Significance
The nuclear pore complex (NPC) consisting of hundreds of proteins embeds into the nuclear envelope and functions as a selectively permeable barrier regulating the bidirectional trafficking of macromolecules between the nucleoplasm and cytoplasm in eukaryotic cells. However, the exact copy number and the specific function of each composite protein within the NPC remain vague. To interrogate this gap in knowledge, in this paper, we utilized a high-speed single-molecule microscopy in conjunction with highly specific auxin-inducible degron strategies to detail the stoichiometric ratio of NPC’s nuclear basket proteins and also reveal their distinct roles in mediating nuclear export of messenger RNAs. The demonstrated techniques might also serve as an effective model for characterizing other large macromolecule complexes.
Keywords: nucleocytoplasmic transport, 3D super-resolution microscopy, NPC stoichiometry
Abstract
The in vivo characterization of the exact copy number and the specific function of each composite protein within the nuclear pore complex (NPC) remains both desirable and challenging. Through the implementation of live-cell high-speed super-resolution single-molecule microscopy, we first quantified the native copies of nuclear basket (BSK) proteins (Nup153, Nup50, and Tpr) prior to knocking them down in a highly specific manner via an auxin-inducible degron strategy. Second, we determined the specific roles that BSK proteins play in the nuclear export kinetics of model messenger RNA (mRNA) substrates. Finally, the three-dimensional (3D) nuclear export routes of these mRNA substrates through native NPCs in the absence of specific BSK proteins were obtained and further validated via postlocalization computational simulations. We found that these BSK proteins possess the stoichiometric ratio of 1:1:1 and play distinct roles in the nuclear export of mRNAs within live cells. The absence of Tpr from the NPC predominantly reduces the probability of nuclear mRNAs entering the NPC for export. Complete depletion of Nup153 and Nup50 results in an mRNA nuclear export efficiency decrease of approximately four folds. mRNAs can gain their maximum successful export efficiency as the copy number of Nup153 increased from zero to only half the full complement natively within the NPC. Lastly, the absence of Tpr or Nup153 seems to alter the 3D export routes of mRNAs as they pass through the NPC. However, the removal of Nup50 alone has almost no impact upon mRNA export route and kinetics.
Large protein complexes perform complex functions critical to the survival and proliferation of cells. A prime example of this phenomenon is one of the largest protein complexes within eukaryotic cells, the nuclear pore complex (NPC). Embedded within the nuclear envelope (NE), the NPC is composed of hundreds of proteins that are classified into approximately 30 different types of proteins. Together, these proteins form a complex that functions as the sole gatekeeper to the nucleus by regulating the bidirectional trafficking of macromolecules between the nucleus and cytoplasm. The structure of vertebrate NPCs, as evaluated by electron microscopy (EM) reconstructions, indicates that vertebrate NPCs are composed of an octagonal central framework that is ∼40 nm in length with an internal and external diameter of ∼50 nm and ∼120 nm, respectively. A cytoplasmic ring moiety with cytoplasmic filaments of ∼50 nm in length is attached to the central framework on the cytoplasmic face of the NPC. Both the nuclear ring moiety and the distal ring together form the nuclear basket of the NPC, a structure that is ∼75 nm in length. This basket is connected to the central framework on the nuclear face of the NPC (1–4). The NPC is composed of component proteins called nucleoporins (Nups). Approximately two-thirds of these Nups are classified as structural or scaffold Nups and together form the predominant physical structure of the pore. The remaining one-third component proteins are classified as phenylalanine-glycine-repeat Nups (FG-Nups) that form the selectively permeable barrier within the nuclear pore (5–8).
The nuclear basket of the NPC is composed of nuclear basket (BSK)-Nups, which, in vertebrates, consists of three FG-Nups: Nup153, Tpr, and Nup50 (9–12). The stoichiometry of these BSK-Nups has been estimated to be 8 for Nup153, 32 for Nup50, and 16 for Tpr in the research of Cronshaw et al. (5) or 32 for Nup153, 16 for Nup50, and 32 for Tpr from the study of Ori et al. (13). Due to conflicting studies, there are two different stoichiometric ratios, 1:4:2 and 2:1:2 (5, 13), which have been obtained via Western blotting, proteomics, and mass spectrometry in isolated nuclei (SI Appendix, Table S1). However, due to the inherent difficulty of determining protein quantities within individual supramolecular complexes in live cells, the reports of BSK-Nups stoichiometry per NPC may represent a best estimate of the average but still show obvious deviation. A more precise count of the FG-Nups copy number within the individual NPCs of live cells would produce a better and more realistic three-dimensional (3D) architectural map of the NPC, thereby further improving our understanding of the function and compositional dynamics of NPCs. Our first attempt to perform a direct count of Nups utilized in situ single-molecule counting technology to directly count the copy number of 24 different Nups within individual NPCs of live yeast. The resulting stoichiometry ratio of Nup1 and Nup60 located at the nuclear basket was determined to be 1:1 (14). In this paper, we expanded this approach to quantify the copies of nuclear basket proteins in live human cells after utilizing CRISPR/Cas9 techniques to fully label targeted Nups within NPCs.
Export of messenger RNA (mRNA) from nucleus to cytoplasm within eukaryotic cells likely occurs through several substeps: trafficking within the nucleus, anchoring at the NPC’s nuclear basket, moving through NPC, and releasing into the cytoplasm for subsequent translation (15–18). Briefly, within the nucleus, nascent mRNAs undergo mRNA processing (19–23) and then assemble mRNA:protein complexes (mRNPs) with protein cofactors prior to the mRNPs docking at the NPC. These cofactors include the transcription-export complex and nuclear transport receptors (24–30). Following docking, the transport receptors of mRNPs (the transport receptor protein Tap forming a heterodimer with cofactor p15 in human cells) enhance the complexes’ movement through the NPC’s selectively permeable barrier to reach the cytoplasm by directly interacting with FG-Nups (31–35). After shuttling through the NPC, mRNPs are directionally dissociated into the cytoplasm by the essential mRNP export factors Gle1, IP6, and DDX at the location of Nup214 (36–41). Previous single-molecule microscopy studies have revealed critical details of the dynamic features of mRNP export through the nuclear pore (42–47). It is important to note that some studies indicate that the nuclear basket could function as the rate-limiting step during mRNP export, thereby suggesting that FG-Nups within the nuclear basket of the NPC could play critical roles for nuclear mRNP export (15, 18, 46, 48). Echoing this finding, anti-Nup153 and anti-Tpr antibodies were found to block mRNP transport from the nucleus to the cytoplasm in vertebrate cells (49–51). Moreover, knockdown of Nup153 or Tpr caused nuclear accumulation of mRNA in eukaryotes (52–54). However, in conflict with these conclusions, knockout studies in yeast showed that depleting all FG-region Nups within the nuclear basket (Nup1, Nup2, Nup60) did not cause an obvious mRNA export defect (55).
In order to further refine the exact number of Nups within native NPCs and to determine the specific roles of BSK-Nups in the nuclear export of mRNPs, we first quantified the precise copy numbers for Nup153, Nup50, and Tpr in live human cells. This was accomplished utilizing the single-molecule counting approach previously employed by our laboratory (14). Next, auxin-inducible degron (AID) strategies were utilized to generate NPCs containing various copies of these BSK FG-Nups. To track model mRNP substrates interacting with NPCs containing different copies of BSK-Nups, high-speed single-particle tracking microscopy was employed. Through the implementation of this technique, we found that only Tpr and Nup153 played critical, but distinct, roles in regulating mRNA as it is exported from the nucleus. Finally, single-point edge-excitation subdiffraction (SPEED) microscopy and its two-dimensional (2D)-to-3D transformation algorithm enabled us to generate a 3D map of the export routes followed by mRNPs as they transport through the NPC in the absence of specific BSK proteins. The combination of these techniques enabled us to detail the structural and functional roles of BSK proteins within the NPC and might also serve as an effective model for further characterizing other large macromolecule complexes, such as ribosomal subunits and the transition zone in primary cilia.
Results
Auxin-Inducible Rapid Degradation of Nuclear Basket Nucleoporins from the NPC.
As reported, Nup153, Nup50, and Tpr are essential intracellular proteins with long half-lives of ∼20 to 30 h within tissue-culture cells (56). While these proteins have a long half-life, RNAi-mediated BSK-Nups degradation requires more than 72 h and multiple rounds of cell division to achieve complete protein depletion (56–58). Thus, prolonged RNAi depletion is permanently disruptive to the pore’s import-export function and partially disruptive to pore disassembly, which, as a consequence, causes a pleiotropic phenotype after nucleoporin loss that cannot be used for studying nucleoporins’ function within the NPC.
To overcome these problems, we employed an AID strategy for selective and rapid degradation of BSK-Nups (SI Appendix, Fig. S1A) described in Aksenova et al. (53). Briefly, we marked endogenous BSK-Nups in vivo utilizing the CRISPR/Cas9 system to integrate both an AID tag (∼25 kDa) and NeonGreen (NG) fluorescent proteins (∼27.5 kDa) in both alleles of the basket nucleoporins genes within the human DLD-1 cell line. Next, a TIR1 gene encoding transport inhibitor response 1 (TIR1) protein was integrated into the regulator of chromosome condensation 1 (RCC1) gene locus of homozygously AID-tagged clones (53). TIR1 acts as a subunit of the Skp1-cullin 1-F-box ubiquitin ligase complex, targeting AID-fused proteins for rapid and selective degradation upon addition of the plant hormone auxin. Concurrently, the RCC1 gene was fused to the infrared fluorescent protein, which we used to follow cells’ nuclei in vivo (SI Appendix, Fig. S2 A, C, and E). A fusion of basket nucleoporins with the fluorescent tag (NG) allowed us to follow nucleoporins degradation in live cells, estimate the precise protein depletion time, and monitor the copy number of nuclear basket nucleoporins (SI Appendix, Fig. S1 E–G). Here, to reflect the orientation of the tag, we named the DLD-1–based AID-tagged BSK-Nups as Nup50-NG-AID, Nup153-NG-AID, and AID-NG-Tpr cell lines, respectively.
First, we confirmed BSK-Nups degradation by Western blot analysis (SI Appendix, Fig. S1 B–D) and live-cell imaging (SI Appendix, Fig. S1 E–G). In addition, the quantification of the fluorescent intensity of NG-tagged BSK-NUPs on the NE shows that Tpr was depleted in 20 min, Nup153 in 40 min, and Nup50 in 60 to 70 min upon auxin treatment (SI Appendix, Fig. S1 E–G). We confirmed that Nup153 and Tpr (SI Appendix, Fig. S2 A–D), but not Nup50 (SI Appendix, Fig. S2 E and F), depleted cells, showing the growth arrest under the constant presence of auxin for more than 15 h in DLD-1 cells. To follow the immediate response of BSK-Nups loss from the basket nucleoporins to the copy number and mRNPs traffic of model mRNA substrate, we used 30 min, 60 min, and 90 min for Tpr, Nup153, and Nup50 depletion, respectively.
Single-Molecule Counting Revealed Copy Numbers for Nuclear Basket Proteins in the NPCs of Live Cells.
To determine the copy number of BSK-Nups in the NPC in vivo, we adapted the single-molecule counting approach detailed in our previous publication (14). The “counting” accuracy of the approach has been previously validated by using molecular complexes of GFP formed by controlled chemical coupling of GFP molecules. The molecular complexes used as standards contain 1 (1xGFP), 2 (2xGFP), 4 (4xGFP), 6 (6xGFP), 12 (12xGFP), and 28 molecules of GFP (28xGFP) (SI Appendix, Fig. S3). The prephotobleaching effect, the transient indistinguishable initial or intermediate photobleaching steps, the background noise from the local environment, and the ability to distinguish photobleaching steps without artificial bias have been fully considered in the methodology (14). The method has enabled us to distinguish 1, 2, 4, 6, 12, and 28 photobleaching steps in the photobleaching curves of the 1xGFP, 2xGFP, 4xGFP, 6xGFP, 12xGFP, and 28xGFP molecular standards, respectively (SI Appendix, Fig. S3). Moreover, to complement the direct-counting approach, we also calculated the total initial number of GFP molecules in each complex, by dividing the initial fluorescence intensity of the complex (at time 0 of its photobleaching curve) by the averaged intensity of an individual photobleaching step (SI Appendix, Fig. S3W).
More detailed technical information can be found in our previous publication (14), and here, we highlighted the key steps in applying the single-molecule counting approach to determine the copy numbers for BSK-Nups in live human DLD-1 cells. By using laser scanning confocal microscopy, we first scanned the NPCs labeled by Nup-NG at both the equator and the bottom of the NE in living DLD-1 cells. As shown in SI Appendix, Fig. S4 A and B, spatially well-separated and unevenly distributed fluorescent spots of Nup-NG were collected at the NE’s bottom. After fitting hundreds of these spots by 2D Gaussian function, the averaged distance between the nearest neighboring NPCs on the NE is ∼633 nm in DLD-1 cells (SI Appendix, Fig. S4 C–E), which allows for convenient illumination of single NPCs on the NE by SPEED microscopy (see Single-Molecule Tracking of Model mRNPs in Live DLD-1 Cells by SPEED Microscopy).
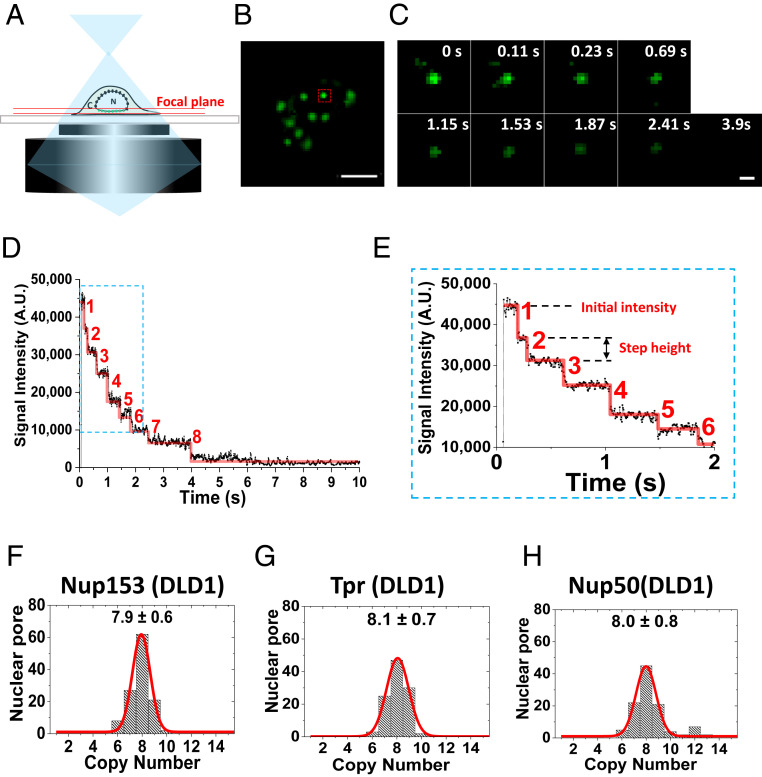
Secondly, to avoid the prephotobleaching of NG fluorescence from the NPCs in living cells, we moved the bottom of the NE to the microscope’s focal plane by using wide-field transmission white light through the cells. Then, we utilized 488-nm-laser narrow-field fluorescence illumination to burst multiple NPCs labeled by Nup-NG into fluorescence (Fig. 1 A and B). Any misalignments between these two imaging modes have been determined and corrected in our data analyses (Materials and Methods). Also, a high-speed charge-coupled device (CCD) camera was turned on to capture the initial fluorescence intensity of NG-NPCs prior to engaging the 488-nm laser to excite the NG fluorescence from NPCs. The recording continued until the NG’s fluorescence was completely photobleached by the laser (Fig. 1C). Finally, the photobleaching curves of single NG-NPCs were obtained by subtracting the integral intensity of NG-NPC by the background fluorescence intensity contributed from local environment at each time interval (Fig. 1 D and E and SI Appendix, Fig. S5 A and B). The photobleaching steps contained in the photobleaching curve were further distinguished by an established computational algorithm (59). We only selected the NPCs with at least three well-resolved photobleaching steps and then averaged these steps to acquire the average step-height fluorescence intensity (Fig. 1E). By dividing the initial fluorescence intensity of an NG-NPC by its step-height intensity, we obtained the copy numbers of BSK-Nups in this NPC. In our experiments, both the initial intensity–based calculation and the direct-counting approach were used in determining the copy number of each BSK-Nup within individual NPCs.
Fig. 1.
Copy number of BSK-NUPs counted by wide-field epi-fluorescent microscopy. (A) NG-labeled NPCs (green) on the bottom of the NE were illuminated at the focal plane (between the double red lines) by wide-field epi-fluorescent microscopy. C, cytoplasm; N, nucleus. (B) A typical image of NPCs harboring NG-fusion Nups (Nup153) proteins at the bottom of the NE in a live DLD-1 cell. Photobleaching of a single NPC (red square) is shown in C. (Scale bar: 5 μm.) (C) A typical sequential photobleaching image of Nup153-NG located in a single NPC over time. (Scale bar: 1 μm.) (D) A typical photobleaching curve for a Nup153-NG–labeled NPC in DLD-1 cells. Eight NG-fusion Nups photobleaching steps were resolved corresponding to eight copies of Nup153 per NPC. The first several transient steps were enlarged for a detailed look in E. (E) The first six transient steps were enlarged for D. (F–H) Histograms of copy numbers of NG-labeling BSK-NUPs per DLD-1 cells’ NPC achieved from the initial intensities per NG-NPC divided by the intensity of a single NG-Nups molecule. Gaussian fittings (red line) of the histograms yielded averaged copies for three BSK-NUPs in the NPC. The inset numbers denote the fitting results as peak ± SD.
From 124 NPCs labeled with Nup153-NG at the NE in 18 live DLD-1 cells, we found that the photobleaching curves of 44 NPCs revealed the maximum 8 well-resolved photobleaching steps, and the average copy number of Nup153-NG per NPC obtained from the initial-intensity calculation is 7.9 ± 0.6 (Fig. 1F). The 111 NPCs fused with NG-Tpr in 15 cells revealed an average copy number of 8.1 ± 0.7, and Nup50-NG was found to have an average copy of 8.0 ± 0.8 in 112 NPCs of 17 cells, in which 37 and 39 NPCs were equipped with the maximum 8 well-resolved photobleaching steps, respectively (Fig. 1 G and H). Additionally, the counting approach was also expanded to quantify singular Nups in the NPCs of DLD-1 cells, yielding 16.4 ± 1.7 copies for Nup88-NG (SI Appendix, Fig. S5 and S6A). Since all BSK-Nups have been fused with NG and there is no prephotobleaching occurring for NG’s fluorescence in the NPCs to obtain the maximum fluorescence, we believe that the aforementioned determined copy numbers are the maximum copies for these BSK-Nups in the NPCs of live DLD-1 cells. Conclusively, our single-molecule counting approach revealed that Nup153, Nup50, and Tpr have an average copy number of primarily 8 in DLD-1 cells and a stoichiometric ratio of 1:1:1 in live DLD-1 cells. Finally, as demonstrated, our approach is to directly determine the copy numbers within individual NPCs in live cells, which differs significantly from previous estimations of abundance ratio among these BSK-Nups based on whole isolated nuclei or NEs (5, 13). Direct scanning of hundreds of individual NPCs in their native status avoids the loss or addition of Nups during the process of isolating nuclei or NEs. Therefore, our determination of the copy numbers and stoichiometric ratio for BSK-Nups are likely to be more accurate.
Besides the aforementioned technical advances in obtaining the copies for BSK-Nups within the native NPCs, we also examined how the NG-AID tag and the Nup’s residence time at the NPC could affect the copies of BSK-Nups in our measurements. We noticed that the AID system leads to a lower abundance of Nups within the TIR1+ cells (cell integrated with TIR1 gene) but not the TIR1− cells (cell not integrated with TIR1 gene) (53). This allowed us to conclude that the NG-AID tagging itself does not influence the total amount of proteins, but TIR1 integration does. Here, to examine whether the AID-NG tag affects the copy numbers of BSK-Nups within the NPC, we quantified the copy numbers of Nup153-NG-AID, Nup50-NG-AID, and AID-NG-TPR in single NPCs of TIR1− DLD-1 cell lines. As shown in SI Appendix, Fig. S6 B–D, we found that these BSK-Nups possess the same copy numbers in both DLD-1 and TIR1− DLD-1 cells, which confirms that the NG-AID tag does not affect the copies of BSK-Nups within the NPC. Previously, BSK-Nups were reported to have distinct dynamics within the NPC (60% of Nup153 with a residence time of 26 s and the remainder with 13 min; 20 s and 3.3 min for 11 and 89% of Nup50, respectively; no data for Tpr) (60). How do these dynamics impact our measures of the copies of these BSK-Nups in live cells? As shown in Fig. 1, with our microscope setup, the eight copies of NG-labeled Nup153 and Nup50 were completely photobleached within ∼5 s, which is much shorter than the reported residence times of Nup153 and Nup50. We can therefore conclude that the dynamics of Nups are exceedingly unlikely to impact the determinations of copy numbers for these Nups.
Single-Molecule Tracking of Model mRNPs in Live DLD-1 Cells by SPEED Microscopy.
Previous studies have suggested that BSK-Nups could play critical roles in mediating mRNP export through NPCs (15, 18, 31, 32, 46, 48). However, it remains obscure whether Nup153, Nup50, and Tpr play equal roles in the export of mRNPs. Furthermore, the question arises whether Nup153, Nup50, and Tpr can compensate for one another’s absence within the nuclear basket during nuclear export of mRNPs. To answer these questions, we measured and compared the nuclear export kinetics of model mRNPs (firefly luciferase mRNA) both in the presence and absence of the nuclear basket FG-Nups in the NPCs. Here, we will focus on three major kinetics including nuclear mRNP’s entrance frequency into the nuclear basket, nuclear export time, and successful nuclear export efficiency. The entrance frequency is defined as the number of mRNPs diffusing into the nuclear basket of NPCs from nucleoplasm per NPC per second. For these entered events, we found that they have three different destinations. First, some of them could successfully export through the NPC to reach cytoplasm. Second, some of them just abort their exports and return to nucleus. The third is that their final destinations could not be detected after entering the nuclear pore due to photobleaching or moving out of the focal plane. As a result, the events in the first two groups were used to calculate the successful and abortive nuclear export efficiency. In detail, the successful (or abortive) nuclear export efficiency is defined as the percentage of successful (or abortive) mRNP export events over the sum of successful and abortive mRNP export events. Finally, the nuclear export time is defined as the dwelling time of mRNPs within the NPCs before they enter the cytoplasm or return to the nucleus, in which the former is the successful nuclear export time and the latter the abortive nuclear export time.
We began the nuclear export experiments by firstly transfecting Nup153-NG-AID DLD-1 cell lines with mCherry-labeled firefly luciferase mRNPs (SI Appendix, Fig. S7A and Materials and Methods). By following the established protocol of labeling mRNPs (43, 46), the 3′ end of mRNA is fused with 24× MS2 loops, and then mCherry-tagged MS2 coating proteins (MCP) linked with nuclear localization signal recognize these loops in dimers (Materials and Methods). We found that the actual labeling ratio for mRNPs in live Nup153-NG-AID DLD-1 cells was determined to be about eight copies of mCherry per mRNP by using the single-molecule counting method (SI Appendix, Fig. S7B). This result agreed well with the labeling ratio of MCP:mRNA in HeLa cells and also reaffirmed the relatively low binding affinity/efficiency between MCP and MS2 loops in live cells (43, 46). Additionally, we estimated that the total number of the model mRNA substrate in a DLD-1 cell is ∼12,000 and about half of them in the nucleus through the application of RNA fluorescence in situ hybridization (SI Appendix, Fig. S8 and Methods and Materials).
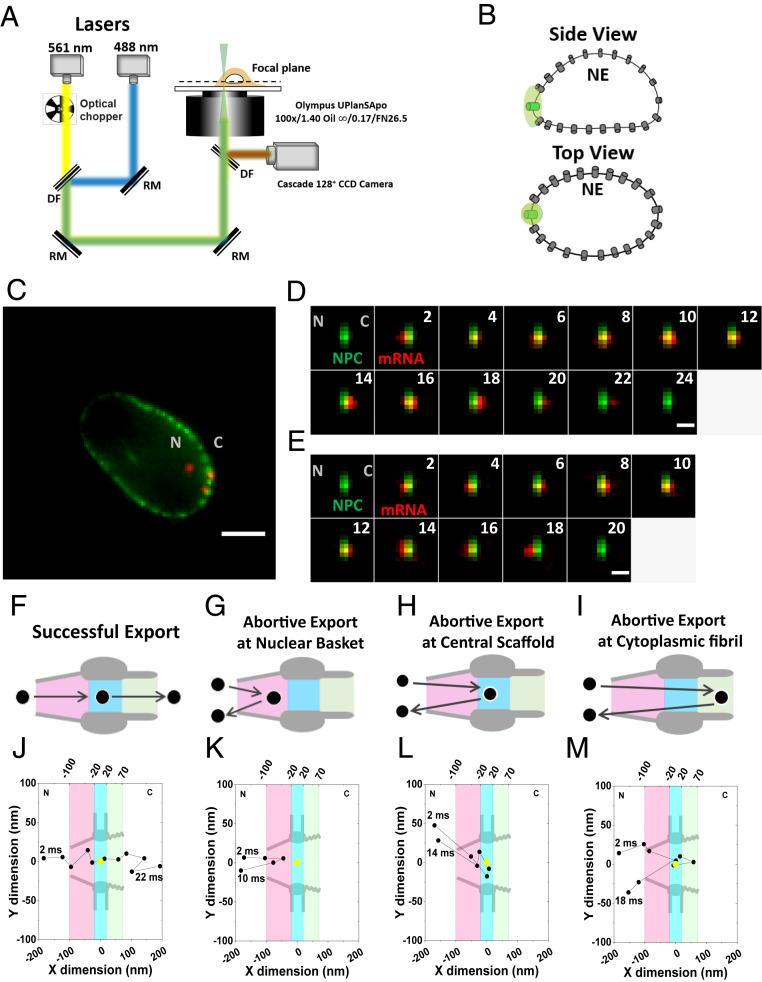
After labeling, we then tracked nuclear export events of mRNPs at the single-molecule level in live DLD-1 cells containing all nuclear basket FG-Nups (Fig. 2 and Movies S1 and S2). The individual mRNPs labeled with eight copies of mCherry can be well distinguished from free single mCherry-MCP molecules due to the significant differences in their fluorescence intensities and nuclear transport kinetics (SI Appendix, Fig. S7B and Table S2). Specifically, we employed SPEED microscopy to illuminate and localize a single NG-labeled NPC on the NE by a 488-nm laser and then tracked the trajectories of individual mCherry-labeled nuclear mRNPs interacting with the NPC via a 561-nm laser (Fig. 2 A and B). The entire interaction process of mRNPs with the NPCs was recorded by a high-speed CCD camera at 500 frames per second (Fig. 2 C–E). We found that the NPCs in the same cell are not equally active in mediating the nuclear export of the firefly luciferase mRNPs in live DLD-1 cells during the detection time, as demonstrated in the SI Appendix, Fig. S9. Specifically, some active NPCs allowed the relatively frequent entrance events of mRNPs (≥0.3 event per pore per second, Table 1), while other less active or inactive NPCs had a much lower frequency (<0.05 event per pore per second). Based upon our observations, including integrated NPCs and Nups-depletion NPCs, a rough estimation of the percentage of active NPCs as a percentage of the total NPCs is ∼18% (SI Appendix, Fig. S9). It is worth noting that this estimation could be underestimated for the following reasons. First, the estimation is not from a large-area scan of NPCs on NE. Instead, it is based on the single-point illumination of SPEED microscopy that enables us to localize an individual NPC per cell while tracking single-molecular mRNPs export through the NPC. Second, the “inactive NPCs” would be omitted particularly from Nup-depletion data where the NG-Nups were fully absent and NPCs cannot be identified by mRNPs export events due to extremely low entrance frequency.
Fig. 2.
Nuclear export of mRNPs tracked by high-speed single-molecule SPEED microscopy. (A) Optical schematic of the high-speed single-molecule SPEED microscope. The 488-nm laser was utilized to excite NG-tagged NPCs at the equatorial plane of NE. The 561-nm laser was chopped by an optical chopper to achieve an on-off laser mode with a laser-on time of 60 ms and a laser-off time of 140 ms. The longer laser-off time gives enough time for fresh fluorescent mRNPs to diffuse from the nucleus into the NPC. (B) Individual NPC was illuminated by a point spread function of high-speed single-molecule SPEED microscopy at the equatorial plane of a DLD-1 cell nucleus in the focal plane. (C) An overlay of a wide-field epi-fluorescence image of the entire NE’s equator labeled with Nup153-NG (green curved line) and a narrow-field microscopy image of several mCherry-mRNPs particles (red spots) within a ∼3-μm area at the equator of the NE. (Scale bar: 5 μm.) C, cytoplasm; N, nucleus. (D) A typical successful single mRNP export event captured by high-speed single-molecule SPEED microscopy. A single mCherry-tagged mRNP (red spot) started from the nucleus, interacted with NPC (green spot), and arrived in the cytoplasm. Numbers denote time in milliseconds. (Scale bar: 1 μm.) C, cytoplasm; N, nucleus. (E) A typical abortive single mRNP export event. A single mCherry-tagged mRNP (red spot) started from nucleus, interacted with NPC (green spot), and returned to the nucleus. Numbers denote time in milliseconds. (Scale bar: 1 μm.) C, cytoplasm; N, nucleus. (F) The model of successful mRNPs export. The mRNP (black circle) sequentially diffuses through nuclear basket (pink region), central scaffold (light blue region), and cytoplasmic fibril (light green region) of the NPC into cytoplasm. (G–I) The models for three types of mRNP’s abortive export. (J) A typical successful export trajectory of mRNA. Single-particle tracks (black dots) were acquired by 2D Gaussian fitting to point spread functions of mRNP–mCherry particles in a series of images for the successful event in D. The centriole of NPC (yellow dot) was acquired by 2D Gaussian fitting to point spread function of single NPC-NG in D. Based on NPC scaffold structure obtained from EM study, nuclear basket (pink region), central scaffold (light blue region), and cytoplasmic fibril (light green region) along the axial dimension are arbitrarily defined as 80 nm, 40 nm, and 50 nm in length, respectively. C, cytoplasm; N, nucleus. (K–M) The examples of three types of mRNA abortive export trajectories. C, cytoplasm; N, nucleus.
Table 1.
Entrance frequency of mRNPs into the nuclear basket under different conditions
| ΔTpr 140 events from 26 cells | ΔNup50 151 events from 14 cells | ΔNup153 163 events from 13 cells | Wild-type control 183 events from 17 cells | |
| Docking frequency (events × [pore × s]−1) | 0.3 ± 0.1 | 1.2 ± 0.2 | 1.4 ± 0.3 | 1.2 ± 0.2 |
| Normalized ratio | 0.25 | 1 | 1.17 | 1 |
Event denotes mRNP nuclear entrance event that mRNPs enter the nuclear basket of the NPC from the nucleus.
The aforementioned observed NPCs’ different levels of activities in mediating the nuclear export of mRNAs are consistent with the previous report that different NPCs in the same cell can differ significantly in handling the numbers of protein complexes importing from the cytoplasm to the nucleus (59). Because over 100 nuclear export events of mRNPs are needed to obtain nuclear transport kinetics and thousands of single-molecule localizations within the NPC for mRNA’s 3D transport routes with high reproducibility, the data reported here were collected from the relatively active NPCs that each had mediated multiple mRNP nuclear export events (≥3) within 10 s. By tracking mRNP export events in 17 integrated NPCs of 17 cells, ∼31% of them successfully moved through the NPC to reach the cytoplasm, and the remaining ∼69% mRNPs that entered the pore returned back to nucleus in live DLD-1 cells, thereby failing to export (Fig. 2 F and J and Table 2). For these abortive mRNP events, ∼16% of them returned to the nucleus after interacting with the nuclear basket, ∼36% of them returned after reaching the central scaffold, and ∼17% returned from the cytoplasmic side of the NPC (Fig. 2 G–I, K–M). We also noticed that these successful nuclear export events and the three types of abortive events have different interaction times with the NPC at 14 ± 2 ms, 7 ± 1 ms, 11 ± 1 ms, and 13 ± 2 ms, respectively (Table 2).
Table 2.
Export time and successful export efficiency of mRNPs under various conditions
| ΔTpr 104 events from 26 cells | ΔNup50 101 events from 14 cells | ΔNup153-ΔNup50 125 events from 13 cells | Wild-type control 183 events from 17 cells | ||
| Efficiency (%) | Successful export | 27.6 | 28.3 | 8.1 | 31.2 |
| Abortive export at nuclear basket | 20.6 | 18.0 | 71.7 | 16.0 | |
| Abortive export at central scaffold | 34.7 | 35.7 | 15.1 | 36.0 | |
| Abortive export at cytoplasmic fibril | 17.1 | 18.0 | 5.1 | 16.6 | |
| Export time (ms) | Successful export | 12.0 ± 1.0 | 12.0 ± 2.0 | 12.0 ± 2.0 | 14.0 ± 2.0 |
| Abortive export at nuclear basket | 6.0 ± 1.0 | 6.0 ± 2.0 | 4.0 ± 1.0 | 7.0 ± 1.0 | |
| Abortive export at central scaffold | 10.0 ± 2.0 | 9.0 ± 1.0 | 8.0 ± 2.0 | 11.0 ± 1.0 | |
| Abortive export at cytoplasmic fibril | 12.0 ± 1.0 | 11.0 ± 2.0 | 12.0 ± 1.0 | 13.0 ± 2.0 |
Event denotes mRNP export event that mRNPs diffuse from the nucleus into the NPC and then move into the cytoplasm or back to the nucleus from the NPC.
Nuclear Export Kinetics of mRNPs in the Absence of Nuclear Basket Proteins.
To study the impact of the absence of BSK-Nups on the nuclear export of mRNPs, we treated Nup153-NG-AID DLD-1 live cells with 1 mM auxin for 1 h to generate Nup153-free NPCs in live cells (as verified in SI Appendix, Fig. S1 C and F). As a result of Nup153-NG-AID being completely removed from the NPCs, we were unable to utilize NG fluorescence to localize the NPC and NE. Instead, transmitted white light was used to visualize and localize the NE in live cells for single-particle tracking of mRNPs across the NE (SI Appendix, Fig. S10). It is important to note that the loss of Nup153, but not Tpr, causes gradual mislocalization of Nup50 in the NPC (SI Appendix, Fig. S11 A and B). Furthermore, the removal of Tpr or Nup50 had no impact on the localization of the other, and neither affected Nup153’s localizations within the NPC (53). These observations agree with previous reports that the deficiency of Nup153 within the NPC causes mislocalization of Nup50 (57, 61). Furthermore, this confirms that Tpr is peripherally attached to the NPC and does not act as an essential anchoring site for other Nups (57), although Tpr might form the major structural scaffold of the nuclear basket (10, 62, 63). For clarification, we used ΔNup153-ΔNup50 to represent both Nup153 and Nup50 removed from the NPC.
By using the aforementioned single-molecule tracking approach, we first determined the nuclear export kinetics of mRNPs in the ΔNup153-ΔNup50 NPCs of live DLD-1 cells. As shown in Table 2, the complete degradation of Nup153 and loss of Nup50 from the nuclear periphery in the cells caused a significant reduction in the successful nuclear export events of mRNPs. However, this degradation only caused slight changes in its export time and almost no changes in its entrance frequency. In detail, the successful nuclear export efficiency of mRNPs through the NPC was reduced to ∼8.1% (a total of 125 export events collected from 14 NPC of 14 cells, Table 2). This is nearly a fourfold decrease when compared to the ∼31% rate of successful nuclear export efficiency of mRNPs through NPCs containing both Nup153 and Nup50. Consequently, the remaining ∼92% of mRNPs aborted nuclear export with different abortive rates in the NPC’s three subregions: ∼71.7% (nuclear basket), ∼15.1% (central scaffold), and ∼5.1% (cytoplasmic fibril). Compared to situations where the NPCs contained both Nup153 and Nup50 (Table 2), these subregion data suggest that the absence of Nup153 and peripheral Nup50 seriously diminishes the role of the nuclear basket in mediating the successful nuclear export of mRNPs. As for the export time, at ΔNup153-ΔNup50, almost no changes were found for the successful (12 ± 2 ms) and abortive (12 ± 2 ms) export events reaching the NPC’s cytoplasmic side. However, shorter dwelling times occurred for the abortive export at the nuclear basket (4 ± 1 ms) and for the abortive events at the central channel (8 ± 2 ms), as expected (Table 2). Lastly, the entrance frequency of mRNPs into the nuclear basket was determined to be 1.4 ± 0.3 per pore per second in ΔNup153-ΔNup50 NPCs. This is highly similar to the entrance frequencies observed when both Nup153 and Nup50 are present in the NPCs, suggesting that ΔNup153-ΔNup50 has minimal impact upon the mRNP’s entrance frequency (Table 1).
To generate ΔNup50 NPCs and ΔTpr NPCs, we treated Nup50-NG-AID DLD-1 cells and NG-AID-Tpr cells with 1 mM auxin for 90 min and 30 min, respectively (SI Appendix, Fig. S1 B, D, E, and G). As shown in Tables 1 and 2, we found that the complete removal of Nup50 from the nuclear basket did not cause significant changes in either the nuclear export time, the successful nuclear export efficiency, or the entrance frequency of mRNPs. The tests performed on Nup50 also clarified that it is Nup153 loss, not loss of Nup50 from the periphery, that significantly reduced the successful nuclear export rate of mRNPs. Moreover, we found that the absence of Tpr from the nuclear basket, although it caused no changes in nuclear export time and efficiency, greatly reduced the entrance frequency of mRNPs from 1.2 ± 0.2 to 0.3 ± 0.1 (Table 1). This decrease is approximately four times lower than observations where Tpr was present within the NPC.
In summary, these results reveal the roles that these nuclear basket FG-Nups play in mediating the docking of mRNPs at the nuclear basket as well as their movement through the nuclear pore channel. Specifically, BSK-Nups impact these functions in the following manner: 1) The successful nuclear export efficiency of mRNPs through the nuclear pore channel appear to rely heavily upon the presence of Nup153 in the NPC; 2) Tpr may only play a critical role in the docking of mRNPs on the nuclear basket; and 3) Nup50 might play negligible roles in both of these processes.
Efficient Nuclear Export of mRNA Relies on the Abundance of BSK-Nups in the NPC.
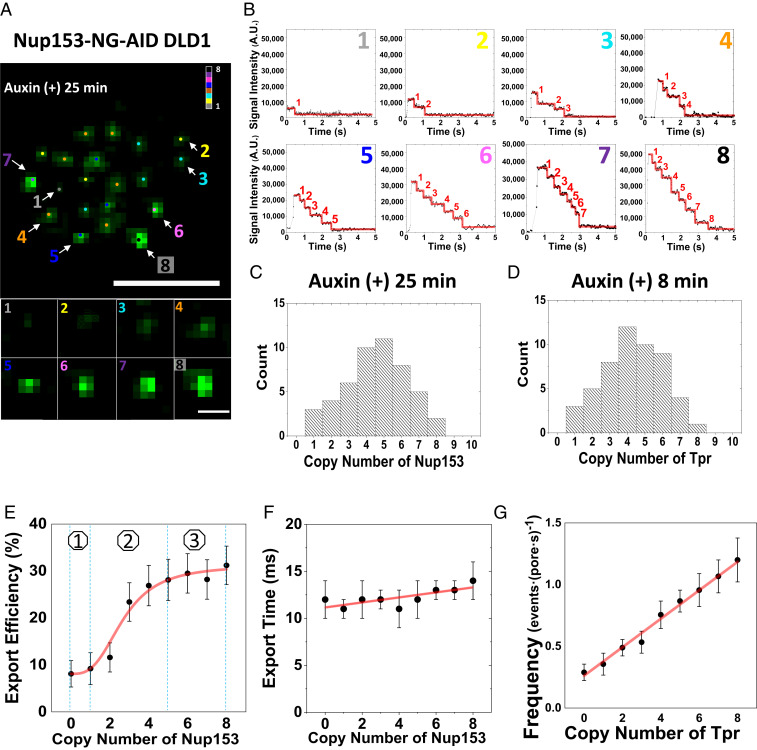
Interestingly, we also found that an earlier termination of the auxin-inducible degradation of BSK-Nups in live cells can generate NPCs containing different copies of BSK-Nups. For example, we treated Nup153-NG-AID DLD-1 live cells with 1 mM auxin for a decreased time of 25 min rather than the standard 60 min (Fig. 3A). Using the aforementioned single-molecule counting approach, we found that cells treated with the shorter incubation time contained NPCs possessing differential copies of Nup153, in which one to eight copies of Nup153 can be identified (Fig. 3 B and C). Similarly, after treating AID-NG-Tpr DLD-1 and Nup50-NG-AID DLD-1 cells for ∼8 and 40 min, respectively, we obtained NPCs containing a copy number from one to eight for either Tpr or Nup50 (Fig. 3D).
Fig. 3.
Export kinetics of mRNPs at various depletion rates of BSK-Nups. (A) The wide-field epi-fluorescent image of NPCs (green spots) harboring Nup153-NG on the bottom of DLD-1 Cell NE after 25-min auxin-inducible degradation. Different NPCs were found to contain different copy numbers of Nup153 after the treatment. (Scale bar: 5 μm.) Pointed NPCs with different copies of Nup153 are enlarged at the bottom. (Scale bar: 1 μm.) Numbers denote the copy number of Nup153-NG. (B) The photobleaching curves show one to eight copy numbers of Nup153 in the pointed eight NPCs shown in A. Numbers denote the copy number of Nup153-NG. (C) The frequency distribution for the copy number of Nup153-NG per NPC in live DLD-1 cells after 25-min auxin-inducible degradation. (D) The frequency distribution for copy number of Tpr-NG per NPC in live DLD-1 cells after 8-min auxin-inducible degradation. (E) Correlation between copy number of Nup153 and mRNP’s nuclear export efficiency in DLD-1 cells. The curve is perfectly fitted by the sigmoid function (red line) yielding the three phases: lag phase, intermediate phase, and plateau phase. (F) Correlation between the copy number of Nup153 and successfully export time of mRNPs in DLD-1 cells. (G) The frequency of mRNPs anchoring on NPCs with the copy number of Tpr from zero to eight, which is fitted with the linear function (red line).
With these NPCs containing different copies of BSK-Nups, we set out to answer a long-standing question: what are the minimum copies of a Nup in an NPC that can maintain its normal functions in mediating nucleocytoplasmic transport? By following the aforementioned imaging and analysis approach used in characterizing the nuclear export of mRNPs through the NPCs, we determined the export kinetics of mRNPs through the NPCs containing one to eight copies of Nup153. When Nup153 increased from zero to four to five copies in the NPC, as shown in Fig. 4 E and F, the successful export efficiency of mRNPs increased notably from ∼8 to ∼28%, while its export time remained almost unchanged. Interestingly, further increasing the copies of Nup153 incrementally from five to eight within the NPC did not further increase the successful nuclear export efficiency of mRNPs; instead, the export efficiency remained at ∼31%, which is functionally the same as what was observed in eight-copy Nup153 NPCs (SI Appendix, Table S3). Moreover, the relationship between the successful export efficiency of mRNPs and the copy number of Nup153 is not linear; rather, the relationship follows a sigmoid function mode including three possible phases (Fig. 3E): 1) lag phase: slight increases in mRNP’s export efficiency as the copy number of Nup153 increases from zero to one or two per NPC; 2) intermediate phase: a logarithmic growth in the successful export efficiency of mRNPs as the copies of Nup153 increased from two to five per NPC; and 3) plateau phase: the successful export efficiency of mRNPs is recovered to its normal level as the copy number of Nup153 increased from five to eight per NPC.
Fig. 4.
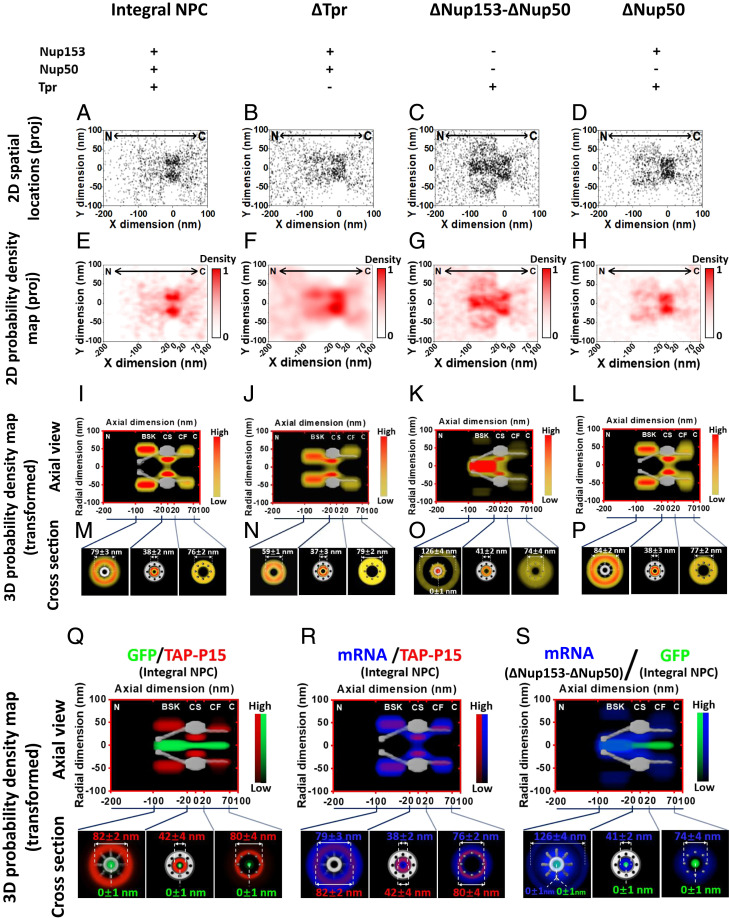
Export routes of mRNPs in the whole NPC under different conditions. (A–D) 2D spatial localizations of mRNP diffusion in and around integral NPCs, ∆Tpr NPCs, ∆Nup153 NPCs, and ∆Nup50 NPCs, respectively. The centroid of the NPC was firstly determined directly from labeled NPC and then further refined by single-molecule tracking data described in Determinations of NPC’s Centroid on page 10 and 11 of the SI Appendix as well as SI Appendix, Fig. S12. Based on the NPC structure obtained by EM study, we defined the axial length of nuclear basket, central scaffold, and cytoplasmic fibril as 80 nm, 40 nm, and 50 nm, respectively. C, cytoplasm; N, nucleus; proj, projection. (E–H) 2D probability density map of mRNP diffusion in and around integral NPCs, ∆Tpr NPCs, ∆Nup153 NPCs, and ∆Nup50 NPCs, respectively. C, cytoplasm; N, nucleus; proj, projection. Color bar shows density from low to high. (I–L) An axial view of 3D probability density maps is shown for mRNPs in NPC, including the nuclear basket, central scaffold, and cytoplasmic fibril under four conditions. C, cytoplasm; CF, cytoplasmic fibril; CS, central scaffold; N, nucleus. Color bar shows density from low to high. (M–P) The corresponding cross section of 3D probability density maps is shown for mRNPs in each subregion of NPCs under four conditions with distances between radial density peaks. (Q) The superimposed 3D probability density maps of GFP (green) and TAP-p15 (red) demonstrating the distinct routes of passive diffusion and facilitated transport in the NPC. The corresponding cross section of 3D probability density maps is shown for each subregions of NPCs with distances between radial density peaks. C, cytoplasm; CF, cytoplasmic fibril; CS, central scaffold; N, nucleus. Color bar indicates density from low to high. (R) The overlapping 3D probability density maps of mRNPs (blue) and TAP-p15 (red) showing their highly overlapped routes. The corresponding cross section of 3D probability density maps is shown for each subregion of the NPC with distances between radial density peaks. C, cytoplasm; CF, cytoplasmic fibril; CS, central scaffold; N, nucleus. Color bar shows density from low to high. (S) The superimposed 3D probability density maps of mRNPs (blue) in ∆Nup153 NPC and GFP (green) indicating mRNPs passively diffuse into the central axial region in the nuclear basket. The corresponding cross section of 3D probability density maps were shown for each subregions of NPCs with distances between radial density peaks. C, cytoplasm; CF, cytoplasmic fibril; CS, central scaffold; N, nucleus. Color bar shows density from low to high.
Similarly, we found that the entrance frequency of mRNPs increased from 0.3 ± 0.1 to 1.2 ± 0.2 per pore per second as the copy number of Tpr changed from zero to eight (SI Appendix, Table S4). Unlike the correlation between the successful export efficiency of mRNPs and the copy number of Nup153, the relationship between the nuclear entrance frequency of mRNPs and the copy number of Tpr within the NPC is a linearly dependent relationship (Fig. 3G).
3D Nuclear Export Routes of mRNPs through DLD-1 NPCs at the Absence of Nuclear Basket Proteins.
To further explore whether the export route of mRNPs through the NPC are affected by the absence of BSK-Nups, we first superimposed the spatial localizations of individual mCherry-mRNPs as they export through the NPC to obtain the 2D spatial localization distributions of mRNPs within, and around, the NPC. Next, the NPC centroid was indirectly determined (SI Appendix, Methods and Materials and Fig. S12) due to the absence of a fluorescent NPC. As shown in Fig. 4 A–D, thousands of mRNP spatial localizations from over 100 mRNP nuclear export events were obtained at different compositions of BSK proteins within the nuclear basket in live DLD-1 cells. This includes the presence of all BSK-Nups (17 NPCs of 17 different cells), ∆Tpr (26 NPCs of 26 different cells), ∆Nup153-∆Nup50 (13 NPCs of 13 different cells), and ∆Nup50 (14 NPCs of 14 different cells). As expected, 2D single-molecule trajectories of mRNPs were mainly located within the NPC and were relatively sparse at the NE (Fig. 4 A–H), a distribution that agrees with previous measurements (46). The 2D spatial distributions of mRNPs are projected from their 3D movements through the NPC. Thus, an accurate understanding of their export routes through the NPC requires a 3D view of their spatial distributions within the NPC. To this end, a previously developed 2D to 3D transformation algorithm for calculating the 3D spatial probability density maps for either passive diffusion of small molecules or transport receptor–facilitated translocation of proteins and mRNAs through the NPC was employed (SI Appendix, Figs. S12–S15 and Table S5) (64, 65). Monte-Carlo simulations previously developed by our laboratory group were employed to verify high reproducibility for all axial dimension subregion 3D transport routes (66) (SI Appendix, Table S5).
Demonstrated in Fig. 4Q, passive diffusion of signal-independent GFP mainly uses the NPC’s central axial channel, while the transport receptor Tap-p15 directly interacts with FG-Nups that mainly occupy the periphery around the central channel. Referencing these two distinct pathways, the 3D spatial locations of mRNPs within the NPC in the presence of all BSK proteins are mainly located at the peripheral regions of the central channel, a distribution pattern that is remarkably similar to the facilitated transport pathway (Fig. 4 I and M and Movie S3). An overlay of the 3D paths of mRNP and Tap-p15 reaffirmed that Tap-p15 functions as a major transport receptor by facilitating mRNP transport through the NPC (Fig. 4R and Movie S7). In the ∆Tpr NPCs, the path of mRNPs clearly changed in the nuclear basket but remained unaltered as they passed through both the central scaffold and the cytoplasmic fibrils (Fig. 4 J and N and Movie S4). In ΔNup153-ΔNup50 NPCs, similarly, the changes in mRNP pathways only occurred in the nuclear basket; however, the absence of Nup153 and peripheral Nup50 caused ∼80% of mRNPs to shift their major dwelling positions to the central axial channel (Fig. 4 K, O, and S and Movies S5 and S8). This suggests that mRNPs may passively diffuse into the central channel due to a lack of FG repeats in the ΔNup153-ΔNup50 nuclear basket. Conversely, the ΔNup50 NPCs exhibited no significant alterations in the mRNP’s nuclear export route, which holds true for all three subregions of the NPC (Fig. 4 L and P and Movie S6).
Discussion
By combining multiple advanced imaging and biological techniques, in this study, we have demonstrated an approach to count and alter copy numbers for BSK-Nups in live cells. With the altered copies of BSK proteins in the NPC, we further studied the relationship between the copy number of the BSK proteins and the nuclear export of mRNPs. Our research aims are to answer several critical questions: first, whether these BSK-Nups play equal roles in the nuclear export of mRNPs; second, whether these FG-Nups could compensate for one another’s absence during the export process of mRNPs; and third, whether the impact of BSK-Nups’ absence on mRNP export is limited to the nuclear basket or extended over the entire nuclear pore. It is worth noting that the model mRNP used in this study is firefly luciferase mRNA fused with 24 MS2 stem loops, which is believed to export through the NPCs with the assistance of transport cofactors of Tap-P15 in human cells. Our studies can only represent the best knowledge for these model mRNAs, and it requires further work if the revealed mechanism here could be expanded to other types of mRNAs or export of mRNAs that are mediated by different transport receptors such as Crm1.
Based on the measured export kinetics and 3D maps of export routes of mRNPs in the ∆Nup153-∆Nup50, ∆Nup50, and ∆Tpr NPCs in live DLD-1 cells, we found that these BSK-Nups played unequal roles and cannot compensate for one another’s absence in the nuclear export process of mRNPs. In our measurements, the absence of Tpr in the NPC directly caused almost 75% of incoming mRNPs from the nucleus to fail to dock at the nuclear basket. Meanwhile, the result also suggested that Tpr’s function was not compensated by Nup153 or Nup50 even if they possessed their full copies when Tpr was depleted in the NPC. This conclusion was further enhanced by observing that the docking frequency was fully recovered only when the full complement of Tpr copies was present in the NPC. Furthermore, we also found that the absence of Tpr in the NPC slightly altered the nuclear export route of model mRNPs after the mRNPs had entered the nuclear basket. These results together support previous conclusions that Tpr is involved in the quality control and retention of unspliced mRNAs in the nucleus (66–68). However, we did not observe changes in successful export efficiency of model mRNPs in the absence of Tpr that evidence to Tpr’s major influence is potentially limited to the docking step in the nuclear basket.
On the other hand, our data showed that Nup153 dominated the transit process for nuclear mRNPs through the NPC following docking at the nuclear basket but did not influence the docking frequency of mRNPs into the nuclear basket. This is based on the following measurements: 1) ∆Nup153 caused the successful nuclear export efficiency for mRNPs to drop from ∼31 to ∼8% but left mRNP’s docking frequency at the nuclear basket unchanged. 2) ∆Nup153 not only altered mRNP’s transit efficiency through the nuclear basket but also extended the changes through the central scaffold and the cytoplasmic side of the NPC. 3) ∆Nup153 caused the export routes of ∼80% of mRNP molecules to shift into the axial central channel of the nuclear basket from the periphery, suggesting the changed diffusion patterns for these mRNPs to be a result of changing from facilitated transport to passive diffusion. The change in mRNP’s diffusion pattern might also explain the reduction in its successful nuclear export efficiency at ∆Nup153, since the facilitated translocation enables large molecules to efficiently move through the NPC comparing to the passive diffusion. 4) The successful nuclear export rate for mRNPs was recovered as the copy of Nup153 increased from zero to approximately half of its full copies. 5) The presences of the full copies of Tpr or Nup50 while Nup153 is absent in the NPC did not rescue the nuclear export efficiency of mRNPs and the export route of mRNPs. 6) Nup50 was found to play negligible roles in the docking frequency, nuclear export rate, and nuclear export route for mRNPs, as its absence from the NPC did not cause any noticeable changes in the aforementioned processes. Together, in light of the experiments of ∆Nup153, these results revealed that it is Nup153, not Nup50, that played the major role in regulating mRNP’s nuclear export rate and export patterns. This conclusion is consistent with previous conclusions that Nup153 plays critical regulation and gating roles in the nucleocytoplasmic transport (46, 49, 52).
The distinct and critical roles that Tpr and Nup153 played in the nuclear export of mRNA could be rooted in the spatial locations of Tpr and Nup153 within the NPC as well as their structural features. First, EM data revealed that Tpr is the FG Nup in the NPC that extends farthest into the nucleus (9, 10). For nuclear mRNPs, Tpr could be an available FG Nup interacting with mRNA’s transport receptors, providing the primary docking sites, and functioning as a quality-control step. Second, based on the structures of these BSK-Nups, Nup153 contains 26 FG repeats. That is much more than Tpr (three FG repeats) and Nup50 (five FG repeats) (69–71). Correspondingly, Nup153 alone and the pair of Nup153 and Nup50 respectively contribute ∼77 and ∼91% of the total FG repeats in the nuclear basket. As a result, ∆Nup153 could jeopardize the majority of binding sites for mRNPs in the nuclear basket, which impedes the facilitated diffusion of mRNPs within the nuclear basket and induces the observed reduced rate of successful nuclear export of mRNPs through the NPC.
Additionally, we found that the nuclear export time of mRNPs was not significantly impacted by the absence of any BSK-Nups in our experiments. Given the facilitated diffusion mode adopted by mRNPs to move through the NPCs, their nuclear export time is mainly determined by their diffusion coefficients and the properties of the local microenvironment in the NPC (SI Appendix, Methods and Materials). Since neither of these factors could be noticeably changed in the absence of these BSK-Nups, the nuclear export time of mRNPs had almost no changes when these BSK-Nups were removed from the NPCs. It is also worth noting that the successful nuclear export rate (product of entrance frequency, successful nuclear export efficiency, and number of NPCs) determined at the single-molecule level might be different from the bulk nuclear export rate for mRNPs measured through other approaches, such as fluorescence recovery after photobleaching, because the former only quantifies the transport through individual NPCs that are relatively active during the detection, while the latter combines the effects of active and inactive NPCs and also the concentration, diffusion, and unspecific bindings of targeted molecules in both the nucleus and the cytoplasm besides the nuclear export behaviors through the NPCs (40–43, 53, 72).
Next, our studies also explored answers to two more questions: 1) what is the minimum required copy number for the export functionality of Tpr and Nup153 to fully recover in the nuclear export of mRNPs? And 2) what is the relationship between the copies of these BSK-Nups and the nuclear export kinetics of mRNPs? Our results revealed that Tpr and Nup153 are again distinct with regard to these questions. First, the entrance/docking frequency of mRNPs increased linearly when the copies of Tpr increased from zero to eight in live DLD-1 cells (Fig. 3G). As shown, Tpr needs to have its full copy number in the NPC to ensure its full function in handling the entrance frequency of mRNPs. However, Nup153 can recover nuclear export efficiency for mRNPs back to the maximal rate when its copy number increased to half of its full complement. Moreover, the relationship between Nup153’s copy number and the successful export efficiency of mRNPs follows a sigmoid function rather than a linear correlation. When this correlation was fitted to a sigmoid function, the relationship curve seems to include three phases: lag phase, intermediate phase, and plateau phase (Fig. 3E). In the lag phase, when Nup153 is completely absent from the nuclear basket, Nup50 cannot be present either, thereby leaving only Tpr within the nuclear basket. As a result, Tpr is the only source for FG-repeat domains in the nuclear basket. However, Tpr’s three FG repeats per copy as well as its spatially distant location, the Tpr protein situated farthest toward the nucleoplasm, would be unlikely to provide enough binding sites to facilitate mRNP movement through the nuclear basket. Interestingly, when Nup153 began to increase its copies from zero to one or two, the 26 FG repeats contained in each copy of Nup153 and the 5 FG repeats per Nup50 together increase the concentration of FG repeats in the nuclear basket. As a result, the successful nuclear export efficiency through the nuclear basket for mRNPs started to increase during the lag phase. In the intermediate phase, Nup153’s copy increased from two to four, and the concentration of FG repeats in the nuclear basket would have been increased rapidly. Notably, we found that the rate of successful nuclear export for mRNPs fully recovered at approximately four to five copies of Nup153. In the plateau phase, as the copy number of Nup153 continued to increase from five to eight, the successful nuclear export rate of mRNPs did not further increase. This suggests that the successful export rate of mRNPs may have already reached its maximum rate. These in vivo observations agree with previous in vitro tests and computational simulations that have shown that Nup153 and other FG-Nups could form an effective selectivity barrier only as its concentration reaches a certain level (73–76). Finally, we noticed that the sigmoid function that fit the relationship between the successful nuclear export rate of mRNPs and the copy number of Nup153 shares much in common with the logistic curve previously used to simulate the correlation between NPC’s effective selectivity barrier and concentrations of FG domains (74).
Materials and Methods
In our experiments, we used CRISPR/Cas9-tagged Nup153-NG-AID, AID-NG-Tpr, and Nup50-NG-AID, TIR1−, and TIR1+ DLD-1 cell lines by following the procedure published in Aksenova et al. study (53). All CRISPR/Cas9-tagged cell lines have been genotyped and tested for tagging and degradation by Western blot analysis and immunofluorescence to prove that all copies Nup gene have been fused with NG and AID tag and degrade properly. Depletion of Nups was achieved by the cultivation of cells in complete media and 5% CO2 atmosphere at 37 °C in the presence of 1 mM auxin (Sigma Aldrich). Firefly luciferase mRNA was extended by 24 MS2 stem loops on the 3′ end, and each loop was the binding target of an MCP dimer fused to mCherry. High-speed single-molecule microscopy was performed with an Olympus IX81 equipped with a 1.4-NA 100× oil-immersion apochromatic objective (UPLSAPO 100×; Olympus), a 50-mW 488-nm semiconductor laser (Coherent OBIS), a 50-mW 561-nm laser (Coherent OBIS), an on-chip multiplication gain CCD camera (Cascade 128+; Photometrics), and the Slidebook software package (Intelligent Imaging Innovations) for data acquisition and processing (77). A more detailed explanation appears in the SI Appendix, Methods and Materials.
Supplementary Material
Acknowledgments
The project was supported by grants from the US NIH (NIH GM116204 and GM22552 to W.Y.). We thank Dr. Eric Schirmer (University of Edinburgh, UK) for providing his critical comments on this manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. L.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015621118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Akey C. W., Radermacher M., Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol. 122, 1–19 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Q., Rout M. P., Akey C. W., Three-dimensional architecture of the isolated yeast nuclear pore complex: Functional and evolutionary implications. Mol. Cell 1, 223–234 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Kiseleva E., et al., Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J. Struct. Biol. 145, 272–288 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Maimon T., Elad N., Dahan I., Medalia O., The human nuclear pore complex as revealed by cryo-electron tomography. Structure 20, 998–1006 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J., Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158, 915–927 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M., Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. U.S.A. 100, 2450–2455 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryan K. J., Wente S. R., The nuclear pore complex: A protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12, 361–371 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Rout M. P., et al., The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 148, 635–651 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frosst P., Guan T., Subauste C., Hahn K., Gerace L., Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 156, 617–630 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krull S., Thyberg J., Björkroth B., Rackwitz H.-R., Cordes V. C., Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell 15, 4261–4277 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fahrenkrog B., et al., Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J. Struct. Biol. 140, 254–267 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Guan T., et al., Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol. 20, 5619–5630 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ori A., et al., Cell type-specific nuclear pores: A case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 9, 648 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mi L., Goryaynov A., Lindquist A., Rexach M., Yang W., Quantifying nucleoporin stoichiometry inside single nuclear pore complexes in vivo. Sci. Rep. 5, 9372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carmody S. R., Wente S. R., mRNA nuclear export at a glance. J. Cell Sci. 122, 1933–1937 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iglesias N., Stutz F., Regulation of mRNP dynamics along the export pathway. FEBS Lett. 582, 1987–1996 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Erkmann J. A., Kutay U., Nuclear export of mRNA: From the site of transcription to the cytoplasm. Exp. Cell Res. 296, 12–20 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Köhler A., Hurt E., Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8, 761–773 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Shatkin A. J., Manley J. L., The ends of the affair: Capping and polyadenylation. Nat. Struct. Biol. 7, 838–842 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Proudfoot N., New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16, 272–278 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Proudfoot N. J., Furger A., Dye M. J., Integrating mRNA processing with transcription. Cell 108, 501–512 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Moore M. J., Proudfoot N. J., Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Bentley D. L., Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15, 163–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng H., et al., Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127, 1389–1400 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Masuda S., et al., Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19, 1512–1517 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guzik B. W., et al., NXT1 (p15) is a crucial cellular cofactor in TAP-dependent export of intron-containing RNA in mammalian cells. Mol. Cell. Biol. 21, 2545–2554 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun I. C., Herold A., Rode M., Conti E., Izaurralde E., Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 276, 20536–20543 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Grüter P., et al., TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1, 649–659 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Segref A., et al., Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16, 3256–3271 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiegand H. L., et al., Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell. Biol. 22, 245–256 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grünwald D., Singer R. H., Rout M., Nuclear export dynamics of RNA-protein complexes. Nature 475, 333–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart M., Nuclear export of mRNA. Trends Biochem. Sci. 35, 609–617 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Braun I. C., Herold A., Rode M., Izaurralde E., Nuclear export of mRNA by TAP/NXF1 requires two nucleoporin-binding sites but not p15. Mol. Cell. Biol. 22, 5405–5418 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katahira J., Straesser K., Saiwaki T., Yoneda Y., Hurt E., Complex formation between Tap and p15 affects binding to FG-repeat nucleoporins and nucleocytoplasmic shuttling. J. Biol. Chem. 277, 9242–9246 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Herold A., Klymenko T., Izaurralde E., NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7, 1768–1780 (2001). [PMC free article] [PubMed] [Google Scholar]
- 36.Dossani Z. Y., Weirich C. S., Erzberger J. P., Berger J. M., Weis K., Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1. Proc. Natl. Acad. Sci. U.S.A. 106, 16251–16256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kendirgi F., Rexer D. J., Alcázar-Román A. R., Onishko H. M., Wente S. R., Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: A conserved mechanism in the export of Hsp70 mRNA. Mol. Biol. Cell 16, 4304–4315 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alcázar-Román A. R., Bolger T. A., Wente S. R., Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J. Biol. Chem. 285, 16683–16692 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alcázar-Román A. R., Tran E. J., Guo S., Wente S. R., Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8, 711–716 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Weirich C. S., et al., Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 8, 668–676 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Tran E. J., Zhou Y., Corbett A. H., Wente S. R., The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 28, 850–859 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Reed R., Hurt E., A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108, 523–531 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Grünwald D., Singer R. H., In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 467, 604–607 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mor A., et al., Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 12, 543–552 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Siebrasse J. P., Kaminski T., Kubitscheck U., Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 109, 9426–9431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma J., et al., High-resolution three-dimensional mapping of mRNA export through the nuclear pore. Nat. Commun. 4, 2414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnell S. J., Ma J., Yang W., Three-dimensional mapping of mRNA export through the nuclear pore complex. Genes (Basel) 5, 1032–1049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beach D. L., Salmon E. D., Bloom K., Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9, 569–578 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Ullman K. S., Shah S., Powers M. A., Forbes D. J., The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell 10, 649–664 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soop T., et al., Nup153 affects entry of messenger and ribosomal ribonucleoproteins into the nuclear basket during export. Mol. Biol. Cell 16, 5610–5620 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibata S., Matsuoka Y., Yoneda Y., Nucleocytoplasmic transport of proteins and poly(A)+ RNA in reconstituted Tpr-less nuclei in living mammalian cells. Genes Cells 7, 421–434 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Umlauf D., et al., The human TREX-2 complex is stably associated with the nuclear pore basket. J. Cell Sci. 126, 2656–2667 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Aksenova V., et al., Nucleoporin TPR is an integral component of the TREX-2 mRNA export pathway. Nat. Commun. 11, 4577 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee E. S., et al., TPR is required for the efficient nuclear export of mRNAs and lncRNAs from short and intron-poor genes. Nucleic Acids Res. 48, 11645–11663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terry L. J., Wente S. R., Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178, 1121–1132 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwanhäusser B., et al., Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Hase M. E., Cordes V. C., Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell 14, 1923–1940 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacinto F. V., Benner C., Hetzer M. W., The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 29, 1224–1238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang W., Gelles J., Musser S. M., Imaging of single-molecule translocation through nuclear pore complexes. Proc. Natl. Acad. Sci. U.S.A. 101, 12887–12892 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rabut G., Doye V., Ellenberg J., Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6, 1114–1121 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Makise M., et al., The Nup153-Nup50 protein interface and its role in nuclear import. J. Biol. Chem. 287, 38515–38522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krull S., et al., Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 29, 1659–1673 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niepel M., et al., The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Biol. Cell 24, 3920–3938 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y., Junod S. L., Ruba A., Kelich J. M., Yang W., Nuclear export of mRNA molecules studied by SPEED microscopy. Methods 153, 46–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruba A., Luo W., Kelich J., Yang W., Obtaining 3D super-resolution information from 2D super-resolution images through a 2D-to-3D transformation algorithm. bioRxiv [Preprint] (2017). 10.1101/188060 (Accessed 11 October 2017). [DOI]
- 66.Rajanala K., Nandicoori V. K., Localization of nucleoporin Tpr to the nuclear pore complex is essential for Tpr mediated regulation of the export of unspliced RNA. PLoS One 7, e29921 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Snow C. J., Paschal B. M., “Roles of the nucleoporin Tpr in cancer and aging” in Cancer Biology and the Nuclear Envelope (Springer, 2014), pp. 309–322. [DOI] [PubMed] [Google Scholar]
- 68.Coyle J. H., Bor Y.-C., Rekosh D., Hammarskjold M.-L., The Tpr protein regulates export of mRNAs with retained introns that traffic through the Nxf1 pathway. RNA 17, 1344–1356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strausberg R. L., et al., Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. U.S.A. 99, 16899–16903 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Collins J. E., et al., A genome annotation-driven approach to cloning the human ORFeome. Genome Biol. 5, R84 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Byrd D. A., et al., Tpr, a large coiled coil protein whose amino terminus is involved in activation of oncogenic kinases, is localized to the cytoplasmic surface of the nuclear pore complex. J. Cell Biol. 127, 1515–1526 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watkins L. P., Yang H., Detection of intensity change points in time-resolved single-molecule measurements. J. Phys. Chem. B 109, 617–628 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Hülsmann B. B., Labokha A. A., Görlich D., The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 150, 738–751 (2012). [DOI] [PubMed] [Google Scholar]
- 74.Ghavami A., Van der Giessen E., Onck P. R., Sol–gel transition in solutions of FG-Nups of the nuclear pore complex. Extreme Mech. Lett. 22, 36–41 (2018). [Google Scholar]
- 75.Frey S., Richter R. P., Görlich D., FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Labokha A. A., et al., Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 32, 204–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y., Tingey M., Ruba A., Yang W., High-speed super-resolution imaging of rotationally symmetric structures using SPEED microscopy and 2D-to-3D transformation. Nat. Protoc. 16, 532–560 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.