Summary
Exorbitant outputs of waste xylose mother liquor (WXML) and corncob residue from commercial‐scale production of xylitol create environmental problems. To reduce the wastes, a Saccharomyces cerevisiae strain tolerant to WXML was conferred with abilities to express the genes of xylose reductase, a xylose‐specific transporter and enzymes of the pentose phosphate pathway. This strain showed a high capacity to produce xylitol from xylose in WXML with glucose as a co‐substrate. Additionally, a simultaneous saccharification and fermentation (SSF) process was designed to use corncob residues and cellulase instead of directly adding glucose as a co‐substrate. Xylitol titer and the productivity were, respectively, 91.0 g l‐1 and 1.26 ± 0.01 g l‐1 h‐1 using 20% WXML, 55 g DCW l‐1 delignified corncob residues and 11.8 FPU gcellulose ‐1 cellulase at 35° during fermentation. This work demonstrates the promising strategy of SSF to exploit waste products to xylitol fermentation process.
Genetically modified yeast produces xylitol from waste xylose mother liquor. Corncob residues can be used as the source of the glucose co‐substrate. The approach of simultaneous saccharification and fermentation achieved high xylitol productivity.
Introduction
Xylitol is a five‐carbon sugar alcohol, which naturally exists in many fruits and vegetables. It has similar sweetness to sucrose but with 40% less calories. Moreover, it has additional advantages over sucrose including protection against tooth enamel softening, osteoporosis and ear infections, as well as anti‐carcinogenic and anti‐inflammatory effects. Most important to diabetics, xylitol metabolism does not require insulin. Because of its many beneficial properties, this sugar alcohol has been widely used in food and pharmaceutical products (Dasgupta et al., 2017; Xu et al., 2019). One market research report estimated the international market value of xylitol to continue to grow beyond its 2019 value of more than US$ 875 million (https://www.imarcgroup.com, 2020).
Commercial‐scale production of xylitol applies the chemical process of xylose hydrogenation (Wang et al., 2016; Dasgupta et al., 2017; Xu et al., 2019). Xylan‐rich lignocellulosic materials, such as corncob, are pretreated with acid at high temperature and high pressure to generate xylose‐rich hydrolysates. A by‐product, corncob residues, is also generated during this process. The lignin in corncob residues can be removed to generate delignified corncob residues (DCCR), which are mainly composed of cellulose. The xylose‐rich hydrolysates are concentrated to precipitate xylose crystals, and the xylose is hydrogenated at 80–140°C under 8–10 MPa of hydrogen pressure in the presence of a nickel catalyst (Xu et al., 2019). In this xylose crystallization process, about 0.6 tons of waste xylose mother liquor (WXML) are produced per ton of xylose. Globally, over 100 000 tons of WXML is generated annually. The WXML is an organic pollutant containing approximately 400 g l‐1 xylose that cannot be extracted by the crystallization process due to the high contents of other sugars (arabinose, glucose, and galactose) in the liquor (Wang et al., 2016; Hua et al., 2019). Moreover, compounds toxic to microorganisms, such as furfural and hydroxymethyl furfural (HMF), are also present in the WXML (Wang et al., 2016). Therefore, finding ways to use the wastes and by‐products (WXML and DCCR) may reduce environmental pollution and bring additional economic benefits from xylitol production.
Bioconversion of xylose to xylitol by microorganisms is a promising strategy to produce xylitol. Some filamentous fungi and yeasts, such as Scheffersomyces (Pichia) stipitis (Rodrigues et al., 2011), Candida tropicalis (Wang et al., 2016), Kluyveromyces marxianus (Hua et al., 2019; Park et al., 2019), and so on, have a complete xylose metabolic pathway. In these microorganisms, xylose is sequentially converted into xylitol, xylulose and xylulose‐5 phosphate by xylose reductase (XR), xylitol dehydrogenase (XDH) and xylulokinase (XK), respectively, and then, it enters the pentose phosphate pathway in the form of xylose‐5‐phosphate (Fig. 1). Unfortunately, these microorganisms are inefficient xylitol producers because quite a lot of the xylitol produced is further metabolized to support the growth of the strain and thus less xylitol can be accumulated (Dasgupta et al., 2017; Xu et al., 2019).
Fig. 1.
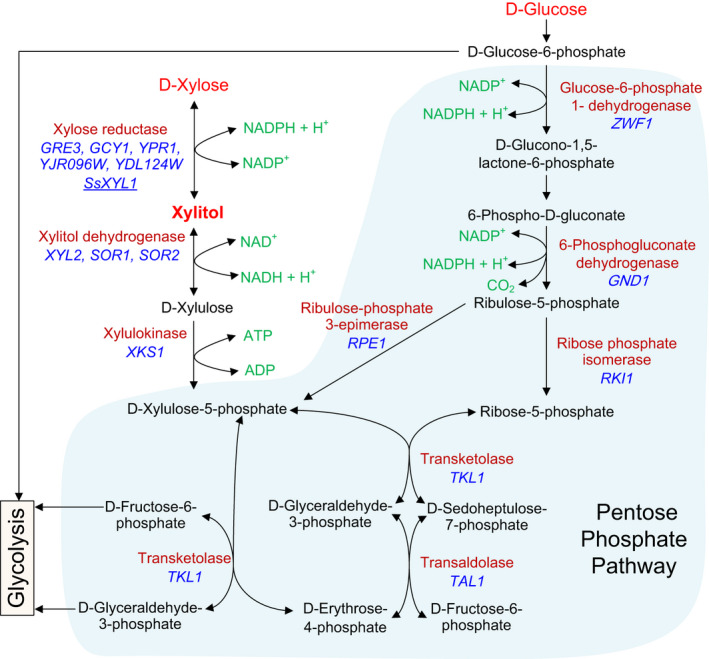
Main metabolic pathways related to xylitol production in filamentous fungi and yeasts. Xylose is sequentially converted into xylitol, xylulose, and xylulose‐5 phosphate by xylose reductase, xylitol dehydrogenase, and xylulokinase, respectively, and then enters the pentose phosphate pathway in the form of xylose‐5 phosphate. The NADPH needed for xylose reduction is mainly generated from the glucose metabolism through the pentose phosphate pathway.
A potentially better candidate as a cell factory for xylitol production from lignocellulosic materials is Saccharomyces cerevisiae because it has been designated as generally recognized as safe by the US Food and Drug Administration, exhibits high tolerance to the toxic compounds in the hydrolysates, and has little to no ability to further metabolize xylitol through xylitol dehydrogenase. Although proteins encoded by the genes XYL2, SOR1 and SOR2 in S. cerevisiae (Fig. 1) have XDH activity, the activity is low and not all S. cerevisiae strains have these genes (Wenger et al., 2010; Li et al., 2015; Wang et al., 2016; Kwak and Jin, 2017). Still, high xylitol yield could be in principle obtained as the produced xylitol is not further metabolized. Furthermore, though the toxic compounds in the lignocellulose hydrolysates may limit the growth and overall metabolic efficiency of yeast strains, S. cerevisiae has higher tolerance than many other microorganisms (Park et al., 2016, Venkateswar Rao et al., 2016; Kwak and Jin, 2017; Xu et al., 2019).
There are some features that need to be improved, however, to use S. cerevisiae to process lignocellulose hydrolysates or WXML for xylitol production. First, xylose reductase (XR) activity of the wild type strain is low, although proteins encoded by several genes (GRE3, GCY1, YPR1, YJR096W, and YDL124W) in S. cerevisiae (Fig. 1) all exhibit XR activity (Wenger et al., 2010; Konishi et al., 2015). Increasing the XR activity is necessary to enhance the xylose reduction capacity of S. cerevisiae strains. Second, it is necessary to increase the supply of NADPH/NADH, which is not sufficient for xylose reduction in wild‐type S. cerevisiae (Jo et al., 2015; Kwak and Jin, 2017). Third, xylose uptake by S. cerevisiae depends on its hexose transporters that have a greater affinity for glucose than xylose (Wang et al., 2015; Kwak and Jin, 2017), and glucose is commonly used as a co‐carbon substrate to support cell growth and generate the coenzyme NADPH/NADH needed for xylose reduction.
The primary purpose of this study was to develop a strategy of using the wastes and by‐products from commercial xylitol production to achieve more xylitol production and reduce waste accumulation. Because of the potential of S. cerevisiae to process WXML for xylitol, we found S. cerevisiae strains with inherently high tolerance to WXML by evaluating their growth in media containing the waste product. Then, using genes related to xylose and glucose metabolism (Fig. 1), we genetically modified these selected strains to overcome their limitations in xylitol production, some of which were described above. The xylose reductase gene of S. stipitis, SsXYL1, was introduced into these strains, and we selected the best of these modified strains for additional modifications. Pentose phosphate pathway (PPP) genes (RKI1, TAL1, TKL1 and RPE1) were overexpressed to increase the flux of PPP, since the first two reactions in PPP are the major source for yeast to generate NADPH that is required for xylose reduction (Jo et al., 2015). The XKS1 (encodes xylulokinase) gene was deleted to block the potential pathway through which xylitol can be consumed. A xylose‐specific transporter, Mgt05196N360F (Wang et al., 2015), was expressed in our selected strain to improve its uptake of xylose. The final optimized strain was then tested for successful growth and xylitol production by fed‐batch fermentation using WXML as the substrate and glucose as the co‐substrate. Additionally, we applied the simultaneous saccharification and fermentation (SSF) method, a process often used in the fermentation of cellulosic ethanol (Loaces et al., 2017) and recently used in xylitol production (Baptista et al., 2018; Baptista et al., 2020), to use DCCR and cellulase to produce the glucose in one system instead of separately adding glucose as a co‐substrate in the xylitol fermentation process. Our work demonstrates a promising strategy of using wastes and by‐products to increase xylitol production while decreasing the pollution resulting from the commercial production of xylitol.
Results and Discussion
The selection of S. cerevisiae strains for xylitol production
To select a suitable strain to construct xylitol‐producing S. cerevisiae, we evaluated five strains (Table 1) for their tolerance to WXML. The results (Fig. 2A) showed that strain RC212, 6508 and GZ‐5 grew better than the other two strains on a YP agar plate with 20% (v/v) WXML. Furthermore, we evaluated growth characteristics of these three strains cultured in YP liquid medium with 25% (v/v) WXML. The results showed that the maximum specific growth rates of strains RC212, 6508 and GZ‐5 were 0.252 ± 0.002, 0.273 ± 0.001 and 0.302 ± 0.002 h‐1, respectively. Moreover, GZ‐5 and 6508 achieved higher cell biomass than RC212 (Fig. 2B).
Table 1.
Strains and plasmids used in this work.
| Strains and Plasmids | Description | Sources |
|---|---|---|
| Strains | ||
| BSIF | Diploid S. cerevisiae, isolated from tropical fruit in Thailand | Li et al. (2015) |
| RC212 | Diploid S. cerevisiae, isolated from grape skin | Li et al. (2015) |
| 6508 | Diploid S. cerevisiae, used in starch‐based ethanol production | Li et al. (2015) |
| GZ‐4 | Diploid S. cerevisiae, isolated from waste materials in sugar‐cane industry | Li et al. (2015) |
| GZ‐5 | Diploid S. cerevisiae, isolated from waste materials in sugar‐cane industry | Li et al. (2015) |
| R3 | Derived from RC212, δ::PsXYL1 | This Work |
| X3 | Derived from 6508, δ:: PsXYL1 | This Work |
| DX7 | Derived from GZ‐5, δ:: PsXYL1 | This Work |
| X3kZ | Derived from X3, xks1::loxP‐PGK1t‐ZWF1‐TEF1p | This Work |
| X3kZM | Derived from X3kZ, gre3::TDH3p‐Mgt05196(N360F)‐CYC1t‐loxP | This Work |
| X3kZPM | Derived from X3kZ, gre3::TPI1p‐RKI1‐RKI1t‐PGK1p‐TAL1‐TAL1t‐FBA1p‐TKL1‐TKL1t‐ADH1p‐RPE1‐RPE1t‐loxP‐Mgt01596(N360F) | This Work |
| Plasmids | ||
| pUG6 | loxP‐KanMX‐loxP | Guldener et al. (1996) |
| YEp‐CH | YEp24 derivative; GAL1p‐Cre‐CYC1t, TEF1p‐hygB‐TEF1t | Li et al. (2016) |
| pXIδ | pUC19 derivative, integration plasmid; targets to δ‐sequence, three tandem expression cassettes of TEF1p‐Ru‐xylA ‐PGK1t, and the selectable marker loxP‐KanMX4‐loxP | Li et al. (2016) |
| P‐K‐δ1δ2‐XR | pUC19 derivative, integration plasmid; targets to δ‐sequence, TEF1p‐SsXYL1‐PGK1t, loxP‐KanMX4‐loxP | This work |
| pUkT13Z | pUG6 derivative, integration plasmid; target to XKS1(−142, +1533), PGK1t‐ZWF1‐TEF1p | This work |
| pUkT23Z | pUG6 derivative, integration plasmid; target to XKS1(530, +1533), PGK1 t‐ZWF1‐TEF1p | This work |
| pUC‐N360F | pUC19 derivative, integration plasmid; targets to GRE3 (+113, +984), TDH3p‐MGT05196N360F‐CYC1 t, loxP‐KanMX4‐loxP | Li et al. (2016) |
| pJPPP4 | pUC19 derivative, integration plasmid; targets to GRE3(−241,+113), TPI1p‐RKI1‐RKI1t‐PGK1p‐TAL1‐TAL1t‐FBA1p‐TKL1‐TKL1t‐ADH1p‐RPE1‐RPE1t, loxP‐KanMX4‐loxP | Li et al. (2016) |
Fig. 2.
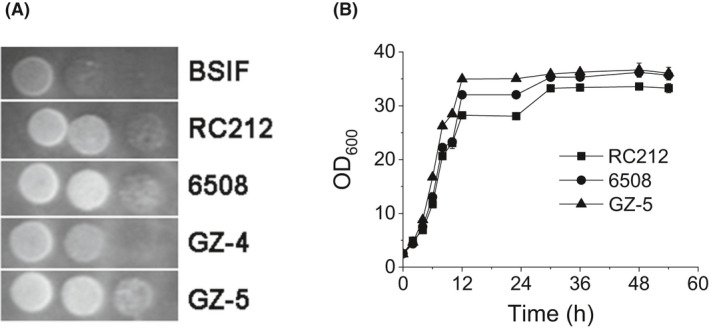
Growth characteristics of industrial S. cerevisiae strains cultivated on YP plates containing 20% xylose mother liquor (A) or cultivated in YP liquid medium containing 25% xylose mother liquor (B), data are mean values of three biological repeats. Symbols: ■, RC212; ●, 6508; ▲ GZ‐5.
Wenger et al., (2010) reported that the enzymes encoding by endogenous genes (XYL2, SOR1 and SOR2) of S. cerevisiae have xylitol dehydrogenase activity, and gene XKS1 encodes xylulokinase (Fig. 1). We endeavoured to detect the existence of these purported endogenous genes by PCR, because if these genes exist, xylitol may be further metabolized through them, and consequently decrease xylitol yield. The results showed that XKS1 and XYL2 were detected in all three strains, while SOR1 and SOR2 were detected in RC212 and GZ‐5 but not in 6508. Strong growth of the strains in the WXML is necessary for them to ferment WXML. Moreover, to produce as much xylitol as possible, xylitol production capacity of the strain should be improved, on the one hand, and further metabolism of xylitol should be prevented, on the other hand. Of the three strains that grew best in WXML, the ones with the most potential to achieve this goal were GZ‐5, because it had the highest tolerance to WXML, and 6508, because it had high tolerance to WXML as well as fewer genes of the xylitol metabolic pathway.
Expression of xylose reductase gene in S. cerevisiae strains
Endogenous aldose reductase activity, which is needed for xylose reduction to xylitol, is very low in wild‐type S. cerevisiae strains (Kogje and Ghosalkar, 2016; Dasgupta et al., 2017). To enhance the xylitol producing capacity of S. cerevisiae strains, the xylose reductase gene of S. stipitis, SsXYL1, was integrated into the delta sequences in the genomes of strains RC212, 6508 and GZ‐5 to produce modified strains R3, X3 and DX7, respectively. The enzyme activity assay showed that the specific XR activities in the cell extracts of R3, X3 and DX7 were 0.87 ± 0.02, 2.88 ± 0.53 and 9.20 ± 0.13 U mg‐1 total protein, respectively. The fermentation of strains in YP medium with 20 g l‐1 glucose and 50 g l‐1 xylose as carbon sources (Fig. 3) showed that xylose conversion and xylitol production rates of the three strains ranked in the following order: DX7 > X3 > R3. This was consistent with the ranking of XR activity levels in each strain. However, the fermentation results of the three strains grown in YP medium with 20% (v/v) WXML showed that X3 grew fastest and attained the highest yield of cell biomass. Furthermore, it consumed the most xylose and produced the most xylitol (Fig. 4). Here we must point out that although what we determined is the total amount of xylose, arabinose and galactose (XAG) in this work, the strain mainly consumed xylose and little arabinose and galactose in WXML were consumed (Fig. S1).
Fig. 3.
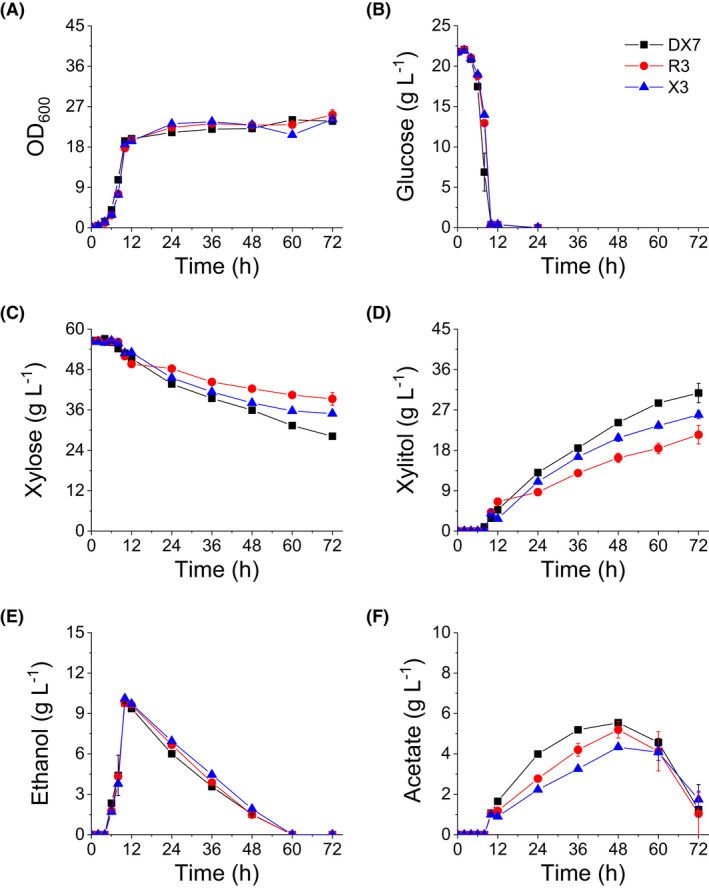
The fermentation characteristics of strains in YP medium with 20 g L‐1 glucose and 50 g L‐1 xylose as carbon sources in shake flasks: (A) growth curve (OD600), (B) glucose consumption, (C) xylose consumption, (D) xylitol accumulation, (E) ethanol accumulation and (F) acetate accumulation. Fermentations were performed at 30° and 200 rpm with an initial OD600 of 0.2 and initial pH of 6.0. Data are mean values of two biological repeats. Symbols: ■, DX7;  , R3;
, R3;  , X3.
, X3.
Fig. 4.
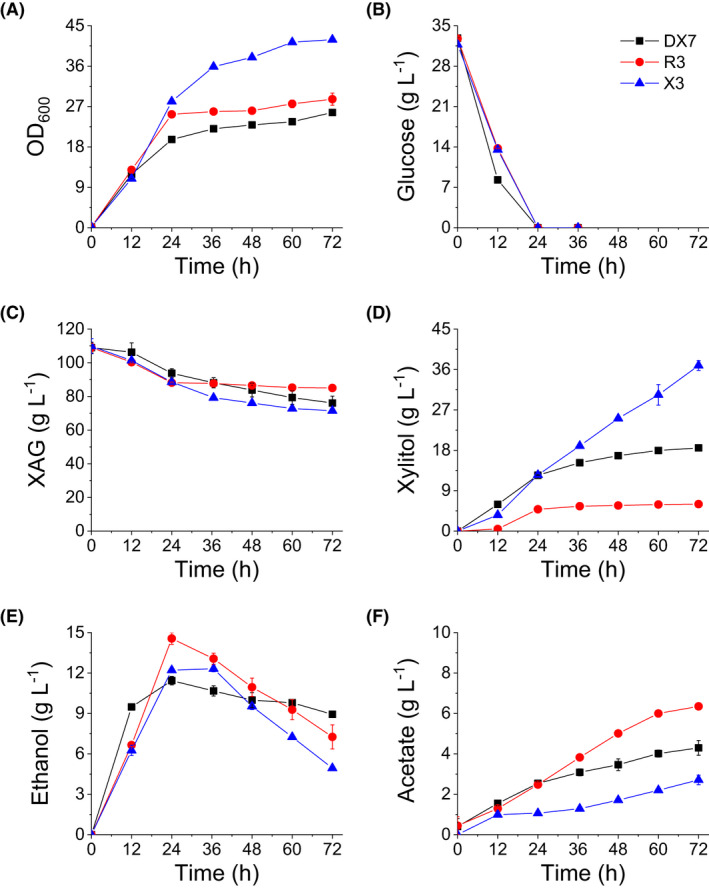
Fermentation characteristics of strains in YP medium with 20% (v/v) xylose mother liquor in shake flasks: (A) growth curve (OD600); (B) glucose; (C) the total amount of xylose, arabinose, and galactose (denoted as XAG); (D) xylitol; (E) ethanol and (F) acetate. Fermentations were performed at 30° and 200 rpm with an initial OD600 of 0.2 and initial pH of 6.0. Data are mean values of two biological repeats. Symbols: ■, DX7;  , R3;
, R3;  , X3.
, X3.
The results of the fed‐batch fermentation of strains DX7 and X3 showed that the X3 culture kept growing over the 72 h of being fed with 1.125 g l‐1 h‐1 glucose. In those 72 h, X3 consumed 78.5 ± 1.7 g l‐1 xylose and produced 70.9 ± 7.4 g l‐1 xylitol (Fig. 5). In contrast, DX7 stopped growing after 24 h of fermentation possibly because too much acetate (7.8 ± 0.2 g l‐1) had accumulated. In 72 h, DX7 only consumed 51.7 ± 0.5 g l‐1 xylose and produced 23.5 ± 0.5 g l‐1 xylitol. Due to the complexity of the raw materials (WXML and DCCR) we used, neither here nor thereafter can we exclude the contribution of xylose additionally released from these raw materials and other nutrient to the production of xylitol. Therefore, both the consumption of xylose (the decrease in xylose in the medium) and the xylose conversion rate we showed were based on the data of HPLC.
Fig. 5.
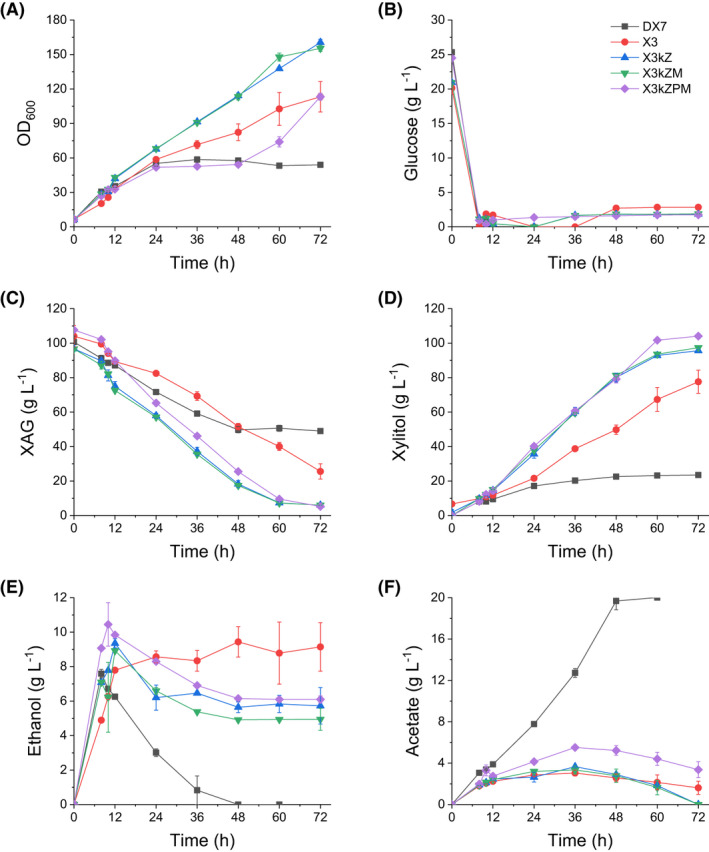
The measured rates during fed‐fermentation of strains: (A) growth curve (OD600); (B) glucose; (C) the total amount of xylose, arabinose and galactose (denoted as XAG); (D) xylitol; (E) ethanol and (F) acetate. Fermentations were performed in 1.4 l fermenters containing 800 mL medium at conditions of 30°, 500 rpm and 1 vvm. The pH was maintained at 5.5 by adding an aqueous ammonia solution and 2 M HCl. The YP medium contained 20% (v/v) xylose mother liquor, and the initial cell biomass was ˜ 1 g l‐1 DCW. After 8 h of fermentation, 1.125 g l‐1 h‐1 of glucose was added over 72 h. Data are mean values of two biological repeats. Symbols: ■, DX7;  , X3;
, X3;  , X3kZ;
, X3kZ;  , X3kZM;
, X3kZM;  , X3kZPM.
, X3kZPM.
Many studies have reported that high XR activity is beneficial to xylitol production (Bae et al., 2004; Jeon et al., 2012; Hong et al., 2014), and our fermentation results from strains cultivated in the medium without toxic compounds (without WXML) also support this viewpoint (Fig. 3). Our data showed that although DX7 had much stronger XR activity than X3, xylitol production capacity of X3 was much greater than that of DX7 in WXML fermentation. We suspect the cause is based on their different metabolic characteristics, particularly the accumulation of acetate (Figs 4 and 5), which is toxic to yeasts (Semchyshyn et al., 2011; Orlandi et al., 2013) and shows negative effect on the yeast‐based xylitol production process (Baptista et al., 2020). The acetate produced by DX7 kept accumulating after 36 h of fermentation, while the acetate produced by X3 did not accumulate and actually decreased after 36 h (Fig. 5F). Our data suggest that WXML initially inhibited acetate metabolism in DX7, causing the accumulation of acetate during fermentation which in turn hampered the growth and metabolism of cells, maybe also the XR activity. The mechanisms underlying the effect of acetate accumulation on cell growth deserve further study.
Modification of the metabolic pathway improved xylitol production capacity
In addition to XR activity, the transport of xylose into microbial cells and the supply of NADPH are also limiting factors of xylitol production (Venkateswar Rao et al., 2016; Xu et al., 2019). To further improve the xylitol production capacity of strain X3, we modified genes in the xylitol metabolic pathway related to xylose transportation and NADPH generation. First, we introduced an expression cassette of the ZWF1 gene (under the control of a strong constitutive promoter PTEF1 ) into the XKS1 loci in the genome of strain X3 to generate the modified strain X3kZ. Then, we introduced the xylose‐specific transporter Mgt05196N360F into the GRE3 loci of X3kZ to produce X3kZM. Furthermore, we introduced the genes of PPP (RKI1, TAL1, TKL1 and RPE1) into other parts of the GRE3 loci in X3kZM to produce X3kZPM.
The fed‐batch fermentation results (Fig. 5, Table 2) revealed that overexpression of ZWF1 with the deletion of XKS1 significantly increased the xylose conversion rate (from 1.14 ± 0.02 to 1.58 ± 0.03 g l‐1 h‐1) and xylitol production rate (from 1.11 ± 0.08 to 1.60 ± 0.02 g l‐1 h‐1) compared to those of the X3 strain. The expression of Mgt05196N360F transporter showed no positive effect on xylitol productivity (X3kZM vs X3kZ). This suggested that the uptake of xylose did not affect glucose in fed‐batch fermentation, which may because under this condition the glucose levels keep low. Overexpression of PPP genes further improved xylose reduction activity: xylose conversion rate (1.78 ± 0.01 g l‐1 h‐1) and xylitol production rate (1.80 ± 0.01 g l‐1 h‐1) in X3kZPM were higher than those of the other strains.
Table 2.
The fermentation characteristics of strains in fed‐fermentation.
| Strains |
Xylose consumption rates a , b (g l‐1 h‐1) |
Xylitol production rates a (g l‐1 h‐1) |
(g l‐1) |
Xylitol producedc (g l‐1) |
|---|---|---|---|---|
| X3 | 1.14 ± 0.02 | 1.11 ± 0.08 | 78.5 ± 6.4 | 70.9 ± 7.4 |
| X3kZ | 1.58 ± 0.03 | 1.60 ± 0.02 | 90.7 ± 0.30 | 93.9 ± 1.2 |
| X3kZM | 1.54 ± 0.01 | 1.61 ± 0.03 | 90.6 ± 0.4 | 97.4 ± 0.6 |
| X3kZPM | 1.78 ± 0.01 | 1.80 ± 0.01 | 102.4 ± 0.2 | 104.1 ± 0.4 |
Data are means ± standard errors calculated from two biological repeats of fermentation.
Rates calculated between 8 h (the initiation of glucose feeding) and 60 h (the nearly full depletion of xylose) in fed‐fermentation.
Calculated according to the decrease in xylose in the medium.
Measured at 72 h when fermentation was halted.
To date, many microorganisms used in xylitol production have been reported. Their features are summarized in a recent report (Hua et al., 2019) and other reviews (Dasgupta et al., 2017; Xu et al., 2019). Saccharomyces cerevisiae is a promising competitor because of its high xylitol yield and status of being generally recognized as safe. However, some other engineered yeasts, such as C. tropicalis (Wang et al., 2016) and K. marxianus (Hua et al., 2019), do have higher xylitol productivity than S. cerevisiae. So, metabolic engineering strategies were performed on S. cerevisiae strains to improve their XR activity and xylose uptake and increase the supply of the reduced coenzyme. As far as we know, the best S. cerevisiae strain was one that co‐expressed two types of xylose reductase (XR) with either NADPH‐dependence or NADH‐preference, and it also co‐overexpressed ZWF1 and ACS1 genes in response to increased intracellular concentrations of NADPH and NADH (Jo et al., 2015). The xylitol productivity and xylitol yield of this strain were ˜ 2 g l‐1 h‐1 and ˜ 1 g g‐1, respectively, when fermented with a glucose feeding rate of 1.8 g l‐1 h‐1. The performance of our strain X3kZPM is second to this strain.
Production of xylitol from xylose mother liquor and delignified corncob residues
To make full use of waste and by‐products and reduce production costs, we designed the SSF process to incorporate DCCR, a by‐product of xylitol production, instead of feeding yeast with glucose. The addition of cellulase was needed to continuously digest the DCCR to generate glucose. The optimum temperature of catalytic activity of cellulase is ˜ 55°, which is much higher than the optimum temperature for yeast growth (30°). Thus, we first tested the glucose release rate at 30 and 35°C with different amounts of cellulase. The results (Table 3) showed that the 18.8 FPU gcellulose ‐1 cellulase at 30°C and 11.8 FPU gcellulose ‐1 cellulase at 35°C used to digest DCCR over 12 h released glucose at rates of 1.120 ± 0.037 and 1.104 ± 0.057 g l‐1 h‐1, respectively. These rates were similar to the glucose feeding rate (1.125 g l‐1 h‐1 glucose) that we used in fed‐batch fermentation.
Table 3.
The glucose release rate from delignified corncob residues saccharified by cellulase obtained from Trichoderma reesei TX.
| Temperature (°C) | Time (h) | Cellulase dosage (FPU gcellulose ‐1) | |||
|---|---|---|---|---|---|
| 11.8 | 18.8 | 23.5 | 35.3 | ||
| 30 | 4 | 1.072 ± 0.041 | 1.653 ± 0.055 | 1.958 ± 0.064 | 2.768 ± 0.154 |
| 12 | 0.740 ± 0.032 | 1.120 ± 0.037 | 1.325 ± 0.009 | 1.827 ± 0.088 | |
| 24 | 0.613 ± 0.015 | 0.866 ± 0.017 | 1.043 ± 0.025 | 1.393 ± 0.096 | |
| 35 | 3 | 1.618 ± 0.112 | 2.527 ± 0.086 | 2.952 ± 0.127 | 4.076 ± 0.093 |
| 12 | 1.104 ± 0.057 | 1.702 ± 0.089 | 1.927 ± 0.089 | 2.671 ± 0.052 | |
| 24 | 0.815 ± 0.054 | 1.248 ± 0.059 | 1.498 ± 0.053 | 1.868 ± 0.042 | |
The results of the SSF performed in 1.4 l fermenters showed that 71.0 ± 0.3 g l‐1 xylose was consumed and 83.4 ± 0.1 g l‐1 xylitol was produced after 72 h of fermentation at 30°, while 83.2 ± 3.6 xylose was consumed and 91.0 ± 0.6 g l‐1 xylitol was produced after 72 h of fermentation at 35° (Fig. 6). The xylitol production rates were 1.16 ± 0.02 and 1.26 ± 0.01 g l‐1 h‐1 at 30 and 35°, respectively. It is worth noting that the cellulose mixture used is a crude fermentation product of T. reesei and has xylanase activity (Bischof et al., 2016). The hemicellulose existing in DDCR may release additional xylose that was not quantified; we were not able to calculate the accurate xylitol yield. The experimental xylitol yields were close to 1 both at 30 and 35°C. Our results showed that the SSF process was an effective method to produce xylitol using WXML and DCCR. Moreover, similarly high rates of xylitol could be produced using less cellulase at the higher temperature (35°) compared to using more cellulase at the lower temperature (30°) in the SSF process.
Fig. 6.
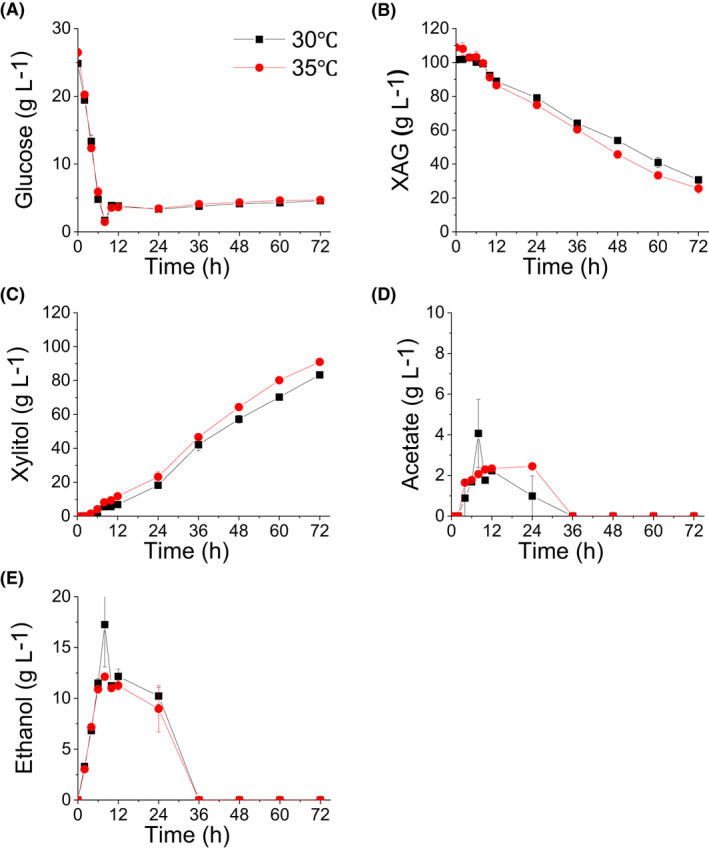
The recorded rates during simultaneous saccharification and fermentation of strain X3kZPM: (A) glucose; (B) the total amount of xylose, arabinose and galactose (denoted as XAG); (C) xylitol; (D) acetate and (E) ethanol. The fermentation was performed in 1.4 l fermenters containing 800 ml YP medium plus 20% (v/v) xylose mother liquor and 55 g DCW l‐1 DCCR. The pH was maintained at 5.5 by adding an aqueous ammonia solution and 2 M HCl. Cellulase was added at a rate of 18.8 or 11.8 FPU gcellulose
‐1 cellulase in fermentations at temperature of 30 or 35°, respectively. Aeration rate was 1 vvm and stirring velocity was 500 rpm. Data are mean values of two biological repeats. Symbols: ■, 30°;  , 35°.
, 35°.
For strains that do not use xylose as a carbon source, fed‐batch cultures with glucose as a substrate is the best way to supply the NADPH that is required for the reduction of xylose (Pratter et al., 2015; Dasgupta et al., 2017; Xu et al., 2019). For strains that can use xylose as a carbon source, feeding with co‐substrates (e.g. glucose or glycerol) because they are cheaper than xylose is also a common strategy to achieve high yields of xylitol with low production cost (Dasgupta et al., 2017; Hua et al., 2019; Park et al., 2019; Xu et al., 2019). Recently, the SSF process was also used in xylitol production from corn cob whole slurry (Baptista et al., 2018; Baptista et al., 2020). The high yield was gotten, while the final xylitol titer was limited by the xylose concentration in the substrate.
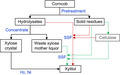
In the present work, we showed that the SSF process is feasible to produce xylitol using the wastes from xylitol‐producing factories. Moreover, the cellulase used in the SSF process also can be produced using DCCR as the substrate (Cheng et al., 2009). Using wastes should theoretically be more economical than using pre‐processed substrates. Another advantage of the SSF process is that it is simpler than the fed‐fermentation method. In the SSF process, cellulosic materials and cellulase are all added at one time point. Thus, glucose is continuously released but only from the cellulose initially added to the batch. In contrast, in the fed‐fermentation method, glucose is consistently being pumped into the fermenter at a certain rate throughout the fermentation process. The simpler mode of operation of the SSF method is more likely to be favoured in industrial production (Dasgupta et al., 2017). In Fig. 7, we illustrate the chemical and potential biotechnological (SSF) routes of xylitol production and indicate where SSF may fit in with the conventional chemical processes.
Fig. 7.
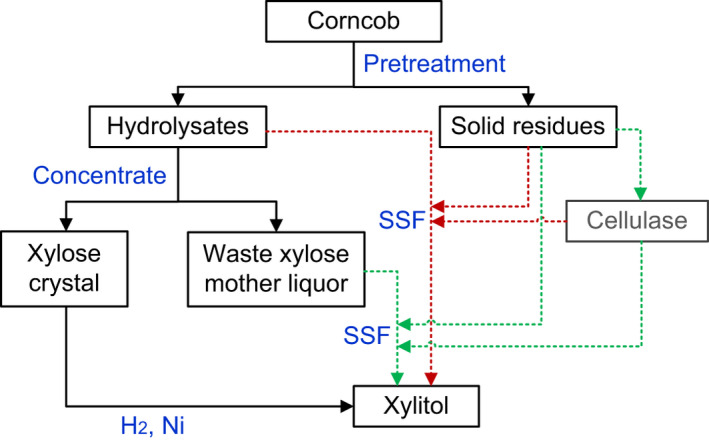
Chemical and biotechnological production routes for the conversion of xylose‐rich lignocellulosic materials to xylitol. SSF: simultaneous saccharification and fermentation. The green and red arrows show the SSF processes to produce xylitol using corncob and the waste xylose mother liquor, respectively.
Conclusion
We engineered a S. cerevisiae strain that efficiently converts xylose to xylitol using xylose‐rich wastes. Furthermore, we demonstrate the successful application of the SSF method for the sustained production of glucose, sourced from the additions of cellulase and DCCR, that was as effective as the conventional method of controlled feeding of glucose in fed‐batch culture for xylitol production. Our work exemplifies a promising, simpler strategy where commercial waste and by‐products can still be of value, which ultimately can help alleviate industrial impacts on natural environments.
Experimental procedures
Strain and culture medium
The strains and plasmids used in this work are listed in Table 1. All the genetic modifications were performed on the chromosomes of strains by homologous recombination. The plasmids with fragments for homologous recombination were constructed, and then, the fragments were cut from the plasmids before transformation into the yeast cells. The open reading frame (ORF) of SsXYL1 was cloned from chromosomal DNA of S. stipitis, and the expression cassette of TEF1p‐SsXYL1‐PGK1t was ligated in plasmid pXIδ (Li et al., 2016), while the expression cassettes of gene Ru‐xylA in pXIδ were deleted. The resulting plasmid P‐K‐δ1δ2‐XR contained the δ‐sequence‐targeting recombinant arms, expression cassette of TEF1p‐SsXYL1‐PGK1t, and selectable marker loxP‐KanMX4‐loxP. The plasmids pUkT13Z and pUkT23Z were constructed to introduce ZWF1 into the XKS1 loci in the vector pUG6 (Guldener et al., 1996), and each contained the fragments of XKS1 as recombinant arms and the expression cassette of TEF1p‐ZWF1‐PGK1t. The plasmids pUC‐N360F and pJPPP4 were, respectively, used to integrate the xylose transporter gene MGT05196N360F and four PPP genes (RPE1, RIK1, TAL1 and TKL1) into different parts of the GRE3 loci, as described in our previous study (Li et al., 2016).
The DNA fragments were introduced into S. cerevisiae cells by the LiAc‐mediated yeast transformation method (Gietz and Woods, 2002). Generally, strains were cultured in YPD medium (10 g l‐1 yeast extract and 20 g l‐1 peptone [YP medium] supplied with 20 g l‐1 glucose as the carbon source) at 30°C. Moreover, different carbon sources (glucose, xylose or WXML) were added as needed to YP medium for strain cultivation and fermentation depending on the specific experiments. The transformants were selected on YPD agar plates supplied with 400 mg l‐1 of the antibiotic G418 (Li et al., 2016). After each step of the genetic engineering process, the loxP‐KanMX4‐loxP fragment in the chromosomes of a recombinant strain was removed by recombinase. First, the plasmid YEp‐CH was introduced into the strains, and the transformants were selected on YPD agar plates supplied with 200 mg l‐1 hygromycin B. Then, the expression of the CreA recombinase gene in plasmid YEp‐CH was induced by galactose to remove the fragment loxP‐KanMX4 as we previously described (Li et al., 2016).
The components of waste xylose mother liquor and delignified corncob residues
The WXML and DCCR were supplied by Shandong Longlive Bio‐technology Co., Ltd (Yucheng, China). The WXML contained about 401.9 g l‐1 xylose, 180.1 g l‐1 arabinose, 151.8 g l‐1 glucose, 136.6 g l‐1 galactose, 15.9 g l‐1 arabitol, 15.7 g l‐1 xylitol, 5.8 g l‐1 glycerol, 4.8 g l‐1 ethanol, 2.9 g l‐1 acetate, 0.5 g l‐1 furfural, 4.1 g l‐1 5‐HMF and 2.8 g l‐1 lignin. The DCCR contained about 0.235 g g‐1 cellulose and 0.011 g g‐1 hemicellulose. There were slight differences between batches of samples.
Yeast spotting and growth assays in waste xylose mother liquor
Yeast strain growth capacity on WXML was determined by the spotting assay (Liang et al., 2021). Overnight cultures were transferred into fresh YPD medium with an initial OD600 of 0.2 and cultured for another 12 h. The cells were then harvested and washed three times with ddH2O and resuspended in ddH2O. The suspension was incubated at 30°C for 9 h and then diluted to an OD600 of 1.0 with ddH2O. Ten‐fold serial dilutions were prepared, and 4‐μL aliquots of each cell suspension were spotted onto YP agar medium with 20% WXML (pH 6.0) and then incubated at 30 ° for one day.
The growth assay of yeast in the liquid medium containing WXML was performed to obtain the growth curves of yeast samples. The cells were pre‐cultured in a 100 ml flask containing 20 ml YPD medium for 12 h at 30° while shaking at 200 rpm. Then, the culture was transferred into 20 ml of fresh YPD medium, starting at an OD600 of 0.2, and cultured for another 12 h. Subsequently, the cells were transferred into 100‐ml flasks containing 40 ml of YP medium with 25% WXML (pH 6.0) at an initial OD600 of 0.5. The growth curve of each strain was determined.
Detection of specific genes in the genome
The cells were harvested by centrifugation after 12 h of cultivation in YPD medium. Then, the genomic DNA was extracted using a Yeast Genomic DNA Extraction Kit (Tiangen Biotech Co., Ltd, Beijing, China) according to the kit’s instructions. The presence of genes was detected by PCR using the genomic DNA of strains as templates. The primers for gene SOR1 were SOR1‐F (5’‐AGTAACCCTGCAGTAGTTCTAG‐3’) and SOR1‐R (5’‐TAGTCTTGACTACCTCTCCACC‐3’). The primers for gene SOR2 were SOR2‐F (5’‐AGTCGGCGATATTGCCATCG‐3’) and SOR2‐R (5’‐GTCTTGACTACCTCTCCACCATG‐3’). The primers for gene XYL2 were XYL2‐F (5’‐CTATTGTTCTAGAGCGACCTGG‐3’) and XYL2‐R (5’‐GCCCTCAATGATCGTCTTG‐3’). The size of expected PCR products of SOR1, SOR2 and XYL2 was 1043, 1015 and 1038 bp, respectively. If the correct‐sized PCR products were obtained, a gene was considered to be present in the genome; otherwise, if not obtained, the gene was absent. Three biological replications were performed for each strain each gene.
Preparation of cell extracts and enzyme assays
The cell extracts were prepared, and the activities of XR were measured as previously described (Eliasson et al., 2000). Briefly, cells were harvested by centrifugation after 8 h of cultivation in YPD medium and then washed with sterile water before lysing them by vortexing the mixture of cells with glass beads (0.5 mm in diameter) in a disintegration buffer. The cell extracts were separated by centrifugation (20 000 g, 5 min, 4°), and the remaining supernatant was used for determination of XR activity levels. Enzyme reactions were monitored based on the oxidation of NADPH at 340 nm. Specific activities of XR were expressed as units per milligram of protein. Units are defined as micromoles of NADPH reduced per minute. The protein concentrations of cell extracts were measured using a BCA protein assay reagent kit (Sangon Biotech Co., Ltd., Shanghai, China) with bovine serum albumin as the standard.
Fermentations
The yeast cells were cultured in 15‐ml tubes with 5 ml YPD medium for 12 h and then transferred into 100 ml shake‐flasks with 20 ml YPD where the initial OD600 was 0.2. After 10 h of cultivation, the cells were harvested by centrifuging them at 5000 g for 10 min. The harvested cells were washed with ddH2O and then used to seed the batch fermentations. The batch fermentation was performed in 100 ml shake‐flasks. The medium was 40 ml YP supplied with 20 g l‐1 glucose and 50 g l‐1 xylose or only 20% (v/v) xylose mother liquor as the carbon source(s). The pH was initially adjusted to 6.0, and the initial OD600 was 0.2 (˜ 0.036 g DCW l‐1). Growth conditions included incubation at 30 ° and shaking of flasks at 200 rpm.
The yeast cells were successively cultured in 5 and 20 ml YPD for activation as described above, and then, cells were transformed to 1 l shake‐flasks with 400 ml YPD medium for another 10 h of cultivation. The cells were harvested and washed with ddH2O and then used as seed cultures for fed‐batch fermentation and SSF. The fed‐batch fermentation was performed in 1.4 l fermenters at 30°. The YP medium with 20% (v/v) xylose mother liquor was used, and the initial cell biomass was ˜1 g l‐1 DCW (OD600 ˜ 6). After 8 h of fermentation, 1.125 g l1 h‐1 glucose was fed to cultures for over 72 h. We selected this rate because it was neither too fast nor too slow for xylitol production. The SSF was also performed in 1.4 l fermenters with 800 ml of the YP medium with 20% (v/v) xylose mother liquor. After 8 h of fermentation, 55 g DCW l‐1 DCCR and 18.8 or 11.8 FPU gcellulose ‐1 cellulase (produced by Trichoderma reesei TX and provided by Dr. Xu Fang) were added when the fermentation temperature was 30 and 35°, respectively. For both fed‐batch fermentation and SSF, the initial volume was 800 ml, pH was maintained at 5.5 by adding a solution of aqueous ammonia and 2 M HCl, aeration rate was 1 vvm, and stirring velocity was 500 rpm.
Analytical methods
The concentrations of glucose, xylose (or XAG, the total amount of xylose, arabinose and the galactose), xylitol, acetate and ethanol were determined as previously described (Li et al., 2016), using the Prominence LC‐20A HPLC (Shimadzu, Japan) equipped with an Aminex HPX‐87H ion‐exchange column (Bio‐Rad, Hercules, CA, USA) and a refractive index detector RID‐10A (Shimadzu, Japan). Samples were eluted from the column at 45°C with 5 M H2SO4 at a flow rate of 0.6 ml min‐1.
Calculation of physiological parameters
Cell density (OD600) was determined with a UV–visible spectrophotometer (Eppendorf, Germany). Cell biomass was estimated according to the correlations of measured OD600 values and dry weights. One unit of OD600 equalled 0.18 g DCW l‐1. The rates of xylose conversion and xylitol production (also referred to as xylitol productivity) are the rates at which xylose is consumed per hour. Xylitol yield is the ratio of the amount of xylitol produced to the amount of xylose consumed.
Funding Information
This work was supported by the National Key Research and Development Program of China (No. 2018YFB1501702), the National Natural Science Foundation of China (No. 31770046), and the Major Program of Shandong Province Natural Science Foundation (No. ZR2018ZB0209).
Conflict of interest
The authors declare no conflict of interests.
Supporting information
Fig. S1. The sugars in a fermentation of X3. Fermentations were performed in 1.4 l fermenters containing 800 ml medium at conditions of 30°, 500 rpm, and 1 vvm. The pH was maintained at 5.5 by adding an aqueous ammonia solution and 2 mol l‐1 HCl. The YP medium contained 20% (v/v) xylose mother liquor and the initial cell biomass was ˜1 g l‐1 DCW. After 8 h of fermentation, 0.75 g l‐1 h‐1 of glucose was added over 72 h. Data are mean values of two biological repeats.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (No. 2018YFB1501702), the National Natural Science Foundation of China (No. 31770046), and the Major Program of Shandong Province Natural Science Foundation (No. ZR2018ZB0209). The authors thank Shandong Longlive Bio‐technology Co., Ltd., for kindly providing samples of their xylose mother liquor and delignified corncob residues and Dr. Xu Fang at State Key Laboratory of Microbial Technology of Shandong University for the cellulase.
Microb. Biotechnol. (2021) 14(5), 2059–2071
References
- Bae, S.‐M., Park, Y.‐C., Lee, T.‐H., Kweon, D.‐H., Choi, J.‐H., Kim, S.‐K., et al. (2004) Production of xylitol by recombinant Saccharomyces cerevisiae containing xylose reductase gene in repeated fed‐batch and cell‐recycle fermentations. Enzyme and Microbial Technology 35: 545–549. [Google Scholar]
- Baptista, S.L., Carvalho, L.C., Romaní, A., and Domingues, L. (2020) Development of a sustainable bioprocess based on green technologies for xylitol production from corn cob. Indust Crops Prod 156: 112867. [Google Scholar]
- Baptista, S.L., Cunha, J.T., Romani, A., and Domingues, L. (2018) Xylitol production from lignocellulosic whole slurry corn cob by engineered industrial Saccharomyces cerevisiae PE‐2. Biores Technol 267: 481–491. [DOI] [PubMed] [Google Scholar]
- Bischof, R.H., Ramoni, J., and Seiboth, B. (2016) Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei . Microb Cell Fact 15: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., Song, X., Qin, Y., and Qu, Y. (2009) Genome shuffling improves production of cellulase by Penicillium decumbens JU‐A10. J Appl Microbiol 107: 1837–1846. [DOI] [PubMed] [Google Scholar]
- Dasgupta, D., Bandhu, S., Adhikari, D.K., and Ghosh, D. (2017) Challenges and prospects of xylitol production with whole cell bio‐catalysis: a review. Microbiol Res 197: 9–21. [DOI] [PubMed] [Google Scholar]
- Eliasson, A., Christensson, C., Wahlbom, C.F., and Hahn‐Hagerdal, B. (2000) Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol 66: 3381–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods, R.A. (2002) Transformation of yeast by lithium acetate/single‐stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96. [DOI] [PubMed] [Google Scholar]
- Guldener, U., Heck, S., Fielder, T., Beinhauer, J., and Hegemann, J.H. (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y., Dashtban, M., Kepka, G., Chen, S., and Qin, W. (2014) Overexpression of D‐xylose reductase (xyl1) gene and antisense inhibition of D‐xylulokinase (xyiH) gene increase xylitol production in Trichoderma reesei . Biomed Res Int 2014: 169705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://www.imarcgroup.com (2020) Xylitol Market: Global Industry Trends, Share, Size, Growth, Opportunity and Forecast 2020‐2025.
- Hua, Y., Wang, J., Zhu, Y., Zhang, B., Kong, X., Li, W., et al. (2019) Release of glucose repression on xylose utilization in Kluyveromyces marxianus to enhance glucose‐xylose co‐utilization and xylitol production from corncob hydrolysate. Microb Cell Fact 18: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, W.Y., Yoon, B.H., Ko, B.S., Shim, W.Y., and Kim, J.H. (2012) Xylitol production is increased by expression of codon‐optimized Neurospora crassa xylose reductase gene in Candida tropicalis. Bioprocess Biosyst Eng 35: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, J.H., Oh, S.Y., Lee, H.S., Park, Y.C., and Seo, J.H. (2015) Dual utilization of NADPH and NADH cofactors enhances xylitol production in engineered Saccharomyces cerevisiae . Biotechnol J 10: 1935–1943. [DOI] [PubMed] [Google Scholar]
- Kogje, A., and Ghosalkar, A. (2016) Xylitol production by Saccharomyces cerevisiae overexpressing different xylose reductases using non‐detoxified hemicellulosic hydrolysate of corncob. 3 Biotech 6: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi, J., Fukuda, A., Mutaguchi, K., and Uemura, T. (2015) Xylose fermentation by Saccharomyces cerevisiae using endogenous xylose‐assimilating genes. Biotechnol Lett 37: 1623–1630. [DOI] [PubMed] [Google Scholar]
- Kwak, S., and Jin, Y.S. (2017) Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective. Microb Cell Fact 16: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Shen, Y.u., Wu, M., Hou, J., Jiao, C., Li, Z., et al. (2016) Engineering a wild‐type diploid Saccharomyces cerevisiae strain for second‐generation bioethanol production. Bioresour Bioprocess 3: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Wu, M., Xu, L., Hou, J., Guo, T., Bao, X., and Shen, Y. (2015) Evaluation of industrial Saccharomyces cerevisiae strains as the chassis cell for second‐generation bioethanol production. Microb Biotechnol 8: 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z., Wang, X., Bao, X., Wei, T., Hou, J., Liu, W., and Shen, Y. (2021) Newly identified genes contribute to vanillin tolerance in Saccharomyces cerevisiae . Microb Biotechnol 14: 503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loaces, I., Schein, S., and Noya, F. (2017) Ethanol production by Escherichia coli from Arundo donax biomass under SSF, SHF or CBP process configurations and in situ production of a multifunctional glucanase and xylanase. Biores Technol 224: 307–313. [DOI] [PubMed] [Google Scholar]
- Orlandi, I., Ronzulli, R., Casatta, N., and Vai, M. (2013) Ethanol and acetate acting as carbon/energy sources negatively affect yeast chronological aging. Oxid Med Cell Longev 2013: 802870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.B., Kim, J.S., Kweon, D.H., Kweon, D.H., Seo, J.H., and Ha, S.J. (2019) Overexpression of endogenous xylose reductase enhanced xylitol productivity at 40 degrees C by thermotolerant yeast Kluyveromyces marxianus . Appl Biochem Biotechnol 189: 459–470. [DOI] [PubMed] [Google Scholar]
- Park, Y.C., Oh, E.J., Jo, J.H., Jin, Y.S., and Seo, J.H. (2016) Recent advances in biological production of sugar alcohols. Curr Opin Biotechnol 37: 105–113. [DOI] [PubMed] [Google Scholar]
- Pratter, S.M., Eixelsberger, T., and Nidetzky, B. (2015) Systematic strain construction and process development: Xylitol production by Saccharomyces cerevisiae expressing Candida tenuis xylose reductase in wild‐type or mutant form. Biores Technol 198: 732–738. [DOI] [PubMed] [Google Scholar]
- Rodrigues, R.C., Kenealy, W.R., and Jeffries, T.W. (2011) Xylitol production from DEO hydrolysate of corn stover by Pichia stipitis YS‐30. J Ind Microbiol Biotechnol 38: 1649–1655. [DOI] [PubMed] [Google Scholar]
- Semchyshyn, H.M., Abrat, O.B., Miedzobrodzki, J., Inoue, Y., and Lushchak, V.I. (2011) Acetate but not propionate induces oxidative stress in bakers' yeast Saccharomyces cerevisiae . Redox Rep 16: 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswar Rao, L., Goli, J.K., Gentela, J., and Koti, S. (2016) Bioconversion of lignocellulosic biomass to xylitol: An overview. Biores Technol 213: 299–310. [DOI] [PubMed] [Google Scholar]
- Wang, C., Bao, X., Li, Y., Jiao, C., Hou, J., Zhang, Q., et al. (2015) Cloning and characterization of heterologous transporters in Saccharomyces cerevisiae and identification of important amino acids for xylose utilization. Metab Eng 30: 79–88. [DOI] [PubMed] [Google Scholar]
- Wang, H., Li, L., Zhang, L., An, J., Cheng, H., and Deng, Z. (2016) Xylitol production from waste xylose mother liquor containing miscellaneous sugars and inhibitors: one‐pot biotransformation by Candida tropicalis and recombinant Bacillus subtilis . Microb Cell Fact 15: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger, J.W., Schwartz, K., and Sherlock, G. (2010) Bulk segregant analysis by high‐throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae . PLoS Genet 6: e1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Chi, P., Bilal, M., and Cheng, H. (2019) Biosynthetic strategies to produce xylitol: an economical venture. Appl Microbiol Biotechnol 103: 5143–5160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The sugars in a fermentation of X3. Fermentations were performed in 1.4 l fermenters containing 800 ml medium at conditions of 30°, 500 rpm, and 1 vvm. The pH was maintained at 5.5 by adding an aqueous ammonia solution and 2 mol l‐1 HCl. The YP medium contained 20% (v/v) xylose mother liquor and the initial cell biomass was ˜1 g l‐1 DCW. After 8 h of fermentation, 0.75 g l‐1 h‐1 of glucose was added over 72 h. Data are mean values of two biological repeats.


