Abstract
Objective
The study sought to conduct an informatics analysis on the National Evaluation System for Health Technology Coordinating Center test case of cardiac ablation catheters and to demonstrate the role of informatics approaches in the feasibility assessment of capturing real-world data using unique device identifiers (UDIs) that are fit for purpose for label extensions for 2 cardiac ablation catheters from the electronic health records and other health information technology systems in a multicenter evaluation.
Materials and Methods
We focused on data capture and transformation and data quality maturity model specified in the National Evaluation System for Health Technology Coordinating Center data quality framework. The informatics analysis included 4 elements: the use of UDIs for identifying device exposure data, the use of standardized codes for defining computable phenotypes, the use of natural language processing for capturing unstructured data elements from clinical data systems, and the use of common data models for standardizing data collection and analyses.
Results
We found that, with the UDI implementation at 3 health systems, the target device exposure data could be effectively identified, particularly for brand-specific devices. Computable phenotypes for study outcomes could be defined using codes; however, ablation registries, natural language processing tools, and chart reviews were required for validating data quality of the phenotypes. The common data model implementation status varied across sites. The maturity level of the key informatics technologies was highly aligned with the data quality maturity model.
Conclusions
We demonstrated that the informatics approaches can be feasibly used to capture safety and effectiveness outcomes in real-world data for use in medical device studies supporting label extensions.
Keywords: informatics analysis, medical device evaluation, cardiac ablation catheters, real-world evidence, RWE, unique device identifier, UDI
INTRODUCTION
With the increasing availability of digital health data and wide adoption of electronic health records (EHRs), there is an opportunity to capture and analyze real-world data (RWD) to generate real-world evidence (RWE) from health information technology (IT) systems for evaluations of medical product safety and effectiveness.1 Under the 21st Century Cures Act, signed into law in 2016,2 the Food and Drug Administration (FDA) has been tasked with developing a program to evaluate the use of RWE to support approval of expanded indications for approved medical products or to meet postmarket surveillance requirements. In the FDA guidance document focused on medical devices, RWE is defined as the clinical evidence regarding the usage and potential benefits and risks of a medical product derived from the analysis of RWD.3 In particular, the FDA and the medical device evaluation community have envisioned a system that can not only promote patient safety through earlier detection of safety signals,4 but also generate, synthesize, and analyze evidence on real-world performance and patient outcomes in situations in which clinical trials are not feasible.5
In this context, the FDA created the National Evaluation System for health Technology Coordinating Center (NESTcc),6 which seeks to support the sustainable generation and use of timely, reliable, and cost-effective RWE throughout the medical device life cycle, using RWD that meet robust methodological standards. As part of the funding commitment to NESTcc from the FDA and the Medical Device User Fee Amendment, some of the pilot projects (or test cases) need to focus on medical devices that are either in the premarket approval (PMA) or 510k phase of the total product life cycle. Test cases that can generate regulatory grade data that are fit for purpose and can support a regulatory submission will prove the strength and reliability of RWD as an effective and alternative method to traditional clinical trials.7 The intent of NESTcc is to provide accurate and detailed information regarding medical devices including the identification of devices that may result in adverse events and act as a neutral conduit reporting on device performance in clinical practice. Notably, the NESTcc has released a Data Quality Framework8 developed by its Data Quality Working Committee to be used by all stakeholders across the NESTcc medical device ecosystem, laying out the foundation for the capture and use of high-quality data for evaluation of medical devices. The framework focuses on the use of RWD generated in routine clinical care, instead of data collected specifically for research or evaluation purposes.
The goal of this article is to conduct an analysis to demonstrate the role of informatics approaches in data capture and transformation in a NESTcc test case, helping to determine if these data are of sufficient relevance, reliability, and quality to generate evidence evaluating the safety and effectiveness of target devices. The NESTcc test case study aimed to explore the feasibility of capturing RWD from the EHRs and other health IT systems of 3 NESTcc Network Collaborators (Mercy Health, Mayo Clinic, and Yale New Haven Hospital [YNHH]), including data from hospital EHRs, and determining whether RWD are fit for purpose for postmarket evaluation of outcomes when 2 ablation catheters were used in new populations and to support submissions to the FDA for indication expansion. The study was proposed to the NESTcc by Johnson & Johnson (New Brunswick, NJ), with the objective of evaluating the safety and effectiveness of 2 cardiac ablation catheters when used in routine clinical practice. The specific catheters of interest are the ThermoCool Smarttouch catheters, initially approved by the FDA in February 2014, and the ThermoCool Smarttouch Surround Flow catheters, initially approved by the FDA in August 2016. The hypotheses of the NESTcc test case are whether the safety and effectiveness of versions of ThermoCool catheters that do not have a labeled indication for ventricular tachycardia (VT) are noninferior to ThermoCool catheters that already have such an FDA approved indication, and similarly versions of ThermoCool catheters that do not have labeled indications for persistent atrial fibrillation (AF) are noninferior to those that do.
Background
Unique device identifiers for device exposure data capture
Data standardization is key for documentation of and linking medical device identification information to diverse data sources.9,10 The FDA has recognized the need to improve the tracking of medical device safety and performance, with implementation of unique device identifiers (UDIs) in electronic health information as a key strategy.11 Notably, the FDA initiated the regulation of the UDI implementation and established a Global Unique Device Identification Database 12 for making unique medical device identification possible. By September 24, 2018, all Class III and Class II devices were required to bear a permanent UDI. Meanwhile, a number of demonstration projects have demonstrated the feasibility of using informatics technology to build a medical device evaluation system and to identify keys to success and challenges of achieving targeted goals.10,11,13,14 These projects served as the proof of concept that UDIs can be used as the index key to combine device and clinical data in a database useful for device evaluation.
Common data models for standardized data capture and analytics
A variety of data models have been developed to provide a standardized approach to store and organize clinical research data.15 These approaches often support query federation, which is the ability to run a standardized query within separate remote data repositories and facilitate the conduct of distributed data analyses where each healthcare system keeps its information, yet a standardized analysis can be conducted across multiple healthcare systems. Examples of these models include the FDA Sentinel Common Data Model (CDM),16 the Observational Medical Outcomes Partnership (OMOP) CDM,17 the National Patient-Centered Research Networks (PCORnet) CDM,18 the Informatics for Integrating Biology and the Bedside (i2b2) Star Schema,19 and the Accrual to Clinical Trials ACT model.20 However, the applicability of CDMs to medical device studies, particularly whether there is sufficient granularity of device identifiers and aggregate codes for procedures, is an unanswered question. We described and analyzed the CDM implementation status at each site and assessed their potential contributions to the data quality maturity model.
NESTcc data quality framework
The NESTcc data quality framework6 focuses primarily on the use of EHR data in the clinical care setting, and is composed of 5 sections which cover data governance, characteristics, capture and transformation, curation, and the NESTcc data quality maturity model. The NESTcc data quality maturity model addresses the varying stages of an organization’s capacity to support these domains, which allows collaborators to indicate progress toward achieving optimal data quality. Supplementary Table S1 shows the description and core principles of the 5 sections in the framework.
MATERIALS AND METHODS
In this study, we analyzed the successes and challenges of acquiring RWD that are fit for purpose for evaluation of outcomes from 2 ablation catheters, focusing on data capture and transformation and the data quality maturity model (as defined in the NESTcc data quality framework) from an informatics analysis perspective, while also highlighting differences between data quality and fit for purpose in RWD studies of medical devices. The informatics analysis included the use of UDIs for identifying device exposure data, the use of standardized codes (eg, International Classification of Diseases [ICD], Current Procedural Terminology [CPT], RxNorm) to define computable phenotypes that could identify study cohorts, covariates and outcome endpoints accurately, the use of natural language processing (NLP) for capturing unstructured data elements from clinical data systems, and the use of CDMs for standardizing data collection and analyses (Supplementary Table S2).
Use of UDIs for identifying device exposure data
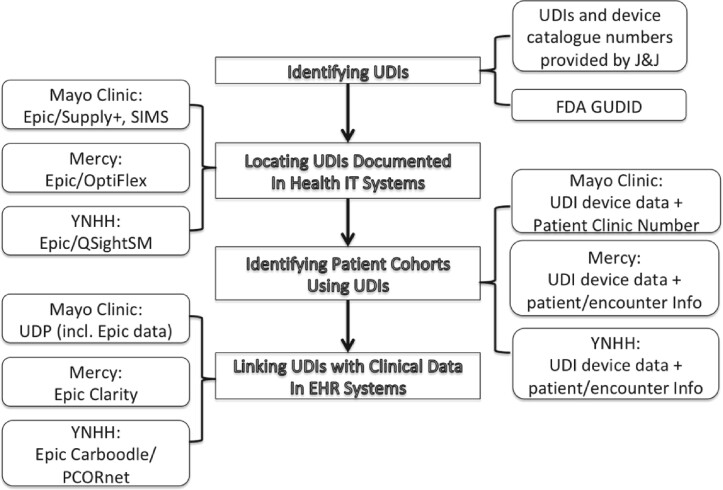
We identified a typical process (see Figure 1) for the use of UDIs for collecting device exposure data and described it as follows.
Figure 1.
A data flow diagram illustrating a typical process for the use of unique device identifiers (UDIs) for collecting device exposure data and clinical data. EHR: electronic health record; GUDID: Global Unique Device Identification Database; IT: information technology; J&J: Johnson & Johnson; PCORnet: Patient-Centered Research Network; YNHH: Yale New Haven Hospital.
Identifying UDIs for target devices. In this study, the UDIs and device catalogue numbers of the target devices were identified and provided by Johnson & Johnson. The FDA Global Unique Device Identification Database was used for the UDI identification. The rationale for relying on UDIs is that target devices are ThermoCool devices, which are brand specific, and the hypotheses to be tested involved comparing 2 different versions of the ThermoCool catheters (ie, those with vs those without the target label). A collection of UDIs for each of brand-specific devices was used to capture related device data.
Locating UDIs documented in the health IT systems in each site. At the Mayo Clinic, UDIs are documented in different health IT systems. As Epic EHR system (Epic Systems, Verona, WI) was introduced as of May 2018, the UDI-linked device data after May 2018 are documented in the Supply+ (Cardinal Health, Dublin, OH) and Plummer (Epic), which have worked together to standardize multiple clinical and business processes to improve efficiency and optimize inventory. Supply+ (Cardinal Health) is an enterprise-wide, integrated inventory management system to implement standardized surgical and procedure inventory management. Historical device data dating back to January 2014 are documented in the Mayo Clinic supply chain management system known as SIMS. SIMS was a Mayo-designed and supported system to improve surgical case management and Mayo Group Practices across Mayo enterprise. At Mercy, manufacturer numbers and UDIs were used to extract the devices of interest from Mercy’s OptiFlex (Omnicell, Mountain View, CA) point of care barcode scanning system for devices used after 2016—the year this system was installed. To pull device-related data prior to 2016 (before OptiFlex was installed), Mercy identified procedures linked to patient information using HCPCS (Healthcare Common Procedure Coding System) codes and Mercy-specific charge codes for device billing. At YNHH, device data elements were captured within the QSightSM (Owens and Minor, Mechanicsville, VA) inventory management system, in use since October 2017, during the normal course of clinical care and administrative activities.
Identifying patient cohorts with device exposure using the UDIs. At the Mayo Clinic, the UDI-linked device data are documented in patient level, and a unique patient clinic number (that is used across enterprise health IT systems, including patient medical records) can be retrieved with this linkage for patient cohort identification. At Mercy, the device data were joined with transaction data to obtain patient and encounter information for each instance of device use. At YNHH, device-related records extracted from QSightSM were linked via transaction data to procedure encounter records within the Epic EHR to verify the specific use of the device and to link with the clinical record.
Linking UDIs with clinical data (eg, procedures of interest) in EHR systems. At the Mayo Clinic, the Unified Data Platform (UDP) has been implemented to provide practical data solutions and creates a combined view of multiple heterogeneous EHR data sources (including Epic) through effective data orchestration, along with a number of data marts based on CDMs. The UDP serves as a data warehouse that contains millions of patient data points for the support of both clinical practice and research. The UDP is updated in real time, data are cleaned, and many of the medical elements are matched with standard medical terminologies such as ICD and SNOMED CT (Systematized Nomenclature of Medicine Clinical Terms) codes. The UDP was used to collect device-related EHR data. We used all of the patient clinic numbers of the device users as the identifiers to extract the data from UDP. At Mercy, device data were joined to ablation procedure data based on patient ID and the dates of procedure in order to examine the device usage during procedures. Mercy utilized Epic Clarity to research the presence of the various diagnosis and procedural codes relevant to the study. At YNHH, clinical data from the EHR are populated by a vendor-provided extract, transform, and load (ETL) process from a nonrelational model into a relational model (Clarity), followed by a second vendor-provided ETL to create a clinical data warehouse (Caboodle). Data were transformed from the Caboodle data warehouse into the PCORnet common data model and analyzed within the YNHH data analytics platform.21 The Caboodle-PCORnet ETL process removes test patients and also standardizes the representation of certain elements such as dates and encounter types.
Use of standard codes for defining computable phenotypes
For the NESTcc test case study, standardized codes were used to define the algorithms to compute phenotypes that could identify target indications (ie, either VT or persistent AF using ICD codes), procedures of interest (ie, cardiac ablation for either VT or persistent AF using CPT codes), outcome endpoints (eg, ischemic stroke, acute heart failure, and rehospitalization with arrhythmia using ICD codes), and covariates of interest (eg, therapeutic drugs using RxNorm codes). Supplementary Table S3 provides a list of phenotype definitions using standardized codes. These standardized codes serve as a common language that would reduce ambiguous interpretation of the algorithm definitions across sites.
Data quality validation
Data quality validation using registry data and chart review is an important component in the study design. In particular, it is well recognized in the research community that the accuracy of phenotype definitions based on simple ICD codes is not optimal, except for markers of healthcare utilization,22 such that these codes cannot be used as a “gold standard.” We found that clinical registry data (if available) constitute a very valuable resource to enable efficient data quality check, if the variables of interest are similar between the real-world study and registry. The Mayo Clinic utilized this internal registry as a data validation source, and the AF cases were classified as paroxysmal, persistent, or longstanding persistent by physicians through a manual review process (note that the physician-based confirmation was done as part of registry-building process, not as a separate effort for this research study).
Use of NLP for unstructured clinical data
In the NESTcc test case, the NLP technology is used in the following aspects.
First, Mercy used a previously validated NLP algorithm to validate AF patient phenotypes. As Mercy does not participate in an AF registry, an NLP tool and validated dataset were used as the gold standard for validation of the extracted data. Specifically, Linguamatics (IQVIA, Danbury, CT) software was utilized within Mercy’s Hadoop warehouse for NLP. This tool was built and validated on a group of patients who were diagnosed with arrhythmia and stroke for a previous Johnson & Johnson project. All EHR notes of those patients were queried and validated for their AF diagnoses. We used this group of patients as our test case to validate ICD codes for the following 3 AF types: paroxysmal, persistent, and chronic. The diagnoses defined by the previously developed NLP tool served as the gold standard for AF subtypes for this project.
Second, left ventricular ejection fraction (LVEF) is one of the covariates of interest to identify. NLP-based methods were used to extract LVEF from echocardiogram reports when it is not available in a structured format.
Use of CDMs for standardizing data collection and analyses
In the NESTcc test case study, we realized that there would be of great value if we could standardize the data collection process across sites, and the infrastructure of CDM-based health IT systems makes this possible. We investigated the CDM implementation status (ie, whether a prevailing CDM such as i2b2, PCORnet, OMOP, Sentinel, and Fast Healthcare Interoperability Resources [FHIR] has been implemented) in the 3 health systems.
Conducting a maturity level analysis
We also conducted a maturity level analysis on the key informatics technologies used in data capture and transformation, highlighting current maturity level (ie, conceptual, reactive, structured, complete, and advanced) of the key technologies and their correlations with the NESTcc data quality domains (ie, consistency, completeness, CDM, accuracy, and automation) as defined in the NESTcc data quality framework. Two representatives from each site assessed the maturity level of the 4 key technologies for their respective system and assigned the maturity level scores.
RESULTS
Initial population and device exposure data
Using standard codes (Supplementary Table S3), we were able to retrieve initial populations of AF and VT patients and their procedure of interest. A total of 337 181 AF patients were identified, including 27 865 patients with persistent AF, and a total of 59 425 VT patients were identified, including 39 092 patients with ischemic VT from 3 sites (Table 1). In addition, a total of 8676 cardiac catheter ablation procedures were identified for AF population and 1865 ablation procedures for VT population (data not shown). Using UDIs, we were able to break down device counts for target population by brand-specific device subtypes (Table 2). Notably, no analyses of safety and effectiveness outcomes by catheter type were conducted in this feasibility study to avoid influencing the second stage study that will test the hypotheses.
Table 1.
AF and VT patient counts by disease subtype (Note that 1 patient may have more than 1 diagnosis)
| AF | Paroxysmal AF | Persistent AF | Permanent AF | Unspecified and other AF | VT | Ischemic VT | Nonischemic VT | |
|---|---|---|---|---|---|---|---|---|
| Mercy (01/01/2014-02/20/2020) | 169 062 | 88 387 | 11 898 | 31 753 | 145 903 | 24 401 | 16 379 | 8022 |
| Mayo Clinic (01/01/2014-12/31/2019) | 133 298 | 60 999 | 12 372 | 21 800 | 98 839 | 20 920 | 13 114 | 7806 |
| YNHH (02/01/2013-08/13/2019) | 54 821 | 15 007 | 3594 | 14 961 | 21 259 | 14 104 | 9599 | 4505 |
| Total | 357 181 | 164 393 | 27 864 | 68 514 | 266 001 | 59 425 | 39 092 | 20 333 |
AF: atrial fibrillation; VT: ventricular tachycardia; YNHH: Yale New Haven Hospital.
Table 2.
Device counts for AF patients by brand-specific subtypes of interest
| Paroxysmal AF |
Persistent AF |
|||
|---|---|---|---|---|
| ThermoCool ST | ThermoCool STSF | ThermoCool ST (treatment catheter) | ThermoCool STSF (control catheter) | |
| Mercy (01/01/2014-02/20/2020) | 377 | 408 | 251 | 492 |
| Mayo Clinic (01/01/2014-12/31/2019) | 625 | 248 | 233 | 100 |
| YNHH (02/01/2013-08/13/2019) | 96 | 135 | 65 | 115 |
| Total | 1098 | 791 | 549 | 707 |
AF: atrial fibrillation; ST: Smarttouch; STSF: Smarttouch Surround Flow; YNHH: Yale New Haven Hospital.
Data quality validation
Table 3 shows cross-validation results for the AF subtype cases identified using ICD codes and registry data at Mayo Clinic. Positive predictive values (PPVs) were calculated as results. For AF cases identified by ICD–Ninth Revision code 427.31, we identified 304 cases of paroxysmal AF and 427 cases of persistent AF from the Mayo Clinic registry, indicating that registry data provide specific subtypes. For 496 cases of paroxysmal AF identified by ICD–Tenth Revision (ICD-10) code I48.0, a total of 260 (PPV = 52.4%) were confirmed as true cases from the registry. For 176 cases of persistent AF identified by ICD-10 code I48.1, 124 cases (PPV = 70.5%) were confirmed as true persistent AF cases from the registry. The results indicated that the case identification algorithms based on ICD-10 codes at Mayo Clinic are not optimal and that the clinical registry had great value in validating the case identification algorithms, though the accuracy of the registry itself has not been validated (and uses retrospective diagnosis based on chart review by a nurse clinician to determine AF type). Note that Mercy used a previously validated NLP algorithm to validate AF patient phenotypes (see details in the following section), and YNHH participates in the National Cardiovascular Data Registry AF Ablation Registry, which is another registry resource used for AF data quality validation in the NESTcc test case study.
Table 3.
Validation of the AF subtype cases identified using ICD codes against the prospective nurse-abstracted registry data at Mayo Clinic
| Code | Vocabulary | Term | Paroxysmal AF in registry | Persistent AF in registry | Total |
|---|---|---|---|---|---|
| 427.31 | ICD-9 | AF | 304 (41.6) | 427 (58.4) | 731 (100) |
| I48.0 | ICD-10 | Paroxysmal AF | 260 (52.4) | 236 (47.6) | 496 (100) |
| I48.1 | ICD-10 | Persistent AF | 52 (29.5) | 124 (70.5) | 176 (100) |
| I48.2 | ICD-10 | Chronic AF | 4 (19.0) | 17 (81.0) | 21 (100) |
| I48.91 | ICD-10 | Unspecified AF | 251 (41.8) | 349 (58.2) | 600 (100) |
Values are n (%). ICD-9 codes were used prior to October 2015 and ICD-10 codes thereafter.
AF: atrial fibrillation; ICD: International Classification of Diseases; ICD-9: International Classification of Diseases–Ninth Revision; ICD-10: International Classification of Diseases–Tenth Revision;
For outcome endpoint validation, a manual chart review process was used to confirm target cases. Owing to time and funding restrictions, the consensus was to focus on 3 primary outcome endpoints: ischemic stroke, acute heart failure, and rehospitalization with arrhythmia. We started the algorithms based on codes obtained from a published literature review and refined this further using consensus clinician review from several practicing electrophysiology physicians, data scientists, epidemiologists, and other team members. Once the code algorithms were finalized, we identified the patient counts for each of the 3 outcome endpoints. We also used the full algorithms that restrict patients within 30 days of postablation (ie, a time window used to identify outcomes) and identified a subset of patients. We then randomly selected 25 cases from the results of the full algorithm for each of the 3 outcome endpoints. Clinicians at each site performed manual chart review to evaluate the clinical outcomes’ algorithms. PPVs were calculated as results (data not shown).
NLP for unstructured data
Mercy does not participate in an AF registry; therefore, a NLP tool was used on a group of patients who were diagnosed with arrythmia and stroke using a collection of ICD codes for a previous Johnson & Johnson project. Table 4 shows the summary of predictive values for ICD codes by AF type.
Table 4.
Summary of predictive values for ICD codes by AF type at Mercy as compared with an natural language processing tool
| Paroxysmal AF (%) | Persistent AF (%) | Chronic AF (%) | |
|---|---|---|---|
| Sensitivity (recall) | 82.80 | 62.70 | 74.80 |
| Specificity | 86.50 | 95.90 | 90.80 |
| Positive predictive value (precision) | 94.20 | 80.40 | 87.60 |
| Negative predictive value | 65.70 | 90.60 | 80.70 |
AF: atrial fibrillation; ICD: International Classification of Diseases.
In addition, we found that LVEF is not readily available in a structured format. At Mercy, LVEF was extracted using an NLP method and at the Mayo Clinic, it was extracted from echocardiogram reports using an open-source NLP program.23 Yale was able to capture ejection fractions available in structured fields.
CDM implementation status
Table 5 shows the CDM implementation status of 3 health systems: the Mayo Clinic, Mercy, and YNNH. Both the Mayo Clinic and YNNH have majority of CDMs (i2b2, PCORnet, OMOP, and FHIR) implemented, whereas Mercy have Sentinel CDM and FHIR implemented. This indicates that the CDM implementation varies across 3 sites.
Table 5.
The CDM implementation status of 3 health systems: Mayo Clinic, Mercy, and YNNH
| CDM Implementation Status | Mayo Clinic | Mercy | YNHH |
|---|---|---|---|
| i2b2 Star Schema | X | X | |
| PCORnet CDM | X | X | |
| OMOP CDM | X (in progress) | X | |
| Sentinel CDM | X | ||
| FHIR | X | X | X |
CDM: common data model; CPT: Current Procedural Terminology; FHIR: Fast Healthcare Interoperability Resources; i2b2: Informatics for Integrating Biology and the Bedside; OMOP: Observational Medical Outcomes Partnership; PCORnet: Patient-Centered Research Network; YNHH: Yale New Haven Hospital.
Maturity level analysis
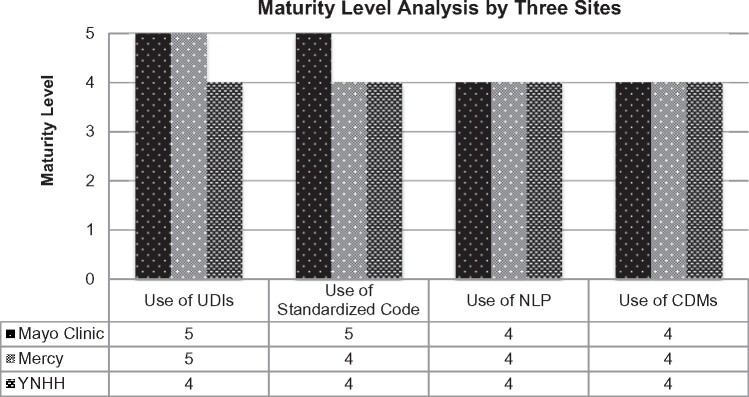
Figure 2 shows the maturity level analysis results for the key technologies used in the data capture and transformation.
Figure 2.
The maturity level analysis results by 3 sites for the key technologies used in the data capture and transformation. Maturity level consists of 5 levels (ie, 1 = conceptual, 2 = reactive, 3 =structured, 4 = complete, and 5 = advanced). CDM: common data model; NLP: natural language processing; UDI: unique device identifier; YNHH: Yale New Haven Hospital.
By design, the maturity model can help researchers identify weaknesses, in terms of the ability to capture data consistently and completely, to represent data via CDMs, to validate the accuracy of data, and to then use the data through automated queries. These are examples of key processes that drive data quality.
A summary of the informatics analysis
The successes and challenges of the informatics analysis are described in detail in Table 6.
Table 6.
Successes and challenges from informatics analysis
| Successes vs Challenges | |
|---|---|
Use of UDIs
|
Successes:
|
Challenges:
| |
Use of Standardized Codes
|
Successes:
|
Challenges:
| |
Use of NLP technology
|
Successes:
|
Challenges:
| |
Use of CDMs
|
Advantages:
|
Challenges:
|
CDM: common data model; CPT: Current Procedural Terminology; EHR: electronic health record; i2b2: Informatics for Integrating Biology and the Bedside; ICD-10: International Classification of Diseases–Tenth Revision; IT: information technology; NC: network collaborator; NESTcc: National Evaluation System for Health Technology Coordinating Center; NLP: natural language processing; OMOP: Observational Medical Outcomes Partnership; PCORnet: Patient-Centered Research Network; UDI: unique device identifier; YNHH: Yale New Haven Hospital.
DISCUSSION
Use of UDIs
We found that, with the UDIs implemented in the health IT systems, the target device exposure data can be effectively identified, particularly for brand-specific devices as targeted in the NESTcc case study. For example, when another device was identified as a potential comparator for a VT ablation study, we needed to assess initial counts of its usage to inform availability of comparator or control data for a potential label extension study for the catheter of interest. The project team at the Mayo Clinic, Mercy, and Johnson & Johnson was able to identify device UDIs and use them to get initial counts of its usage in a short turnaround.
One of key challenges is that the UDI implementation is uneven across sites. For example, Mercy implemented UDIs in its health IT systems in 2016. As mentioned previously, to pull device-related data prior to 2016 (before OptiFlex was installed), Mercy identified procedures linked to patient information using HCPCS codes and Mercy-specific charge codes for device billing. These codes were reviewed and confirmed by Johnson & Johnson before data extraction. Device data were joined together with those UDI-linked data to create a final dataset after duplicates were removed. Supplementary Table S4 shows the UDI implementation status of 3 health systems (Mayo Clinic, Mercy, and YNHH).
Use of standardized codes
We found that coming to an agreement on standard computable covariate and outcome definitions took more time than we foresaw. In particular, this consensus process involved input from clinicians to ensure algorithm definitions were clinically meaningful and precise. For example, to define cardiac ablation as a procedure of interest, we used CPT procedure codes. The initial list of the CPT codes included 93650 (atrioventricular node ablation), and through discussion with the clinical group, the CPT code was questioned as not representing AF ablation and consensus was achieved to remove the CPT code 93650 from the definition list.
Use of NLP technology
The use of NLP in this study was limited to a number of specific tasks. The main challenges for using NLP technology include (1) requiring advanced expertise in using existing NLP tools or developing fit-for-purpose NLP probes to search clinical text and notes, (2) lack of NLP solutions that are portable across sites, and (3) challenges in validating NLP probes. In addition, we also noticed that NLP, in general, has its own challenges including accuracy and maintenance issues, and potential for accidental privacy breaches.24,25
Use of CDMs
The advantages of using CDM-based research repositories are described in Table 6. Note that if there is no CDM, the researcher still must understand the source data and convert it to a usable form that is consistent across the multiple healthcare systems participating in the study, so they have similar work with or without the data model. But a CDM can provide significant benefit when provided by a coordinating center for use by individual researchers, helping make language consistent related to queries developed, thus saving the investigator significant work. One of the main challenges is that the implementation of CDMs often requires significant time and effort to extract and convert data from clinical data information systems, such as EHRs and laboratory information systems, to the format required to load into each CDM. Fortunately, this challenge can be alleviated with the advancement of mature ETL technology involved in the CDM implementation. Moreover, the processing and transformation of data into CDMs provides a logical pathway for enabling standardized analyses that are portable and consistent across sites, the benefit of which can help make a decision for the investment on the CDM implementation.
Data quality vs fit for purpose
Fit for purpose is defined as a conclusion that the level of validation associated with a medical product tool is sufficient to support its context of use.26 The NESTcc Data Quality Framework6 has made clear that useful data must be both reliable (high quality) and relevant (fit for purpose) across a broad and representative population based on the experimental, approved, or real-world use of a medical device.
The nature of capturing RWD from health IT systems for device evaluation is the secondary use of the data for a research purpose. The underlying data can have quality issues (eg, typed in wrong value, only captured a portion of UDI when a standard operating procedure calls for identifier capture in its entirety, manual data entry instead of barcode scanning). However, it is important to separate those issues from data that may not be present because the data weren’t needed (or needed in structured formats) for direct clinical care.
In addition, lacking a gold standard, the reports of detected data quality rely heavily on the quality of the evaluation plan. We found that different modalities such as ablation registries, NLP tools and chart reviews were required for validating data quality of the phenotypes.
Clinical aspects of the NESTcc test case
The focus of this article is on the informatics approaches used in the NESTcc test case. A separate clinical article reports on the feasibility of using the informatics approaches to capture RWD from the EHRs and other health IT systems at 3 health systems that are fit for purpose for postmarket evaluation of outcomes for label extensions of 2 cardiac ablation catheters. In brief, such evaluation was preliminarily determined feasible based on (1) the finding of adequate sample size of device of interest and control device use; (2) the presence of sufficient in-person encounter follow-up data (up to 1 year); (3) the availability of adequate data quality validation modalities, including clinician chart reviews; and (4) the potential use of CDMs for distributed data analytics. Reporting the detailed findings of the project’s clinical aspects and feasibility assessments is beyond the scope of the article.
CONCLUSION
We demonstrated that the informatics approaches can be feasibly used to capture RWD that are fit for purpose for postmarket evaluation of outcomes for label extensions of 2 ablation catheters from the EHRs and other health IT systems in a multicenter evaluation. While variations of such systems in each institution caused some data quality issues on data capture and transformation, we argue that development of coordination across otherwise perfectly fit-for-use (for other purposes) systems would be required for the device data integration needs of the postmarket surveillance study. However, we also identified a number of challenging areas for future improvements, including integrating UDI-linked device data with clinical data into a research repository; improving the accuracy of phenotyping algorithms with additional data points such as timing, medication use, and data elements extracted from unstructured clinical notes using NLP; specifying a chart review guideline to standardize the chart review process; and using CDM-based research repositories to standardize the data collection and analysis process.
FUNDING
This project was supported by a research grant from the Medical Device Innovation Consortium as part of the National Evaluation System for Health Technology (NEST), an initiative funded by the U.S. Food and Drug Administration (FDA). Its contents are solely the responsibility of the authors and do not necessarily represent the official views nor the endorsements of the Department of Health and Human Services or the FDA. While the Medical Device Innovation Consortium provided feedback on project conception and design, the organization played no role in collection, management, analysis, and interpretation of the data, nor in the preparation, review, and approval of the manuscript. The research team, not the funder, made the decision to submit the manuscript for publication. Funding for this publication was made possible, in part, by the FDA through grant 1U01FD006292. Views expressed in written materials or publications and by speakers and moderators do not necessarily reflect the official policies of the Department of Health and Human Services; nor does any mention of trade names, commercial practices, or organization imply endorsement by the U.S. government. In the past 36 months, JSR received research support through Yale University from the Laura and John Arnold Foundation for the Collaboration for Research Integrity and Transparency at Yale, from Medtronic and the FDA to develop methods for postmarket surveillance of medical devices (U01FD004585) and from the Centers of Medicare and Medicaid Services (CMS) to develop and maintain performance measures that are used for public reporting (HHSM-500-2013-13018I); JSR currently receives research support through Yale University from Johnson & Johnson to develop methods of clinical trial data sharing, from the FDA for the Yale-Mayo Clinic Center for Excellence in Regulatory Science and Innovation program (U01FD005938); from the Agency for Healthcare Research and Quality (R01HS022882); from the National Heart, Lung, and Blood Institute of the National Institutes of Health (R01HS025164, R01HL144644); and from the Laura and John Arnold Foundation to establish the Good Pharma Scorecard at Bioethics International. SSD reports receiving research support from the National Heart, Lung, and Blood Institute of the National Institutes of Health (K12HL138046), the Greenwall Foundation, Arnold Ventures, and the NEST Coordinating Center. PAN reports receiving research support from the National Institute of Aging (R01AG 062436-1) and the Heart, Lung, and Blood Institute (R21HL 140205-2, R01HL 143070-2, R01HL 131535-4) of the National Institutes of Health and the NEST Coordinating Center.
AUTHOR CONTRIBUTIONS
GJ, JPD, SSD and WLS developed the initial drafts of the manuscript. JC, YY, PT, EB, SZ, WLS, GJ contributed to the data collection and analysis. AAD, PAN, SSD and JPD contributed their clinical expertise required for the study. JPD, NSS, JSR, PC and KRE led the conception, and provided oversight and interpretation, of the project and the manuscript. All authors reviewed and approved of the submitted manuscript and have agreed to be accountable for its contents.
ETHICS STATEMENT
This study was approved by the institutional review boards (IRBs) of Mercy Health (IRB Submission No. 1349229-1), Mayo Clinic (IRB Application No. 19-001493), and Yale New Haven Hospital (IRB Submission No. 2000024523).
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kim Collison-Farr for her work as the project manager, Ginger Gamble for her work as Yale project manager, Lindsay Emmanuel for her work as Mayo Clinic project manager, and Robbert Zusterzeel, MD, at the NEST Coordinating Center for his support throughout the project.
DATA AVAILABILITY STATEMENT
The data underlying this article cannot be shared publicly due to ethical/privacy reasons (ie, they are patient-level device data).
CONFLICT OF INTEREST STATEMENT
WLS was an investigator for a research agreement, through Yale University, from the Shenzhen Center for Health Information for work to advance intelligent disease prevention and health promotion; collaborates with the National Center for Cardiovascular Diseases in Beijing; is a technical consultant to HugoHealth, a personal health information platform, and cofounder of Refactor Health, an AI-augmented data management platform for healthcare; and is a consultant for Interpace Diagnostics Group, a molecular diagnostics company. PC and SZ are employees of Johnson & Johnson and own stock in the company; Johnson & Johnson’s cardiac ablation catheters were the research topic of the NESTcc test case, although this study was a feasibility study to evaluate the quality of the data to support regulatory decisions.
REFERENCES
- 1.Corrigan-Curay J, Sacks L, Woodcock J.. Real-world evidence and real-world data for evaluating drug safety and effectiveness. JAMA 2018; 320 (9): 867–8. [DOI] [PubMed] [Google Scholar]
- 2.21st Century Cures Act. 2020. https://www.fda.gov/regulatory-information/selected-amendments-fdc-act/21st-century-cures-act. Accessed March 29, 2020.
- 3.FDA Guidance on RWE for medical devices. 2020. https://www.fda.gov/media/99447/download. Accessed October 19, 2020.
- 4.Gottlieb S, Shuren J. Director of the Center for Devices and Radiological Health, on FDA’s updates to Medical Device Safety Action Plan to enhance post-market safety. 2020. https://www.fda.gov/news-events/press-announcements/statement-fda-commissioner-scott-gottlieb-md-and-jeff-shuren-md-director-center-devices-and-2. Accessed April 10, 2020.
- 5.Krucoff MW, Sedrakyan A, Normand SL.. Bridging unmet medical device ecosystem needs with strategically coordinated registries networks. JAMA 2015; 314 (16): 1691–2. [DOI] [PubMed] [Google Scholar]
- 6.National Evaluation System for Health Technology Coordinating Center (NESTcc). 2020. https://nestcc.org/. Accessed March 29, 2020.
- 7.MDUFA Performance Goals and Procedures Fiscal Years 2018 Through 2022. 2020. https://www.fda.gov/media/100848/download. Accessed January 13, 2021.
- 8.NESTcc Data Quality Framework. 2020. https://nestcc.org/data-quality-and-methods/. Accessed March 29, 2020.
- 9.Jiang G, Yu Y, Kingsbury PR, Shah N.. Augmenting medical device evaluation using a reusable unique device identifier interoperability solution based on the OHDSI common data model. Stud Health Technol Inform 2019; 264: 1502–3. [DOI] [PubMed] [Google Scholar]
- 10.Zerhouni YA, Krupka DC, Graham J, et al. UDI2Claims: planning a pilot project to transmit identifiers for implanted devices to the insurance claim. J Patient Saf 2018. Nov 21 [E-pub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 11.Drozda JP Jr, Roach J, Forsyth T, Helmering P, Dummitt B, Tcheng JE.. Constructing the informatics and information technology foundations of a medical device evaluation system: a report from the FDA unique device identifier demonstration. J Am Med Inform Assoc 2018; 25 (2): 111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Global Unique Device Identification Database (GUDID). 2020. https://www.fda.gov/medical-devices/unique-device-identification-system-udi-system/global-unique-device-identification-database-gudid. Accessed March 30, 2020.
- 13.Drozda J, Zeringue A, Dummitt B, Yount B, Resnic F.. How real-world evidence can really deliver: a case study of data source development and use. BMJ Surg Interv Health Technologies 2020; 2 (1): e000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tcheng JE, Crowley J, Tomes M, et al. ; MDEpiNet UDI Demonstration Expert Workgroup. Unique device identifiers for coronary stent postmarket surveillance and research: a report from the Food and Drug Administration Medical Device Epidemiology Network Unique Device Identifier demonstration. Am Heart J 2014; 168 (4): 405–13.e2. [DOI] [PubMed] [Google Scholar]
- 15.Weeks J, Pardee R.. Learning to share health care data: a brief timeline of influential common data models and distributed health data networks in U.S. health care research. EGEMS (Wash DC) 2019; 7 (1): 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.FDA Sentinel Common Data Model. 2020. https://www.sentinelinitiative.org/sentinel/data/distributed-database-common-data-model. Accessed April 4, 2020.
- 17.OHDSI OMOP Common Data Model. 2020. https://github.com/OHDSI/CommonDataModel/wiki. Accessed April 4, 2020.
- 18.PCORnet Common Data Model Specification 2020. https://pcornet.org/wp-content/uploads/2019/09/PCORnet-Common-Data-Model-v51-2019_09_12.pdf. Accessed April 4, 2020.
- 19.I2B2 Star Schema. 2020. https://i2b2.cchmc.org/faq. Accessed April 4, 2020.
- 20.Visweswaran S, Becich MJ, D'Itri VS, et al. Accrual to Clinical Trials (ACT): a clinical and translational science award consortium network. JAMIA Open 2018; 1 (2): 147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McPadden J, Durant TJS, Bunch DR, et al. Health care and precision medicine research: analysis of a scalable data science platform. J Med Internet Res 2019; 21 (4): e13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guimaraes PO, Krishnamoorthy A, Kaltenbach LA, et al. Accuracy of medical claims for identifying cardiovascular and bleeding events after myocardial infarction: a secondary analysis of the TRANSLATE-ACS study. JAMA Cardiol 2017; 2 (7): 750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adekkanattu P, Jiang G, Luo Y, et al. Evaluating the portability of an NLP system for processing echocardiograms: a retrospective, multi-site observational study. AMIA Annu Symp Proc 2019; 2019: 190–9. [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C, Ta CN, Rogers JR, et al. Ensembles of natural language processing systems for portable phenotyping solutions. J Biomed Inform 2019; 100: 103318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Carrell D, Aberdeen J, et al. Optimizing annotation resources for natural language de-identification via a game theoretic framework. J Biomed Inform 2016; 61: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.FDA‐NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, & other Tools) Resource. Silver Spring, MD: U.S. Food and Drug Administration; 2016. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly due to ethical/privacy reasons (ie, they are patient-level device data).