Summary
Lactiplantibacillus plantarum (formerly Lactobacillus plantarum) is a lactic acid bacteria species found on plants that is essential for many plant food fermentations. In this study, we investigated the intraspecific phenotypic and genetic diversity of 13 L. plantarum strains isolated from different plant foods, including fermented olives and tomatoes, cactus fruit, teff injera, wheat boza and wheat sourdough starter. We found that strains from the same or similar plant food types frequently exhibited similar carbohydrate metabolism and stress tolerance responses. The isolates from acidic, brine‐containing ferments (olives and tomatoes) were more resistant to MRS adjusted to pH 3.5 or containing 4% w/v NaCl, than those recovered from grain fermentations. Strains from fermented olives grew robustly on raffinose as the sole carbon source and were better able to grow in the presence of ethanol (8% v/v or sequential exposure of 8% (v/v) and then 12% (v/v) ethanol) than most isolates from other plant types and the reference strain NCIMB8826R. Cell free culture supernatants from the olive‐associated strains were also more effective at inhibiting growth of an olive spoilage strain of Saccharomyces cerevisiae. Multi‐locus sequence typing and comparative genomics indicated that isolates from the same source tended to be genetically related. However, despite these similarities, other traits were highly variable between strains from the same plant source, including the capacity for biofilm formation and survival at pH 2 or 50°C. Genomic comparisons were unable to resolve strain differences, with the exception of the most phenotypically impaired and robust isolates, highlighting the importance of utilizing phenotypic studies to investigate differences between strains of L. plantarum. The findings show that L. plantarum is adapted for growth on specific plants or plant food types, but that intraspecific variation may be important for ecological fitness and strain coexistence within individual habitats.
The phenotypic and genetic properties were examined for 13 Lactiplantibacillus plantarum (formerly Lactobacillus plantarum) strains from (fermented) plant sources. The findings broadly show that strains obtained from the same or similar plant environments tend to be more genetically related and share similar carbohydrate utilization and stress tolerance capacities. However, there were still significant differences between all strains, irrespective of their source, indicating that intraspecific variation ‐is important for ecological fitness of ‐L. plantarum within individual habitats.
Introduction
Certain lactic acid bacteria (LAB) required for food fermentations are recognized for their genetic and phenotypic diversity and have been classified as ‘nomadic’ or ‘generalist’ because of their broad habitat range (Duar et al., 2017; Choi et al., 2018; Yu et al., 2020). Lactiplantibacillus plantarum [formerly Lactobacillus plantarum (Zheng et al., 2020)] is included among those nomadic LAB (Duar et al., 2017) and is known for its significant intraspecific versatility (Molenaar et al., 2005; Siezen et al., 2010; Martino et al., 2016; Cen et al., 2020). L. plantarum is frequently isolated from fresh and fermented plant, meat and dairy foods and is an inhabitant of the gastrointestinal and vaginal tracts of humans and animals (Delgado et al., 2005; Aquilanti et al., 2007; Di Cagno et al., 2008; Yang et al., 2010; Ciocia et al., 2013; Jose et al., 2015; Zago et al., 2017; Parichehreh et al., 2018; Barache et al., 2020). This species is frequently required for or is involved in the production of numerous fermented foods (e.g. fermented olives, sauerkraut, salami, and sourdough), and certain strains are effective probiotics (Marco, 2010; Seddik et al., 2017; Crakes et al., 2019). Consistent with its host and environmental range, L. plantarum strains have larger genomes compared with LAB with narrow host ranges and also carry strain‐specific genes, often located on lifestyle adaptation islands (Molenaar et al., 2005; Sun et al., 2015; Zheng et al., 2015; Duar et al., 2017; Salvetti et al., 2018).
Despite the robust growth of L. plantarum in different host‐associated and food environments, L. plantarum genomes and cell properties have thus far shown limited correlations with isolation source across disparate habitats (Siezen et al., 2010; Martino et al., 2016). These findings indicate that either intraspecific variation of L. plantarum within individual sources is fortuitous and members of this species have not evolved for growth in specific habitats (Martino et al., 2016) or that this observed variation is the result of adaptive evolution of the L. plantarum species within certain habitats with the outcome of maximizing co‐occurrence by niche complementarity (Bolnick et al., 2011; Ehlers et al., 2016).
To begin to address these two hypotheses, we examined the intraspecies variation of a collection of L. plantarum strains isolated from fermented olives and other plant food types. L. plantarum is typically highly abundant in olive fermentations (Hurtado et al., 2012). Assessments of the population sizes of individual L. plantarum strains in olive fermentations over time have shown how these fermentations are highly dynamic, likely undergoing succession processes at both the species and strain levels (Zaragoza et al., 2017). These findings are notable because although LAB have received considerable attention for their contributions to plant fermentations, the diversity, abundance and importance of L. plantarum and other LAB in plant microbiomes are not well understood (Yu et al., 2020). It has been found that LAB in spontaneous (wild) plant food fermentations are subject to dispersal and selection constraints (Miller et al., 2019). However, adaptations expressed by these bacteria that are specific to plant environments and interactions between the same or highly related LAB species remain to be determined.
Lactiplantibacillusplantarum was isolated from olive fermentations (AJ11, BGM55, BGM37, BGM40 and EL11), tomato fermentations (T2.5 and WS1.1), teff injera fermentations (W1.1, B1.1 and B1.3), wheat sourdough starter (K4), wheat boza (8.1) and prickly pear cactus fruit (1B1) (Table 1). Some isolates were collected from the same source either at the same time (strain B1.1 and B1.3) or on different days over the course of fermentation (strains AJ11, BGM37 and BGM40). The strains were selected without considering special criteria or selective pressure. A reference strain from saliva (NCIMB8826R) was used for comparison. To investigate their phenotypic range, the L. plantarum strains were evaluated for growth on a variety of plant‐associated carbohydrates and during exposure to high NaCl [4% (v/v)], ethanol [8% and 12% (v/v)] or surfactant [sodium dodecyl sulfate (SDS, 0.03% (w/v)] stress. The isolates were measured for the capacity to grow at a low pH (pH 3.5) as well as survive (pH 2) and tolerate a high temperature (50°C) incubation. Biofilm formation and growth inhibition of Saccharomyces cerevisiae UCDFST 09‐448, a pectinolytic spoilage yeast (Golomb et al., 2013), were also tested. Lastly, to establish the genetic basis for the observed strain differences, multi‐locus sequence typing (MLST) and comparative genomics were performed.
Table 1.
Lactiplantibacillusplantarum strains used in this study.
| Strain name | Isolation source | Isolation datea | References |
|---|---|---|---|
| AJ11 | Fermented olives; commercial fermentation | 12/02/2010 | Golomb et al. (2013) |
| BGM55 | Fermented olives; pilot‐scale fermentation inoculated with S. cerevisiae 09‐448 | 03/07/2011 | Golomb et al. (2013) |
| BGM37 | Olive fermentation brine; commercial fermentation | 01/04/2011 | Golomb et al. (2013) |
| BGM40 | Fermented olives; commercial fermentation | 01/26/2011 | Golomb et al. (2013) |
| EL11 | Fermented olives; commercial fermentation | 12/04/2009 | Golomb et al. (2013) |
| K4 | Wheat sourdough starter | 09/15/2014 | This study |
| 8.1 | Wheat boza | 09/15/2014 | This study |
| W1.1 | White flour teff injera | 04/04/2015 | This study |
| B1.1 | Brown flour teff injera | 04/04/2015 | This study |
| B1.3 | Brown flour teff injera | 04/04/2015 | This study |
| T2.5 | Fermented tomatoes | 08/20/2015 | This study |
| WS1.1 | Fermented tomatoes (spoiled) | 08/20/2015 | This study |
| 1B1 | Ripe cactus fruit (Opuntia ficus‐indicia) | 10/25/2011 | Tyler et al. (2016) |
| NCIMB8826R | Human saliva | N/A | Yin et al. (2018) |
Month/Day/Year. N/A, not available.
Results
Strain differentiation and phylogenetic analysis
The isolates were identified as L. plantarum by 16S rRNA gene sequence analysis and differentiated from the closely related species Lactiplantibacillus pentosus [formerly Lactobacillus pentosus (Zheng et al., 2020)] and Lactiplantibacillus paraplantarum [formerly Lactobacillus paraplantarum (Zheng et al., 2020)] by multiplex PCR targeting recA (Torriani et al., 2001).
The strains were also found to have unique allelic multi‐locus sequence typing (MLST) sequence types (ST) (Table S1), thus confirming that they are genetically distinct and not derived from the same clonal populations. Among the eight genes tested by MLST, between 6 (uvrC) and 12 (pyrG) different alleles were found (Table S1). Phylogenetic analysis of the ST showed that the L. plantarum strains clustered into two clades (Fig. 1A). The isolates from fermented olives were contained in one clade, suggesting they are more closely related to each other and to the teff injera strain B1.3 than those retrieved from other sources. The two other strains from teff injera (B1.1 and W1.1) clustered together in the other clade which also contained NCIMB8826R and the strains isolated from wheat boza, sourdough, cactus fruit and fermented tomatoes (Fig. 1A). When examined in a MLST phylogenetic tree containing 264 other L. plantarum strains (Fig. S1), the L. plantarum isolates collected from fermented olives remained clustered closely together, whereas the others were distributed across the tree.
Fig. 1.
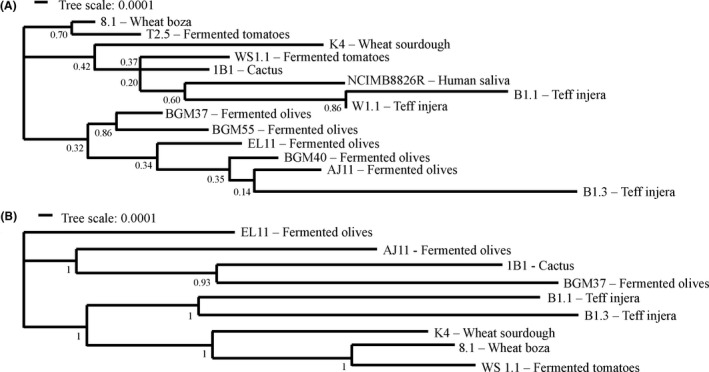
Phylogenetic relationships between Lactiplantibacillus plantarum strains.
A. Phylogenetic relationships of 14 strains of L. plantarum based on MLST profiles with pheS, pyrG, uvrC, recA, clpX, murC, groEL and murE (Table S8).
B. Relationships of nine L. plantarum strains based on concatenated core protein sequences using the maximum likelihood method with bootstrap values calculated from 500 replicates using Mega (7.0) (Kumar et al., 2016).
Carbohydrate utilization capacities
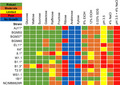
The capacity of the L. plantarum strains to use different sugars for growth was measured using MRS, a complete medium commonly used for cultivation of LAB (De Man et al., 1960). To exclude metabolizable carbon sources, the MRS was modified (mMRS) to remove beef extract and dextrose. In mMRS containing glucose, maltose or sucrose, all L. plantarum strains except B1.3 (teff injera) and 8.1 (wheat boza) were found to have robust growth according to area under the curve (AUC) rankings (Figs 2 and 3; Table S2). Those strains which grew robustly reached maximum OD600 values within 12 h (Fig. 3; Table S3) and displayed growth rates ranging from a low of 0.31 ± 0.01 h−1 (strain W1.1 (teff injera) in maltose) to a high of 0.45 ± 0.01 h−1 [BGM37 (fermented olives) in glucose] (Table S4). By comparison, the growth rate of B1.3 was lower in glucose (0.20 ± 0.00 h−1) and maltose (0.15 ± 0.00 h−1) compared to the other strains (Fig. 3; Table S4). In mMRS‐sucrose, both B1.3 and 8.1 exhibited poor growth (Figs 2 and 3; Table S2).
Fig. 2.
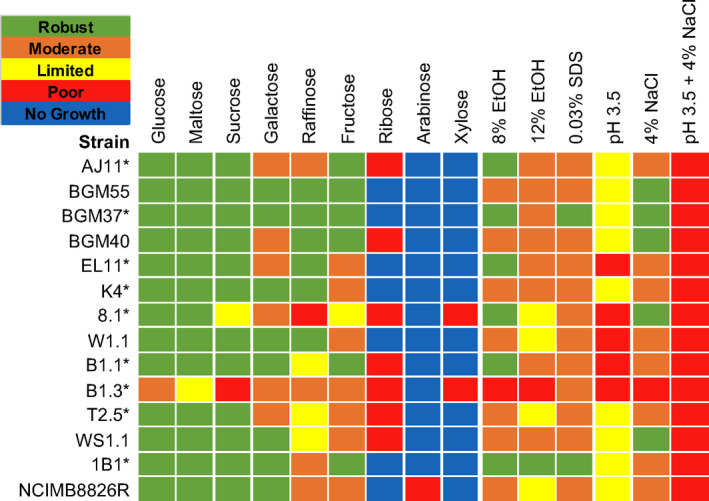
Lactiplantibacillusplantarum phenotype profiles. Area under the curve (AUC) values were used to illustrate L. plantarum capacities to grow in mMRS containing different sugars and in mMRS‐glucose in the presence of 8% (v/v) ethanol (EtOH), 8% (v/v) ethanol and then 12% (v/v) ethanol (12% ethanol), 0.03% (w/v) SDS, 4% (w/v) NaCl or set at pH 3.5 without or with 4% (w/v) NaCl. AUC values for the growth curves were ranked as ‘robust’ (AUC between 150 and 115), ‘moderate’ (AUC between 114 and 80), ‘limited’ (AUC between 79 and 45), ‘poor’ (AUC < 45) or ‘no growth’ (AUC was equivalent to the strain growth in mMRS lacking a carbohydrate source). L. plantarum growth in mMRS‐glucose supplemented with an equal volume of water instead of ethanol, NaCl or SDS was not significantly different compared to growth in mMRS‐glucose (P > 0.05). * indicates strains examined by whole genome sequencing.
Fig. 3.
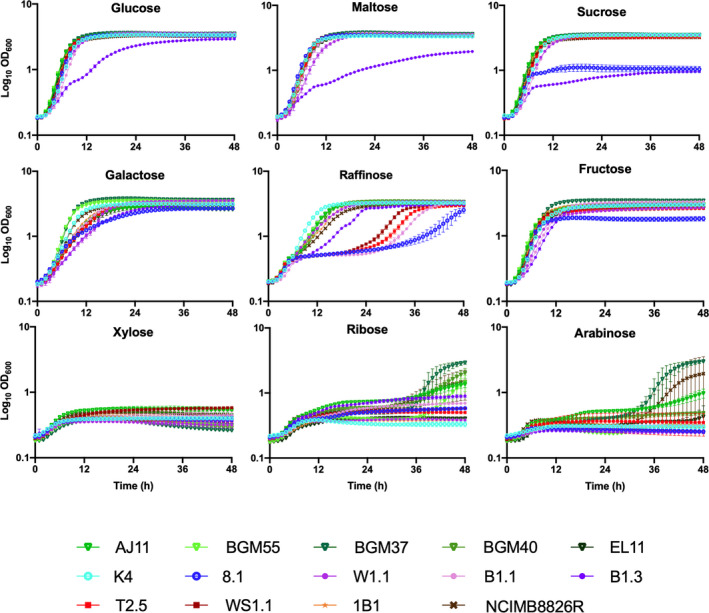
Growth of Lactiplantibacillus plantarum in mMRS containing different mono‐, di‐ and tri‐saccharides. L. plantarum was incubated in mMRS containing 2% (w/v) of each sugar at 30°C for 48 h. The avg ± stdev OD600 values of three replicates for each strain are shown.
All strains grew moderately to robustly when galactose was provided as the sole carbon source in mMRS (Figs 2 and 3; Table S2). Growth rates ranged from a low of 0.16 ± 0.003 h−1 [B1.3 (teff injera)] to a high of 0.42 ± 0.01 h−1 [BGM37 (fermented olives)] (Table S4). Final OD600 values measured after 24 h incubation ranged from 2.58 ± 0.05 [BGM40 (fermented olives)] to 3.62 ± 0.03 (BGM37) (Table S3). Because incubation in glucose‐containing MRS prior to exposure to mMRS‐galactose might result in carbon catabolite repression (Kremling et al., 2015), several strains with only moderate growth in that culture medium [AJ11, BGM40 and EL11 (fermented olives), 8.1 (wheat boza), B1.3 (teff injera) and T2.5 (fermented tomatoes)] were inoculated in succession into mMRS‐galactose. However, prior exposure to mMRS‐galactose did not result in higher AUC values (data not shown).
In mMRS with raffinose, all five L. plantarum strains isolated from fermented olives (BGM37, BGM55, BGM40, AJ11 and EL11) exhibited either moderate or robust growth (Figs 2 and 3; Tables S2–S4). Although strain W1.1 (teff injera) also grew robustly, the other strains isolated from teff and wheat fermentations (8.1, B1.1 and B1.3) and both strains isolated from fermented tomatoes (T2.5 and WS1.1) displayed limited or poor growth (Figs 2 and 3; Tables S2–S4). To address whether the poor growth of those isolates was due to carbon catabolite repression, serial passage in mMRS‐raffinose was performed. Notably, growth of four out of the five strains (B1.1, 8.1, T2.5 and WS1.1) was improved by successive cultivation in mMRS‐raffinose (Fig. S2).
When fructose was provided, all L. plantarum isolates except for strain 8.1 (wheat boza) exhibited either moderate or robust growth (Figs 2 and 3; Table S2). Similar to incubation in glucose and galactose, strain BGM37 (fermented olives) reached the highest OD600 (OD600 = 3.44 ± 0.03) (Table S3). Notably, growth of B1.3 (teff injera) was improved in mMRS‐fructose compared to the other sugars tested, as demonstrated by a higher growth rate (Table S4) and final OD600 (Table S3). Similar to the lack of effect on AUC values found after successive passage in the presence of mMRS‐galactose, no significant differences in growth were found for any of the 14 strains after multiple passages in mMRS‐fructose (data not shown).
Growth of L. plantarum was poor in mMRS containing xylose, ribose or arabinose. Only four olive‐associated strains (AJ11, BGM55, BGM37 and BGM40) and NCIMB8826R grew in the presence of mMRS‐ribose or mMRS‐arabinose and none grew in mMRS‐xylose (Figs 2 and 3; Tables S2–S4). After 38 h in mMRS‐ribose, the OD600 values for those strains ranged from a low of 1.36 ± 0.15 [AJ11 (fermented olives)] to a high of 2.93 ± 0.17 [BGM37 (fermented olives)] (Fig. 3; Table S3). In mMRS with arabinose, only NCIMB8826R and BGM37 grew, reaching an OD600 of 1.94 ± 1.56 and 2.99 ± 0.14 respectively (Fig. 3; Table S3). To investigate whether growth could be improved by prior exposure to those pentose sugars, strains AJ11, BGM37, 8.1 and NCIMB8826R were incubated with successive passages in mMRS‐ribose or mMRS‐arabinose. This resulted in shorter lag phase times and higher final OD600 values for AJ11, BGM37 and NCIMB8826R in both media (Figs S3 and S4). By comparison, no difference in growth was observed for strain 8.1 (wheat boza) in mMRS‐ribose or mMRS‐arabinose irrespective of the adaptation period (Figs S3 and S4).
Growth in the presence of ethanol
Because mMRS‐glucose resulted in robust growth of the majority of L. plantarum strains investigated here, that culture medium was used for investigation of stress tolerance properties. In mMRS‐glucose containing 8% (v/v) (174 mM) ethanol (EtOH), the AUCs for all strains except B1.3 (teff injera) were either moderate or robust (Figs 2 and 4; Table S2). Although lag phase times were longer (data not shown) and growth rates were reduced when ethanol was included in the culture medium (Table S5), the growth curves of six strains [AJ11, BGM37 and EL11 (fermented olives), 8.1 (wheat boza), B1.1 (teff injera) and 1B1 (cactus fruit)] were still regarded as robust according to AUC assessments (Fig. 2). Surprisingly, two strains, BGM37 (fermented olives) and 1B1 (cactus fruit), reached a higher final OD600 in mMRS‐glucose with 8% (v/v) ethanol than in mMRS‐glucose alone (Student’s t‐test, P < 0.05; Table S3).
Fig. 4.
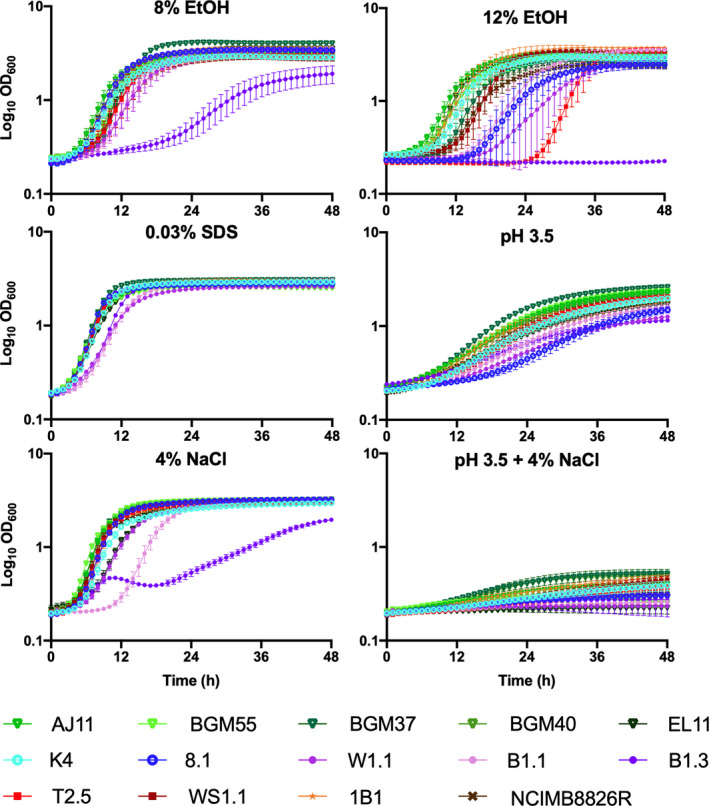
Growth of Lactiplantibacillus plantarum in mMRS‐glucose exposed to different environmental stresses. L. plantarum was incubated in mMRS‐glucose containing 8% (v/v) ethanol (EtOH), 12% (v/v) ethanol, 0.03% (w/v) SDS or 4% (w/v) NaCl with or without adjustment to pH 3.5 and incubated at 30°C for 48 h. The avg ± stdev OD600 values of three replicates for each strain are shown.
None of the L. plantarum strains tested here were able to grow over a 48 h period when incubated directly in mMRS‐glucose with 12% (v/v) (260 mM) ethanol (data not shown). To determine whether a more gradual exposure to high ethanol concentrations would change this outcome, the strains were incubated in mMRS‐glucose containing 8% (v/v) ethanol overnight prior to inoculation into mMRS‐glucose with 12% (v/v) ethanol. This modification resulted in robust growth of 1B1 (cactus fruit; Figs 2 and 4; Tables S2, S3 and S5). Eight other strains [AJ11, BGM55, BGM37, BGM40 and EL11 (fermented olives), K4 (wheat sourdough), B1.1 (teff injera) and WS1.1 (fermented tomatoes)] exhibited moderate growth according to AUC values as a result of the stepwise transfer to the higher [12% (v/v)] ethanol conditions (Figs 2 and 4; Tables S2, S3 and S5).
Growth in the presence of detergent (SDS) stress
While most of the L. plantarum strains exhibited moderate growth when sodium dodecyl sulfate (SDS) [0.03% (w/v) (0.10 mM)] was included in mMRS‐glucose, two strains BGM37 (fermented olives) and 1B1 (cactus fruit) grew robustly (Figs 2 and 4; Tables S2, S3 and S5). Remarkably, the growth rate of strain B1.3 (teff injera) was higher in the presence of SDS (0.32 ± 0.003 h−1) (Table S5) as opposed to its absence (0.20 ± 0.003 h−1) (Table S4) and it reached a higher AUC (107 ± 0.17) (Table S2).
Growth at pH 3.5 and in the presence of 4% NaCl
Growth of L. plantarum was reduced in mMRS‐glucose adjusted to a pH of 3.5 (Figs 2 and 4; Table S2). However, the strains isolated from brine‐based, fruit fermentations [AJ11, BGM55, BGM37, BGM40 and EL11 (fermented olives) and T2.5 and WS1.1 (fermented tomatoes)] grew significantly better under those conditions compared to the L. plantarum isolated from grain fermentations (Student’s t‐test, P < 0.05). The strains from grain‐based fermentations [K4 (wheat sourdough), 8.1 (wheat boza), W1.1, B1.1 and B1.3 (teff injera)] grew poorly in the acidified mMRS (pH 3.5) (Figs 2 and 4; Table S2), yielding low growth rates (0.06 ± 0.01 h−1) (Table S5) and final OD600 values (1.50 ± 0.19) (Table S3).
When 4% (w/v) NaCl was included in mMRS‐glucose, five strains isolated from different sources [BGM55, BGM37 and BGM40 (fermented olives), 8.1 (wheat sourdough) and WS1.1 (fermented tomatoes)] were classified as robust according to their AUC values (Fig. 2; Table S2). The growth of strain B1.3 (teff injera) was the most negatively impacted by the addition of salt into the laboratory culture medium (Fig. 4; Tables S2, S3 and S5).
All L. plantarum strains were inhibited in mMRS‐glucose containing 4% (w/v) NaCl and a starting pH of pH 3.5 (Figs 2 and 4; Table S2). The final OD600 values ranged from a low of 0.23 ± 0.00 (W1.1, teff injera) to a high of 0.52 ± 0.06 (BGM37, fermented olives) (Table S3). Although the AUCs of all strains were regarded to be poor, growth rates of those isolated from brine‐based, fruit fermentations [AJ11, BGM55, BGM37, BGM40 and EL11 (fermented olives) and T2.5 and WS1.1 (fermented tomatoes)] were significantly higher than those isolated from grain‐based fermentations [K4 (wheat sourdough), 8.1 (wheat boza), W1.1, B1.1, and B1.3 (teff injera)] (P < 0.05, Student’s t‐test).
Survival at pH 2
Within 15 min incubation in physiological saline adjusted to pH 2, a 104‐ to 106‐fold reduction in cell viability was observed (Fig. 5A). After 30 min exposure to pH 2, strains B1.3 (teff injera), BGM40 (fermented olives) and NCIMB8826R (saliva, reference strain) were no longer detectable by colony enumeration. BGM37 (fermented olives), B1.1 (teff injera) and T2.5 (fermented tomatoes) were no longer viable by 60 min (Fig. 5A). L. plantarum AJ11, BGM55 and EL11 (fermented olives), 8.1 (wheat boza) and WS1.1 (fermented tomatoes) exhibited the highest acid tolerance and were still viable according to colony enumerations performed on cells collected after 60 min incubation. Unlike the findings for growth under acidic conditions (pH 3.5) (Fig. 4), there were no obvious isolation‐source dependent trends in L. plantarum strain survival.
Fig. 5.
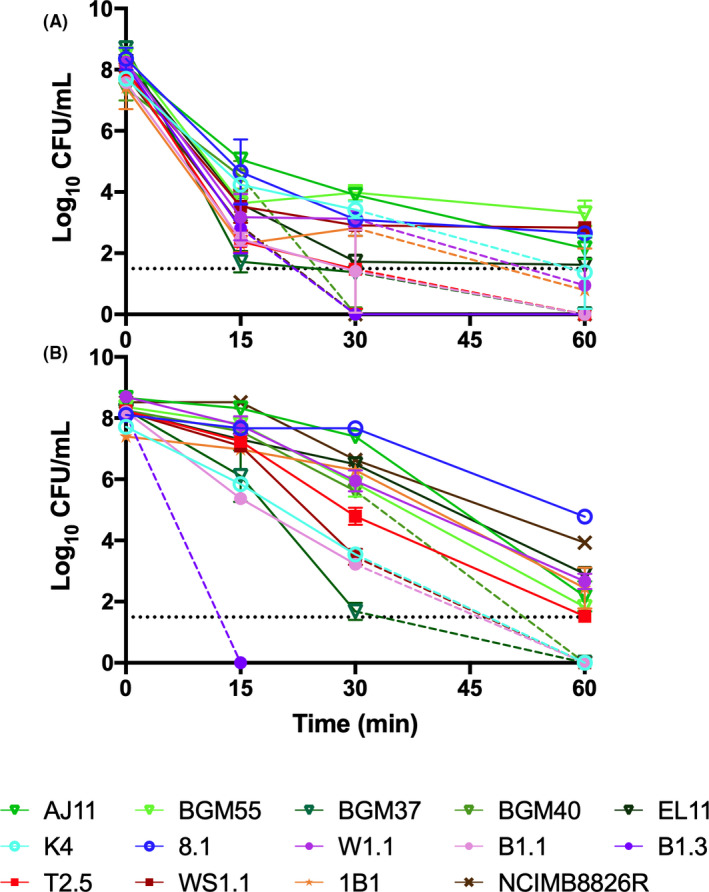
Survival of Lactiplantibacillus plantarum at (A) pH 2 and at (B) 50°C. (A) Viable cells were enumerated after 0, 15, 30 and 60 min of incubation in physiological saline at pH 2 or (B) in PBS at 50°C. The dashed lines indicate when the number of viable cells was below the detection limit (34 CFU ml−1). The avg ± stdev CFU ml−1 values of three replicates for each strain are shown.
Survival at 50°C
Survival of L. plantarum at 50°C spanned a 106‐ fold range (Fig. 5B). Viable B1.3 (teff injera) cells were no longer detected after incubation at 50°C for 15 min (1 × 108 cells ml−1 present in the inoculum). After 60 min, AJ11 and EL11 (fermented olives), 8.1 (wheat boza), W1.1 (teff injera), 1B1 (cactus fruit) and NCIMB8826R (saliva, reference strain) were still culturable in a range from 5 × 104 (8.1) to 1.5 × 102 (AJ11) CFU ml−1, spanning a 103‐ to 106‐fold reduction in viable cell numbers (Fig. 5B). Similar to survival to pH 2, no obvious isolation‐source dependent differences in survival were observed.
Biofilm forming capacity
Because biofilm formation is an indicator of bacterial capacities to tolerate environmental stress (Yin et al., 2019) and L. plantarum biofilm formation is partially dependent on carbon source availability (Fernández Ramírez et al., 2015), we examined the capacity of L. plantarum to produce biofilms during growth in mMRS with glucose, fructose or sucrose. Only BGM55 and BGM37 (fermented olives), 8.1 (wheat boza), W1.1 and B1.1 (teff injera), T2.5 and WS1.1 (fermented tomatoes) formed robust biofilms after growth in at least one of those laboratory culture media (Fig. 6). Whereas injera strain W1.1 only developed a biofilm when grown in mMRS‐fructose, the other isolates formed robust biofilms in the presence of at least two different sugars (Fig. 6). Both strains isolated from fermented tomatoes, T2.5 and WS1.1, formed extensive biofilms when grown in the presence of either glucose or fructose. Notably, biofilm formation was not associated with robust strain growth. Strain 8.1 formed a biofilm in mMRS‐sucrose (Fig. 6) despite showing poor growth (Fig. 2) and reaching a low final OD600 (Table S3) in that culture medium. Conversely, strain K4 grew well in mMRS‐sucrose but did not produce a biofilm.
Fig. 6.
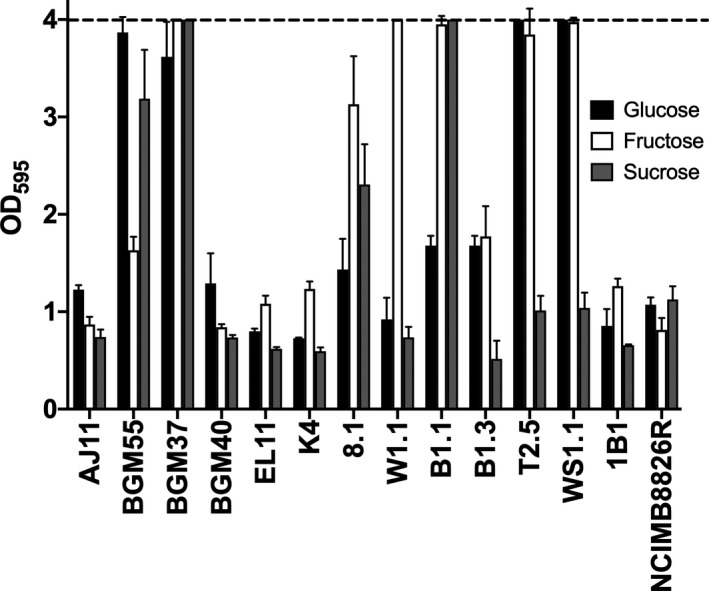
Lactiplantibacillusplantarum biofilm formation during growth in mMRS with glucose, fructose or sucrose. L. plantarum was incubated in mMRS‐glucose, mMRS‐fructose and mMRS‐sucrose in 96‐well, polystyrene microtiter plates at 30°C for 48 h. The non‐adherent cells were removed by washing with PBS. The remaining cells were stained with 0.05% crystal violet (CV). OD595 values of wells without cells did not exceed 0.22. The upper detection limit as indicated by the stippled line was an OD595 of 4.0. The avg ± stdev OD595 values of three replicate wells after CV staining are shown.
Antifungal activity of L. plantarum cell‐free culture supernatant (CFCS)
Growth rates and final OD600 values of S. cerevisiae UCDFST 09‐448 were reduced when incubated in the presence of the L. plantarum CFCS (Table S6). All L. plantarum CFCSs inhibited S. cerevisiae growth; however, there were some strain‐specific differences (Fig. 7; Table S6). Collectively, the CFCSs from strains isolated from fermented olives (AJ11, BGM55, BGM37, BGM40, EL11) and fermented tomatoes (WS1.1 and T2.5) were significantly (P < 0.05, Student’s t‐test) more inhibitory than those isolated from fermented grains (K4, 8.1, W1.1, B1.1 and B1.3). Among the strains isolated from fermented olives, growth inhibition resulting from exposure to the CFCS ranged between 29.8% ± 4.87 (BGM55) and 34.1% ± 9.4 (BGM40). By comparison, growth inhibition with CFCS from most L. plantarum isolated from grain fermentations was only between 20.1% ± 1.06 (B1.1) and 22.68% ± 1.46 (8.1). Interestingly, the growth pattern of S. cerevisiae in the presence of teff injera strain B1.3 CFCS (31.4 ± 1.27) was more similar to strains from fermented olives than grains.
Fig. 7.
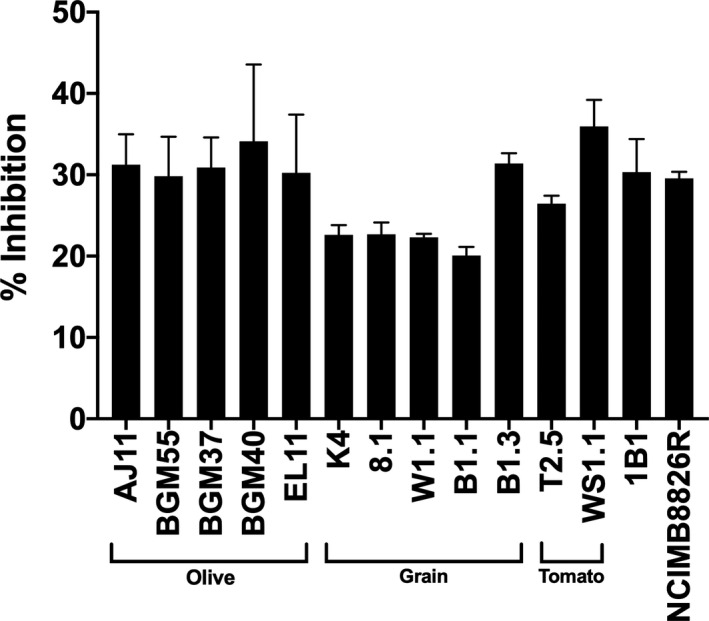
Saccharomycescerevisiae growth inhibition in the presence of Lactiplantibacillus plantarum CFCS. S. cerevisiae UCDFST‐09‐448 was incubated in a 1:1 ratio of 2X YM and pH adjusted L. plantarum CFCS from cMRS (pH 3.8). Growth was measured by monitoring the change in OD600 over 24 h. Per cent inhibition was determined by comparing the final OD600 of S. cerevisiae incubated in the presence of CFCS to cells incubated in a 1:1 ratio of 2X YM and pH adjusted (pH 3.8) cMRS (pH 3.8).
Comparisons of L. plantarum genomes
Nine of the fourteen strains were selected for genome sequencing (PacBio or Illumina platforms) based on the variations in their phenotypic profiles (Fig. 2). Genome assembly for strains sequenced using PacBio resulted in fewer contigs (min of 3 and max of 9) and higher coverage (min of 140X and max of 148X) compared to Illumina (contigs: min of 29 and max of 120; coverage (min of 27X and max of 128X) (Table 2). Genome sizes ranged from 3.09 Mbp [B1.3 (teff injera)] to 3.51 Mbp [WS1.1 (fermented tomatoes)], and total numbers of predicted coding sequences ranged from 3088 [K4 (wheat sourdough)] to 3613 (WS1.1) (Table 2).
Table 2.
Lactiplantibacillusplantarum genome coverage and assembly statistics.
| Straina | Accession No. | Genome size (Mb) | # of Contigs | Coverage | N50 | L50 | % GC content | # of CDS |
|---|---|---|---|---|---|---|---|---|
| AJ11 | WWDD00000000 | 3.27 | 29 | 27X | 252487 | 6 | 44.54 | 3214 |
| BGM37 | WWDC00000000 | 3.46 | 46 | 66X | 155998 | 7 | 44.15 | 3467 |
| EL11 | WWDB00000000 | 3.28 | 29 | 128X | 1944449 | 5 | 44.31 | 3231 |
| K4 | WWDF00000000 | 3.16 | 3 | 148X | 3157988 | 1 | 44.60 | 3088 |
| 8.1 | WWDE00000000 | 3.37 | 9 | 140X | 3066287 | 1 | 44.40 | 3366 |
| B1.1 | WWCZ00000000 | 3.17 | 120 | 76X | 59472 | 19 | 44.55 | 3242 |
| B1.3 | WWCY00000000 | 3.09 | 5 | 145X | 2939357 | 1 | 44.50 | 3157 |
| WS1.1 | WWDA00000000 | 3.51 | 99 | 30X | 78600 | 12 | 44.11 | 3613 |
| 1B1 | WWDG00000000 | 3.34 | 60 | 28X | 109565 | 11 | 44.34 | 3371 |
The genomes of AJ11, EL11, BGM37, WS1.1 and 1B1 were sequenced by Illumina MiSeq V2 (2 × 250). The genomes of strains K4, 8.1 and B1.3 were sequenced by PacBio RSII (P6‐C4 sequencing chemistry).
The core‐ and pan‐genomes of the nine strains consisted of 2222 and 6277 genes, respectively (Fig. S5), numbers consistent with previous comparisons examining larger collections of L. plantarum strains (Siezen et al., 2010; Martino et al., 2016; Choi et al., 2018). Alignments of the predicted amino acid sequences for the genes in the core genomes indicated that strains isolated from grain fermentations [K4 (wheat sourdough), 8.1 (wheat boza) and B1.1, and B1.3 (teff injera)] and strain WS1.1 from fermented tomatoes are more closely related to each other than isolates from olives and cactus fruit (Fig. 1B). B1.1 and B1.3, two strains originating from the same sample of teff injera, were also shown to share similar core genomes (Fig. 1B).
Just as strains 8.1 (wheat boza) and WS1.1 (fermented tomatoes) were found to have similar core genomes (Fig. 1B), those two strains are similar according to hierarchical clustering based on the numbers of genes in individual cluster of orthologous group (COG) categories (Fig. 8). The three strains isolated from olives formed a separate clade from those recovered from other sources and were shown to have higher numbers of genes in the carbohydrate metabolism and transport (G) and transcription (K) COGs. L. plantarum BGM37, a strain from olives that exhibited the most robust growth on the different carbohydrates compared tested here (Fig. 2), also contained the highest numbers of gene clusters annotated to the carbohydrate metabolism and transport COG (256 gene clusters, Fig. 8; Table S7).
Fig. 8.
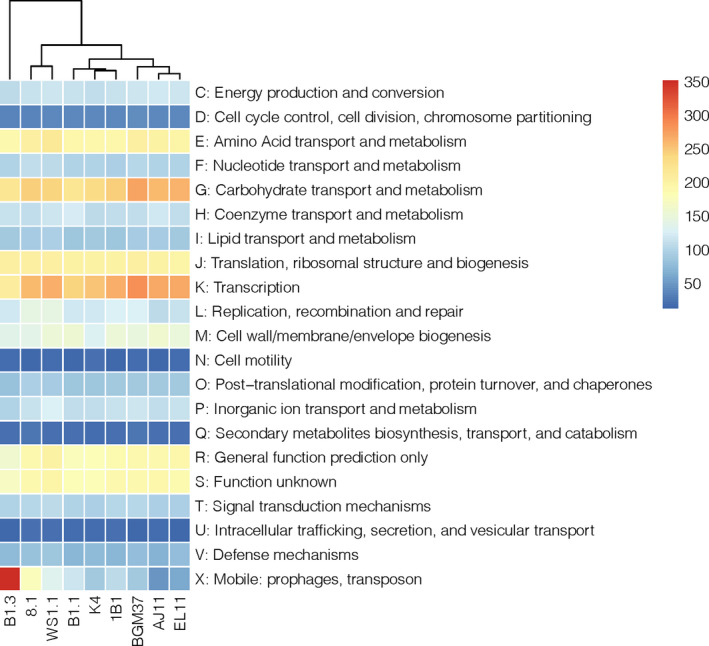
Distribution of COG Categories across Lactiplantibacillus plantarum genomes. Hierarchical clustering of L. plantarum based on the number of gene clusters assigned to each functional COG category. Number of gene clusters present in each strain is indicated by the colour gradient.
Strain B1.3 was found to contain the lowest number of gene clusters in the carbohydrate metabolism and transport COG (206 gene clusters, Fig. 8; Table S7) and is specifically lacking numerous genes required for sugar metabolism and sugar‐importing phosphotransferase (PTS) systems (data not shown). Conversely, the genome of B1.3 harbours at least twofold higher numbers of genes and genetic elements in the mobilome (X) COG compared to the other strains examined (352 gene clusters, Table S7). These genomic features include prophages, insertion sequence elements and transposases that are interspersed throughout the genome and frequently located between genes with known function. For example, a transposon (3.8 kb) is located between the glucose‐6‐phosphate isomerase (lp_2502) and glucose/ribose porter family sugar transporter (lp_2503) genes that are annotated to be associated with glucose metabolism. Other genes were not present in the B1.3 genome such as the sucrose‐associated PTS (lp_3819; pts24BCA), possibly indicating why this strain exhibited poor growth in mMRS‐sucrose. Strain 8.1, the only other L. plantarum strain tested here that grew to a limited extent on sucrose (Fig. 2), lacks the first 650 bp of pts1BCA (lp_0185), a gene in the sucrose phosphoenolpyruvate (PEP)‐dependent phosphotransferase system (PTS; Saulnier et al., 2007; Yin et al., 2018).
The number of gene clusters in the other COG categories was largely conserved between strains (Fig. 8; Table S7). These COG categories encode pathways required energy metabolism (glycolysis), synthesis of macromolecules (proteins, nucleotides and lipids) and stress response. The genomes of all nine strains contain genes encoding chaperones (DnaJK, GroEL, GroES, GrpE, ClpB, ClpL), proteases (ClpX, ClpP, ClpE), DNA repair proteins (RecA, UvrABC) and transcriptional regulators (HrcA, CtsR) critical for L. plantarum tolerance to numerous environmental stresses (Papadimitriou et al., 2016). Although genes required for citrate metabolism (citCDEF) were previously found to be associated with ethanol tolerance (van Bokhorst‐van de Veen et al., 2011) and that locus was flanked by mobile elements in several of the L. plantarum strains examined here, the presence of those mobile elements was not correlated with ethanol sensitivity.
Discussion
This study investigated the phenotypic and genetic properties of L. plantarum strains from (fermented) plant sources. The findings broadly show that strains obtained from the same or similar plant environments tend to be more genetically related and share similar carbohydrate utilization and stress tolerance capacities. However, there were still significant differences between all strains, irrespective of their source, a result which suggests that L. plantarum has adapted for growth in specific habitats (e.g. olive fermentations) but that intraspecific variation of this generalist species may afford the opportunity for L. plantarum strain coexistence by niche differentiation.
Our use of growth curve area under the curve (AUC) rankings and the monitoring of growth rates and final OD600 values provided a detailed view of L. plantarum carbohydrate utilization capacities. The majority of strains exhibited robust growth on glucose, maltose, sucrose and galactose, moderate growth on raffinose and fructose, and only limited to no growth on ribose, arabinose and xylose. The moderate or poor growth observed for a few strains when incubated the presence galactose or fructose was likely not due to carbon catabolite repression (Görke and Stülke, 2008; Kremling et al., 2015), but rather a lack of enzymatic capacity to utilize those sugars. These conserved carbohydrate consumption patterns are consistent with prior reports on L. plantarum isolated from plants and other host‐associated sources (Westby et al., 1993; Saulnier et al., 2007; Siezen et al., 2010; Filannino et al., 2014; Siragusa et al., 2014). The strains tested here were also able to grow in the presence of 0.03% (w/v) SDS and were severely impaired when incubated in mMRS at pH 3.5 with 4% (w/v) NaCl or inoculated directly into mMRS with 12% (v/v) ethanol.
Other findings were strain‐specific and similarly consistent with reported phenotypic (Parente et al., 2010; Siezen et al., 2010; Guidone et al., 2014; Ferrando et al., 2015, 2016; Gheziel et al., 2019; Fuhren et al., 2020; Prete et al., 2020) and genomic variations (Molenaar et al., 2005; Siezen et al., 2010; Siezen and van Hylckama Vlieg, 2011; Martino et al., 2016; Choi et al., 2018; Cen et al., 2020; Pérez‐Díaz et al., 2021) observed for the L. plantarum species. We found that L. plantarum growth was highly variable following the sequential incubation in 8% (v/v) and then 12% (v/v) ethanol. Strain growth rates in mMRS with 8% (v/v) ethanol were correlated with those observed for mMRS containing 0.03% SDS (r = 0.561, P < 0.05), thereby indicating overlapping mechanisms in L. plantarum strain tolerance to membrane‐disruptive compounds (Seddon et al., 2004; Bravo‐Ferrada et al., 2015; Mukhopadhyay, 2015). High temperature tolerance also differed between the L. plantarum isolates, such that incubation at 50°C for 60 min resulted in over a 105‐fold range in strain survival. Survival at pH 2 followed a similar trend, such that some strains were no longer culturable after 15 min, while other strains still formed colonies after prolonged (60 min) incubation. Notably, only two strains from olive fermentations (AJ11 and EL11) and 8.1 from boza survived well under both high temperature and low pH conditions. Although, the genomes were found contain chaperones and proteases known to be involved in L. plantarum heat and acid shock responses (Corcoran et al., 2008; Mills et al., 2011), the unique proteins or pathways expressed by those strains which confer heightened stress tolerance remain to be determined.
Despite the conserved and variable aspects of L. plantarum carbohydrate utilization and environment stress tolerance phenotypes, there were other remarkable trends associated with strain isolation source. For example, the isolates from acidic, brine‐containing ferments (olives and tomatoes) were more resistant to acidic pH (pH 3.5) and high NaCl (4% w/v) concentrations than those recovered from grain fermentations (wheat boza, wheat sourdough and teff injera). Genome comparisons using concatenated core gene amino acids showed that strains isolated from grain fermentations are more related to each other than those from other sources. Genetic conservation between olive fermentation‐associated strains was observed by MLST and COG gene numbers. These results are in agreement with a recent comparative genomics study of 140 strains of L. plantarum isolated from variable environments, which found that strains isolated from similar ecological niche share similar functional profiles (Cen et al., 2020).
The strains from fermented olives also showed the greatest capacity to consume raffinose (a tri‐saccharide composed of galactose, glucose and fructose). It is also notable that two of those isolates (BGM37 and BGM55) grew equally well in mMRS‐galactose as in mMRS‐glucose. These results are consistent with the findings that olives leaves and roots contain both raffinose (2.7 ± 0.1 µmol) and galactose (4.8 ± 0.3 µmol) (Cataldi et al., 2000) and that the fruits contain galactose along with higher concentrations of glucose, mannitol and fructose (Gómez‐González et al., 2010). All strains from olive fermentations also exhibited at least moderate or robust growth in mMRS in the presence of 8% (v/v) ethanol, and the CFCSs from those strains resulted in greater inhibition of S. cerevisiae UCDFST‐09‐448 compared to the CFCSs from L. plantarum isolated from other environments. Because yeast are normal members of olive fermentation microbiota, the inhibitory capacity may indicate the presence of shared mechanisms required to prevent yeast overgrowth.
Several strains also showed unique properties illustrative of the phenotypic range of the L. plantarum species. Among those strains was BGM37 isolated from the brine of fermented olives. This strain exhibited the most robust growth on the carbohydrates tested here, showed the highest tolerance to 8% ethanol and 0.03% SDS and was able to form biofilms in the presence of glucose, fructose and sucrose. Compared to the other strains for which genome sequences were obtained, BGM37 was found to have the second largest genome size (3.46 Mbp) after WS1.1 (3.51 Mbp), a magnitude comparable to the other L. plantarum strains with large (complete) genomes published at NCBI (maximum of 3.70 Mbp as of Jan 2021).
Lactiplantibacillusplantarum 1B1, a strain isolated from ripe cactus fruit, is notable because of its robust growth in the presence of either SDS or ethanol. Although other studies reported growth of L. plantarum in the presence of ethanol (van Bokhorst‐van de Veen et al., 2011; Brizuela et al., 2019; Chen et al., 2020), the capacity to grow well at 12% ethanol is an unusual trait even among oenological‐associated L. plantarum (Succi et al., 2017). Thus, the unique properties of this single isolate from a fresh fruit source may indicate the presence of a broader diversity of LAB present in the carposphere (Yu et al., 2020).
Lastly, strain B1.3 from teff injera exhibited the most restrictive carbon utilization capacities and the lowest levels of environmental stress tolerance among all isolates tested. B1.3 grew poorly on glucose and most other carbohydrates, whereas the other strains from teff injera B1.1 and W1.1 exhibited robust growth on a variety of sugars. Limitations in the ability of B1.3 to consume different sugars were also shown by the lower numbers of gene clusters in the B1.3 genome that are responsible for carbohydrate transport and metabolism. The overall smaller genome size of this strain (3.09 Mbp) and high numbers of genes in the mobilome COG potentially indicates that this strain is undergoing genome reduction for habitat specialization as found for other LAB [e.g. Lactobacillus bulgaricus (yoghurt) (van de Guchte et al., 2006), Lactobacillus iners (vagina) (France et al., 2016) and Apilactobacillus apinorum (honeybee) (Endo et al., 2018)]. Remarkably, the higher growth rate of B1.3 in mMRS‐fructose and in the presence of SDS indicates it may be fructophilic and capable of withstanding the presence of membrane disrupting compounds in teff flour. The finding that the CFCS from B1.3 inhibited S. cerevisiae UCDFST 09‐448 growth also suggests that B1.3 may be adapted to compete with yeast in teff injera. This result is consistent with the proximity of B1.3 to the olive‐associated strains in the MLST phylogenetic tree. However, it is also noteworthy that B1.3 shares genetic similarity with the teff injera isolate (B1.1) and other grain‐associated L. plantarum according to core genome comparisons.
Although disruptions in sucrose PTS systems may indicate why neither strain B1.3 nor 8.1 was able to grow in the presence of sucrose, the specific genes and pathways conferring the phenotypic variations observed in this study remain to be determined. To this regard, identification of the genome composition alone is insufficient to understand the full metabolic and functional potential of this species. For example, there still remains a lack of resolution in some PTS and other carbohydrate transport and metabolic pathways among lactobacilli (Gänzle and Follador, 2012; Zheng et al., 2015) and stress response mechanisms frequently involve numerous pathways with overlapping cell functions (e.g. membrane synthesis, protein turnover and energy metabolism pathways; Papadimitriou et al., 2016).
The genetic and phenotypic variation observed for the L. plantarum isolates indicate this species has evolved towards specialization in different plant‐associated habitats (e.g. fruit vs cereal grains), but at the same time is under selective pressure for sustaining intraspecific diversity within those habitats, possibly as a mechanism promoting L. plantarum species stability through co‐occurrence in those ecosystems (Maynard et al., 2019). This can be investigated using L. plantarum strains possessing shared and variable traits in plant and fermented plant food colonization assays. Overall, these efforts will be useful for understanding bacterial interactions and habitat partitioning in other complex host‐associated (e.g. Lloyd‐Price et al., 2017; Truong et al., 2017; Bongrand and Ruby, 2019; Ma et al., 2020) and environmental (e.g. Ellegaard et al., 2015; Koch et al., 2020; Props and Denef, 2020) sites wherein significant intraspecies diversity has been found but not yet understood. These findings may also be used to guide the selection of robust, multi‐strain starter cultures that are suited to inter‐ and intraspecies selection pressures in fruit and vegetable fermentations to result in optimal sensory and safety characteristics.
Experimental procedures
Bacterial strains and growth conditions
Lactiplantibacillusplantarum strains used in this study are shown in Table 1. The isolates from olive fermentations and cactus fruit were described previously (Golomb et al., 2013; Tyler et al., 2016), and NCIMB8826R, a rifampicin‐resistant variant (Yin et al., 2018) of strain NCIMB8826 (Hayward and Davis, 1956), was used as a reference. For L. plantarum isolation from injera batter, the batter was mixed with phosphate buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4‐7H2O, 1.4 mM KH2PO4) (pH 7.2) at a ratio of 1:10. For isolation from boza and sourdough, the batter was mixed with physiological saline (145 mM NaCl) (pH 7.0) at a ratio of 1:10. For isolation from fermented tomatoes, three tomatoes were placed in sterile bags containing mesh filters (Nasco, Modesto, CA) with 1 ml of PBS (pH 7.2) and macerated by hand. Serial dilutions of the injera, boza, sourdough and tomato suspensions were then plated on de Man, Rogosa and Sharpe (MRS) agar from a commercial source (BD, Franklin Lakes, NJ) (cMRS). Natamycin (25 μg ml−1) (Dairy Connection, Wisconsin, WI) was included in the cMRS agar to inhibit fungal growth. The cMRS agar plates were incubated at 30°C under aerobic or anaerobic conditions [BD BBL GasPak system (BD, Franklin Lakes, NJ)] for 48 h. Single colony isolates were repeatedly streaked for isolation on cMRS prior to characterization. For phenotypic and genotypic analysis, the L. plantarum strains were routinely grown in cMRS without aeration at 30°C.
Strain identification and typing
Lactiplantibacillusplantarum 16S rRNA genes were amplified from individual colonies using the 27F and 1492R primers (Lane et al., 1991; Table S8) with ExTaq DNA polymerase (TaKaRa, Shiga, Japan). Thermal cycling conditions were as follows: 95°C for 3 min, 30 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 90 s, and a final elongation step of 72°C for 5 min. The PCR products were purified [Wizard SV Gel and PCR Clean‐Up System (Promega, Madison, WI)] and sequenced at the UC Davis DNA Sequencing Facility http://dnaseq.ucdavis.edu/. The DNA sequences were compared against the National Center for Biotechnology Information (NCBI) database using the nucleotide Basic Local Alignment Search Tool (Blastn; https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the Ribosomal Database Project (RDP; http://rdp.cme.msu.edu/). Multiplex PCR targeting the recA gene was also used to confirm L. plantarum at the species level according to methods described by (Torriani et al., 2001; Table S8). The 16S rRNA sequencing data for the strains in this study can be found National Center for Biotechnology Information (BankIt) under accession numbers MT937284‐MT937296.
For multi‐locus sequence typing (MLST), genomic DNA was isolated with the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. PCR was then performed using primers targeting the variable regions of L. plantarum pheS, pyrG, uvrC, recA, clpX, murC, groEL and murE (Table S8; Xu et al., 2014). PCR amplification was preformed using ExTaq DNA polymerase (TaKaRa, Shiga, Japan) as previously described (Xu et al., 2014). The PCR products were sequenced in both directions using the forward and reverse primers at the UC Davis DNA Sequencing Facility (http://dnaseq.ucdavis.edu/) and Genewiz (South Plainfield, NJ). DNA sequences were aligned, trimmed and analysed using the Mega 7.0 software package (Kumar et al., 2016). Based on the findings, unique nucleotide sequences for a gene were defined as an allele and unique allelic profiles were defined as a sequence type. The concatenate sequences in the order of pheS, pyrG, uvrC, recA, clpX, murC, groEL and murE was used for phylogenetic tree analysis with maximum likelihood supported with a multi‐locus bootstrap approach using Mega 7.0 (Kumar et al., 2016). For comparisons to other strains of L. plantarum, the sequences of 264 strains of L. plantarum were downloaded from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/), and a minimum spanning tree of the 278 strains was made using Phyloviz Online (Ribeiro‐Gonçalves et al., 2016). The MLST DNA sequences can be found in the National Center for Biotechnology Information (BankIt) under gene accession numbers MT864201–MT864291 and MT880889–MT880901.
Genome sequencing, assembly, annotation and analysis
Nine strains were selected for genome sequencing by either Illumina MiSeq (Illumina, San Diego, CA) (B1.1, WS1.1, 1B1, AJ11, BGM37, EL11) or Pacific Biosciences (PacBio, Menlo Park, CA) (B1.3, 8.1, K4) DNA sequencing methods. For the Illumina MiSeq, approximately 3 − 109 cells were suspended in lysis buffer containing 200 mM NaCl, 20 mM (ethylenediaminetetraacetic acid) EDTA, 500 µl of 793 mM sodium dodecyl sulfate (SDS) and 300 mg of zirconium beads (0.1 mm, BioSpec Products, Bartlesville, OK). The cells were then mechanically lysed by bead‐beating at 6.5 m/s for 1 min with a FastPrep‐24 (MP Biomedical, Santa Ana, CA). To obtain larger DNA fragments appropriate for PacBio DNA sequencing, total genomic DNA was extracted from each strain by incubating approximately 3 × 109 cells in the presence of 20 mg ml−1 lysozyme (Sigma‐Aldrich, St. Louis, MO) at 37°C for 60 min. After extraction by either mechanical or enzymatic lysis, DNA was purified using phenol‐chloroform and ethanol precipitation methods (Sambrook and Russell, 2006).
Illumina libraries were prepared for paired‐end 250‐bp sequencing (2 × 250 bp) using the Nextera DNA Flex Library kit (Illumina, San Diego, CA). The libraries were sequenced at the UC Davis Genome Center (Davis, CA) (https://genomecenter.ucdavis.edu/) on an Illumina Miseq V2 according to the manufacturer’s protocol. Genomes were assembled with Spades (v3.12.0, using k‐mers 31, 51, 71), and Quast (v 4.6.3) was used to confirm assembly quality. The assembled genome sequences were then annotated with Rasttk (https://rast.nmpdr.org/) and PATRIC (Wattam et al., 2017). PATRIC comprehensive genome analysis was run using default auto parameters. This program encompasses BayesHammer for read error correction, Velvet, IDBA, and Spades for assembly, and ARAST to verify assembly quality (Wattam et al., 2017).
PacBio libraries were prepared and sequenced at the UC Davis Genome Center (Davis, CA) (https://genomecenter.ucdavis.edu/) on a Pacific Biosciences RSII instrument using P6‐C4 sequencing chemistry. Sequence SMRTcell files were imported into the PacBio SMRT portal graphical interface unit (https://www.pacb.com/) for de novo assembly using the hierarchical genome‐assembly process (HGAP) protocol (Chin et al., 2013) and RS HGAP Assembly 2 in Smart analysis version 2.3 software. The resulting assemblies were used for subsequent annotation with Rasttk (https://rast.nmpdr.org/) and Patric (Wattam et al., 2017). The whole genome sequencing data for this study can be found in the National Center for Biotechnology Information under the BioProject PRJNA598971.
Edgar 2.0 was used to evaluate the size of the pangenome and identify the number of genes shared between all nine sequenced strains as well as to identify the phylogenetic relationships between the different strains (Blom et al., 2016). The pan and core genomes were identified, and the results were presented as ortholog sets. To evaluate phylogenetic relationships, concatenate core amino acid sequences were aligned using Muscle (Edgar, 2004). The resulting alignment was used to construct a phylogenetic tree using a maximum likelihood method with bootstrapping in Mega 7.0 (Kumar et al., 2016). Anvi’o (v6.1) was used to group orthologous protein sequences into gene clusters for cluster of orthologues group (COG) functional assignments using the program ‘anvi‐pan‐genome’ (Eren et al., 2015; Delmont and Eren, 2018) with the flags ‘‐use‐ncbi‐blast’ (Altschul et al., 1990) and parameters ‘‐minibit 0.5’ (Benedict et al., 2014) and ‘mcl‐inflation 10’. COG frequency heat map with hierarchical clustering was generated using RStudio with the package ‘pheatmap’ (https://www.rstudio.com/). To confirm the truncation of pts1BCA in L. plantarum 8.1, the pts1BCA gene was amplified from genomic DNA from strains B1.3, K4, 8.1, and NCIMB8826R using the pts1BCA_trunF (5′‐ TCGTCACCGAGTGTTCGTTT) and pts1BCA_trunR (5′‐ AGTTGCTGGCCACTGTTCAT) primers (Table S8) and ExTaq DNA polymerase (TaKaRa, Shiga, Japan). Thermal cycling conditions were as follows: 95°C for 3 min, 30 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 90 s, and a final elongation step of 72°C for 5 min. PCR products were visualized on a 1% agarose gel.
Carbohydrate utilization
Lactiplantibacillusplantarum strains were first incubated in cMRS for 24 h at 30°C. The cells were then collected by centrifugation at 5000× g for 5 min, washed twice in PBS to remove residual nutrients (pH 7.2) and then suspended in a modified MRS (mMRS) without beef extract or dextrose (pH 6.5) (De Man et al., 1960). The cell suspensions were then distributed into 96‐well microtiter plates (Thermo Fisher Scientific, Waltham, MA) at an optical density (OD) at 600 nm (OD600) of 0.2. To test the capacity to grow on different sugars, mMRS was amended to contain 2% (w/v) of d‐glucose (111 mM) (Fisher Scientific, Fair Lawn, NJ), d‐maltose monohydrate (55 mM) (Amresco, Solon, OH), sucrose (58 mM) (Sigma, St. Louis, MO), d‐galactose (111 mM) (Fisher Scientific, Fair Lawn, NJ), d‐raffinose pentahydrate (40 mM) (VWR International, Solon, OH), D‐fructose (55 mM) (Fisher Scientific, Fair Lawn, NJ), d‐xylose (133 mM) (Acros Organics, Morris Plains, NJ), d‐ribose (133 mM) (Acros Organics, Morris Plains, NJ) or l‐arabinose (133 mM) (Acros Organics, Morris Plains, NJ). The OD600 values were measured hourly for 48 h in a Synergy 2 microplate reader (Biotek, Winooski, VT) set at 30°C without aeration.
Growth during exposure to ethanol, SDS, NaCl and pH 3.5
Lactiplantibacillusplantarum was incubated in cMRS for 24 h at 30°C. The cells were then collected by centrifugation at 5000× g for 5 min, washed twice in PBS (pH 7.2) and then suspended in mMRS‐glucose (2% (w/v) (111 mM) d‐glucose) (pH 6.5). The cell suspensions were then distributed into 96‐well microtiter plates containing mMRS‐glucose amended to contain ethanol [8% (v/v) (174 mM) or 12% (v/v) (260 mM)], SDS [0.03% (w/v) (0.10 mM)] or NaCl [4% (w/v) (68 mM)]. For measuring the effects of low pH, mMRS‐glucose was adjusted to pH 3.5 with 1 M HCl. For measuring the effect of both low pH and high NaCl concentration, mMRS‐glucose (pH 3.5) was supplemented with 4% (w/v) (68 mM) NaCl. Each strain was also incubated in mMRS diluted with water between [4 and 12% (v/v)] to control for dilution of mMRS due to amendment addition. The OD600 was used to monitor growth during incubation at 30°C for 48 h without aeration using a Synergy 2 microplate reader (Biotek, Winooski, VT).
Survival at pH 2 or 50°C
For assessing acid tolerance, L. plantarum was incubated in cMRS for 24 h at 30°C prior to collection by centrifugation at 5000× g for 5 min and washing twice in physiological saline (145 mM NaCl) (pH 7.0). L. plantarum was then inoculated at a concentration of 1 × 108 cells ml−1 in physiological saline adjusted to pH 2 with 5 M HCl in 1.5 ml tubes. Survival was measured after 0, 15, 30 and 60 min incubation at 30°C. At each time point, three tubes were retrieved per stain for centrifugation at 10 000× g for 1 min. The supernatant was discarded, and the resulting cell pellet was suspended in 1mL physiological saline (pH 7.0). Serial dilutions were then plated on cMRS agar and incubated at 30°C for 48 h prior to colony enumeration.
Survival at 50°C
To measure thermal tolerance, L. plantarum was incubated in cMRS for 24 h at 30°C prior to collection by centrifugation at 5000× g for 5 min and washing twice in PBS (pH 7.2). The suspensions were then distributed into 0.2 ml tubes at approximately 1 × 108 CFU ml−1 and incubated in a C1000 Thermal Cycler (Bio‐Rad Laboratories, Foster City, CA) at 50°C for 0, 15, 30 and 60 min. At each time point, three tubes were retrieved per strain. Serial dilutions of the cell suspensions were plated onto cMRS agar and incubated at 30°C for 48 h prior to colony enumeration.
Biofilm formation assay
The potential for L. plantarum to form biofilms was assessed by measuring adherence to polystyrene according to previously described methods (Kopit et al., 2014) with several modifications. Briefly, 96‐well polystyrene plates (Thermo Fisher Scientific, Waltham, MA) containing either mMRS‐glucose, mMRS‐fructose or mMRS‐sucrose were inoculated with L. plantarum to a starting OD600 of 0.2 and the plates were incubated at 30°C for 48 h. The wells were then rinsed with PBS (pH 7.2), stained with 0.05% (w/v) crystal violet (CV), dried in an inverted position for 30 min and then rinsed again three times with PBS (pH 7.2). Absorbance at OD595 was measured with a Synergy 2 microplate reader (Biotek, Winooski, VT) to determine adherence. Wells containing mMRS with the corresponding sugar without L. plantarum inoculum were included as controls.
Yeast inhibition assay
Lactiplantibacillusplantarum cell‐free culture supernatants (CFCS) were prepared from the spent media collected after L. plantarum incubation in cMRS for 24 h at 30°C. CFCS was collected by centrifugation at 4000× g for 10 min at 4°C followed by filtration of the supernatant through a 0.45 μm polyethersulfone (PES) filter (Genesee Scientific, San Diego, CA). To eliminate the effects of differences of pH on yeast inhibition, the CFCS was adjusted with lactic acid (1.3 M) to pH 3.8, the lowest pH reached by L. plantarum after incubation in cMRS (data not shown). S. cerevisiae UCDFST 09‐448 (Golomb et al., 2013), a strain shown to cause olive tissue damage and spoilage during olive fermentations, was grown in yeast mould (YM) broth (BD, Franklin Lakes, NJ) for 24 h at 30°C with aeration at 250 rpm. Cells were collected by centrifugation at 20 000× g for 5 min at 4°C and then washed twice with PBS. S. cerevisiae UCDFST 09‐448 was then inoculated into 96‐well microtiter plates containing 1:1 ratio of 2X YM and CFCS at a starting OD600 of 0.05. OD600 was measured in a Synergy 2 microplate reader (Biotek, Winooski, VT) set at 30°C for 24 h aerated every hour by shaking for 10 s before each read. Controls included S. cerevisiae UCDFST 09‐448 incubated in YM and YM supplemented with cMRS (pH 3.8).
Statistical analysis
Area under the curve (AUC) was used to examine the growth and survival of L. plantarum under different conditions (Sprouffske and Wagner, 2016). The AUC was calculated with GraphPad Prism 8 (Graph Pad Software, San Diego, CA). Hierarchical clustering was generated using RStudio with the package ‘pheatmap’ based on AUC values (https://www.rstudio.com/). Unpaired, two‐tailed Student’s t‐tests were used to compare between the different L. plantarum groups (e.g. brine‐ and grain‐based fermentations). P values of < 0.05 were considered significant.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting information
Table S1. Allelic profiles of L. plantarum strains according to MLSTa.
Table S2. Average AUC values of L. plantarum under different growth conditions.
Table S3. Final OD600 of L. plantarum under different growth conditions.
Table S4. Growth rates of L. plantarum in mMRS containing different sugars.
Table S5. Growth rates of L. plantarum exposed to different environmental stresses.
Table S6. Growth characteristics of S. cerevisiae UCDFST 09‐448 incubated in L. plantarum cell free supernatant (CFCS).
Table S7. Distribution of gene clusters across L. plantarum genomes based on COG categories.
Table S8. PCR primers used in this study.
Fig. S1. Minimum spanning tree of L. plantarum using MLST.
Fig. S2. Growth of L. plantarum in mMRS containing 2% (w/v) raffinose after repeated incubation in that culture medium.
Fig. S3. Growth of L. plantarum in mMRS containing 2% (w/v) ribose after repeated‐incubation in that culture medium.
Fig. S4. Growth of L. plantarum in mMRS containing 2% (w/v) arabinose after repeated‐incubation in that culture medium.
Fig. S5. The core‐genome and pan‐genome of nine L. plantarum strains examined in this study.
Acknowledgements
We would like to thank Nathan Lee and Brendan McCarthy‐Sinclair for their assistance with conducting these assays. We would like to thank Menkir Tamrat for providing the injera from which we isolated L. plantarum B1.1 and B1.3. This work was supported by the USDA National Institute of Food and Agriculture (Grant No. 2015‐67017‐23116).
Microb. Biotechnol. (2021) 14(5), 1990–2008
Funding information
This work was supported by the USDA National Institute of Food and Agriculture (Grant No. 2015‐67017‐23116).
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Aquilanti, L., Santarelli, S., Silvestri, G., Osimani, A., Petruzzelli, A., and Clementi, F. (2007) The microbial ecology of a typical Italian salami during its natural fermentation. Int J Food Microbiol 120: 136–145. [DOI] [PubMed] [Google Scholar]
- Barache, N., Ladjouzi, R., Belguesmia, Y., Bendali, F., and Drider, D. (2020) Abundance of Lactobacillus plantarum strains with beneficial attributes in blackberries (Rubus sp.), fresh figs (Ficus carica), and prickly pears (Opuntia ficus‐indica) grown and harvested in Algeria. Probiot Antimicro Protein 12: 1514–1523. [DOI] [PubMed] [Google Scholar]
- Benedict, M.N., Henriksen, J.R., Metcalf, W.W., Whitaker, R.J., and Price, N.D. (2014) ITEP: An integrated toolkit for exploration of microbial pan‐genomes. BMC Genom 15: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom, J., Kreis, J., Spänig, S., Juhre, T., Bertelli, C., Ernst, C., and Goesmann, A. (2016) EDGAR 2.0: an enhanced software platform for comparative gene content analyses. Nucleic Acids Res 44: W22–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick, D.I., Amarasekare, P., Araújo, M.S., Bürger, R., Levine, J.M., Novak, M., et al. (2011) Why intraspecific trait variation matters in community ecology. Trends Eco Evol 26: 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongrand, C., and Ruby, E.G. (2019) Achieving a multi‐strain symbiosis: strain behavior and infection dynamics. ISME J 13: 698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo‐Ferrada, B.M., Gonçalves, S., Semorile, L., Santos, N.C., Tymczyszyn, E.E., and Hollmann, A. (2015) Study of surface damage on cell envelope assessed by AFM and flow cytometry of Lactobacillus plantarum exposed to ethanol and dehydration. J Appl Microbio 118: 1409–1417. [DOI] [PubMed] [Google Scholar]
- Brizuela, N., Tymczyszyn, E.E., Semorile, L.C., Valdes La Hens, D., Delfederico, L., Hollmann, A., and Bravo‐Ferrada, B. (2019) Lactobacillus plantarum as a malolactic starter culture in winemaking: a new (old) player? Electron J Biotech 38: 10–18. [Google Scholar]
- Cataldi, T.R.I., Margiotta, G., Iasi, L., Di Chio, B., Xiloyannis, C., and Bufo, S.A. (2000) Determination of sugar compounds in olive plant extracts by anion‐exchange chromatography with pulsed amperometric detection. Anal Chem 72: 3902–3907. [DOI] [PubMed] [Google Scholar]
- Cen, S., Yin, R., Mao, B., Zhao, J., Zhang, H., Zhai, Q., and Chen, W. (2020) Comparative genomics shows niche‐specific variations of Lactobacillus plantarum strains isolated from human, Drosophila melanogaster, vegetable and dairy sources. Food Bioscience 35: 100581. [Google Scholar]
- Chen, X., Wang, T., Jin, M., Tan, Y., Liu, L., Liu, L., et al. (2020) Metabolomics analysis of growth inhibition of Lactobacillus plantarum under ethanol stress. Int J Food Sci Technol 55: 3441–3454. [Google Scholar]
- Chin, C.‐S., Alexander, D.H., Marks, P., Klammer, A.A., Drake, J., Heiner, C., et al. (2013) Nonhybrid, finished microbial genome assemblies from long‐read SMRT sequencing data. Nat Methods 10: 563–569. [DOI] [PubMed] [Google Scholar]
- Choi, S., Jin, G.‐D., Park, J., You, I., and Kim, E.B. (2018) Pan‐genomics of Lactobacillus plantarum revealed group‐specific genomic profiles without habitat association. J Microbiol Biotechnol 28: 1352–1359. [DOI] [PubMed] [Google Scholar]
- Ciocia, F., McSweeney, P.L.H., Piraino, P., and Parente, E. (2013) Use of dairy and non‐dairy Lactobacillus plantarum, Lactobacillus paraplantarum and Lactobacillus pentosus strains as adjuncts in cheddar cheese. Dairy Sci Technol 93: 623–640. [Google Scholar]
- Corcoran, B.M., Stanton, C., Fitzgerald, G., and Ross, R.P. (2008) Life under stress: the probiotic stress response and how it may be manipulated. Curr Pharm Des 14: 1382–1399. [DOI] [PubMed] [Google Scholar]
- Crakes, K.R., Santos Rocha, C., Grishina, I., Hirao, L.A., Napoli, E., Gaulke, C.A., et al. (2019) PPARα‐targeted mitochondrial bioenergetics mediate repair of intestinal barriers at the host–microbe intersection during SIV infection. Proc Natl Acad Sci USA 116: 24819–24829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Man, J.C., Rogosa, M., and Sharpe, M.E. (1960) A medium for the cultivation of Lactobacilli. J Appl Bacteriol 23: 130–135. [Google Scholar]
- Delgado, S., Flórez, A.B., and Mayo, B. (2005) Antibiotic Susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr Microbiol 50: 202–207. [DOI] [PubMed] [Google Scholar]
- Delmont, T.O., and Eren, A.M. (2018) Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ 6: e4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cagno, R., Surico, R.F., Siragusa, S., De Angelis, M., Paradiso, A., Minervini, F., et al. (2008) Selection and use of autochthonous mixed starter for lactic acid fermentation of carrots, French beans or marrows. Int J Food Microbiol 127: 220–228. [DOI] [PubMed] [Google Scholar]
- Duar, R.M., Lin, X.B., Zheng, J., Martino, M.E., Grenier, T., Pérez‐Muñoz, M.E., et al. (2017) Lifestyles in transition: evolution and natural history of the genus Lactobacillus . FEMS Microbiol Rev 41: S27–S48. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers, B.K., Damgaard, C.F., and Laroche, F. (2016) Intraspecific genetic variation and species coexistence in plant communities. Biol Lett 12: 20150853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegaard, K.M., Tamarit, D., Javelind, E., Olofsson, T.C., Andersson, S.G., and Vásquez, A. (2015) Extensive intra‐phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genom 16: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, A., Maeno, S., Tanizawa, Y., Kneifel, W., Arita, M., Dicks, L., and Salminen, S. (2018) Fructophilic lactic acid bacteria, a unique group of fructose‐fermenting microbes. Appl Environ Microbiol 84: e01290‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren, A.M., Esen, Ö.C., Quince, C., Vineis, J.H., Morrison, H.G., Sogin, M.L., and Delmont, T.O. (2015) Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3: e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández Ramírez, M.D., Smid, E.J., Abee, T., and Nierop Groot, M.N. (2015) Characterization of biofilms formed by Lactobacillus plantarum WCFS1 and food spoilage isolates. Int J Food Microbio 207: 23–29. [DOI] [PubMed] [Google Scholar]
- Ferrando, V., Quiberoni, A., Reinhemer, J., and Suárez, V. (2015) Resistance of functional Lactobacillus plantarum strains against food stress conditions. Food Microbiol 48: 63–71. [DOI] [PubMed] [Google Scholar]
- Ferrando, V., Quiberoni, A., Reinheimer, J., and Suárez, V. (2016) Functional properties of Lactobacillus plantarum strains: a study in vitro of heat stress influence. Food Microbiol 54: 154–161. [Google Scholar]
- Filannino, P., Cardinali, G., Rizzello, C.G., Buchin, S., Angelis, M.D., Gobbetti, M., and Cagno, R.D. (2014) Metabolic responses of Lactobacillus plantarum strains during fermentation and storage of vegetable and fruit juices. Appl Environ Microbiol 80: 2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- France, M.T., Mendes‐Soares, H., and Forney, L.J. (2016) Genomic comparisons of Lactobacillus crispatus and Lactobacillus iners reveal potential ecological drivers of community composition in the vagina. Appl Environ Microbiol 82: 7063–7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhren, J., Rösch, C., ten Napel, M. , Schols, H.A., and Kleerebezem, M. (2020) Synbiotic matchmaking in Lactobacillus plantarum: substrate screening and gene‐trait matching to characterize strain‐specific carbohydrate utilization. Appl Environ Microbiol 86: e01081‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gänzle, M.G., and Follador, R. (2012) Metabolism of oligosaccharides and starch in Lactobacilli: a review. Front Microbiol 3: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheziel, C., Russo, P., Arena, M.P., Spano, G., Ouzari, H.‐I., Kheroua, O., et al. (2019) Evaluating the probiotic potential of Lactobacillus plantarum strains from Algerian infant feces: towards the design of probiotic starter cultures tailored for developing countries. Probiot Antimicro Protein 11: 113–123. [DOI] [PubMed] [Google Scholar]
- Golomb, B.L., Morales, V., Jung, A., Yau, B., Boundy‐Mills, K.L., and Marco, M.L. (2013) Effects of pectinolytic yeast on the microbial composition and spoilage of olive fermentations. Food Microbiol 33: 97–106. [DOI] [PubMed] [Google Scholar]
- Gómez‐González, S., Ruiz‐Jiménez, J., Priego‐Capote, F., and Luque de Castro, M.D. (2010) Qualitative and quantitative sugar orofiling in olive fruits, leaves, and stems by gas chromatography−tandem mass spectrometry (GC‐MS/MS) after ultrasound‐assisted leaching. J Agric Food Chem 58: 12292–12299. [DOI] [PubMed] [Google Scholar]
- Görke, B., and Stülke, J. (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6: 613–624. [DOI] [PubMed] [Google Scholar]
- Guidone, A., Zotta, T., Ross, R.P., Stanton, C., Rea, M.C., Parente, E., and Ricciardi, A. (2014) Functional properties of Lactobacillus plantarum strains: a multivariate screening study. LWT ‐ Food Sci and Technol 56: 69–76. [Google Scholar]
- Hayward, A.C., and Davis, G.H.G. (1956) The isolation and classification of Lactobacillus strains from Italian saliva samples. Br Dent J 101: 2733–2741. [Google Scholar]
- Hurtado, A., Reguant, C., Bordons, A., and Rozès, N. (2012) Lactic acid bacteria from fermented table olives. Food Microbiol 31: 1–8. [DOI] [PubMed] [Google Scholar]
- Jose, N.M., Bunt, C.R., and Hussain, M.A. (2015) Comparison of microbiological and probiotic characteristics of Lactobacilli isolates from dairy food products and animal rumen contents. Microorganisms 3: 198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, H., Germscheid, N., Freese, H.M., Noriega‐Ortega, B., Lücking, D., Berger, M., et al. (2020) Genomic, metabolic and phenotypic variability shapes ecological differentiation and intraspecies interactions of Alteromonas macleodii . Sci Rep 10: 809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopit, L.M., Kim, E.B., Siezen, R.J., Harris, L.J., and Marco, M.L. (2014) Safety of the surrogate microorganism Enterococcus faecium NRRL B‐2354 for use in thermal process validation. Appl Environ Microbiol 80: 1899–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremling, A., Geiselmann, J., Ropers, D., and de Jong, H. (2015) Understanding carbon catabolite repression in Escherichia coli using quantitative models. Trends in Microbiol 23: 99–109. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Stecher, G., and Tamura, K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, D. J. (1991) 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt, E., and Goodfellow, M. (eds). Chichester, UK: John Wiley and Sons, pp. 115–175. [Google Scholar]
- Lloyd‐Price, J., Mahurkar, A., Rahnavard, G., Crabtree, J., Orvis, J., Hall, A.B., et al. (2017) Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550: 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, B., France, M.T., Crabtree, J., Holm, J.B., Humphrys, M.S., Brotman, R.M., and Ravel, J. (2020) A comprehensive non‐redundant gene catalog reveals extensive within‐community intraspecies diversity in the human vagina. Nature Commun 11: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco, M. (2010) Lactobacillus plantarum in foods. In Encyclopedia of Biotechnology in Agriculture and Food. 1st edition, Boca Raton, FL, USA: CRC Press, pp. 360–362. [Google Scholar]
- Martino, M.E., Bayjanov, J.R., Caffrey, B.E., Wels, M., Joncour, P., Hughes, S., et al. (2016) Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ Microbiol 18: 4974–4989. [DOI] [PubMed] [Google Scholar]
- Maynard, D.S., Serván, C.A., Capitán, J.A., and Allesina, S. (2019) Phenotypic variability promotes diversity and stability in competitive communities. Ecol Lett 22: 1776–1786. [DOI] [PubMed] [Google Scholar]
- Miller, E.R., Kearns, P.J., Niccum, B.A., O’Mara Schwartz, J., Ornstein, A., and Wolfe, B.E. (2019) Establishment Limitation constrains the abundance of lactic acid bacteria in the Napa cabbage phyllosphere. Appl Environ Microbiol 85: e00269‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, S., Stanton, C., Fitzgerald, G.F., and Ross, R.P. (2011) Enhancing the stress responses of probiotics for a lifestyle from gut to product and back again. Microb Cell Fact 10: S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar, D., Bringel, F., Schuren, F.H., de Vos, W.M. , Siezen, R.J., and Kleerebezem, M. (2005) Exploring Lactobacillus plantarum genome diversity by using microarrays. J Bacteriol 187: 6119–6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, A. (2015) Tolerance engineering in bacteria for the production of advanced biofuels and chemicals. Trends in Microbiol 23: 498–508. [DOI] [PubMed] [Google Scholar]
- Papadimitriou, K., Alegría, Á., Bron, P.A., de Angelis, M., Gobbetti, M., Kleerebezem, M., et al. (2016) Stress physiology of lactic acid bacteria. Microbiol Mol Biol Rev 80: 837–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente, E., Ciocia, F., Ricciardi, A., Zotta, T., Felis, G.E., and Torriani, S. (2010) Diversity of stress tolerance in Lactobacillus plantarum, Lactobacillus pentosus and Lactobacillus paraplantarum: a multivariate screening study. Int J Food Microbiol 144: 270–279. [DOI] [PubMed] [Google Scholar]
- Parichehreh, S., Tahmasbi, G., Sarafrazi, A., Imani, S., and Tajabadi, N. (2018) Isolation and identification of Lactobacillus bacteria found in the gastrointestinal tract of the dwarf honeybee, Apis florea Fabricius, 1973 (Hymenoptera: Apidae). Apidologie 49: 430–438. [Google Scholar]
- Pérez‐Díaz, I.M., Johanningsmeier, S.D., Anekella, K., Pagán‐Medina, C.G., Méndez‐Sandoval, L., Arellano, C., et al. (2021) Genotypic and phenotypic diversity among Lactobacillus plantarum and Lactobacillus pentosus isolated from industrial scale cucumber fermentations. Food Microbiol 94: 103652. [DOI] [PubMed] [Google Scholar]
- Prete, R., Long, S.L., Joyce, S.A., and Corsetti, A. (2020) Genotypic and phenotypic characterization of food‐associated Lactobacillus plantarum isolates for potential probiotic activities. FEMS Microbiol Lett 367: 76. [DOI] [PubMed] [Google Scholar]
- Props, R., and Denef, V.J. (2020) Temperature and nutrient levels correspond with lineage‐specific microdiversification in the ubiquitous and abundant freshwater genus Limnohabitans . Appl Environ Microbiol 86: e00140‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro‐Gonçalves, B., Francisco, A.P., Vaz, C., Ramirez, M., and Carriço, J.A. (2016) PHYLOViZ Online: web‐based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res 44: W246–W251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti, E., Harris, H.M.B., Felis, G.E., and O’Toole, P.W. (2018) Comparative genomics of the genus Lactobacillus reveals robust phylogroups that provide the basis for reclassification. Appl Environ Microbiol 84: e00993‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2006) Purification of nucleic acids by extraction with Phenol:Chloroform. Cold Spring Harb Protoc 2006: pdb.prot4455. [DOI] [PubMed] [Google Scholar]
- Saulnier, D.M.A., Molenaar, D., de Vos, W.M. , Gibson, G.R., and Kolida, S. (2007) Identification of prebiotic fructooligosaccharide metabolism in Lactobacillus plantarum WCFS1 through microarrays. Appl Environ Microbiol 73: 1753–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddik, H.A., Bendali, F., Gancel, F., Fliss, I., Spano, G., and Drider, D. (2017) Lactobacillus plantarum and its probiotic and food potentialities. Probiot Antimicro Protein 9: 111–122. [DOI] [PubMed] [Google Scholar]
- Seddon, A.M., Curnow, P., and Booth, P.J. (2004) Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta – Biomembranes 1666: 105–117. [DOI] [PubMed] [Google Scholar]
- Siezen, R.J., and van Hylckama Vlieg, J.E. (2011) Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb Cell Fact 10: S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen, R.J., Tzeneva, V.A., Castioni, A., Wels, M., Phan, H.T.K., Rademaker, J.L.W., et al. (2010) Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol 12: 758–773. [DOI] [PubMed] [Google Scholar]
- Siragusa, S., De Angelis, M., Calasso, M., Campanella, D., Minervini, F., Di Cagno, R., and Gobbetti, M. (2014) Fermentation and proteome profiles of Lactobacillus plantarum strains during growth under food‐like conditions. J Proteom 96: 366–380. [DOI] [PubMed] [Google Scholar]
- Sprouffske, K., and Wagner, A. (2016) Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Succi, M., Pannella, G., Tremonte, P., Tipaldi, L., Coppola, R., Iorizzo, M., et al. (2017) Sub‐optimal pH preadaptation improves the survival of Lactobacillus plantarum strains and the malic acid consumption in wine‐like medium. Front Microbiol 8: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z., Harris, H.M.B., McCann, A., Guo, C., Argimón, S., Zhang, W., et al. (2015) Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6: 8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani, S., Felis, G.E., and Dellaglio, F. (2001) Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene‐derived primers. Appl Environ Microbiol 67: 3450–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong, D.T., Tett, A., Pasolli, E., Huttenhower, C., and Segata, N. (2017) Microbial strain‐level population structure and genetic diversity from metagenomes. Genome Res 27: 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, C.A., Kopit, L., Doyle, C., Yu, A.O., Hugenholtz, J., and Marco, M.I. (2016) Polyol production during heterofermentative growth of the plant isolate Lactobacillus florum 2F. J Appl Microbiol 120: 1336–1345. [DOI] [PubMed] [Google Scholar]
- van Bokhorst‐van de Veen, H., Abee, T., Tempelaars, M., Bron, P.A., Kleerebezem, M., and Marco, M.L. (2011) Short‐ and long‐term adaptation to ethanol stress and its cross‐protective consequences in Lactobacillus plantarum . Appl Environ Microbiol 77: 5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Guchte, M., Penaud, S., Grimaldi, C., Barbe, V., Bryson, K., Nicolas, P., et al. (2006) The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc Natl Acad Sci USA 103: 9274–9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam, A.R., Davis, J.J., Assaf, R., Boisvert, S., Brettin, T., Bun, C., et al. (2017) Improvements to PATRIC, the all‐bacterial bioinformatics database and analysis Resource Center. Nucleic Acids Res 45: D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westby, A., Nuraida, L., Owens, J.D., and Gibbs, P.A. (1993) Inability of Lactobacillus plantarum and other lactic acid bacteria to grow on D‐ribose as sole source of fermentable carbohydrate. J Appl Bacteriol 75: 168–175. [Google Scholar]
- Xu, H., Sun, Z., Liu, W., Yu, J., Song, Y., Lv, Q., et al. (2014) Multilocus sequence typing of Lactococcus lactis from naturally fermented milk foods in ethnic minority areas of China. J Dairy Sci 97: 2633–2645. [DOI] [PubMed] [Google Scholar]
- Yang, J., Cao, Y., Cai, Y., and Terada, F. (2010) Natural populations of lactic acid bacteria isolated from vegetable residues and silage fermentation. J Dairy Sci 93: 3136–3145. [DOI] [PubMed] [Google Scholar]
- Yin, W., Wang, Y., Liu, L., and He, J. (2019) Biofilms: the microbial “protective clothing” in extreme environments. Int J Mol Sci 20: 3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, X., Heeney, D.D., Srisengfa, Y.T., Chen, S.‐Y., Slupsky, C.M., and Marco, M.L. (2018) Sucrose metabolism alters Lactobacillus plantarum survival and interactions with the microbiota in the digestive tract. FEMS Microbiol Ecol 94: fiy084. [DOI] [PubMed] [Google Scholar]
- Yu, A.O., Leveau, J.H.J., and Marco, M.L. (2020) Abundance, diversity and plant‐specific adaptations of plant‐associated lactic acid bacteria. Environ Microbiol Rep 12: 16–29. [DOI] [PubMed] [Google Scholar]
- Zago, M., Scaltriti, E., Bonvini, B., Fornasari, M.E., Penna, G., Massimiliano, L., et al. (2017) Genomic diversity and immunomodulatory activity of Lactobacillus plantarum isolated from dairy products. Benef Microbes 8: 597–604. [DOI] [PubMed] [Google Scholar]
- Zaragoza, J., Bendiks, Z., Tyler, C., Kable, M.E., Williams, T.R., Luchkovska, Y., et al. (2017) Effects of exogenous yeast and bacteria on the microbial population dynamics and outcomes of olive fermentations. mSphere 2: e00315‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J., Ruan, L., Sun, M., and Gänzle, M. (2015) A genomic view of Lactobacilli and Pediococci demonstrates that phylogeny matches ecology and physiology. Appl Environ Microbiol 81: 7233–7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J., Wittouck, S., Salvetti, E., Franz, C.M.A.P., Harris, H.M.B., Mattarelli, P., et al. (2020) A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 70: 2782–2858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Allelic profiles of L. plantarum strains according to MLSTa.
Table S2. Average AUC values of L. plantarum under different growth conditions.
Table S3. Final OD600 of L. plantarum under different growth conditions.
Table S4. Growth rates of L. plantarum in mMRS containing different sugars.
Table S5. Growth rates of L. plantarum exposed to different environmental stresses.
Table S6. Growth characteristics of S. cerevisiae UCDFST 09‐448 incubated in L. plantarum cell free supernatant (CFCS).
Table S7. Distribution of gene clusters across L. plantarum genomes based on COG categories.
Table S8. PCR primers used in this study.
Fig. S1. Minimum spanning tree of L. plantarum using MLST.
Fig. S2. Growth of L. plantarum in mMRS containing 2% (w/v) raffinose after repeated incubation in that culture medium.
Fig. S3. Growth of L. plantarum in mMRS containing 2% (w/v) ribose after repeated‐incubation in that culture medium.
Fig. S4. Growth of L. plantarum in mMRS containing 2% (w/v) arabinose after repeated‐incubation in that culture medium.
Fig. S5. The core‐genome and pan‐genome of nine L. plantarum strains examined in this study.


