Abstract
Objective
This study investigated the epidemiology, virulence and drug resistance of invasive Klebsiella pneumoniae (K. pneumoniae) isolates at a children’s medical center in eastern China in order to obtain epidemiologic, virulence, and antimicrobial resistance data that can guide for the selection and development of anti-infection treatments.
Methods
A total of 94 invasive K. pneumoniae strains were isolated from children between January 2016 and December 2020 at the Children’s Hospital of Soochow University. The strains were identified by mass spectrometry. The Kirby–Bauer method and VITEK 2 Compact system were used to analyze the antimicrobial susceptibility. Polymerase chain reaction (PCR) and sequencing was performed to detect the capsular serotypes, virulence-associated genes, β-lactam antibiotic resistance genes and multilocus sequence typing.
Results
The PCR results showed that 87 strains (92.55%) of invasive K. pneumoniae were hypervirulent capsular serotypes, with K57 as the dominant capsular serotype (62.77%). All strains carried virulence-associated genes. Among them, 84 strains (89.36%) carried hypervirulence genes, with iroB (86.17%) being the predominant; meanwhile, other virulence genes, including wabG (100.00%), mrkD (98.94%), ycfM (96.81%), fimH (95.74%) and Uge (88.30%), were detected in most strains. All strains carried β-lactam antibiotic resistance genes; the main extended-spectrum β-lactamase gene was blaSHV-11 (86.17%) and the major AmpC cephalosporinase genes were blaFOX-1 (86.17%) and blaACT-1 (70.21%). Carbapenemase genes were detected in only a few isolates. Notably, 12 invasive K. pneumoniae isolates were identified as carbapenem-resistant and hypervirulent K. pneumoniae (CR-HVKP), and 14 other multidrug resistance (MDR) isolates were also detected.
Conclusion
The results of this study reveal the epidemiology, virulence and antimicrobial resistance of invasive K. pneumoniae in pediatric patients. Both CR-HVKP and MDR strains were identified, which should be of great concern to clinicians.
Keywords: Klebsiella pneumoniae, invasive infection, children, virulence factors, resistance genes
Introduction
Invasive bacterial infection caused by Klebsiella pneumoniae (K. pneumoniae) is one of the most common diseases in children.1,2 The infection can involve the respiratory tract, blood, nervous system and surgical site, and lead to pneumonia, bacteremia/septicemia, meningitis and abscesses.2 In severe clinical cases, it can also lead to multiple organ failure, or even death.3
In recent years, infection with hypervirulent K. pneumoniae (HVKP) has been reported in many countries and has become a global public health concern.4–6 The virulence of HVKP is associated with capsular polysaccharides, siderophores and other virulence factors.7–9 Previous studies of HVKP mainly screened for aerobactin, which is encoded by iucA.9 However, more hypervirulence genes such as prmpA, rmpA2, crmpA, peg-344, terB, iroB and irp2 have recently been identified.7 These HVKP strains warrant closer attention as they have the ability to cause severe and life-threatening infections in healthy individuals through these virulence factors.6
The development of drug resistance by K. pneumoniae and the emergence of carbapenem-resistant K. pneumoniae (CRKP) caused by the misuse and overuse of antibiotics is also a major threat to public health.10,11 The isolation rate of CRKP is higher among children than in adults and is increasing in China.12,13 Meanwhile, the multidrug-resistant (MDR) is more likely to occur than CRKP,14–16 and MDR pathogens can be transmitted to humans via the food chain or through direct contact with infected animals,17–19 which should be of concern as well.
HVKP and CRKP were not traditionally overlapped.13 However, cases of infection with carbapenem-resistant and hypervirulent K. pneumoniae (CR-HVKP) have recently been reported, which have a poor prognosis and are challenging to treat.20–23 To date, there have been limited studies of CR-HVKP isolated from pediatric patients. On this basis, the present study investigated the virulence and antibiotic resistance characteristics of invasive K. pneumoniae at a children’s medical center in eastern China.
Materials and Methods
Study Site
This study was conducted in Suzhou, a major city with a population of 12.7 million in the southeast area of Jiangsu Province in eastern China. The population of children aged under 15 years old was 1.7 million in 2020. The Children’s Hospital of Soochow University (CHSU), located in the central area of Suzhou, is a children’s medical center in eastern China and the only provincial tertiary children’s hospital in Jiangsu Province. CHSU has 1500 beds and serves >70,000 inpatients and >2 million outpatients annually. This study had no impact on patients and was approved by the Ethics Committee of CHSU (No. 2020CS099).
Bacterial Strains
Nonduplicated invasive K. pneumoniae isolates were collected at CHSU from January 1, 2016 to December 31, 2020. Invasive K. pneumoniae was defined as K. pneumoniae isolated from a normally sterile site, namely blood, cerebrospinal fluid (CSF), pleural fluid or joint fluid. All invasive K. pneumoniae strains were confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, Bruker, Mannheim, Germany).24
Detection of Capsular Serotypes and Virulence-Associated Genes
The common hypervirulent capsular serotypes (K1, K2, K5, K20, K54 and K57), hypervirulence genes (iucA, prmpA, prmpA2, crmpA, peg-344, terB, iroB and irp2) and other virulence genes (ybtS, mrkD, entB, kfu, allS, iutA, k2A, wabG, Uge, fimH, wcaG, Kpn, ycfM, iroN, Hly and cnf-1) of invasive K. pneumoniae isolates were screened by PCR as previously described.7,13,25 The PCR products were analyzed by 1.5% agarose gel electrophoresis, and positive products were sequenced and aligned with sequences in GenBank using the Basic Local Alignment Search Tool (BLAST).
Antimicrobial Susceptibility Test
The antimicrobial susceptibility of invasive K. pneumoniae isolates was determined by the Kirby–Bauer (K-B) method and Vitek 2 Compact system (bioMérieux, Marcy-l’Etoile, France) according to the manufacturer’s instructions. The results were analyzed according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI).24,26
Detection of β-Lactam Antibiotic and Colistin Resistance Genes
PCR was performed to detect carbapenemase genes (blaKPC, blaOXA, blaNDM, blaVIM, blaIMP, blaAIM, blaGIM, blaSIM, blaSPM, blaBIC and blaDIM), extended-spectrum β-lactamase (ESBL) genes (blaTEM, blaSHV, blaCTX-M, blaVEB and blaPER) and AmpC cephalosporinase genes (blaCMY, blaACT, blaACC, blaDHA, blaFOX, blaMOX, blaCIT and blaEBC).13,27 Colistin resistance genes (MCR-1 and MCR-2) were also examined in parallel.28,29 The primers used to screen the abovementioned genes have been previously reported and the PCR products were analyzed as described above.
Multilocus Sequence Typing (MLST)
MLST was performed using the primers and protocol available from the Pasteur MLST website (https://bigsdb.pasteur.fr/klebsiella/primers_used.html). Seven housekeeping genes of invasive K. pneumoniae isolates, namely rpoB, gapA, mdh, pgi, phoE, infB and tonB, were analyzed and the sequence types (STs) were compared with those available in the Pasteur MLST database.
Data Analysis
The chi-squared test was used to analyze the data and p<0.05 was considered statistically significant. Antibiotic resistance data were analyzed with WHONET v5.6 software (WHO Collaborating Centre for Surveillance of Antimicrobial Resistance, Boston, MA, USA).
Results
Clinical Characteristics and Epidemiology of Invasive K. pneumoniae Infection
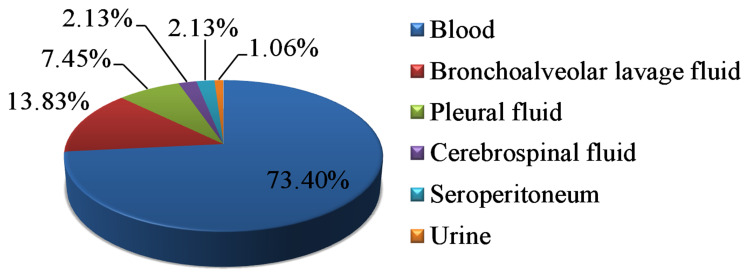
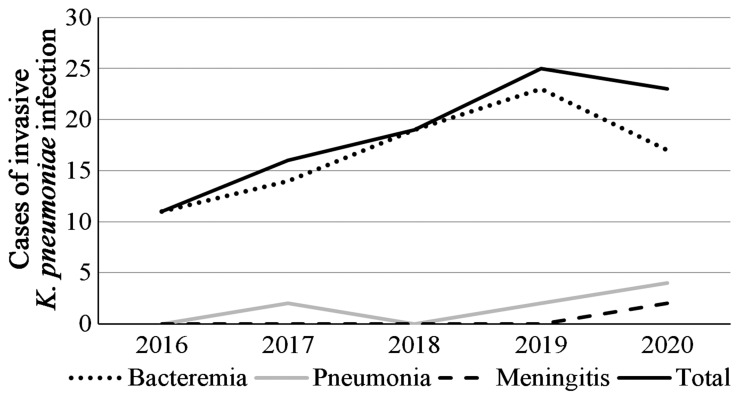
Over the 5-year study period, a total of 94 non-duplicated invasive K. pneumoniae strains were collected and identified at our hospital. The clinical characteristics of the study population are summarized in Table 1. Of the 94 children, 59 (62.77%) were male. The median age at the time of diagnosis was 24 months (range: 3 days-180 months); at the time of infection, 62 children (65.96%) were aged <5 years and 32 (34.04%) were aged ≥ 5 years. Most of the invasive K. pneumoniae strains were isolated from the department of hematology (52.13%) and neonatology (25.53%). The clinical syndromes recognized in children with invasive K. pneumoniae infection were bacteremia (89.36%, 84/94), pneumonia (8.51%, 8/94) and meningitis (2.13%, 2/94). Based on the first isolates, the primary sites of infection were blood (73.40%, 69/94), bronchoalveolar lavage fluid (13.83%, 13/94), pleural fluid (7.45%, 7/94), cerebrospinal fluid (2.13%, 2/94), seroperitoneum (2.13%, 2/94) and urine (1.06%, 1/94) (Figure 1). The annual trends of invasive K. pneumoniae cases by clinical diagnosis are shown in Figure 2. There was an increase in the overall number of cases between 2016 and 2019, primarily driven by an increase in K. pneumoniae bacteremia although there were also marginal increases in the number of cases of invasive K. pneumoniae pneumonia and meningitis. Additionally, 12 strains of invasive K. pneumoniae were CR-HVKP.
Table 1.
Clinical Characteristics and Epidemiology of Invasive K. pneumoniae Infection at Children’s Hospital of Soochow University from 2016 to 2020
| Bacteremia | Pneumonia | Meningitis | Total | |
|---|---|---|---|---|
| (n=84) | (n=8) | (n=2) | (n=94) | |
| Gender, n (%) | ||||
| Male | 51 (60.71) | 6 (75.00) | 2 (100.00) | 59 (62.77) |
| Female | 33 (39.29) | 2 (25.00) | 0 (0) | 35 (37.23) |
| Age* | ||||
| Median | 30 m | 17 d | 6 m | 24 m |
| Range | 3 d-168 m | 7 d-180 m | 2–10 m | 3 d-180 m |
| Age*, n (%) | ||||
| <5 y | 53 (63.10) | 7 (87.50) | 2 (100.00) | 62 (65.96) |
| <1 m | 14 (16.67) | 5 (62.50) | 0 (0) | 19 (20.21) |
| 1–5 m | 14 (16.67) | 0 (0) | 1 (50.00) | 15 (15.96) |
| 6–11 m | 5 (5.95) | 0 (0) | 1 (50.00) | 6 (6.38) |
| 12–59 m | 20 (23.81) | 2 (25.00) | 0 (0) | 22 (23.40) |
| 5–18 y | 31 (36.90) | 1 (12.50) | 0 (0) | 32 (34.04) |
| Wards, n (%) | ||||
| Haematology | 49 (58.33) | 0 (0) | 0 (0) | 49 (52.13) |
| Neonatology | 19 (22.62) | 5 (62.50) | 0 (0) | 24 (25.53) |
| ICU | 5 (5.95) | 3 (37.50) | 1 (50.00) | 9 (9.57) |
| Infectious disease | 3 (3.57) | 0 (0) | 0 (0) | 3 (3.19) |
| Others | 8 (9.52) | 0 (0) | 1 (50.00) | 9 (9.57) |
Notes: Others: including the departments of neurosurgery, cardiovasology, general surgery, gastroenterology, nephrology and emergency ward.
Abbreviations: *d, days; m, months; y, years; ICU, intensive care unit.
Figure 1.
The first isolated samples of invasive K. pneumoniae strains in this study.
Figure 2.
The annual trends of invasive K. pneumoniae cases by clinical diagnosis from 2016 to 2020 in this study.
Distribution of Capsular Types and Virulence-Associated Genes
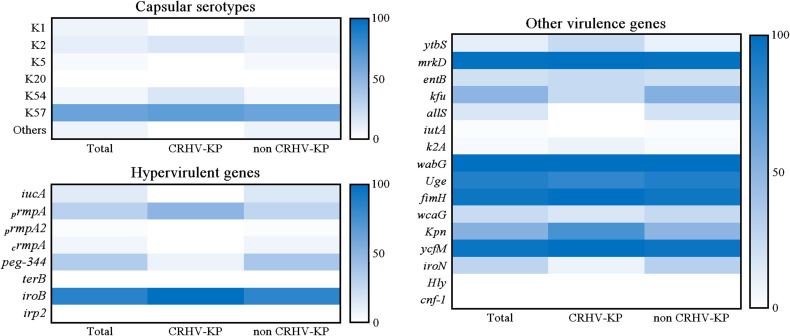
Previous studies have shown that some capsular serotypes of K. pneumoniae (eg, K1, K2, K5, K20, K54, and K57) contribute to hypervirulence.13 In this study, common hypervirulent capsular serotypes were detected in 87 strains (92.55%) of invasive K. pneumoniae; the main capsular serotype was K57 (62.77%, 59/94) and the remaining ones were K2 (11.70%, 11/94), K1 (7.45%, 7/94), K54 (6.38%, 6/94), K5 (4.26%, 4/94) and others (7.45%, 7/94). The K20 capsular serotype was not detected (Table 2 and Figure 3). All invasive K. pneumoniae strains carried virulence-associated genes; among them, 84 (89.36%) carried hypervirulence genes, which were iroB (86.17%, 81/94), peg-344 (34.04%, 32/94), prmpA (30.85%, 29/94), iucA (13.83%, 13/94), crmpA (6.38%, 6/94) and prmpA2 (2.13%, 2/94). The other main virulence genes were wabG (100%, 94/94), mrkD (98.94%, 93/94), ycfM (96.81%, 91/94), fimH (95.74%, 90/94) and Uge (88.30%, 83/94) (Table 2 and Figure 3). There were no significant differences between CR-HVKP and non CR-HVKP strains in terms of capsular types and virulence-associated genes. These data suggest that the isolated invasive K. pneumoniae strains, including CR-HVKP and non CR-HVKP, all carried a high proportion of virulence genes that may contribute to disease.
Table 2.
The Capsular Serotypes and Virulence-Associated Genes of Invasive K. pneumoniae Strains in This Study
| Total | CRHV-KP | non CRHV-KP | P value | |
|---|---|---|---|---|
| (n=94) | (n=12) | (n=82) | ||
| Capsular serotypes, n (%) | ||||
| K1 | 7 (7.45) | 0 (0) | 7 (8.54) | 0.5895 |
| K2 | 11 (11.70) | 2 (16.67) | 9 (10.98) | 0.9266 |
| K5 | 4 (4.26) | 0 (0) | 4 (4.88) | 1.0000 |
| K20 | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
| K54 | 6 (6.38) | 2 (16.67) | 4 (4.88) | 0.1678 |
| K57 | 59 (62.77) | 8 (66.67) | 51 (62.20) | 0.9837 |
| Others | 7 (7.45) | 0 (0) | 7 (8.54) | 0.5895 |
| Hypervirulent genes, n (%) | ||||
| iucA | 13 (13.83) | 0 (0) | 13 (15.85) | 0.2992 |
| prmpA | 29 (30.85) | 6 (50.00) | 23 (28.05) | 0.2289 |
| prmpA2 | 2 (2.13) | 0 (0) | 2 (2.44) | 1.0000 |
| crmpA | 6 (6.38) | 0 (0) | 6 (7.32) | 1.0000 |
| peg-344 | 32 (34.04) | 1 (8.33) | 31 (37.80) | 0.0918 |
| terB | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
| iroB | 81 (86.17) | 12 (100.00) | 69 (84.15) | 0.2992 |
| irp2 | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
| Other virulence genes, n (%) | ||||
| ytbS | 11 (11.70) | 3 (25.00) | 8 (9.76) | 0.2921 |
| mrkD | 93 (98.94) | 12 (100.00) | 81 (98.78) | 1.0000 |
| entB | 20 (21.28) | 3 (25.00) | 17 (20.73) | 0.9680 |
| kfu | 47 (50.00) | 3 (25.00) | 44 (53.66) | 0.0637 |
| allS | 16 (17.02) | 0 | 16 (19.51) | 0.2046 |
| iutA | 2 (2.13) | 0 | 2 (2.44) | 1.0000 |
| k2A | 4 (4.26) | 1 (8.33) | 3 (3.66) | 0.4264 |
| wabG | 94 (100.00) | 12 (100.00) | 82 (100.00) | 1.0000 |
| Uge | 83 (88.30) | 10 (83.33) | 73 (89.02) | 0.9266 |
| fimH | 90 (95.74) | 12 (100.00) | 78 (95.12) | 1.0000 |
| wcaG | 23 (24.47) | 2 (16.67) | 21 (25.61) | 0.7538 |
| Kpn | 50 (53.19) | 9 (75.00) | 41 (50.00) | 0.1050 |
| ycfM | 91 (96.81) | 12 (100.00) | 79 (96.34) | 1.0000 |
| iroN | 27 (28.72) | 1 (8.33) | 26 (31.71) | 0.1836 |
| Hly | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
| cnf-1 | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
Figure 3.
The heat-map of capsular serotypes and virulence-associated genes of invasive K. pneumoniae strains in this study.
Antimicrobial Susceptibility
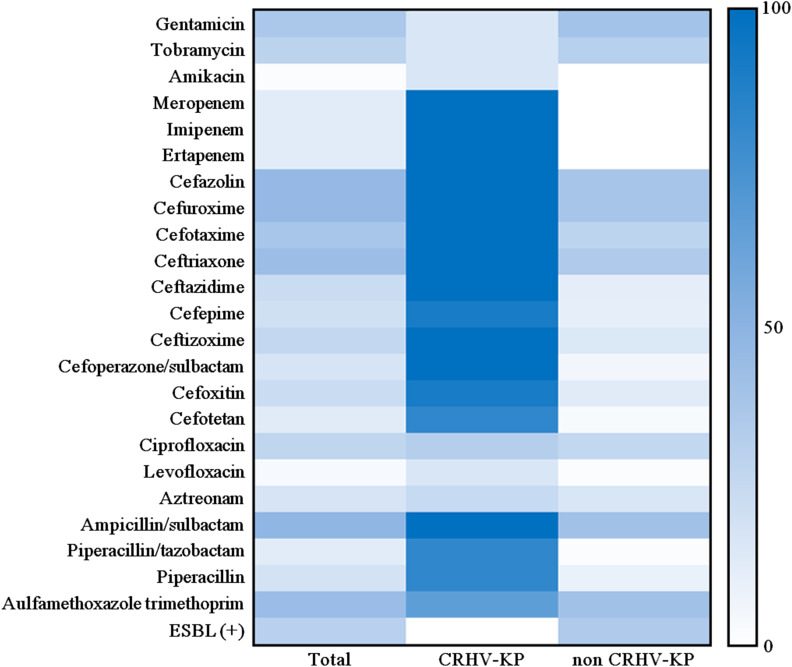
The invasive K. pneumoniae strains exhibited different patterns of resistance to various antibiotics, but in general, they showed low resistance to aminoglycosides (amikacin and tobramycin), carbapenems (meropenem, imipenem and ertapenem), cephalosporins (ceftazidime, cefepime and ceftizoxime), cephalosporins+β-lactamase inhibitors (cefoperazone/sulbactam), cephamycins (cefoxitin and cefotetan), fluoroquinolones (ciprofloxacin and levofloxacin), monobactams (aztreonam), penicillins+β-lactamase inhibitors (piperacillin/tazobactam) and penicillins (piperacillin) (Table 3 and Figure 4). Notably, 29 strains (30.85%) of non CR-HVKP produced ESBLs. The resistance rates of CR-HVKP strains were much higher than those of non–CR-HVKP strains (Table 3 and Figure 4). Moreover, there were significant differences (p<0.05) between CR-HVKP and non CR-HVKP strains in terms of susceptibility to aminoglycosides (amikacin), carbapenems, cephalosporins, cephalosporins+β-lactamase inhibitors, cephamycins, penicillins+β-lactamase inhibitors, and penicillins (Table 3 and Figure 4).
Table 3.
The Antimicrobial Resistance of Invasive K. pneumoniae Strains in This Study
| Antibiotics | Total | CRHV-KP | non CRHV-KP | P value |
|---|---|---|---|---|
| (n=94) | (n=12) | (n=82) | ||
| R (n, %) | R (n, %) | R (n, %) | ||
| Aminoglycosides | ||||
| Gentamicin | 35 (37.23) | 2 (16.67) | 33 (40.24) | 0.2083 |
| Amikacin | 2 (2.13) | 2 (16.67) | 0 (0) | 0.0151 |
| Tobramycin | 28 (29.79) | 2 (16.67) | 26 (31.71) | 0.4677 |
| Carbapenems | ||||
| Meropenem | 12 (12.77) | 12 (100.00) | 0 (0) | 2.6496E-20 |
| Imipenem | 12 (12.77) | 12 (100.00) | 0 (0) | 2.6496E-20 |
| Ertapenem | 12 (12.77) | 12 (100.00) | 0 (0) | 2.6496E-20 |
| Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins | ||||
| Cefazolin | 44 (46.81) | 12 (100.00) | 32 (39.02) | 7.6944E-05 |
| Cefuroxime | 44 (46.81) | 12 (100.00) | 32 (39.02) | 7.6944E-05 |
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins | ||||
| Cefotaxime | 36 (38.30) | 12 (100.00) | 24 (29.27) | 1.1345E-05 |
| Ceftriaxone | 41 (43.62) | 12 (100.00) | 29 (35.37) | 2.4768E-05 |
| Ceftazidime | 22 (23.40) | 12 (100.00) | 10 (12.20) | 2.2286E-10 |
| Cefepime | 20 (21.28) | 11 (91.67) | 9 (10.98) | 1.9556E-09 |
| Ceftizoxime | 25 (26.60) | 12 (100.00) | 13 (15.85) | 6.1744E-09 |
| Cephalosporins + β-lactamase inhibitors | ||||
| Cefoperazone/sulbactam | 17 (18.09) | 12 (100.00) | 5 (6.10) | 6.7846E-14 |
| Cephamycins | ||||
| Cefoxitin | 22 (23.40) | 11 (91.67) | 11 (13.41) | 1.9690E-08 |
| Cefotetan | 13 (13.83) | 10 (83.33) | 3 (3.66) | 2.2231E-12 |
| Fluoroquinolones | ||||
| Ciprofloxacin | 26 (27.66) | 4 (33.33) | 22 (26.83) | 0.9006 |
| Levofloxacin | 4 (4.26) | 2 (16.67) | 2 (2.44) | 0.0780 |
| Monobactams | ||||
| Aztreonam | 17 (18.09) | 3 (25.00) | 14 (17.07) | 0.7911 |
| Penicillins + β-lactamase inhibitors | ||||
| Ampicillin/sulbactam | 46 (48.94) | 12 (100.00) | 34 (41.46) | 0.0002 |
| Piperacillin/tazobactam | 12 (12.77) | 10 (83.33) | 2 (2.44) | 1.5841E-13 |
| Penicillins | ||||
| Piperacillin | 18 (19.15) | 10 (83.33) | 8 (9.76) | 1.5374E-08 |
| Sulfonamides | ||||
| Aulfamethoxazole trimethoprim | 42 (44.68) | 8 (66.67) | 34 (41.46) | 0.1010 |
| ESBL (+) | 29 (30.85) | 0 (0) | 29 (35.37) | 0.0321 |
Figure 4.
The heat-map of the different degrees of antimicrobial resistance of invasive K. pneumoniae strains in this study.
Distribution of β-Lactam Antibiotic and Colistin Resistance Genes
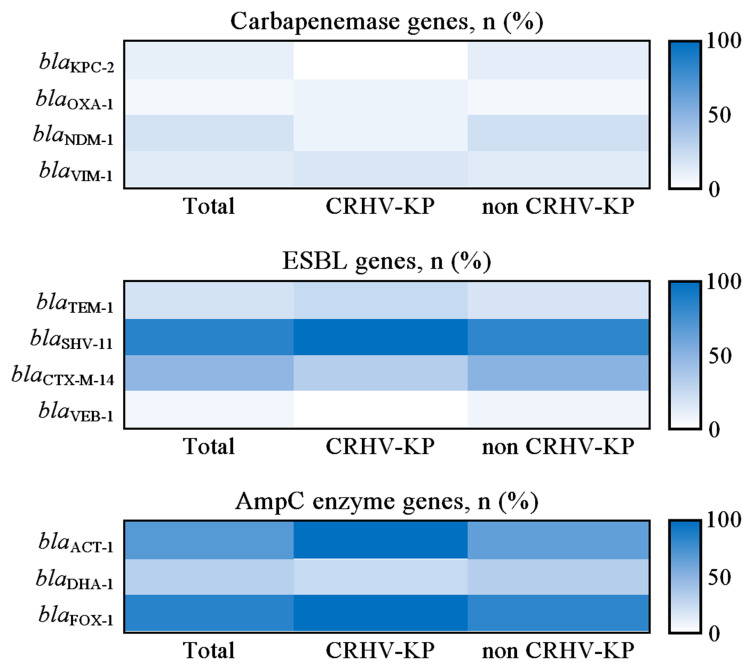
Carbapenemase, ESBL and AmpC cephalosporinase genes are the main β-lactam antibiotic resistance genes in Enterobacteriaceae.13,27 In this study, all invasive K. pneumoniae strains carried β-lactam antibiotic resistance genes. The carbapenemase genes were blaNDM-1 (19.15%, 18/94), blaVIM-1(13.83%, 13/94), blaKPC-2 (10.64%, 10/94) and blaOXA-1 (5.32%, 5/94); the ESBL genes were blaSHV-11 (86.17%, 81/94), blaCTX-M-14 (48.94%, 46/94), blaTEM-1 (19.15%, 18/94) and blaVEB-1 (6.38%, 6/94); the AmpC cephalosporinase genes were blaFOX-1 (86.17%, 81/94), blaACT-1 (70.21%, 66/94) and blaDHA-1 (31.91%, 30/94). Colistin resistance genes (MCR-1 and MCR-2) were not detected (Table 4 and Figure 5). There were no significant differences between CR-HVKP and non CR-HVKP strains in terms of the rates of carbapenemase, ESBL and AmpC cephalosporinase genes (with the exception of blaACT-1). These data suggest that invasive K. pneumoniae strains including CR-HVKP and non CR-HVKP harbor a high proportion of β-lactam antibiotic resistance genes simultaneously.
Table 4.
The β-Lactam Antibiotic and Colistin Resistance Genes of Invasive K. pneumoniae Strains in This Study
| Total | CRHV-KP | non CRHV-KP | P value | |
|---|---|---|---|---|
| (n=94) | (n=12) | (n=82) | ||
| Carbapenemase genes, n (%) | ||||
| blaKPC-2 | 10 (10.64) | 0 (0) | 10 (12.20) | 0.4363 |
| blaOXA-1 | 5 (5.32) | 1 (8.33) | 4 (4.88) | 0.5029 |
| blaNDM-1 | 18 (19.15) | 1 (8.33) | 17 (20.73) | 0.5308 |
| blaVIM-1 | 13 (13.83) | 2 (16.67) | 11 (13.41) | 0.8864 |
| blaIMP, blaAIM, blaGIM, blaSIM, blaSPM, blaBIC, blaDIM | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
| ESBL genes, n (%) | ||||
| blaTEM-1 | 18 (19.15) | 3 (25.00) | 15 (18.29) | 0.8738 |
| blaSHV-11 | 81 (86.17) | 12 (100.00) | 69 (84.15) | 0.2992 |
| blaCTX-M-14 | 46 (48.94) | 4 (33.33) | 42 (51.22) | 0.2470 |
| blaVEB-1 | 6 (6.38) | 0 (0) | 6 (7.32) | 1.0000 |
| blaPER | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
| AmpC enzyme genes, n (%) | ||||
| blaACT-1 | 66 (70.21) | 12 (100.00) | 54 (65.85) | 0.0377 |
| blaDHA-1 | 30 (31.91) | 3 (25.00) | 27 (32.93) | 0.8269 |
| blaFOX-1 | 81 (86.17) | 12 (100.00) | 69 (84.15) | 0.2992 |
| blaCMY, blaACC, blaMOX, blaCIT, blaEBC | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
| Colistin resistance genes, n (%) | ||||
| MCR-1 | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
| MCR-2 | 0 (0) | 0 (0) | 0 (0) | 1.0000 |
Figure 5.
The heat-map of β-lactam antibiotic genes of invasive K. pneumoniae strains in this study.
Characteristics of CR-HVKP Strains
A total of 12 invasive K. pneumoniae strains were identified as CR-HVKP, which carried at least 3 β-lactam antibiotic resistance genes. Meanwhile, all 12 CR-HVKP strains carried hypervirulence and other virulence genes, and had hypervirulent capsular serotypes. Additionally, 7 distinct STs were observed among these CR-HVKP strains: ST29 (25.00%, 3/12), ST14 (16.67%, 2/12), ST17 (16.67%, 2/12), ST37 (16.67%, 2/12), ST45 (8.33%, 1/12), ST101 (8.33%, 1/12) and ST234 (8.33%, 1/12) (Table 5). The 12 CR-HVKP strains had low resistance to gentamicin, amikacin, tobramycin, levofloxacin and aztreonam, but were resistant to most other antimicrobial drugs (Table 3 and Figure 4).
Table 5.
The Capsular Serotypes, Genotypes and MLST of CR-HVKP Strains Identified in This Study (n=12)
| STs | Capsular Serotypes | Virulence Genes | β-Lactam Antibiotics Resistance Genes |
|---|---|---|---|
| ST14 (n=2) | K2 | prmpA, iroB, mrkD, kfu, K2A, wabG, Uge, fimH, ycfM | blaSHV-11, blaCTX-M-14, blaACT-1, blaDHA-1, blaFOX-1 |
| K2 | prmpA, iroB, mrkD, kfu, wabG, Uge, fimH, ycfM, iroN | blaSHV-11, blaACT-1, blaDHA-1, blaFOX-1 | |
| ST17 (n=2) | K57 | iroB, mrkD, wabG, Uge, fimH, Kpn, ycfM | blaSHV-11, blaNDM-1, blaVIM-1, blaACT-1, blaFOX-1 |
| K57 | prmpA, iroB, mrkD, wabG, Uge, fimH, Kpn, ycfM | blaSHV-11, blaACT-1, blaFOX-1 | |
| ST29 (n=3) | K57 | iroB, ytbS, mrkD, entB, wabG, Uge, fimH, Kpn, ycfM | blaTEM-1, blaSHV-11, blaOXA-1, blaACT-1, blaFOX-1 |
| K54 | iroB, mrkD, entB, wabG, Uge, fimH, wcaG, Kpn, ycfM | blaTEM-1, blaSHV-11, blaACT-1, blaFOX-1 | |
| K54 | prmpA, iroB, peg-344, ytbS, mrkD, wabG, Uge, fimH, wcaG, Kpn, ycfM | blaSHV-11, blaACT-1, blaFOX-1 | |
| ST37 (n=2) | K57 | prmpA, iroB, mrkD, entB, wabG, Uge, fimH, Kpn, ycfM | blaSHV-11, blaCTX-M-14, blaVIM-1, blaACT-1, blaFOX-1 |
| K57 | iroB, mrkD, wabG, fimH, Kpn, ycfM | blaSHV-11, blaCTX-M-14, blaACT-1, blaFOX-1 | |
| ST45 (n=1) | K57 | prmpA, iroB, ytbS, mrkD, wabG, Uge, fimH, Kpn, ycfM | blaTEM-1, blaSHV-11, blaACT-1, blaFOX-1 |
| ST101 (n=1) | K57 | iroB, mrkD, kfu, wabG, Uge, fimH, ycfM | blaSHV-11, blaACT-1, blaFOX-1 |
| ST234 (n=1) | K57 | iroB, mrkD, wabG, fimH, Kpn, ycfM | blaSHV-11, blaCTX-M-14, blaACT-1, blaDHA-1, blaFOX-1 |
Correlation Between Phenotypic and Genotypic MDR Patterns in Invasive K. pneumoniae
Besides the 12 CR-HVKP strains, other MDR strains of invasive K. pneumoniae were also observed in this study. A total of 14 strains (14.89%) were MDR (defined as resistant to ≥ 1 agent in ≥ 3 antimicrobial classes30); these strains exhibited MDR to 7 antimicrobial classes (n=3), 6 antimicrobial classes (n=2), 5 antimicrobial classes (n=6), 4 antimicrobial classes (n=2), and 3 antimicrobial classes (n=1). Additionally, 8 of the strains harbored 5 β-lactam antibiotic resistance genes, 5 strains had 4 of these genes, and 1 strain had 3 of the genes (Table 6).
Table 6.
The Correlation Between Phenotypic and Genotypic Resistance Patterns Among the MDR K. pneumoniae Strains in This Study (n=14)
| Strain Number | In-vitro Phenotypic Resistance | The Antimicrobial Resistance Genes |
|---|---|---|
| 2 | Aminoglycosides: Gentamicin | blaSHV-11, blaACT-1, blaFOX-1, |
| Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | ||
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Ceftriaxone, Ceftizoxime | ||
| Monobactams: Aztreonam | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| Penicillins: Piperacillin | ||
| Sulfonamides: Aulfamethoxazole trimethoprim | ||
| 6 | Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | blaTEM-1, blaSHV-11, blaOXA-1, blaACT-1, blaFOX-1 |
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Ceftriaxone, Ceftazidime, Cefepime, Ceftizoxime | ||
| Monobactams: Aztreonam | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| Penicillins: Piperacillin | ||
| Sulfonamides: Aulfamethoxazole trimethoprim | ||
| 8 | Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | blaSHV-11, blaOXA-1, blaACT-1, blaFOX-1 |
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Ceftriaxone, Ceftizoxime | ||
| Fluoroquinolones: Ciprofloxacin | ||
| Monobactams: Aztreonam | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| Penicillins: Piperacillin | ||
| Sulfonamides: Aulfamethoxazole trimethoprim | ||
| 9 | Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | blaSHV-11, blaCTX-M-14, blaVIM-1, blaACT-1, blaFOX-1 |
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Ceftriaxone | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| Penicillins: Piperacillin | ||
| Sulfonamides: Aulfamethoxazole trimethoprim | ||
| 15 | Aminoglycosides: Gentamicin, Tobramycin | blaSHV-11, blaCTX-M-14, blaACT-1, blaDHA-1, blaFOX-1 |
| Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | ||
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Ceftriaxone, Ceftazidime, Cefepime | ||
| Monobactams: Aztreonam | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| Penicillins: Piperacillin | ||
| Sulfonamides: Aulfamethoxazole trimethoprim | ||
| 16 | Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | blaSHV-11, blaCTX-M-14, blaACT-1, blaDHA-1, blaFOX-1 |
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Ceftriaxone, Ceftizoxime | ||
| Cephamycins: Cefoxitin | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| Penicillins: Piperacillin | ||
| 22 | Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | blaTEM-1, blaSHV-11, blaVIM-1, blaACT-1, blaFOX-1 |
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Cefotaxime, Ceftriaxone | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| 24 | Aminoglycosides: Gentamicin | blaTEM-1, blaSHV-11, blaACT-1, blaFOX-1 |
| Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | ||
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Cefotaxime, Ceftriaxone | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| Sulfonamides: Aulfamethoxazole trimethoprim | ||
| 37 | Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | blaTEM-1, blaSHV-11, blaACT-1, blaFOX-1 |
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Cefotaxime, Ceftriaxone | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| Sulfonamides: Aulfamethoxazole trimethoprim | ||
| 48 | Aminoglycosides: Gentamicin | blaTEM-1, blaSHV-11, blaACT-1, blaFOX-1 |
| Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | ||
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Cefotaxime, Ceftriaxone | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| Sulfonamides: Aulfamethoxazole trimethoprim | ||
| 65 | Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | blaSHV-11, blaNDM-1, blaVIM-1, blaVEB-1, blaFOX-1 |
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Cefotaxime, Ceftriaxone | ||
| Monobactams: Aztreonam | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| Sulfonamides: Aulfamethoxazole trimethoprim | ||
| 73 | Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | blaSHV-11, blaCTX-M-14, blaDHA-1, blaFOX-1 |
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Cefotaxime, Ceftriaxone | ||
| Cephamycins: Cefoxitin | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam | ||
| Sulfonamides: Aulfamethoxazole trimethoprim | ||
| 79 | Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | blaSHV-11, blaNDM-1, blaVIM-1, blaVEB-1, blaFOX-1 |
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Cefotaxime, Ceftriaxone, Ceftizoxime | ||
| Monobactams: Aztreonam | ||
| Sulfonamides: Aulfamethoxazole trimethoprim | ||
| 94 | Aminoglycosides: Gentamicin | blaTEM-1, blaSHV-11, blaKPC-2, blaACT-1, blaFOX-1 |
| Non-extended spectrum cephalosporins; 1st and 2nd generation cephalosporins: Cefazolin, Cefuroxime | ||
| Extended-spectrum cephalosporins; 3rd and 4th generation cephalosporins: Cefotaxime, Ceftriaxone | ||
| Fluoroquinolones: Ciprofloxacin | ||
| Monobactams: Aztreonam | ||
| Penicillins + β-lactamase inhibitors: Ampicillin/sulbactam |
Discussion
K. pneumoniae is one of the most common pathogens responsible for nosocomial and community-acquired infections in children, which is associated with high morbidity and mortality.1–3 In recent years, the increased prevalence of HVKP has become a serious global threat to public health.4 HVKP can cause many types of invasive infection, such as severe pneumonia, bacteremia/septicemia and meningitis, which are often associated with severe symptoms and high mortality rates.4–6 The capsular serotype of K. pneumoniae is one of the factors responsible for its hypervirulence, protecting the bacteria from phagocytosis. There are at least 78 capsular serotypes, with K1, K2, K5, K20, K54 and K57 being the most common hypervirulent serotypes.8,31 In addition, K. pneumoniae also carries several genes that were shown to contribute to hypervirulence, such as iucA (involved in aerobactin siderophore biosynthesis), the plasmid-borne rmpA gene (prmpA), prmpA2, chromosomal gene rmpA (crmpA) (which regulates of the mucoid phenotype via increased capsule production), peg-344 (a putative transporter), terB (involved in tellurite resistance), iroB (involved in salmochelin siderophore biosynthesis), and irp2 (involved in yersiniabactin siderophore biosynthesis).7 K1 and K2 are the most common hypervirulent serotypes of K. pneumoniae.13,31,32 But in our study, K57 was the main capsular serotype of invasive K. pneumoniae strains, which may be explained by differences in serotype distribution according to geographic areas and across populations. Moreover, all invasive K. pneumoniae strains carried virulence-associated genes and the proportion with hypervirulence genes was significantly higher than that reported in previous studies, indicating that these factors play an important role in the pathogenicity of invasive K. pneumoniae in children.
The prevalence of CRKP is also a global public health problem and the clinical isolation rate of CRKP has been increasing in recent years.10–13 In China, the rate of detection of CRKP in children increased from 3% to > 20% according to a surveillance program conducted from 2005 to 2017. Notably, this rate was higher than that recorded in adults.11,13,22 CRKP is resistant to most antimicrobial drugs and there are currently limited therapeutic options available, making infections difficult to control and resulting in high mortality rates.12,13 The main mechanism of CRKP resistance is the production of enzymes, such as carbapenemases, ESBLs and AmpC, that hydrolyze antimicrobial drugs.13,27 blaKPC-2 is the most common carbapenemase in adults in China,13,33 while the predominant carbapenemases in children were blaIMP-4, blaNDM-1 and blaOXA-232 in different periods.13,34 In our study, carbapenemase genes were detected in only a few invasive K. pneumoniae strains; however, most strains harbored ESBL and AmpC enzyme genes, mainly blaSHV-11, blaFOX-1 and blaACT-1. This diverges from previous findings that blaTEM-1 was the main ESBL gene while the proportion of AmpC genes was low.27,35 An evaluation of drug susceptibility showed that the 94 invasive K. pneumoniae strains identified in this study had low drug resistance rates but high carriage rates of ESBL and AmpC enzyme genes. Moreover, we found 14 MDR K. pneumoniae strains that carried at least 3 β-lactam antibiotic resistance genes, representing potential threats. Therefore, the drug resistance of invasive K. pneumoniae in children warrants further attention.
In this study, 12 CR-HVKP strains were identified belonging to 7 STs. ST29 was the most common ST in these strains, whereas ST11 was previously reported as the predominant ST among CRKP isolated from both adults and children in China.27,36 The predominant ST of CRKP and CR-HVKP may differ. However, there have been limited studies of CR-HVKP isolated from pediatric patients and further investigations are required. CR-HVKP has high pathogenicity in children and is resistant to most antimicrobial drugs, which presents a challenge for the clinical management of infections. More research is needed to develop strategies to overcome these challenges.
Conclusions
In summary, our findings reveal the epidemiology, virulence, and antimicrobial resistance of invasive K. pneumoniae at a children’s medical center in eastern China. Both CR-HVKP and MDR strains were identified. HVKP and CRKP were found to overlap in pediatric patients; their accurate identification can aid diagnosis and effective treatment.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (82002106), the High-level Innovative and Entrepreneurial Talents Introduction Program of Jiangsu Province (2020-30186 and 2020-30191), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (20KJB310009 and 20KJB310012), the Medical Research Project of Jiangsu Commission of Health (M2020027), the Science and Technology Program of Suzhou (SYS2020163, SYSD2019120, SLC201904 and SS201867).
Ethical Approval
The authors are accountable for all aspects of the work and ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Children’s Hospital of Soochow University (No. 2020CS099). The investigation was conducted in accordance with the guidelines of the Declaration of Helsinki.
This was a retrospective study, and the clinical isolates were collected and stored during routine diagnostic laboratory examination. The Review Board exempted the study from the requirement informed consent as it was focused on the bacteria and did not have an impact on the patients.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Viau RA, Hujer AM, Marshall SH, et al. “Silent” dissemination of Klebsiella pneumoniae isolates bearing K. pneumoniae carbapenemase in a long-term care facility for children and young adults in Northeast Ohio. Clin Infect Dis. 2012;54(9):1314–1321. doi: 10.1093/cid/cis036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. doi: 10.3389/fcimb.2018.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bengoechea JA, Sa Pessoa J. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev. 2019;43(2):123–144. doi: 10.1093/femsre/fuy043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3):e00001–19. doi: 10.1128/CMR.00001-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cubero M, Grau I, Tubau F, et al. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin Microbiol Infect. 2016;22(2):154–160. doi: 10.1016/j.cmi.2015.09.025 [DOI] [PubMed] [Google Scholar]
- 6.Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae-clinical and molecular perspectives. J Intern Med. 2020;287(3):283–300. doi: 10.1111/joim.13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9):e00776–18. doi: 10.1128/JCM.00776-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker KA, Miller VL. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr Opin Microbiol. 2020;54:95–102. doi: 10.1016/j.mib.2020.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo TA, Olson R, Macdonald U, et al. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun. 2014;82(6):2356–2367. doi: 10.1128/IAI.01667-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Mathema B, Chavda KD, et al. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686–696. doi: 10.1016/j.tim.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Y, Liu C, Shen Z, et al. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg Microbes Infect. 2020;9(1):1771–1779. doi: 10.1080/22221751.2020.1799721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu F, Zhu D, Wang F, et al. Current status and trends of antibacterial resistance in China. Clin Infect Dis. 2018;67(suppl2):S128–S134. doi: 10.1093/cid/ciy657 [DOI] [PubMed] [Google Scholar]
- 13.Wang B, Pan F, Wang C, et al. Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in a paediatric hospital in China. Int J Infect Dis. 2020;93:311–319. doi: 10.1016/j.ijid.2020.02.009 [DOI] [PubMed] [Google Scholar]
- 14.Makharita RR, El-Kholy I, Hetta HF, et al. Antibiogram and genetic characterization of carbapenem-resistant gram-negative pathogens incriminated in healthcare-associated infections. Infect Drug Resist. 2020;13:3991–4002. doi: 10.2147/IDR.S276975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Algammal AM, Hetta HF, Elkelish A, et al. Methicillin-resistant Staphylococcus aureus (MRSA): one health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect Drug Resist. 2020;13:3255–3265. doi: 10.2147/IDR.S272733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Algammal AM, El-Kholy AW, Riad EM, et al. Genes encoding the virulence and the antimicrobial resistance in enterotoxigenic and Shiga-toxigenic E. coli isolated from diarrheic calves. Toxins (Basel). 2020;12(6):383. doi: 10.3390/toxins12060383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Algammal AM, Mabrok M, Sivaramasamy E, et al. Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes. Sci Rep. 2020;10(1):15961. doi: 10.1038/s41598-020-72264-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Algammal AM, Enany ME, El-Tarabili RM, et al. Prevalence, antimicrobial resistance profiles, virulence and enterotoxins-determinant genes of MRSA isolated from subclinical bovine mastitis in Egypt. Pathogens. 2020;9(5):362. doi: 10.3390/pathogens9050362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abolghait SK, Fathi AG, Youssef FM, et al. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from chicken meat and giblets often produces staphylococcal enterotoxin B (SEB) in non-refrigerated raw chicken livers. Int J Food Microbiol. 2020;328:108669. doi: 10.1016/j.ijfoodmicro.2020.108669 [DOI] [PubMed] [Google Scholar]
- 20.Yao H, Qin S, Chen S, et al. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2018;18(1):25. doi: 10.1016/S1473-3099(17)30628-X [DOI] [PubMed] [Google Scholar]
- 21.Karlsson M, Stanton RA, Ansari U, et al. Identification of a carbapenemase-producing hypervirulent Klebsiella pneumoniae isolate in the United States. Antimicrob Agents Chemother. 2019;63(7):e00519-19. doi: 10.1128/AAC.00519-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a Molecular Epidemiological Study. Lancet Infect Dis. 2018;18(1):37–46. doi: 10.1016/S1473-3099(17)30489-9 [DOI] [PubMed] [Google Scholar]
- 23.Fasciana T, Gentile B, Aquilina M, et al. Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy. BMC Infect Dis. 2019;19(1):928. doi: 10.1186/s12879-019-4565-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang H, Zheng W, Kong Z, et al. Disease burden and molecular epidemiology of carbapenem-resistant Klebsiella pneumonia infection in a tertiary hospital in China. Ann Transl Med. 2020;8(9):605. doi: 10.21037/atm.2020.03.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Candan ED, Aksöz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62(4):867–874. doi: 10.18388/abp.2015_1148 [DOI] [PubMed] [Google Scholar]
- 26.Gu B, Bi R, Cao X, et al. Clonal dissemination of KPC-2-producing Klebsiella pneumoniae ST11 and ST48 clone among multiple departments in a tertiary teaching hospital in Jiangsu Province, China. Ann Transl Med. 2019;7(23):716. doi: 10.21037/atm.2019.12.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao M, Wen H, Xu P, et al. Genetic diversity of carbapenem-resistant Enterobacteriaceae (CRE) clinical isolates from a tertiary hospital in eastern China. Front Microbiol. 2019;9:3341. doi: 10.3389/fmicb.2018.03341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a Microbiological and Molecular Biological Study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 29.Moosavian M, Emam N. The first report of emerging mobilized colistin-resistance (mcr) genes and ERIC-PCR typing in Escherichia coli and Klebsiella pneumoniae clinical isolates in southwest Iran. Infect Drug Resist. 2019;12:1001–1010. doi: 10.2147/IDR.S192597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Algammal AM, Hashem HR, Alfifi KJ, et al. atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci Rep. 2021;11(1):9476. doi: 10.1038/s41598-021-88861-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee CR, Lee JH, Park KS, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. doi: 10.3389/fcimb.2017.00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Q, Zhou JW, Qiu CN, et al. Antimicrobial susceptibility and microbiological and epidemiological characteristics of hypermucoviscous Klebsiella pneumoniae strains in a tertiary hospital in Hangzhou, China. J Glob Antimicrob Resist. 2018;15:61–64. doi: 10.1016/j.jgar.2018.05.025 [DOI] [PubMed] [Google Scholar]
- 33.Wang Q, Wang X, Wang J, et al. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis. 2018;67(suppl2):S196–S205. doi: 10.1093/cid/ciy660 [DOI] [PubMed] [Google Scholar]
- 34.Dong F, Zhang Y, Yao K, et al. Epidemiology of carbapenem-resistant Klebsiella pneumoniae bloodstream infections in a Chinese children’s hospital: predominance of New Delhi metallo-β-lactamase-1. Microb Drug Resist. 2018;24(2):154–160. doi: 10.1089/mdr.2017.0031 [DOI] [PubMed] [Google Scholar]
- 35.Tian D, Pan F, Wang C, et al. Resistance phenotype and clinical molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Shanghai. Infect Drug Resist. 2018;11:1935–1943. doi: 10.2147/IDR.S175584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]