Abstract
Objective
: Developing clinical natural language processing systems often requires access to many clinical documents, which are not widely available to the public due to privacy and security concerns. To address this challenge, we propose to develop methods to generate synthetic clinical notes and evaluate their utility in real clinical natural language processing tasks.
Materials and Methods
: We implemented 4 state-of-the-art text generation models, namely CharRNN, SegGAN, GPT-2, and CTRL, to generate clinical text for the History and Present Illness section. We then manually annotated clinical entities for randomly selected 500 History and Present Illness notes generated from the best-performing algorithm. To compare the utility of natural and synthetic corpora, we trained named entity recognition (NER) models from all 3 corpora and evaluated their performance on 2 independent natural corpora.
Results
: Our evaluation shows GPT-2 achieved the best BLEU (bilingual evaluation understudy) score (with a BLEU-2 of 0.92). NER models trained on synthetic corpus generated by GPT-2 showed slightly better performance on 2 independent corpora: strict F1 scores of 0.709 and 0.748, respectively, when compared with the NER models trained on natural corpus (F1 scores of 0.706 and 0.737, respectively), indicating the good utility of synthetic corpora in clinical NER model development. In addition, we also demonstrated that an augmented method that combines both natural and synthetic corpora achieved better performance than that uses the natural corpus only.
Conclusions
: Recent advances in text generation have made it possible to generate synthetic clinical notes that could be useful for training NER models for information extraction from natural clinical notes, thus lowering the privacy concern and increasing data availability. Further investigation is needed to apply this technology to practice.
Keywords: natural language processing, neural language model, text generation, clinical notes, named entity recognition
INTRODUCTION
Natural language processing (NLP) is an important technology for unlocking unstructured patient information from clinical notes in electronic health records (EHRs) to support clinical research or practice. Currently, a few clinical NLP systems, eg, cTAKES (clinical Text Analysis and Knowledge Extraction System),1 MetaMap,2 and CLAMP,3 have been developed and applied to different clinical applications such as clinical decision support and observational studies.4–6 While developing high-performance clinical NLP systems, especially machine learning–based ones, it often requires a large number of clinical documents,7 which are often not widely available to the public due to privacy and security concerns. Current practice is usually to develop programs to remove personal identifiers in clinical notes (called de-identification). Different manual and automatic de-identification methods and systems have been developed to address this issue in the past few years.8–13 For example, the MIMIC-III (Medical Information Mart for Intensive Care) has developed hybrid approaches to de-identify textual documents in critical care settings in accordance with Health Insurance Portability and Accountability Act (HIPAA) standards, and it has been widely shared by large communities.11,12,14
Although de-identification techniques show promising performance and can alleviate the concerns regarding protected patient information to some extent, they are still insufficient to enable free sharing of data to ensure privacy guarantees.15 For example, reidentification risks still exist due to unique combinations of clinical events of a single patient. A complementary approach (ie, can be applied after de-identification) is to generate fully synthetic notes for NLP method development. Although text generation for clinical documents has been explored,16,17 none of the previous studies has evaluated the utility of these synthetic notes for clinical NLP development for common tasks such as named entity recognition (NER), one of the most fundamental and critical NLP information extraction tasks in the medical domain.
The goal of this study is 2-fold: (1) to systematically assess and compare 4 state-of-the-art text generation algorithms in the medical domain by applying them to the task of generating History and Present Illness (HPI) sections in discharge summaries; and (2) to build an annotated corpus from synthetic documents generated by the best-performing algorithm; train NER models using the synthetic corpus for identifying clinical problems, treatments, and tests; and evaluate its performance on additional annotated natural clinical corpora collected from independent sources, and thus to assess its utility in real clinical NER tasks. To the best of our knowledge, this is the first study that systematically compares different algorithms for HPI text generation and assesses the utility of synthetic clinical notes on real NLP information extraction tasks such as NER. The manually labeled synthetic corpus, together with the codes used in study, is publicly available at https://github.com/UTHealth-CCB/synthetic_hpi_ner.
RELATED WORK
Recent advances in neural network technology have greatly improved performance on text generation in the open domain. Bengio et al18 proposed the first neural network language model to explore text generation. Subsequently, a recurrent neural network language model (RNNLM) was proposed by Mikolov et al19 to solve the problem of long-distance contexts. The classical RNNLM with its improved variants, eg, long short-term memory (LSTM)20 and gated recurrent unit,21 gained popular attention and yielded promising results. However, the well-known exposure bias problem generated by maximum likelihood estimation (MLE) in RNNLM made it hard to generate satisfactory results. Bengio et al22 tried to alleviate the exposure bias problem using scheduled sampling. Another trending solution is to make use of the generative adversarial network (GAN) 23 framework together with the REINFORCE algorithm.24 In addition to RNN- and GAN-based language models and their variants, most recently, models based on various transformer architectures with attention mechanisms have achieved state-of-the-art performance in many text generation tasks.25–27
Several studies have investigated synthetic text generation in the medical domain. Guan et al16 proposed a medical text GAN (mtGAN) to generate synthetic text of electronic medical records. The mtGAN model is based on the GAN framework trained with the REINFORCE algorithm and is evaluated on the micro, macro, and application levels. For the application-level evaluation, it designed a classification experiment by comparing the models trained on the natural and synthetic data. Liu et al17 introduced a Transformer-based language modeling task 25,26 to generate clinical notes based on EHR data. In Liu et al,17 the trained language model was evaluated using different evaluation metrics and showed the utility in supporting assistive note-writing features. Ive et al28 presented a neural Transformer model to generate artificial mental health records, which were used to train a downstream text classification model that obtained comparable results as to those obtained from using the original data. Nevertheless, all these efforts were focused on specific types of neural language models (NLMs)—GAN models compared with MLE in Guan et al,16 and transformer models in Liu et al17 and Ive et al28—and lacked the systematic comparisons of state-of-the-art NLMs in the medical domain.
Furthermore, none of the previously mentioned text generation studies has investigated the utility of generated documents for the development of the NLP information extraction tasks such as NER. NER tasks such as recognizing important clinical entities (eg, problems, treatments, tests) are one of the fundamental and critical NLP tasks in the medical domain. Diverse methods and tools have already been developed for clinical entity recognition, including a number of shared tasks. For example, the 2010 i2b2/VA clinical NLP challenge on concept extraction from clinical discharge summaries is an NER task that has been widely studied by many research groups.29 So far, almost all clinical NER works are based on natural clinical notes (either de-identified or not). The use of synthetic clinical documents for medical NER tasks has not been explored, which is one of the goals of this study.
MATERIALS AND METHODS
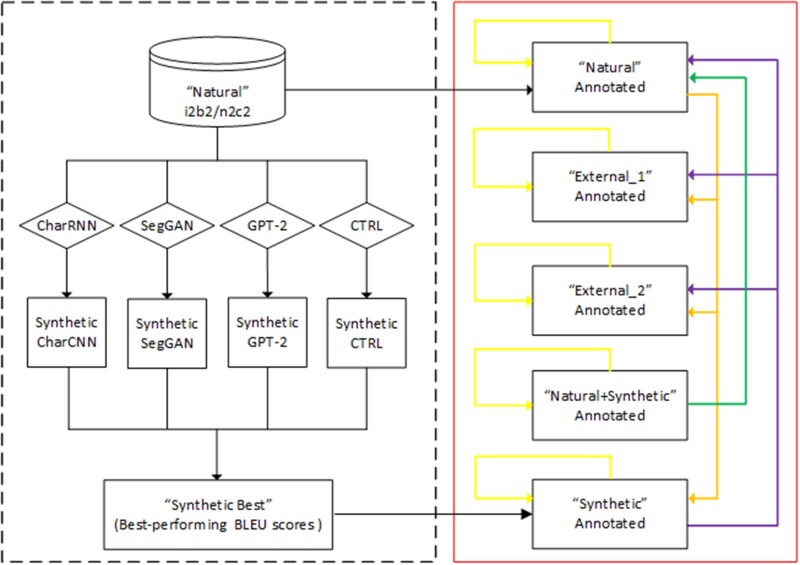
Figure 1 shows an overview of the proposed study, which consists of 2 parts. First, we implemented and compared 4 state-of-the-art text generation models, namely CharRNN (Character Recurrent Neural Network),30,31 SegGAN (Sub-sequence Generative Adversarial Network),32 GPT-2 27 (Generative Pre-Training), and CTRL (Conditional Transformer Language),33 for the task of generating text of the HPI sections. A corpus of 570 HPI sections from natural clinical notes in the i2b2 and n2c2 challenges were used to train different text generation models. The BLEU (bilingual evaluation understudy) metrics were used to evaluate different text generation methods, and the best-performing algorithm was used to generate a synthetic corpus of 500 HPI sections, which were then manually annotated for clinical entities including problems, treatments, and tests. Second, we trained NER models using the bidirectional LSTM with a conditional random field algorithm34 on the 292 HPI sections in the i2b2 challenge, with the same annotations of problems, treatments, and tests (corpus—natural), and the 500 annotated synthetic HPI sections (corpus—synthetic), and evaluated their performances on these 2 corpora, as well as on 2 external natural clinical corpora from 2 independent sources (named external_1 and external_2 respectively), to assess the utility of synthetic corpora on real clinical NER tasks. Furthermore, we trained NER models for the augmented corpus (natural training set+synthetic) and evaluated its performance on the test set of the natural corpus.
Figure 1.
Overall framework of the synthetic text generation and evaluation of clinical named entity recognition tasks. (Left box) Compare different text generation language algorithms and generate the synthetic corpus of History and Present Illness sections (BLEU [bilingual evaluation understudy] measures reported in Table 1); (right box) train named entity recognition models and evaluate their performance across different corpora: synthetic, natural, external_1, and external_2. Yellow arrows indicate 10-fold cross validation for each corpus (performance is reported in Table 3); purple arrows indicate train on the synthetic corpus and predict on test sets of natural, external_1, and external_2 (performance is reported in Table 4); orange arrows indicate train on the natural corpus and predict on test sets of synthetic, external_1, and external_2 (performance is reported in Table 5); and green arrows indicate train on the natural+synthetic corpus and predict on test sets of natural corpus (performance is reported in Table 6).
Synthetic text generation
Four state-of-the-art NLM, namely CharRNN, SegGAN, GPT-2, and CTRL, were implemented to generate clinical text for the HPI section. The following sections will describe the dataset we used for training and the 4 text generation language models that we implemented here.
HPI training data for text generation
To train the NLMs for text generation, we used discharge summaries from the 2010 i2b2/VA NLP challenge.29 That corpus contains 826 available clinical notes (after excluding notes from the University of Pittsburgh Medical Center as by the updated data user agreement), of which 292 HPI sections were extracted and used in this study. In addition, to increase the training data, we also included clinical notes from the 2018 n2c2 Shared-Task,35 which contributed additional 278 HPI sections. We combined both datasets, which resulted in 570 HPI sections from natural clinical notes, containing 9159 sentences and 149 920 tokens in total (with the average numbers of sentences and tokens per file as 16 and 263, respectively). These 570 HPI sections were used to train text generation models.
Neural language models
NLMs18 play an important role in many NLP tasks, such as machine translation, text summarization, speech recognition, and text generation. The NLMs learn to predict a probability distribution over the vocabulary given some linguistic context, ie, , where is the t-th word in the vocabulary and context can be words before and after . For our task of generating HPI sections, we have a corpus of natural HPI sections, where each document contains a sequence of words and each word comes from a vocabulary of tokens. Our goal is to generate a set of synthetic HPI sections by training a NLM to learn the underlying distribution of the natural data.
CharRNN
RNNs19 and their improved variants, eg, LSTM 20,36 and gated recurrent unit,21,37 have shown impressive results in the text generation task due to their ability to capture long-term dependencies, ie, to remember the preceding inputs using internal memory. The RNN language models are usually trained by MLE using teacher forcing.38 During the training time, RNN encodes preceding inputs to a hidden vector and makes use of the hidden vector to conduct the inference of the next word at each iteration. Character-level RNN language models30,31 have proven to work successfully in text generation, with better capability to handle the out-of-vocabulary problem (https://www.youtube.com/watch?v=B4v545V3Dq0&t=12s; Lecture 17: Issues in NLP and Possible Architectures for NLP, Stanford CS224N: NLP with Deep Learning). To study the classic RNN model in clinical text generation as a baseline framework, we apply a character-level RNN based on multilayer recurrent neural networks to train a CharRNN (https://github.com/sherjilozair/char-rnn-tensorflow) NLM for synthetic clinical text generation.
SegGAN
RNN NLMs try to predict the next word given the preceding ground-truth words and language models are usually only exposed to the training data distribution instead of their own prediction, which leads to the well-known exposure bias problem.22 Recently, the GAN 23 framework has attracted a lot of attention for text generation due to its success in image generation. The GAN model contains 2 neural networks that compete with one another: a generator G tries to generate fake data, and a discriminator D tries to classify the natural data from the fake ones. However, it’s difficult to apply GANs in text generation because the gradient from the discriminator cannot be back-propagated to the generator due to discrete text outputs. To tackle the drawback of GANs, variant GANs are developed. The SegGAN (https://github.com/liyzcj/seggan) proposed by Chen et al32 demonstrated significant improvements over state-of-the-art GAN models—SeqGAN,24 LeakGAN,39 and RelGAN40—by making the adversarial learning not only on the entire sequence, but also on the subsequences. Here, we apply the SegGAN mechanism (an improvement based on RelGAN) to train a GAN-based NLM for synthetic clinical text generation.
GPT-2
GPT-2 (https://github.com/openai/gpt-2) is a large transformer-based pretrained language model (1.5 billion parameters) published by OpenAI that shows the unprecedented capability to generate synthetic text,27 when compared with other contextual embedding models, eg, BERT41–43 and XLNet.44,45 Here, we fine-tune (https://github.com/minimaxir/gpt-2-simple) the GPT-2 model (1.5 billion parameters) on our natural clinical HPI corpus to train a GPT-2 NLM for synthetic clinical text generation. During fine-tuning, all the notes in the natural HPI corpus are concatenated with blank line, which also provides a customized character to indicate the start and end of a note. After the new model has been trained for the natural HPI corpus based on the pretrained GPT-2 language model, synthetic samples are generated, and each note is identified using the customized blank line character. Although the latest GPT-series model GPT-346 has shown improved performance on text generation, currently it is not publicly available. We plan to include GPT-3 models in our future work as soon as they become available.
CTRL
CTRL (https://github.com/salesforce/ctrl) is a 1.63-billion-parameter conditional transformer language model that was trained to condition on control codes that govern the content and task-specific behavior.33 The control codes were derived from the structure that naturally co-occurs with raw text, preserving the advantages of unsupervised learning while providing more explicit control over text generation. Like GPT-2, we fine-tune the CTRL pretrained model (256 sequence length) on the HPI corpus to train a CTRL NLM for synthetic clinical text generation.
Parameter settings
For CharRNN, we set the hyperparameter of length of sequence as 100, the number of sequences in batch as 32, and the learning rate as 0.001. For SegGAN, we set word embedding dimensions of LSTM cell as 32 and 64 for generator and discriminator, respectively, and the batch_size as 16. For GPT-2, we set the number of training epochs as 1000 and the learning rate as 0.0001; and for CTRL, we create a new control code and set the iterations for training steps as 256.
Evaluation
BLEU aims to evaluate how similar 2 sentences are and is widely used for text generation evaluation.47 To automatically evaluate the performance of the 4 text generation algorithms, we generated 500 synthetic HPI sections using each algorithm and calculated BLEU scores for n-grams of size 1 to 4 (denoted as BLEU-1, BLEU-2, BLEU-3, and BLEU-4, respectively) for each generated HPI corpus.
Evaluation of synthetic corpus on the clinical NER task
We evaluated the utility of the synthetic corpus using the clinical NER task in the 2010 i2b2/VA challenge, which is to recognize clinical problems, treatments, and tests. For the 500 HPI sections (the synthetic corpus) generated by the best-performing algorithm, we manually annotated problem, treatment, and test entities following the same guideline used in the challenge. In addition, we included 2 existing clinical corpora with the same annotations. Detailed information about these datasets and evaluation experiments is provided in the following sections.
The NER task and annotated datasets
In the 2010 i2b2/VA clinical NLP challenge, one task is to extract important clinical entities including clinical problems, treatments, and lab tests.29 This is a general clinical information extraction task, and it has been used as a benchmark for comparing different clinical NER methods in many studies.48–50 Here, we proposed to evaluate the utility of the synthetic corpus using this widely studied task.
Four annotated corpora (following the same annotation guideline as in the challenge) were included in this study, including (1) the annotated synthetic corpus: 500 HPI sections generated by the best-performing algorithm were manually annotated by experienced annotators in our group, following the guidelines used in the 2010 i2b2/VA challenge; (2) the annotated natural corpus: as described in the previous section, the raw natural corpus contains HPI sections from both the i2b2 and the n2c2 challenges, of which the 292 HPI sections from the i2b2 challenge were already annotated and were used in this evaluation here; (3) the annotated external_1 corpus: in a previous study,3 a corpus of outpatient clinic visit notes from the University of Texas Health Science Center at Houston (1351 notes) were already annotated using the same guidelines, and after we limited it to HPI sections, it resulted in an annotated corpus of 805 HPI sections as the first external corpus (external_1); and (4) the annotated external_2 corpus: a corpus of mock clinical documents from MTSamples (https://www.mtsamples.com/) (338 notes) were also annotated using the same guidelines in the previous work,3 and from that, 153 annotated HPI sections were included here as the second external corpus (external_2).
NER model training and evaluation experiments
All NER models were trained using the bidirectional LSTM with a conditional random field algorithm34 in the TensorFlow Named Entity Recognition (tf-ner) package (https://github.com/guillaumegenthial/tf_ner), which has been reported to have good performance in several clinical NER studies.51–53 We set the epochs as 25 for training all the models. As shown in Figure 1 (right box), we first evaluated the performance of models trained on each corpus itself using 10-fold cross-validation performed by dividing each dataset into train, development, and test subsets with a ratio of 80%:10%:10%, respectively. We then evaluated the performance of the NER model trained on the annotated synthetic corpus using the 3 other corpora: natural, external_1, and external_2. Models from 10-fold cross-validation experiments have different sets of optimized hyperparameters, and it is difficult to determine the best set of hyperparameters when predicting on external corpora. Our approach is to build 10 models (each with optimized hyperparameters) from 10-fold cross-validation settings and apply all of them to the external corpora via an ensemble approach, which implements a simple voting strategy to combine predicted labels from 10 models. To compare the performance between natural and synthetic, we also evaluated the performance of the NER model trained on the natural corpus using the synthetic, external_1, and external_2 corpora.
There are scenarios in which the number of available natural clinical notes is limited, which causes low performance of NER models. In that case, generating and annotating additional synthetic notes would be very helpful, if we can approve that combining annotated natural and synthetic notes can further improve the performance of NER models. Therefore, we conducted an additional experiment to evaluate an augment method: we trained NER models by combining the synthetic corpus and the training set of the natural corpus (synthetic + natural) and evaluated its performance on the test set of the natural corpus.
Strict and relaxed precision, recall, and F1 measures54 are reported for each entity type as well as for the overall performance for each NER model.
RESULTS
Table 1 shows the BLEU-1, -2, -3, and -4 scores of different text generation methods. Results show that GPT-2 achieved the best performance among 4 methods, with the highest BLEU-2 score of 0.92. Table 2 shows 2 examples of HPI sections generated by GPT-2. Both read well, although not all sentences make sense semantically (eg, admission due to medical bills in the second example).
Table 1.
Synthetic clinical notes generation performance
| Metric | CharRNN | SegGAN | GPT-2 | CTRL |
|---|---|---|---|---|
| BLEU-1 | 87.75 | 94.89 | 97.69a | 91.73 |
| BLEU-2 | 69.16 | 87.77 | 92.39a | 68.94 |
| BLEU-3 | 48.56 | 79.73 | 85.17a | 49.65 |
| BLEU-4 | 32.29 | 72.37 | 77.28a | 35.62 |
BLEU: bilingual evaluation understudy.
indicates the highest score among different text generation methods for BLEU-1, -2, 3- AND -4 respectively.
Table 2.
Sample excerpts from 2 synthetic History and Present Illness sections generated by GPT-2
| Sample excerpts | |
|---|---|
| On the day of admission, the patient was found to be unresponsive at home with recent unresponsiveness first noticed at 6 am. EMS was called to the residence and found a 75-year-old woman unresponsive with no obvious signs of intra-respiratory hemorrhage. She was given IV fluids and antibiotics and intravenous antibiotics for a possible PNA. Her temperature was 100.4. Breath sounds were not affected. Blood pressure was noted to be down in the 30s. | This is a 39 year-old female with a history of diabetes mellitus, coronary artery disease, who presents with shortness of breath and cough. It is a drought-stressed female with a history of adult-use diabetes mellitus, tobacco abuse, who presents with acute onset of chest pain since nine in the morning with chest pressure x 3 days. She is admitted now with increasing radiation damage to her home and extensive medical bills . |
Table 3 shows the performance of NER models trained and tested on each individual corpus using 10-fold cross-validation. For the same NER task, the NER model on external_1 achieved the highest strict overall F1 score of 0.859, while the model on external_2 achieved the lower strict overall F1 score of 0.767, indicating intrinsic differences among corpora from different sources.
Table 3.
Named entity recognition performances on synthetic and natural clinical corpora: synthetic refers to model trained on training dataset and tested on test dataset of synthetic corpus; natural, external_1, and external_2 all similarly refer to models trained and tested on the same corpus’ train and test datasets
| Entity | synthetic |
natural |
||||
|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Precision | Recall | F1 | |
| Problem | 0.811 (0.907) | 0.818 (0.917) | 0.814 (0.911) | 0.825 (0.921) | 0.841 (0.938) | 0.833 (0.929) |
| Test | 0.785 (0.863) | 0.779 (0.855) | 0.780(0.857) | 0.838 (0.901) | 0.821 (0.877) | 0.829 (0.888) |
| Treatment | 0.804 (0.889) | 0.789 (0.874) | 0.796 (0.881) | 0.828 (0.914) | 0.794 (0.881) | 0.810(0.896) |
| Overall | 0.804 (0.894) | 0.803 (0.893) | 0.803 (0.893) | 0.829 (0.914) | 0.827 (0.913) | 0.828 (0.914) |
| Entity | external_1 | external_2 | ||||
| Precision | Recall | F1 | Precision | Recall | F1 | |
| Problem | 0.867 (0.927) | 0.881 (0.947) | 0.874 (0.937) | 0.779 (0.899) | 0.776 (0.905) | 0.777 (0.902) |
| Test | 0.821 (0.887) | 0.779 (0.842) | 0.798 (0.863) | 0.773 (0.880) | 0.769 (0.881) | 0.770(0.879) |
| Treatment | 0.782 (0.853) | 0.809 (0.886) | 0.795 (0.869) | 0.728 (0.848) | 0.742 (0.874) | 0.734 (0.859) |
| Overall | 0.847 (0.910) | 0.859 (0.926) | 0.852 (0.918) | 0.768 (0.885) | 0.768 (0.894) | 0.767 (0.889) |
Numbers in the parentheses are results based on relaxed matching criteria.
Tables 4 and 5 show the performance of NER models that were trained on either the synthetic or natural corpus and were evaluated on the 3 remaining corpora. The synthetic corpus actually achieved slightly higher performance than that of the natural corpus on both external_1 and external_2 corpora: strict and relaxed overall F1 scores for synthetic vs natural are 0.709 (0.854) vs 0.706 (0.857) and 0.748 (0.871) vs 0.737 (0.859) on external_1 and external_2, respectively, indicating the great utility of the synthetic corpus in real NLP tasks.
Table 4.
NER performances on synthetic and natural clinical corpora: synthetic_for_natural, synthetic_for_external_1, and synthetic_for_external_2 refer to models trained from synthetic and tested on natural, external_1, and external_2 test datasets, respectively
| Entity | synth_for_natural |
synth_for_external_1 |
synth_for_external_2 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 | |
| Problem | 0.807 (0.930) | 0.784 (0.899) | 0.795 (0.914) | 0.705 (0.842) | 0.752 (0.909) | 0.727 (0.874) | 0.750 (0.869) | 0.773 (0.904) | 0.761 (0.886) |
| Test | 0.825 (0.911) | 0.724 (0.793) | 0.770 (0.848) | 0.674 (0.807) | 0.642 (0.768) | 0.657 (0.786) | 0.780 (0.882) | 0.750 (0.852) | 0.763 (0.865) |
| Treatment | 0.802 (0.893) | 0.782 (0.872) | 0.791 (0.882) | 0.682 (0.830) | 0.632 (0.770) | 0.656 (0.798) | 0.685 (0.811) | 0.708 (0.858) | 0.695 (0.832) |
| Overall | 0.809 (0.918) | 0.772 (0.872) | 0.790 (0.894) | 0.698 (0.837) | 0.720 (0.871) | 0.709 (0.854) | 0.741 (0.859) | 0.755 (0.884) | 0.748 (0.871) |
Numbers in the parentheses are results based on relaxed matching criteria.
Table 5.
NER performances on synthetic and natural clinical corpora: natural_for_synthetic, natural_for_external_1, and natural_for_external_2 refer to models trained from natural and tested on synthetic, external_1, and external_2 test datasets, respectively
| Entity | natural_for_synth |
natural_for_external_1 |
natural_for_external_2 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 | |
| Problem | 0.772 (0.866) | 0.841 (0.950) | 0.805 (0.906) | 0.703 (0.849) | 0.749 (0.920) | 0.725 (0.883) | 0.707 (0.820) | 0.800 (0.939) | 0.751 (0.875) |
| Test | 0.768 (0.841) | 0.762 (0.841) | 0.764 (0.840) | 0.605 (0.703) | 0.682 (0.803) | 0.640 (0.749) | 0.726 (0.818) | 0.736 (0.835) | 0.730 (0.826) |
| Treatment | 0.749 (0.852) | 0.787 (0.897) | 0.767 (0.874) | 0.658 (0.791) | 0.666 (0.827) | 0.662 (0.808) | 0.660 (0.778) | 0.750 (0.914) | 0.701 (0.838) |
| Overall | 0.765 (0.858) | 0.812 (0.915) | 0.788 (0.886) | 0.686 (0.825) | 0.728 (0.892) | 0.706 (0.857) | 0.700 (0.810) | 0.778 (0.915) | 0.737 (0.859) |
Numbers in the parentheses are results based on relaxed matching criteria.
Table 6 shows the performance of NER models that were trained on the augmented corpus (natural+synthetic) vs NER models that were trained on the natural corpus alone. NER models trained on the augmented corpus achieved better performance than that trained on the natural corpus only: strict (relaxed) F1 scores of 0.851 (0.927) vs 0.828 (0.914), which indicate another use of synthetic notes—to augment the natural corpus to further improve NER performance.
Table 6.
NER performances on natural+synthetic and natural corpora: natural+synthetic_for_natural and natural_for_natural refer to models trained from the augmented corpus of natural+synthetic and the natural corpus (training set), respectively, and then tested on natural test set
| Entity | natural+synthetic_for_natural |
natural_for_natural |
||||
|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Precision | Recall | F1 | |
| Problem | 0.862 (0.948) | 0.848 (0.930) | 0.855 (0.939) | 0.825 (0.921) | 0.841 (0.938) | 0.833 (0.929) |
| Test | 0.857 (0.913) | 0.850 (0.899) | 0.853 (0.905) | 0.838 (0.901) | 0.821 (0.877) | 0.829 (0.888) |
| Treatment | 0.849 (0.930) | 0.828 (0.909) | 0.838 (0.919) | 0.828 (0.914) | 0.794 (0.881) | 0.810 (0.896) |
| Overall | 0.858 (0.936) | 0.844 (0.919) | 0.851 (0.927) | 0.829 (0.914) | 0.827 (0.913) | 0.828 (0.914) |
Numbers in the parentheses are results based on relaxed matching criteria.
DISCUSSION
In this study, we first systematically investigated 4 state-of-the-art algorithms for the task of generating HPI sections and demonstrated that GPT-2 achieved the highest BLEU scores. We then annotated GPT-2–generated HPI corpus, trained deep learning–based NER models, and evaluated their performance on external clinical corpora. Our results show that the automatically generated synthetic corpus is useful for developing clinical NER models, indicating its real utility in clinical NLP development. To the best of our knowledge, this is the first work on systematic evaluation of text generation methods and its utility on NLP NER development in the medical domain.
Among the 4 different text generation methods, GPT-2 achieved the best scores on all 4 BLEU-1, -2, -3, and -4 metrics, indicating the advantages of transformer-based NLMs for text generation. NER models trained from the synthetic corpus generated by GPT-2 obtained comparable results, eg, for the natural dataset, the synthetic model achieved a strict F1 measure of 0.790, which is lower than the performance of the NER model trained with the natural corpus itself (F1 of 0.828). When applying both synthetic and natural models to external corpora (external_1 and external_2), both dropped performance significantly, probably owing to different note types in the external corpora. However, the synthetic model actually achieved slightly higher performance than that of the natural model on both external corpora, which greatly demonstrates the utility of the synthetic notes. One possible reason that synthetic model performed better than natural model on the external corpora is that the annotated synthetic corpus contains more samples than the annotated natural corpus.
Another interesting use of synthetic corpora would be to augment an existing natural clinical corpus that is with limited samples. There are scenarios in which a natural clinical corpus has limited samples (ie, hundreds of clinical documents from a shared NLP task) and end users want to train models with more samples. In that case, users can consider combining the natural dataset with the synthetically generated dataset for model training, thus achieving better performance, just as what we have demonstrated in this study, ie, natural+synthetic corpus improved F1 score by 2.3% compared with natural corpus only (F1 scores of 85.1% vs 82.8%).
We also conducted error analyses on predictions for the external_1 corpus, by both synthetic and natural models. We randomly collected 100 false positives and 100 false negatives from each model and manually reviewed those errors and categorized them into 3 classes: (1) the predicted boundary is not correct, (2) the predicted semantic type is wrong, and (3) manual annotation errors. The results of the error analysis are (1) for false positives, errors for boundary, semantic type, and annotation are (40%, 57%, 3%) and (35%, 61%, 4%) for synthetic and natural models, respectively; and (2) for false negatives, errors for boundary, semantic type, and annotation are (43%, 55%, 2%) and (47%, 50%, 3%) for synthetic and natural models, respectively. There were no obvious differences between the synthetic and natural models, in terms of error patterns.
In this study, we assume that a synthetic corpus does not contain original orders of clinical events of patients, thus avoiding potential adversarial attacks on reidentification when sharing them. However, current text generation metrics such as BLEU do not measure this aspect. To demonstrate that a synthetically generated HPI section is different from any original HPI section in natural clinical notes, in terms of the sequential pattern of mentioned clinical events, we conducted an additional analysis to compare event sequences in synthetic notes with those in the original notes. Based on annotation, each HPI section was converted into a sequence of events (problems, treatments, and tests) and an event sequence similarity (ESS) metric was introduced to measure the similarity of event sequences between a synthetic and a natural HPI section. Basically, the ESS metric is a modified BLEU without brevity-penalty, and it is based on clinical events only. Our results show that all 500 generated HPI sections have ESS scores close to zero, indicating that their event sequences are not similar to the original text at all, thus reducing reidentification risk. We are aware that the proposed analysis is not a strong measurement to ensure that the synthetic notes are not identifiable, as it is based on the order of events only. However, as the synthetic notes can be generated based on the already de-identified natural notes, we would argue that this measure provides additional insights about how synthetic notes can further reduce potential reidentification risks. Nevertheless, more in-depth investigation is necessary to further validate such advantages.
Recently, the utility of synthetic clinical data (eg, EHRs) has gained great attention. Choi et al55 proposed to synthesize EHRs data using a deep learning model called a medical GAN (medGAN). By training one neural network to generate synthetic records and another to discriminate those synthetic records from the natural records, the model can learn the distribution of both count- and binary-valued variables in the EHRs, which can then be used to produce patient-level records that preserve the analytic properties of the data. As their research is focused on discrete variable records and does not address the wealth of information embedded in clinical notes, our approach could be complementary to their work by introducing synthetic clinical notes. Of course, generating synthetic notes with correct semantic meanings that can be used for health analytics would be more challenging than that for NLP development, but it is worth exploring and pursuing.
Our current study has several limitations. Text generation algorithms have evolved rapidly. During the development of our work, the latest GPT-series model GPT-346 has been reported, with a capacity of 175 billion parameters (compared with 1557 million parameters in GPT-2) and improved performance on text generation. Nevertheless, GPT-3 is not publicly available at this time. We have submitted a request to OpenAI for accessing GPT-3, and we plan to further develop our text generation methods using this new model later. In addition, we studied the utility of synthetic clinical texts on the NER tasks only. As shown in Table 2, the GPT-2 model may not be able to generate semantically meaningful text with coherent discourse information, which may limit its usage in other NLP tasks such as timeline extraction, relation extraction, and discourse analysis. We will investigate the use of synthetic corpus in those additional NLP tasks in the future.
CONCLUSION
Recent advances in text generation have made it possible to generate synthetic clinical notes that could be used to train NER models for information extraction from natural clinical notes, thus lowering the privacy concern and increasing data availability. Further investigations are required to apply this technology into practice.
FUNDING
This research was partially supported by the National Institutes of Health under Award Numbers U01TR002062, R01AI130460, R01GM103859, and U24CA194215.
AUTHOR CONTRIBUTIONS
HX, XJ, HL, SVP, and JL conceived and designed the research; JL and YZ participated in dataset construction and annotation; JL and YZ performed the experiments and analyzed the data; JL and HX drafted the manuscript; HX, XJ, HL, SVP, and YZ offered insights and guidance for manuscript revision; all authors checked and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
HX and The University of Texas Health Science Center at Houston have financial related interests at Melax Technologies Inc.
DATA AVAILABILITY
The manually annotated synthetic data underlying this article are available at https://github.com/UTHealth-CCB/synthetic_hpi_ner.
REFERENCES
- 1.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson AR, Lang FM. The evolution of MetaMap, a concept search program for biomedical text. AMIA Annu Symp Proc2009; 2009: 22.
- 3.Soysal E, Wang J, Jiang M, et al. CLAMP–a toolkit for efficiently building customized clinical natural language processing pipelines. J Am Med Inform Assoc 2018; 25 (3): 331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ford E, Carroll JA, Smith HE, Scott D, Cassell JA.. Extracting information from the text of electronic medical records to improve case detection: a systematic review. J Am Med Inform Assoc 2016; 23 (5): 1007–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perera S, Sheth A, Thirunarayan K, Nair S, Shah N. Challenges in understanding clinical notes: Why NLP engines fall short and where background knowledge can help. In: Proceedings of the 2013 International Workshop on Data Management & Analytics for Healthcare; 2013: 21–6.
- 6.Wu Y, Warner JL, Wang L, Jiang M, Xu J, Chen Q. Discovery of noncancer drug effects on survival in electronic health records of patients with cancer: a new paradigm for drug repurposing. JCO Clin Cancer Inform 2019; 3: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spasic I, Nenadic G.. Clinical text data in machine learning: Systematic review. J Med Internet Res 2020; 8 (3): e17984. doi:10.2196/17984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douglass M, Clifford GD, Reisner A, Moody GB, Mark RG.. Computer-assisted de-identification of free text in the MIMIC II database. In: Computers in Cardiology, 2004. New York: IEEE; 2004: 341–4. [Google Scholar]
- 9.Gupta D, Saul M, Gilbertson J.. Evaluation of a Deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am J Clin Pathol 2004; 121 (2): 176–86. [DOI] [PubMed] [Google Scholar]
- 10.Gobbel GT, Garvin J, Reeves R, et al. Assisted annotation of medical free text using RapTAT. J Am Med Inform Assoc 2014; 21 (5): 833–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dernoncourt F, Lee JY, Uzuner O, Szolovits P.. De-identification of patient notes with recurrent neural networks. J Am Med Inform Assoc 2017; 24 (3): 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3: 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich M, Köhn A, Wiedemann G, Biemann C. Adversarial learning of privacy-preserving text representations for de-identification of medical records. In: Proceedings of the 57th Annual Meeting of the Association of Computational Linguistics. Florence, Italy: Association for Computational Linguistics; 2019: 5829–39. doi:10.18653/v1/p19-1584.
- 14.Beaulieu-Jones BK, Wu ZS, Williams C, et al. Privacy-preserving generative deep neural networks support clinical data sharing. Circ Cardiovasc Qual Outcomes 2019; 12 (7): 1–10. doi:10.1161/CIRCOUTCOMES.118.005122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shokri R, Stronati M, Song C, Shmatikov V. Membership inference attacks against machine learning models. In: 2017 IEEE Symposium on Security and Privacy. New York: IEEE; 2017: 3–18.
- 16.Guan J, Li R, Yu S, Zhang X. Generation of synthetic electronic medical record text. In: Proceedings - 2018 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2018; 2019: 374–80. doi:10.1109/BIBM.2018.8621223
- 17.Liu PJ. Learning to write notes in electronic health records. arXiv, doi: http://arxiv.org/abs/1808.02622, 8 Aug 2018.
- 18.Bengio Y, Ducharme R, Vincent P, Jauvin C.. A neural probabilistic language model. J Mach Learn Res 2003; 3 (Feb): 1137–55. [Google Scholar]
- 19.Mikolov T, Karafiát M, Burget L, Černocký J, Khudanpur S. Recurrent neural network based language model. In: Eleventh Annual Conference of the International Speech Communication Association. 2010: 1045–8.
- 20.Hochreiter S, Schmidhuber J.. Long short-term memory. Neural Comput 1997; 9 (8): 1735–80. [DOI] [PubMed] [Google Scholar]
- 21.Cho K, Van Merriënboer B, Gulcehre C, et al. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv, doi: http://arxiv.org/abs/1406.1078, 3 Sep 2014, preprint: not peer reviewed.
- 22.Bengio S, Vinyals O, Jaitly N, Shazeer N. Scheduled sampling for sequence prediction with recurrent neural networks. In: NIPS ’15: Proceedings of the 28th International Conference on Neural Information Processing Systems – Volume 1; 2015: 1171–9.
- 23.Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. In: NIPS ’14: Proceedings of the 27th International Conference on Neural Information Processing Systems – Volume 2; 2014: 2672–80.
- 24.Yu L, Zhang W, Wang J, Yu Y. Seqgan: Sequence generative adversarial nets with policy gradient. In: AAAI ’17: Proceedings of the Thirty-First Conference on Artificial Intelligence; 2017: 2852–8.
- 25.Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. In: NIPS ’17: Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017: 6000–10.
- 26.Liu PJ, Saleh M, Pot E, et al. Generating Wikipedia by summarizing long sequences. In: 6th International Conference on Learning Represent ICLR 2018 - Conference Track Proceedings; 2018.
- 27.Radford A, Wu J, Child R, Luan D, Amodei D, Sutskever I.. Language models are unsupervised multitask learners. OpenAI Blog 2019; 1 (8): 9. [Google Scholar]
- 28.Ive J, Viani N, Kam J, et al. Generation and evaluation of artificial mental health records for natural language processing. NPJ Digit Med 2020; 3 (1): 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uzuner Ö, South BR, Shen S, DuVall SL.. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18 (5): 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung J, Ahn S, Bengio Y. Hierarchical multiscale recurrent neural networks. arXiv, doi: http://arxiv.org/abs/1609.01704, 9 Mar 2017, preprint: not peer reviewed.
- 31.Ling W, Luís T, Marujo L, et al. Finding function in form: compositional character models for open vocabulary word representation. In: Conference Proceedings - EMNLP 2015 Conf Empir Methods Nat Lang Process. 2015: 1520–30. doi:10.18653/v1/d15-1176
- 32.Chen X, Li Y, Jin P, et al. Adversarial sub-sequence for text generation. arXiv, doi: http://arxiv.org/abs/1905.12835, 30 May 2019, preprint: not peer reviewed.
- 33.Keskar NS, McCann B, Varshney LR, Xiong C, Socher R. CTRL: A Conditional Transformer Language Model for Controllable Generation. arXiv, doi: http://arxiv.org/abs/1909.05858, 20 Sep 2019, preprint: not peer reviewed.
- 34.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural architectures for named entity recognition. In: 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies NAACL HLT 2016 - Proceedings Conference; 2016: 260–70. doi:10.18653/v1/n16-1030
- 35.Henry S, Buchan K, Filannino M, Stubbs A, Uzuner O.. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J Am Med Inform Assoc 2020; 27 (1): 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sundermeyer M, Schlüter R, Ney H. LSTM neural networks for language modeling. In: INTERSPEECH 2012: 13th Annual Conference of the International Speech Communication Association; 2012; 194–7.
- 37.Chung J, Gulcehre C, Cho K, Bengio Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv, doi: http://arxiv.org/abs/1412.3555, 11 Dec 2014, preprint: not peer reviewed.
- 38.Williams RJ, Zipser D.. A learning algorithm for continually running fully recurrent neural networks. Neural Comput 1989; 1 (2): 270–80. [Google Scholar]
- 39.Guo J, Lu S, Cai H, Zhang W, Yu Y, Wang J. Long text generation via adversarial training with leaked information. In: 32nd AAAI Conference on Artificial Intelligence AAAI 2018; 2018: 5141–8.
- 40.Nie W, Narodytska N, Patel AB. Relgan: Relational generative adversarial networks for text generation. In: 7th International Conference on Learning Representations, ICLR 2019; 2019.
- 41.Devlin J, Chang MW, Lee K, Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers); 2019: 4171–86.
- 42.Wang A, Cho K. BERT has a mouth, and it must speak: BERT as a Markov random field language model. arXiv, doi: http://arxiv.org/abs/1902.04094, 11 Feb 2019, preprint: not peer reviewed.
- 43.Cho K. BERT has a mouth and must speak, but it is not an MRF. 2019. https://sites.google.com/site/deepernn/home/blog/amistakeinwangchoberthasamouthanditmustspeakbertasamarkovrandomfieldlanguagemodel. Accessed May 1, 2020.
- 44.Yang Z, Dai Z, Yang Y, Carbonell J, Salakhutdinov R, Le QV. XLNet: Generalized autoregressive pretraining for language understanding. Adv Neural Inf Process Syst 2019; 32 (NeurIPS): 1–18. [Google Scholar]
- 45.Rusia A. XLNet speaks. Comparison with GPT-2. 2019. https://amanrusia.medium.com/xlnet-speaks-comparison-to-gpt-2-ea1a4e9ba39e. Accessed May 1, 2020.
- 46.Brown TB, Kaplan J, Ryder N, et al. Language models are few-shot learners. arXiv preprint arXiv:2005.14165. 2020.
- 47.Papineni K, Roukos S, Ward T, Zhu W-J. BLEU: a method for automatic evaluation of machine translation. In: Proceedings of the 40th Annual Meeting on Association for Computational Linguistics; 2002: 311–8.
- 48.Jiang M, Chen Y, Liu M, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc 2011; 18 (5): 601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang B, Wu Y, Jiang M, Chen Y, Denny JC, Xu H.. A hybrid system for temporal information extraction from clinical text. J Am Med Inform Assoc 2013; 20 (5): 828–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Yang M, Wang X, et al. Entity recognition from clinical texts via recurrent neural network. BMC Med Inform Decis Mak 2017; 17 (2): 53–61. doi:10.1186/s12911-017-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chalapathy R, Borzeshi EZ, Piccardi M. Bidirectional LSTM-CRF for clinical concept extraction. In: Proceedings of the Clinical Natural Language Processing Workshop, ClinicalNLP 2016; 2016: 7–12. [Google Scholar]
- 52.Zhu H, Paschalidis IC, Tahmasebi A. Clinical concept extraction with contextual word embedding. arXiv, http://arxiv.org/abs/1810.10566, 26 Nov 2018, preprint: not peer reviewed.
- 53.Jauregi Unanue I, Zare Borzeshi E, Piccardi M.. Recurrent neural networks with specialized word embeddings for health-domain named-entity recognition. J Biomed Inform 2017; 76 (September): 102–9. [DOI] [PubMed] [Google Scholar]
- 54.Stubbs A, Kotfila C, Uzuner Ö.. Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/UTHealth shared task Track 1. J Biomed Inform 2015; 58: S11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi E, Biswal S, Malin B, Duke J, Stewart WF, Sun J.. Generating multi-label discrete patient records using generative adversarial networks. Proc Mach Learn Res 2017: 68: 286–305. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The manually annotated synthetic data underlying this article are available at https://github.com/UTHealth-CCB/synthetic_hpi_ner.