Summary
Med‐ORF10, a single‐domain protein with unknown function encoded by a gene located in a gene cluster responsible for the biosynthesis of a novel antitumour antibiotic medermycin, shares high homology to a group of small proteins widely distributed in many aromatic polyketide antibiotic pathways. This group of proteins contain a nuclear transport factor‐2 (NTF‐2) domain and appear to undergo an evolutionary divergence in their functions. Gene knockout and interspecies complementation suggested that Med‐ORF10 plays a regulatory role in medermycin biosynthetic pathway. Overexpression of med‐ORF10 in its wild‐type strain led to significant increase of medermycin production. It was also shown by qRT‐PCR and Western blot that Med‐ORF10 controls the expression of genes encoding tailoring enzymes involved in medermycin biosynthesis. Transcriptome analysis and qRT‐PCR revealed that Med‐ORF10 has pleiotropic effects on more targets. However, there is no similar conserved domain available in Med‐ORF10 compared to those of mechanistically known regulatory proteins; meanwhile, no direct interaction between Med‐ORF10 and its target promoter DNA was detected via gel shift assay. All these studies suggest that Med‐ORF10 regulates medermycin biosynthesis probably via an indirect mode.
Med‐ORF10 homologues are widely distributed and functionally divergent in Streptomyces. Med‐ORF10 plays a positive regulatory role in medermycin biosynthesis and also shows pleiotropic effects on other biosynthetic pathways in an unknown indirect mode.

Introduction
Streptomycetes are soil‐dwelling filamentous bacteria and notable for their ability to produce an impressive range of secondary metabolites, including many natural antibiotics with interesting biological activities and potential pharmacological and agricultural applications (Liu et al., 2018). Biosynthesis of natural products derived from streptomycetes is often coordinately controlled by both pleiotropic regulators and pathway‐specific regulators at different levels (Jones and Elliot, 2018). To date, several families of pathway‐specific regulators that have been identified are absolutely required for specific antibiotic production, while the pleiotropic regulators often regulate the onset of more than one antibiotic production and morphological differentiation of streptomycetes (Xia et al., 2020). Additionally, there still exist numerous regulatory candidate genes in certain antibiotic clusters with unknown mechanism. These mechanism‐unknown genes are becoming of great interest for their critical influence on the production of antibiotics and potential applications in the breeding of high‐yielding strains for antibiotic production.
Medermycin (MED, 1), as a member of Streptomyces‐derived aromatic polyketide antibiotics known as benzoisochromanequinones (BIQs, also called pyranonaphthoquinones, PNQs) (Fig. 1A) (Ichinose et al., 2003; Jiang et al., 2018; Zhou et al., 2019), exhibits prominent antitumour activities through a novel alkylation mechanism (Toral‐Barza et al., 2007; Salaski et al., 2009). In 2003, a 30‐kb complete medermycin biosynthetic gene cluster (med cluster) consisting of 29 genes was cloned from Streptomyces sp. AM‐7161 (AM‐7161) and sequenced. Most of med genes are functionally assigned to MED biosynthesis (Ichinose et al., 2003; Li et al., 2005; He et al., 2015). However, some genes, including med‐ORF10 encoding a small protein composed of 147 amino acid residues, have yet to be annotated (Fig. 1B). Thus far, a few complete BIQ gene clusters have been fully cloned and sequenced, which are responsible for the production of MED, actinorhodin (ACT), granaticin, alnumycin and qinimycin (Ichinose et al., 1998; Oja et al., 2008; Wu et al., 2017) (Fig. 1B). Frenolicin biosynthetic gene cluster was also partially cloned (Bibb et al., 1994). Bioinformatic analysis of gene clusters for BIQ/PNQ antibiotics described above revealed a common existence of Med‐ORF10 homologs with unassigned functions in their corresponding pathways, such as ActVI‐ORFA for ACT, Gra‐ORF31 for granaticin, FrnO for frenolicin and Aln‐ORF2 for alnumycin. This supported that they might play essential roles in the pathways of BIQ antibiotics through unknown mechanisms of action. However, functional characterization of these BIQ‐related proteins has not been conducted yet, except for ActVI‐ORFA that was verified to be involved in the regulation of ACT accumulation by controlling the expression of several tailoring‐step genes in ACT biosynthetic pathway via unknown mode of action (Ozawa et al., 2003; Taguchi et al., 2007).
Fig. 1.
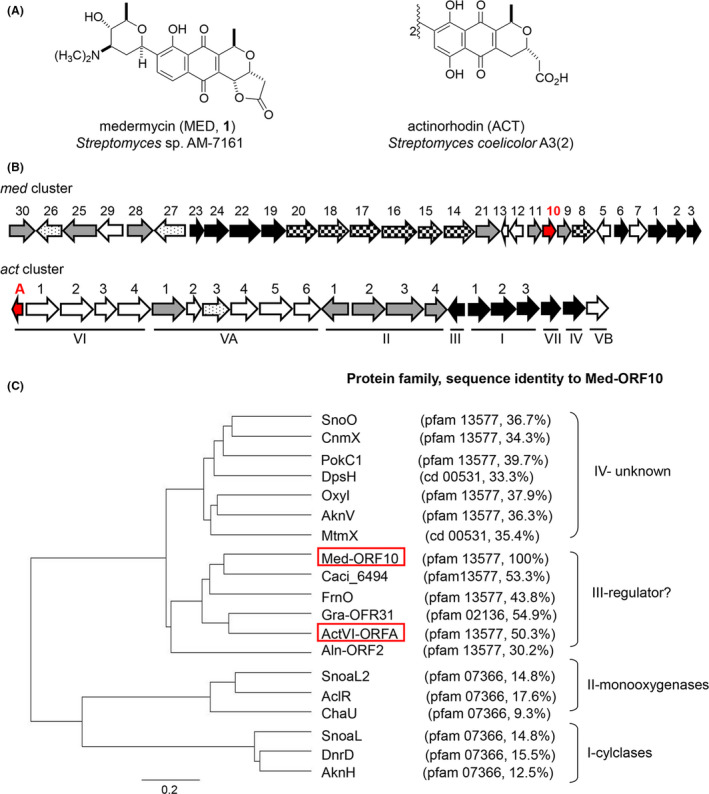
Structures and gene clusters of medermycin (MED, 1) and actinorhodin (ACT) and phylogenetic tree of Med‐ORF10 homologues.
A. Structures and producers of medermycin and actinorhodin.
B. Gene clusters of medermycin and actinorhodin. med10 (med‐ORF10) and actA (actVI‐ORFA) were indicated in red.
C. Phylogenetic tree of med‐ORF10‐homologs in antibiotic pathways. Their protein families and amino acid sequence identities to Med‐ORF10 were shown as well. Their accession numbers in GenBank are AknV (AAF73458.1), SnoO (AAF01807.1), CnmX (CAE17525.1), PokC1 (ACN64848.1), DpsH (AAD04719.1), OxyI (AAZ78332.2), Med‐ORF10 (BAC79038.1), Gra‐ORF31 (CAA09658.1), ActVI‐ORFA (NP_629222.1), FrnO (AAC18110.1), SnoaL (AAF01813), DnrD (AAA99000.1), AknH (AAF70112.1), SnoaL2 (2GEX_A), AclR (CAJ87106.1), DpsH (AD04719.1), ChaU (CAH10171.1), MtmX (CAA61988.1), Aln‐ORF2 (ACI88858.1) and Caci_6494 (ACU75344.1).
In order to investigate the function and mechanism of med‐ORF10, here we performed functional analysis of med‐ORF10 via phylogenetic analysis, complementation, overexpression and gene knockout experiments. Furthermore, qRT‐PCR and transcriptome analysis were conducted to determine the target genes of Med‐ORF10 followed by gel shift assay to detect whether Med‐ORF10 could directly bind to the promoter DNA of its targets. All results suggest that Med‐ORF10 acts as a positive regulator regulating MED biosynthesis in an as‐yet‐unknown indirect manner.
Results
Comparative analysis of Med‐ORF10 homologs
BLAST analysis revealed that med‐ORF10 homologs are located within the biosynthetic gene clusters for many aromatic polyketides besides BIQs/PNQs, such as aknH for aclacinomycins (Kallio et al., 2006), snoaL and snoaL2 for nogalamycin (Sultana et al., 2004; Bechthold and Yan, 2012; Siitonen et al., 2012), aclR for cinerubin (Beinker et al., 2006) and so on (Fig. 1C). It was shown that Med‐ORF10 homologous proteins share a nuclear transport factor‐2 (NTF‐2) domain in common by conserved domain search using CCD program. These homologs constitute a NTF‐2‐like superfamily (cl09109). Phylogenetic analysis suggested that this group of proteins encoded by Streptomyces secondary metabolite gene clusters can be divided into at least four subgroups (Fig. 1C). Among them, there are two subgroups (I and II) with predictable function since SnoaL and AknH in subgroup I were characterized as polyketide cyclases and SnoaL2 and AclR in subgroup II as polyketide monooxygenases (Sultana et al., 2004; Bechthold and Yan, 2012). ActVI‐ORFA within subgroup III was identified as a regulator controlling the expression of tailoring enzymes involved in ACT biosynthesis. Besides ActVI‐ORFA, Med‐ORF10 along with other homologs derived from different BIQ/PNQ biosynthetic pathways were also grouped into subgroup III, which occupied a distinctive position. A higher number of proteins constitute a subgroup IV, but none of which has been clarified functionally.
Interspecies complementation using med‐ORF10
Given the high sequence similarity and close evolutionally relationship between med‐ORF10 and actVI‐ORFA, we proposed that med‐ORF10 could complement actVI‐ORFA deficiency in ACT production. Subsequently, we tested such a possibility by delivering a med‐ORF10‐containing expression plasmid, pIJ8600‐med10, into the actVI‐ORFA‐deficient strain S. coelicolor J1501/ΔactVI‐ORFA (Taguchi et al., 2007) to generate a complementation strain J1501/ΔactVI‐ORFA/med10 for detection of the change in ACT production. Expression of med‐ORF10 in the complement strain was under the control of a strong inducible promoter (tipA promoter) induced by thiostrepton. The typical pH indicator property of ACT (red under acidic conditions and blue under basic ones) (Li et al., 2005; Taguchi et al., 2007) allowed us to carry out a simple complementation test using pigmentation to detect the ACT production in S. coelicolor. The significant increase of ACT production from J1501/ΔactVI‐ORFA/med10 was observed by visualizing a stronger blue pigmentation on R4 agar plate under basic conditions, compared to that in the deficient strain J1501/ΔactVI‐ORFA (Fig. 2A). This result supported our hypothesis that med‐ORF10 can complement the deficiency of actVI‐ORFA.
Fig. 2.
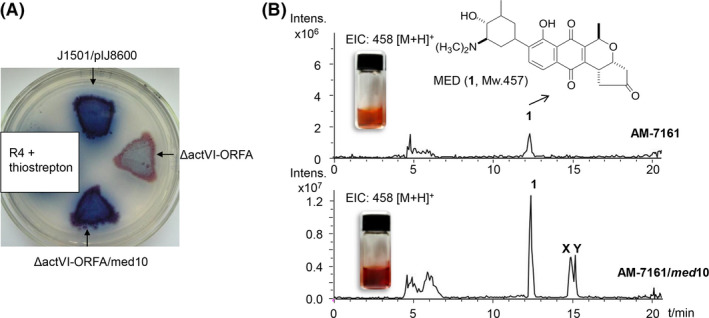
Complementation of med‐ORF10 in the actVI‐ORFA‐deficient mutant and overexpression of med‐ORF10 in AM‐7161.
A. J1501/pIJ8600, J1501/ΔactVI‐ORFA and J1501/(ΔactVI‐ORFA+med10) grown on R4 agar plate.
B. Methanol crude extracts of AM‐7161 and AM‐7161/med10 in glass vials and extracted ion chromatogram (EIC) of 1 (458.1, [M + H]+) from crude extracts of AM‐7161 and AM‐7161/med10; the peaks at 12.2 (1, MED) and 15 min (X and Y, MED‐related unknown compounds) were indicated respectively.
Enhancement of MED production by overexpression of med‐ORF10
To interrogate the regulatory role of med‐ORF10 in MED biosynthesis, we constructed a med‐ORF10 overexpression strain AM‐7161/med‐10 to check its effects on MED production by introduction of a free replicating med‐ORF10 expression plasmid pWHM4*‐med10 to AM‐7161. MED displays a typical red‐brown pigmentation in R4 medium (Ichinose et al., 2003), allowing us to easily observe MED production with the naked eyes. We then separately cultivated the recombinant strain AM‐7161/med10 and wild‐type (WT) strain AM‐7161 used as a control in R4 liquid medium, followed by extraction. Based upon the pigmentation observed in crude extracts, the MED production in AM‐7161/med10 was apparently higher than that in AM‐7161 (Fig. 2B). LC/MS analysis further showed that the MED production in AM‐7161/med10 was increased by approximately fourfold, compared to the WT strain. Additionally, two unknown compounds X and Y in AM‐7161/med10 at retention time of 15 min were increased as well (Fig. 2B).
Knockout of med‐ORF10
To further investigate the function of med‐ORF10, we attempted to knock out med‐ORF10 to check its effects on the production of MED. It has been still difficult to bring suicide plasmids into the WT strain for knockout experiments, though either integrative or auto‐replicating plasmids could be delivered into this strain (Deng et al., 2010). However, pIK340 carrying the entire med cluster has been heterologously expressed successfully for MED in a Streptomyces coelicolor CH999 (CH999) that is unable to produce PNQ‐related compounds (McDaniel et al., 1993; Ichinose et al., 2003). Therefore, we decided to delete med‐ORF10 from pIK340 in E. coli to generate a med‐ORF10‐deficiency plasmid pIK340‐Δmed10. pIK340‐Δmed10 was subsequently delivered into CH999 to obtain a CH999/pIK340‐Δmed10. It was shown obviously that the red‐brown pigmentation and sporulation of CH999/pIK340‐Δmed10 grown on R4 agar plate were reduced relative to those of CH999/pIK340 (Fig. 3A). LC/MS analysis of metabolite production from both strains revealed that MED (1) was dramatically decreased (Fig. 3B), but not completely abolished in CH999/pIK340‐Δmed10, compared to that in CH999/pIK340 (Fig. 3B and C).
Fig. 3.
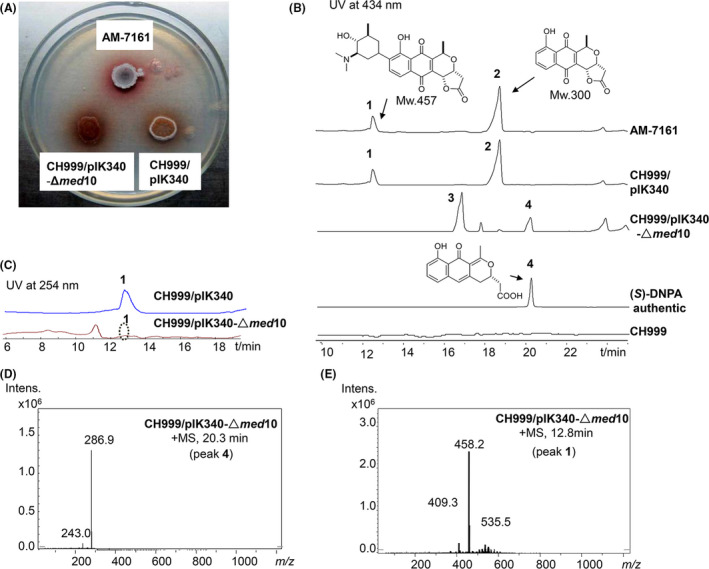
Medermycin production in med‐ORF10 knockout mutant strain.
A. AM‐7161 (native and wild type for med cluster), CH999/pIK340 and CH999/pIK340‐Δmed10 grown on R4 agar plate.
B. Comparison of UV spectra of crude extracts from AM‐7161 and CH999/pIK340 and CH999/pIK340‐Δmed10 at 434 nm.
C. Comparison of UV spectra at 254 nm, medermycin (1), kalafungin (2), (S)‐NHAB (3) and (S)‐DNPA (4).
D. The mass spectra of 4 ((S)‐DNPA) (286.9, [M + H]+) at 20 min.
E. The mass spectra of 1 (MED) (458.2, [M + H]+) at 12 min.
Meanwhile, the deficiency of med‐ORF10 also led to considerable reduction of kalafungin (2) with antitumour activity that is an intermediate of MED prior to C‐glycosylation (Salaski et al., 2009; Lv et al., 2015) (Fig. 3B). Additionally, obvious accumulation of two compounds 3 and 4 were detected in CH999/pIK340‐Δmed10 (Fig. 3B). By comparison of UV absorption, retention time and exact mass, we deduced that compound 4, also as an intermediate in the pathway of MED, is 4‐dihydro‐9‐hydroxy‐1‐methyl‐10‐oxo‐3‐naphtho‐[2,3‐c]‐pyran‐3‐(S)‐acetic acid ((S)‐DNPA, Mw 286) and compound 3 might be a shunt product, 1,4‐naphthoquinone‐8‐hydroxy‐3‐[(3S)‐acetoxybutyric acid] ((S)‐NHAB, Mw 318) (Taguchi et al., 2007) (Fig. 3B and D). Both of them have been also detected in the actVI‐ORFA‐deficiency strain of S. coelicolor (Ozawa et al., 2003; Taguchi et al., 2007). This further supported that MED and ACT share the same earlier and middle steps during their biosynthetic pathways (Ichinose et al., 2003; Li et al., 2005). All these results indicated that med‐ORF10 is a regulatory gene rather than a structural gene involved in the biosynthesis of MED. Moreover, no second copy or homologue from the genome of the CH999/pIK340‐Δmed10 or AM‐7161 could be found to complement its function.
Determination of the possible targets of med‐ORF10
Based on the results obtained above, we assumed that Med‐ORF10 regulates the biosynthesis of MED probably by controlling the expression level of structural genes in med gene cluster. med‐ORF12 in med gene cluster encodes a stereospecific ketoreductase responsible for the conversion of the bicyclic intermediate into (S)‐DNPA (4) in the pathway of MED (Li et al., 2005; He et al., 2015). As Med‐ORF10 deficiency can result in (S)‐DNPA (4) accumulation (Fig. 3B), we decided to check whether the expression of med‐ORF12 was controlled by Med‐ORF10. The result of qRT‐PCR showed that the transcription level of med‐ORF12 was reduced significantly by 70% in the med‐ORF10‐deficient strain (CH999/pIK340‐Δmed10), compared to CH999/pIK340 (Fig. 4A).
Fig. 4.
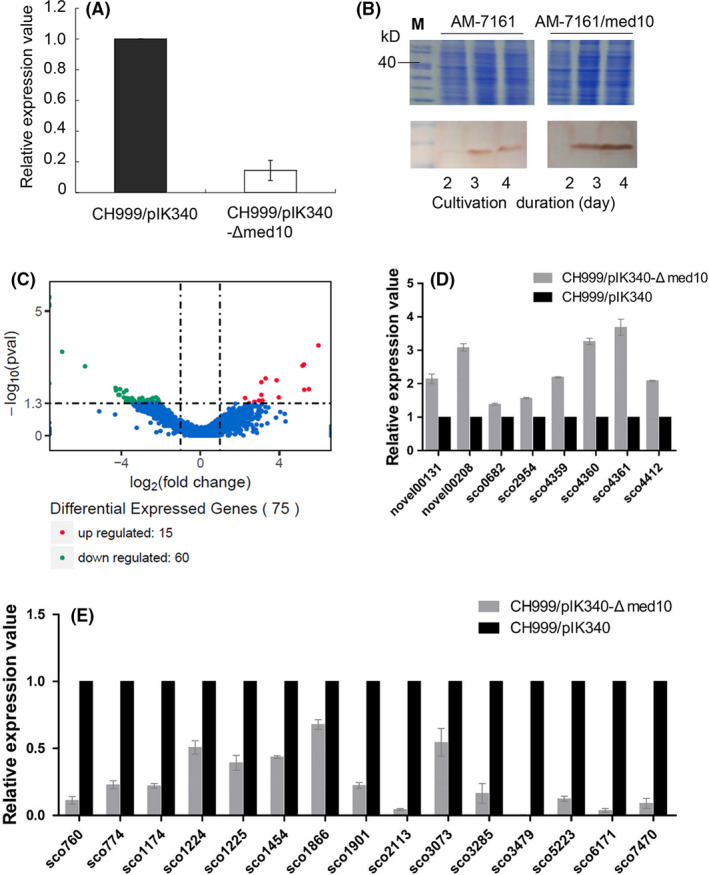
Regulatory effects of med‐ORF10 on expression levels of proposed targets.
A. Transcriptional analysis of med‐ORF12 in CH999/pIK340 and CH999/pIK340‐Δmed10) by qRT‐PCR.
B. Translation analysis of med‐ORF29 in AM‐7161 and AM‐7161/med10 was analysed by SDS‐PAGE (upper panel) and Western blot (bottom panel). M, protein marker. Total protein samples were isolated from 2‐, 3‐ and 4‐day cultures respectively.
C. The volcano plot of transcriptomics assay for differentially expressed native genes (non‐med genes) between CH999/pIK340‐Δmed10 versus CH999/pIK340 (mapped against the genome sequence of S. coelicolor A(3)2). The transcriptional level of different genes from CH999/pIK340 was normalized into 1. Differentially expressed genes are represented by red dots (upregulated) or green dots (downregulated) while blue dots represent genes without significantly differential expression. The x‐axis indicated the expression difference (P < 0.05 and log2(fold‐change) > 1 or < −1) of genes between CH999/pIK340 and CH999/pIK340‐Δmed10 while y‐axis represents the statistical significance (−log10(pval)) of expression difference.
D. qRT‐PCR analysis for partial upregulated genes in C.
E. qRT‐PCR analysis for partial downregulated genes in C.
Accordingly, it has been shown in our previous study that the translational level of Med‐ORF12 was also remarkably increased in AM‐7161/med10 (Sun et al., 2012). The med‐ORF29 in med gene cluster was annotated to encode an enoylreductase for reduction of (S)‐DNPA (4) (Ichinose et al., 2003). We then checked the expression of med‐ORF29 at protein level in AM‐7161/med10 and AM‐7161. Western blot analysis demonstrated that the protein expression of Med‐ORF29 was obviously increased in the AM‐7161/med10, compared to AM‐7161 (Fig. 4B).
Since Med‐ORF10 may positively regulate not only MED production and the sporulation in Streptomyces (Fig. 3A), but the production of unknown compounds X and Y (Fig. 2B), we next conducted transcriptomic analysis to find out whether Med‐ORF10 has pleiotropic effect as a global regulator. Therefore, CH999/pIK340 and CH999/pIK340‐Δmed10 were subsequently cultivated in biological replicates for each strain. The cultures from two technical replicates for each strain were collected and followed by RNA extraction and RNA‐Seq. A total of 11–14 million reads per sample were generated by RNA‐Seq and mapped to the genome sequence of the S. coelicolor A3(2) (A3(2)) in NCBI. As a derivative stain of A3(2), the host CH999 used here lacks ACT gene cluster (McDaniel et al., 1993; Kieser et al., 2000). These reads showed 75 native genes (non‐med genes) in total were significantly differentially expressed (P < 0.05 and log2(fold‐change) > 1 or < −1) between CH999/pIK340‐Δmed10 and CH999/pIK340 (Fig. 4C) and assigned for different physiological pathways (Figs S1–S3). Among them, 60 genes showed significant downregulation in CH999/pIK340‐Δmed10, compared to that in CH999/pIK340 (Tables S2 and S3), including several genes located in the gene clusters for secondary metabolites (polyketide, terpene and indole alkaloid): SCO1224 (annotated as sugar‐phosphate isomerase/Type 3‐PKS: herboxidiene), SCO1225 (osmoprotectant transporter/Type 3‐PKS: herboxidiene), SCO5223 (cytochrome P450/Terpene: albaflavenone) and SCO7470 (phenylacetic acid degradation protein PaaI/Indole alkaloid:5‐isoprenylindole‐3‐carboxylate β‐D‐glycosyl ester) (Table S2 and S3). Additionally, we also detected differential expression of some med genes for MED listed in Table S2, including med‐ORF12 and med‐ORF29.
We further confirmed the pleiotropic role of Med‐ORF10 by qRT‐PCR, which showed consistency with the data from transcriptomic analysis (Fig. 4D and E). Overall, these results suggested that Med‐ORF10 could control more targets apart from MED‐related biosynthetic genes, probably in a pleiotropic manner.
Interaction between Med‐ORF10 and a target promoter DNA in med cluster
Though the regulatory role of Med‐ORF10 was verified as above, it is difficult to predict how it controls the transcription of its target genes because bioinformatic analysis of Med‐ORF10 using various prediction tools showed that med‐ORF10 lacks an obvious DNA‐binding domain and cannot be classified into any regulatory family with known mechanism. Nevertheless, we performed EMSA for Med‐ORF10 with the intention of detecting the promoter DNA bound to med‐ORF12.
To this end, Med‐ORF10 was heterologously expressed in E. coli BL21(DE3), purified in vitro and then analysed by SDS‐PAGE and Native‐PAGE. It was shown that Med‐ORF10 appears to be a homogeneous multimer owing to its slower mobility on Native‐PAGE than SDS‐PAGE (Fig. 5A). A 182‐bp DNA region in the upstream of med‐ORF12 was searched using an online promoter prediction server (http://www.fruitfly.org/seq_tools/promoter.html) (Figs 5B and S3). Then, we incubated the biotin‐labelled promoter DNA with purified Med‐ORF10, followed by analysis of their binding ability via Native‐PAGE. After several times of repetitive experiments using recommended systems in the kit as positive control, there was no detectable binding between Med‐ORF10 and the target promoter DNA (Fig. 5C). Chromatin immunoprecipitation analysis also showed no binding between Med‐ORF10 and the genome DNA of AM‐7161 (data not shown).
Fig. 5.
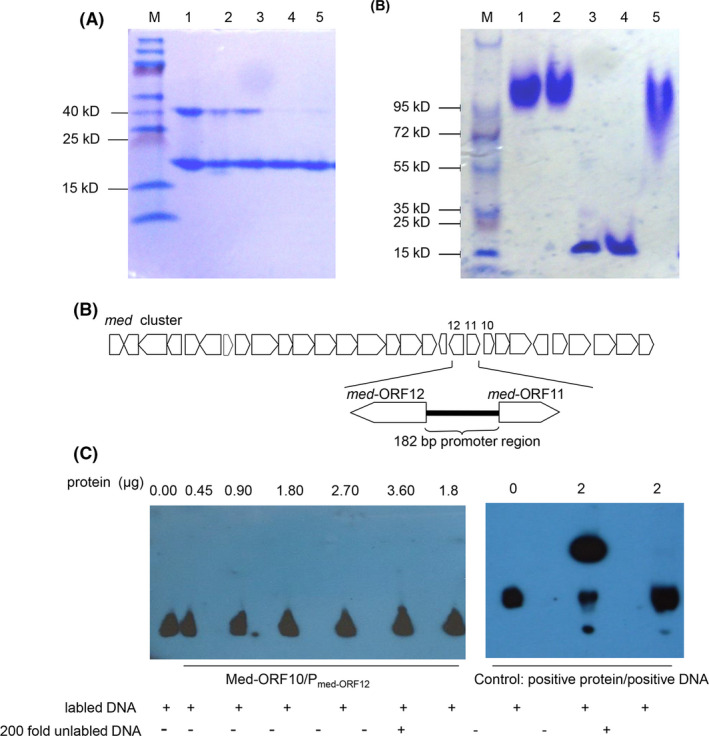
Analysis of purified Med‐ORF10 by electrophoresis and gel shift assay
A. Med‐ORF10 pre‐treated with different denaturing agents and analysed by SDS‐PAGE (left panel). M: Protein marker; lane 1: Med‐ORF10 without addition of loading buffer; lane 2: Med‐ORF10 with loading buffer; lane 3: Med‐ORF10 mixed with loading buffer and 2% SDS; lane 4: Med‐ORF10 mixed with loading buffer and 2% SDS and 250 mM DTT; and lane 5: Med‐ORF10 mixed with loading buffer and 250 mM DTT.
B. Med‐ORF10 pre‐treated with different denaturing agents and then analysed by Native‐PAGE (right panel). M: Protein marker; lane 1: Med‐ORF10 without addition of loading buffer; lane 2: Med‐ORF10 mixed with loading buffer containing 100 mM DTT; lanes 3‐4: Med‐ORF10 mixed with loading buffer containing 2% SDS and 250 mM DTT; and lane 5: Med‐ORF10 mixed with loading buffer containing 2% SDS.
C. Location of a 182‐bp promoter region of med‐ORF12 in med gene cluster for MED biosynthesis.
D. EMSA including biotin‐labelled 182‐bp promoter DNA incubated with different amounts of Med‐ORF10 (left panel) and positive control (right panel).
Discussion
It has been reported that NTF‐2‐domain‐containing proteins are widely distributed in bacteria, archaea and eukaryotes with a great diversity in their functions, which can be divided into several protein families (such as, pfam 07366, pfam 13577, pfam02136 and cd00531) (Yamada et al., 2004; Sone et al., 2018). Med‐ORF10 and its homologs containing a single NTF‐2 domain are extensively found in Streptomyces polyketide biosynthetic pathways, such as anthracycline and PNQ chemical families, triggering our interest into their functional roles in the biosynthetic pathways of these antibiotics (Duan et al., 2018). Combined with previous reports (Sultana et al., 2004; Beinker et al., 2006; Kallio et al., 2006; Taguchi et al., 2007), our phylogenetic analysis supports convincingly that these Med‐ORF10 homologs within aromatic polyketide antibiotic biosynthetic pathways undergo evolutionary divergence in their functions. For example, SnoaL (cyclase) and SnoaL2 (monooxygenase) play completely different roles in antibiotic pathways, although they share quite high sequence similarity and similar folds in their crystal structures (Sultana et al., 2004; Bechthold and Yan, 2012; Siitonen et al., 2012). In subgroup III, ActVI‐ORFA is an only functionally known example reported to regulate the expression of tailoring enzymes in ACT pathway (Taguchi et al., 2007). The close distance between ActVI‐ORFA and Med‐ORF10 in the phylogenetic tree suggested that they may share a similar function.
Besides ActVI‐ORFA and Med‐ORF10, Caci_6494 from an uncharacterized biosynthetic cluster in Catenulispora acidiphila and Aln2 from Streptomyces sp. CM020 in the pathway of alnumycin are also assigned to subgroup III. In a recent report about the X‐ray crystal structure elucidation of three NTF‐2‐like proteins (ActVI‐ORFA, Caci_6494 and Aln2) (Vuksanovic et al., 2020), the authors proposed that these three proteins are enzymes in polyketide biosynthesis only because of the presence of a solvent‐accessible cavity and the conservation of the His/Asp dyad in their structures. However, by far, no direct evidences support such a speculation. On the contrary, our previous (Taguchi et al., 2007) and present data strongly demonstrated that ActVI‐ORFA and Med‐ORF10 act as regulators and no second copy or homolog in the native genomes was found to complement their roles in the gene knockout mutants.
In both previous and present study, we observed the deletion of ActVI‐ORFA and Med‐ORF10 resulted in increase of accumulation of (S)‐DNPA (4) and shunt product NHAB (3) derived from earlier stages prior to (S)‐DNPA, implying that more than one target in MED and ACT pathways were downregulated. It was shown previously that the ActVI‐ORFA controls the expression of ActVI‐ORF1 (ketoreductase) and ActII‐ORF4 (regulator) in ACT pathway (Taguchi et al., 2007). Our previous studies revealed that Med‐ORF12, as a homologue of ActVI‐ORF1, acts as ketoreductase in MED pathway (Li et al., 2005; He et al., 2015). Med‐ORF29 shares high similarity to ActVI‐ORF2 for post‐modification of ACT (Ichinose et al., 2003). Here, we also proved that expression of Med‐ORF12 and Med‐ORF29 was regulated by Med‐ORF10 at transcriptional and translational levels. As expected, besides med‐ORF12 and med‐ORF29, more med genes, especially located in the operons for middle and later steps of MED biosynthesis, showed obvious differential expression at the absence of Med‐ORf10 using transcriptomics (Table S2 and Fig. S3), in accordance with the speculation from the data in Fig. 3 that Med‐ORF10 deficiency caused accumulation of some shunt or intermediate products in middle or later stages of MED biosynthesis.
Also as expected, the transcriptomic and qRT‐PCR data further supported the regulatory role of Med‐ORF10 but rather as a pleiotropic regulator because many genes involved in different physiological pathways were regulated obviously in the Med‐ORF10‐deficient strain (Table S2‐S3). Although we proved that Med‐ORF10 positively regulate MED biosynthesis, it is impossible to identify its target promoter DNA and explain its regulatory mechanism of action. This is probably resulted from the lack of DNA‐binding domain in Med‐ORF10.
What is more, overexpression of Med‐ORF10 in AM‐761 led to not only an increase of MED, but also an obvious appearance of unknown compounds X and Y which exact masses are very close to MED. Our transcriptomic data suggested some genes related to other secondary metabolite pathways might be controlled by Med‐ORF10. So, we will further try to isolate these compounds and elucidate their chemical structures, as well confirm the pleiotropic role of Med‐ORF10, especially its possible potential to enhance the production of more natural products.
In conclusion, herein, we reported that a function‐unknown small protein Med‐ORF10 positively regulates MED biosynthesis. Overexpression of Med‐ORF10 led to an increase of MED production. The qRT‐PCR and transcriptomic analysis further revealed its regulatory role, but rather as a pleiotropic regulator. The fact that there is no obvious DNA‐binding domain and detectable interaction between Med‐ORF10 and the DNA promoter region of its target implies its indirect participation in antibiotic biosynthesis.
Experimental procedures
Bacterial strains and plasmid vectors
Streptomyces strains used in this study include Streptomyces coelicolor CH999 (CH999, an act‐cluster‐deficient mutant) as a host for heterologous expression (McDaniel et al., 1993) and Streptomyces sp. AM‐7161 (AM‐7161) as MED‐producing strain (Ichinose et al., 2003) and S. coelicolor J1501/ΔactVI‐ORFA (actVI‐ORFA‐deficient strain) (Taguchi et al., 2007). E. coli 25113/pIJ790 was used as the host for λRED‐mediated PCR‐targeted gene inactivation (Gust et al., 2003; Abbasi et al., 2020). E. coli ET12567/pUZ8002 was used for intergeneric conjugation (Kieser et al., 2000). pIK340 is an integrative supercosmid containing the entire med gene cluster, which was used as a template for amplification of med genes and used for med‐ORF10 gene inactivation (Ichinose et al., 2003). pT7Blue and pET28a (+) obtained from Novagen were vectors for routine DNA cloning and prokaryotic expression in E. coli respectively. pIJ8600 containing an inducible tipA promoter and pWHM4* containing a strong constitutive ermE promoter were used as Streptomyces vectors for med‐ORF10 complementation and overexpression experiments respectively (Kieser et al., 2000). pIJ773 was used as the template for the amplification of apramycin resistance gene cassette (Gust et al., 2003).
Culture conditions and general genetic manipulations
Streptomyces strains were cultivated on R4 or GYM agar medium and grown in YEME or TSB liquid medium (Kieser et al., 2000). E. coli strains were cultivated in LB agar or liquid medium (Sambrook and Russel, 2001). When needed, antibiotics were added at a final concentration of 25 μg ml−1 for thiostrepton, 50 μg ml−1 for apramycin, 25 μg ml−1 for kanamycin, 25 μg ml−1 chloramphenicol, 100 μg ml−1 for nalidixic acid or 100 μg ml−1 for ampicillin. DNA isolation and manipulation from E. coli and Streptomyces were carried out according to standard protocols (Kieser et al., 2000; Sambrook and Russel, 2001).
Bioinformatic analysis of DNA and protein sequences
MEGA (version 7.0.26) was used to perform sequence alignment and phylogenetic analysis of proteins. BLAST programs online were used for similarity search. Conserved domains of proteins were predicted and analysed with Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/).
Gene knockout of med‐ORF10
Inactivation of med‐ORF10 in the entire med gene cluster on the plasmid pIK340 was performed as described in the λRED‐mediated PCR‐targeted REDIRECT technology kit (Gust et al., 2003). The apramycin resistance gene cassette for replacing med‐ORF10 was amplified from pIJ773 using two long primers (med10‐A‐Red and med10‐B‐Red) (Table S1). The resulting plasmid containing the mutant med cluster was designated as pIK340‐Δmed10. Subsequently, pIK340 and pIK340‐Δmed10 were introduced individually into S. coelicolor CH999 by intergeneric conjugation (Kieser et al., 2000). Apramycin‐resistant conjugants CH999/pIK340‐Δmed10 and CH999/pIK340 were picked out and further confirmed by PCR.
To detect the production of MED, intermediate or shunt products by CH999/pIK340‐Δmed10, its spore suspension was firstly inoculated into the seed medium and then transferred into R4 liquid medium as described in the previous studies (Li et al., 2005). Extraction procedures and LC/APCI/MS (Agilent 1100 HPLC/Brucker Esquire HCT) analytic conditions of the culture supernatant were performed according to those described previously (Ichinose et al., 2003).
Interspecies gene complementation
med‐ORF10 was amplified from pIK340 by PCR using primers med10‐A‐PIJ and med10‐B‐PIJ (Table S1). The 441‐bp PCR product of med‐ORF10 was inserted into the NdeI‐BamHI sites downstream of tipA promoter on pIJ8600 to generate a recombinant plasmid, pIJ8600‐med10, which was subsequently introduced into E. coli ET12567/pUZ8002, followed by intergeneric conjugation between E. coli and Streptomyces (S. coelicolor J1501/ΔactVI‐ORFA) (Kieser et al., 2000; Taguchi et al., 2007). Apramycin‐resistant exconjugants (J1501/ΔactVI‐ORFA/med10) were selected and confirmed by PCR using the genomic DNA as the templates isolated from the corresponding exconjugants. Subsequently, colonies of J1501/ΔactVI‐ORFA/med10 grown on R4 agar medium supplemented with thiostrepton (6.25 μg ml−1) for inducible gene expression was overlaid with 1N NaOH for blue pigmentation indicating ACT production.
Overexpression of med‐ORF10 in the MED‐producing strain
PCR amplification of med‐ORF10 gene was conducted using primers med10‐A‐WHM and med10‐B‐WHM (Table S1). The PCR product composed of med‐ORF10 gene and a native ribosome binding site upstream of it was inserted into the downstream of ermE promoter on pWHM4*, generating a recombinant plasmid, pWHM4*‐med10. This auto‐replicative expression plasmid was delivered into a medermycin‐producing strain AM‐7161 via intergeneric conjugation to obtain an overexpressing strain AM‐7161/med10. Subsequently, MED production in the broth of AM‐7161/med10 cultivated in R4 liquid medium was measured by LC/MSn using the system 1100 LC/MSD Trap (Agilent Technologies, Santa Clara, CA, USA) under the specific analytic conditions described by Ichinose et al., (2003).
qRT‐PCR analysis
Total RNA was isolated with the RNApure kit (BioTeke) from the mycelia of CH999/pIK340 and CH999/pIK340‐Δmed10 (med‐ORF10‐deficient strain) after cultivation on R4 plates for 5 d. RNasin (Takara)‐ and DNase I (Takara)‐treated RNA (1 mg) was used as the template for reverse transcription with random hexamers and reverse transcriptase M‐MLV (Promega). The qRT‐PCR using resultant cDNA samples was performed with the Premix DimerEraser® (Perfect Real Time) (TaKaRa, Japan) according to the manufacturer’s instructions on Applied Bio systems QuantStudio™ 3 Real‐Time PCR System (Applied Biosystems Inc., Waltham, MA, USA). Primer pairs for amplification of proposed targets and 23S rDNA (Table S1) were used to produce PCR products ranging from 250 to 400 bp. The experiment was independently conducted in triplicates. The relative mRNA expression of med‐ORF12 was calculated using the 2‐∆∆CT method (Livak and Schmittgen, 2001).
Prokaryotic expression in E. coli and purification of proteins
The med‐ORF10 gene was amplified from pIK340 by PCR using primers med10‐A‐PET and med10‐B‐PET (Table S1). The 441‐bp amplicon was ligated into pET28a (+) to generate pET28a‐med10 and followed by introduction into E. coli BL21(DE3) (Liu et al., 2015). Expression of Med‐ORF10 was induced by 0.1 mM IPTG at 16 °C for 25 h, followed by purification using Ni‐NTA column (Qiagen) and visualization on SDS‐PAGE. The same procedures were taken for Med‐ORF29 expression, purification and visualization (PCR primers: med29‐A‐PET and med29‐B‐PET; Table S1). To analyse Med‐ORF10 multimers using SDS‐PAGE and Native‐PAGE, 5 μL of Med‐ORF10 (0.68 mg ml−1) was pre‐incubated with 5 μl of 2 × protein loading buffer (Sambrook and Russel, 2001) for 5 min at room temperature, then loaded directly onto 6.5% polyacrylamide gel as a control, while the same amount of protein Med‐ORF10 was pre‐incubated with 5 μl loading buffer supplemented with 2% SDS and/or 250 mM DTT.
Preparation of polyclonal antibodies
400–600 μg purified protein Med‐ORF29 was mixed with Freund's complete or incomplete adjuvant and then used for immunizing rabbits for three times to prepare polyclonal antiserum at Wuhan Institute of Virology, Chinese Academy of Sciences. The titre and specificity of polyclonal antibodies were analysed with enzyme‐linked immunosorbent assay and Western blot (Sambrook and Russel, 2001).
Expression of Med‐ORF29 in Streptomyces
Total proteins were isolated from AM‐7161 and AM‐7161/med10 after cultivation for 2, 3 and 4 days in R4 medium, respectively, then analysed by SDS‐PAGE (15% polyacrylamide), and then transferred from PAGE onto PVDF membrane (Kieser et al., 2000). Subsequently, Western blotting with polyclonal antibodies against Med‐ORF29 was performed to detect the expression level of Med‐ORF29 (Kieser et al., 2000; Sambrook and Russel, 2001).
Electrophoretic mobility shift assay (EMSA) between Med‐ORF10 and DNA
A 182‐bp promoter DNA region upstream of med‐ORF12 was amplified from pIK340 by PCR using primers Pmed12A and Pmed12B (Table S1), followed by labelling of 50 pmol promoter DNA with biotin as described in the Biotin 3' End DNA Labeling Kit (Thermo, Waltham, MA, USA). Purified Med‐ORF10 was mixed with labelled DNA and then incubated at room temperature for approximately 20 min. Subsequently, the mixture was loaded onto a 6.5% polyacrylamide gel for Native‐PAGE and then followed by the band shift detection using LightShift® Chemiluminescent (Thermo, Waltham, MA, USA).
Transcriptomics analysis
Total RNA samples were isolated from 3‐day cultures CH999/pIK340 and CH999/pIK340‐Δmed10 using RNAprep pure Cell/Bacteria Kit DP430 (Tiangen, Beijing) and then sent to Novogene Technology Co., Ltd (Beijing, China) for RNA‐Seq. Sequencing libraries were generated using NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA). Complementary DNA (cDNA) library was verified using Agilent Bioanalyzer 2100 system (Agilent Technologies) and then sequenced using NovaSeq 6000 (Illumina, San Diego, CA, USA).
RNA‐Seq data were analysed using Bowtie2‐2.2.3 for mapping reads to the reference genome. Rockhopper was used for novel genes, operons and transcription start points. TDNN was employed to predict the promoters, RBSfinder and TransTermHP for predicting SD sequence and terminator sequence, IntaRNA for predicting sRNA targets, RNAfold for RNA secondary structures. Quantification of gene expression level and differential expression analysis were analysed by HTSeq v0.6.1 and DESeq R package respectively. GOseq R package was used to analyse gene enrichment, KOBAS software for the statistical enrichment of differential expression genes in KEGG pathways. Picard‐tools v1.96 and samtools v0.1.18 were employed to sort, mark duplicated reads and reorder the bam alignment results of each sample and GATK2 software performed SNP calling.
All sequences were trimmed, and forward and reverse sequenced reads were generated for each sample and then mapped against genome sequences of S. coelicolar A3(2) in NCBI sequence databases to annotate and quantify gene expression levels for each strain. The expression value was measured in RPKM (reads per kilo base per million mapped reads). Genes were considered as differentially expressed genes (DGEs) when log2(fold‐change) was above 1 or below −1 with statistical significance (P‐value < 0.05, false‐discovery rate (FDR < 0.001)). In order to find out functional relationship between DGEs in two strains, pathway enrichment analysis was applied to identify biological themes in the complex lists of DGEs. After analysis, results were visualized using volcano plot, heat map and GO enrichment histogram to analyse the pathway enrichment of differentially expressed genes between two strains.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Table S1. Primers used in this study.
Table S2. The downregulated genes in CH999/pIK340‐Δmed10 (log2(fold‐change) >1 or <−1 and P‐value < 0.05) using transcriptomics.
Table S3. The upregulated genes in CH999/pIK340‐Δmed10 (log2(fold‐change) >1 or <−1 and P‐value < 0.05) using transcriptomics.
Fig. S1. The overall FPKM hierarchical clustering diagram.
Fig. S2. The ordinate is the enriched GO term, and the abscissa is the number of differential genes in the term.
Fig. S3. The proposed operons in med gene cluster. (A) and differential expression of med genes using transcriptomics (B). Ω: terminators; arrow: promoters. Genes lower than that the dashed line indicated the differential expression. The y‐axis indicated the expression difference (P<0.05 and log2 (fold‐change) >1 or <−1) of med genes between CH999/pIK340 and CH999/pIK340‐Δmed10. The transcriptional level of different genes from CH999/pIK340 was normalized into 1.
Acknowledgements
This study was supported by the National Key R&D Program of China (No. 2018YFA0900400 and 2019YFA0905700) and the Shandong Province Natural Science Foundation (ZR2017MC031). We are thankful to National Natural Science Foundation of China (31170050), the Open Project Program of the State Key Laboratory of Bio‐based Material and Green Papermaking (KF201825) and the 111 Project (B16030) for financial support. All authors are grateful to Prof. Satoshi Ōmura for the generous gift of Streptomyces sp. AM‐7161 and thank Dr. Micheal C. Wilson (Global Scientific Communications Manager – Alzheimer's Disease at Roche) for refining the manuscript.
Microb. Biotechnol. (2021) 14(5), 1918–1930
Funding information
This study was supported by the National Key R&D Program of China (No. 2018YFA0900400 and 2019YFA0905700) and the Shandong Province Natural Science Foundation (ZR2017MC031). We are thankful to National Natural Science Foundation of China (31170050), the Open Project Program of the State Key Laboratory of Bio‐based Material and Green Papermaking (KF201825) and the 111 Project (B16030) for financial support.
References
- Abbasi, M.N., Fu, J., Bian, X., Wang, H., Zhang, Y., and Li, A. (2020) Recombineering for genetic engineering of natural product biosynthetic pathways. Trend Biotechnol 38: 715–728. [DOI] [PubMed] [Google Scholar]
- Bechthold, A., and Yan, X. (2012) SnoaW/SnoaL2: a different two‐component monooxygenase. Chem Biol 19: 549–551. [DOI] [PubMed] [Google Scholar]
- Beinker, P., Lohkamp, B., Peltonen, T., Niemi, J., Mäntsälä, P., and Schneider, G. (2006) Crystal structures of SnoaL2 and AclR: two putative hydroxylases in the biosynthesis of aromatic polyketide antibiotics. J Mol Biol 359: 728–740. [DOI] [PubMed] [Google Scholar]
- Bibb, M.J., Sherman, D.H., Omura, S., and Hopwood, D.A. (1994) Cloning, sequencing and deduced functions of a cluster of Streptomyces genes probably encoding biosynthesis of the polyketide antibiotic frenolicin. Gene 142: 31–39. [DOI] [PubMed] [Google Scholar]
- Deng, H., Cai, X., Peng, J., Hong, H., Ichinose, K., and Li, A. (2010) Practical procedures for genetic manipulation systems for medermycin‐producing Streptomyces sp. AM‐7161. J Basic Microbiol 50: 299–301. [DOI] [PubMed] [Google Scholar]
- Duan, Y., Liu, Y., Huang, T., Zou, Y.i., Huang, T., Hu, K., et al. (2018) Divergent biosynthesis of indole alkaloids FR900452 and spiro‐maremycins. Org Biomol Chem 16: 5446–5451. [DOI] [PubMed] [Google Scholar]
- Gust, B., Challis, G.L., Fowler, K., Kieser, T., and Chater, K.F. (2003) PCR‐targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100: 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Q., Li, L., Yang, T., Li, R., and Li, A. (2015) Functional characterization of a ketoreductase‐encoding gene med‐ORF12 involved in the formation of a stereospecific pyran ring during the biosynthesis of an antitumor antibiotic medermycin. PLoS One 10: e0132431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose, K., Bedford, D.J., Tornus, D., Bechthold, A., Bibb, M.J., Peter Revill, W., et al. (1998) The granaticin biosynthetic gene cluster of Streptomyces violaceoruber Tü22: sequence analysis and expression in a heterologous host. Chem Biol 5: 647–659. [DOI] [PubMed] [Google Scholar]
- Ichinose, K., Ozawa, M., Itou, K., Kunieda, K., and Ebizuka, Y. (2003) Cloning, sequencing, and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM‐7161 towards comparative analysis of the benzoisochromanequionone gene clusters. Microbiology 149: 1633–1645. [DOI] [PubMed] [Google Scholar]
- Jiang, Y.J., Zhang, D.S., Zhang, H.J., Li, J.Q., Ding, W.J., Xu, C.D., et al. (2018) Medermycin‐type naphthoquinones from the marine‐derived Streptomyces sp. XMA39. J Nat Prod 81: 2120–2124. [DOI] [PubMed] [Google Scholar]
- Jones, S.E., and Elliot, M.A. (2018) 'Exploring' the regulation of Streptomyces growth and development. Curr Opin Microbiol 42: 25–30. [DOI] [PubMed] [Google Scholar]
- Kallio, P., Sultana, A., Niemi, J., Mäntsälä, P., and Schneider, G. (2006) Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity. J Mol Biol 357: 210–220. [DOI] [PubMed] [Google Scholar]
- Kieser, T., Bibb, M.J., Butter, M.J., Chater, K.F., and Hopwood, D.A. (2000) Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation. [Google Scholar]
- Li, A., Itoh, T., Taguchi, T., Xiang, T., Ebizuka, Y., and Ichinose, K. (2005) Functional studies on a ketoreducatse gene from Streptomyces sp. AM‐7161 to control the stereochemistry in medermycin biosynthesis. Bioorg Med Chem 13: 6856–6863. [DOI] [PubMed] [Google Scholar]
- Liu, R., Deng, Z., and Liu, T. (2018) Streptomyces species: Ideal chassis for natural product discovery and overproduction. Metab Eng 50: 74–84. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Liu, S., Yang, T., Guo, X., Jiang, Y., Zahid, K.R., et al. (2015) Expression, crystallization and preliminary X‐ray diffraction analyses of Med‐ORF10 in the biosynthetic pathway of an antitumor antibiotic medermycin. Protein J 34: 404–410. [DOI] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lv, J., He, Q., Huang, L., Cai, X., Guo, W., He, J., et al. (2015) Accumulation of a bioactive benzoisochromanequinone compound kalafungin by a wild type antitumor‐medermycin‐producing streptomycete strain. PLoS One 10: e0117690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel, R., Ebert‐Khosla, S., Hopwood, D.A., and Khosla, C. (1993) Engineered biosynthesis of novel polyketides. Science 262: 1546–1550. [DOI] [PubMed] [Google Scholar]
- Oja, T., Palmu, K., Lehmussola, H., Leppäranta, O., Hännikäinen, K., Niemi, J., et al. (2008) Characterization of the alnumycin gene cluster reveals unusual gene products for pyran ring formation and dioxan biosynthesis. Chem Biol 15: 1046–1057. [DOI] [PubMed] [Google Scholar]
- Ozawa, M., Taguchi, T., Itoh, T., Ebizuka, Y., Booker‐Milburn, K.I., Stephenson, G.R., and Ichinose, K. (2003) Structure and biosynthetic implication of (S)‐NHAB, a novel shunt product, from a disruptant of the actVI‐ORFA gene for actinorhodin biosynthesis in Streptomyces coelicolor A3(2). Tetrahedron 59: 8793–8798. [Google Scholar]
- Salaski, E.J., Krishnamurthy, G., Ding, W.D., Zu, K., Insaf, S.S., Eid, C., et al. (2009) Pyranonaphthoquinone lactones: a new class of AKT selective kinase inhibitors alkylates a regulatory loop cysteine. J Med Chem 52: 2181–2184. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russel, D. (2001) Molecular Cloning: A Laboratory Manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Siitonen, V., Blauenburg, B., Kallio, P., Mäntsälä, P., and Metsä‐Ketelä, M. (2012) Discovery of a two‐component monooxygenase SnoaW/SnoaL2 involved in nogalamycin biosynthesis. Chem Biol 19: 638–646. [DOI] [PubMed] [Google Scholar]
- Sone, Y., Nakamura, S., Sasaki, M., Hasebe, F., Kim, S.Y., and Funa, N. (2018) Bacterial enzymes catalyzing the synthesis of 1,8‐dihydroxynaphthalene, a key precursor of dihydroxynaphthalene melanin, from Sorangium cellulosum . Appl Environ Microbiol 84: e00258‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana, A., Kallio, P., Jansson, A., Wang, J.‐S., Niemi, J., Mäntsälä, P., and Schneider, G. (2004) Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation. EMBO J 23: 1911–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, R., Liu, M., Gong, C., Wang, W., Zeng, A., and Li, A. (2012) Expression detection of med‐ORF12 encoding a stereochemical ketoreductase possibly involved in medermycin biosynthesis. Acta Microbiol Sinica 52: 60–68. [PubMed] [Google Scholar]
- Taguchi, T., Okamoto, S., Lezhava, A., Li, A., Ochi, K., Ebizuka, Y., and Ichinose, K. (2007) Possible involvement of ActVI‐ORFA in transcriptional regulation of actVI tailoring‐step genes for actinorhodin biosynthesis. FEMS Microbiol Lett 269: 234–239. [DOI] [PubMed] [Google Scholar]
- Toral‐Barza, L., Zhang, W.‐G., Huang, X., McDonald, L.A., Salaski, E.J., Barbieri, L.R., et al. (2007) Discovery of lactoquinomycin and related pyranonaphthoquinones as potent and allosteric inhibitors of AKT/PKB: mechanistic involvement of AKT catalytic activation loop cysteines. Mol Cancer Ther 6: 3028–3038. [DOI] [PubMed] [Google Scholar]
- Vuksanovic, N., Zhu, X., Serrano, D.A., Siitonen, V., Metsä‐Ketelä, M., Melançon, C.E., and Silvaggi, N.R. (2020) Structural characterization of three noncanonical NTF2‐like superfamily proteins: implications for polyketide biosynthesis. Acta Crystallogr F Struct Biol Commun 76: 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., Du, C., Ichinose, K., Choi, Y.H., and van Wezel, G.P. (2017) Discovery of C‐glycosyl pyranonaphthoquinones in Streptomyces sp. MBT76 by a combined NMR‐based metabolomics and bioinformatics workflow. J Nat Prod 80: 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, H., Li, X., Li, Z., Zhan, X., Mao, X., and Li, Y. (2020) The application of regulatory cascades in Streptomyces: yield enhancement and metabolite mining. Front Microbiol 11: 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, N., Motoyama, T., Nakasako, M., Kagabu, S., Kudo, T., and Yamaguchi, I. (2004) Enzymatic characterization of scytalone dehydratase Val75Met variant found in melanin biosynthesis dehydratase inhibitor (MBI‐D) resistant strains of the rice blast fungus. Biosci Biotechnol Biochem 68: 615–621. [DOI] [PubMed] [Google Scholar]
- Zhou, B., Jiang, Y.J., Ji, Y.Y., Zhang, H.J., and Shen, L. (2019) Lactoquinomycin C and D, two new medermycin derivatives from the marine‐derived Streptomyces sp. SS17A. Nat Prod Res 26: 1–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used in this study.
Table S2. The downregulated genes in CH999/pIK340‐Δmed10 (log2(fold‐change) >1 or <−1 and P‐value < 0.05) using transcriptomics.
Table S3. The upregulated genes in CH999/pIK340‐Δmed10 (log2(fold‐change) >1 or <−1 and P‐value < 0.05) using transcriptomics.
Fig. S1. The overall FPKM hierarchical clustering diagram.
Fig. S2. The ordinate is the enriched GO term, and the abscissa is the number of differential genes in the term.
Fig. S3. The proposed operons in med gene cluster. (A) and differential expression of med genes using transcriptomics (B). Ω: terminators; arrow: promoters. Genes lower than that the dashed line indicated the differential expression. The y‐axis indicated the expression difference (P<0.05 and log2 (fold‐change) >1 or <−1) of med genes between CH999/pIK340 and CH999/pIK340‐Δmed10. The transcriptional level of different genes from CH999/pIK340 was normalized into 1.


