Significance
We find that the coronin family protein POD-1 debranches the actin network in vitro. Our results reveal that POD-1 regulates actin organization and cell polarity in migrating neuroblasts and one-cell−stage embryos of C. elegans. We provide evidence that F-actin debranching is critical for establishing cell polarity during cell migration and asymmetric cell division.
Keywords: directional cell migration, actin debranching, cell polarity, coronin
Abstract
The formation of the branched actin networks is essential for cell polarity, but it remains unclear how the debranching activity of actin filaments contributes to this process. Here, we showed that an evolutionarily conserved coronin family protein, the Caenorhabditis elegans POD-1, debranched the Arp2/3-nucleated actin filaments in vitro. By fluorescence live imaging analysis of the endogenous POD-1 protein, we found that POD-1 colocalized with Arp2/3 at the leading edge of the migrating C. elegans neuroblasts. Conditional mutations of POD-1 in neuroblasts caused aberrant actin assembly, disrupted cell polarity, and impaired cell migration. In C. elegans one-cell−stage embryos, POD-1 and Arp2/3, moved together during cell polarity establishment, and inhibition of POD-1 blocked Arp2/3 motility and affected the polarized cortical flow, leading to symmetric segregation of cell fate determinants. Together, these results indicate that F-actin debranching organizes actin network and cell polarity in migrating neuroblasts and asymmetrically dividing embryos.
Cell polarity is a fundamental feature of virtually all eukaryotic cells and plays crucial roles in a wide range of cellular processes, including cell motility, asymmetric cell division, and cell signaling (1). The establishment of cell polarity involves the asymmetric assembly of distinct cellular components to perform specialized functions. The actin-related protein (Arp) 2/3 complex-dependent branched actin networks and the pushing force they produce provide the principal means for cells to remodel the plasma membrane during cellular polarization (2). For example, in the leading edge of a migrating cell, the local Arp2/3-nucleated actin polymerization powers asymmetric projections of the plasma membrane (3). During asymmetric cell division of the Caenorhabditis elegans zygote, an actomyosin flow is central to the transport of the polarity PAR proteins into defined subcellular domains (4).
Actin filaments' continuous assembly must be balanced by actin depolymerization to ensure a constant supply of actin monomers for new growth. The Arp2/3 complex potency in actin nucleation empowers this complex as an essential regulator to organize the actin cytoskeleton. While Arp2/3 by itself is biochemically inactive, interactions with nucleation-promoting factors (NPFs) such as the Wiskott Aldrich syndrome protein (WASP)/WASP family verproline-homologous (WASP/WAVE) family proteins shift the Arp2/3 complex from its open, inactive conformation to a closed, active conformation (5, 6). The conformationally activated Arp2/3 complex then binds to the side of preexisting actin filaments to nucleate a branch from the mother filament (7–12). Conversely, nucleation by Arp2/3 can be inhibited by several binding partners, including glia maturation factor (GMF), Gadkin, Arpin, and Coronin, whose activities replenish available pools of actin monomers and Arp2/3 complexes for sustained actin assembly (13–18).
The coronin family proteins are conserved actin regulators (19). The phylogenetic analysis grouped coronin genes into three types (19, 20). The best-characterized coronin is the Type I coronin (e.g., Coronin 1B) that binds to actin filaments through the β-propeller structure and to the Arp2/3 complex via its N terminus. These interactions block the docking of Arp2/3 onto actin filaments or facilitate debranching the existing actin network (20). Coronin 1B simultaneously interacts with the Slingshot phosphatase to dephosphorylate and activate ADF/Cofilin proteins that sever actin filaments, thereby promoting the actin network disassembly (13). Despite significant progress on Type I coronin, the activity and function of other coronins remain unclear. In particular, Type III coronins, known as POD-1 in C. elegans and Drosophila or Coronin7 in Dictyostelium and humans, contain two tandem coronin repeats, making them distinct from other coronins (19–21). POD-1 was biochemically isolated from C. elegans oocytes (22), and its mutations disrupted the polarity and architecture in early C. elegans embryos and impaired midlife touch sensitivity of the nematode (21, 23). However, it remains unclear how the Type III coronin functions. The Drosophila homolog of POD-1 is required for correct axon guidance, and the purified Dpod-1 cross-links the actin and microtubule cytoskeletons (24), whereas the mammalian Coronin7 was implicated in the Golgi morphology and function (25, 26), demonstrating the functional divergence of this family of coronin. Here, we show that the C. elegans POD-1 debranches Arp2/3-nucleated actin filaments in vitro and that POD-1 regulates cell polarity by remodeling the actin cytoskeleton during cell migration and asymmetric cell division.
Results
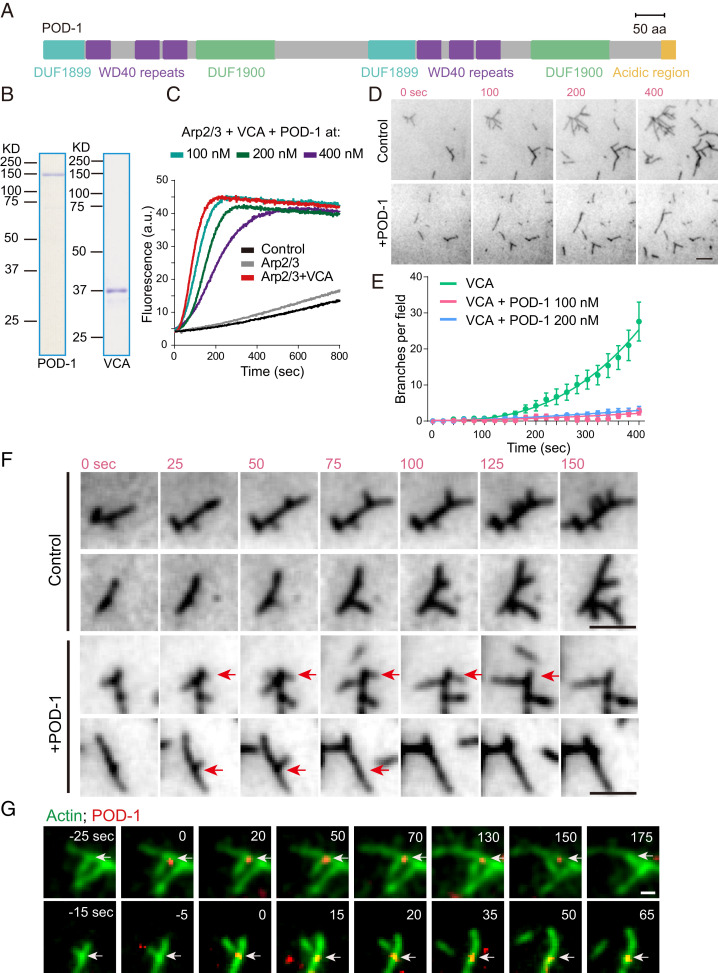
POD-1 Localizes to Actin Branches and Induces Debranching In Vitro.
To determine whether POD-1 inhibits Arp2/3 nucleation, we performed in vitro spectrofluorimetry and total internal reflection fluorescence (TIRF) microscopy assays to examine the impact of POD-1 on Arp2/3 activity. First, we prepared the full-length POD-1 protein from human HEK293F cells and the recombinant C. elegans WASP protein homolog/WSP-1-VCA domain from bacteria (Fig. 1 A and B). By adding these proteins into the pyrene actin polymerization reactions, we showed that the VCA domain activated the Arp2/3-dependent actin polymerization and that the POD-1 protein inhibited the activation in a dose-dependent manner (Fig. 1C). The acidic (A) domain of SCAR/WASP proteins interacts with the Arp2/3 complex, and, unlike the Type I coronins, the Type III coronins have such a highly acidic (A) region at the C terminus (19) (SI Appendix, Fig. S1A). To understand the role of this region, we prepared the POD-1ΔA truncated protein lacking the acidic region (SI Appendix, Fig. S1B) and found that POD-1ΔA inhibited actin polymerization like POD-1 full-length protein (SI Appendix, Fig. S1C). Consistently, the acidic peptide POD-1A alone neither inhibited nor promoted actin polymerization (SI Appendix, Fig. S1C), suggesting that this region may be dispensable for POD-1 to regulate actin polymerization in vitro.
Fig. 1.
POD-1 inhibits Arp2/3-mediated actin nucleation in vitro. (A) Schematics of the full-length protein of POD-1. Protein is 1,057 residues long. DUF1899: a putative actin-binding site; WD40 repeats: act as a site for protein−protein or protein−DNA interaction; DUF1900: predominantly found in the structural protein coronin with unknown function; acidic region: homologous to the most C-terminal acidic motif of SCAR/WASP proteins which contacts the Arp2/3 complex. (B) Sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis of strep-tagged full-length POD-1 and GST-tagged WSP-1-VCA domain. Full-length POD-1 was expressed in HEK293F cells, and the WSP-1-VCA domain was expressed in E. coli. These proteins were used for in vitro actin polymerization assays. (C) Spectrofluorimetry assay using pyrene-labeled actin to monitor polymerization. The reactions contain 4 μM actin (10% pyrene labeled), 20 nM Arp2/3 complex, and 10 nM VCA. The concentrations of full-length POD-1 are as indicated; a.u., arbitrary units. See SI Appendix, Fig. S1C for POD-1ΔA and POD-1A. (D) Actin (1.2 μM, 50% Oregon green labeled) filaments growing in the presence of 13.5 nM Arp2/3 and 10 nM VCA ± 100 nM POD-1 observed by TIRF microscopy. Time is represented in seconds, with t = 0 corresponding to the frame from where the imaging was initiated. (Scale bar, 5 μm.) (E) The number of actin branches/field (1,600 μm2) was plotted as a function of time. Actin (1.2 μM, 50% Oregon green labeled), 13.5 nM Arp2/3, and 10 nM VCA, and indicated amounts of POD-1 were mixed immediately before the reactions started. Data are presented as mean ± SEM from five fields for each condition. See SI Appendix, Fig. S1E for POD-1ΔA and POD-1A. (F) High magnification view of filament debranching. Reactions contained actin (1.2 μM, 50% Oregon green labeled), 13.5 nM Arp2/3, and 5 nM VCA ± 100 nM POD-1. Red arrows indicate debranching events. (Scale bar, 3 μm.) (G) Actin (1.2 μM, 50% Oregon green labeled) filaments growing in the presence of 13.5 nM Arp2/3 complex, 10 nM VCA, and 200 nM Alexa Fluor-568 labeled POD-1 in the flow chamber observed by TIRF microscopy. White arrows indicate the localization of POD-1. (Scale bar, 1 μm.) See a larger field view in SI Appendix, Fig. S1G.
To explore the mechanism of how POD-1 blocks Arp2/3-dependent actin assembly, we monitored actin polymerization and branching using time-lapse TIRF microscopy. In the presence of the full-length POD-1 or the truncated POD-1ΔA, we observed a significant reduction of total F-actin polymers and a fewer generations of actin branched junctions (Fig. 1D and SI Appendix, Fig. S1D). Our quantification of actin branch formation over time showed that POD-1 and POD-1ΔA suppressed the rate of actin branch formation (Fig. 1E and SI Appendix, Fig. S1E). Under identical experimental conditions, our addition of POD-1A protein did not affect actin debranching, which also serves as a negative control to rule out the underflow in microfluidics (SI Appendix, Fig. S1 D and E). From TIRF recording, most of the F-actin branches stayed stable in reactions without POD-1; however, the presence of POD-1 induced dissociation of branches from the preexisting actin filaments (Fig. 1F and Movie S1). Furthermore, we assembled the Arp2/3-based actin network first and then added the POD-1 protein or the buffer as the control (SI Appendix, Fig. S1F and Movie S2). By determining the debranching rate compared to the untreated sample, we found a marked increase of actin filament debranching compared to the control (SI Appendix, Fig. S1G). Next, we directly visualized fluorescence-labeled POD-1 and actin branches in one reaction. We found that 53% of POD-1 spots target actin branches (n = 42), and, in some cases, filament branches disassembled after binding POD-1 (Fig. 1G, SI Appendix, Fig. S1H, and Movie S3). We revealed the sequential events: 1) Actin branches formed (−25 s, upper; −15 s, lower); 2) POD-1 localized to branch junctions (0 s); and 3) debranching occurs (175 s upper or 35 s lower) (Fig. 1G). These results indicate that POD-1, at least to a certain extent, directly impacts branch stability; however, we do not rule out that POD-1 may have other biochemical functions. Using the established method to determine POD-1 debranching efficiency (27), we found that POD-1 accelerated the rate of debranching, and it had debranching activity similar to the known debranchers Coronin-1 and GMF (SI Appendix, Fig. S1 G and I) (15, 27, 28). Thus, our results show that POD-1 localizes to actin branches and induces debranching, thereby inhibiting Arp2/3-dependent actin nucleation and branching in vitro.
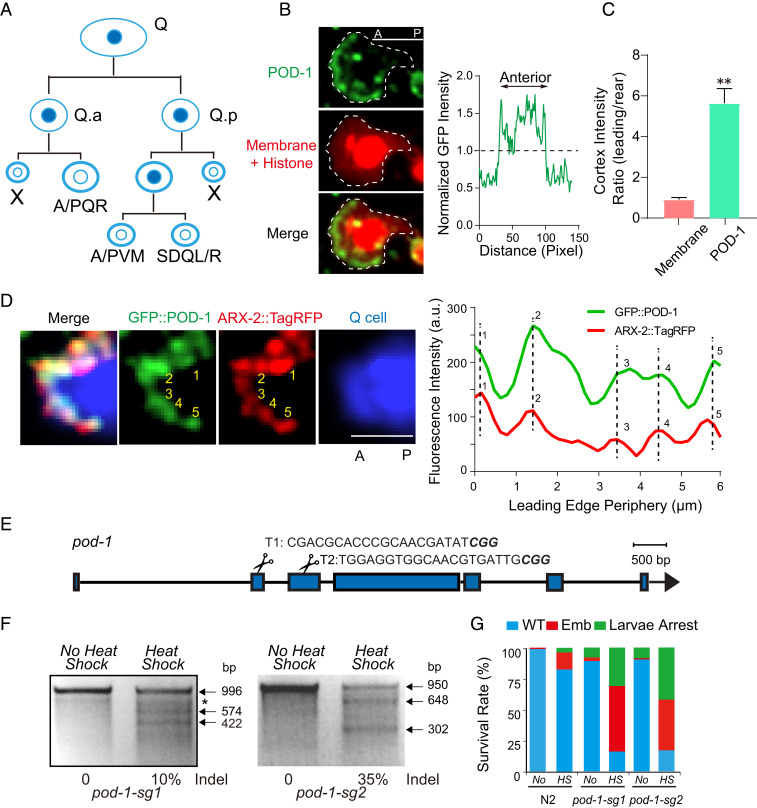
POD-1 and Arp2/3 Colocalize at the Leading Edge of Migrating Neuroblasts.
We studied the in vivo function of POD-1 during the C. elegans Q neuroblast migration. Q neuroblasts on the right (QR) or the left (QL) undergo three rounds of asymmetric cell division during the first larval stage to generate three distinct neurons (29) (Fig. 2A). The Q-cell descendants move to well-defined positions along the anterior/posterior (A/P) axis but in opposite directions, with QR and descendants (abbreviated as QR.x) migrating anteriorly and QL and descendants (QL.x) migrating posteriorly (30) (Fig. 3B). To examine the subcellular localization of endogenous POD-1 protein during Q-cell migration, we employed the CRISPR-Cas9−based knock-in (KI) technique to construct a GFP::POD-1 KI strain. We did not detect any defects in cell migration or animal development in the KI animals (n > 100 animals), suggesting that GFP fusion may not affect the functionality of POD-1. We depicted the plasma membrane and nucleus using mCherry tagged with a myristoylation signal and a histone H2B. By examining GFP fluorescence in the migrating Q cells, we revealed enrichment of POD-1 at the leading edge (SI Appendix, Fig. S2A and Movie S4). Because GFP-POD-1 KI reporter illuminates all the endogenous POD-1 proteins in the larvae, including the neighboring cells of Q cells, we constructed a transgenic C. elegans strain that expressed GFP-tagged POD-1 under the control of a Q-cell specific promoter Pegl-17. Essentially, we confirmed the asymmetric localization of POD-1 in the leading edge of migrating Q cells (Fig. 2 B and C and Movie S5). By performing the triple fluorescence labeling, we showed that the green fluorescence of POD-1 and the red fluorescence of the Arp2/3 complex colocalized in the leading edge in animals carrying ARX-2::TagRFP KI, GFP::POD-1 KI, and the Q-cell marker Plin-32::TagBFP (Fig. 2D and Movie S6). By plotting ARX-2 and POD-1 intensity distribution as a function of distance from the leading edge, we found that ARX-2::TagRFP and GFP::POD-1 overlapped (Fig. 2D). To further understand the function of the A domain of POD-1, we generated a transgenic animal expressing POD-1ΔA::GFP in Q-cell lineages. Like the full-length POD-1 protein, POD-1ΔA::GFP localized at the leading edge and rescued the cell migration defects in pod-1 conditional knockout (KO) animals (SI Appendix, Fig. S2 B and C, Fig. 3 D and E, and Movie S5). In agreement with our biochemical data on POD-1ΔA, our results suggest that the acidic domain may not be essential for the function of POD-1 in actin debranching and cell migration.
Fig. 2.
Distribution of POD-1 in migrating neuroblasts and generation of pod-1 conditional KO strain. (A) Schematic of Q neuroblast lineages. QL and QR neuroblasts divide three times and generate three neurons (QL: PQR, PVM, and SDQL; QR: AQR, AVM, and SDQR) and two apoptotic cells (X). (B) (Left) Representative fluorescence images of GFP-tagged POD-1 and mCherry tagged plasma membrane and histone in migrating QR.ap cell. The dashed lines indicate the cell periphery; (Right) the normalized GFP fluorescence distribution plot around the periphery of the representative image shown on Left. The trace starts from the posterior of QR.ap and moves counterclockwise along the cell periphery to the anterior and back to the rear. (Scale bar, 5 μm.) See SI Appendix, Fig. S2A for POD-1 localization in Q cell of GFP::POD-1 KI animals. (C) Quantification of POD-1::GFP fluorescence intensity ratio of the leading edge to the rear part of QR.ap cell. The corresponding mCherry-membrane intensity ratio was used as an internal control. Data are presented as mean ± SEM; **P < 0.01 by Student’s t tests. The number of examined animals indicated by N, n = 5. (D) (Left) Triple fluorescence images of AQR. (Right) The corresponding normalized fluorescence KI intensity distribution along the leading edge. Individual puncta of GFP::POD-1 or ARX-2::TagRFP are labeled and numbered in representative images and the corresponding plots. (Scale bar, 3 μm.) (E) Schematic of pod-1 gene model and single guide RNA (sgRNA) sequences (sgRNA1 and sgRNA2 target the second and third exon, respectively). (F) Representative gels of the T7 endonuclease I (T7EI) assay for pod-1 PCR products amplified from the genomic DNA of worms expressing Phsp::Cas9 and PU6::pod-1-sgRNA1 (Left) or PU6::pod-1-sgRNA2 (Right) with or without heat shock treatment. (G) Quantification of embryo survival rates in WT and pod-1 conditional knockouts; n = 50 to 100 from three different generations.
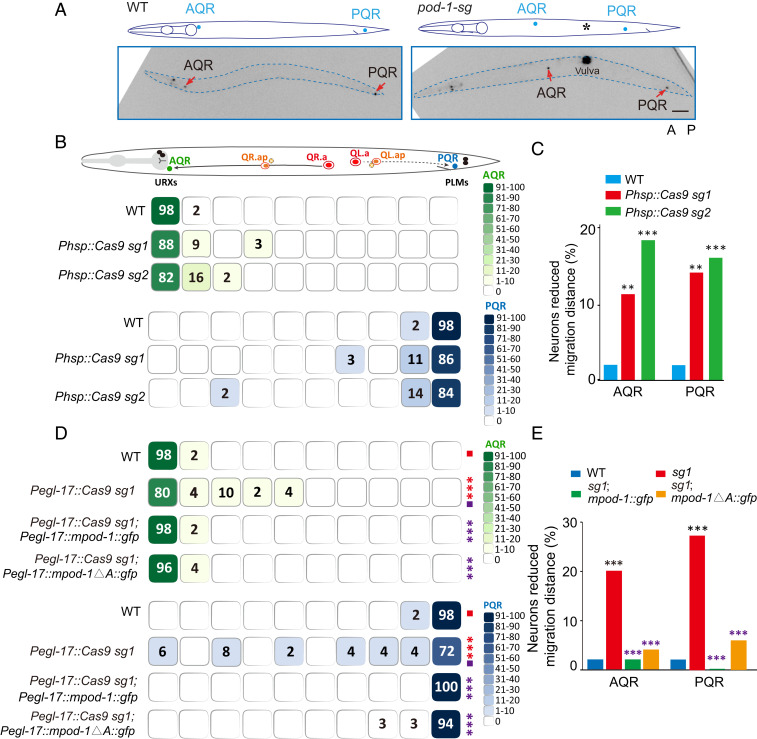
Fig. 3.
Depletion of POD-1 causes Q-cell migration defects. (A) Schematics (Top) and fluorescence inverted images (Bottom) of the A/PQR position in WT and pod-1 conditional knockouts. A/PQR neurons were visualized using Pgcy-32::mCherry. The image is inverted so that high mCherry fluorescence intensity is black. The cell identities are denoted adjacent to the cells. Dotted blue lines show the periphery of C. elegans. (Scale bar: 50 μm.) (B) (Bottom) A color-coded heat map scores the A/PQR position in L4 animals of the indicated genotypes. (Top) As shown in the schematic overview of Q-cell migration, the QR descendant (QR.x), AQR, migrates anteriorly, whereas the QL descendant (QL.x), PQR, migrates posteriorly. The total length between the URX and the tail is divided into 10 blocks, and the percent of A/PQRs that stopped within each block is listed. The color darkness of the blocks symbolizes the range of percentage values; n ≥ 50 for all genotypes. (C) Quantification of defects in Q-cell migration of A/PQR in pod-1 conditional knockouts; n ≥ 50. **P < 0.01, ***P < 0.001 by the χ2 test. (D) A color-coded heat map scores the A/PQR position in L4 animals of the indicated genotypes. The quantization method used is the same as in E; n ≥ 50 for all genotypes. Statistical significance compared to the control (purple square) with matching color codes, ***P < 0.001 by the Fisher’s exact tests. (E) Quantification of defects in Q-cell migration in pod-1 conditional knockouts and rescued animals; n ≥ 50. Black stars represent the comparisons between the WT and mutants; purple stars represent the comparisons between the mutants and rescued animals. ***P < 0.001 by the χ2 test.
To explore the interaction between POD-1 and Arp2/3, we performed affinity purification and mass spectrometry to identify interacting proteins of POD-1. Using an anti-GFP antibody, we purified GFP-tagged POD-1 with its associated proteins from the lysate of GFP::POD-1 KI larvae (SI Appendix, Fig. S2D). Protein constituents were determined by employing liquid chromatography−tandem mass spectrometry. We detected four subunits in the Arp2/3 complex (SI Appendix, Fig. S2 E and F). Using TBA-1(an α-tubulin subunit)::GFP KI animals as the negative control, we did not identify any Arp2/3 subunits (SI Appendix, Fig. S2 E and F), validating the specificity of the purification scheme. Together, these results indicate that POD-1 interacts with Arp2/3 at the leading edge of migrating neuroblasts.
POD-1 Regulates Cell Polarity during Neuroblast Migration.
The germline deletion of POD-1 leads to embryonic lethality (21). To investigate how POD-1 regulates neuronal cell migration, we created conditional mutations of pod-1. Using the somatic CRISPR-Cas9 technique (31), in which the Cas9 endonuclease is expressed under the control of a heat shock−inducible promoter Phsp, we generated conditional mutants of pod-1 during Q-cell development (Fig. 2E). T7 endonuclease I (T7EI)-based assays demonstrated that these transgenic animals produce molecular lesions with the expected sizes at the target loci of pod-1 after heat shock induction of Cas9 expression (Fig. 2F). Consistent with the reported phenotypes of the pod-1(ye11) animals (21), the pod-1 conditional mutant embryos, but not wild-type (WT) embryos, exhibited embryonic lethality and larval arrest phenotypes (Fig. 2G), demonstrating the success in generating conditional mutations in the pod-1 locus. We next examined the final positions of QR.ap and QL.ap in these animals, which will become the AQR and PQR neurons after long-distance migration toward the anterior or posterior, respectively. We showed that AQR was localized at a more posterior position, and PQR was confined at a more anterior region in pod-1 conditional mutants (Fig. 3 A–C) or Q-cell specific pod-1 mutants (Fig. 3 D and E) when compared with WT animals, indicating an essential role of POD-1 in cell migration. Considering that animals expressing Phsp::Cas9 and pod-1-sg caused animal lethality by mutating pod-1 in the majority of cells, we suspect that the strong loss-of-function mutant animals did not survive to the young adult stage when migration phenotypes were quantified. By contrast, Pegl-17::Cas9 mutated pod-1 within Q-cell lineages and did not cause lethality, allowing us to quantify the phenotypes from both strong and weak mutants, thereby generating the higher penetrance of cell migration defects than Phsp::Cas9. The cell migration defects from Q-cell−specific pod-1 mutants suggested a cell-autonomous contribution of POD-1 to cell migration.
To dissect the cellular mechanism for which the cell migration is reduced in pod-1 mutants, we performed fluorescence time-lapse analysis of cell morphology and the actin cytoskeleton in migrating Q cells (32). The plasma membrane and histone were marked with mCherry, and the actin cytoskeleton was depicted using GFP tagged with the actin-binding domain of moesin (GFP::moesin ABD). In WT animals, QR.a or QR.ap polarizes toward the anterior and extends a lamellipodium with F-actin enrichment at the leading edge (Fig. 4 A and C and Movie S7). In the pod-1 conditional knockouts, ectopic actin bundles assemble behind the leading edge, and the GFP fluorescence of GFP::moesinABD accumulates in the lagging area, causing an even distribution of F-actin in QR.a and QR.ap (Fig. 4 B, D, and E and Movie S8). The fluorescence intensity ratio of GFP::moesinABD between the leading edge and the rear was reduced from 2.0-fold in WT to 0.9-fold in pod-1 conditional KO animals (Fig. 4F), indicating the loss of F-actin asymmetry in the migrating cells. To examine the localization of Arp2/3 in the POD-1 KO cells, we introduced GFP-tagged ARX-2 KI marker into the pod-1-sg animals. Compared to WT Q cells, Arp2/3 becomes less abundant at the leading edge in the POD-1 KO cell (SI Appendix, Fig. S3 A and B), resembling reduced F-actin localization in this area, suggesting that branched F-actin may not be accumulating at the leading edge in the absence of POD-1. We speculate that the Arp2/3-dependent actin branching and the POD-1−mediated debranching events are highly dynamic and balanced. Inhibition of actin debranching can disrupt the branched actin network, affecting Arp2/3 localization in the leading edge.
Fig. 4.
Depletion of POD-1 in Q-cell lineages disrupts actin network at the leading edge of migrating Q neuroblasts. (A and B) Fluorescence time-lapse images of F-actin (GFP:: ABDmoesin) in the QR.a cell of WT (A) and pod-1 conditional KO (B) animals. The white arrows indicate the leading edge, and the asterisks denote nuclei. The blue arrows in B indicate ectopic F-actin. The GFP image (Top) is inverted, and Bottom shows the merged images. (Scale bar: 5 μm.) The image of A is also shown in Fig. 5B. (C and D) Fluorescence time-lapse images of F-actin (GFP:: ABDmoesin) in the QR.ap cell of WT (C) and pod-1 conditional KO (D) animals. The white arrows indicate the leading edge, and the asterisks denote nuclei. The blue arrows in D indicate ectopic F-actin. The GFP image (Top) is inverted, and Bottom shows merged images. (Scale bar: 5 μm.) (E) Quantification of the formation of ectopic actin filaments (longer than 2 μm) in Q neuroblasts of WT and pod-1 conditional mutant animals (n = 14 to 20). **P < 0.01 by the χ2 test. (F) Quantification of the F-actin fluorescence intensity ratio of the leading edge to the rear part of QR.ap cell in WT or pod-1 mutant animals (n = 10). The corresponding mCherry-membrane intensity ratio was used as an internal control. Data are presented as mean ± SEM; n.s., not significant, ***P < 0.001 by Student’s t tests. (G) Schematic of a migrating Q cell. The anterior, posterior, and total length or area of the Q cell was quantified as indicated. (H and I) Quantification of the length (H) and area (I) ratios of the leading end to the lagging end of Q cell in the WT and pod-1 conditional KO animals (n = 12 to 14). **P < 0.01, ***P < 0.001 by Student’s t tests. Error bars indicate SEM.
Strikingly, Q-cell morphology was altered in pod-1 conditional knockouts, including the formation of an oversized larger leading area and the lack of a persistent migration direction (Figs. 4 B and D and 5C). By measuring the ratio of the area or the length of the leading to lagging portions of the Q neuroblasts, we show that these ratios in the pod-1 conditional mutants were from 1.5- to 1.7-fold longer and from 1.7- to 1.8-fold larger, respectively, than those in WT cells (Fig. 4 H and I). To quantify the migration direction, we measured the angle between the C. elegans A−P body axis and the Q-cell longitudinal axis (Fig. 5A). In WT animals, QR.ap only changed their angles within 45° during a 40-min time-lapse recording, and the migration angle within 20° occupied 98% of the recorded migration events. However, the pod-1 conditional KO QR.ap dramatically altered the migration direction, varying the angles from 0° to ±125°, and the percentage of QR.ap cells whose migration angles are within 20° was reduced to 54% (Fig. 5 B–F and Movie S9). Consequently, QR.ap failed to translocate the cell body toward the anterior continuously, reducing the migration speed from 27.1 μm/h to 16.1 μm/h (Fig. 5 G and H). Together, these data indicate that POD-1 promotes neuronal migration by regulating an asymmetric F-actin network at the leading edge.
Fig. 5.
Depletion of POD-1 disrupts the directionality of neuronal migration and slows whole-cell migration. (A) Quantification scheme of the angle (α) between the longitudinal axis of a migrating cell and the AP body axis. A, anterior; P, posterior; D, dorsal; V, ventral. (B and C) Fluorescence time-lapse images of F-actin, plasma membrane, and histones during AQR/QR.ap migration in WT (B) or pod-1 mutant (C) animals. Merged images are on Left; inverted fluorescence images of F-actin are on Right; white arrows, migration direction; asterisks, nucleus; double-headed arrows, migration distance. F-actin is labeled with the GFP-tagged actin-binding domain of Moesin (GFP::Moesin ABD), and the plasma membrane and histones are labeled with mCherry-tagged myristoylation signal or histone H2B, respectively. The dotted lines indicate the cell periphery in C. Time is presented in minutes. (Scale bars, 5 μm.) (D) Quantification of the QR.ap migration angle. Each line represents the measurement from one time-lapse movie; n = 10. (E) A color-coded heat map scores the QR.ap migration angle. The migration angles were recorded from 40-min migration tracks in WT and pod-1 mutant animals (n = 10 for each strain). The color darkness of the blocks symbolizes the range of percentage values. Animals used were the same as in D. (F) Quantification of the QR.ap migration angle. ***P < 0.001 by Student’s t test. Error bars indicate SD; n = 10. Animals used were the same as in D. (G and H) Quantification of the QR.ap migration distance (G) and speed (H). Each line in G represents the measurement from one time-lapse movie; **P < 0.01 by Student’s t test. Error bars indicate SD; n = 10. Animals used were the same as in D. (I and J) A proposed model for POD-1 in debranching the Arp2/3-based actin network (I) during neuronal migration (J). POD-1 limits the Arp2/3-based branches by promoting debranching in vitro (I). Depletion of POD-1 in vivo disrupts dynamic actin network formation during neuronal migration (J).
Discussion
In this study, we show that the coronin family protein POD-1 debranches the actin network in vitro. Our genetic and imaging results revealed that POD-1 is essential for actin organization and cell polarity in migrating neuroblasts of C. elegans (Fig. 5 I and J).
POD-1 Induces F-Actin Debranching In Vitro.
POD-1 was identified by isolating actin-associated protein from C. elegans oocytes extracts (22). The past research focused on biochemical activities of POD-1 homologs across species: The Dictyostelium Coronin7 binds directly to F-actin, and the CRN7NPST (N-terminal WD repeat including the PST domain) polypeptide stabilizes actin filaments and protects filaments from depolymerization (33), whereas recombinant Drosophila POD-1 can cross-link both actin and microtubules (24). Our results support the notion that Coronin7/POD-1 functions in actin-related processes. Importantly, this study demonstrated the activity of POD-1 in debranching the Arp2/3-nucleated actin network in vitro.
We noticed that POD-1 generates a weaker effect on kinetic actin polymerization curves than that in the TIRF assay. The distinct sensitivity between the TIRF assay and Pyrene assay may explain the difference. In TIRF assays, the addition of POD-1 removed Arp2/3-generated nascent branched actin filaments, likely generating short filaments beyond the detection limit of fluorescence light microscopy. In pyrene assays, the increase of pyrene fluorescence results from actin monomer polymerization, and the formation of short actin filaments, beyond TIRF detection, can be monitored by the changes in pyrene fluorescence. Considering that POD-1 debranches actin branches, it is likely that the formation of short actin filaments in the presence of POD-1 can be monitored as the pyrene fluorescence changes; however, these filaments might be too short to be visualized by fluorescence light microscopy.
We suggest that POD-1’s inhibition of Arp2/3-dependent actin branching occurs mostly through its β-propeller units but may not require its C-terminal acidic region. Consistently, the Type I coronins also interact with actin filaments through their β-propeller (8). Recent observations showed that the N-terminal β-propeller domain of the Type I coronin positioned near the p35/ARPC2 subunit in the Arp2/3 complex and induced a standard open/inactive conformation of the complex (18). The Arp2/3 inhibitory proteins such as Arpin contain an acidic motif that binds to Arp2/3 and induces its inactive conformation (14, 18). However, the conserved C-terminal acidic domain of POD-1 did not affect Arp2/3-based actin polymerization in vitro (SI Appendix, Fig. S1 C–E). A possible explanation is that the binding of POD-1's acidic domain to Arp2/3 did not induce an open conformation, and, alternatively, the acidic domain in POD-1 might not interact with Arp2/3. While the β-propeller units of POD-1 are sufficient to inhibit Arp2/3 nucleation, we cannot exclude the possibility that the C-terminal acidic domain perhaps assists this inhibition. Future studies will elucidate the structural basis of POD-1's inhibition of the Arp2/3 complex. Considering that the daughter filaments seemingly sprang down from mother filaments (Movies S1–S3), we speculate that POD-1's debranching activity might involve the conformational changes in Arp2/3.
POD-1 Regulates Cell Polarity during Asymmetric Cell Division.
We wondered how POD-1 regulates actin organization and polarity establishment in the C. elegans one-cell−stage embryos. We showed that POD-1 was enriched in the anterior cortex of WT embryos (SI Appendix, Fig. S3C; 20/20 embryo examined), which is consistent with an early result based upon immunofluorescence (22). ARX-2 is asymmetrically localized in the C. elegans zygote with a large portion of ARX-2 and POD-1 coclustering (SI Appendix, Fig. S3C; 20/20 embryo examined). The pod-1 RNAi perturbed the localization of ARX-2 protein (SI Appendix, Fig. S3C; 8/10 embryo examined), but the asymmetric localization of POD-1 was not affected by eliminating ARX-2 (SI Appendix, Fig. S3C; 9/10 embryo examined). By imaging double fluorescence KI embryos, we showed that GFP-tagged POD-1 puncta colocalized with the mCherry-tagged Arp2/3 complex and that the green and red fluorescence puncta move together during polarity establishment (SI Appendix, Fig. S3C and Movie S10). Considering that the Arp2/3 positive structures are distinct in size, we speculate that they may contain a variable number of the Arp2/3 complexes, likely ranging from one to multiple Arp2/3 complexes. Virtually, RNA interference (RNAi) in Arp2/3 eliminated the red fluorescence but did not markedly change the motility of POD-1. By contrast, inhibition of POD-1 by RNAi blocked the movement of Arp2/3 puncta.
To quantify the motility of POD-1 and Arp2/3 in embryos, we performed the mean-square displacement (MSD) analysis of multiple trajectories of both proteins in WT and RNAi embryos. According to the previously described method (34), we calculated time-averaged MSD curves. The MSD analysis of the transport paths showed that POD-1 and Arp2/3 underwent identical and directional movement in WT embryos. While the loss of Arp2/3 slightly perturbs the directionality of POD-1 motility, the inhibition of POD-1 almost completely blocked any directional movement of Arp2/3. Together, these results indicate that POD-1 and Arp2/3 move together and that POD-1 controls the motility of Arp2/3 but not vice versus (SI Appendix, Fig. S3D).
Next, we determined whether the lack of POD-1−dependent F-actin debranching affects the early embryos' cortical contractility and polarity establishment. We monitored the cortical distribution of GFP-tagged nonmuscle myosin II (NMY-2) over time. In comparison with control embryos, our kymography analysis revealed that pod-1 (RNAi) embryos did not undergo the stereotypical cortical flow from posterior to anterior, and GFP-tagged NYM-2 remained stationary (SI Appendix, Fig. S3 E and F and Movie S11). Because the cortical flow is essential for the embryonic anterior-posterior polarity establishment, we assessed the impact of POD-1 inhibition on the localization of polarity proteins. In WT embryos, PAR-3 protein became enriched at the anterior cell cortex during the polarity establishment; however, GFP-tagged PAR-3 signal appeared at both the anterior and posterior cell cortex in pod-1-depleted embryos (SI Appendix, Fig. S4 A and B and Movie S12). The cell fate determinants such as P-granule components undergo polarized segregation into the posterior portion of the one-cell−stage WT embryos, leading to an asymmetric inheritance of P-granule in the P1 cell. In contrast, P-granules were evenly distributed in pod-1 RNAi embryos (SI Appendix, Fig. S4 D and E and Movie S13). These results show that the POD-1-based actin debranching activity regulates embryonic cell polarity by promoting Arp2/3 dynamics and NYM-2-based cortical contractility (SI Appendix, Fig. S4F).
POD-1 Organizes Actin Network and Establishes Cell Polarity In Vivo.
Our results establish that POD-1 is a crucial regulator of neuronal migration. Depletion of POD-1 perturbs the lamellipodial actin network and impaired the C. elegans neuroblast migration. In the one-cell−stage embryos, the loss of POD-1 blocks Arp2/3 motility and NMY-2−dependent cortical flow, disrupting polarity establishment. POD-1 depletion may alter lamellipodial morphology or embryonic polarity by maintaining existing branched filaments. These results support the view that a dynamic branching and debranching of actin filaments in a living cell is essential for cell motility and polarity.
POD-1/Coronin7 appears to perform diverse functions that are associated with the actin cytoskeleton. In Dictyostelium, Coronin7 colocalized with the actin cytoskeleton in the pseudopod to regulate cell migration (33). The Drosophila POD-1 is localized to the growth cone to regulate axon guidance (24). While the mammalian Coronin7 is ubiquitously distributed in different tissues throughout development, Coronin7 is strongly up-regulated in the brains during embryonic development and in adult animals (35). Immunolocalization of Coronin7 in NIH 3T3 and HeLa cells described their localization in the Golgi apparatus and showed that Coronin7 was involved in Golgi morphology and membrane trafficking (25). However, these cells do not undergo directional cell migration or polarization, and it remains unclear how the mammalian Coronin7 acts in the polarized cells. POD-1/Coronin7 may regulate various cellular processes that require the debranching of the actin cytoskeleton. Indeed, the C. elegans POD-1 also localized in the growth cone when these neuroblasts grew their dendrites (SI Appendix, Fig. S5A), implying its role in dendrite formation. This observation is consistent with the role of its Drosophila homolog in axon development. Therefore, a systematic investigation of Coronin7 in the mammalian nervous system or other tissues may expand their functional reservoir and reveal the broad significance of actin debranching activity during development and physiology.
Our previous studies described the cell migration defects in the Coronin-1 conditional KO animals, and the current work showed similar cell migration phenotype in pod-1 conditional KO animals, including abnormally long F-actin bundles, reduced actin in the leading edge, and the perturbed cell morphology, consistent with their similar biochemical activities in promoting filament disassembly. Despite the similar contribution to neuroblast migration, COR-1 and POD-1 appear to behave differently in the early embryos. We showed that loss of POD-1 caused a loss of Arp2/3 motility when other redundant mechanisms (e.g., Coronin-1) exist to promote debranching and filament disassembly in the C. elegans embryo. By constructing a GFP KI strain for Coronin-1 (GFP::COR-1), we confirmed that COR-1 localizes in the early embryos. Distinct from POD-1, COR-1 does not colocalize with Arp2/3 and does not show any apparent asymmetric distribution during embryonic divisions (SI Appendix, Fig. S5B and Movie S14; 10/10 embryo examined), suggesting that POD-1 may be the primary actin debrancher in the embryo.
Our previous work showed that the C. elegans Type I coronin COR-1 acts with cofilin to modulate F-actin organization during neuroblast migration (31). The current study revealed that POD-1 limits the generation of Arp2/3-based branches in vitro and organizes the actin cytoskeleton and cell polarity in migrating cells and embryos. We suggest that distinct coronin proteins may work in concert to tune the actin network in vivo. Recent studies demonstrated that Coronin7 interacts with Rac/Cdc-42 GTPase and that Coronin7 may respond to environmental cues (36, 37). The debranching activity of POD-1 may be modulated by the Rac family proteins, such as MIG-2 that is essential for Q neuroblast migration, or CDC-42 that regulates embryonic cell polarity. Q cells sense environmental cues to direct cell migration in a developing larva. The canonic Wnt signaling activates the transcription factor MAB-5, which specifies cell fate and migration directionality (38). POD-1 may not be directly recruited or affected by Wnt signaling.
On the other hand, a transmembrane protein MIG-13 guides the anterior cell migration of QR descendants by directly recruiting WAVE and WASP to the leading edge (39, 40). Although the signal molecule that activates MIG-13 remains unknown, the MIG-13−mediated signaling pathway may recruit POD-1 to the leading edge of migrating Q cells. Future studies on the regulation of POD-1 by cell signaling will advance our understanding of how the actin debranching events are controlled and provide new perspectives for elucidating the physical mechanisms underlying polarity establishment. While this study focuses on the contribution of POD-1 to the actin debranching mechanism during cell migration, POD-1 may regulate cell migration through other means. For example, the POD-1 homolog showed the activity of cross-linking actin and microtubules in Drosophila (24), which suggests that POD-1 may organize the microtubule cytoskeleton during neuronal cell migration.
Materials and Methods
C. elegans Strains and Genetics.
We raised C. elegans strains on nematode growth medium (NGM) plates that were seeded with Escherichia coli strain OP50. Worms were cultured at 20 °C. SI Appendix, Tables S1–S3 summarize the primers, plasmids, and strains.
Molecular Biology for Generating Knock-in or Transgenic Animals.
We inserted CRISPR-Cas9 targets into the pDD162 vector (Addgene, #47549) by linearizing the vector with primers in SI Appendix, Table S1. We retreated PCR products with DpnI digestion overnight. PCR products contain 15 base pairs with overlapped double-strand DNA ends. We cyclized the linearized PCR products to generate plasmids by spontaneous recombination in E. coli. To generate fluorescence tag KI strains, we constructed the homology recombination templates, cloning the ∼2 kb 5′ and 3′ homology arms into the pPD95.77 plasmid. We used the In-Fusion Advantage PCR cloning kit (Clontech; catalog no. 639621). The CRISPR design tool (https://zlab.bio/guide-design-resources) was used to select the target sequence. We inserted the GFP tag into the N terminus of POD-1. We confirmed KI strains using PCR and Sanger sequencing. We generated conditional KO strains using our previously described protocols (31, 41).
We used a PCR fusion-based method to produce Pegl-17::pod-1 constructs. We placed the Q-cell−specific promoter Pegl-17 adjacent to the complementary DNA fragment of modified pod-1 that contains synonymous mutations in the target sequence. We used the unc-54 3′ untranslated region that was amplified using the C. elegans genomic DNA as the template. We created transgenic lines carrying extrachromosomal arrays of the pod-1 expression constructs by microinjecting ∼50 ng/μL DNA with coinjection markers into C. elegans germlines.
To perform RNAi of pod-1 or arx-2, we produced the corresponding double-stranded RNA (dsRNA) molecules using the T7 RiboMAX Express RNAi System. We diluted the synthesized dsRNAs 100 times to a final concentration of 30 ng/μL to 50 ng/μL. We microinjected dsRNA into young adult worms and performed live imaging for one-cell−stage embryos 12 h after microinjection.
Mass Spectrometry Analysis.
Using our previously established protocol (42), we identified the binding proteins of POD-1. We first raised the GFP::POD-1 KI animals on ∼100 90-mm NGM plates. We collected worms and washed them three times with M9 buffer. Using FastPrep-24 (MP Biomedicals), we made worm lysates from 1 mL to ∼2 mL of packed worms in lysis buffer [25 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 10% glycerol, 1× mixture of the complete and (ethylenedinitrilo)tetraacetic acid−free protease inhibitors from Roche, 40 mM NaF, 5 mM Na3VO4] and 3 mL to ∼4 mL of 0.5-mm-diameter glass beads. We used GFP-Trap A beads (Chromoteck) to pull down proteins and eluted them with 300 mL of 0.1 M glycine-HCl, pH 2.5 into 15 mL of 1.5 M Tris⋅HCl pH 8.8. We further precipitated proteins with 100 mL of trichloroacetic acid and redissolved them in 60 mL of 8 M urea, 100 mM Tris⋅HCl, pH 8.5. We treated samples with 5 mM TCEP for reduction and 10 mM iodoacetamide for alkylation, and then diluted them fourfold with 100 mM Tris⋅HCl 8.5. After adding 1 mM CaCl2 and 20 mM methylamine, we further digested proteins with 0.2 mg trypsin at 37 °C overnight. Resultant peptides were desalted with Zip Tip pipette tips (Merck Millipore). We searched tandem mass spectrometry spectra against the C. elegans proteome database using Proteome Discoverer (version PD1.4; Thermo Fisher Scientific). Protein components in SI Appendix, Fig. S2 were reproducibly identified from three biological repeats.
Live-Cell Imaging.
We performed live-cell imaging for Q-cell migration using our previously described methods (43, 44). We first anesthetized L1 larvae with 0.1 mmol/L levamisole in M9 buffer and mounted them on 3% agarose pads at 20 °C. We imaged the larvae on an Axio Observer Z1 microscope (Carl Zeiss) equipped with a 100×, 1.49 numerical aperture (N.A.) objective, an electron-multiplying (EM) charge-coupled device (CCD) camera (Andor iXon+ DU-897D-C00-#BV-500), and the 488- and 561-nm lines of a Sapphire CW CDRH USB Laser System attached to a spinning disk confocal scan head (Yokogawa CSU-X1 Spinning Disk Unit). To image worm embryos, we dissected gravid worms in the M9 buffer. We mounted embryos on 3% agarose pads at 20 °C (43). We collected images of live embryos using an Olympus IX83 microscope equipped with a 150×, 1.45 N.A. oil objective, an EM CCD camera (Andor iXon+ DU-897D-C00-#BV-500), and the 405-, 488-, and 568-nm lines of a Sapphire CW CDRH USB Laser System attached to a spinning disk confocal scan head (Yokogawa CSU-X1 Spinning Disk Unit). We used μManager (https://micro-manager.org) to acquire time-lapse images. The exposure time is 200 ms. We processed and quantified images using the ImageJ software.
Quantifications and Statistical Analysis.
We used the previous established methods to quantify our images (45). In brief, we used the ImageJ software to circumscribe the fluorescence field and measure fluorescence intensity. In all the intensity quantifications, we subtracted the background. When we measure the F-actin marker GFP::moesinABD or KI fluorescence protein signal intensity along the cortex, “leading” refers to the circumference of the lamellipodium (i.e., the widespread area) whereas “rear” refers to the circumference around the rest of the cell. We calculated the intensity ratio between the two using the corresponding mCherry fluorescence intensity as a control. We quantified the migration distances as the cell body movement along the A/P body axis. We measured the migration angle between the protrusion and the A/P body axis. The anterior was defined as degree 0, and the posterior was defined as degree 180. We presented the quantifications by the mean value ± SD for each group. N is the number of animals or cells used for the corresponding quantification. As indicated in figure legends, we used the one-sample or two-tailed Student’s t test or χ2 analysis, or Fisher’s exact tests to determine statistical differences.
Protein Preparation.
We purified actin from rabbit skeletal muscle acetone powder by Sephacryl S-300 chromatography and stored it on ice in buffer G (5 mM Tris HCl, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT, and 0.01% NaN3) using the established methods (46). We labeled actin with pyrene iodoacetamide for fluorometry assay and Oregon Green-488 iodoacetamide. We directly visualized actin filaments using the TIRF microscopy (TIRFM) as described (47).
We used HEK293F cells to express 2×Strep-Flag-POD-1 and 2×Strep-Flag-POD-1ΔA (1 to 1,036 aa proteins lacking the acidic motif). We performed transient transfection using polyethyleneimine at 37 °C, and harvested cells 48 h after transfection. We purified these proteins via Twin-Strep-tag affinity chromatography using gravity flow columns (IBA; catalog no. 2-0910-001). We expressed GST-tagged POD-1A (acidic domain: 1,037 to 1,057 aa) and GST-tagged WSP-1-VCA in E. coli strain BL21 (DE3) by inducing them with 0.3 mM isopropyl β-D-thiogalactoside at 16 °C overnight. We purified the proteins using glutathione-affinity chromatography (GE Healthcare). We then dialyzed proteins in 5 mM Tris HCl, pH 8.0, and flash froze them in liquid nitrogen and stored them with 10% glycerol at −80 °C.
Actin Polymerization Assays.
We adapted the previously described protocols to assay actin polymerization (48, 49). We mixed 4 μM actin (10% pyrene labeled), 20 nM bovine brain Arp2/3 complex (Cytoskeleton. Inc.), and 10 nM VCA with increasing concentrations of full-length POD-1 (100 nM to 400 nM) or POD-1ΔA (100 nM to 400 nM) or POD-1A (1 μM to 30 μM) in G buffer. We added a one-tenth volume of 10×KMEI (1× contains 50 mM KCl, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid (EGTA), and 10 mM imidazole-HCl, pH 7.0). Using a QuantaMaster Luminescence QM 3 PH fluorometer (Photo Technology International, Inc.), we measured these reactions by monitoring the changes in pyrene fluorescence.
TIRFM Assays and Analysis.
Using the previously developed protocols, we performed the TIRFM-based actin-branching assay (42, 45, 47). In brief, we first nucleated actin (1.2 μM, 50% Oregon green labeled) in the presence of 13.5 nM Arp2/3 and 5 nM VCA with or without full-length POD-1 or POD-1ΔA or POD-1A in freshly prepared 1×TIRFM buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 50 mM CaCl2, 15 mM glucose, 20 mg/mL catalase, 100 mg/mL glucose oxidase, and 0.5% methylcellulose) in the dark. We then injected the samples into an N-ethylmalaeimide (NEM)-myosin−treated flow chamber and imaged them at 3- or 5-s intervals using objective-based TIRF microscopy. To visualize the localization of POD-1 to actin filaments, we labeled POD-1 proteins with Alexa Fluor-568 by following the manufacturer’s protocol. We incubated POD-1 proteins with Alexa Fluor-568 conjugate of streptavidin (Invitrogen; catalog no. S11226) at 20 °C for 20 min in the dark. We then added the AlexaFluor-568−labeled POD-1 to the nucleation reactions. The mixtures in the TIRF imaging buffer were injected into a NEM-myosin−treated flow chamber and imaged at 5-s intervals using objective-based TIRF microscopy. As previously described (27), we analyzed the debranching efficiency of POD-1. To determine branch lifetimes, we tracked the branches that were formed at the beginning until they debranched or the end of the movie. We assumed a constant probability per unit time of debranching (single-exponential kinetics) and calculated the debranching rate by a Bayesian likelihood analysis (50).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 32070706, 31730052, 31525015, 31861143042, 31561130153, 31671444, 31871352, and 31991190) and the National Key R&D Program of China (Grants 2017YFA0503501, 2019YFA0508401, and 2017YFA0102900).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100805118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.St Johnston D., Establishing and transducing cell polarity: Common themes and variations. Curr. Opin. Cell Biol. 51, 33–41 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Inagaki N., Katsuno H., Actin waves: Origin of cell polarization and migration? Trends Cell Biol. 27, 515–526 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Krause M., Gautreau A., Steering cell migration: Lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 15, 577–590 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Gubieda A. G., Packer J. R., Squires I., Martin J., Rodriguez J., Going with the flow: Insights from Caenorhabditis elegans zygote polarization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodal A. A., et al., Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat. Struct. Mol. Biol. 12, 26–31 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Xu X. P., et al., Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 31, 236–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchoin L., Pollard T. D., Mullins R. D., Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10, 1273–1282 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Molinie N., Gautreau A., The Arp2/3 regulatory system and its deregulation in cancer. Physiol. Rev. 98, 215–238 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Mullins R. D., Heuser J. A., Pollard T. D., The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U.S.A. 95, 6181–6186 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodnick-Smith M., Luan Q., Liu S. L., Nolen B. J., Role and structural mechanism of WASP-triggered conformational changes in branched actin filament nucleation by Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3834–E3843 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouiller I., et al., The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 180, 887–895 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volkmann N., et al., Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science 293, 2456–2459 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Cai L., Marshall T. W., Uetrecht A. C., Schafer D. A., Bear J. E., Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell 128, 915–929 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang I., et al., Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 503, 281–284 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Gandhi M., et al., GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr. Biol. 20, 861–867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S. L., Needham K. M., May J. R., Nolen B. J., Mechanism of a concentration-dependent switch between activation and inhibition of Arp2/3 complex by coronin. J. Biol. Chem. 286, 17039–17046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maritzen T., et al., Gadkin negatively regulates cell spreading and motility via sequestration of the actin-nucleating ARP2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 109, 10382–10387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokolova O. S., et al., Structural basis of Arp2/3 complex inhibition by GMF, coronin, and arpin. J. Mol. Biol. 429, 237–248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uetrecht A. C., Bear J. E., Coronins: The return of the crown. Trends Cell Biol. 16, 421–426 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Chan K. T., Creed S. J., Bear J. E., Unraveling the enigma: Progress towards understanding the coronin family of actin regulators. Trends Cell Biol. 21, 481–488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rappleye C. A., Paredez A. R., Smith C. W., McDonald K. L., Aroian R. V., The coronin-like protein POD-1 is required for anterior-posterior axis formation and cellular architecture in the nematode Caenorhabditis elegans. Genes Dev. 13, 2838–2851 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aroian R. V., Field C., Pruliere G., Kenyon C., Alberts B. M., Isolation of actin-associated proteins from Caenorhabditis elegans oocytes and their localization in the early embryo. EMBO J. 16, 1541–1549 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melentijevic I., et al., C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature 542, 367–371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothenberg M. E., Rogers S. L., Vale R. D., Jan L. Y., Jan Y. N., Drosophila pod-1 crosslinks both actin and microtubules and controls the targeting of axons. Neuron 39, 779–791 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Rybakin V., et al., Crn7 interacts with AP-1 and is required for the maintenance of Golgi morphology and protein export from the Golgi. J. Biol. Chem. 281, 31070–31078 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Rybakin V., et al., Molecular mechanism underlying the association of coronin-7 with Golgi membranes. Cell. Mol. Life Sci. 65, 2419–2430 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ydenberg C. A., et al., GMF severs actin-Arp2/3 complex branch junctions by a cofilin-like mechanism. Curr. Biol. 23, 1037–1045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai L., Makhov A. M., Schafer D. A., Bear J. E., Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell 134, 828–842 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou G., Stuurman N., D’Ambrosio M., Vale R. D., Polarized myosin produces unequal-size daughters during asymmetric cell division. Science 330, 677–680 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sulston J. E., Horvitz H. R., Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156 (1977). [DOI] [PubMed] [Google Scholar]
- 31.Shen Z., et al., Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev. Cell 30, 625–636 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Ou G., Vale R. D., Molecular signatures of cell migration in C. elegans Q neuroblasts. J. Cell Biol. 185, 77–85 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shina M. C., et al., A coronin7 homolog with functions in actin-driven processes. J. Biol. Chem. 285, 9249–9261 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manzo C., Garcia-Parajo M. F., A review of progress in single particle tracking: From methods to biophysical insights. Rep. Prog. Phys. 78, 124601 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Rybakin V., et al., Coronin 7, the mammalian POD-1 homologue, localizes to the Golgi apparatus. FEBS Lett. 573, 161–167 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharya K., et al., Novel coronin7 interactions with Cdc42 and N-WASP regulate actin organization and Golgi morphology. Sci. Rep. 6, 25411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swaminathan K., et al., Coronin7 regulates WASP and SCAR through CRIB mediated interaction with Rac proteins. Sci. Rep. 5, 14437 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Middelkoop T. C., Korswagen H. C., Development and migration of the C. elegans Q neuroblasts and their descendants. WormBook, 10.1895/wormbook.1.173.1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z., et al., Spatial confinement of receptor activity by tyrosine phosphatase during directional cell migration. Proc. Natl. Acad. Sci. U.S.A. 117, 14270–14279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Z., et al., Functional coordination of WAVE and WASP in C. elegans neuroblast migration. Dev. Cell 39, 224–238 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y., et al., Spectraplakin induces positive feedback between fusogens and the actin cytoskeleton to promote cell-cell fusion. Dev. Cell 41, 107–120.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Chai Y., et al., Live imaging of cellular dynamics during Caenorhabditis elegans postembryonic development. Nat. Protoc. 7, 2090–2102 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Jia R., et al., Spectrin-based membrane skeleton supports ciliogenesis. PLoS Biol. 17, e3000369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian D., et al., Anillin regulates neuronal migration and neurite growth by linking RhoG to the actin cytoskeleton. Curr. Biol. 25, 1135–1145 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Pollard T. D., Polymerization of ADP-actin. J. Cell Biol. 99, 769–777 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amann K. J., Pollard T. D., Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 98, 15009–15013 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang S., et al., Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17, 486–501 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ojala P. J., et al., The two ADF-H domains of twinfilin play functionally distinct roles in interactions with actin monomers. Mol. Biol. Cell 13, 3811–3821 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ensign D. L., Pande V. S., Bayesian single-exponential kinetics in single-molecule experiments and simulations. J. Phys. Chem. B 113, 12410–12423 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.