Abstract
Objective
Clinical notes contain an abundance of important, but not-readily accessible, information about patients. Systems that automatically extract this information rely on large amounts of training data of which there exists limited resources to create. Furthermore, they are developed disjointly, meaning that no information can be shared among task-specific systems. This bottleneck unnecessarily complicates practical application, reduces the performance capabilities of each individual solution, and associates the engineering debt of managing multiple information extraction systems.
Materials and Methods
We address these challenges by developing Multitask-Clinical BERT: a single deep learning model that simultaneously performs 8 clinical tasks spanning entity extraction, personal health information identification, language entailment, and similarity by sharing representations among tasks.
Results
We compare the performance of our multitasking information extraction system to state-of-the-art BERT sequential fine-tuning baselines. We observe a slight but consistent performance degradation in MT-Clinical BERT relative to sequential fine-tuning.
Discussion
These results intuitively suggest that learning a general clinical text representation capable of supporting multiple tasks has the downside of losing the ability to exploit dataset or clinical note-specific properties when compared to a single, task-specific model.
Conclusions
We find our single system performs competitively with all state-the-art task-specific systems while also benefiting from massive computational benefits at inference.
Keywords: multitask learning; natural language processing; clinical natural language processing, named entity recognition, textual entailment, semantic text similarity
INTRODUCTION
Electronic health records (EHRs) contain a wealth of actionable patient information in the form of structured fields and unstructured narratives within a patient’s clinical note. While structured data such as billing codes provide a coarse-grained signal pertaining to common conditions or treatments a patient may have experienced, a large quantity of vital information is not directly accessible due to being stored in unstructured, free-text notes. The task of automatically extracting structured information from this free-form text is known as information extraction and has been an intensely studied line of research over the past 2 decades. While the primary objective of information extraction is to gather fine-grained information about patients, such as problems experienced, treatments underwent, tests conducted, and drugs received, auxiliary tasks such as the automatic identification and subsequent removal of personal health information (PHI) are also of pragmatic interest to the functioning of the health system controlling the EHR.
To support this diverse set of information extraction challenges, several community-led shared tasks have annotated datasets for the construction and evaluation of automated information extraction systems. These include the identification of problems, treatments and tests;1,2 the identification of drugs, adverse drug events, and drug-related information;3 and the deidentification of PHI.4 While these shared tasks have produced well-performing solutions, the resulting systems are disjoint, meaning that no information is shared between systems addressing each individual information extraction task. Notably, this means that each task requires a separate engineering effort to solve, narrow technical expertise to construct, and disjoint computational resources to apply in clinical practice. Recently, this gap has been narrowed by advances in large-scale self-supervised text pretraining.5,6 This paradigm has resulted in well-known language representation systems, such as BERT, which can easily be adapted to any single domain-specific task and achieve state-of-the-art performance. In the clinical space, researchers have similarly leveraged large clinical note repositories such as MIMIC-III7 to pretrain Clinical BERT8 instances, achieving large performance gains on several clinical natural language proceeding (NLP)-related tasks. While well-performing, a single fine-tuned Clinical BERT instance requires significant resources to deploy into a clinical informatics workflow, thus limiting its practical applicability. This fact is amplified by the observation that an isolated 110 million parameter model is required for each clinical task; scaling linearly the required hardware resources.
This article introduces Multitask-Clinical-BERT: a single, unified deep learning-based clinical multitask learning system that concurrently addresses 8 datasets across 3 distinct NLP tasks. MT-Clinical BERT augments the BERT5 deep learning architecture with a novel round robin task fine-tuning schema that allows the learning of features for multiple clinical tasks simultaneously. As a result, our system decreases the hardware and computational requirements of deploying BERT into clinical practice by successfully condensing eight 110 million parameter BERT instances into a single model while retaining nearly all BERT-associated task performance gains.
Our main contributions are summarized as follows:
We develop a single deep learning model that concurrently achieves competitive performance over 8 clinical tasks spanning named entity recognition, entailment, and semantic similarity. As a result, we achieve an 8-fold computational speed-up at inference compared to traditional per-task, sequentially fine-tuned models.
We demonstrate the feasibility of multitask learning towards developing a universal clinical information extraction system that shares information among disjointly annotated datasets.
We release and benchmark against a new and more competitive BERT fine-tuning baseline for 8 clinical tasks by performing extensive hyperparameter tuning for each task’s dataset.
RELATED WORK
Multitask learning has been an integral subfield of the machine learning community for many decades.9–11 In the context of deep learning, programs in several domains spanning drug discovery, computer vision, and NLP have continued to achieve successes by sharing supervised signal and data between machine learning tasks.12–15 Within the biomedical and clinical domain, much of the work has been focused on information extraction tasks. Previous works can be divided into 2 categories: multitask learning on a single task across multiple datasets and multitask learning across related tasks within a single domain. Multitask learning over a single task has primarily centered around named entity recognition. In the biomedical domain, these systems focused on extracting biomedical entities (eg, chemical, genes, diseases, and species) from scientific literature.16–20 Within the clinical domain, these systems focused on extracting clinical concepts (eg, problems, treatments, and tests) from clinical notes.13 These works focused on data efficiency with the hypothesis that multitask learning on existing named entity regression (NER) datasets would reduce the annotated data required to extract new entity types.
Much of the multitask learning across tasks has focused on learning both entity and relations. Within the biomedical domain, Li et al21 evaluated their approach over 2 entity/relation tasks within scientific literature: adverse drug events and bacteria biotopes. Within the clinical domain, Shi et al22 focused on family history entities and relations in clinical records. These works, focused on jointly processed correlated tasks, increased the overall performance of the model. Peng et al23 evaluated joint learning entities and relations across both the clinical and biomedical texts to evaluate the transfer of knowledge not only between tasks but also domains. Their findings showed information can be shared across the 2 domains, improving the overall results for information extraction tasks.
These previous works perform multitask learning via a sum of losses objective in which, given the losses on a batch from each dataset, the model updates the weights with a backpropagation against the sum of all the losses. In this work, we explore utilizing a round robin technique comparing the results to utilizing the sum of the loss.
Within clinical multitask learning, these previous works focused on single task and related tasks. However, Li et al24 evaluated multitask learning across 8 distinct tasks within the general English domain. Their findings showed that related tasks may not always help each other. However, unrelated tasks are not correlated tasks, and therefore the sharing of the input features and hidden units can benefit each other during training creating a more generalizable system. Collobert et al25 evaluated multitasking across 6 distinct NLP tasks showing that multitask learning improved generalization even across possibly unrelated tasks. In this work, we explore multitask learning across 3 distinct NLP tasks: entity recognition, semantic text similarity, and natural language inference, across 8 datasets.
MATERIALS AND METHODS
This section begins with description of the clinical text benchmarks and then a description of our clinical multitask learning system. Our goal is to investigate the effect of multitask learning across a set of diverse clinical tasks.
Data
We use 8 clinical tasks to evaluate our multitasking system. Table 1 describes the tasks, the predefined train and evaluation splits used in our experiments, and the corresponding task evaluation metric.
Table 1.
Clinical information extraction benchmarks with reported performance metric
| Task | Dataset | Metric | Description | # Train Inst. | # Test Inst. |
|---|---|---|---|---|---|
| STS | n2c2-201926 | Pearson Rho | Sentence pair semantic similarity | 1641 | 410 |
| Entailment | MedNLI27 MedRQE28 |
|
|
12 627 8588 | 1422 302 |
| NER | n2c2-20183 i2b2-20144 i2b2-20122 |
|
|
|
|
| i2b2-20101 | Micro-F1 | Problems, treatments and tests | 27 837 | 45 009 | |
| quaero-201429 | Micro-F1 | UMLS semantic groups (French) | 2695 | 2260 |
The Semantic Textual Similarity (STS) task is to assign a numerical score to sentence pairs, indicating their degree of semantic similarity. Our system includes 1 STS dataset:
-
The n2c2-2019 dataset consists of deidentified pairs of clinical text snippets from the Mayo Clinic that were ordinally rated from 0 to 5 with respect to their semantic equivalence where 0 indicates no semantic overlap and 5 indicates complete semantic overlap. The training dataset contains 1642 sentence pairs, while the test dataset contains 412 sentence pairs.
Textual entailment is the task of determining if 1 text fragment is logically entailed by the previous text fragment. We utilize 2 entailment datasets:
The MedNLI27 dataset consists of the sentence pairs developed by physicians from the Past Medical History section of MIMIC-III clinical notes annotated for Definitely True, Maybe True, and Definitely False. The dataset contains 11 232 training, 1395 development, and 1422 test instances. We combined the training and development instances for our work.
-
The MedRQE28 dataset consists of question–answer pairs from the National Institutes of Health’s National Library of Medicine clinical question collection (FAQ). The positive examples were drawn explicitly from the dataset while the negative pairs were collected by associating a randomly combined question–answer pair as having at least 1 common keyword and at least 1 different keyword from the original question. The dataset contains 8588 training pairs and 302 test pairs with approximately 54.2% as positive instances.
NER is the task of automatically identifying mentions of specific entity types within unstructured text. In this work, we utilize 5 NER datasets:
The n2c2-2018 dataset3 consists of 505 deidentified discharge summaries drawn from the MIMIC-III clinical care database and annotated for adverse drug events and the drug that caused them; reason for taking the drug and the associated dosage, route, and frequency information. The training and test sets contain 303 and 202 instances, respectively.
The i2b2-2014 dataset2 consists of 28 772 deidentified discharge summaries provided from Partners HealthCare annotated for PHI including, patient names, physician names, hospital names, identification numbers, dates, locations, and phone numbers. The training and test sets contain 17 310 and 11 462 instances, respectively.
The i2b2-2012 dataset consists of deidentified discharge summaries provided by Partners HealthCare and MIMIC-II. The dataset was annotated for 2 entity types: 1) clinical events, including both clinical concepts, departments, evidentials, and occurrences; and 2) temporal expressions, referring to the dates, times, durations, or frequencies. In this work, we evaluated only the event annotations. The training and test sets contain 16 468 and 13 594 instances, respectively.
The i2b2-2010 dataset1 consists of deidentified discharge summaries, provided by Partners HealthCare and MIMIC-II, and deidentified discharge and progress notes from the University of Pittsburgh Medical Center. The dataset was annotated for 3 entity types—clinical concepts, clinical tests, and clinical problems. These entities overlap with the i2b2-2010 event annotations. The training and test sets contain 27 837 and 45 009 instances, respectively.
The quaero-2014 dataset29 consists of a French medical corpus containing 3 document types: 1) the European Medicines Agency drug information; 2) MEDLINE research article titles; and 3) European Patent Office patents. The dataset was annotated for 10 types of clinical entities from the Unified Medical Language System Semantic Groups:30 Anatomy, Chemical and Drugs, Devices, Disorders, Geographic Areas, Living Beings, Objects, Phenomena, Physiology, and Procedures. The training and test sets contain 2695 and 2260 instances, respectively.
Multitasking clinical BERT
In this section, we describe our multitask learning framework which aims to adapt the weights of a base pretrained model into a feature encoder capable of generating text representations suitable for multiple tasks simultaneously. In a non-multitask learning environment, the standard practice of learning from pretrained transformers, such as BERT, is a method known as sequential fine-tuning. During sequential fine-tuning, a BERT encoder is initialized with self-supervised pretraining and then fine-tuned with loss signal from a task-specific head. This procedure adapts the weights of the base pretrained model into a task-specific feature encoder capable of representing the input text, such that the task objective is easily discernible by the task-specific head (eg, linearly separable in the case of classification).
In contrast, in a multitask learning environment the weights of the base pretrained model into a feature encoder are tuned for multiple tasks simultaneously. In our case, this is achieved by treating the BERT transformer encoder as a feature encoder that feeds into multiple lightweight task-specific architectures, each implementing a different task objective. Traditionally, hard parameter multitask learning is achieved through loss summation,21,22 where the encoder is updated to minimize the sum of all losses across tasks. While effective, this approach does not scale to larger numbers of tasks due to the linearly increasing device memory requirements and tuning necessary to prevent a single task loss signal from dominating during learning. In turn, we propose and validate a hard parameter multitask learning model that learns a multitask encoder with memory requirements independent of the number of tasks present during training.
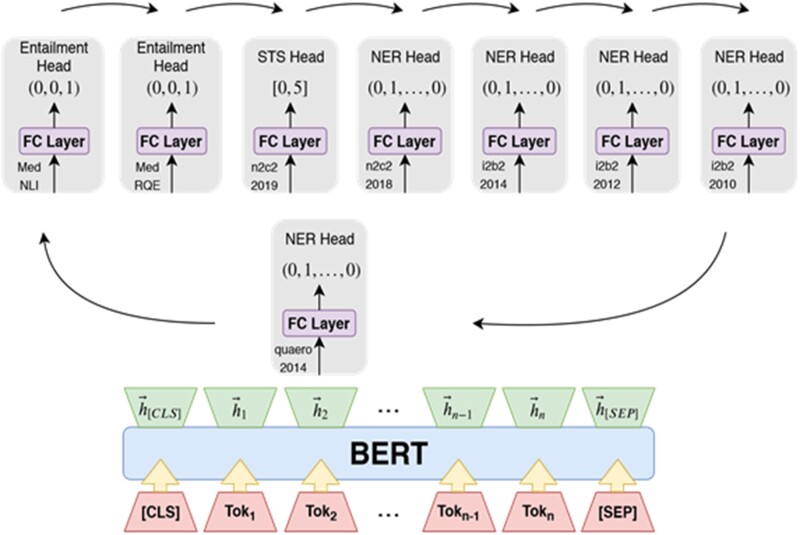
Our multitasking model (Figure 1) comprises of a BERT feature encoder with weights initialized from Bio + Clinical BERT8 and 8 per-dataset task-specific heads. The head architectures are as follows:
Figure 1.
Eight-headed MT Clinical BERT with a round robin training schedule. Each Entailment head predicts a one-hot class indicator. The Semantic Text Similarity (STS) head predicts a similarity score in [0, 5] representing the semantic similarity of the 2 input sentences. Each Named Entity Recognition head predicts a one-hot entity indicator for each input sub-word token.
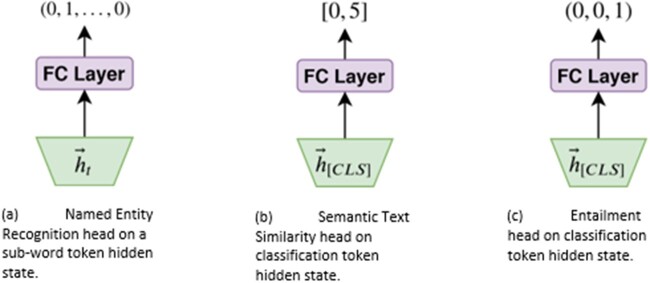
Named Entity Recognition (Figure 2a): token classification via a per-entity linear classifier on subword tokens providing loss signal with cross entropy loss.
Semantic Text Similarity (Figure 2b): sentence pair semantic similarity scoring via a linear regression on the sequence representation (sentence classification [CLS]) token providing loss signal via the mean squared error.
Natural Language Inference (Figure 2c): sentence pair logical entailment via a linear classifier on the sequence representation (CLS) token providing loss signal with cross entropy loss.
Figure 2.
Task-specific heads with corresponding input representations from the BERT hidden state sequence.
To train our multitasking model, the feature encoder was adapted to support all tasks simultaneously. There are several established methods of adapting the feature encoder parameters by combining loss signal from each head during training31 (eg, averaging/adding losses); however, most assume that the loss function is constant across all of the heads. In general, this is not necessarily true. When different loss functions are present, the standard solution is to subsample instances from each dataset proportional to the dataset size, compute a weighted sum of all the per-task losses, and then proceed with batch stochastic gradient descent against the total sum of losses.31 Instead, we propose a simpler and less involved training scheme that additionally reduces the device memory constraints imposed by methods such as loss summation. We train our multitask learning model as follows. During each data epoch, batches are randomly sampled from each NLP task and paired with the task’s corresponding linear prediction head. Each linear head takes turns having its parameters adjusted towards executing its designated NLP task, given the BERT encoder features. Each time a linear heads’ parameters update, the shared BERT encoders’ parameters receive an update. Heads are cycled on the encoder in a round robin fashion to ensure that the encoder does not become overly specialized in producing features for any specific task head. This training schedule is formally described in Algorithm 1.
Algorithm 1 MT-Clinical BERT Training Schedule
Require: θE: pretrained Transformer encoder.
Require: θH = {θh1,..,θhn}: n task-specific heads.
1: Randomly initialize θhi ∀i ∈{1,..,n}
2: while all batches from largest task dataset are not sampled do
3: Sample a batch Di for each θhi ∈ θH
4: for each (θhi, Di) do → One round robin iteration
5: Let θ = θE θhi → Output of encoder into head θh
6: θ0 = θ − α∇θL(θ,Di)
7: Update θ with θ0
8: end for
9: end while
Experimental details and reproducibility
We base our implementation on the well-known HuggingFace “Transformers” implementation of BERT. During hyperparameter tuning, we reinitialize with random seeds in the set {1,…,5}. All fine-tuning is performed with constant learning rate 5e−5. The NER heads train with a batch size of 25 length 512 subword sequences, while the STS and entailment training is performed with a batch size of 40. All training and evaluation are conducted on a single Nvidia V100 32GB GPU. It takes roughly 3 hours of sampled training to achieve the results in MT-Clinical BERT results in Table 2. It takes roughly 8 hours of training to achieve the multitask learning (MTL) Loss Summation results in Table 2. In addition to our pretrained models, we support reproducibility by including all preprocessing necessary to replicate our results in the code release.
Table 2.
Clinical information extraction performance of MT-Clinical BERT vs hyperparameter searched Clinical BERT fine-tuning runs. All span level metrics are exact match. Task performances showcased in the column MT-Clinical BERT represent a single multitask round robin trained feature encoder with individual task-specific heads. Task performances showcased in the column MTL Loss Summation represent a multitask feature encoder trained with loss summation. All other reported results are generated from task-specific BERT models. Higher is better
| MT-Clinical BERT | MTL Loss Summation | Optimized Clinical BERT | Clinical BERT8 | |
|---|---|---|---|---|
| n2c2-2019 | 86.7 (−0.5) | 84.5 | 87.2 | – |
| MedNLI | 80.5 (−2.3) | 80.2 | 82.8 | 82.7 |
| MedRQE | 76.5 (−3.6) | 77.5 | 80.1 | – |
| n2c2-2018 | 87.4 (−0.7) | 85.5 | 88.1 | – |
| i2b2-2014 | 91.9 (−3.6) | 94.2 | 95.5 | 92.7 |
| i2b2-2012 | 84.1 (+0.2) | 84.8 | 83.9 | 78.9 |
| i2b2-2010 | 89.5 (−0.3) | 90.6 | 89.8 | 87.8 |
| quaero-2014 | 49.1 (−6.4) | 52.2 | 55.5 | – |
Evaluation
To demonstrate that round-robin trained MT-Clinical BERT retains task-specific performance, we evaluate the model alongside BERT fine-tuning, the state-of-the-art single task approach, and a multitask BERT encoder trained with traditional loss summation. To insure a competitive and fair comparison with existing state-of-the-art solutions, we perform a hyperparameter search for each individual Clinical BERT task fine-tuning run and report the best performing model on each task. Recent work32 has found negligible performance differences between random seed reinitialization and more complex methods of hyperparameter search during BERT fine-tuning, so we opt for the former. Specifically, for each task, a Clinical BERT instance is initialized and fine-tuned for 20 training data epochs over 5 unique random seeds resulting in 100 unique task-specific models. We report the top performing model at evaluation. We do not utilize a development set for training MT-Clinical BERT, as the multitasking paradigm itself largely removes the ability for a model to overfit any specific task (for if it did, performance would degrade on other tasks). Additionally, we do not perform hypothesis testing due to the significant computational resources required.
RESULTS AND DISCUSSION
We compare the performance of our multitasking information extraction system to state-of-the-art BERT sequential fine-tuning baselines and multitask loss summation in Table 2. The reported multitask results are based on the epoch that provided the greatest overall performance of the system, rather than the individual best per task. Evaluations reported in the column (Optimized) Clinical BERT represent individually fine-tuned, per-task BERT models. Evaluations reported in the column MT-Clinical BERT represent lightweight task-specific heads over a single multitask trained BERT feature encoder. We find that the performances reported in the Clinical BERT8 paper can be substantially improved via hyperparameter search. While this is not surprising (the authors specify that performance was not their goal), it is important to compare improvements or degradations against a competitive baseline. All further discussion compares the multitasking model to the hyperparameter Optimized Clinical BERT baseline.
When comparing the 2 evaluated approaches for hard parameter multitask learning, we found no discernable patterns or trends in performance metrics. Each training method, round robin and loss summation, performs better for some tasks and worse for others. Notably, we found that round robin trained MT-Clinical BERT requires significantly less computational resources and training time on the fixed compute (Nvidia V100 32GB) to achieve the reported performance.
Overall, our results show that information sharing exists in our multitasking model as all task predictions depend only on the representations produced by a single encoder and a task-specific linear probing MLP over these representations. If information from all tasks were not present in the encoders hidden state embeddings, performance would degrade for 1 or all tasks. We observe a slight but consistent performance degradation in MT-Clinical BERT relative to sequential fine-tuning. Intuitively, this suggests that learning a general clinical text representation capable of supporting multiple tasks has the downside of losing the ability to exploit dataset or clinical note-specific properties when compared to a single, task-specific model. This phenomenon can best be illustrated among the English token classification tasks, where the deidentification task, i2b2-2014, suffered the greatest performance degradation. Clinical BERT is pretrained over MIMIC-III. As MIMIC-III is deidentified, all PHI markers in the original notes are replaced with special PHI tokens that do not linguistically align with the surrounding text (eg, an instance of a hospital name would be replaced with the token [HOSPITAL]). Due to this, no PHI tokens are present in MIMIC-III, and, thus the pretraining procedure of Clinical BERT over the MIMIC-III corpus provides little signal pertaining to PHI tokens. Alsentzer et al8 observes and discusses this property in depth. These results suggest that a lack of PHI-related information during pretraining can be overcome by the encoder during sequential fine-tuning. However, this is not as successful when regularized by the requirement of supporting multiple tasks due to the mixture of PHI and special PHI tokens across the datasets.
Surprisingly, MT-Clinical BERT confers a slight performance increase in the problem, treatment, and test extraction task i2b2-2012 relative to the hyperparameter-tuned Clinical BERT baseline. This suggests that multitask regularization with the related problem, treatment, and test extraction task in i2b2-2010 may be inducing features more suited to generalizability for these entity types. These are the only NER tasks with overlapping entity definitions.
Our final observation reinforces the commonly laid out claim in the multitasking community related to task orthogonality/overlap. In the supervised multitask setup, 2 tasks are said to have overlap when some characteristics of a given task (eg, data domain, task objective, target label space, etc) should intuitively help with performance on a different but related task. Otherwise, tasks are said to be orthogonal along that characteristic.31 The majority of the tasks (5/8) in this study are token classification objectives. Unlike the 3 segment level tasks, these require the BERT feature encoder to learn task-robust contextual token representations that, due to their prevalence during training, may negatively harm the formation of segment level representations. This objective orthogonality is suggested by consistent and large performance decreases in the entailment tasks (MedNLI and MedRQE). We speculate that this could be aided by including additional clinical-related segment level objectives during training or by incorporating the original next sentence prediction pretraining objective into the multitasking mix. Similarly, the quaero 2014 corpus is entirely in French. This naturally induces a lingual orthogonality relative to the other 7 English corpora. This orthogonality manifests by inducing the largest loss in competitiveness (−6.4%) to fine-tuning baselines across all tasks. Again, we suspect that the inclusion of additional non-English token level tasks could close this performance gap.
To summarize, the main insights from our analysis are:
A general trend of degradation in MT-Clinical BERT task-specific performance over individual task-specific models. This is a direct tradeoff to the 8-fold reduction in parameters and computational speed up at inference provided by MT-Clinical BERT.
The observation of the task-specific performance increase on i2b2-2012 by MT-Clinical BERT. This is potentially due to the quader regularization provided during multitask learning.
The observation that the greatest relative reduction in multitasking performance occurs on datasets (MedNLI, MedRQE and quaero-2014) with orthogonal characteristics to the predominately English token classification (NER) tasks considered.
Limitations
We foresee the following limitations for both the implementation and scaling of our proposed system. First, the datasets considered are annotated over patient discharge summaries. Naturally, different types of notes may have differing underlying data distribution that can lead to performance degradation. Second, we have observed from experiments in other domains that scaling the number of tasks (> 40) during training inversely correlates with per-task performance. This means that multitask training with a large number of tasks may require careful ablation experiments to gauge the net benefit of adding any given task.
FUTURE WORK
There are several directions for future work. We describe them and provide insights below.
Adding more tasks and datasets. Is adding more tasks feasible and beneficial? There is strong evidence12 suggesting that including a greater number of overlapping tasks may increase task-specific predictive performance. This comes with the additional benefit of increasing computational performance at inference as described in this work.
Learning from limited data. Do the representations obtained via multitask learning serve as a better initialization for learning from limited data resources? Work in this direction would benefit from the inclusion of instance ablation studies.
Unifying NLP pipelines into end-to-end systems. Many common NLP tasks build upon the output of previous tasks. This naturally results in phenomena such as error propagation. Can the shared representations produced by a multitask encoder construct an effective joint NER and relation identification system? Recent work33 suggests this is possible, but can it be accomplished in the multitasking framework?
Incorporating pretraining objectives during multitasking. In low, annotated data domains, such as clinical text, we suspect it may be useful to incorporate the self-supervised masked language modeling and next sentence prediction objectives during multitask training. During preliminary experiments, we find that this does not harm system performance.
Task-specific analysis. This work evaluated the effectiveness of multitask learning across tasks. In the future, we would like to explore how the different tasks affect the extraction of the different clinical entity types within our NER datasets.
CONCLUSION
In this work, we evaluated multitask learning over 8 datasets across 3 distinct NLP tasks. We compare our model to a competitive single-task learning baseline. We found that multitask learning is an effective mechanism to distill information from multiple clinical tasks into a single system. Our results show that information sharing exists in our multitasking model, as all task predictions depend only on the representations produced by a single encoder. This has the main benefit of significant hardware and computational reductions at inference with the trade-off of a small performance degradation. Our system directly increases the potential for the use of recent state-of-the-art NLP methods in clinical applications. In addition, we contribute new state-of-the-art baselines for several clinical information extraction tasks. The data repositories and resources of the clinical NLP community have grown steadily over the past two decades—the doors have been opened to consolidate, cross-leverage and jointly build on these expensive annotation efforts. We make our implementation and pretrained models publicly accessible at https://github.com/AndriyMulyar/multitasking_transformers.
FUNDING
This study was supported in part by the National Library of Medicine under award number R15LM013209.
AUTHOR CONTRIBUTIONS
AM and BTM are the primary authors of the manuscript; AM developed the software and performed experimentation. All authors contributed to writing and editing the manuscript.
DATA AVAILABILITY
The n2c2 and i2b2 data were provided by the n2c2 National NLP Clinical Challenges (https://portal.dbmi.hms.harvard.edu/). The quaero-2014 data were provided by the Laboratoire d'Informatique pour la Mécanique et les Sciences de l'Ingénieur (https://quaerofrenchmed.limsi.fr/). The MedRQE and MedNLI datasets were provided by the National Library of Medicine (https://github.com/abachaa/MEDIQA2019 and https://jgc128.github.io/mednli/).
ACKNOWLEDGMENTS
The authors would like to thank Nick Rodriguez for his valuable commentary and suggestions on the final draft of this article.
CONFLICT OF INTEREST STATEMENT
None declared.
References
- 1.Uzuner Ö, South BR, Shen S, DuVall SL.. 2010 i2b2/va challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18 (5): 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun W, Rumshisky A, Uzuner O.. Evaluating temporal relations in clinical text: 2012 i2b2 challenge. J Am Med Inform Assoc 2013; 20 (5): 806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry S, Buchan K, Filannino M, Stubbs A, Uzuner O.. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J Am Med Inform Assoc 2020; 27 (1): 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stubbs A, Kotfila C, Uzuner Ö.. Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/uthealth shared task track 1. J Biomed Inform 2015; 58: S11–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devlin J, Chang M-W, Lee K, Toutanova K.. BERT: Pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; June 3–5, 2019; Minneapolis, MN. [Google Scholar]
- 6.Yang Z, Dai Z, Yang Y, Carbonell J, Salakhutdinov RR, Le QV. Xlnet: Generalized autoregressive pretraining for language understanding. Adv Neural Inf Process Syst2019; 32: 5753–63. [Google Scholar]
- 7.Johnson AE, Pollard TJ, Shen L, et al. Mimic-iii, a freely accessible critical care database. Scientific Data 2016; 3: 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alsentzer E, Murphy J, Boag W, et al. Publicly available clinical BERT embeddings. In: Proceedings of the 2nd Clinical Natural Language Processing Workshop; June 7, 2019; Minneapolis, MN. [Google Scholar]
- 9.Caruana R.Multitask learning. Mach Learn 1997; 28 (1): 41–75. [Google Scholar]
- 10.Worsham J, Kalita J.. Multi-task learning for natural language processing in the 2020s: where are we going? Pattern Recogn Lett 2020; 136: 120–6. [Google Scholar]
- 11.Zhang Y, Yang Q.. A survey on multi-task learning. arXiv preprint arXiv:1707.08114, 2017. [Google Scholar]
- 12.Ramsundar B, Kearnes S, Riley P, Webster D, Konerding D, Pande V.. Massively multitask networks for drug discovery. arXiv preprint arXiv:1502.02072, 2015. [Google Scholar]
- 13.Yan K, Tang Y, Peng Y, et al. Mulan: multitask universal lesion analysis network for joint lesion detection, tagging, and segmentation In: International Conference on Medical Image and Computing and Computer-Assisted Invervention; 2019: 194–202. Springer, Cham.
- 14.Liu X, He P, Chen W, Gao J.. Multi-task deep neural networks for natural language understanding. In: Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics; July 28–August 2, 2019;Florence, Italy. [Google Scholar]
- 15.Raffel C, Shazeer N, Roberts A, et al. Exploring the limits of transfer learning with a unified text-to-text transformer, J Mach Learn Res 2020; 21: 1–67.
- 16.Crichton G, Pyysalo S, Chiu B, Korhonen A.. A neural network multi-task learning approach to biomedical named entity recognition. BMC Bioinform 2017; 18 (1): 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Zhang Y, Ren X, et al. Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics 2019; 35 (10): 1745–52. [DOI] [PubMed] [Google Scholar]
- 18.Khan MR, Ziyadi M, AbdelHady M.. Mt-bioner: Multi-task learning for biomedical named entity recognition using deep bidirectional transformers. arXiv preprint arXiv:2001.08904; 2020. [Google Scholar]
- 19.Mehmood T, Gerevini A, Lavelli A, Serina I. Multi-task learning applied to biomedical named entity recognition task. In: CLIC-IT Italian Conference on Computational Linguistics; November 13–15, 2019; Bari, Italy.
- 20.Akdemir A, Shibuya T.. Analyzing the effect of multi-task learning for biomedical named entity recognition. arXiv Preprint arXiv:2011.00425; 2020. [Google Scholar]
- 21.Li D, Xie Z, Zand M, Fogg T, Dye T.. A neural joint model for entity and relation extraction from biomedical text+. BMC Bioinform 2017; 18 (1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi X, Jiang D, Huang Y, et al. Family history information extraction via deep joint learning. BMC Med Inform Decis Mak 2019; 19 (S10): 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng Y, Chen Q, Lu Z.. An empirical study of multi-task learning on BERT for biomedical text mining. arXiv:2005.02799 [cs] May 2020. [Google Scholar]
- 24.Li J, Liu X, Yin W, Yang M, Ma L, Jin Y.. Empirical evaluation of multi-task learning in deep neural networks for natural language processing. Neural Comput Appl 2020: 1–12. [Google Scholar]
- 25.Collobert R, Weston J. A unified architecture for natural language processing: deep neural networks with multitask learning. In: Proceedings of the 25th International Conference on Machine Learning; Jul 5–9, 2008; New York, NY.
- 26.Henry S, Buchan K, Filannino M, Stubbs A, Uzuner O. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J Am Med Inform Assoc 2020; 27 (1): 3–12. [DOI] [PMC free article] [PubMed]
- 27.Romanov A, Shivade C.. Lessons from natural language inference in the clinical domain. arXiv preprint arXiv:1808.06752; 2018. [Google Scholar]
- 28.Ben Abacha A, Demner-Fushman D.. Recognizing question entailment for medical question answering. AMIA Annu Symp Proc 2016; 2016: 310–8. [PMC free article] [PubMed] [Google Scholar]
- 29.Névéol A, Grouin C, Leixa J, Rosset S, Zweigenbaum P. The Quaero French medical corpus: a resource for medical entity recognition and normalization. In: Proceedings of the Fourth Workshop on Building and Evaluating Ressources for Health and Biomedical Text Processing; May 31, 2014; Reykjavik, Iceland.
- 30.McCray AT, Burgun A, Bodenreider O.. Aggregating UMLs semantic types for reducing conceptual complexity. Stud Health Technol Inform 2001; 84 (01): 216. [PMC free article] [PubMed] [Google Scholar]
- 31.Ruder S.Neural Transfer Learning for Natural Language Processing [doctoral thesis]. Galway, Ireland, National University of Ireland; 2019.
- 32.Dodge J, Ilharco G, Schwartz R, Farhadi A, Hajishirzi H, Smith N. Fine-tuning pretrained language models: weight initializations, data orders, and early stopping. arXiv:2002.06305; 2020.
- 33.Giorgi J, Wang X, Sahar N, Young Shin W, Bader GD, Wang B. End-to-end named entity recognition and relation extraction using pre-trained language models. arXiv:1912.13415; 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The n2c2 and i2b2 data were provided by the n2c2 National NLP Clinical Challenges (https://portal.dbmi.hms.harvard.edu/). The quaero-2014 data were provided by the Laboratoire d'Informatique pour la Mécanique et les Sciences de l'Ingénieur (https://quaerofrenchmed.limsi.fr/). The MedRQE and MedNLI datasets were provided by the National Library of Medicine (https://github.com/abachaa/MEDIQA2019 and https://jgc128.github.io/mednli/).