Fv Region (pIFv_3D).
The pI of the Fv region of an antibody influences its solution properties (27) in vitro and PK/PD in vivo (19). Both sequence- and structure-based methods can calculate pI, and both have been shown to correlate well with experimental pI measurements (39). Fig. 2B shows the distribution of structure-based pI values for Fv regions of antibody-based biotherapeutics currently available in the market. The pIFv_3D values range from 4.28 to 9.50 (average = 7.9 ± 1.2, Table 2), and the pIFv_3D of trastuzumab is 8.4 (red and green dotted lines in Fig. 2B). The distribution for pIFv_3D is long tailed, with a small population of molecules exhibiting low pIFv_3D values. The lowest pIFv_3D values are shown by abciximab and brentuximab (pIFv_3D = 4.2 and 4.6, respectively), and the greatest pIFv_3D values are shown by alemtuzumab and erenumab (pIFv_3D = 9.5 for both). Values of the formulation buffer pH range from 4.8 to 8 for the 77 marketed biotherapeutics (Dataset S2). The difference between pIFv_3D and formulation buffer pH has been shown to determine whether antibody solutions are repulsive or attractive at high concentrations (27).
The Ratio of Dipole Moment to Hydrophobic Moment.
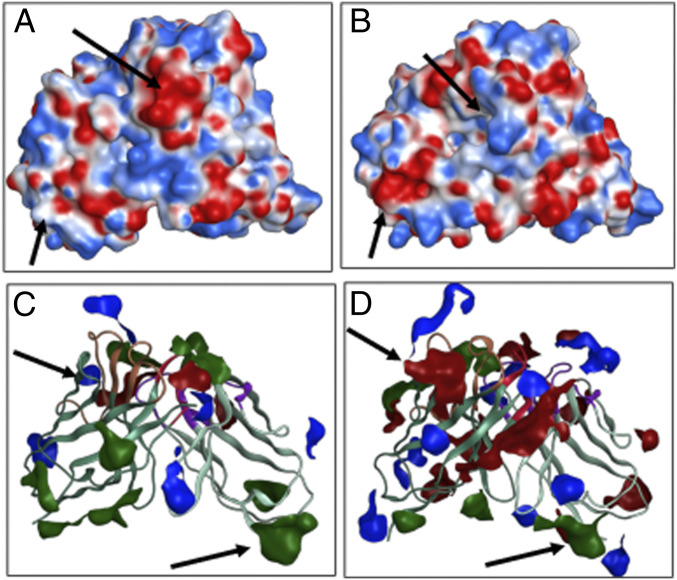
Concentration-dependent solution behavior of a biologic molecule in a given solvent is driven by both solute–solute and solute–solvent interactions (40). At the molecular level, these interactions are both polar and nonpolar. Therefore, distributions of charged and nonpolar residues in a biologic molecule can influence its solution behavior. The descriptors, dipole moment (µD) (11) and hydrophobic moment (µH) (41, 42), inform us about distributions of charged and hydrophobic residues in the Fv regions of marketed antibody-based biotherapeutics. Dipole moment quantifies separation between positively and negatively charged residues in a biologic molecule, while hydrophobic moment quantifies the separation between hydrophobic and hydrophilic residues in it (11). The ratio of dipole to hydrophobic moments (RM) therefore denotes a balance between electrostatic and hydrophobic attributes of the biologic molecules. Average magnitudes of µD and µH for the 79 Fv regions are 316.7 ± 129.6 D (range = 45.5 to 693.8) and 339.9 ± 143.4 (range = 88.3 to 794.9), respectively, and the ratio (RM = µD/µH) has an average of 1.1 ± 0.6 D (range = 0.09 to 3.29, Table 2). Fig. 2C shows the distribution plot for RM. RM of trastuzumab is 0.48 D (µD = 196.3 D and µH = 405.1). Efalizumab shows the lowest RM with a value of 0.09 D (µD = 45.5D and µH = 480.8), while galcanezumab displays the highest RM of 3.29 D (µD = 290.5 D and µH = 88.2). These differences in RM lead to considerable differences in Poisson Boltzmann electrostatic surfaces for the Fv regions of efalizumab and galcanezumab (Fig. 3 A and B).
Fig. 3.
Poisson Boltzmann electrostatic surfaces are shown for (A) efalizumab (RM = 0.09 D) and (B) galcanezumab (RM = 3.60 D). The molecular surface of galcanezumab shows significantly greater electrostatic polarization than efalizumab. Charged (blue for positive and red for negative) and hydrophobic (green) patches on molecular surfaces of (C) cemiplimab (RP = 0.98) and (D) emicizumab_anti-FX (RP = 3.32). A greater portion of the molecular surface of emicizumab is covered by the charged patches, while the hydrophobic patches cover a greater part of the molecular surface of cemplimab. The arrows indicate regions of significant differences in the molecular surfaces.
The Ratio of Surface Areas of Charged Patches to Hydrophobic Patches.
Characteristics of molecular surface patches also influence solution behavior of an antibody. For example, large charged and hydrophobic surface patches have been linked to undesirable aggregation as well as high viscosity in antibody formulations (4, 15, 16, 43). Moreover, large positively charged patches vicinal to the CDRs can lead to nonspecific binding (4, 16, 44, 45). It has been also shown that disrupting the charged and hydrophobic patches can improve solution properties of antibodies (43, 45). A descriptor, RP, was devised to quantify the balance between molecular surface areas covered by charged (sum of the areas of positively charged and negatively charged patches) and hydrophobic patches (Materials and Methods). The distribution of RP is shown in Fig. 2D. The average value of RP for Fv regions of marketed antibody-based biotherapeutics is 1.8 ± 0.5 (range = 0.98 to 3.32, Table 2), and the RP of trastuzumab is 1.57 (green dotted line in Fig. 2D). The lowest RP is found for cemiplimab with a value of 0.98, whereas the highest RP is observed as 3.32 for emicizumab_anti-FX. Therefore, charged and hydrophobic patches on molecular surfaces for the two Fvs show significant differences (Fig. 3 C and D).
Average Hydrophobic Imbalance.
The average hydrophobic imbalance (Avg_HI) measures anisotropy in the distribution of hydrophobic residues on a protein’s surface. It was devised to explain chromatographic behaviors of small proteins by Salagado et al. (46). A small value for Avg_HI suggests that hydrophobic residues are distributed evenly over a protein’s surface, whereas a large Avg_HI value shows that the hydrophobic residues may be localized in a region of its surface. In this work, average HI value for Fv regions in 77 marketed antibody-based biotherapeutics is 1.0 ± 0.4 (range = 0.30 to 1.92, Table 2), and the Avg_HI value for trastuzumab is 0.97 (Fig. 2E). Guselkumab shows the lowest Avg_HI of 0.30, whereas benralizumab has the greatest Avg_HI of 1.92.
Potential Applications of the Intrinsic Physicochemical Profile.
As stated in the introduction, the intrinsic physicochemical profile derived by analyzing Fv regions of 77 antibody-based biotherapeutics can be used in multiple ways. In this section, four potential examples are described. First, Fv regions from 271 CST antibodies (Phase I to III) were analyzed for similarity of their intrinsic physicochemical characteristics to those of the 79 Fv regions found in marketed biotherapeutics from a perspective of portfolio risk evaluation. Second, 14,037 antibodies from human next-generation sequencing (NGS) repertoires, studied by Raybould et al. (15), were studied here to deepen our understanding of the similarity between natural human antibodies and marketed biotherapeutics (47). Third, 3,120 hits from our internal antibody discovery campaigns (2015 to early 2019) were evaluated for their physicochemical similarity to the marketed antibody-based biotherapeutics from the perspective of hit selection during initial stages of drug discovery. Fourth, we analyze specific examples of mAbs from the perspective of lead identification and optimization. Intrinsic physicochemical profiles of the Fv regions from two mAbs are compared as a worked example for lead identification. We then describe two well-known case studies of mAbs that faced challenges in their product development stages. These challenges were later shown to be mitigated via mutations at a single or a few positions in their amino acid sequences (48–50).
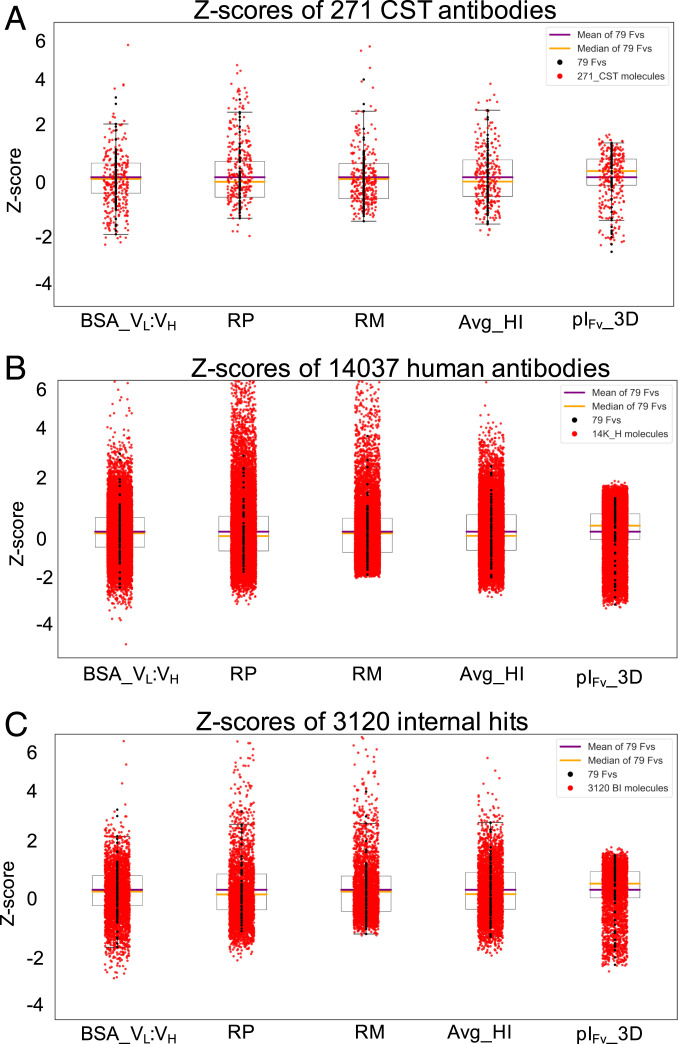
In the first three of these analyses, two different statistical measures, namely, flags and Z-distances, were used. These measures are described in Materials and Methods. Briefly, for each Fv region in the above mentioned sets of antibody sequences, its homology-based model was used to compute the values for five nonredundant descriptors identified in the previous section. Each of these five descriptor values was used to compute Z-scores by comparing them with the average and SD values of the corresponding descriptors for the 79 Fv regions as described in SI Appendix, Eq. 5. The distributions of Z-scores of the five nonredundant descriptors are shown as box plots in Fig. 4 A–C. Each descriptor with Z-score > 1.96 or < −1.96 contributes a flag for an Fv region. Therefore, an Fv region can collect up to five flags. Furthermore, Z-scores for the five nonredundant descriptors of an Fv region were combined as shown in SI Appendix, Eq. 5 to compute its Z-distance. The greater the Z-distance of an Fv, the further it is from the average physicochemical properties of the 79 Fvs from the 77 marketed biotherapeutics.
Fig. 4.
Boxplots showing distributions of Z-scores for (A) 271 CST, (B) 14,037 human, and (C) 3,120 internal hit antibodies with respect to the five noncorrelated descriptors derived from the 79 Fv regions from 77 marketed biotherapeutics. The individual Z-scores for each Fv region were combined to obtain its Z-distance using SI Appendix, Eq. 5.
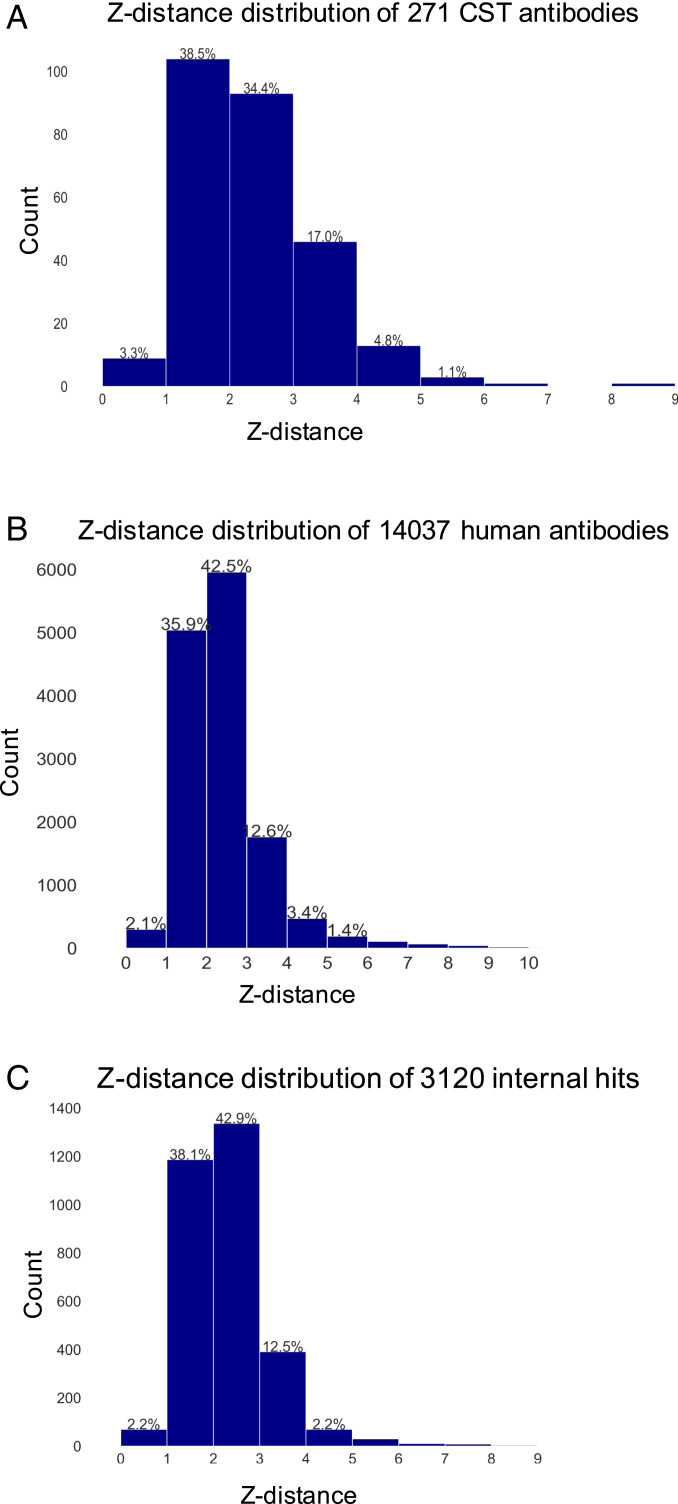
Table 3 summarizes statistics on flags for Fv regions of 271 CST antibodies, 14,037 human antibodies, and 3,120 internal hits. Data on 79 Fvs from the marketed biotherapeutics is also included for reference. Note that no Fv region in these three antibody-sequence sets is flagged more than four times. A majority of Fv regions in all the three datasets do not have any flags, that is, Fv regions in 177 (∼65%) of the 271 CST antibodies, 10,054 (72%) of the 14,037 human antibodies, and 2,285 (∼73%) of the 3,120 internal hits possess average physicochemical properties that are similar to those of the 79 Fvs. Furthermore, 81 (30%) CST antibodies, 3,466 (25%) human antibodies, and 727 (23%) of internal hits possess a single flag, while two flags were found for 10 (4%), 490 (3%), and 99 (3%) of CST, human, and our internal hit antibodies, respectively. Finally, less than 1% of antibodies in these three sets of antibody sequences possess three or more flags (Table 3). A small number of flags among antibodies in these three datasets suggests that the 77 marketed antibody-based biotherapeutics may possess broad ranges for the five physicochemical descriptors. A second reason could be the use of a Z-score cutoff value of >|1.96|, which covers 95% of the distributions (average ±2 SD). Nonetheless, these results show that 271 CST antibodies contain fewer Fvs whose physicochemical properties are similar to those of the marketed biotherapeutics than the other two datasets.
Table 3.
Flags for the Fv regions of 271 CST, 14037 human, and 3120 internal hit antibodies
| No. of flags | 79 Fvs in 77 marketed biotherapeutics | Fvs in 271 CST antibodies | Fvs in 14,037 human antibodies | Fvs in 3,120 internal antibodies |
| 0 | 58 (73%) | 174 (64%) | 9,861 (70%) | 2,240 (72%) |
| 1 | 18 (23%) | 82 (30%) | 3,609 (26%) | 756 (24%) |
| 2 | 3 (4%) | 11 (4%) | 539 (4%) | 115 (4%) |
| 3 | 0 | 3 (1%) | 27 (∼0%) | 9 (∼0%) |
| 4 | 0 | 1 (0%) | 1 (0%) | 0 |
Data on 79 Fvs is provided for the sake of reference.
The distribution of flags among Fv regions of the 271 CST antibodies was further investigated for their incidence in Phase I (86 antibodies), Phase II (129 antibodies), and Phase III (56 antibodies) (Dataset S1). A breakdown analysis shows that 62 (72%), 79 (61%), and 33 (59%) Fv regions in antibodies in Phases I, II and III do not have any flag. This observation suggests that greater proportions of antibody-based biotherapeutic candidates discovered in recent years (Phase I) have physicochemical properties that are like those of the marketed antibodies in comparison to those discovered several years ago (Phase III). This is consistent with improvements in antibody discovery technologies and emphasis on developability in recent years. Fv regions of 20 (23%) Phase I, 43 (33%) Phase II, and 19 (34%) Phase III antibodies have a single flag, while 3 (3%), 5 (4%) and 3(5%) of antibodies show two flags for Phase I, II, and III, respectively. As anticipated, Fv regions for only one antibody in Phase I and two in Phase II possess three flags. Interestingly, one antibody, lampalizumab, in the Phase III of clinical trials at the time of this analysis, is flagged four times. Note that several antibodies may have progressed to the next phases and others might have been approved or discontinued since the collection of this data.
Table 4 compares average values and ranges of the five nonredundant physicochemical descriptors seen for Fv regions in 271 CST, 14,037 human, and 3,120 internal hit antibodies with those of 79 Fvs from 77 marketed antibody-based biotherapeutics. The physicochemical descriptors for 79 Fvs show smaller variations (ranges) than those seen for the other three datasets. Z-distance values for Fv regions in 271 CST, 14,037, human and 3,120 internal hit antibodies were also calculated to quantitatively assess their similarity to the marketed antibodies. The average Z-distance values are given in Table 4, and histograms showing the Z-distance distributions are plotted in Fig. 5 A–C. In addition to these, a quartile analysis of the distribution of Z-distances observed for all the 79 Fvs was performed. The Z-distance values at the first and the fourth quartile intervals observed for the 79 Fvs were used to obtain cutoff values for classifying the 271 CST, 14,037 human, and 3,120 internal hit antibodies as having physicochemical properties very similar (Z-distance < 1.57), similar (1.57 ≤ Z-distance ≤ 2.67), or dissimilar (Z-distance > 2.67) to those of the marketed antibody-based biotherapeutics (Table 5). Note that high Z-distance values do not imply that such antibodies cannot be developed into biotherapeutic products. Instead, these values imply that such drug candidates may require greater attention during their sequence optimizations and/or drug product development. In the case of 271 CST antibodies, the intrinsic physicochemical properties of the Fv regions from 53 (∼20%) of them are very similar to those of the marketed antibody-based biotherapeutics. Additionally, 130 (48%) of them have similar physicochemical properties as those of the marketed biotherapeutics. However, approximately one-third of 271 CST antibodies (88, 32.5%) possess physicochemical properties that are dissimilar from those seen for the antibodies-based biotherapeutics (Z-distance > 2.67, Table 5). Interestingly, physicochemical properties of 2,362 (16.8%) human antibodies are very similar, and those of another 7,581 (54%) human antibodies are similar to physicochemical properties of the marketed antibodies. These observations agree with those of Deane and coworkers (15, 47), who reported that human NGS repertoires contain antibodies with sequences highly similar to those of the marketed antibody-based biotherapeutics. Taken together, these observations suggest that ideal antibody-based drug products are likely to be human antibodies that possess good manufacturability and physicochemical stability characteristics. Furthermore, our internal hits analyzed in this work were obtained from antibody generation campaigns that involved either transgenic mice expressing human antibodies or phage display libraries constructed using human germlines. These hits also show comparable results. Three-fourths of them (2,280 out of 3,120, 73%) have Z-distance values ≤ 2.67, and the remaining one-fourth (840, 27%) possess physicochemical properties that are different from those of 77 marketed biotherapeutics (Z-distance > 2.67, Table 5). Note that the 271 CST antibodies contain a greater proportion of Fvs (32.5%) with physicochemical properties dissimilar to those of the 79 Fvs found in marketed biotherapeutics in comparison to 27% in our internal hits and 29% of human antibodies. This trend is consistent with the increased representation of humanized or fully human antibodies among the approved biotherapeutics.
Table 4.
Average values and ranges for five nonredundant descriptors and Z-distances for the Fv regions of 77 marketed antibody-based biotherapeutics, 271 CST, 14,037 human, and 3,120 internal hit antibodies
| Descriptor | 79 Fvs in 77 marketed biotherapeutics* | 271 CST antibodies | 14,037 human antibodies | 3,120 internal hits |
| BSA_VL: VH (Å2) | 797 ± 81 (618 to 1,046) | 781 ± 93 (585 to 1,211) | 796 ± 83 (436 to 1,456) | 778 ± 82 (521 to 1,318) |
| pIFv_3D | 7.9 ± 1.2 (4.3 to 9.5) | 7.7 ± 1.3 (4.6 to 9.9) | 8 ± 1.3 (4.1 to 10.3) | 7.8 ± 1.3 (3.9 to 9.8) |
| RM(µD/µH) (D) | 1.1 ± 0.6 (0.1 to 3.6) | 1.07 ± 0.7 (0.1 to 6.1) | 0.9 ± 0.8 (0 to 22.9) | 1.08 ± 1.07 (0.1 to 42.3) |
| RP | 1.8 ± 0.5 (1.0 to 3.3) | 2 ± 0.9 (0.6 to 11.1) | 1.9 ± 0.9 (0.3 to 16) | 1.7 ± 0.7 (0.4 to 7.8) |
| Avg_HI | 1 ± 0.4 (0.3 to 1.9) | 0.9 ± 0.4 (0.14 to 2.3) | 1 ± 0.4 (0 to 3.1) | 0.9 ± 0.4 (0.03 to 3.2) |
| Z-distance | 2.1 ± 0.7 (0.6 to 4.2) | 2.5 ± 1.4 (0.7 to 18.6) | 2.5 ± 1.4 (0.3 to 37.5) | 2.4 ± 1.7 (0.5 to 71.5) |
Note that the ranges for the five nonredundant descriptors are smaller for the Fvs from marketed biotherapeutics.
Fig. 5.
Histograms showing distributions of Z-distances for (A) 271 CST antibodies, (B) 14,037 human antibodies, and (C) 3,120 internal antibodies. Z-distance values greater than 10 are not shown in the histograms.
Table 5.
The physicochemical similarity of the Fv regions in 271 CST antibodies, 14,037 human antibodies, and 3,120 internal hit antibodies with those found in marketed biotherapeutics
| Dataset | Total number of Fv regions | Highly similar (Z-distance < 1.57) | Similar (1.57 ≤ Z-distance ≤ 2.67) | Dissimilar (Z-distance > 2.67) |
| 77 biotherapeutics | 79 | 15 (19%) | 43 (54%) | 21 (27%) |
| 271 CST antibodies | 271 | 54 (20%) | 129 (48%) | 88 (32%) |
| 14,037 human antibodies | 14,037 | 2,293 (16%) | 7,547 (54%) | 4,197 (30%) |
| 3,120 internal hits | 3,120 | 563 (18%) | 1,697 (54%) | 860 (28%) |
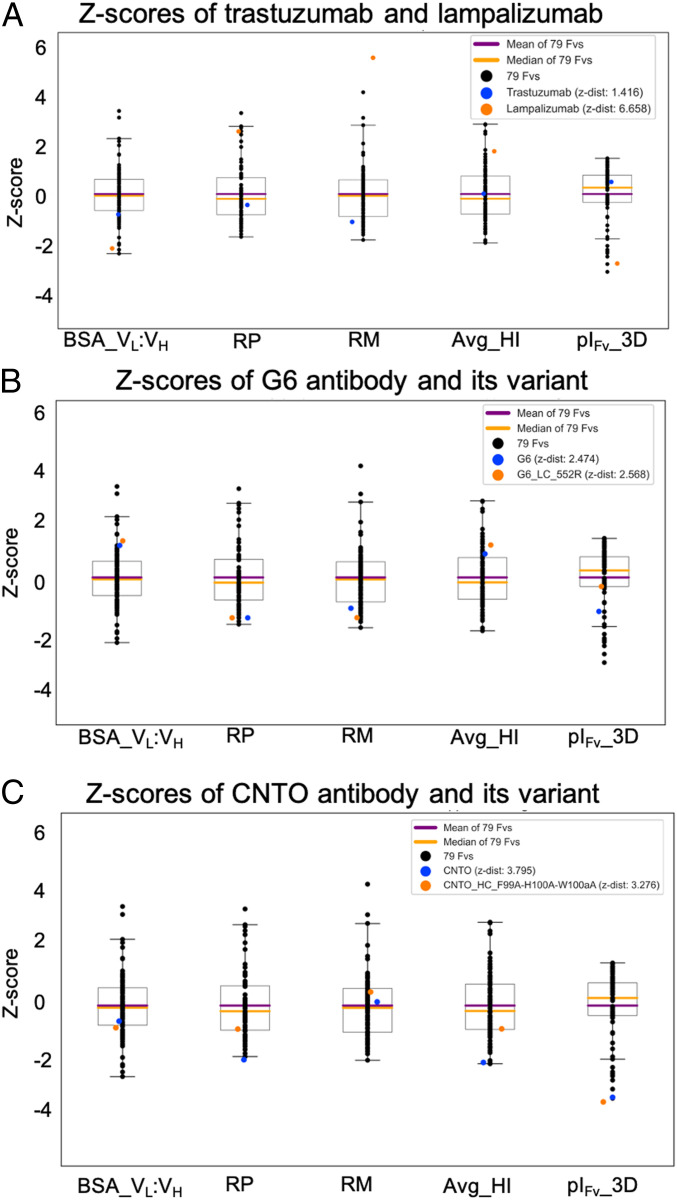
In the fourth application of this work, we first study intrinsic physicochemical profiles of the Fv regions of two mAbs, namely, trastuzumab and lampalizumab, and then compare two poorly behaving parent mAbs with their rationally optimized variants from previously reported case studies (48, 49). The boxplots shown in Fig. 6A compare intrinsic physicochemical profiles of the Fv regions from trastuzumab and lampalizumab, a Phase III antibody that has been flagged four times in this analysis (Table 3). In a thought experiment, let us consider that these two antibodies have been identified as lead candidates in a hypothetical biologic drug discovery program and are equivalent function wise. Now, Fig. 6A shows that intrinsic physicochemical parameters of the Fv region of the lead candidate with a Z-distance of 1.4 (trastuzumab) are more similar to those of the 79 Fvs from the marketed biotherapeutics than the one with a Z-distance of 6.7 (lampalizumab, Fig. 6A); therefore, it should be prioritized for further optimization and drug development. The availability of such information during early discovery can be crucial toward mitigating attrition at the later stages. The boxplots in Fig. 6 B and C extend this thought experiment to the considerations during lead candidate optimization via two different case studies. In the first case study, the presence of an aggregation-prone region (APR) in light chain complementarity determining region 2 (LCDR2) of an anti-VEGF antibody [G6 mAb, Protein Data Bank (PDB) entry 2FJF (51)] contributed toward aggregation when expressed in a transient system (48). Bauer et al. (48) used an in silico tool, Solubis (52), to identify the APR and disrupted it by introducing a single point mutation, Ser-52 → Arg, in LCDR2. This mutation led to significantly improved productivity, decreased self-association, reduced opalescence, better resistance to heat-induced aggregation, and improved colloidal stability while maintaining target binding affinity. We note that the parent mAb G6 and its optimized variant exhibit very similar properties for four of the five descriptors, with more than a one unit increase in pI of the Fv region because of a polar to charged residue substitution (SI Appendix, Table S2). This is reflected in the Z-scores plot comparing the physicochemical profiles of the parent and the variant G6 antibodies in reference to the 79 Fvs from 77 marketed biotherapeutics (Fig. 6B). In the second case study, we assessed multiple variants generated to establish an aggregation model for an anti-IL13 antibody (CNTO607) which displayed poor solubility in physiological formulation conditions (49). Again, a crystal structure of the Fab portion of CNTO607 [PDB entry 3G6A (53)] was used to identify an aggregation hotspot containing three contiguous amino acid residues, 99-F-H-W-100a, in the amino acid sequence of its HCDR3. This hotspot was hypothesized to cause self-association, leading to precipitation of CNTO607 antibody, and the three residues were mutated to Ala singly and all together. We have calculated five nonredundant descriptors for all the four variants (F99A, H100A, W100aA, and F99A-H100A-W100aA) and the parent CNTO607 (SI Appendix, Table S2). The descriptors RP and Avg_HI show significant differences in their values for the variants carrying the mutations F99A, W100aA, and F99A-H100A-W100aA compared to the parent CNTO607 but not for the variant H100A, in agreement with the experimental results (49, 50). Fig. 6C compares physiochemical profiles for the parent CNTO607 and the triple point variant F99A-H100A-W100aA. The triple mutant F99A-H100A-W100aA showed the greatest degree of improvements in the solution behavior of CNTO607 antibody (49, 50). This agrees with the largest changes observed for the ratio of charged to hydrophobic patches and average hydrophobic imbalance (Fig. 6C and SI Appendix, Table S2).
Fig. 6.
Applications of the intrinsic physicochemical profiles in lead candidate identification and optimization. This figure utilizes boxplots showing Z-scores to compare intrinsic Fv region physicochemical profiles for (A) trastuzumab and lampalizumab, (B) an anti-VEGF antibody (G6) and its variant S52R in the light chain CDR2, and (C) an anti-IL13 antibody (CNTO607) and its triple mutant (F99A-H100A-W100aA). All these profiles are made in reference to the 79 Fvs from 77 marketed biotherapeutics.
Discussion
The translation of a biotherapeutic drug candidate into a marketed biotherapeutic drug product requires that the candidate can withstand various stresses during manufacturing, shipping, and storage. In addition, the drug product needs to possess acceptable pharmacology, safety, immunogenicity, and toxicology attributes in vivo. Now, different pharmaceutical companies follow different drug development and manufacturing processes. Clinical trial designs and patient populations also show significant variations. Furthermore, the final decision to approve or decline a given biotherapeutic candidate is commonly made on a case-by-case basis. Pharmacology, safety, immunogenicity and toxicology, CMC attributes as well as risk versus benefit profile in relation to patient indication(s) of a biotherapeutic drug candidate are important considerations during the development (technical as well as clinical) and the approval processes. While all of this is true, we argue that the following common threads unite all marketed antibody-based biotherapeutics: 1) all of them are efficacious in vivo and possess acceptable pharmacological attributes, 2) all of them can be reliably manufactured in enormous quantities repeatedly over many years, and 3) all of them are generally safe. We hypothesize that structural and physicochemical attributes, derived from primary sequences of marketed biotherapeutics, may contribute toward each of these common threads. Therefore, insights gained by studying these intrinsic factors of the marketed biotherapeutics could be useful toward enhancing pipeline productivity by prioritizing drug candidates that possess physicochemical attributes similar to those of the marketed biotherapeutics. This systematic analysis of intrinsic physicochemical properties of the marketed biotherapeutics could not have been performed without the availability of their primary sequences in public databases. A similar comprehensive analysis of multiple attributes measured via standardized experiments that inform all aspects of developability (manufacturability, safety, efficacy, and pharmacology) shall also be very useful. As far as we know, there are no publicly available databases that track such multidimensional experimental data for biotherapeutics. In addition to intrinsic sequence and structural elements, there are a myriad of extrinsic factors that may have also significantly contributed toward the success of a biotherapeutic product. Examples of such extrinsic factors include target drugability, target biology and its implications for therapeutic intervention, disease mechanism(s) and prevalence, biomarkers used for patient population stratification, manufacturing process details, business decisions, and so on. These factors may not be easily linkable to the sequence and/or structural characteristics of a marketed biotherapeutic product. However, again, it is currently difficult to study these extrinsic elements systematically because there are no comprehensive public databases that capture such information.
This research has sought to capture a “holistic” physicochemical profile, which is intrinsic to molecular sequences and structures of the Fv regions of marketed biotherapeutics, by distilling numerous aspects of in vitro and in vivo behaviors, analogous to therapeutic antibody profile for candidates in clinical trials (15). This report has established the boundary conditions around variations of the physicochemical attributes of Fv regions in marketed antibody-based biotherapeutics since the five nonredundant descriptors show smaller variations for the 79 Fvs from the marketed biotherapeutics in comparison to the 271 CST drug candidates, the 14,037 human antibodies, and our 3,120 internal hits (Table 4). This profile is a phenomenological model, and it may not directly correlate with individual aspects of developability.
The intrinsic physicochemical profile described in this theoretical work has several practical uses as described in the introduction and shown via examples in Results. This profile can be very impactful at the earliest stage of biologic drug discovery, namely, selection of hits generated via antibody discovery campaigns for experimental testing. With recent methodological advancements in the single B cell repertoire sequencing and analyses, typical antibody discovery campaigns against a given target can yield thousands of hits (54, 55), and testing each hit experimentally can be cost and time prohibitive. Along with the diversity in antibody germlines and epitopes, similarity of their physicochemical properties to biotherapeutics already in the market can potentially help select functional as well as easily developable hits for experimental testing. This can help save costs and improve efficiency by eliminating the need to express and purify hits that may be difficult to develop. The availability of multiple functional hits with good developability attributes at the very beginning of a discovery process can also help shorten the time and reduce costs associated with the experiments needed to move on to the next stages, namely, lead identification and optimization. Another aspect of biologic drug discovery and development that can benefit significantly from the availability of this profile is about making informed business decisions. The intrinsic physicochemical profile described in this work can be used as a risk assessment tool for biotherapeutic candidates already in product and/or clinical development. Inclusion of this profile in due diligence exercises for in-licensing purposes can also add to the success of industrial collaborations. This profile is clearly not intended to predict the function of a given biotherapeutic drug candidate and therefore should not be used for such activities.
Limitations of the Intrinsic Physicochemical Profile.
The intrinsic physicochemical profile reported in this work has several limitations. First of all, the number of biotherapeutic drug products available in the market is still small in comparison to the small molecule drug products. Therefore, this profile is expected to be updated as more antibody-based biotherapeutic candidates become marketed and more data on them (experimental as well as computational) becomes available. This is our first attempt, and the profile is limited to Fv regions of the marketed biotherapeutics. It does not account for the effect of sequence structural diversity in the constant regions or formats of the marketed biotherapeutics. Currently marketed antibody-based biotherapeutics come in several different molecular formats (e.g., IgG1, IgG2, IgG4, bispecific, Fabs, ScFvs, and Fvs), formulations (e.g., lyophilized powders, high concentration liquid formulations, and so on), and presentations optimized for different routes of administration (e.g., intravenous, intramuscular, subcutaneous injections, and so on). The profile developed in this work does not consider the physicochemical characteristics specific to individual biotherapeutic product classes, such as high concentration liquid formulations suitable for subcutaneous administration versus low concentration formulations suitable for intravenous administration, or old versus new therapeutic antibodies, and so on. However, the data in Table 2 shows that dividing the 79 Fvs into such classes does not significantly change the average values of these descriptors. This observation supports our hypothesis that the intrinsic physicochemical profile of the Fv region is the key to estimating which biotherapeutic candidates can potentially pass all stages of manufacturing, product development, clinical development, regulatory approval, and become drug products available in the market to serve unmet medical needs. Additionally, this profile is focused on physicochemical descriptors computed using the homology-based structural models. It does not provide information on incidence of potential aggregation-prone regions, T cell immune epitopes, or chemical degradation motifs found in biotherapeutic candidates. Along with the structure-based physicochemical attributes, these sequence characteristics are also important aspects of developability assessments guiding the optimization of the lead candidates. Finally, marketed biotherapeutic products span a broad spectrum of disease indications, patient populations, mechanisms of action, molecular formats, and sequence—structural characteristics. Therefore, data analysis studies involving them are inherently subjective. Our study has attempted to mitigate this subjectivity by focusing on variable regions of the marketed biotherapeutics. Despite the above-described limitations, this work has important implications toward devising rational biopharmaceutical informatics (9) approaches to biologic drug discovery and development. This is one step forward in learning from biotherapeutics already in the market to improve the selection and engineering of newly discovered biotherapeutic candidates.
Materials and Methods
Full details of methods followed in this work are described in SI Appendix. Briefly, homology-based models of variable regions of the marketed antibody-based biotherapeutics were used to derive a large number of physicochemical descriptors. Clustering these in silico descriptors has yielded a set of five nonredundant descriptors that show no significant correlations among themselves. These nonredundant descriptors constitute an intrinsic physicochemical profile for variable regions found in the marketed biotherapeutics. Potential uses of this profile are discussed based on physicochemical similarity of variable regions found in three different antibody-sequence datasets with those of the marketed biotherapeutics.
Supplementary Material
Acknowledgments
L.A. and K.P.M. thank Boehringer Ingelheim Pharmaceutical, Inc. for supporting their postdoctoral research. We thank Kenny Tsang for helping with scripting. Drs. Thomas Fox, Nels Thorsteinson, Alexander Jung, Michael Marlow, Giuseppe Licari, Anne Karow-Zwick, and Joschka Bauer are thanked for numerous discussions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020577118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Smietana K., Siatkowski M., Møller M., Trends in clinical success rates. Nat. Rev. Drug Discov. 15, 379–380 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Tomar D. S., et al., In-silico prediction of concentration-dependent viscosity curves for monoclonal antibody solutions. MAbs 9, 476–489 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiCara D. M., et al., High-throughput screening of antibody variants for chemical stability: Identification of deamidation-resistant mutants. MAbs 10, 1073–1083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma V. K., et al., In silico selection of therapeutic antibodies for development: Viscosity, clearance, and chemical stability. Proc. Natl. Acad. Sci. U.S.A. 111, 18601–18606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar S., Singh S. K., Developability of Biotherapeutics: Computational Approaches (CRC Press, 2015). [Google Scholar]
- 6.Xu Y., et al., Structure, heterogeneity and developability assessment of therapeutic antibodies. MAbs 11, 239–264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomar D. S., Singh S. K., Li L., Broulidakis M. P., Kumar S., In silico prediction of diffusion interaction parameter (kD), a key indicator of antibody solution behaviors. Pharm. Res. 35, 193 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Chennamsetty N., Voynov V., Kayser V., Helk B., Trout B. L., Design of therapeutic proteins with enhanced stability. Proc. Natl. Acad. Sci. U.S.A. 106, 11937–11942 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S., Plotnikov N. V., Rouse J. C., Singh S. K., Biopharmaceutical informatics: Supporting biologic drug development via molecular modelling and informatics. J. Pharm. Pharmacol. 70, 595–608 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Lauer T. M., et al., Developability index: A rapid in silico tool for the screening of antibody aggregation propensity. J. Pharm. Sci. 101, 102–115 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Singh S. N., Yadav S., Shire S. J., Kalonia D. S., Dipole-dipole interaction in antibody solutions: Correlation with viscosity behavior at high concentration. Pharm. Res. 31, 2549–2558 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Lobo S. A., et al., Stability liabilities of biotherapeutic proteins: Early assessment as mitigation strategy. J. Pharm. Biomed. Anal. 192, 113650 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Dumas J., et al., Developability assessment with case studies highlighting the decision taking. Med. Sci. (Paris) 35, 1171–1174 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Jain T., et al., Biophysical properties of the clinical-stage antibody landscape. Proc. Natl. Acad. Sci. U.S.A. 114, 944–949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raybould M. I. J., et al., Five computational developability guidelines for therapeutic antibody profiling. Proc. Natl. Acad. Sci. U.S.A. 116, 4025–4030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X., Singh S. K., Kumar S., Potential aggregation-prone regions in complementarity-determining regions of antibodies and their contribution towards antigen recognition: A computational analysis. Pharm. Res. 27, 1512–1529 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S., Singh S. K., Wang X., Rup B., Gill D., Coupling of aggregation and immunogenicity in biotherapeutics: T- and B-cell immune epitopes may contain aggregation-prone regions. Pharm. Res. 28, 949–961 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Kumar S., Mitchell M. A., Rup B., Singh S. K., Relationship between potential aggregation-prone regions and HLA-DR-binding T-cell immune epitopes: Implications for rational design of novel and follow-on therapeutic antibodies. J. Pharm. Sci. 101, 2686–2701 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Boswell C. A., et al., Effects of charge on antibody tissue distribution and pharmacokinetics. Bioconjug. Chem. 21, 2153–2163 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Shah D. K., Pharmacokinetic and pharmacodynamic considerations for the next generation protein therapeutics. J. Pharmacokinet. Pharmacodyn. 42, 553–571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg Y., et al., Pharmacokinetics and immunogenicity of broadly neutralizing HIV monoclonal antibodies in macaques. PLoS One 10, e0120451 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauna Z. E., Richards S. M., Maillere B., Jury E. C., Rosenberg A. S., Editorial: immunogenicity of proteins used as therapeutics. Front. Immunol. 11, 614856 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datta-Mannan A., et al., Influence of physiochemical properties on the subcutaneous absorption and bioavailability of monoclonal antibodies. MAbs 12, 1770028 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laptoš T., Omersel J., The importance of handling high-value biologicals: Physico-chemical instability and immunogenicity of monoclonal antibodies. Exp. Ther. Med. 15, 3161–3168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuster J., et al., In vivo stability of therapeutic proteins. Pharm. Res. 37, 1–17 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Singh S. K., Impact of product-related factors on immunogenicity of biotherapeutics. J. Pharm. Sci. 100, 354–387 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Li L., et al., Concentration dependent viscosity of monoclonal antibody solutions: Explaining experimental behavior in terms of molecular properties. Pharm. Res. 31, 3161–3178 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Li B., et al., Framework selection can influence pharmacokinetics of a humanized therapeutic antibody through differences in molecule charge. MAbs 6, 1255–1264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y., et al., Minipig as a potential translatable model for monoclonal antibody pharmacokinetics after intravenous and subcutaneous administration. MAbs 4, 243–255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., et al., Physicochemical rules for identifying monoclonal antibodies with drug-like specificity. Mol. Pharm. 17, 2555–2569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starr C. G., Tessier P. M., Selecting and engineering monoclonal antibodies with drug-like specificity. Curr. Opin. Biotechnol. 60, 119–127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finlay W. J. J., Coleman J. E., Edwards J. S., Johnson K. S., Anti-PD1 ‘SHR-1210’ aberrantly targets pro-angiogenic receptors and this polyspecificity can be ablated by paratope refinement. MAbs 11, 26–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kingsbury J. S., et al., A single molecular descriptor to predict solution behavior of therapeutic antibodies. Sci. Adv. 6, eabb0372 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipinski C. A., Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 44, 235–249 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Lipinski C. A., Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today. Technol. 1, 337–341 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J., Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 64, 4–17 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Stanfield R. L., Takimoto-Kamimura M., Rini J. M., Profy A. T., Wilson I. A., Major antigen-induced domain rearrangements in an antibody. Structure 1, 83–93 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Warszawski S., et al., Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces. PLoS Comput. Biol. 15, e1007207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jarasch A., et al., Developability assessment during the selection of novel therapeutic antibodies. J. Pharm. Sci. 104, 1885–1898 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Tomar D. S., Kumar S., Singh S. K., Goswami S., Li L., Molecular basis of high viscosity in concentrated antibody solutions: Strategies for high concentration drug product development. MAbs 8, 216–228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisenberg D., Weiss R. M., Terwilliger T. C., The helical hydrophobic moment: A measure of the amphiphilicity of a helix. Nature 299, 371–374 (1982). [DOI] [PubMed] [Google Scholar]
- 42.Eisenberg D., Weiss R. M., Terwilliger T. C., The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. U.S.A. 81, 140–144 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nichols P., et al., Rational design of viscosity reducing mutants of a monoclonal antibody: Hydrophobic versus electrostatic inter-molecular interactions. MAbs 7, 212–230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Datta-Mannan A., et al., The interplay of non-specific binding, target-mediated clearance and FcRn interactions on the pharmacokinetics of humanized antibodies. MAbs 7, 1084–1093 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datta-Mannan A., et al., Balancing charge in the complementarity-determining regions of humanized mAbs without affecting pI reduces non-specific binding and improves the pharmacokinetics. MAbs 7, 483–493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salgado J. C., Rapaport I., Asenjo J. A., Predicting the behaviour of proteins in hydrophobic interaction chromatography. 1: Using the hydrophobic imbalance (HI) to describe their surface amino acid distribution. J. Chromatogr. A 1107, 110–119 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Krawczyk K., Raybould M. I. J., Kovaltsuk A., Deane C. M., Looking for therapeutic antibodies in next-generation sequencing repositories. MAbs 11, 1197–1205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bauer J., et al., Rational optimization of a monoclonal antibody improves the aggregation propensity and enhances the CMC properties along the entire pharmaceutical process chain. MAbs 12, 1787121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bethea D., et al., Mechanisms of self-association of a human monoclonal antibody CNTO607. Protein Eng. Des. Sel. 25, 531–537 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Wu S. J., et al., Structure-based engineering of a monoclonal antibody for improved solubility. Protein Eng. Des. Sel. 23, 643–651 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Fuh G., et al., Structure-function studies of two synthetic anti-vascular endothelial growth factor Fabs and comparison with the Avastin Fab. J. Biol. Chem. 281, 6625–6631 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Van Durme J., et al., Solubis: A webserver to reduce protein aggregation through mutation. Protein Eng. Des. Sel. 29, 285–289 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Teplyakov A., et al., Epitope mapping of anti-interleukin-13 neutralizing antibody CNTO607. J. Mol. Biol. 389, 115–123 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Goldstein L. D., et al., Massively parallel single-cell B-cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies. Commun. Biol. 2, 304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parola C., Neumeier D., Reddy S. T., Integrating high-throughput screening and sequencing for monoclonal antibody discovery and engineering. Immunology 153, 31–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.