Significance
HIV-1 replication is critically dependent on the selective incorporation of viral RNA genomes into assembling virions. Although RNA elements that promote packaging have been identified, the determinants of authentic packaging fidelity and efficiency have until now remained unknown. The present studies show that genome selection is achieved by a bipartite mechanism that requires both dimerization-dependent exposure of packaging signals within the 5′ leader of the viral RNA and structural sequestration of the 5′ cap. Cap sequestration likely prevents capture by the cellular RNA processing and translation machinery, a mechanism that may help explain why some cellular RNA polymerase III transcripts that lack 5′ caps can parasitize virions. Cap sequestration may be commonly employed among RNA viruses that package 5′-capped genomes.
Keywords: HIV-1, RNA, genome, 5′ cap, packaging
Abstract
HIV-1 selectively packages two copies of its 5′-capped RNA genome (gRNA) during virus assembly, a process mediated by the nucleocapsid (NC) domain of the viral Gag polyprotein and encapsidation signals located within the dimeric 5′ leader of the viral RNA. Although residues within the leader that promote packaging have been identified, the determinants of authentic packaging fidelity and efficiency remain unknown. Here, we show that a previously characterized 159-nt region of the leader that possesses all elements required for RNA dimerization, high-affinity NC binding, and packaging in a noncompetitive RNA packaging assay (ΨCES) is unexpectedly poorly packaged when assayed in competition with the intact 5′ leader. ΨCES lacks a 5′-tandem hairpin element that sequesters the 5′ cap, suggesting that cap sequestration may be important for packaging. Consistent with this hypothesis, mutations within the intact leader that expose the cap without disrupting RNA structure or NC binding abrogated RNA packaging, and genetic addition of a 5′ ribozyme to ΨCES to enable cotranscriptional shedding of the 5′ cap promoted ΨCES-mediated RNA packaging to wild-type levels. Additional mutations that either block dimerization or eliminate subsets of NC binding sites substantially attenuated competitive packaging. Our studies indicate that packaging is achieved by a bipartite mechanism that requires both sequestration of the 5′ cap and exposure of NC binding sites that reside fully within the ΨCES region of the dimeric leader. We speculate that cap sequestration prevents irreversible capture by the cellular RNA processing and translation machinery, a mechanism likely employed by other viruses that package 5′-capped RNA genomes.
HIV-1 replication is critically dependent on selective and efficient incorporation of two copies of the unspliced, 5′-capped RNA genome (gRNA) into assembling virus particles (1–5). Genomes are recruited to plasma membrane assembly sites from a cytoplasmic milieu that contains a substantial excess of cellular RNAs, spliced viral messenger RNAs (mRNAs) that encode for viral accessory proteins, and unspliced viral mRNAs that encode the viral Gag and Gag-Pol proteins (6). gRNA selection is mediated by the nucleocapsid (NC) domain of Gag, which interacts with cis-acting RNA “packaging signals” located within the 5′ leader (5′-L) of the unspliced RNA (7–12). Genomes are initially anchored to the plasma membrane by a small number of Gag proteins, possibly two dozen or fewer (13, 14), forming a ribonucleoprotein nucleation site that recruits additional Gag proteins and cellular factors required for virus assembly and budding (15). Genomes are packaged as dimers, and there is considerable evidence that dimerization and packaging are mechanistically coupled (4, 5, 7, 16–18). Although gRNA versus mRNA functions were originally thought to be controlled by dimerization-dependent structural modulation of a single viral transcript (19, 20), it now appears that RNA fates are established by a heterogeneous transcriptional start site mechanism, in which two functionally distinct pools of RNAs containing one or three 5′ guanosines are transcribed from the single integrated provirus (21, 22). RNAs containing a single 5′ guanosine preferentially form dimers in vitro and are selected for packaging as gRNA whereas those with three (and to a lesser extent, two) 5′ guanosines adopt a monomeric leader structure in vitro and are retained in cells as mRNAs (22, 23).
Efforts to identify the specific RNA residues and structures that promote packaging have relied primarily on biophysical studies of recombinant RNAs and in situ packaging experiments involving transiently transfected or infected cells (8, 24–27). The dimeric gRNA leader that promotes packaging adopts a branched multihairpin structure, in which residues important for transcriptional activation, initiation of reverse transcription, dimerization, and packaging (TAR, PBS, DIS, and ΨHP, respectively), or that contain a polyadenylation sequence (polyA), adopt hairpin structures that decorate a central tandem three-way junction structure (Fig. 1A) (8, 17, 24, 28, 29). The structure exposes approximately two dozen high-affinity NC binding sites under physiological-like conditions (27). A variant of the 5′-L that lacks the TAR and polyA hairpins and contains a GAGA tetraloop (A, adenosine; G, guanosine) substituted for the PBS loop exhibits dimerization and NC-binding properties similar to those of the intact 5′-L (24, 27), and vectors containing 5′-Ls that lack these elements can be readily packaged into virus-like particles (24). This region of the leader has therefore been called the “core encapsidation signal” (ΨCES) (Fig. 1 A and C) (24, 27, 29).
Fig. 1.
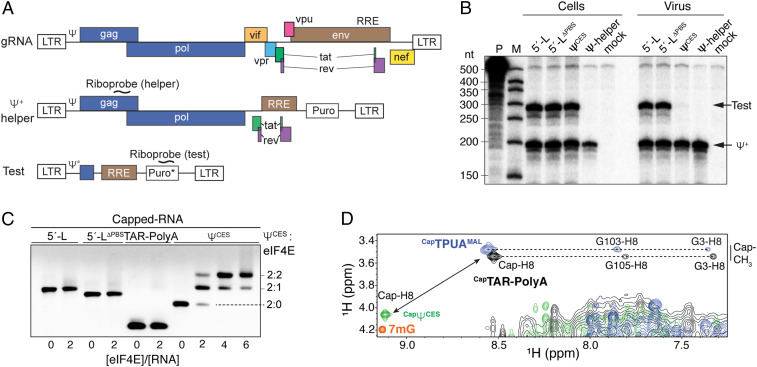
Secondary structure of the HIV-1NL4-3 gRNA 5′-L and constructs used to probe RNA packaging. (A) Native 5′-L. The PBS region is shaded in gray; nucleotides of TAR, polyA, U5, DIS, SD (splice donor site), ΨHP, and AUG are color-coded. The two three-way junctions are labeled as Ψ3WJ-1 and Ψ3WJ-2. (B) Schematic representation of the 5′-LΔPBS RNA (PBS substituted by a GAGA tetraloop). (C) Schematic representation of ΨCES (5′-LΔTAR-ΔPolyA-ΔPBS). Native residues U106 and G345 are substituted by G and C, respectively.
Packaging efficiencies have generally been assessed by quantifying extracellular viral RNA levels (virion-associated RNA) relative to extracellular Gag protein levels or copackaged host RNA levels (24, 30–32) and, in prior studies, the packaging efficiency of ΨCES was measured indirectly by comparisons with the noncoding host 7SL RNA that is enriched in virions (24). However, in the absence of authentic genomes, Gag proteins can assemble into virus-like particles that incorporate nongenomic mRNAs, many in proportions that roughly match their intracellular concentrations (33–36), and this promiscuity can confound assessment of packaging efficiencies relative to that of the authentic gRNA. To better understand the contributions of specific leader elements to packaging, we have now employed a competitive RNA packaging approach that allows quantitative comparison of packaging efficiencies from cells cotransfected with vectors containing native and modified 5′-Ls. Unexpectedly, the ability of ΨCES to direct packaging is severely attenuated compared with that of the native leader, despite having similar structural, dimerization, and NC-binding properties. Our studies further show that the PBS loop does not contribute to packaging but that the TAR and polyA helices are indispensable for competitive packaging even though these elements do not contribute to high-affinity NC binding or dimerization. Recent NMR studies revealed that the TAR and polyA helices adopt an end-to-end stacking arrangement in which the 5′ cap (5′-5′-triphosphate–linked 7-methylguanosine) is base-paired with, and sandwiched between, the TAR and polyA helices (23). We now show that cap sequestration by the TAR–polyA cassette is an essential determinant of packaging and that all other elements required for authentic packaging selectivity and efficiency reside fully within the ΨCES region of the leader. Mechanistic implications of cap sequestration for genome packaging by HIV-1 and other viruses that encapsidate 5′-capped RNAs are discussed.
Results
RNA Elements Required for Competitive Packaging.
To identify leader elements that contribute to wild-type packaging efficiency, competitive packaging experiments were performed using human 293T cells cotransfected with plasmid DNAs encoding native (Ψ+) and modified (test) HIV-1 5′-L sequences (27) (Fig. 2A). The Ψ+ vector contains the HIV-1NL4-3 genome with deletions of the vpu, vif, and part of env genes and with nef replaced by a puromycin-resistance expression cassette (20). Test plasmids contain native or modified forms of the initial 688 nt of the HIV-1NL4-3 genome followed by the Rev responsive element (RRE) and a puromycin-resistance gene (24) (Fig. 2A). Only cells expressing similar levels of Ψ+ and test RNAs, as assessed by an RNase protection assay (RPA) (37), were used for comparative packaging analyses. Experiments were conducted with test constructs containing the native 5′-L as a control, and with leaders that lack the primer binding site loop (5′-LΔPBS) (Fig. 1B) or the PBS, TAR, and polyA hairpins (5′-LΔTAR-ΔPolyA-ΔPBS: ΨCES) (Fig. 1C). Packaging data are reported throughout as the mean ± SD from three individual packaging experiments.
Fig. 2.
RNA packaging signal in the HIV-1 5′-L. (A, Top) The proviral DNA of HIV-1NL4-3. (A, Middle) The Ψ+-helper vector is a replication-defective version of the HIV-1NL4-3 proviral DNA. (A, Bottom) The test vector’s 5′-L includes the 5′ untranslated region plus 353 nt from the gag-coding region. In the test vectors, the puromycin cassette (Puro*) has an extra CMV promoter region, for which the riboprobe is targeted. LTR, long terminal repeat. (B) Packaging of test RNAs containing 5′-L, 5′-LΔPBS, and ΨCES in competition with Ψ+, assayed by ribonuclease protection. M, RNA size marker; P, undigested probe. (C) Native gel electrophoresis showing that eIF4E binds the dimeric CapΨCES but not capped RNAs containing TAR–polyA (Cap5′-L, Cap5′-LΔPBS, and CapTAR–polyA). (D) Portion of 2D NOESY spectra overlay showing cap chemical shifts and NOEs indicative of a sequestered 5′ cap in CapTAR–polyA (black) and exposed 5′ cap in CapΨCES (green); ppm, parts per million. Corresponding spectra for 7mG (orange) and CapTPUAMAL [TAR, polyA, and U5:AUG regions of the HIV-1MAL leader (23)] (blue) are also shown. The chemical shifts and NOEs observed for the cap-CH3 and -H8 protons of CapTPUAMAL were similar to those of the HIV-1MAL 5′-L (23). Residue G105 of HIV-1NL4-3 gRNA corresponds to G103 in the HIV-1MAL leader (SI Appendix, Fig. S1).
Test RNAs containing the authentic 5′-L were packaged as efficiently as Ψ+, indicating that differences in residues downstream of the 5′-L do not affect packaging (Fig. 2B) (38). Test RNAs containing the 5′-LΔPBS were also efficiently packaged relative to Ψ+ (104 ± 11%) (Fig. 2B), as expected since 5′-LΔPBS exhibits dimerization and NC-binding properties similar to those of the intact leader (27). However, test RNAs containing the ΨCES leader were poorly packaged relative to Ψ+ (Fig. 2B) even though ΨCES efficiently promotes packaging in noncompetitive packaging assays (24). ΨCES differs from 5′-LΔPBS only by the absence of the TAR and polyA hairpins (Fig. 1), revealing that this 5′-tandem hairpin element is essential for competitive packaging. Neither the TAR nor polyA hairpins exhibit significant affinity for the NC in vitro (27, 39) and removal of these two hairpins does not affect 5′-L dimerization (24), suggesting that factors other than NC binding or dimerization are responsible for TAR–polyA–dependent packaging. Recent NMR studies revealed that the TAR and polyA helices of the MAL strain of the HIV-1 (subtype A; HIV-1MAL) gRNA leader are coaxially stacked in a manner that structurally sequesters the 5′ cap and prevents binding by eukaryotic translation initiation factor 4E (eIF4E) in vitro (23). Cap sequestration has been proposed to inhibit cap-dependent degradation of the gRNA and is likely to interfere with translation and splicing (23), but has not been previously considered important for packaging. Experiments were therefore conducted to test this possibility.
State of the 5′ Cap in Native and Mutant HIV-1NL4-3 Leader RNAs.
NMR and cap accessibility titration experiments were performed to determine if native and mutant HIV-1NL4-3 gRNA leader constructs adopt a cap-sequestered structure similar to that observed for the HIV-1MAL leader (23). Titrations of HIV-1NL4-3 5′-L and 5′-LΔPBS–capped RNAs with recombinant cap-binding protein eIF4E did not give rise to detectable band shifts in native electrophoretic mobility-shift assays (Fig. 2C), indicating that these RNAs do not bind eIF4E. An RNA construct comprising only the capped TAR and polyA hairpins (CapTAR–polyA) was also unable to bind eIF4E (Fig. 2C), indicating that the CapTAR–polyA tandem hairpin functions as an independent cassette for cap sequestration. Two-dimensional (2D) nuclear Overhauser effect spectroscopy (NOESY) data obtained for CapTAR–polyA exhibited NMR chemical shifts and NOE cross-peaks similar to those observed for the HIV-1MAL 5′-L (Fig. 2D) (23), indicating that the 5′-capped HIV-1NL4-3 gRNA leader also adopts a cap-sequestered structure with end-to-end stacking of the TAR (G3) and polyA (G105) hairpins. In contrast, 1H NMR chemical shifts and NOEs observed for the cap-CH3 and -H8 protons of 5′-capped ΨCES (CapΨCES) were similar to those of 7-methylguanosine triphosphate (7mG; Fig. 2D), indicating that the 5′ cap of CapΨCES exists in an exposed conformation. eIF4E titration experiments confirmed that the dimeric CapΨCES RNA readily binds to eIF4E molecules (Fig. 2C). These findings suggest a possible link between 5′-cap exposure and the inability of CapΨCES to promote competitive packaging (Fig. 2), and also that the TAR and polyA helices of the native leader may promote packaging by sequestering the 5′ cap.
Cap Exposure in the Full-Length Leader Aabrogates RNA Packaging.
To test the hypothesis that cap exposure inhibits RNA packaging, we prepared a test vector with a stretch of AAGG residues inserted immediately downstream of the transcriptional start site (Fig. 3A). This insertion affords RNA transcripts with a leader sequence containing a nonnative A(1*)-A(2*)-G(3*)-G(4*) segment inserted immediately after the 5′-capped G3 residue (Cap5′-LAAGG) (Fig. 3A). The nonnative residues were engineered to substitute the native Cap:C58 and G3:C57 base pairs by G(3*):C58 and G(4*):C57 pairs and extend the cap away from the TAR hairpin (Fig. 3B). Under physiological-like ionic strength conditions (PI buffer: 10 mM Tris⋅HCl, 140 mM KCl, 10 mM NaCl, and 1 mM MgCl2, pH 7.5), Cap5′-LAAGG forms dimers and retains NC-binding properties of the native leader (Fig. 3 C and D). However, the 5′ cap of Cap5′-LAAGG exhibits NMR chemical shifts and NOEs consistent with a nonstacked and disordered conformation (Fig. 3E). Titration experiments confirmed that the 5′ cap of the dimeric Cap5′-LAAGG is accessible for eIF4E binding (Fig. 3F). Test RNAs containing the 5′-AAGG–modified leader were unable to detectably promote packaging when competing with Ψ+ (Fig. 3G). These findings collectively indicate that cap exposure is detrimental to packaging, even though the RNA exhibits dimerization and NC-binding properties similar to those of the native leader.
Fig. 3.
Cap sequestration is required for competitive gRNA packaging. (A) The AAGG sequence was inserted after the transcription start site. (B) The nonnative AAGG residues extend the cap away from the TAR hairpin. (C and D) The inserted AAGG residues do not affect dimerization (C) and NC binding (D) (D, dimer; M, monomer). (E) Portion of 2D NOESY spectra overlay showing cap chemical shifts and NOEs indicative of the exposed 5′ cap for CapΨCES (green) and Cap5′-LAAGG (purple). NOEs for 7mG are shown in orange. ppm, parts per million. (F) eIF4E binds the Cap5′-LAAGG dimer. (G) Packaging of test leader RNA with an AAGG insertion under competitive conditions as measured by ribonuclease protection.
Cotranscriptional Shedding of the 5′ Cap Enables Authentic Packaging by ΨCES.
To determine if cap exposure accounts for the poor competitive packaging of ΨCES test RNAs, we generated chimeric RNA constructs in which active and inactive forms of the self-cleaving hepatitis delta virus (HDV) ribozyme were appended to the 5′ end of ΨCES (δ-ΨCES and δmut-ΨCES, respectively) (Fig. 4A). The HDV element undergoes rapid self-cleavage to liberate its capped 5′ hairpin both in vitro (Fig. 4B) and within the nucleus in vivo (40), whereas the HDV ribozyme with a single cytidine-to-uridine substitution remains intact. Both RNAs with 5′ HDV sequences exhibit in vitro dimerization properties similar to those of the parent ΨCES RNA (Fig. 4C). However, whereas test RNAs containing the cap-shedding δ-ΨCES leader exhibited packaging efficiencies similar to those of Ψ+ (106 ± 11%), δmut-ΨCES RNAs that retain the 5′ cap were poorly packaged (9 ± 6% compared with the authentic 5′-L) (Fig. 4D). These findings further confirm that cap sequestration is required for competitive packaging and indicate that all other elements required for native-like RNA encapsidation reside within the ΨCES region of the leader.
Fig. 4.
Cap-dependent modulation of ΨCES packaging. (A) Construct design of δ-ΨCES and δmut-ΨCES. (B) The wild-type HDV ribozyme cotranscriptionally cleaves the capped 5′ fragment in vitro but the C-to-U mutant does not. (C) δ-ΨCES and δmut-ΨCES exhibit the same dimerization properties as the parent ΨCES RNA. M, monomer; D, dimer; W, water; PI, PI buffer. (D) Packaging of test RNAs containing δ-ΨCES and δmut-ΨCES in competition with Ψ+ as measured by ribonuclease protection.
ΨCES Is the Minimal Gag Recognition Signal.
We also asked if efficient packaging could be directed by cap-sequestered RNAs containing smaller regions of ΨCES. The highest-affinity NC binding sites (Kd ∼30 nM) known to be critical for packaging are located within the base of the Ψ-hairpin (ΨHP), where it joins the proximal three-way junction (3WJ-1; Fig. 1A) (27). Test RNAs containing the cap-sequestering TAR–polyA cassette and either the proximal three-way junction or an extended Ψ-hairpin (TP–Ψ3WJ-1 and TP–ΨHP, respectively; Fig. 5A) were not detectably packaged (Fig. 5B). In addition, test RNAs containing GAGA tetraloops substituted for the apical loop of the DIS hairpin, which prevents dimerization without altering the structure or NC-binding properties of the RNA (29), were also poorly packaged (36 ± 6% and 38 ± 8% for 5′-LDISm and 5′-LΔPBS-DISm, respectively; Fig. 5 B–E). These findings indicate that authentic packaging efficiency requires the entire dimeric ΨCES element.
Fig. 5.
Dimeric ΨCES is required for authentic packaging fidelity and efficiency. (A) TP-Ψ3WJ-1 and TP-ΨHP contain the proximal three-way junction and the extended Ψ-hairpin, respectively. TP, cap-sequestering TAR–polyA cassette. (B) Test RNAs containing TP-Ψ3WJ-1 and TP-ΨHP were not detectably packaged in competition with Ψ+. (C–E) Substitution of the native DIS loop by GAGA (C) prevents dimerization (D) and attenuates packaging of test RNAs containing 5′-LDISm and 5′-LDPBS-DISm under competitive conditions (E). (F) Working model for HIV-1 RNA packaging. Cap-dependent capture by the RNA processing or translation machinery precludes packaging of host and viral mRNAs. gRNAs are selected by a bipartite mechanism that involves structural sequestration of the 5′ cap and exposure of a cluster of high-affinity Gag binding sites. CBP, cap-binding protein; ORF, open reading frame.
Discussion
Understanding the molecular mechanism of HIV-1 genome packaging has been challenging for several reasons. Residues important for packaging sometimes have multiple functions that can complicate interpretation of mutagenesis outcomes. In addition, Gag can efficiently assemble into RNA-containing virus-like particles from cells lacking viral gRNAs, and mutant RNAs with packaging defects can therefore be packaged at substantial levels if overexpressed in transfected cells in the absence of a competing authentic genome. As a result, nearly every portion of the HIV-1 5′-L, and even stretches of the gag-coding region (31, 41, 42), have been controvertibly proposed to play roles in genome packaging (38, 43). Prior studies of the TAR element illustrate these challenges. Early studies showed that base pairing near the 5′ end of the TAR hairpin is required for optimal viral replication (44), and studies implicating TAR in translation, dimerization, packaging, and reverse transcription were complicated by the dominant-negative effects of TAR mutations on transcription (45). Berkhout and coworkers conducted a series of studies with HIV-1 variants that do not require TAR for transcription. These studies revealed that, although transcriptionally competent variants lacking the TAR hairpin can replicate (albeit less efficiently than the wild-type virus), long-term replication leads to restoration of a 5′–stem-loop structure (45). These and other studies showed that a stable 5′ hairpin, but not necessarily TAR, is required for efficient genome packaging, and that disruption of base pairing can alter the folding and dimerization properties of the 5′-L (46). Similar studies revealed that a stable hairpin at the polyA position is required for efficient replication and RNA packaging (47). These results can be explained by our findings that cap sequestration, mediated by end-to-end stacking of the TAR and polyA helices, is required for efficient genome packaging. Mutations that disrupt base pairing near the bases of the TAR and polyA helices are likely to disrupt coaxial helical stacking and consequently expose the 5′ cap.
Our finding that 5′-LAAGG and ΨCES test RNAs are not packaged, despite possessing wild-type NC binding and dimerization behaviors, indicates that cap exposure is a dominant-negative determinant of packaging. How does cap exposure prevent packaging? The inability of HIV-1 Gag proteins to package cap-exposed viral RNAs is not likely due to altered intracellular trafficking since live-cell imaging studies have shown that both translating and nontranslating pools of HIV RNAs actively transit sites of virus assembly (48, 49). Retroviral transcripts involved in translation and packaging exist in separate, noninterconverting pools (22, 48–51), and splicing of HIV-1 RNAs transcribed with three 5′ guanosines, which adopt a cap-exposed conformation, occurs at significantly greater levels than those transcribed with a single 5′ guanosine that are known to adopt a cap-sequestered conformation (52). These findings are collectively consistent with a mechanism in which HIV-1 transcripts containing an exposed 5′ cap are irreversibly sequestered by the cellular processing and translation machinery prior to becoming accessible for Gag-dependent packaging, and that gRNAs avoid this capture by structural sequestration of their 5′ cap (Fig. 5F).
The negative influence of cap exposure on packaging may also help explain why certain host noncoding RNAs synthesized by RNA polymerase III, including 7SL, U6 small nuclear RNA, and transfer RNAs, are enriched in retroviruses (53–56). These RNAs lack a 7-methylguanosine cap, and their expression and processing pathways are independent of the cap-dependent RNA processing and translation machinery (53, 56) (Fig. 5F). Cap sequestration may also be important for RNA packaging by other viruses; for example, the 5′ cap of rotavirus plus-strand RNA associates with eIF4E when functioning as mRNA but is sequestered by the copackaged viral polymerase VP1 when functioning as gRNA (57, 58). Suppression of plus-strand viral RNA translation appears to be coupled with genome packaging by hepatitis B virus and flaviviruses and is also mediated by the binding of the viral polymerase to sites proximal to the 5′ cap (59–61).
In summary, our studies reveal that cap sequestration is an essential determinant of HIV-1 genome packaging, that the TAR and polyA hairpins indirectly promote packaging by sequestering the cap, and that all additional RNA elements required for authentic packaging fidelity and efficiency reside fully within the intact ΨCES region of the dimeric 5′-L. Attenuated packaging of cap-exposed RNAs likely results from cap-dependent sequestration by cellular RNA processing and translation machinery. Since cap exposure can be induced at relatively modest energetic cost (a few kcal/mol) (23), the TAR–polyA cassette could serve as an attractive antiviral target.
Materials and Methods
Plasmid Construction and DNA Template Preparation.
The construction of plasmids for 5′-L, 5′-LΔPBS, and ΨCES has been described previously (24, 29). Plasmids for 5′-LDISm were generated by site-directed mutagenesis on the 5′-L using forward primer 5′-CGC AGG ACT CGG CTT GCT GGA GAC GGC AAG AGG CGA GGG GCG G-3′ and reverse primer 5′-CCG CCC CTC GCC TCT TGC CGT CTC CAG CAA GCC GAG TCC TGC G-3′. Plasmids for 1G5′-L and 1G5′-LΔPBS were generated by site-directed mutagenesis using forward primer 5′-CTA ATA CGA CTC ACT ATA GTC TCT CTG GTT AGA CCA G-3′ and reverse primer 5′-CTG GTC TAA CCA GAG AGA CTA TAG TGA GTC GTA TTA G-3′ on parent plasmids 5′-L and 5′-LΔPBS, respectively. The plasmid for 1GΨCES was prepared by site-directed mutagenesis using forward primer 5′-CTA ATA CGA CTC ACT ATA GTG CCC GTC TGT TGT GTG-3′ and reverse primer 5′-CAC ACA ACA GAC GGG CAC TAT AGT GAG TCG TAT TAG-3′ on ΨCES. The plasmid for GGAAG5′-L was prepared by site-directed mutagenesis using forward primer 5′-CTA ATA CGA CTC ACT ATA GGA AGG TCT CTC TGG TTA GAC C-3′ and reverse primer 5′-GGT CTA ACC AGA GAG ACC TTC CTA TAG TGA GTC GTA TTA G-3′ on plasmid 5′-L. The plasmid for GAAG5′-L was generated by site-directed mutagenesis on the 5′-L using forward primer 5′-CTA ATA CGA CTC ACT ATA GAA GGT CTC TCT GGT TAG ACC-3′ and reverse primer 5′-GGT CTA ACC AGA GAG ACC TTC TAT AGT GAG TCG TAT TAG-3′. DNA fragments of δ-ΨCES and δmut-ΨCES were synthesized and inserted into pUC57 plasmids by Genewiz. The T7 promoter sequence was included at the 5′ end of the RNA-encoding sequences in all the plasmids. All the site-directed mutagenesis was performed with the QuikChange Lightning Mutagenesis Kit (Agilent Technologies).
The DNA templates for RNA in vitro transcription were obtained by standard PCR amplification using the above-mentioned plasmids (EconoTaq PLUS 2X Master Mix; Lucigen). A common forward amplification primer targeting a vector region ∼80 nt upstream of the T7 promoter sequence was used for all PCR reactions (5′-GGG ATG TGC TGC AAG GCG ATT AAG TTG GG-3′). The reverse amplification primers are 5′-mCmGC ACC CAT CTC TCT CCT TCT AGC CTC C-3′ for 5′-L, 5′-LΔPBS, 1G5′-L, 1G5′-LΔPBS, GGAAG5′-L, GAAG5′-L, δ-ΨCES, and δmut-ΨCES; 5′-mGmGC ACC CAT CTC TCT CCT TCT AGC CTC C-3′ for ΨCES and 1GΨCES; and 5′-mCmAC TAC TTT GAG CAC TCA AGG CAA GC-3′ for TAR–polyA; “m” denotes 2′-O-methyl modification to reduce nontemplated nucleotide addition by T7 RNA polymerase (62). All the DNA oligos were synthesized by Integrated DNA Technologies.
In Vitro Transcription.
RNAs were prepared by in vitro transcription as described previously (27). The reactions (15 to 30 mL) each contained ∼0.5 mg of PCR-amplified DNA template, 20 mM MgCl2, 3 to 6 mM nucleoside triphosphates (NTPs), 2 mM spermidine, 2 mM dithiothreitol (DTT), 20% (volume [vol]/vol) dimethyl sulfoxide, 80 mM Tris⋅HCl (pH 9.0), and 0.3 mg T7 RNA polymerase (purified in-house). The reaction was quenched after a 5-h incubation at 37 °C by addition of an ethylenediaminetetraacetate (EDTA)/urea mixture (250 mM EDTA and 7 M urea, pH 8.0), followed by boiling for 5 min and snap cooling on ice for 5 min; 50% glycerol was added to the sample to a final concentration of 6% (vol/vol). RNAs were purified by electrophoresis on urea-containing polyacrylamide denaturing gels (SequaGel; National Diagnostics) at 20 W overnight, visualized by ultraviolet shadowing, and eluted using the Elutrap Electroelution System (Whatman) at 150 V overnight. The eluted RNAs were concentrated and washed twice with 2 M high-purity NaCl followed by washing eight times with ultrapure water using an Amicon Ultra Centrifugal Filter Device (Millipore).
NTPs for In Vitro Transcription.
Fully protonated NTPs were purchased from Sigma. Perdeuterated NTPs were purchased from Cambridge Isotope Laboratories. A2r was obtained by selective deuteration of the C8 position of fully protonated adenosine triphosphate (ATP). Briefly, 140 µL triethylamine (TEA) was added to 0.2 g ATP dissolved in 8 mL D2O (99.8%; Cambridge Isotope Laboratories); 1-mL aliquots were incubated at 60 °C for 5 d. TEA was subsequently removed by lyophilization. Gr was obtained by analogous treatments for guanosine triphosphate (GTP) with a shorter incubation time at 60 °C (24 h).
Purification of Vaccinia Virus Capping Enzyme.
The expression vector containing the coding sequences of the His-tagged D1 and D12 subunits of vaccinia virus capping enzyme was a kind gift from the Stephen Cusack laboratory, European Molecular Biology Laboratory, Grenoble, France (63). The coexpressed D1 and D12 proteins from the plasmid were purified from BL21 (DE3) pLysS cells (Life Technologies). Briefly, the cells were cultured in Terrific broth at 37 °C with 250 rpm shaking until the OD reached ∼1.0, followed by protein induction with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 20 °C overnight. The cells were harvested by centrifugation at 7,000 rpm at 4 °C for 20 min and resuspended in lysis buffer (20 mM Tris base, 200 mM NaCl, 20 mM imidazole, 10% glycerol, and 5 mM Tris[2-carboxyethyl]phosphine [TCEP], pH 8.0), and then lysed by freezing and thawing, followed by microfluidization. After centrifugation at 12,000 rpm at 4 °C for 30 min, the supernatant containing the His-tagged proteins was applied to an Ni-NTA affinity column (Qiagen) and eluted by elution buffer (20 mM Tris base, 200 mM NaCl, 250 mM imidazole, 10% glycerol, and 5 mM TCEP, pH 8.0). Proteins were dialyzed overnight at 4 °C in storage buffer (20 mM Tris⋅HCl, 100 mM NaCl, 10% glycerol, and 1 mM DTT, pH 8.0). The activity of the capping enzyme was assessed in a trial capping reaction using a 20-nt RNA. Capped RNA can be resolved from uncapped RNA on a 20% denaturing acrylamide gel (SequaGel; National Diagnostics) run at 220 V for 3 h. Only one species of capped RNA was generated by vaccinia virus capping enzyme with the cap moiety appended to the 5′ end of the in vitro transcribed RNA. The appropriate amount of capping enzyme was chosen to achieve near-100% capping efficiency.
Preparation of Capped RNA.
The capping reaction contained 16 mL of 20 µM in vitro T7-transcribed and purified RNA in a buffer with 50 mM Tris base, 5 mM KCl, 1 mM MgCl2, and 1 mM DTT (pH 8.0). RNA was boiled and snap-cooled in water followed by the addition of 10× capping buffer and 0.5 mM GTP, 0.1 mM S-adenosyl methionine, and various amounts of vaccinia virus capping enzyme (determined in a 20-µL trial reaction with a 20-nt RNA). The reaction was incubated at 37 °C for 2 h and then quenched with the addition of the EDTA/urea mixture (250 mM EDTA and 7 M urea, pH 8.0) followed by boiling for 3 min and snap cooling on ice. The capped RNA was purified the same way as described above for in vitro transcribed RNAs.
Purification of Human eIF4E.
The pUC57 plasmid containing the codon-optimized DNA sequence for human eIF4E (UniProt ID code P06730) with an N-terminal His6 and GB1 tag was synthesized by Genewiz. The protein sequence is MGHHH HHHSS GGMQY KLILN GKTLK GETTT EAVDA ATAEK VFKQY ANDNG VDGEW TYDDA TKTFT VTEIP TTENL YFQGA MATVE PETTP TPNPP TTEEE KTESN QEVAN PEHYI KHPLQ NRWAL WFFKN DKSKT WQANL RLISK FDTVE DFWAL YNHIQ LSSNL MPGCD YSLFK DGIEP MWEDE KNKRG GRWLI TLNKQ QRRSD LDRFW LETLL CLIGE SFDDY SDDVC GAVVN VRAKG DKIAI WTTEC ENREA VTHIG RVYKE RLGLP PKIVI GYQSH ADTAT KSGST TKNRF VV. The coding sequence was PCR-amplified and inserted into the pLATE11 expression vector (aLICator LIC Cloning & Expression System; Thermo Fisher Scientific). BL21 (DE3) cells harboring the plasmid were cultured in Luria–Bertani broth (LB) medium at 35 °C with 250 rpm shaking until the OD reached ∼0.6, and protein was overexpressed by induction with 1 mM IPTG at 28 °C overnight. The pelleted cells were lysed by freeze–thawing and microfluidization in lysis buffer containing 50 mM Tris base, 500 mM NaCl, 20 mM imidazole, and 2 mM TCEP (pH 8.0). The supernatant after centrifugation was applied to an Ni-NTA affinity column (Qiagen) and eluted by elution buffer (50 mM Tris base, 500 mM NaCl, 250 mM imidazole, and 2 mM TCEP, pH 8.0). The eluted protein was further purified by size-exclusion chromatography on a Superdex 75 column (Amersham) in 20 mM Tris base, 100 mM KCl, and 2 mM DTT (pH 7.5).
Native Gel Electrophoresis.
RNA samples (0.4 to 2 µM) were prepared in PI buffer (10 mM Tris⋅HCl, pH 7.5, 140 mM KCl, 10 mM NaCl, and 1 mM MgCl2), heat-denatured for 3 min, and slowly cooled down to room temperature. For cap-binding gel shift with eIF4E, the protein was diluted into PI buffer and incubated at 37 °C for 1 h. The complex samples were prepared by mixing appropriate amounts of protein and RNA stocks, which were further incubated at 37 °C for 1 h; 10× native agarose gel loading solution containing 0.17% bromophenol blue and 40% (vol/vol) sucrose was added to each sample and mixed. The final samples containing ∼500 ng RNA were loaded onto 1% agarose gels prestained with ethidium bromide and run at 115 V in 1× TB buffer (44.5 mM Tris⋅boric acid, pH 7.5) on ice to prevent thermal denaturation during electrophoresis.
Purification of HIV-1NL4-3 NC.
The NC protein was overexpressed and purified from BL21 (DE3) pLysE cells with a pET-3a plasmid containing the coding sequence of the HIV-1NL4-3 NC (amino acid sequence: MQKGN FRNQR KTVKC FNCGK EGHIA KNCRA PRKKG CWKCG KEGHQ MKDCT ERQAN). The detailed purification steps have been described previously (27).
Isothermal Titration Calorimetry.
Isothermal titration calorimetry (ITC) experiments were carried out using an ITC200 (MicroCal). Both the protein and RNA samples were dialyzed into ITC buffer (10 mM Tris⋅HCl, pH 7.0, 140 mM KCl, 10 mM NaCl, 5 mM MgCl2, and 5 mM TCEP) overnight before use. The injection syringe contained 40 μL ∼240 µM NC, and the calorimetry cell was loaded with 200 μL ∼1.0 µM RNA. After thermal equilibration at 30 °C and the initial 60-s delay, a single injection of 0.2 μL followed by 19 serial injections of 2 μL was made into the calorimetry cell.
NMR Spectroscopy.
NMR data were collected with Bruker AVANCE 600- or 800-MHz spectrometers equipped with cryoprobes. Samples for NMR studies of the capped RNAs were prepared in 90% H2O/10% D2O with 10 mM Tris⋅d11 (pH 7.5), 50 mM KCl, and 1 mM MgCl2 in standard 5-mm NMR tubes (500 μL ∼100 μM). The NMR sample for 7mG (Sigma-Aldrich) was 5 mM. Two-dimensional NOESY spectra were collected at 35 °C with a 300-ms NOE mixing time and 3.2-s relaxation delay using a jump–return pulse sequence for water suppression. NMR data were processed with NMRFx (64) and analyzed with NMRViewJ (65).
Ψ+-Helper and Test Vector Plasmid Construction.
The Ψ+-helper NL4-3 GPP was described previously and consists of an HIV-1NL4-3 genome with vpu, vpr, and partial env deletions and with nef replaced by a puromycin-resistance expression cassette (20). The native test vector plasmid (aka L688-RRE-Puro-LTR) contains 688 base pairs of the parental HIV-1NL4-3 leader, the RRE, and a puromycin-resistance cassette as described previously (24). All changes in test vector 5′-L sequences were introduced by replacement of the corresponding region in the native test vector with a fragment generated using overlap extension PCR.
Virus Production.
Human 293T cells were grown at 37 °C under 5% CO2 in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and 50 µg/mL gentamicin. HIV-1 particles were produced by 293T cells after cotransfection with plasmid DNA mixtures that included helper (NL4-3 GPP) and one test vector plasmid at an ∼1:1 molar ratio using polyethylenimine, as described previously (27). Forty-eight hours later, transfection media were collected and filtered through 0.22-μm filters. Cells were directly lysed with TRIzol reagent (Ambion) and viral particles were first concentrated by ultracentrifugation (25,000 rpm) through a 20% sucrose cushion and then lysed with TRIzol. RNA was extracted from TRIzol lysates according to the manufacturer’s protocol, treated with DNase, and stored at −80 °C.
RNA Content Analysis.
The RNA content of transfected cells and viral particles was analyzed by RPA as previously described (37). RNA samples were hybridized with riboprobe in 1× hybridization buffer (80% formamide, 40 mM piperazine-N,N′-bis[2-ethanesulfonic acid], pH 6.4, 400 mM NaCl, and 1 mM EDTA) by denaturing at 95 °C for 5 min followed by a 16-h incubation at 65 °C. Samples were then digested in a mixture containing 300 mM NaCl, 10 mM Tris (pH 7.5), 10 mM EDTA, 10 µg/µL salmon sperm DNA, 40 µg/mL RNase A (Roche), and 10 U RNase T1 (Ambion) at 37 °C for 30 min. RNase-digested samples were further processed by the addition of sodium dodecyl sulfate (SDS) to a final concentration of 1.3% and 0.04 mg/µL proteinase K (Invitrogen) before incubation at 37 °C for 15 min, followed by phenol/chloroform extraction and ethanol precipitation. Pellets were resuspended in 2× loading dye (80% formamide, 10 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol, and 0.1% SDS) and denatured at 95 °C for 5 min before loading onto a prewarmed 8% polyacrylamide/8 M urea gel and running at 500 V for 1.5 h. The gel was dried and exposed to a phosphorimager screen overnight. Imaging was performed using a Typhoon FLA 9500 (GE Healthcare) instrument, and radiolabeled bands were quantified using ImageQuant software (GE Healthcare). Packaging efficiency was quantified by determining the molar ratio of test vector to Ψ+-helper RNAs present in viral particles as compared with their ratio in cells, normalized to packaging efficiency for a test vector with a native NL4-3 strain 5′-L. Packaging efficiencies are reported as the mean ± SD from three individual packaging experiments. The chimeric riboprobe used in this study, HIVgag/CMV (cytomegalovirus), was described previously and protects a 201-nt fragment unique to the NL4-3 GPP helper and 289 nt of sequence unique to test vectors (27). The riboprobe was transcribed from linearized plasmid templates using T7 RNA polymerase (Promega) and [α-32P]rCTP (PerkinElmer).
Supplementary Material
Acknowledgments
We thank the Howard Hughes Medical Institute (HHMI) staff at the University of Maryland, Baltimore County (UMBC) and C. Burnett at the University of Michigan Medical School for technical assistance, and J. Marchant at UMBC for helpful suggestions. This research was supported by research grants from the NIH (National Institute of Allergy and Infectious Diseases [NIAID] 8R01 AI50498 to M.F.S. and A.T. and NIAID U54 AI150470 to A.T.). E.C. and H.F. were supported by a National Institute of General Medical Sciences grant for enhancing minority access to research careers (MARC U*STAR 2T34 GM008663); R.C. was supported by an HHMI undergraduate education grant and the Meyerhoff Scholars Program at UMBC.
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112475118/-/DCSupplemental.
Data Availability
All data generated and analyzed during this study are included in this published article and its SI Appendix.
References
- 1.Coffin J. M., Hughes S. H., Varmus H. E., Retroviruses (Cold Spring Harbor Laboratory Press, Plainview, NY, 1997). [PubMed] [Google Scholar]
- 2.Berkowitz R., Fisher J., Goff S. P., RNA packaging. Curr. Top. Microbiol. Immunol. 214, 177–218 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Chen J., et al., High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U.S.A. 106, 13535–13540 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell R. S., Liang C., Wainberg M. A., Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology 1, 23 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paillart J.-C., Shehu-Xhilaga M., Marquet R., Mak J., Dimerization of retroviral RNA genomes: An inseparable pair. Nat. Rev. Microbiol. 2, 461–472 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Kuzembayeva M., Dilley K., Sardo L., Hu W.-S., Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 454–455, 362–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Souza V., Summers M. F., How retroviruses select their genomes. Nat. Rev. Microbiol. 3, 643–655 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Lu K., Heng X., Summers M. F., Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 410, 609–633 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson E. D., Musier-Forsyth K., Retroviral Gag protein-RNA interactions: Implications for specific genomic RNA packaging and virion assembly. Semin. Cell Dev. Biol. 86, 129–139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaddis Maldonado R. J., Parent L. J., Orchestrating the selection and packaging of genomic RNA by retroviruses: An ensemble of viral and host factors. Viruses 8, 257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comas-Garcia M., et al., Dissection of specific binding of HIV-1 Gag to the ‘packaging signal’ in viral RNA. eLife 6, e27055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rein A., RNA packaging in HIV. Trends Microbiol. 27, 715–723 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jouvenet N., Simon S. M., Bieniasz P. D., Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. U.S.A. 106, 19114–19119 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jouvenet N., Simon S. M., Bieniasz P. D., Visualizing HIV-1 assembly. J. Mol. Biol. 410, 501–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganser-Pornillos B. K., Yeager M., Sundquist W. I., The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18, 203–217 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miele G., Mouland A., Harrison G. P., Cohen E., Lever A. M., The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 70, 944–951 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbink T. E. M., Berkhout B., A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 278, 11601–11611 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Greatorex J., The retroviral RNA dimer linkage: Different structures may reflect different roles. Retrovirology 1, 22 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ooms M., Huthoff H., Russell R., Liang C., Berkhout B., A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 78, 10814–10819 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu K., et al., NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 334, 242–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda T., et al., Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci. Rep. 5, 17680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kharytonchyk S., et al., Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc. Natl. Acad. Sci. U.S.A. 113, 13378–13383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown J. D., et al., Structural basis for transcriptional start site control of HIV-1 RNA fate. Science 368, 413–417 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heng X., et al., Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 417, 224–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lever A., Göttlinger H., Haseltine W., Sodroski J., Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 63, 4085–4087 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeffman A., Hassard S., Varani G., Lever A., The major HIV-1 packaging signal is an extended bulged stem loop whose structure is altered on interaction with the Gag polyprotein. J. Mol. Biol. 297, 877–893 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Ding P., et al., Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc. Natl. Acad. Sci. U.S.A. 117, 17737–17746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damgaard C. K., Andersen E. S., Knudsen B., Gorodkin J., Kjems J., RNA interactions in the 5′ region of the HIV-1 genome. J. Mol. Biol. 336, 369–379 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Keane S. C., et al., RNA structure. Structure of the HIV-1 RNA packaging signal. Science 348, 917–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Didierlaurent L., et al., Role of HIV-1 RNA and protein determinants for the selective packaging of spliced and unspliced viral RNA and host U6 and 7SL RNA in virus particles. Nucleic Acids Res. 39, 8915–8927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luban J., Goff S. P., Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 68, 3784–3793 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson J. H., Child L. A., Lever A. M. L., Packaging of human immunodeficiency virus type 1 RNA requires cis-acting sequences outside the 5′ leader region. J. Virol. 67, 3997–4005 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S.-W., Aldovini A., RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 76, 11853–11865 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muriaux D., Mirro J., Harvin D., Rein A., RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U.S.A. 98, 5246–5251 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muriaux D., Mirro J., Nagashima K., Harvin D., Rein A., Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 76, 11405–11413 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rulli S. J. Jr, et al., Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 81, 6623–6631 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyazaki Y., et al., An RNA structural switch regulates diploid genome packaging by Moloney murine leukemia virus. J. Mol. Biol. 396, 141–152 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kharytonchyk S., et al., Influence of gag and RRE sequences on HIV-1 RNA packaging signal structure and function. J. Mol. Biol. 430, 2066–2079 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clever J. L., Miranda D. Jr, Parslow T. G., RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 76, 12381–12387 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fong N., Ohman M., Bentley D. L., Fast ribozyme cleavage releases transcripts from RNA polymerase II and aborts co-transcriptional pre-mRNA processing. Nat. Struct. Mol. Biol. 16, 916–922 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchschacher G. L. Jr, Panganiban A. T., Human immunodeficiency virus vectors for inducible expression of foreign genes. J. Virol. 66, 2731–2739 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., et al., HIV-1 sequence necessary and sufficient to package non-viral RNAs into HIV-1 particles. J. Mol. Biol. 429, 2542–2555 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mailler E., et al., The life-cycle of the HIV-1 Gag-RNA complex. Viruses 8, 248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klaver B., Berkhout B., Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 13, 2650–2659 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das A. T., Harwig A., Vrolijk M. M., Berkhout B., The TAR hairpin of human immunodeficiency virus type 1 can be deleted when not required for Tat-mediated activation of transcription. J. Virol. 81, 7742–7748 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das A. T., Vrolijk M. M., Harwig A., Berkhout B., Opening of the TAR hairpin in the HIV-1 genome causes aberrant RNA dimerization and packaging. Retrovirology 9, 59 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das A. T., Klaver B., Klasens B. I., van Wamel J. L., Berkhout B., A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 71, 2346–2356 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J., et al., Visualizing the translation and packaging of HIV-1 full-length RNA. Proc. Natl. Acad. Sci. U.S.A. 117, 6145–6155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J., et al., Impact of nuclear export pathway on cytoplasmic HIV-1 RNA transport mechanism and distribution. mBio 11, e01578-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levin J. G., Rosenak M. J., Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: Evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 73, 1154–1158 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boeras I., et al., The basal translation rate of authentic HIV-1 RNA is regulated by 5′UTR nt-pairings at junction of R and U5. Sci. Rep. 7, 6902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esquiaqui J. M., Kharytonchyk S., Drucker D., Telesnitsky A., HIV-1 spliced RNAs display transcription start site bias. RNA 26, 708–714 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Onafuwa-Nuga A. A., King S. R., Telesnitsky A., Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J. Virol. 79, 13528–13537 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eckwahl M. J., et al., Analysis of the human immunodeficiency virus-1 RNA packageome. RNA 22, 1228–1238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Telesnitsky A., Wolin S. L., The host RNAs in retroviral particles. Viruses 8, 235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eckwahl M. J., Telesnitsky A., Wolin S. L., Host RNA packaging by retroviruses: A newly synthesized story. mBio 7, e02025-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDonald S. M., Patton J. T., Assortment and packaging of the segmented rotavirus genome. Trends Microbiol. 19, 136–144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu X., et al., Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure 16, 1678–1688 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryu D. K., Ahn B. Y., Ryu W. S., Proximity between the cap and 5′ epsilon stem-loop structure is critical for the suppression of pgRNA translation by the hepatitis B viral polymerase. Virology 406, 56–64 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Khromykh A. A., Varnavski A. N., Sedlak P. L., Westaway E. G., Coupling between replication and packaging of flavivirus RNA: Evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J. Virol. 75, 4633–4640 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fajardo T., et al., The flavivirus polymerase NS5 regulates translation of viral genomic RNA. Nucleic Acids Res. 48, 5081–5093 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kao C., Zheng M., Rüdisser S., A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5, 1268–1272 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De la Peña M., Kyrieleis O. J., Cusack S., Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 26, 4913–4925 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Norris M., Fetler B., Marchant J., Johnson B. A., NMRFx Processor: A cross-platform NMR data processing program. J. Biomol. NMR 65, 205–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson B. A., Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed during this study are included in this published article and its SI Appendix.