Significance
The failure of β cells to secrete sufficient amounts of insulin is a key feature of diabetes mellitus. Each β cell secretes only a small amount of insulin upon stimulation in a highly regulated fashion: young insulin is preferentially released, whereas old insulin is mainly degraded within the β cell. How this process is regulated in vivo and likely altered in diabetes is currently unknown. We present here a transgenic pig model that allows the in vivo fluorescent labeling of age-distinct insulin secretory granule pools, hence providing a close-to-life readout of insulin turnover. This will enable the study of alterations in β cell function in an animal model close to humans.
Keywords: β cell, insulin turnover, diabetes mellitus, pig model
Abstract
β cells produce, store, and secrete insulin upon elevated blood glucose levels. Insulin secretion is a highly regulated process. The probability for insulin secretory granules to undergo fusion with the plasma membrane or being degraded is correlated with their age. However, the molecular features and stimuli connected to this behavior have not yet been fully understood. Furthermore, our understanding of β cell function is mostly derived from studies of ex vivo isolated islets in rodent models. To overcome this translational gap and study insulin secretory granule turnover in vivo, we have generated a transgenic pig model with the SNAP-tag fused to insulin. We demonstrate the correct targeting and processing of the tagged insulin and normal glycemic control of the pig model. Furthermore, we show specific single- and dual-color granular labeling of in vivo–labeled pig pancreas. This model may provide unprecedented insights into the in vivo insulin secretory granule behavior in an animal close to humans.
Dysfunction of pancreatic islet β cells is a key contributor to type 2 diabetes mellitus (T2DM) (1, 2), starting in the early onset of the disease (3). Each β cell contains several thousand insulin secretory granules (SGs) (4, 5). However, only a small percentage of insulin SGs undergo exocytosis upon glucose stimulation (6). Insulin is secreted in two phases: a rapid first and a sustained second phase (7–9). On the level of insulin SGs, our understanding of insulin secretion has been shaped by two basic concepts: 1) the recruitment of SG pools defined by their spatial confinement in the cell and 2) the higher probability of young SGs for exocytosis. In model 1), the so-called, readily releasable pool consists of SGs that are already docked with the plasma membrane and are released immediately upon glucose stimulation, thereby creating the first rapid phase of insulin secretion (10). The second prolonged phase is then caused by the recruitment of SGs from the reserve pool, which is located deeper inside the β cell (6). The detailed properties of SGs of the different pools have been debated and refined recently (11). Additionally, data obtained by radio-labeling experiments suggest that young insulin SGs are preferentially secreted (12, 13). A method that allows for the visualization of age-defined pools of the desired protein is to fuse it with the SNAP-tag, a 20-kDa protein tag that reacts covalently in a bioorthogonal manner with fluorescent benzylguanine (BG)-fused substrates in living cells and organisms (14). By using a pulse–chase-labeling approach to track SGs containing an insulin-SNAP chimera, we could confirm the preferential exocytosis of young SGs and also show the preferential intracellular degradation of old SGs (15–17). Furthermore, we found that in insulinoma INS-1 cells a pool of young SGs travels fast on microtubules, while this property is lost for old SGs (18). Young insulin SGs additionally have a more acidic luminal pH compared to old ones (19). Addressing the heterogeneity of insulin SGs and their differential reaction to stimuli and pharmaceutical intervention poses possibilities for the treatment of T2DM. Genetically modified mouse models have been the method of choice to investigate intracellular signaling, as well as metabolism, in diabetes research. Recently, transgenic pigs have been made available that allow for conducting β cell research in a context even closer to humans (20).
Here, we describe the generation and characterization of a transgenic pig model with the SNAP-tag fused to insulin, called the Study OF Insulin granule Aging (SOFIA) pig. We demonstrate the correct targeting and processing of insulin-SNAP to insulin SGs. Finally, we show successful in vivo labeling with one and two SNAP-substrates staining pancreatic islets and distinct insulin SG pools. In summary, our pig model is a valuable system enabling the imaging-based investigation of insulin SG turnover in a large living mammal.
Results
Generation of a Transgenic Pig Model (SOFIA Pig) Expressing Insulin-SNAP.
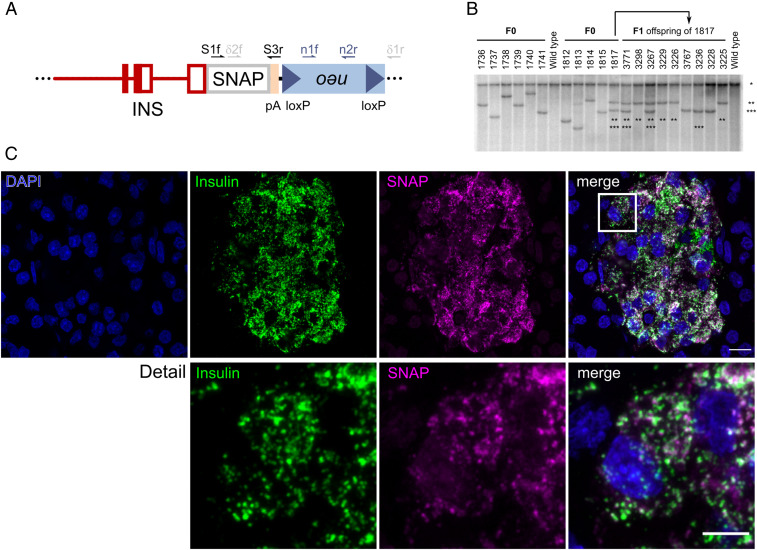
To monitor the intracellular trafficking and turnover of the insulin SGs, we designed an expression vector of porcine insulin containing the porcine INS core promoter and genomic fragments from the porcine INS gene for proper splicing and polyadenylation of the transgene (Fig. 1A). The SNAP-tag was cloned in frame at the 3′-end of the INS coding sequence. After the transfection of the vector into porcine kidney cells, their positive selection and depletion of the floxed neomycin resistance (neo) cassette, we used a mixed population of genetically modified cell clones in somatic cell nuclear transfer (SCNT) experiments and transferred cloned embryos to estrus-synchronized gilts. In total, 11 INS-SNAP transgenic (SOFIA) founder piglets with 10 different integration patterns were obtained (Fig. 1B). The highest transgene expression was detected specifically in β cells of founder 1,817 (SI Appendix, Fig. S1A). This founder pig had two segregating transgene integration sites: one with full Cre-mediated deletion of the floxed neo cassette exhibiting medium transgene expression levels and another with two INS-SNAP copies, one with and the other without neo deletion. The latter integration site resulted in the highest INS-SNAP expression (Fig. 1B and SI Appendix, Fig. S1A) and was used to set up a pig line for further experiments. Immunostaining and confocal microscopy for the SNAP-tag and insulin showed the colocalization of SNAP with insulin in the islets of Langerhans of transgenic offspring, whereas wild-type (WT) pig pancreas was negative for SNAP (Fig. 1C and SI Appendix, Fig. S2).
Fig. 1.
Generation and characterization of SOFIA founder pigs and offspring. (A) Cartoon of the β cell–specific INS-SNAP expression vector with 1.3-kb upstream regions, exon 1 to 3 and intron 1 of the porcine INS gene, in frame SNAP-tag sequence and a polyadenylation (pA) cassette of the bovine growth hormone gene, linked to a floxed neomycin resistance cassette. The sites of primers used for genotyping are indicated. (B) Southern blot analysis for the evaluation of integration pattern. Integration patterns of 11 F0 founder pigs and nine F1 offspring of founder 1,817 are shown. Genomic DNA was digested with the “null cutter” restriction endonuclease enzyme NdeI. *, the band representing the endogenous INS promoter; **, neo/δ-neo transgene integration site with two integrants where neo cassette was deleted only in one INS-SNAP integrant; and ***, δ-neo transgene integration site. (C) Immunofluorescence labeling against SNAP-tag and insulin in transgenic offspring in the SOFIA neo/δ-neo F2 generation. (Scale bar, 20 µm.) Detail shows the magnified boxed region. (Scale bar, 5 µm.)
To determine the transgene integration site(s), we performed targeted locus amplification (TLA) with subsequent next-generation sequencing (NGS). The results demonstrate that the transgene is integrated at a single locus on chromosome 11 in a noncoding, genomic region with a nearest distance of >0.4 Mb to the next coding gene (SI Appendix, Fig. S1B). In detail, integration of the transgene resulted in the duplication of the genomic locus chr11: 56,928,515 to 56,937,394. These duplicated regions flank the transgene, which is present at either of two possible orientations (SI Appendix, Fig. S1C). As observed by Southern blot (SI Appendix, Fig. S1A), the sequencing confirmed the presence of one INS-SNAP cassette with, and another without, the floxed neo cassette (SI Appendix, Fig. S1C). Taken together, these data show the successful integration of the INS-SNAP transgene in the porcine genome in a single locus on chromosome 11.
Characterization of Insulin-SNAP Expression and Targeting.
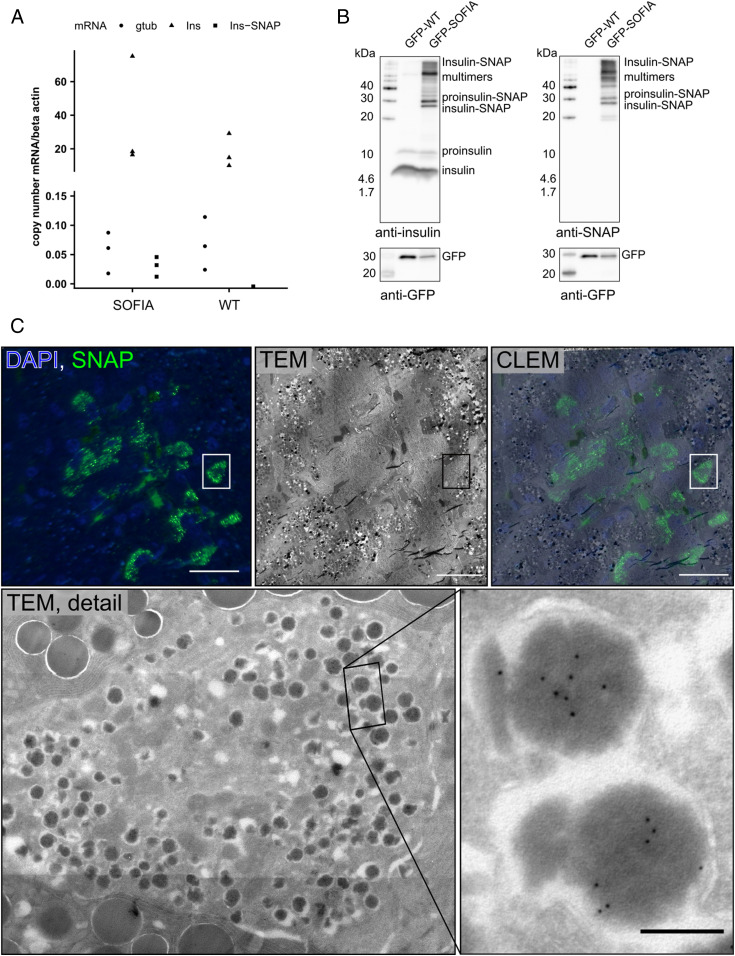
Next, we characterized the expression levels and localization of INS-SNAP at the cellular level by real-time qPCR (RT-qPCR), immunoblotting, and microscopy. To this aim, we isolated messenger RNA (mRNA) from total pancreas for RT-qPCR. As expected, the transgene was not detected in WT animals (Fig. 2A) but at comparable expression levels to γ-tubulin in SOFIA pig pancreata. However, in both SOFIA and WT pigs, endogenous insulin was ∼600-fold higher than the transgene or γ-tubulin. Furthermore, we crossed SOFIA pigs with INS-eGFP animals expressing GFP under the porcine insulin promoter (21) to allow for fluorescence-activated cell sorting (FACS) of β cells from pancreas. Extracts of the sorted cells were then used for immunoblotting. Both SOFIA/INS-eGFP and INS-eGFP animals showed comparable levels of 9 to 12 kDa proinsulin and 6 kDa insulin in FACS-sorted β cells (Fig. 2B). When probing for SNAP, however, only β cells of the SOFIA pig showed a robust signal, shifted about ∼20 kDa, resulting from the additional 19.4 kDa SNAP-tag. In addition, higher–molecular weight bands were detectable in cells of SOFIA pigs. Since they may result from the nonreducing conditions used in tricine gels, we ran the samples also under reducing conditions. In this case, the higher–molecular weight bands disappeared together with endogenous insulin because of the disruption of the intramolecular disulfide bridges within insulin (SI Appendix, Fig. S3).
Fig. 2.
Expression and targeting insulin-SNAP to insulin SGs in SOFIA pig islets. (A) RT-qPCR of SOFIA pig and WT pig pancreas. (B) Western blots of nonreducing tricine gels against insulin, SNAP, and GFP on FACS sorted GFP-SOFIA or GFP-WT β cells. GFP-WT: β cells from INS-eGFP animal and GFP-SOFIA: β cells from SOFIA/INS-eGFP animal. (C) CLEM of SOFIA pig pancreas. The fluorescence image shows anti-SNAP labeling (green) with SNAP+ β cells and DAPI (blue). Corresponding transmission EM (TEM) image shows the pancreatic islet surrounded by exocrine tissue. CLEM overlay shows the SNAP signal to be within β cells. (Scale bars, 20 µm.) TEM detail shows the SNAP+ β cell, with the inset showing immunogold labeling for SNAP (10 nm gold). (Scale bar, 200 nm.)
Finally, we applied correlative light and electron microscopy (CLEM) to fixed pancreas tissue obtained from adult SOFIA pigs. Immunolabeling with a primary antibody against SNAP followed by a secondary Alexa 488–coupled antibody and protein A–gold 10 nm showed granular subcellular staining (Fig. 2 C, Upper Left and Middle). Overlay with the electron microscopy (EM) image allowed for the identification of the fluorescent signal within insulin SGs in the islets of Langerhans (Fig. 2 C, Upper Right). This fluorescent signal further correlated with protein A–gold labeling. The ultrastructure of insulin-SNAP+ SGs appeared to be normal, with insulin SGs containing one or more insulin crystals surrounded by a translucent halo (Fig. 2 C, Lower).
Overall, the ultrastructure of the β cells of SOFIA pigs was comparable to that of pigs not expressing insulin-SNAP and to the published EM data of WT pigs (22, 23), with the normal appearance of insulin SGs, mitochondria, and endoplasmic reticulum without any signs of stress or structural alterations (SI Appendix, Fig. S4). Furthermore, all insulin SGs in SNAP+ β cells contained insulin-SNAP (Fig. 2 C, Lower Right).
SOFIA Pigs Have Normal Glucose Tolerance.
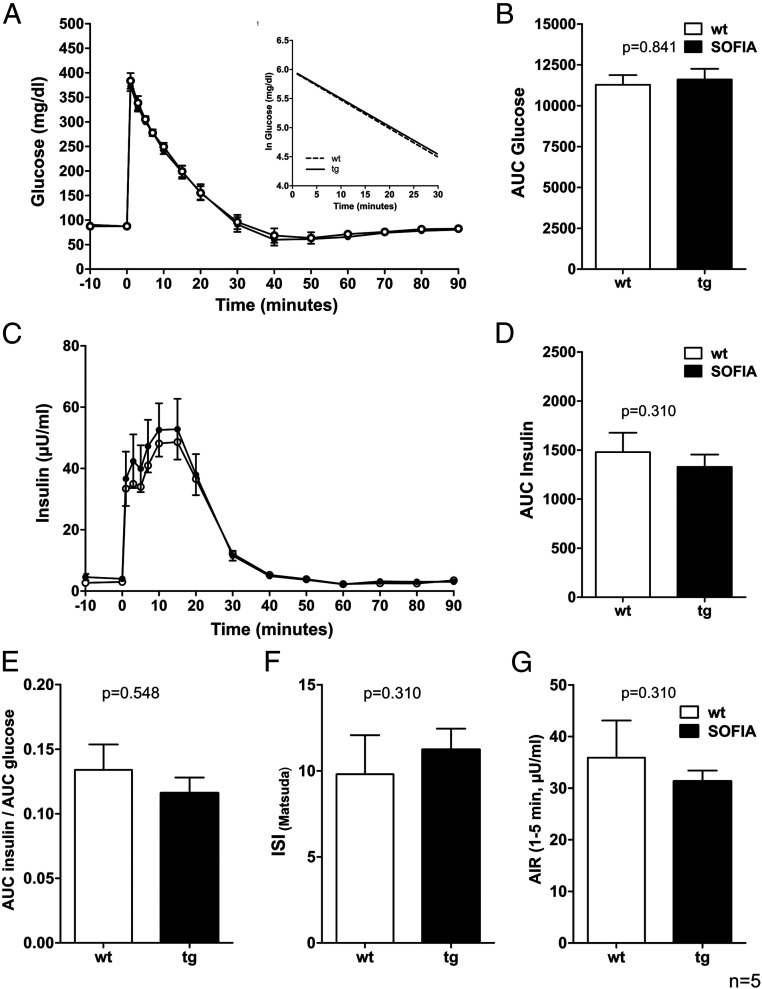
To assess metabolic control of insulin secretion on the organism level, we performed intravenous glucose tolerance tests (IVGTTs) in SOFIA and WT pigs (Fig. 3). After administration, glucose was rapidly cleared from the blood of either pig strain without any obvious difference (Fig. 3 A and B). Simultaneously, plasma insulin levels were comparable between the two groups without any significant difference (Fig. 3 C and D). Accordingly, further parameters, such as the quotient of insulin and glucose, the insulin sensitivity index, and the acute insulin response were not significantly altered in SOFIA pigs in comparison to WT pigs (Fig. 3 E–G). In conclusion, these data indicate that the insertion of the INS-SNAP transgene in this SOFIA pig strain does not interfere with glucose homeostasis in vivo.
Fig. 3.
IVGTT and related indices of 15- to 16-wk-old SOFIA pigs and WT controls. (A) Plasma glucose levels. (Inset) Glucose elimination rate. (B) AUC glucose. (C) Plasma insulin levels. (D) AUC insulin. (E) Quotient of AUC insulin and AUC glucose. (F) Insulin sensitivity index according to Matsuda (ISI Matsuda). (G) Acute insulin response (AIR). Data are represented as means ± SEM n = 5 per GT.
SNAP-Labeling of Insulin SGs In Vivo.
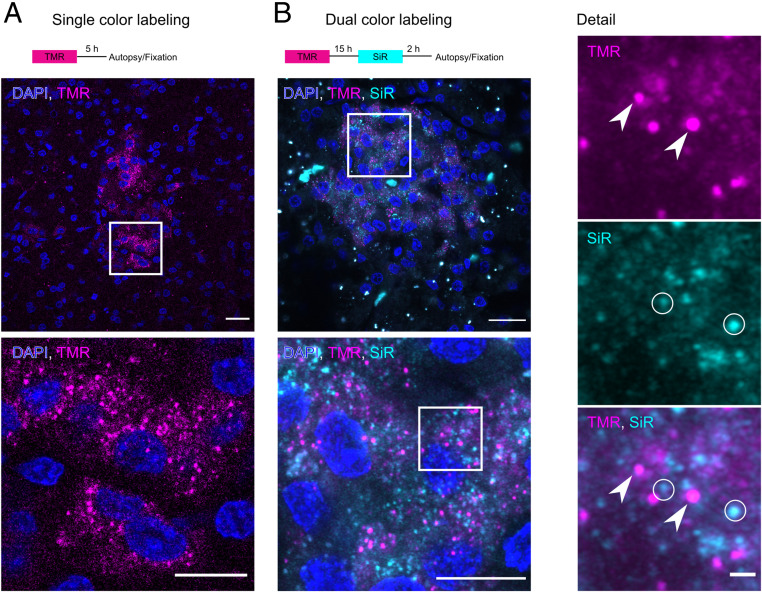
Next, we investigated the functionality and suitability of the SNAP-tag for the age-dependent labeling of insulin SGs in living pigs. To this aim, we injected 0.6 to 2 µmol/100 kg body weight (BW) of fluorescent BG SNAP substrates intravenously in SOFIA and WT pigs. Injection of BG-coupled tetramethylrhodamine (BG-TMR), followed by euthanasia of the animal and fixation of the pancreas, resulted in a clearly detectable TMR signal in cryosections of the pancreas (Fig. 4A). The signal was restricted to the pancreatic islets and was colocalized with the insulin antibody staining (SI Appendix, Fig. S5). Although a fluorescent TMR signal could be selectively imaged after in vivo application of 0.6 µmol TMR per 100 kg BW, we decided to apply 1.8 to 2 µmol SNAP substrate per 100 kg BW for in vivo SNAP labeling to obtain a more robust imaging signal. For comparison, SOFIA mice received 15 nmol SNAP substrate per mouse (15 nmol/25 g) (15) (i.e., 60 µmol/100 kg BW). Hence, for the in vivo labeling of SOFIA pigs, we applied 30-fold less SNAP substrate per kilogram of BW. In order to demonstrate the suitability of the approach for labeling distinct insulin SG pools, we performed a dual-color labeling experiment with a first application of BG-TMR followed by BG–silicon rhodamine (SiR) after 15 h and autopsy and fixation of the pancreas 2 h later. Again, we could detect the red and far-red fluorescence signals of both SNAP substrates in the islets and detect granular labeling at high magnification (Fig. 4B). Furthermore, there was only a modest colocalization of TMR+ and SiR+ insulin SGs (Pearson’s coefficient: 0.36 0.11), whereas SGs labeled with only a single substrate were the majority, indicating the segregation of SG pools over time. Taken together, these data show that INS-SNAP can be efficiently labeled in vivo by the injection of fluorescently modified SNAP substrates and further shows suitability for the age-dependent labeling of distinct SG pools.
Fig. 4.
In vivo labeling of SOFIA pigs. (A) Scheme for single-color BG-TMR labeling. (Below) A confocal microscopy image of a cryosection showing TMR fluorescence in the islets of Langerhans. (Scale bar, 20 µm.) The magnified view shows granular TMR fluorescence (magenta) and nuclei (blue). (Scale bar, 10 µm.) (B) Scheme for dual-color labeling with BG-TMR and BG-SiR. (Below) A confocal image of a cryosection of a SOFIA pig showing TMR+ and SiR+ granular staining with nuclei (blue). (Scale bar, 20 µm.) Detailed views show the magnified boxed area with split TMR and SiR channels. Arrowheads point to exclusively TMR+ SGs, and circles show SiR-positive SGs. (Scale bar, 10 and 1 µm.)
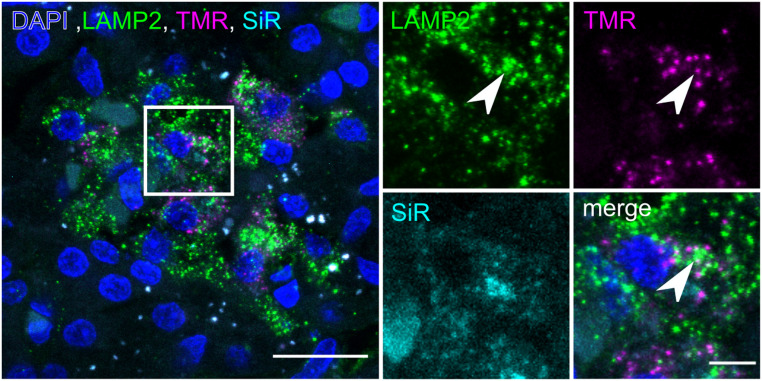
To investigate the intracellular degradation of age-defined insulin SGs, we performed immunolabeling for lysosome-associated protein 2 (LAMP2). This allowed us to assess the colocalization of insulin SGs with lysosomes, hence, the relationship between SG aging and their intracellular degradation in vivo (Fig. 5). While the colocalization of older TMR+ SGs with LAMP2 was low (Pearson’s coefficient: 0.14 ± 0.08), we could, however, observe TMR+ SGs colocalizing with LAMP2+ compartments (Fig. 5, arrowheads). In contrast, the colocalization of younger SiR+ SGs with lysosomes was close to zero (Pearson’s coefficient: 0.04 ± 0.06), consistent with the preferential degradation of older SGs observed in isolated islets in vitro (16, 24).
Fig. 5.
Detection of age-defined insulin SGs in LAMP2+ compartments. Confocal image of cryosection of double-labeled SOFIA pig pancreas with LAMP2 staining. (Scale bar, 20 µm.) The magnified region shows single channels for LAMP2, TMR, and SiR. The arrowhead points to an object positive for LAMP2 and TMR. (Scale bar, 5 µm.)
Discussion and Outlook
In the present study, we have successfully generated and characterized a transgenic pig expressing INS-SNAP and demonstrate its suitability for the in vivo labeling of insulin SGs. We provide a strategy to overcome two major limitations of β cell research: the translational gap between rodents and humans and ex vivo experiments for insulin turnover using isolated pancreatic islets. We generated a transgenic pig as a model system, since it closes this translational gap. Pigs are akin to humans in the anatomy of their gastrointestinal tract and the morphology and function of the pancreas (20). Furthermore, it has been demonstrated that the structure and composition of pig pancreatic islets is much closer to human than to rodent islets (25), although some morphological and functional differences in islets between humans and pigs were reported (26, 27). Moreover, the molecular and developmental signatures of pig and human islets and β cells are also more similar to each other than those of human and rodents (28). This makes the pig a compelling model organism not only for the study of systemic metabolism but specifically also for that of islets and β cell function.
The age of insulin SGs has been associated with the likelihood of exocytosis, and recently, a connection of the dysregulation of SG age with metabolic stress has been made (29). We chose the SNAP-tag fused with insulin in our pig model, since it allows for the conditional and flexible labeling of age-defined insulin SG pools with fluorophores of different colors over longer time spans, compared to fluorescent timer proteins (30). We used this technology successfully in isolated mouse islets to map age-distinct SG pools (16). However, this knock-in mouse showed signs of impaired glucose tolerance (15), a problem that did not occur in the transgenic SOFIA pig. Notably, the SOFIA mouse is a knock-in model with SNAP fused in frame at the C terminus of Ins2, leading to a high expression of insulin2-SNAP. This may delay proinsulin2-SNAP folding and processing and thus could account for the impaired glucose tolerance of these mice. Therefore, we chose to generate the pig as a transgenic model, hoping to overcome the aforementioned problems, which turned out to be successful. Our strategy for the pig model was based on a random integration approach, selection of the most suitable founder animal (based on transgene expression level and absence of side effects), and subsequent characterization of the transgene integration site. For the selected founder animal, TLA sequencing revealed a single-transgene integration site in a noncoding region of chromosome 11 that is unlikely to affect the function of neighboring genes. This locus may turn out to be a safe harbor for the faithful expression of transgenes after targeted integration using CRISPR/Cas9 (31).
So far, our knowledge on insulin SG trafficking is derived exclusively from ex vivo experiments using isolated islets or in vitro work with insulin-producing cell lines. Proteins tagged with self-labeling enzymes have been successfully used for in vivo studies in the mouse brain and in small animals (32–35) by the injection of the substrates directly into the tissue. Administration of a single-color SNAP-tag or Halo-tag substrate via tail–vein injection has resulted in specific labeling in mice (15, 36). This approach is less invasive and overcomes the substantial hurdles and potential severe complications (e.g., pancreatitis) associated with injections into a retroperitoneal and sensitive organ as the pancreas. Nevertheless, the high specificity of the SNAP substrates to their tag and the covalent binding upon contact enabled the specific labeling of insulin SGs, even in adult pigs. We found that a dual-color sequential labeling, with only a relatively short time interval between the labeling steps, results in partially distinct insulin SG pools. Future studies will aim to optimize the delivery of substrate directly to the β cells, as well as to better control the age-defined labeling. Furthermore, the possibility to quantify intracellular degradation of aged SGs by immunolabeling for lysosomal markers provides a further readout to investigate SG turnover. Our preliminary data suggest that the intracellular degradation of insulin occurs already at a relatively young SG age of ∼18 h compared to 2.7 d, as estimated in isolated mouse islets (16). Ultimately, the SOFIA pig may be crossed to diabetic pig models (37, 38) to gain insights into the changes in insulin SG turnover in T2DM.
Methods
Generation of SOFIA Pig.
All animal procedures in this study were approved by the responsible animal welfare authority (Regierung von Oberbayern) and were performed according to the German Animal Welfare Act and Directive 2010/63/EU on the protection of animals used for scientific purposes.
The SNAP-tag sequence (New England Biolabs [NEB]) was cloned into a β cell–specific expression vector with a porcine INS gene promoter, exon and intron sequence (39), leading to an SNAP-tagged insulin protein product. The INS-SNAP vector was combined with a floxed neo cassette. Porcine kidney cells were transfected with the linearized and excised expression vector, and pools of stable, transfected male porcine kidney cell clones were generated. For the Cre-mediated removal of floxed neo cassette, kidney cells were lipofected with a CAG-Cre expression vector directly before being used for SCNT (40). Cloned embryos were laparoscopically transferred to recipient gilts. Genotyping of offspring was performed by PCR using the following primers: SNAP_1_for (5′- ACC AGA GCC ACT GAT GCA G -3′), SNAP_3_rev (5′- GGA GTG GCA CCT TCC AG -3′), δneo_2_for (5′- CCT ACT TTC ACC AGC CTG AG -3′), δneo_1_rev (5′- AGC TTG ATA TCG AAT TCC TGC AG -3′), neo_1_for (5′- ACA ACA GAC AAT CGG CTG CTC TG -3′), and neo_2_rev (5′- TGC TCT TCG TCC AGA TCA TCC TG -3′).
Transgene integration patterns were analyzed by Southern blot analysis as described previously (41). Genomic DNA was extracted from skin using Wizard Genomic DNA Purification Kit (Promega). A total of 10 µg DNA each were digested with NdeI or PshAI, fractionated on 0.7% Tris-Borate-EDTA agarose gel, and blotted under neutral conditions to Hybond-XL nylon membrane (GE Healthcare). Probes were synthesized by PCR from DNA of a transgenic animal using primers INS (5′- TCG TTA AGA CTC TAA TGA CCT C -3′) and INS-SNAP_5_rev (5′- ATC CCA GTT GCA GTA GTT CTC CAG C -3′) for comprising the 3′-region of the porcine INS promoter and the 5′-region of the INS-SNAP sequence, microdialyzed and 32P-labeled using Prime-a-Gene Labeling System (Promega). Filters were prehybridized for 2 h at 65 °C in Rapid-hyb solution (GE Healthcare): Hybridization was performed overnight in the same buffer containing a 32P-labeled probe. After washing the membranes (once for 20 min at room temperature in 2× standard saline citrate containing 0.1% sodium dodecyl sulfate [SDS] and twice for 15 min at 65 °C in 1× standard saline citrate containing 0.1% SDS), the exposition of membranes was done in a Phosphor-Imager cassette overnight. Imaging plates were scanned with a Phosphor-Imager (Typhoon FLA9000; GE Healthcare).
TLA Sequencing.
Commercially available TLA with subsequent NGS was performed by Cergentis.
FACS Sorting of β Cells and Western Blot.
For the FACS sorting of GFP-positive β cells, the SOFIA pig was crossed with the INS-eGFP pig line expressing eGFP specifically in their β cells (21). Dual transgenic (SOFIA/INS-eGFP) and single INS-eGFP transgenic pigs were euthanized at an age of 8 to 10 wk, their pancreata were cut in small pieces, collagenase digested, sieved through 500-μm mesh, and washed as described in ref. 21. Afterward, single-cell suspension by TrypLE Express enzyme digestion was prepared for FACS sorting, as described elsewhere (42). Between 22,109 and 26,665 FACS-sorted β cells were then lysed, boiled in (non)reducing sample buffer, and loaded on tricine gels for the detection of insulin (Sigma, I2018), SNAP-tag (NEB, P9310S), or GFP (Max Planck Institute of Molecular Cell Biology and Genetics [MPI-CBG], self-made; or Takara, 632381).
RT-qPCR.
For mRNA isolation, small pieces of pancreas were homogenized in RLT buffer (Qiagen) at 4 °C using a TissueLyser (Qiagen) with a 5-mm steel bead for 5 min at 50 Hz. The mRNA was then isolated from the homogenate using an RNeasy Mini Kit (Qiagen) according to manufacturer’s recommendations. Reverse transcription (RT) of mRNA into complementary DNA (cDNA) was carried out using an Moloney Murine Leukemia Virus RT RNase kit (Promega). RT-qPCR was performed using the GoTaq qPCR Master Mix (Promega) according to manufacturer’s instructions. Briefly, cDNA from RT reactions was diluted 1:2 in RNase-free water, and reactions were set up as triplicates in semiskirted 96-Well PCR Plates (0.2 mL) with optical strip caps (Agilent). The PCR reactions were run in an AriaMx RT-PCR System (Agilent). For absolute quantification, the serial dilutions of the target sequence cloned into pCRII vectors were used. The results were then normalized by the parallel amplification of porcine β actin mRNA. The following primers were used for the detection of porcine: β-actin (fwd: 5′- CAA CGG CTC CGG CAT GTG -3′; rev: 5′- TCT TCT CCA TGT CGT CCC AGT TG -3′), γ-tubulin (fwd: 5′- CCA CGG TCC TGG ACG TCA T -3′; rev: 5′- GGT GTG GTT GGC CAT CAT GAG C -3′), insulin (fwd: 5′- GAG AAC CCT CAG GCA GGT G -3′; rev: 5′- CCG CAC CCC AAA ACC CAA T -3′), and INS-SNAP (fwd: 5′- AAC CCT CAG GCA GGT GCC -3′; rev: 5′- CTG GAC AAA GAC TGC GAA AT -3′).
Immunohistochemistry and Confocal Microscopy.
Fixed pancreatic tissue was embedded in TissueTek, snap frozen, and stored at −80 °C for cryosectioning. Immunolabeling on cryosections was performed with anti-insulin (Sigma), anti-SNAP (NEB), and LAMP2 (PLID, self-made) antibodies. Labeled cryosections were imaged with a Zeiss LSM 780 (DFG FZT 111) and LSM 980 of the Light Microscopy Facility, a Core Facility of the Center for Molecular and Cellular Bioengineering (CMCB) Technology Platform at Technische Universität (TU) Dresden, and a Nikon C2+ confocal microscope with 20× air and 40× and 63× oil immersion objectives. The colocalization of fluorescent signals was analyzed with the Coloc2 plugin in FIJI (43).
Transmission EM.
Pancreatic tissue was fixed with 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M Sörensen’s phosphate buffer (pH 7.4) at room temperature. After dehydration in a graded series of ethanol, the specimens were embedded in epoxy resin, as described before (4). Ultrathin sections were cut with a Leica EM UC6 μL tramicrotome. After postcontrasting with uranyl-acetate and lead-citrate, the sections were observed with an FEI Morgagni electron microscope running at 80 kV.
CLEM.
For CLEM, pancreatic tissue was fixed with 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M Sörensen’s phosphate buffer (pH 7.4). The specimens were processed for CLEM, as described in ref. 44. In brief, they were embedded in 12% gelatin and immersed in 2.3 M sucrose at 4 °C overnight. The samples were mounted on metal pins and immersed in liquid nitrogen. Ultrathin Tokuyasu sections were cut with a Leica EM UC6 μL tramicrotome equipped with an FC6 cryounit. Sections were stained with the anti-SNAP antibody, followed by an Alexa Fluor–conjugated secondary antibody. For immunogold-labeling protein A–gold 10 nm was applied, followed by DAPI. Fluorescence imaging was done prior to EM with a Keyence Biozero 8000 fluorescence microscope, as described in ref. 45.
IVGTT.
An IVGTT was performed in 15- to 16-wk-old SOFIA and WT control pigs. For stress-free, frequent blood sampling, central venous catheters (Careflow 3 Fr, 200 mm, Merit Medical Systems) were surgically inserted into the external jugular vein via the vena auricularis under anesthesia, as described previously (46). After an 18-h fasting period, a bolus injection of concentrated 50% glucose solution (0.5 g/kg BW) was administered through the central venous catheter. Blood samples were collected at the indicated time points (Fig. 3). Plasma glucose levels were determined using an AU480 autoanalyzer (Beckman Coulter) and adapted reagent kits from Beckman Coulter. Plasma insulin levels were determined by radioimmunoassay (Millipore).
The net glucose elimination rate after glucose injection was calculated as the slope for the interval 1 to 30 min after the glucose injection of the logarithmic transformation of the individual plasma glucose values. Insulin sensitivity indices were calculated according to Matsuda (47). Acute insulin responses were calculated as the difference of mean insulin levels at 1, 3, and 5 min following intravenous glucose load and basal insulin levels. Longitudinal data (glucose/insulin values during IVGTT) were statistically evaluated by ANOVA (linear mixed models; PROC MIXED; and SAS 8.2), taking the fixed effects of genotype (GT; transgenic and control), time (relative to glucose application), and the interaction GT*Time, as well as the random effect of the individual animal into account. Area under the curve (AUC) insulin/glucose was calculated using GraphPad Prism software (version 5.02). AUCs and indices were tested for significance by Mann–Whitney U test using SPSS (version 21) software.
In Vivo Labeling of INS-SNAP in Pigs with SNAP Substrates for Imaging Studies.
BG-TMR and BG-SiR were synthesized as described previously (33, 48). Central venous catheters (Careflow 3 Fr, 200 mm, Merit Medical Systems) were surgically inserted into the external jugular vein via the vena auricularis at least 1 d before SNAP substrate or vehicle application. SNAP substrates were resuspended at a concentration of 1 µmol per 50 µL DMSO and diluted directly prior to intravenous injection with a 50-fold volume of 0.9% NaCl.
For single-SNAP substrate in vivo labeling, 0.6 µmol BG-TMR per 100 kg BW or 1.8 to 2.0 µmol TMR-Star per 100 kg BW were intravenously injected in three overnight-fasted SOFIA pigs and one WT pig. Around 5 h after BG-TMR injection, an autopsy took place for the sampling of pancreas for histological analyses. For dual-SNAP substrate in vivo labeling in two SOFIA pigs, 1.8 to 2.0 µmol per 100 kg BW BG-TMR, as the first SNAP substrate, were intravenously injected at 5:15 PM during the last meal. Second SNAP substrate intravenous injection using BG-SiR at an amount of 1.8 to 1.9 µmol per 100 kg BW was done in overnight-fasted animals at 8:15 AM, 15 h after first SNAP substrate application. Around 2 h after BG-SiR injection, an autopsy took place. As a negative control to demonstrate the specificity of SNAP substrate labeling of INS-SNAP protein on insulin granules in β cells, one SOFIA pig received DMSO without substrate and one WT pig received BG-TMR substrate, as performed for the single labelings of SOFIA pigs.
Supplementary Material
Acknowledgments
We thank Christina Blechinger, Eva Jemiller, Anne Richter, and Tatiana Schroeter (all Ludwig Maximilian University of Munich, Germany) as well as Carla Münster, Daniela Friedland, Eyke Schöniger, Katharina Ganß, Carolin Wegbrod (all Paul Langerhans Institute), Cordula Andreé (MPI-CBG), and Bettina Mathes (MPI for Medical Research) for their excellent technical support. We thank Susanne Kretschmar (CMCB) for EM processing. We are further grateful to Katja Pfriem for administrative assistance. We acknowledge the antibody facility of the MPI-CBG for antibody generation. This work was supported by the Light Microscopy Facility, a Core Facility of the CMCB Technology Platform at TU Dresden. This work was supported by the Grant 82DZD00802 from the German Federal Ministry of Education and Research (BMBF) to the German Center for Diabetes Research (DZD) to E.W. and in part by the German Research Council (TRR127) to E.W. and E.K. M.S. received funding from the DZD by the BMBF, from the German Research Foundation (DFG) jointly with the Agence Nationale de la Recherche (Grant SO 818/6-1), and from the Innovative Medicines Initiative 2 Joint Undertaking under Grant Agreements 115881 (RHAPSODY), which includes financial contributions from the European Union’s Framework Program Horizon 2020, the European Federation of Pharmacological Industries and Associations (EFPIA), and the Swiss State Secretariat for Education, Research, and Innovation under Contract 16.0097, as well as the Juvenile Diabetes Research Foundation (JDRF) International and the Leona M. and Harry B. Helmsley Charitable Trust. A.I. and A.M. were the recipients of MeDDrive Grants (60.372, 60.295, and 60417) from the Carl Gustav Carus Faculty of Medicine at TU Dresden. M.N. was the recipient of a predoctoral fellowship from the Dresden International Graduate School for Biomedicine and Bioengineering in the context of the Excellence Initiative of the DFG. T.K. and the EM facility of the Center for Molecular and Cellular Bioengineering are supported by the European Fund for Regional Development.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107665118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Solimena M., et al., Systems biology of the IMIDIA biobank from organ donors and pancreatectomised patients defines a novel transcriptomic signature of islets from individuals with type 2 diabetes. Diabetologia 61, 641–657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saeedi P.et al., IDF Diabetes Atlas Committee , Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 157 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Cohrs C. M., et al., Dysfunction of persisting β cells is a key feature of early type 2 diabetes pathogenesis. Cell Rep. 31, 107469 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Fava E., et al., Novel standards in the measurement of rat insulin granules combining electron microscopy, high-content image analysis and in silico modelling. Diabetologia 55, 1013–1023 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Müller A., et al., 3D FIB-SEM reconstruction of microtubule-organelle interaction in whole primary mouse β cells. J. Cell Biol. 220, e202010039 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rorsman P., Renström E., Insulin granule dynamics in pancreatic beta cells. Diabetologia 46, 1029–1045 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Cerasi E., Luft R., The plasma insulin response to glucose infusion in healthy subjects and in diabetes mellitus. Acta Endocrinol. (Copenh.) 55, 278–304 (1967). [DOI] [PubMed] [Google Scholar]
- 8.Curry D. L., Bennett L. L., Grodsky G. M., Dynamics of insulin secretion by the perfused rat pancreas. Endocrinology 83, 572–584 (1968). [DOI] [PubMed] [Google Scholar]
- 9.Jaffredo M., et al., Dynamic uni- and multicellular patterns encode biphasic activity in pancreatic islets. Diabetes 70, 878–888 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Barg S., Eliasson L., Renström E., Rorsman P., A subset of 50 secretory granules in close contact with L-type Ca2+ channels accounts for first-phase insulin secretion in mouse β-cells. Diabetes 51(suppl. 1), S74–S82 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Ohara-Imaizumi M., et al., Imaging analysis reveals mechanistic differences between first- and second-phase insulin exocytosis. J. Cell Biol. 177, 695–705 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schatz H., Nierle C., Pfeiffer E. F., (Pro-) insulin biosynthesis and release of newly synthesized (pro-) insulin from isolated islets of rat pancreas in the presence of amino acids and sulphonylureas. Eur. J. Clin. Invest. 5, 477–485 (1975). [DOI] [PubMed] [Google Scholar]
- 13.Halban P. A., Differential rates of release of newly synthesized and of stored insulin from pancreatic islets. Endocrinology 110, 1183–1188 (1982). [DOI] [PubMed] [Google Scholar]
- 14.Keppler A., et al., A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Ivanova A., et al., Age-dependent labeling and imaging of insulin secretory granules. Diabetes 62, 3687–3696 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller A., et al., A global approach for quantitative super resolution and electron microscopy on cryo and epoxy sections using self-labeling protein tags. Sci. Rep. 7, 23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller A., Mziaut H., Neukam M., Knoch K.-P., Solimena M., A 4D view on insulin secretory granule turnover in the β-cell. Diabetes Obes. Metab. 19 (suppl. 1), 107–114 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Hoboth P., et al., Aged insulin granules display reduced microtubule-dependent mobility and are disposed within actin-positive multigranular bodies. Proc. Natl. Acad. Sci. U.S.A. 112, E667–E676 (2015). Correction in: Proc. Natl. Acad. Sci. U.S.A. 112, E2114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neukam M., Sönmez A., Solimena M., FLIM-based pH measurements reveal incretin-induced rejuvenation of aged insulin secretory granules. bioRxiv [Preprint] (2017). https://www.biorxiv.org/content/10.1101/174391v2.full (Accessed 19 February 2021).
- 20.Wolf E., Braun-Reichhart C., Streckel E., Renner S., Genetically engineered pig models for diabetes research. Transgenic Res. 23, 27–38 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Kemter E., et al., INS-eGFP transgenic pigs: A novel reporter system for studying maturation, growth and vascularisation of neonatal islet-like cell clusters. Diabetologia 60, 1152–1156 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Vantyghem M. C., et al., Immunohistochemical and ultrastructural study of adult porcine endocrine pancreas during the different steps of islet isolation. Histochem. Cell Biol. 106, 511–519 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Lukinius A., Korsgren O., The transplanted fetal endocrine pancreas undergoes an inherent sequential differentiation similar to that in the native pancreas. An ultrastructural study in the pig-to-mouse model. Diabetes 50, 962–971 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Orci L., et al., Insulin, not C-peptide (proinsulin), is present in crinophagic bodies of the pancreatic B-cell. J. Cell Biol. 98, 222–228 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoang D.-T., et al., A conserved rule for pancreatic islet organization. PLoS One 9, e110384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim A., et al., Islet architecture: A comparative study. Islets 1, 129–136 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper D. K. C., et al., Progress in clinical encapsulated islet xenotransplantation. Transplantation 100, 2301–2308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S., et al., Molecular and genetic regulation of pig pancreatic islet cell development. Development 147, dev186213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yau B., et al., A fluorescent timer reporter enables sorting of insulin secretory granules by age. J. Biol. Chem. 295, 8901–8911 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duncan R. R., et al., Functional and spatial segregation of secretory vesicle pools according to vesicle age. Nature 422, 176–180 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Ruan J., et al., Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Sci. Rep. 5, 14253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masch J.-M., et al., Robust nanoscopy of a synaptic protein in living mice by organic-fluorophore labeling. Proc. Natl. Acad. Sci. U.S.A. 115, E8047–E8056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poc P., et al., Interrogating surface versus intracellular transmembrane receptor populations using cell-impermeable SNAP-tag substrates. Chem. Sci. 11, 7871–7883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campos C., Kamiya M., Banala S., Johnsson K., González-Gaitán M., Labelling cell structures and tracking cell lineage in zebrafish using SNAP-tag. Dev. Dyn. 240, 820–827 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Yang G., et al., Genetic targeting of chemical indicators in vivo. Nat. Methods 12, 137–139 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Chen S., et al., In vivo ZIMIR imaging of mouse pancreatic islet cells shows oscillatory insulin secretion. Front. Endocrinol. (Lausanne) 12, 613964 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renner S., et al., Permanent neonatal diabetes in INS(C94Y) transgenic pigs. Diabetes 62, 1505–1511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renner S., et al., Glucose intolerance and reduced proliferation of pancreatic β-cells in transgenic pigs with impaired glucose-dependent insulinotropic polypeptide function. Diabetes 59, 1228–1238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klymiuk N., et al., Xenografted islet cell clusters from INSLEA29Y transgenic pigs rescue diabetes and prevent immune rejection in humanized mice. Diabetes 61, 1527–1532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurome M., Kessler B., Wuensch A., Nagashima H., Wolf E., “Nuclear transfer and transgenesis in the pig” in Nuclear Reprogramming: Methods and Protocols, Methods in Molecular Biology, Beaujean N., Jammes H., Jouneau A., Eds. (Springer, 2015), pp. 37–59. [DOI] [PubMed] [Google Scholar]
- 41.Klose R., et al., Expression of biologically active human TRAIL in transgenic pigs. Transplantation 80, 222–230 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Böttcher A., et al., Non-canonical Wnt/PCP signalling regulates intestinal stem cell lineage priming towards enteroendocrine and Paneth cell fates. Nat. Cell Biol. 23, 23–31 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Schindelin J., et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Völkner M., et al., Mouse retinal organoid growth and maintenance in longer-term culture. Front. Cell Dev. Biol. 9, 645704 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fabig G., et al., Labeling of ultrathin resin sections for correlative light and electron microscopy. Methods Cell Biol. 111, 75–93 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Renner S., et al., Metabolic syndrome and extensive adipose tissue inflammation in morbidly obese Göttingen minipigs. Mol. Metab. 16, 180–190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radikova Z., Assessment of insulin sensitivity/resistance in epidemiological studies. Endocr. Regul. 37, 189–194 (2003). [PubMed] [Google Scholar]
- 48.Lukinavičius G., et al., A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat. Chem. 5, 132–139 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.