Significance
Plant architecture is highly plastic and known to respond sensitively to nutritional changes. Although of great agronomic importance, the underlying molecular mechanisms that sense and transduce these cues into plant development and growth are poorly understood. Applying diverse genetic, biochemical, and microscopic approaches, we disclosed that signaling via the central, evolutionarily conserved fuel-sensor kinase Snf1-RELATED KINASE1 (SnRK1) initiates lateral root (LR) primordia formation in response to transient metabolic perturbations. This is accomplished by SnRK1-mediated activation of a signaling cascade involving the pivotal LR regulator AUXIN RESPONSE FACTOR19 (ARF19). We propose that this developmental priming strategy represents a cost-efficient approach to ensure rapid growth recovery after stress release, providing in competitive ecosystems a clear advantage in terms of Darwinian fitness.
Keywords: lateral root, metabolic homeostasis, SnRK1, ARF19, bZIP63
Abstract
Plants adjust their energy metabolism to continuous environmental fluctuations, resulting in a tremendous plasticity in their architecture. The regulatory circuits involved, however, remain largely unresolved. In Arabidopsis, moderate perturbations in photosynthetic activity, administered by short-term low light exposure or unexpected darkness, lead to increased lateral root (LR) initiation. Consistent with expression of low-energy markers, these treatments alter energy homeostasis and reduce sugar availability in roots. Here, we demonstrate that the LR response requires the metabolic stress sensor kinase Snf1-RELATED-KINASE1 (SnRK1), which phosphorylates the transcription factor BASIC LEUCINE ZIPPER63 (bZIP63) that directly binds and activates the promoter of AUXIN RESPONSE FACTOR19 (ARF19), a key regulator of LR initiation. Consistently, starvation-induced ARF19 transcription is impaired in bzip63 mutants. This study highlights a positive developmental function of SnRK1. During energy limitation, LRs are initiated and primed for outgrowth upon recovery. Hence, this study provides mechanistic insights into how energy shapes the agronomically important root system.
Plants display a tremendous plasticity in their overall growth and architecture. Environmental factors, such as ambient light and temperature, abiotic stress factors, or biotic interactions, as well as endogenous cues provided by the circadian clock or metabolite levels reflecting energy availability, need to be integrated into plant growth and developmental programs (1). This is in part mediated by a eukaryotic system of two counteracting kinases that are evolutionarily conserved in plants (2–6). TARGET OF RAPAMYCIN (TOR) kinase signaling supports anabolic, energy-demanding processes frequently linked to cell cycle and growth. On the other hand, SUCROSE NONFERMENTING1 (Snf1) kinase in yeast, Snf1-RELATED PROTEIN KINASE1 (SnRK1) in plants, or AMP-ACTIVATED PROTEIN KINASE (AMPK) in mammals typically stimulate a catabolic or energy-preserving metabolism. The active AMPK/Snf1/SnRK1 kinase complexes consist of three subunits comprising a catalytic α-subunit together with regulatory β- and -subunits (2, 6). Plant SnRK1 subunits are encoded by small gene families, which in part differ in number and composition from their animal counterparts (2). In Arabidopsis, two partially redundant catalytic α-subunits (SnRK1α1 and SnRK1α2, also known as KIN10 or KIN11) are active (7). Whereas mammalian AMPK is regulated by competitive binding of adenosine nucleotides (AMP, ADP [adenosine mono- and diphosphate], and ATP), with increasing AMP and ADP levels reflecting low energy charge, this does not appear to be the case for SnRK1 (8). Accumulating evidence rather suggests a model in which the low abundant metabolite trehalose 6-phosphate (T6P), which mirrors sucrose availability in plants, acts as an inhibitor of SnRK1 activity (9, 10). Moreover, the catalytic SnRK1α1 subunit has been shown to be tethered in the cytosol by the β-subunits. Upon energy starvation, SnRK1α1 is translocated to the nucleus to interact with the chromatin and activate transcription (11, 12).
SnRK1 controls enzymatic activities as well as the transcription of a multitude of genes (7, 13). With respect to the latter, SnRK1-dependent phosphorylation of the basic leucine zipper (14) transcription factor (TF) bZIP63 leads to induction of genes involved in metabolic adaptation during the starvation response (11, 15). bZIP63 participates in a network of nine group C and group S1 bZIP TFs known to form heterodimers and to mediate low-energy responses downstream of SnRK1 (16). SnRK1 has been linked to the regulation of diverse developmental processes such as hypocotyl elongation (17) or flowering (18, 19). How SnRK1 exactly tunes these processes is, however, still poorly understood.
In response to environmental conditions, the root system displays a pronounced plasticity, which is crucial for resource foraging and water uptake as well as anchoring in soil. In angiosperms, the primary root is established during embryogenesis, whereas branching occurs postembryogenically through the formation of lateral roots (LRs) (20, 21). In Arabidopsis, a subset of pericycle cells at the xylem poles are initiated to develop into LR primordia. These xylem pole pericycle cells (XPP) are specified from pericycle initials in the root-apical meristem (RAM). Via anticlinal cell divisions and elongations, XPPs leave the RAM and are activated by various signals including the plant hormone auxin. An oscillating pattern of auxin maxima along the root axes in the prebranch zone (22) controls LR spacing and density (23). In consequence, two adjacent XPPs undergo radial swelling, repolarize, and show migration of the nuclei toward the common anticlinal cell wall (20). These are the earliest microscopically visible events in LR initiation. As a common molecular marker, temporary and localized expression of GATA23 has been established (24). After initiation in one cell file, a group of approximately 8 to 11 founder cells can be detected, which further proliferate to form an LR primordium, establishing a functional meristem. After further proliferation, these cells burst through the concentric root cell layers to produce a novel LR. Auxin signaling is decisive for LR initiation, as demonstrated by the impact of several essential AUXIN RESPOSE FACTOR (ARF) TFs such as ARF7 and ARF19, which have partly redundant functions. Accordingly, the arf7/arf19 double mutant is devoid of LRs when grown on agar plates (25). These ARFs are under the control of auxin-degradable repressors, including INDOLE-3-ACETIC ACID PROTEINS14/28 (IAA14 and IAA28), and exert their function in LR formation via LATERAL ORGAN BOUNDARIES16/29 (LBD16/29) (20).
Under natural conditions, plants are confronted with constantly changing environmental conditions and hence need to steadily balance energy supply and growth. Therefore, a dynamic, energy-sensing system is required to repress growth under low-energy conditions but allow rapid growth recovery upon stress release. Here, we focus on Arabidopsis LR development as an easy-to-quantify output to study how minor perturbations in energy homeostasis are transmitted into developmental plasticity. Using microscopic, genetic, and molecular tools, we disclose that short-term energy deprivation provokes SnRK1-mediated phosphorylation of bZIP63 and its direct binding to the ARF19 promoter. The resulting increase in starvation-triggered expression of the central LR regulator ARF19 is vital for the enhanced LR initiation. As these primed LR initials grow out only after recovery of photosynthesis, we propose a regulatory system that primes development during starvation, which is then executed upon restored photosynthetic energy supply.
Results
Low-Light or Short-Term Unexpected Darkness Increases LR Density without Changing Primary Root Length.
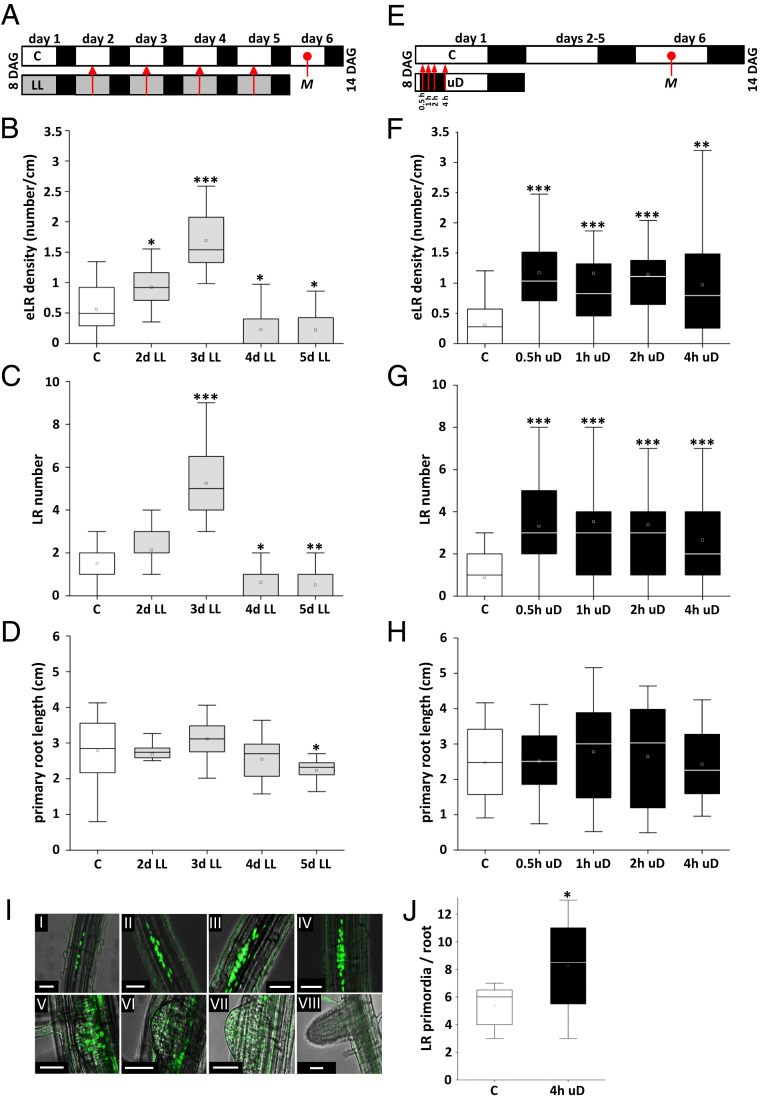
To assess the impact of energy homeostasis on root architecture, we tested several experimental growth conditions that should lead to moderate but controlled perturbations of the photosynthetic energy metabolism and therefore mimic naturally occurring fluctuations in resource availability. Following the setup depicted in Fig. 1A, seedlings were cultivated on Murashige and Skoog (MS) (26) medium without sugars under control light conditions (70 μmol ⋅ m−2 ⋅ s−1, 16-h/8-h–long day regime). At 8 d after germination (DAG), these plants were shifted to low-light conditions close to the light compensation point (15 μmol ⋅ m−2 ⋅ s−1). Under these conditions, cryptochrome and phytochrome signaling is still active (27). After 1 to 5 d of low light, plants were transferred back to control light conditions and root architecture was analyzed 14 DAG (Fig. 1 B–D). In comparison to plants grown under control conditions, an increase in emerged LR density (eLRD) (defined as the number of LRs per primary root length) was observed in conditions with up to 3 d of low-light treatment. However, plants grown in low light for longer times displayed a reduced eLRD compared to control conditions. Importantly, this response was independent of primary root length, which remained constant up to 4 d but showed slightly reduced growth with prolonged low-light treatments (Fig. 1D).
Fig. 1.
Low light or short-term uD increased LR density without changing primary root length. (A) Schematic view describing the experimental setup for low-light (LL) treatment. Arabidopsis (Col-0) seedlings were grown in a long day regime (16 h light/8 h dark) on solidified 1/2 MS media at 70 μmol ⋅ m−2 ⋅ s−1 (control, C, white). After 8 d, plants were cultivated under control or LL (gray, 15 μmol ⋅ m−2 ⋅ s−1) conditions for 1 up to 4 d (as indicated by red arrows). Root parameters were assayed at 14 DAG (M, time of measurement). Given are eLRD (B), LR number (C), and primary root length (D). (E) Schematic view describing the experimental setup for uD treatment. During the 16-h light period, 0.5 up to 4 h of darkness were given (as indicated by red arrows), starting 2 h after onset of the light phase. Cultivation was continued under control conditions, and root parameters were assayed at 14 DAG (M, time of measurement) and given as eLRD (F), LR number (G), and primary root length (H). Data from three independent experiments are presented in the respective boxplots. Statistically significant differences between control and treated samples were determined by Mann–Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001; n = 15 to 30. (I) uD resulted in an increase in the number of cells showing expression of the early-stage LR marker pGATA23::NLS-GFP (24). Confocal microscopy exhibits nuclear GFP signals throughout all stages of LR development. (Scale bar: 50 µm.) (J) Analysis of 5-d-old seedlings under control and 4-h uD conditions. GFP signals from pGATA23::NLS-GFP lines were counted 16 h after treatment and presented as boxplots. Student’s t test. *P < 0.05; n = 8.
We continued testing further perturbation schemes to assess whether the phenotype was more generally observed upon reduced energy (light) supply. Extended night, brought about by prolonging the night for 6 h, was found to increase eLRD, which was, however, due to decreased primary root length (28) and not caused by an increase in eLR number (SI Appendix, Fig. S1 A–D). In contrast, short-term unexpected darkness (uD) during the day period, starting 2 h after onset of light (Fig. 1 E–H), resulted in a significant increase in eLRD, already after 0.5 h of treatment, while primary root length was not affected, even after 4 h of uD. The phenotype was highly reproducible (SI Appendix, Fig. S1 E–H) and did not lead to altered shoot fresh weight. Moreover, quantification of uD-induced eLRD was very robust, as it was observed with three Arabidopsis ecotypes, Columbia-0 (Col-0), Wassilewskija (WS), and Landsberg erecta (Ler) (SI Appendix, Fig. S1I) and found to be independent of root light perception (SI Appendix, Fig. S1J). Finally, this phenotype is not generally stress related, as exemplified by cultivation at high temperatures (28 to 42 °C) (SI Appendix, Fig. S1 K and L). Taken together, LR plasticity rapidly and transiently responds to moderate perturbations in photosynthetic activity and thus serves as a quantitative readout to study low-energy responses on plant development.
To substantiate the phenotypical analysis, we followed LR development using molecular markers. Transcription of GATA23 is specifically and transiently induced in XPPs (24), and so far, monitoring GATA23:GFP expression provides the best approximation of a founder cell specification marker (20). As LR specification is proposed to start in 3- to 5-d-old seedlings (29), we treated 5-d-old seedlings with 4 h of uD and counted GATA23:NLS-GFP expression sites after 16 h (Fig. 1 I and J and SI Appendix, Fig. S2). In line with the phenotypic analysis, a significant increase of the number of GFP sites was observed, supporting the notion that uD treatment increases LR initiation events.
Short-Term uD Leads to Lower Sugar and Trehalose 6-Phosphate Levels and Expression of Low-Energy Stress Markers.
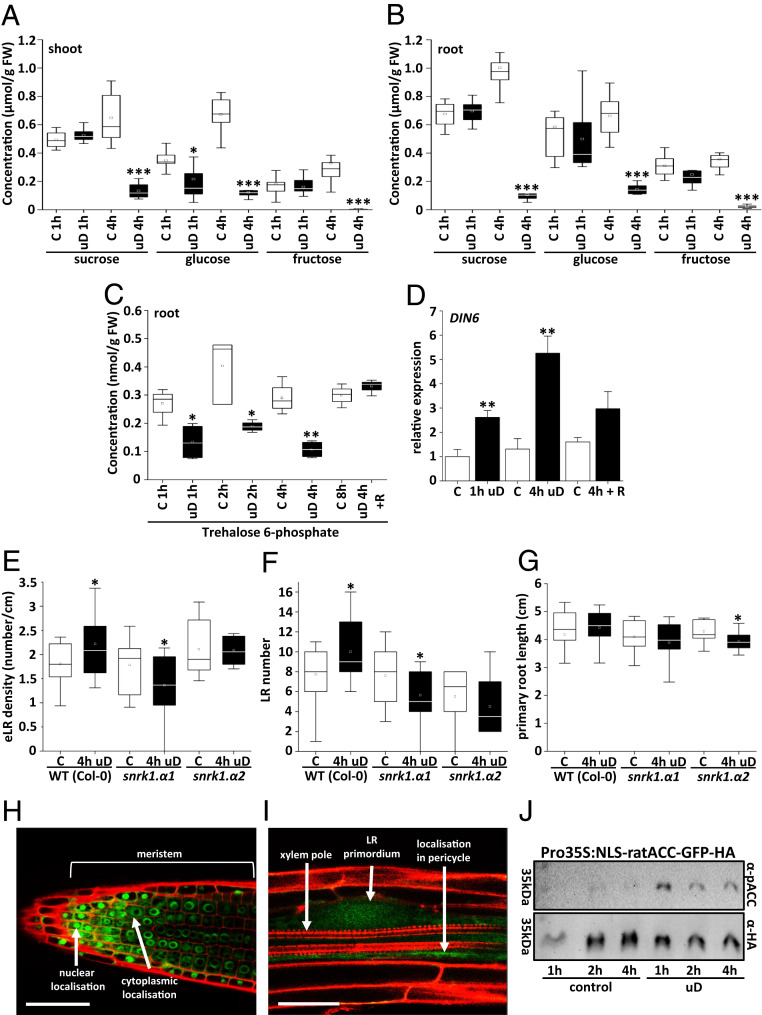
Perturbation of photosynthesis should affect metabolic homeostasis, primarily in photosynthetic tissues. We reasoned that these changes should be reflected in soluble sugar content. Hence, we analyzed sucrose, glucose, and fructose levels directly after the uD treatment or, as control, at the respective daytime in untreated plants and separately in leaves (Fig. 2A) and roots (Fig. 2B). In young, 8-d-old plants, a significant decrease of glucose content was observed in photosynthetic tissues already after a short-term perturbation of 1 h of uD. However, 4 h of uD resulted in a dramatic drop for all sugars under investigation. In roots, the concentration of the important transport sugar Suc decreased to only 10% of that in control conditions. This correlative evidence indicated that access to energy resources is a potential cue affecting LR architecture. The low-abundance sugar phosphate T6P has been proposed to function as a major signal in plant resource management and development (9, 10). Accordingly, T6P levels rapidly decreased by about 50% in roots after 1 h of uD and remained at this low level for up to 4 h of treatment. However, a fast recovery to initial levels could be observed after 4 h of light recovery (Fig. 2C). Moreover, the shift in carbon metabolism correlated with the activation of the well-established energy stress marker gene DARK-INDUCED6/ASPARAGINE SYNTHETASE1 (DIN6/ASN1) (7, 30), as determined by RT-qPCR (Fig. 2D). Taken together, molecular marker analysis in roots as well as metabolite analysis in both shoot and root tissues support the hypothesis that the tested experimental setup transiently perturbs seedling energy metabolism.
Fig. 2.
Enhanced LR density upon uD correlates with reduced hexose levels and activation of low-energy stress markers and requires SnRK1. (A–C) The 8-d-old Arabidopsis (Col-0) seedlings were cultured under control (“C,” white bars) or 1-h and 4-h uD conditions (black bars) (see Fig. 1E). The presented soluble sugars from shoots (A) and roots (B) or T6P from roots (C) were quantified. Data are presented as boxplots, and significance was determined by Mann–Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001; n = 6 to 10 (A and B) or n = 4 (C). (D) Relative expression of the DIN6/ASN1 marker gene in roots under control (white bars), 1 h and 4 h uD conditions (black bars), or after 4 h uD and 8 h light recovery (R) (4 h + R). Transcript abundance was quantified by qRT-PCR. Given are mean values ± SD. Significance relative to control was calculated by Student’s t test; *P < 0.05, **P < 0.01, ***P < 0.001; n = 3. (E–G) Root parameters (E, eLRD; F, LR number; G, primary root length) quantified for WT (Col-0) and snrk1.α1 and snrk1.α2 knockout mutants cultivated in control (white bars) or uD (black bars) according to Fig. 1E. Data from three independent experiments are presented in the respective boxplots. Student’s t test compares control and treated samples. *P < 0.05; n = 10 to 15. (H and I) Confocal microscopy of an SnRK1α1:GFP fusion protein expressed under the native SnRK1α1 promotor in transgenic plants (32). SnRK1α1 was found to be ubiquitously expressed in roots (SI Appendix, Fig. S3). (H) Nuclear staining was observed in the meristematic zone at the root tip. (I) GFP fluorescence is observed in developing LRs, particularly in the pericycle. Counterstaining with propidium iodide. (Scale bar: 50 µm.) (J) Analysis of root SnRK1 kinase activity according to the setup in Fig. 1E. A nuclear rat ACC-GFP-HA reporter protein was expressed in transgenic plants, and its in vivo phosphorylation was assayed by immunodetection using a P-dependent ACC-specific antibody (αP-ACC) and an α-HA antibody for normalization.
The LR Response upon uD Requires SnRK1, a Central Kinase in Energy Homeostasis.
DIN6/ASN1 is a well-known downstream response gene of the central metabolic kinase SnRK1, which activates catabolic processes and pathways for alternative ATP generation upon energy starvation (7, 11, 16). Moreover, T6P has been demonstrated to inhibit SnRK1, at least under in vitro conditions (9, 10, 31). To evaluate SnRK1’s contribution to LR establishment in response to metabolic perturbation, we employed a mutant approach. In Arabidopsis, two catalytic α-units are functionally important and perform in a partially redundant manner (7, 11). Whereas knockout of the SnRK1α2 catalytic subunit (snrk1α2) only had a minor effect on uD-induced eLRD, the snrk1α1 mutant showed a significant reduction in eLRD upon uD treatment (Fig. 2 E–G), suggesting a SnRK1α1 function in maintaining LR initiation after stress recovery. In contrast, snrk1α2 appears to impact particularly primary root length in response to uD, whereas primary root length was unaffected in snrk1α1. This loss-of-function approach demonstrates that SnRK1α1 is required to adjust LR density during photosynthetic perturbations.
Following the assumption that SnRK1 affects LR development upon energy perturbations, we assessed SnRK1α1 localization in roots using a SnRK1α1:GFP fusion expressed under the native promoter. In line with previous findings (32, 33), SnRK1α1:GFP expression was observed rather ubiquitously in many root cell types (SI Appendix, Fig. S3 A–F), predominantly perinuclear or in the nucleus of actively dividing cells at the root tip (Fig. 2H). Whereas strong SnRK1α1:GFP expression was found at all stages of LR development, a weak signal was already observed in LR primordia as well as in pericycle cells (Fig. 2I). This localization is in line with a proposed function of SnRK1α1 in uD-triggered LR formation.
As the SnRK1 catalytic subunit was found to translocate to the nucleus to induce target gene expression (12), we more directly assessed nuclear SnRK1 activity by expressing a reporter harboring a well-described AMPK1 phosphorylation target peptide obtained from rat ACETYL-COA CARBOXYLASE (ACC) with an SV40 nuclear localization sequence (NLS), fused to GFP and a double HA-tag (34). Using commercial phospho (P)-ACC antibodies, phosphorylation of the peptide was detected and normalized to the HA-signal of the reporter. This system enables a quantitative evaluation of SnRK1 phosphorylation activity in the nucleus, as it has been previously demonstrated in vitro and in yeast (34). In transgenic roots, we observed a rapid increase in nuclear SnRK1 activity already 1 h after uD treatment (Fig. 2J and SI Appendix, Fig. S4 A–D), further supporting the role of SnRK1 (particularly the α-1 catalytic subunit) in mediating the LR response to uD.
Increased LR Density upon uD Requires the SnRK1 Target TF bZIP63.
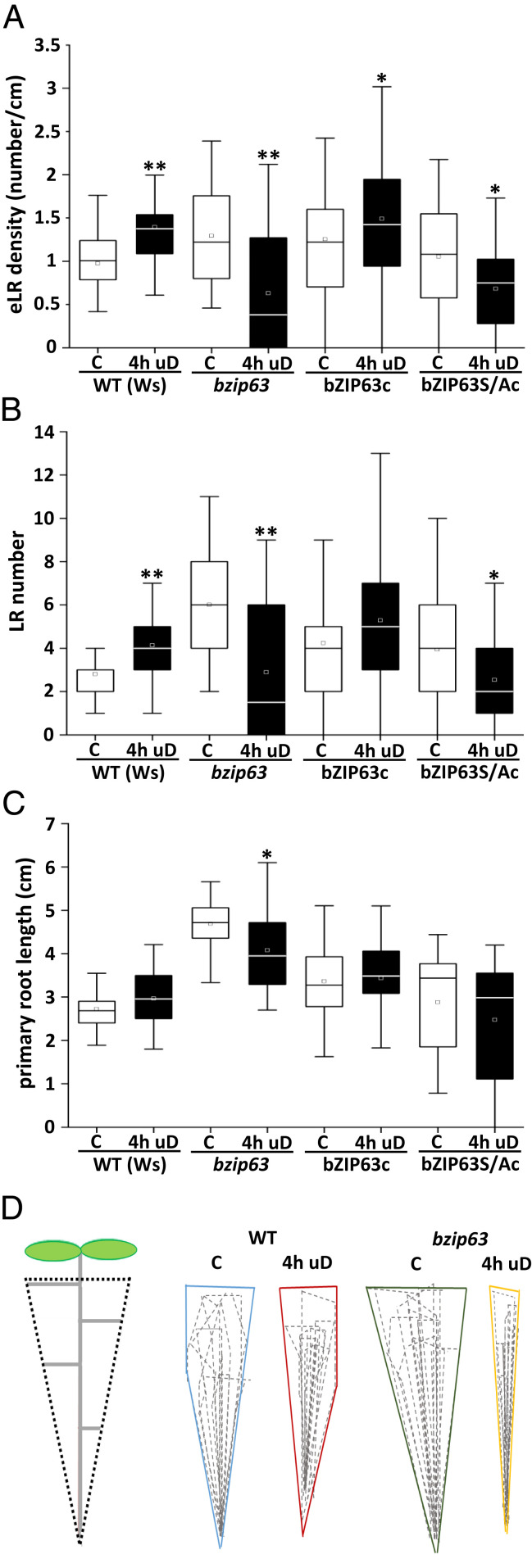
Several bZIPs of the C/S1 TF network have been proposed to function as homo- or heterodimers downstream of the SnRK1 kinase (16). In particular, group C bZIP63 was identified as an in vivo kinase target of SnRK1 (15). Hence, we studied bZIP63 as a potential SnRK1 downstream TF in the LR response. Similar to the snrk1α1 mutant (Fig. 2 E–G), a decreased eLRD was observed upon 4 h of uD in bzip63 transfer DNA knockout seedlings in the WS background (Fig. 3 A–C) or in CRISPR-derived bzip63 mutant seedlings in the Col-0 background (SI Appendix, Figs. S5 A and B and S6A). It should be noted that in comparison to wild type (WT), bzip63 mutants showed increased primary root (PR) length and eLRD under control conditions. Besides the response to uD, low light–induced LR formation was also reduced in the bzip63 mutant (SI Appendix, Fig. S6B). We thus conclude that bZIP63 is required for the observed increased eLRD phenotype in response to short-term perturbations in energy homeostasis.
Fig. 3.
Increased LR density upon uD requires the SnRK1 downstream TF bZIP63. (A–C) Root parameters (eLRD [A]; LR number [B]; and primary root length [C]) quantified for WT (Ws), bzip63 knockout mutant, bzip63 complemented with bZIP63:YFP (bZIP63c) or bZIP63Ala:YFP (bZIP63S/Ac; Ala exchange derivative of bZIP63) (11) grown under control (“C,” white bars) or uD (black bars) conditions according to Fig. 1E. Statistical significant differences between control and treated samples were determined by Mann–Whitney U test. *P < 0.05; **P < 0.01; n = 18 to 35. Data from three independent experiments are presented in the respective boxplots. (D) The bzip63 loss-of-function mutant is affected in overall LR architecture, specifically upon uD treatment. The 8-d-old seedlings were cultured on solidified 1/2 MS under control and 4-h uD conditions and analyzed 14 DAG. The overall root system dimensions were imaged by a maximum projection of 10 individual roots per genotype (WT, bzip63) and condition (uD and control). The outline projections are shown for the indicated conditions.
Three serine residues (S) have been identified in bZIP63 as in vivo SnRK1 phosphorylation sites (15). Triple alanine (A) exchange mutations (S29A, S294A, and S300A) and nonmutated versions were expressed as YFP fusions under control of the native promoter to complement the bzip63 knockout mutant. In contrast to seedlings expressing the WT bZIP63:YFP protein (bZIP63c), seedlings expressing the mutant protein (bZIP63S/Ac) are impaired in SnRK1-mediated phosphorylation and did not display enhanced eLRD upon uD (Fig. 3 A–C). These data strongly support a key role for SnRK1-bZIP63 signaling in the starvation-induced LR response.
In addition, we assessed the impact of bZIP63 on overall root architecture by analyzing root system dimensions of WT and bzip63 mutants under control or uD conditions. To depict the entire root system, we overlayed roots of 10 individual plants to create a maximum root outline projection. By these means, we found that compared to WT, bzip63 mutants exhibited a slightly expanded root system under control conditions, while the root system dimension was strikingly reduced in response to a uD treatment (Fig. 3D). Altogether, these findings strongly support the view that bZIP63 controls LR density, especially under conditions of low energy. Moreover, bZIP63 requires a posttranslational activation via SnRK1-mediated phosphorylation.
bZIP63 Is Expressed throughout LR Development and Impacts Its Initiation.
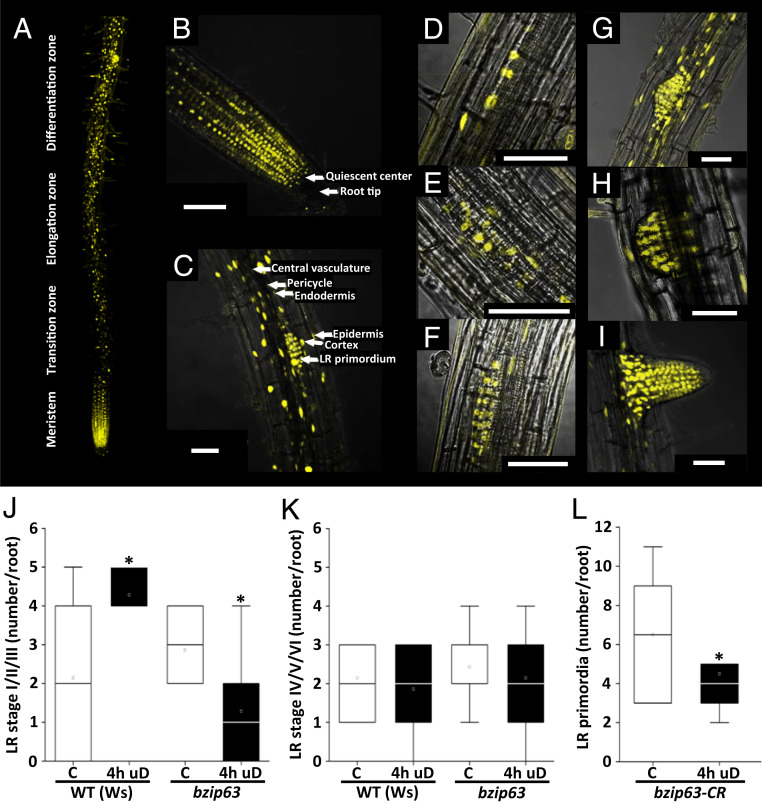
Localization of bZIP63 in the root remains an important prerequisite to further assess its functional impact. Hence, we used confocal fluorescence microscopy to study a transgenic line expressing bZIP63:YFP under the control of its native promoter in a bzip63 mutant background. Periodical clusters of high and low YFP-expressing cells were observed along the root axes (Fig. 4A), whereas strong expression and nuclear localization were obvious in the root meristem (Fig. 4B). In particular, we detected strong YFP signals in areas of LR emergence (Fig. 4C). Imaging at higher magnification revealed nuclear localization of bZIP63:YFP in the cortex, endodermis, and pericycle cells but not in xylem or phloem cells. Moreover, strong bZIP63 expression is visible throughout all developmental stages of LR development (35) (Fig. 4 D–I).
Fig. 4.
bZIP63 is expressed in PRs and LRs and is required for early LR initiation. (A–C) Overview and close-up confocal scanning microscope images of 10-d-old Arabidopsis roots expressing bZIP63:YFP under control of the native promoter in a bzip63 mutant background. (A) Overview panel of the primary root (developmental zones from the meristem at the root tip to differentiation zone [size ∼1.5 cm; magnification 25×] show periodically occurring YFP maxima). Strong nuclear YFP signals were observed at the root tip (B) and LR primordia (C). Magnification 40×. (Scale bar: 50 µm.) (D–I) bZIP63:YFP was detected throughout LR development: stage II (D), stage III (E), stage IV (F), stage V (G), stage VI (H), and emerged LR (I). The plane was adjusted to visualize the xylem pole at each stage; magnification 40×. (Scale bar: 50 µm.) (J and K) Number of early (stages I through III) (J) and late (stages IV through VI) (K) LRs in 5-d-old WT (Ws) and bzip63 mutant seedlings, as determined by DIC microscopy (SI Appendix, Fig. S7), revealed an impact of bZIP63 on early LR development. Student’s t test; *P < 0.05, n = 7. Data from three independent experiments are presented in the respective boxplots. (L) Enhanced expression of the early-stage LR marker pGATA23::NLS-GFP (24) upon uD depends on bZIP63. Using CRISPR/Cas9 technology, a bzip63 knockout (Col-0, bzip63CR) was generated in the pGATA23::NLS-GFP (24) reporter line. The 5-d-old seedlings were treated with 4 h of uD. After treatment, the seedlings were transferred to long day conditions and recovered for 16 h under control (C) conditions. Primordia events expressing GFP were quantified along the primary root with and without uD treatment. Student’s t test relative to the control; *P < 0.05 for n = 8 samples. Data from four (n = 2 per condition) independent experiments are presented in the boxplot.
Based on the observed expression profiles, bZIP63 has the potential to interfere at several stages in LR initiation, specification, or emergence (20). To further evaluate the exact impact of bZIP63 on LR development, we studied the appearance of LR primordia in cleared roots applying differential interference contrast (DIC) imaging (35). At 16 h after uD treatment, LR stages were counted in WT and bzip63 mutant seedlings and compared to the respective control conditions (Fig. 4 J and K and SI Appendix, Fig. S7). These analyses revealed that uD treatment led to more early LR primordia (stages I through III) in the WT but less in bzip63 (Fig. 4J). This finding was corroborated when we assayed the GATA23:NLS-GFP reporter in a bzip63 CRISPR knockout background (Fig. 4L). Again, fewer microscopically quantified GFP sites - reflecting early LR primordia - were found after uD in comparison to the control treatment. In contrast, numbers of LRs classified as stages IV through VI were similar in WT and the mutant (Fig. 4K). Taken together, we conclude that bZIP63 mediates the priming of early LR initiation, particularly during short-term perturbation of energy homeostasis.
bZIP63 Directly Binds the Promoter of ARF19 and Is Required for Increased ARF19 Expression in Response to uD.
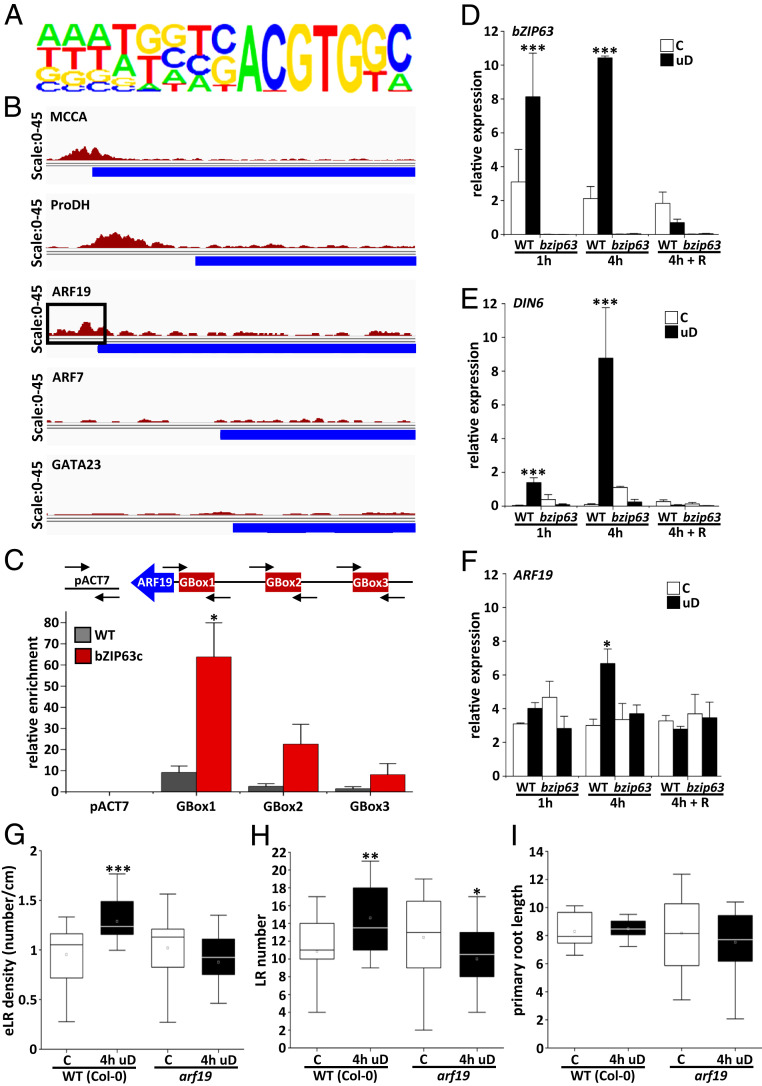
To define direct target genes of bZIP63 in this response, we performed ChIPseq (chromatin immunoprecipitation DNA sequencing) using roots treated with 4 h of uD. For the IP (immunoprecipitation) with a commercial GFP antibody, the bzip63 mutant and the complementation line expressing a bZIP63:YFP fusion protein under the native bZIP63 promoter were used. These experimental settings are important for studying cell-type–specific localization, natural expression levels, and inductive conditions. Data analysis detected 821 signals (peaks) significantly enriched in comparison to the control (Dataset S1). The identified sites correspond to promoters (51.2%), intergenic regions (19%), exons (15.8%), transcription termination sites (11.3%), and introns (2.7%). In line with the well-defined bZIP63 binding site, G-box related sequences (C/GACGTG) (36, 37) were enriched in the promoters detected by the ChIPseq approach (Fig. 5A). Among the genes bound by bZIP63, several previously confirmed target genes were detected, including METHYLCROTONYL-COA CARBOXYLASE (MCCA), ELECTRON-TRANSFER FLAVOPROTEIN: UBIQUINONE OXIDO-REDUCTASE (ETFQO) BRANCHED CHAIN AMINO ACID TRANSAMINASE2 (BCAT2), PROLINE DEHYDROGENASE (ProDH), and DIN6/ASN1 (11) (Fig. 5B and Dataset S1), underlining the quality of the analysis. Interestingly, we identified the promoter of the ARF19 gene as a target bound by bZIP63. Both ARF19 and its homolog ARF7 represent crucial auxin-dependent TFs with established roles in LR development (38, 39). However, ARF7, as well as other well-established LR development genes such as GATA23 or LBD16/29, were not detected in our ChIPseq analysis (Fig. 5B and Dataset S1). Using ChIPPCR, we further confirmed significant binding of bZIP63 to the ARF19 promoter in a region harboring a G-box cis-element (G-box1) (Fig. 5C and SI Appendix, Fig. S8).
Fig. 5.
bZIP63 directly binds the ARF19 promoter and controls ARF19 transcription. (A and B) ChIPseq was performed with roots of 10-d-old seedlings upon 4 h of uD comparing bzip63 and a complementation line expressing a functional bZIP63:YFP under control of the native promoter. Chromatin was immunoprecipitated using anti-GFP antibodies, and genomic fragments were subjected to high-throughput DNA sequencing. A total of 821 enriched binding fragments (peaks) corresponding to 500 target genes were identified (Dataset S1). (A) Nucleotide logo displaying the predicted, enriched cis-element matching the experimentally defined bZIP63 specific binding site (G/C-box; C/GACGTG). (B) Reads from bZIP63:YFP bound DNA fragments mapped against selected known bZIP63 target promoters as controls (MCCA and ProDH) and ARF19 as a potential target in LR development. No strong binding was observed for ARF7 or GATA23. Blue color bars represent the 5′ end of the respective open reading frames. The ARF19 promoter is marked for G-Box-1 binding region (black box). (C) ChIPPCR of roots treated with 4-h uD was used to verify binding of bZIP63:YFP to the ARF19 promoter. Using the primer pairs indicated, significant binding was determined around G-Box-1, whereas no significant enrichment was observed for the nonbinding control (ACTIN7 and ACT7) or G-Box-2 and -3. Enrichment of promoter sequences derived from WT (gray bars) and bZIP63:YFP (red bars) are indicated. Presented are mean values ± SD from three independent plant pools relative to input (determined by ProACTIN8 abundance). Student’s t test, *P < 0.05. (D–F) bZIP63 loss-of-function mutants are impaired in induced ARF19 transcription upon uD. The 8-d-old Arabidopsis WT and bzip63 seedlings were cultivated under control conditions or treated with 1 and 4 h of uD or 4 h uD plus 8 h of light recovery (R) before harvesting. RT-qPCR of roots at the time points indicated for bZIP63 (D), DIN6/ASN1 (E), and ARF19 (F). Given are mean values ± SEM derived from roots of three independent plant pools relative to EF1A. Student’s t test, *P < 0.05, ***P < 0.001. (G–I) ARF19 is required for uD-induced LR initiation. eLRD of WT and arf19 mutant analyzed in the setup described in Fig. 1E. (eLRD [G]; LR number [H]; and primary root length [I]). Seedlings were grown in long day regime (16 h light/8 h dark at 100 μmol ⋅ m−2 ⋅ s−1) on solidified 1/2 MS media. Statistically significant differences between control (C) and treated (uD) samples were determined by Mann–Whitney U test. *P < 0.05, **P < 0.01, and ***P < 0.001.
To study ARF19 gene expression, a RT-qPCR time-course experiment was performed, determining its transcript abundance in roots of WT and bZIP63 mutant plants after 1 and 4 h of uD and after recovery (Fig. 5 D–F). In 8-d-old WT seedlings, bZIP63 and its target gene DIN6/ASN1 were found to be significantly induced after 4 h of uD, while normal transcript levels were reestablished after shifting plants back to light. In line with largely missing bZIP63 expression, and as expected based on previous findings (15), DIN6/ASN1 induction was impaired in the bZIP63 mutant, which therefore serves as control. Importantly, whereas the WT showed a significant twofold induction of ARF19 expression upon 4 h of uD, basal ARF19 expression was found to be independent of bZIP63. Altogether, these data propose a specific input of SnRK1-bZIP63-ARF19 signaling on LR development during perturbed energy homeostasis.
To further support ARF19 as a potential bZIP63 downstream target in this response, arf19 mutants were analyzed. Importantly, the mutant line behaved like WT under control conditions (as ARF7 is still present) but no longer induced eLRD upon uD (Fig. 5 G–I), indicating that ARF19 is required for this response. These data identify ARF19 as a target of SnRK1-bZIP63 signaling and further suggest a role of this auxin-dependent TF in priming LR initiation during energy deprivation.
Discussion
This study was designed to identify molecular players that integrate information on fluctuations in energy availability into developmental plasticity. In order to characterize plant responses to energy-limiting conditions, frequently relatively harsh experimental treatments are applied, which interfere with plants’ photosynthetic activity. In this respect, night extension, limitation of CO2, or treatment with photosynthesis inhibitors are used (11, 27, 28). Recently, photosynthetic inhibitors and extended night treatments were found to strongly interfere with root meristematic activity and affect both primary root and LR development (28). However, as severe treatments impact overall plant physiology, mechanistic aspects of the regulatory circuits are difficult to dissect. Here, we observed that several mild metabolic perturbations caused by short-term uD or low-light treatment led to a consistent increase in eLRD, whereas primary root growth was not affected. Importantly, this developmental output was robust, easy to quantify, and was observed in several Arabidopsis ecotypes. On the other hand, it was specifically related to metabolic perturbations and not a general stress response. To conclude, the employed mild and controlled experimental setup was well suited to mimic fluctuating energy situations regularly occurring in plant life.
The temporary uD treatment resulted in a rapid activation of the DIN6/ASN1 starvation response marker (7, 15, 30) and depletion of soluble sugars as well as the sugar-signaling molecule T6P. These correlative data support the view that the mild perturbation treatments lead to fast and significant alterations in energy homeostasis both in shoots and roots. SnRK1 has been established as an evolutionary conserved metabolic stress-sensor kinase, which responds to limiting energy conditions (2–6). According to the nexus model, the low-abundance signaling metabolite T6P is proposed to mirror and control plant sucrose levels and was found to exert its effects - at least in part - through negative regulation of SnRK1 activity (9, 10). In line with this, transiently reduced T6P levels and increased SnRK1 activity upon uD treatment as well as an impact of the snrk1α1 loss-of-function approach support the importance of this central kinase in stimulating LR development upon metabolic perturbations. Although the two SnRK1 α-subunits have been proposed to exert partially redundant functions, a mutant in the α2-subunit showed only minor effects on LR development, supporting a more pronounced function of SnRK1α1. It needs to be stressed that under standard laboratory growth conditions, WT and snrk1α1 mutants did not differ with respect to primary root and LR architecture. These data propose a developmental function of SnRK1α1, which is executed only upon metabolic perturbation.
Recently, we have established a mechanistic link between SnRK1 and its phosphorylation target protein bZIP63, which functions as a downstream transcriptional regulator (11, 15). Loss-of-function approaches and specific alanine exchange mutations with respect to in vivo bZIP63 phosphorylation sites demonstrated that bZIP63 is required to establish the SnRK1-dependent LR phenotype. In contrast to the snrk1α1 mutant, bzip63 plants showed increased primary root length and a slightly enlarged overall root system under standard growth conditions, indicating additional bZIP63 functions beyond LR development. As bZIP63 is part of the complex C/S1 bZIP network (16), it is conceivable that other bZIPs may perform as heterodimerization partners. Along this line, the poplar ortholog of Arabidopsis bZIP1, which interacts with AtbZIP63 (15, 40), has been implicated in controlling LR formation (41).
As the short uD treatment resulted in decreased sugar and T6P levels and increased SnRK1 activity in roots, it is most likely that the low-energy stimulus is perceived in roots. In line with this, GFP studies demonstrated that SnRK1 and bZIP63 expression domains particularly overlapped in the pericycle and cells crucial to early LR formation, which would enable direct SnRK1-mediated phosphorylation of bZIP63. Nevertheless, perception of metabolic perturbations in photosynthetic leaves and subsequent signaling to the roots cannot be excluded. In particular, we recently observed a reduced polar auxin transport to the root tip upon starvation by an extended night treatment (11), resulting in auxin accumulation in the LR zone. Because of its prime importance in LR initiation (20), auxin very likely contributes to this response. In summary, these hypotheses are not mutually exclusive, and further research is needed to gain insight into long-distance communications in plant energy homeostasis.
XPPs are specified in the pericycle initials of the meristem (20). However, only a subset of them develop into LR founder cells and finally establish an LR primordium. Applying DIC microscopy and a GATA23:GFP reporter, early LR initiation events triggered by uD were found to be significantly reduced in the bzip63 mutant in comparison to WT. These findings support the notion that signals related to metabolic imbalance are transmitted via SnRK1-bZIP63 signaling into early events in LR development. The auxin-regulated TFs ARF19 and ARF7 have been demonstrated to be crucial in early LR specification (38). Importantly, unbiased ChIPseq and ChIPPCR fine mapping support direct binding of bZIP63:YFP to the ARF19 promoter at/in vicinity of G-box1, a well-described binding site for bZIP63 (11). In agreement with these findings, G-box cis-elements were observed to be significantly enriched in the promoters bound by bZIP63. Whereas ARF19 transcription was induced by uD, this response was impaired in the bzip63 mutant. Moreover, ARF19 was found to be essential for the increased LR phenotype upon metabolic perturbations, as it was not observed in the arf19 knockout mutant. Altogether, these data strongly support our hypothesis that the SnRK1-bZIP63-ARF19 module signals information on the metabolic status to a central regulatory hub in LR initiation (Fig. 6A). TFs generally bind and/or regulate hundreds of target genes. Along this line, promoters of several well-known LR regulators, such as PUCHI (42) or MYB77 (43), are directly bound by bZIP63. It is therefore conceivable that bZIP63 mediates its function via several direct target genes. However, ARF7 or the GATA23 promoters were not found to be bound by bZIP63, indicating that these genes are regulated in an indirect manner.
Fig. 6.
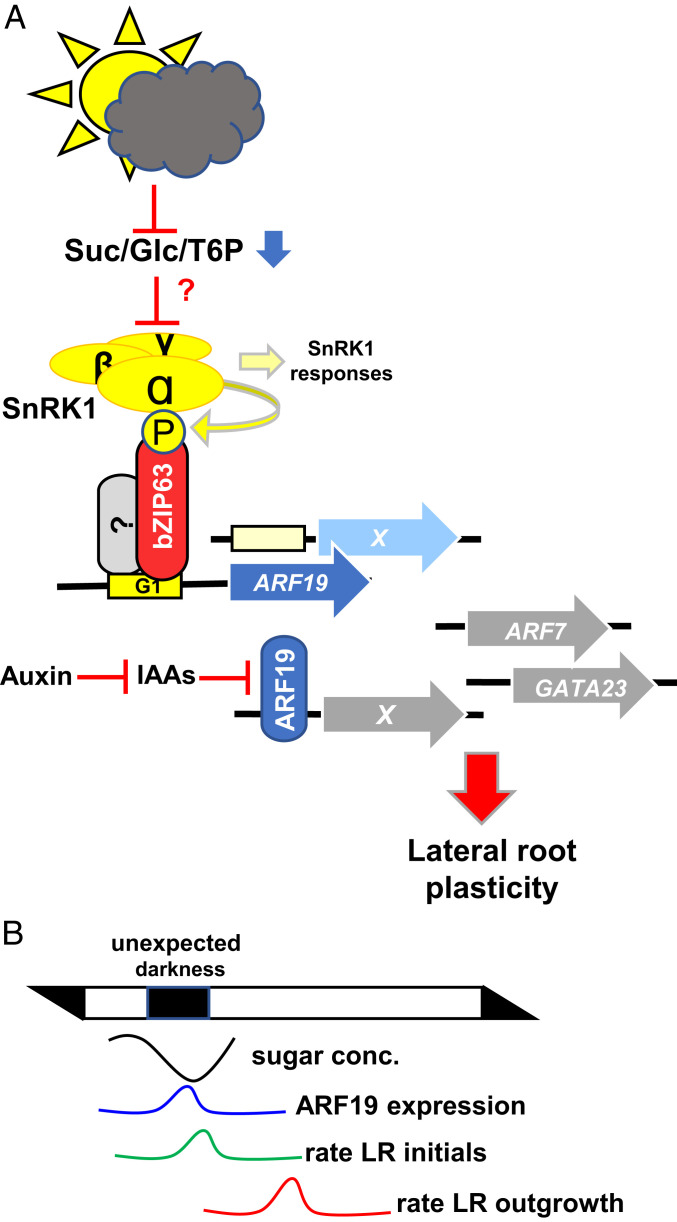
Working model summarizing SnRK1-bZIP63-ARF19 signaling in metabolic control of LR development. (A) Metabolic perturbations activate the SnRK1 kinase, which phosphorylates the bZIP63 TF (15). Via direct promoter binding, bZIP63 activates ARF19 transcription. Being de-repressed by auxin-mediated inactivation of IAA repressors, ARF19 controls auxin-responsive gene expression related to LR initiation (20). In this respect, SnRK1-mediated, metabolic signaling is proposed to tune auxin responses and, consequently, LR plasticity. Further regulators in LR development (ARF7 and GATA23) (20) are not direct targets of the proposed signaling cascade. Localization and metabolic signals triggering SnRK1 activity, as well as potential bZIP heterodimerization partners, remain unresolved. (B) Sketch describing the proposed timing of events leading to low-energy–mediated priming of LR initiation.
Overall, the observation of an increased eLRD upon metabolic perturbations was unexpected and, at first view, counterintuitive. However, our analyses disclosed that under these conditions, primarily early LR development was initiated via SnRK1-bZIP63-ARF19 signaling, while LR outgrowth was deferred until stress release. This highlights a yet uncharacterized positive function of SnRK1 signaling besides the well-established metabolic “brake”, namely the priming of prospective developmental processes, anticipating an upcoming resource supply.
In order to ensure optimal plant growth, shoot and root propagation are highly coordinated. While the photosynthetically active shoot produces carbohydrates, the root system exploits water and mineral resources. In this light, developmental priming (44, 45) of LR initiation under energy-deprived conditions can be interpreted as a cost-efficient strategy to prepare plants for efficient mineral and water uptake required for a rapid restart of overall plant growth, once metabolic (carbon) homeostasis is restored. Upon recovery, the sugar-depleted LR initials act as a strong sink, and sugar levels are rapidly normalized and provide the crucial resources for LR outgrowth. We therefore propose the working model summarized in Fig. 6B, which, however, needs to be challenged experimentally. Importantly, a rapid growth recovery after stress may be essential under fluctuating environmental conditions and in natural (competitive) ecosystems to ensure the plant`s reproductive success and, hence, its Darwinian fitness.
Methods
Plant Material and Culture.
The Arabidopsis thaliana WT accessions Col-0, WS, and Ler, as well as transgenic lines, are listed in SI Appendix, Table S1. For all experiments, seeds were surface sterilized with chlorine gas and stratified for 48 h in darkness at 4 °C. For LR phenotyping approaches, seedlings were grown vertically in square (12 cm × 12 cm) Petri dishes containing half-strength MS (26) medium solidified with 8 g/L phytoagar (Duchefa) under long day conditions (16 h light at 23 °C/8 h darkness at 16 °C) illuminated with 70 (all experiments except Fig. 5 G–I) or 100 (Fig. 5 G–I) µmol ⋅ m−2 ⋅ s−1 light and a relative humidity of 60%. The strongest effect on uD-mediated LR formation was observed when plants were cultivated under 70 µmol ⋅ m−2 ⋅ s−1. At 7 DAG, plants of similar root length (∼2 cm) were transferred to new plates with a spacing of around 1 cm between plants. At 8 DAG, seedlings were transferred to specific low-energy or control conditions. Energy perturbation assays were performed according to the schemes in Fig. 1A (low light), Fig. 1E (uD), or SI Appendix, Fig. S1A (extended night). Seedlings used for low-light treatment were subjected to an irradiance of 15 μmol ⋅ m−2 ⋅ s−1. For the uD experiments, seedlings were treated with complete darkness for 0.5 h to 4 h starting 2 h (ZT2) after onset of the light phase. At 14 DAG, LR number, primary root length, and eLRD were determined for each plant. For laser scanning microscopy (LSM) imaging (Figs. 1 I and J and 4 J–L and SI Appendix, Fig. S2), seedlings were transferred to low-energy conditions already 5 DAG. A detailed description on the root phenotyping procedure and microscopic imaging of root-localized GATA23 and bZIP63 expression can be found in SI Appendix, Supplementary Methods.
Molecular Biological Techniques.
Total plant RNA was prepared from 5 to 10 mg root material using an RNeasy Mini Kit (Qiagen) following the manufacturer’s protocol. Complementary DNA (cDNA) was synthesized from 1 µg total RNA using random nonamer and oligo-dT primers with reverse transcriptase RevertAid H Minus (Thermo Fisher Scientific) as previously described (46). SYBR green was used to visualize the amplified products. Threshold cycle (Ct) values were calculated from three biological replicates employing the 2-ΔCT method (47) using ELONGATION FACTOR 1-ALPHA 1 (EF1A) for normalization. Primers are given in SI Appendix, Table S1.
CRISPR/Cas9 technology was used to generate a bzip63 mutant in the pGATA23::NLS-GFP (24) reporter line using the system described (48). An efficiently binding and target gene–specific single guide RNA targeting exon 1 of bZIP63 was designed using ChopChop (49) (SI Appendix, Fig. S5 A and B). Primers are given in SI Appendix, Table S1. A detailed description of ChIPPCR and ChIPseq can be found in SI Appendix, Supplementary Methods.
Mass Spectrometric Analysis.
Shoots and roots (∼40 mg fresh weight) were separated and frozen in liquid N2. Ground tissue was extracted in 300 µL 80% ethanol, containing 2 µg 1,1-d2-trehalose and 8 µg 6,6-d2-glucose as internal standards. Samples were incubated at 80 °C for 20 min and centrifuged for 10 min at 14,000 rpm. The supernatant was transferred to a new reaction tube, and the residue was reextracted twice using, first, 300 µL 50% (volume/volume) ethanol and, subsequently, 300 µL 80% ethanol (volume/volume) both at 80 °C for 20 min. The extracts were pooled, and the solvent was completely evaporated using a vacuum concentrator at 55 °C. The obtained pellet was redissolved in 25 µL 50% methanol (volume/volume). Samples (5 µL) were analyzed using a Waters Acquity ultrahigh-performance liquid chromatograph coupled to a Waters Micromass Quattro Premier triple quadrupole mass spectrometer (Milford) with electrospray interface (ESI). Chromatographic separation was performed according to application note WA60126 with a modified flow rate of 0.2 mL/min. Sugars were detected in negative electrospray mode (ESI−) at 120 °C source temperature and 3.25 kV capillary voltage. Nitrogen served as a desolvation and a cone gas with flow rates of 800 L ⋅ h−1 at 350 °C and 25 L ⋅ h−1. The mass spectrometer operated in the multiple reaction monitoring mode using argon as a collision gas at a pressure of ∼3 × 10−3 bar. Cone voltage and collision energy were optimized for maximum signal intensity of each individual compound during ionization and collision-induced dissociation with a dwell time of 0.025 per transition. T6P was quantified according to ref. 50, with modifications as in ref. 51.
SnRK1 Kinase Activity Assay.
For stable transformation of the SnRK1 activity reporter in the WT Col-0 background, the coding sequence of a GFP- and double HA–tagged double rat ACC1 peptide with N-terminal SV40 NLS (33) was subcloned in a pCB302-derived mini binary vector with 35SC4PPDK promoter (35S enhancer and maize C4PPDK basal promoter), nopaline synthase terminator, and bar resistance marker (52). Extraction and immunoblotting were performed as previously described (46).
Statistical Analysis.
Statistical tests were performed with the built-in statistical analyzer of Origin software. Student’s t test or the Mann–Whitney U test were used for significance testing in normally and not normally distributed data, respectively.
Supplementary Material
Acknowledgments
We thank Elena Baena-González (Instituto Gulbenkian de Ciência Lisbon) for providing Arabidopsis seeds and Theresa Damm for excellent technical assistance. This work was supported by the Deutsche Forschungsgesellschaft (DR273/10-6), the Jeff Schell Scholarship from the Bayer Foundation provided to P.M., and by the Max Planck Society (R.F. and J.E.L.) and the Austrian Science Fund with Project P-28491 (to M.T.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106961118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Chaiwanon J., Wang W., Zhu J. Y., Oh E., Wang Z. Y., Information integration and communication in plant growth regulation. Cell 164, 1257–1268 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crepin N., Rolland F., SnRK1 activation, signaling, and networking for energy homeostasis. Curr. Opin. Plant Biol. 51, 29–36 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Wu Y., et al., Integration of nutrient, energy, light, and hormone signalling via TOR in plants. J. Exp. Bot. 70, 2227–2238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margalha L., Confraria A., Baena-González E., SnRK1 and TOR: Modulating growth-defense trade-offs in plant stress responses. J. Exp. Bot. 70, 2261–2274 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Shi L., Wu Y., Sheen J., TOR signaling in plants: Conservation and innovation. Development 145, dev160887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broeckx T., Hulsmans S., Rolland F., The plant energy sensor: Evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot. 67, 6215–6252 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Baena-González E., Rolland F., Thevelein J. M., Sheen J., A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Emanuelle S., et al., SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 82, 183–192 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Zhai Z., et al., Trehalose 6-phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell 30, 2616–2627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueroa C. M., Lunn J. E., A tale of two sugars: Trehalose 6-phosphate and sucrose. Plant Physiol. 172, 7–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedrotti L., et al., Snf1-RELATED KINASE1-controlled C/S1-bZIP signaling activates alternative mitochondrial metabolic pathways to ensure plant survival in extended darkness. Plant Cell 30, 495–509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramon M., et al., Default activation and nuclear translocation of the plant cellular energy sensor SnRK1 regulate metabolic stress responses and development. Plant Cell 31, 1614–1632 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nukarinen E., et al., Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 6, 31697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dröge-Laser W., Snoek B. L., Snel B., Weiste C., The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 45, 36–49 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Mair A., et al., SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. eLife 4, e05828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dröge-Laser W., Weiste C., The C/S1 bZIP network: A regulatory hub orchestrating plant energy homeostasis. Trends Plant Sci. 23, 422–433 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Simon N. M. L., et al., The energy-signaling hub SnRK1 Is important for sucrose-induced hypocotyl elongation. Plant Physiol. 176, 1299–1310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong E.-Y., Seo P. J., Woo J. C., Park C.-M., AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in Arabidopsis. BMC Plant Biol. 15, 110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai A. Y.-L., Gazzarrini S., AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 69, 809–821 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Motte H., Vanneste S., Beeckman T., Molecular and environmental regulation of root development. Annu. Rev. Plant Biol. 70, 465–488 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Du Y., Scheres B., Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 69, 155–167 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Moreno-Risueno M. A., et al., Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xuan W., et al., Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 351, 384–387 (2016). [DOI] [PubMed] [Google Scholar]
- 24.De Rybel B., et al., A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20, 1697–1706 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Okushima Y., et al., Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skoog M., Murashige T., Skoog F., A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497 (1962). [Google Scholar]
- 27.Lauxmann M. A., et al., Reproductive failure in Arabidopsis thaliana under transient carbohydrate limitation: Flowers and very young siliques are jettisoned and the meristem is maintained to allow successful resumption of reproductive growth. Plant Cell Environ. 39, 745–767 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Weiste C., et al., The Arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PLoS Genet. 13, e1006607 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kircher S., Schopfer P., Priming and positioning of lateral roots in Arabidopsis. An approach for an integrating concept. J. Exp. Bot. 67, 1411–1420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank A., et al., Circadian entrainment in Arabidopsis by the sugar-responsive transcription factor bZIP63. Curr. Biol. 28, 2597–2606.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., et al., Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 149, 1860–1871 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bitrián M., Roodbarkelari F., Horváth M., Koncz C., BAC-recombineering for studying plant gene regulation: Developmental control and cellular localization of SnRK1 kinase subunits. Plant J. 65, 829–842 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Belda-Palazón B., et al., A dual function of SnRK2 kinases in the regulation of SnRK1 and plant growth. Nat. Plants 6, 1345–1353 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Deroover S., Ghillebert R., Broeckx T., Winderickx J., Rolland F., Trehalose-6-phosphate synthesis controls yeast gluconeogenesis downstream and independent of SNF1. FEMS Yeast Res. 16, 1–15 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Malamy J. E., Benfey P. N., Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Kang S. G., Price J., Lin P.-C., Hong J. C., Jang J.-C., The Arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol. Plant 3, 361–373 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Kirchler T., et al., The role of phosphorylatable serine residues in the DNA-binding domain of Arabidopsis bZIP transcription factors. Eur. J. Cell Biol. 89, 175–183 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Wilmoth J. C., et al., NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43, 118–130 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M., ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ehlert A., et al., Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: Establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J. 46, 890–900 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Dash M., et al., Poplar PtabZIP1-like enhances lateral root formation and biomass growth under drought stress. Plant J. 89, 692–705 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Kang N. Y., Lee H. W., Kim J., The AP2/EREBP gene PUCHI Co-Acts with LBD16/ASL18 and LBD18/ASL20 downstream of ARF7 and ARF19 to regulate lateral root development in Arabidopsis. Plant Cell Physiol. 54, 1326–1334 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Shin R., et al., The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19, 2440–2453 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaturvedi P., Ghatak A., Weckwerth W., Pollen proteomics: From stress physiology to developmental priming. Plant Reprod. 29, 119–132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pantin F., et al., Developmental priming of stomatal sensitivity to abscisic acid by leaf microclimate. Curr. Biol. 23, 1805–1811 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Weiste C., Dröge-Laser W., The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 5, 3883 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Wang Z. P., et al., Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Labun K., et al., CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47 (W1), W171–W174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lunn J. E., et al., Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J. 397, 139–148 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Figueroa C. M., et al., Trehalose 6-phosphate coordinates organic and amino acid metabolism with carbon availability. Plant J. 85, 410–423 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Hwang I., Sheen J., Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.