Abstract
Coronavirus disease is communicable and inhibits the infected person’s immune system. It belongs to the Coronaviridae family and has affected 213 nations and territories so far. Many kinds of studies are being carried out to filter advice and provide oversight to monitor this outbreak. A comparative and brief review was carried out in this paper on research concerning the early identification of symptoms, estimation of the end of the pandemic, and examination of user-generated conversations. Chest X-ray images, abdominal computed tomography scan, tweets shared on social media are several of the datasets used by researchers. Using machine learning and deep learning methods such as K-means clustering, Random Forest, Convolutional Neural Network, Long Short-Term Memory, Auto-Encoder, and Regression approaches, the above-mentioned datasets are processed. The studies on COVID-19 with machine learning and deep learning models with their results and limitations are outlined in this article. The challenges with open future research directions are discussed at the end.
Introduction
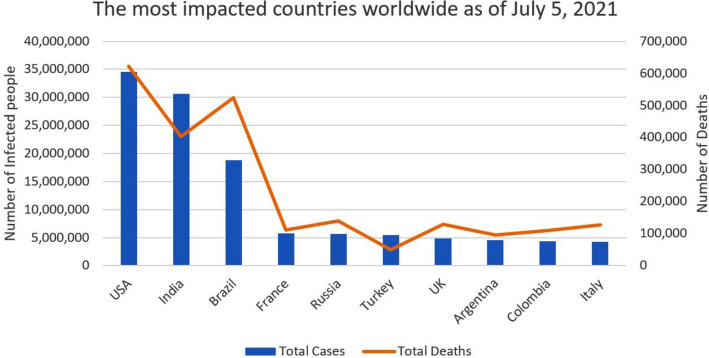
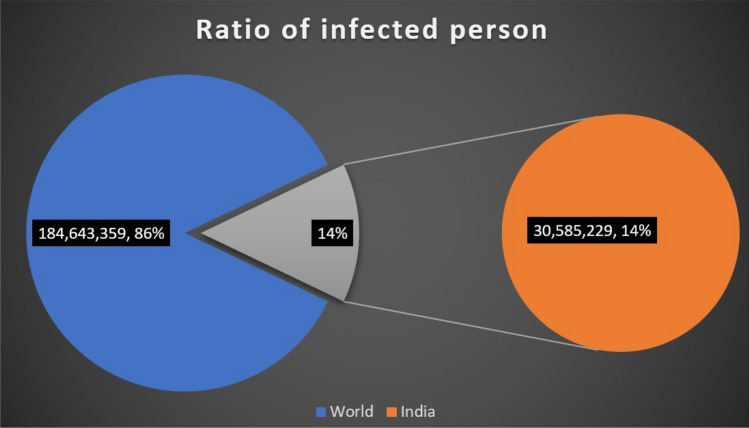
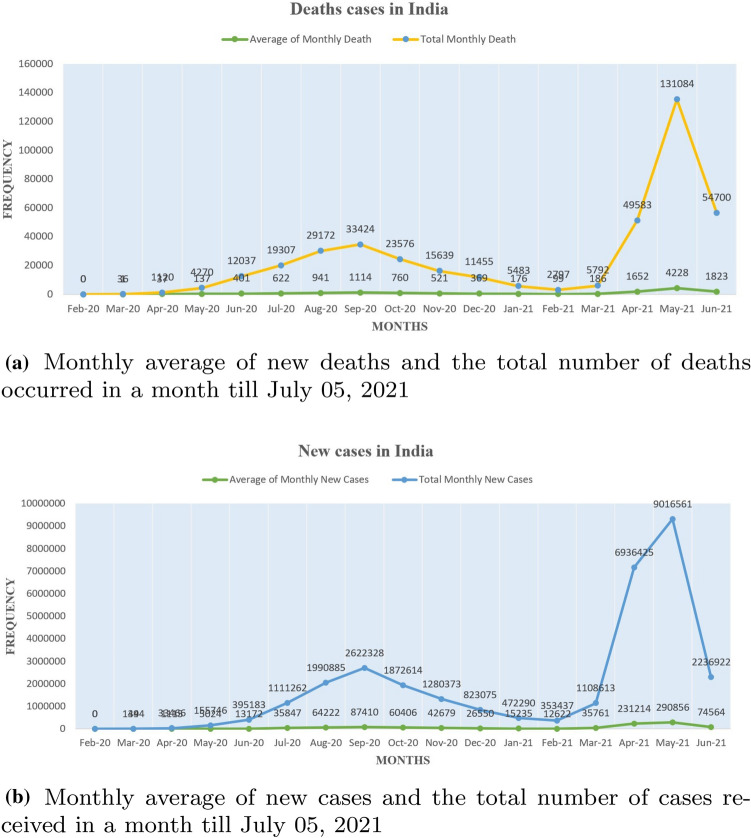
In the fight against the COVID-19 pandemic, humanity’s Achilles’ heel has been the inability to evaluate at scale. The coronavirus pandemic has spread quickly, infecting millions of people in many countries and bringing economic activity to a halt as countries placed stringent travel restrictions to halt the virus’s spread. The upsurge in the spreading of covid-19 has resulted in nearly 184 million cases worldwide until July 20211. The virus has been named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by the International Committee on Virus Taxonomy (ICTV), which is spreading coronavirus infection. As shown in Fig. 1, presently, the United States of America is the worst-hit country with a total number of cases reaching 34 million, while India is the second worst-hit country with a total of 30 million cases. Fig. 2 depicts the total number of India’s cases with 30,585,229 counts till 5th July 2021. Fig. 3 shows India as the most affected country by the exponential growth of death cases due to Coronavirus. This pandemic has left many people and families in torment. Fig. 3 (a) shows the average death count in India, and Fig. 3 (b) an average number of cases till 5th July 2021.
Fig. 1.
Top ten impacted countries in the world as of July 05,2021
Fig. 2.
Status of covid-19 in India with respect to the world till July 05, 2021
Fig. 3.
The number of new COVID -19 cases and deaths in India
The crisis emphasizes the need for immediate action to mitigate the pandemic’s health and economic effects, protect vulnerable communities, and lay the groundwork for a long-term recovery [1, 2]. Different people have different symptoms due to this virus. Various guidelines have been issued by World Health Organization (WHO). For example, compulsory protective masks, lockdown in the country, frequently washing hands, maintaining a minimum distance of two feet, and Personal Protective Equipment (PPE) for doctors and nursing staff to tackle this virus.
According to the guidelines [3], covid-19 is primarily transmitted human-to-human through oral and respiratory aerosols and droplets ejected when an infected person coughs. Other sources include touching an infected object or surface. Signs and symptoms of covid-19 may appear 2 to 14 days after exposure to the virus. Some of the symptoms are respiratory problems, sore throat, cough, and fever. The spread of coronavirus can be controlled by early detection of these symptoms so that they can be treated on time. Many researchers have conducted different experiments that supported the clinical tests in diagnosing the disease at a very early stage [4]. Various approaches such as Big data, Artificial Intelligence, Deep Learning, and Machine learning are being applied to a colossal amount of data related to covid-19 [5–7].
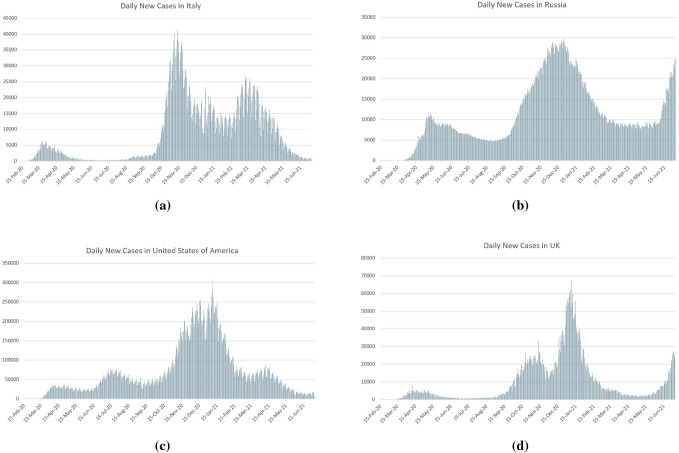
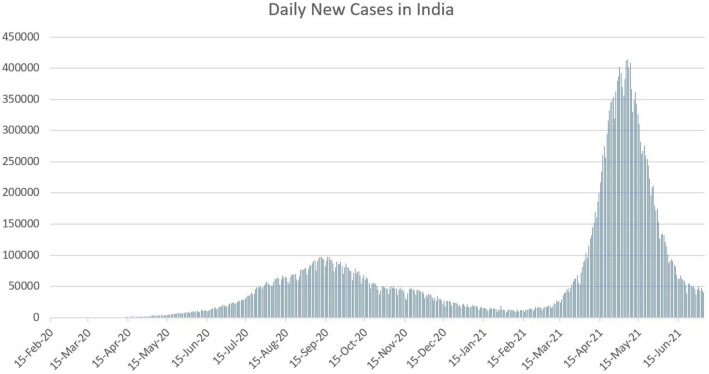
Coronavirus has the propensity to infect people in multiple waves. Some countries which initially successfully curbed the spread of the covid-19 outbreak are now reporting a new resurgence of cases. Fig. 4 shows the countries like Italy, Russia, the USA, and the UK are facing multiple waves of covid-19 outbreak. Italy and Russia have faced much stronger second and third waves as can be seen from Fig. 4 (a) and (b). Russia is still in its peak of the third wave. Fig. 4 (c) shows that US is just coming out of its fourth wave of the pandemic right now, and Fig. 4 (d) shows that the UK is in its fourth wave’s peak. The subsequent multiple waves in the US Fig. 4 (c) and Russia Fig. 4 (b) came instantly after the daily new cases from the previous wave stopped dropping. In Italy Fig. 4 (a) and the UK Fig. 4 (d), a longer gap was observed between the first and the second wave of the outbreak. From Fig. 5, we can see that India has faced one of the deadliest second waves in the world. The trajectory of the graph of the daily new cases in India (initially rose to a peak during the second wave, then decreased rapidly) is similar to the other countries that have seen subsequent pandemic waves. This may signal the start of a new wave for India.
Fig. 4.
Multiple waves of covid-19 outbreak in different countries
Fig. 5.
Covid-19 waves in India by July 05, 2021
This paper comprises key research done using deep learning, machine learning, and other approaches to provide solutions to tackle this situation. Researchers have classified between infected and healthy people by processing images of chest X-ray or abdominal computed tomography (CT) images gathered from various hospitals, universities GitHub, and Kaggle [8–11]. Researches were efficacious in using AI technology to develop on-device mobile applications that can track a person’s travel history and give them a self-assessment test. This would help people analyze their health condition all by themselves and the impact of coronavirus on their body based on the guidelines by the World Health Organisation [3]. Few works anticipated the count of positive coronavirus cases at the end of this outbreak in various countries utilizing the covid-19 data [12–15]. In order to do this, they have used people’s sentiment collected through user-generated comments on social media sites [16]. Editorials mentioning preventive measures that a nation should incorporate and various challenges that people face because of this outbreak are also listed. They advised to compare previous pandemics such as SARS and studying them, and adopt different solutions to tackle this pandemic.
Affluent countries should help the impoverished countries to improve their medical facilities. All these measures and research will reinforce the medical teams and doctors standing as a front-line warrior in this situation and the country in finding efficient prevention methods and stabilizing economic growth. Through these measures, the governments could get additional time in creating policies and guidelines helping both people and medical teams of the nation. The research works that were developed using Artificial Intelligence, Machine learning, and deep learning are summarized in this article. This study also includes the works that provided advantageous information for safety measures and other essential pandemic parameters. This study helps future researchers to identify the shortcomings in the current research and give them details on existing datasets.
Motivation
Since the initial report of Coronavirus Disease (covid-19) in Wuhan, China, in December 2019, it has spread to almost every nation and territory worldwide, resulting in 184 million illnesses and around 4 million fatalities as of July 05, 2021. With the rising issue, firms and researchers all around the globe are seeking methods to address the virus’s difficulties, reduce its spread, and discover a treatment. Science and technology are crucial in this perplexing conflict. For example, when China first started responding to the virus, it relied on facial recognition technology to track infected patients, foods and medicines were delivered to infected patients by robots, drones were used to disinfect public places. On the road to finding a cure for covid-19, AI has been heavily employed to uncover novel compounds.
Many researchers utilize artificial intelligence to discover novel medications and therapies for cures, with computer scientists concentrating on detecting infected individuals using medical image processing such as X-rays and CT scans. AI is being used extensively to discover, track, and anticipate outbreaks and assist in viral diagnosis. To the best of our knowledge, there is no current review paper available that covers the present research on the application of both machine learning and deep learning for covid-19 analysis. Shoeibi et al. [17] published a review article focused solely on deep learning models for covid-19 detection and forecasting. A few other surveys have recently been published, but they didn’t cover many insights into the use of deep learning and how it can be used for both detection and forecasting of covid-19. This inspires us to construct an up-to-date, state-of-the-art study, which aims to examine and cover all the aspects of machine learning and deep learning in the context of the present pandemic.
Contributions
The following are the major contributions:
Gathered articles from various digital libraries and categorized them into Deep Learning and Machine Learning.
To provide the evaluated works and important information in a clear, simple, and accessible manner by taking into account several essential factors such as the data utilized in experiments, the data splitting technique, the suggested architecture for diagnosis, and performance assessment metrics.
Using CT and X-Ray medical imaging samples, thoroughly examine the state-of-the-art advances of deep learning-based covid-19 diagnostic systems.
To present a taxonomy of the examined literature in order to have a better understanding of the advances.
To highlight and debate the difficult elements of existing covid-19 diagnostic systems that are based on deep learning.
To propose future research prospects for the development of covid-19 detection systems that are both efficient and dependable.
The rest of the papers are structured as follows: Section 2 introduces the article selection process and discusses CNN and LSTM model. Section 3 addresses the findings using deep learning models. Section 4 addresses work on models focused on machine learning. Section 5 discusses the open issues and future research directions. Finally, Section 6 concludes this work.
Methods
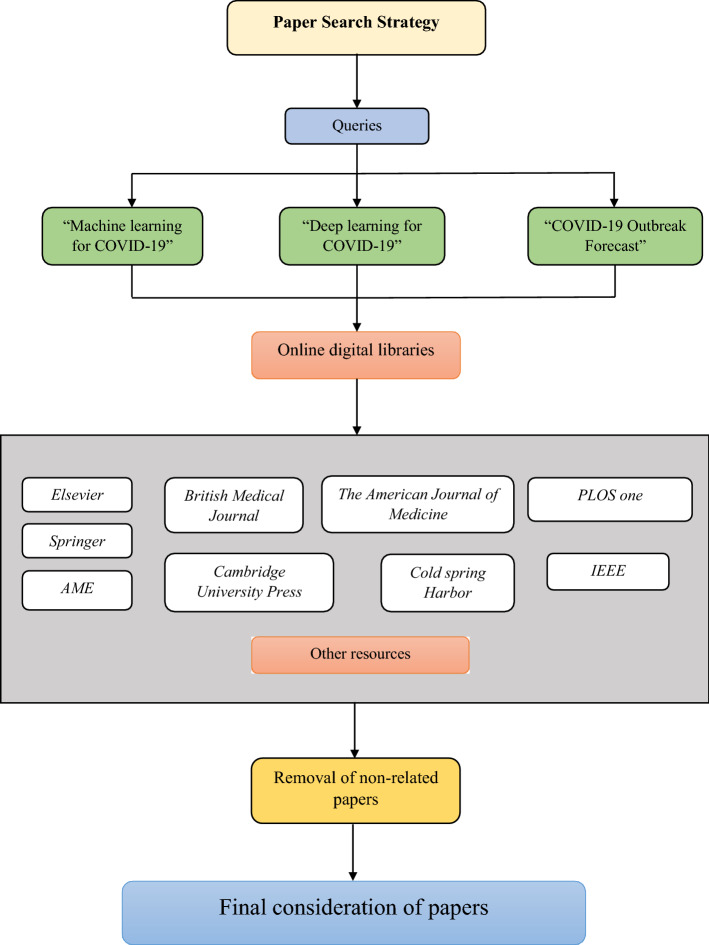
This section highlights the process of collecting the relevant research articles. Since the problem started, much research was published to address the covid-19 issues by worldwide researchers. But, after analyzing the articles, it has been found that there were a large number of articles which are only sharing the prevention mechanism, personal thoughts, and similar ones. Hence, it is a brainstorming task to find the articles from the pools which uses machine learning or deep learning-based framework to address the covid-19 issues. The steps followed to filter out the relevant articles are shown in Fig. 6.
Fig. 6.
Steps to extract the relevant articles
Models used for Predictions
The researchers used many models to address the covid-19 issues. The popular machine learning models include- Support Vector Machine (SVM), Random Forest (RF), K-nearest neighbor (KNN), Gradient Boosting (GB), Logistic Regression (LR), Naive Bayes (NB), and others. The deep learning models such as Convolutional Neural Networks (CNN), Long-Short Term Memory (LSTM), Recurrent Neural Network (RNN), Bidirectional-LSTM, and the hybrid of CNN-LSTM-RNN models are the most preferred deep learning models. Especially, the CNN model yielded promising prediction accuracies with X-ray or CT-scan images dataset. The majority of researchers applied CNN models on various image datasets to address the covid-19 prediction or forecasting purposes.
The CNN model is helpful to extract the hidden contextual features from images with the help of various kernel sizes and hidden units. Moreover, the pooling layers and the dense layer at the end helps to get better accuracies. On the other hand, the LSTM and Bi-LSTM models have achieved good accuracies with the sequential dataset. The working of these models can be studied from the existing literature [18–21].
Convolutional Neural Network
One of the popular deep learning models for covid-19 predictions is Convolutional Neural Network (CNN). The working of the CNN model is discussed by [18]. The CNN model mainly works in three phases: (i) Embedding layer, (ii) Features extractions using kernels, and (iii) Pooling essential features.
The input sample is preprocessed and passed to either a pre-trained embedding or directly fetch to the embedding layer to create the matrix. The different n-gram filters are used to convolve over the matrix formed with the help of embedding. By convolutional operations, the essential contextual features are extracted from the input samples. However, all extracted features may not be equally important; hence, only essential features are pooled out using the pooling layers to reduce the processing overhead. Further, the pooled features are concatenated and passed to a fully connected dense layer to process it. The CNN model uses loss functions such as Binary cross-entropy or categorical cross-entropy to monitor the training losses, whereas tanh and Adam are the popularly used optimizer. The metrics such as accuracy, precision, recall, F1-score, root mean square value evaluate the trained model performance [22].
Long-Short Term Memory
The LSTM model is the most preferred model for predictions with sequential data. The sequential data means the data consisting of the information of previous timestamps as well with the current one [21]. Meaning if the prediction depends on the past knowledge along with the current information, the LSTM model is the preferred network for predictions. For example, to predict the stock market price of a particular stock, the current stock price is not helpful; instead, the last few months price is also needed. Many researchers utilize the LSTM models to forecast the covid-19 disease in various countries and to predict whether a person is infected or not. The summary of the researches using the deep learning-based frameworks are listed in Table 1. The research works on covid-19 using machine learning and deep learning frameworks are separately discussed in consecutive sections.
Table 1.
Research findings and limitations using Deep Learning frameworks
| Objective | Dataset | Time Frame | Source of Dataset | Methods | Model performance | Limitation |
|---|---|---|---|---|---|---|
| To forecast of the future of covid-19 cases [26] | Total 346 people from five countries | January 20, 2020 to April 4, 2020 | Not available | i) ARIIMA and wavelet-based technique ii) Regression Tree | Predictive model | – |
| To Predict the trends and stopping time of covid-19 outbreak [27] | Not available | Till march 31,2020 | Two universities | LSTM networks | End by June 2020 | – |
| Predicting trend of covid-19 in China [5] | Current Covid-19 and 2003 SARS epidemic data | Not available | National Health Commision of China | SEIR and LSTM | Peak of 4,000 daily infections between February 4 to 7, 2020 in China | Limited number of factors are considered |
| Pandemic prediction [29] | Confirmed covid-19 cases | Not available | (i) Johns Hopkins University, (ii) WHO, (iii) Ding Xiang Yuana | SEIR | Reach peak in late May 2020 | – |
| Forecasting of covid-19 in China [31] | Confirmed covid-19 cases | (i) News Networks (ii) WHO | Modified stacked auto-encoder | Predictive model | – | |
| Detection of covid-19 cases using chest X-ray images [11] | 13,975 CXR images | Not available | COVIDx dataset | COVID-Net | Predictive model | Not a production ready solution |
| To determine the uncertainity and interpretability[6] | 5,941 PA chest radiography images | Not available | (i) Dr. Joseph Cohen’s Github repository and (ii) Kaggle’s Chest X-Ray Images | Bayesian Deep Learning | Predictive model | Study is only limited to estimating uncertainty in already developed models |
| Detection of coronavirus disease using X-ray images [32] | 100 chest X-ray images | Not available | GitHub | Convolutional neural networks | Accuracy : 98% | Small dataset |
| covid-19 outbreak prediction in India [13] | Covid-19 cases in India | January 30th 2020 to March 30th 2020 | Johns Hopkins University | SEIR and Regression Model | RMSLE : 1.75 | Limited data |
| Prediction of country wise risk of covid-19 [33] | Confirmed covid-19 cases | January 22 2020 to March 10 2020 | Not available | LSTM networks | Accuracy: 78% | Model achieved less accuracy |
| Forecasting of covid-19 pandemic [82] | Not available | January 21, 2020 to April 02, 2020 | Sourceb | Machine Learning | 97% confidence interval | Small Dataset was taken for Senegal |
| Screening for covid-19 disease using CT images [1] | 453 CT images | Not available | Not available | Deep Learning | AUC: 0.90 | Training dataset is small |
| Detection for covid-19 from chest CT using weak label [3] | 630 CT images | December 13, 2019 to February 6, 2020 | Not available | DeCoVNet | AUC: 0.959 | Dataset from single hospital |
| Automatic detection of covid-19 from X-ray images [34] | 2,870 X-ray images | Not available | Public medical repositories | Deep Learning, Transfer Learning | Accuracy : 96.78% | Small sample of positive cases |
| Prediction of the pandemic trend of covid-19 in Italy [35] | Daily reports of covid-19 | Jan 22,2020 to Apr 02,2020 | Johns Hopkins University | Extended susceptible-infected-removed | 95% CI | Asymptomatic and unconfirmed cases may be ignored. |
| Identification of covid-19 through mobile phone based survey [12] | Not available | Not available | Not available | AI algorithms | Difficult to collect data that is used for this model | |
| Detection of covid-19 using Artificial Intelligence [36] | 260 chest X-ray images | Not available | (i) GitHub, and (ii) Kaggle | Convolutional neural network | Accuracy: 100% | Small dataset was taken to validate model |
| Predicting outbreak trend of coronavirus disease in India [37] | China’s covid-19 cases | Jan 22, 2020 to April 3, 2020 | Kaggle | Machine Learning | Forecasting prediction for India | Limited to Indian context |
| Predicting the trends in covid-19 outbreak in Iran [38] | Iran’s covid-19 data | Feb 15, 2020 to Mar 18, 2020 | (i) WorldOmeter website, and (ii) Google trends | LSTM and Linear Regression | RMSE : 7.562 | Limited Google Search data |
| Analysis of confirmed cases using AI [83] | Not available | Not available | Binary Classification and Regression model | Accuracy : 95.7% | Study only takes weather parameters in consideration | |
| AI model to distinguish covid-19 cases [39] | 4,356 3D chest CT images | Not available | Six medical centers | Transfer Learning | AUC : 0.96 | Train and Test from same dataset. |
| covid-19 case detection [40] | 127 X-ray | Not available | ChestX-ray8 database | Deep Learning | Accuracy : 98.09% | Limited number of covid-19 X-ray images. |
| AI System for covid-19 [43] | 6,752 CT scans images | Not available | CC-CCII | Deep Learning | Accuracy : 92.49% | |
| Diagnosis of coronavirus disease from X-ray images [7] | 5,949 posteroanterior chest radiography images | Not available | COVIDx | COVIDiagnosis-Net | Accuracy : 98.3% | Model require Fine-tuned hyperparameters for accurate predictions |
| Accurate model for covid-19 prediction [44] | 605 real-world data | Not available | Not available | Deep Learning | Accuracy : 94.5% | Limited dataset |
| Time dependent SIR model for covid-19 [45] | Not available | Jan 15,2020 to Mar 2,2020 | NHC | SIR model | Day-wise prediction | Limited domain |
| Lung infection quantification [46] | 549 CT images | Not available | Not available | Deep Learning | Similarity coefficients : 91.6% | Validation was conducted on same dataset |
| Large scale screening method [47] | 2,685 CT images | Not available | Three medical source from China | Random Forest | Accuracy :87.9% | only baseline CT findings of covid-19 patients were included in study |
| To diagnose COIVD-19 patient [48] | 50 chest X-ray images | Not available | Not available | Transfer Learning | F1-score : 0.89 | Small dataset |
| Pneumonia screening [49] | 43,583 chest CT images | Not available | (i) X-VIRAL, and (ii) X-COVID | Deep Learning | AUC : 0.836 | High false negative rate |
| Classification of covid-19 cases [50] | 196 samples of CXRs | Not available | Japanese Society of Radiological Technology | Deep Learning | Accuracy : 95.12% | Limted training data. |
| Diagnosis of covid-19 [51] | 349 covid-19 CT images | Not available | COVID-CT-Dataset | Multi-task learning | F1-score : 0.90 | Limited number of CT images |
| Classification model [52] | 5,856 CT images | Not available | Not available | Transfer Learning | Accuracy :96% | |
| covid-19 diagnosis method using X-ray [54] | 170 X-ray images and 361 CT images | Not available | Multiple Sources | Deep Learning | Accuracy : 98% | Small dataset. |
| Detection of covid-19 cases [56] | 5,863 X-ray images | Not available | Not available | Transfer Learning | Accuracy : 99% | Used only 624 images |
| Identification of abnormal CT patterns [57] | 9,749 chest CT images | Not available | Not available | Deep Learning | Pearson correlation coefficient : 0.92 | Model was trained only with specific abnormalities data |
| Identification of covid-19 using chest X-ray [58] | 455 chest X-ray images | Not available | Multiple Sources | Transfer Learning | Accuracy : 91.24% | Small number of cases are considered |
| covid-19 patient detection using CT images [59] | 109 confirmed cases from China | Not available | Not available | Transfer Learning | AUC : 0.948 | Small dataset |
| Identification of covid-19 cases from X-ray images [60] | 94,323 frontal view chest X-ray images | Not available | NIH CXR dataset | Deep Learning | Accuracy : 95.7% | Pre-training is required for the model to achieve high accuracy |
| Estimation of global covid-19 spread [62] | Not available | Not available | Chinese National Health Commission | Neural network | Analysis based model | |
| To evaluate pneumonia cases during the covid-19 [2] | 5,863 chest X-ray images | Not available | Not available | Transfer Learning | False negatives : 0.7% | low number of covid-19 chest X-ray images in dataset |
| covid-19 diagnosis based on chest X-ray [63] | 15,959 CXR images | Not available | Multiple Sources | Transfer Learning | F1-score : 0.945 | Limited amount of CXR images |
| covid-19 patterns detection through X-ray images [64] | 13,800 X-ray images | Not available | Multiple Sources | Deep artificial neural networks | Accuracy : 93.9% | Less heterogeneous dataset was used |
| covid-19 infection detection [65] | 60 3D CT lung scans | Not available | TCIA dataset | Convolutional neural network | Accuracy : 96.2% | Small dataset |
| To develop covid-19 diagnosis system [66] | 144,167 CT images | Not available | COVID-CS | Transfer Learning | Sensitivity : 95.0% | – |
| covid-19 Detection using CXRs [10] | Not available | Not available | Multiple Sources | Transfer Learning | Accuracy : 99.01% | Limited number of covid-19 pneumonia CXR data |
| To predict covid-19 from chest X-ray images [14] | 5,071 chest X-ray images | Not available | (i) Chestxray-Dataset, and (ii) ChexPert dataset | Transfer Learning | Specificity : 97.8% | Dataset contained less than 100 covid-19 X-ray images |
| covid-19 detection [68] | 100 axial CT images | Not available | covid-19 CT segmentation dataset | Deep Learning | Analysis based model | Small dataset |
| Diagnosis of covid-19 with chest X-ray images | Transfer Learning [70] | 1,076 posteroanterior CXR (PCXR) images | Not available | i) covid-19 Image, and ii) RSNA dataset | Accuracy : 96% | – |
| Estimating covid-19 trend in Spain [71] | Not available | Not available | Sourcec | Bayesian Poison-Gamma model | Predictive model | – |
| covid-19 pneumonia severity predicting model [72] | 94 posteroanterior CXR images | Not available | Multiple Sources | DenseNet model | MAE: 0.78 | Small number of samples |
| Diagnosis of coronavirus using CT images [74] | 88 chest CT scans images | Not available | Not available | Details Relation Extraction neural network | AUC : 0.99 | Small dataset |
| AI system for covid-19 diagnosis [75] | 10,250 CT images | Not available | Multiple Sources | Deep Learning based AI system | AUC : 0.971 | Some limitations arises when unbalanced dataset is used |
| covid-19 diagnosis based on CT scans [76] | 349 CT scan images | Jan 19th to Mar 25th | (i) medRxiv, and (ii) bioRxiv | Transfer Learning | F1-score : 0.85 | Not robust |
| CT imaging classification and segmentation for covid-19 [77] | 1,044 CT images | Not available | (i) COVID-CT data, and (ii) Sourced | MTL architecture | AUC : 0.93 | Limited covid-19 images |
| covid-19 lung infection segmentation model using CT images [78] | 100 axial CT images | Not available | CT Segmentation dataset | Transfer Learning | Sensitivity : 0.870 | Limited dataset |
| covid-19 diagnostic and prognostic analysis [79] | 5,372 CT images | Not available | Not available | Deep Learning | AUC : 0.90 | Many factors are not considered |
| CT images feature analysis to screen covid-19 [80] | 51 CT images | Not available | Kaggle database | Naive Bayes | Accuracy : 96.07% | Small dataset |
| covid-19 predicitons using X-ray Images [81] | 3,905 X-ray images | Not available | Not available | Convolutional neural network | Accuracy : 99.18% | Used limited dataset |
aChinese government authorized website
bhttps://www.tableau.com/covid-19-coronavirus-data-resources
chttps://covid19.isciii.es/resources/serie_historica_acumulados.csv
Metrics
Researchers uses many popular metrics for the model evaluations including- precision, recall, F1-score, accuracy, auc-roc curve [19, 23]. These metrics can be defined as follows:
Precision is defined as the number of samples truly predicted among the retrieved samples for a particular class. If the dataset contains multiple classes, then for each class, a separate precision value is predicted by the model.
Recall is defined as the number of samples truly predicted among the total samples for a particular class. If the dataset contains multiple classes, then a separate recall value is predicted by the model for each class.
F1-Score: the F1-score is the harmonic mean of precision and recall. The F1-score is also different as like precision and recall for different classes present in the dataset.
Accuracy: the samples of the different classes truly predicted among the total sample is termed as accuracy. If a dataset consists of five classes, then accuracy is the number of correctly predicted samples from each class to the total number of samples.
AUC-ROC: the true positive rate and the false positive rate is used to plot the area under the receiver operative curve. The ROC value nearer to 1 is an indicator of a good predictive model.
Research Contributions with Deep Learning
This section discusses the works that were reported by the researcher using the various deep learning models such as, LSTM network [23, 24], GRU [12], CNN [13, 22, 25] and numerous others. The summary of the reported works with their findings and limitation are presented in Table 1.
Chakraborty and Ghosh [26] focused on two problems i) generating short term forecasts and ii) risk assessment of the novel coronavirus. Following are the analysis they did for the first problem i.e., to forecast confirmed cases of covid-19: The dataset contained data from five different countries i) 64 people under observation from India, ii) 65 people under observations from the UK, iii) 70 people under observations for Canada, iv) 71 people under observations from France and v) 76 people under from south Korea. The data was collected from the mentioned countries between January to April 2020. They used hybrid time series forecasting approaches combining ARIIMA and wavelet-based forecasting approach. For risk evaluation of covid-19 cases: they computed the case fatality ratio (CFR) estimates for 50 countries from the known date of starting this coronavirus outbreak until 4 April 2020. The CFR modeling dataset contained 50 observations. They used the Regression Tree (RT) model for processing the CFR dataset for risk assessment. The experimental results suggested four control variables that will significantly impact controlling CFR, i) the number of cases ii) 65 years or more age group people iii) lockdown period, and iv) hospital per 1000 people.
Chimmula and Zhang [27] proposed a deep learning-based model to predict the coronavirus outbreak trend and its possible stopping time. They collected the dataset from Johns Hopkins University and the Canadian Health authority, which included the number of confirmed cases, number of fatalities, and number of recovered patients until March 31, 2020. They utilized long short-term memory (LSTM) networks to predict the future of covid-19. Based on LSTM networks results, the model anticipated that the coronavirus outbreak’s possible ending point would be around June 2020. Tomar and Gupta [28] proposed a data-driven model on the prediction for the spread of covid-19 in India. They utilized data that was collected from 30th January 2020 to 4th April 2020. They also used LSTM and curve fitting method to estimate the number of recovered cases, daily positive cases, deceased cases for the next 30 days in India.
Yang et al. [5] suggested a modified prediction approach for finding the trend of covid-19 in China. The study was done for three provinces: (i) Hubei, (ii) Guangdong, and (iii) Zhejiang. They created a dataset by collecting the data based on daily covid-19 outbreak numbers reported by China’s National Health Commission (NHC). They also collected 2003 SARS epidemic data between April and June 2003 across China from an archived new website (SOHU). They used two models: (i) dynamic SEIR and (ii) LSTM. The SEIR and LSTM model anticipated a peak of 4,000 daily coronavirus infections between February 4 to 7, 2020, with the SEIR model anticipating various smaller peaks of new infections in the middle to late February. Hamzah et al. [29] proposed world-wide covid-19 pandemic data analysis and prediction using SEIR modeling. They collected data of confirmed covid-19 cases from Johns Hopkins University, WHO and DingXiangYuan (Chinese government authorized website). They used SEIR for forecasting the trajectory of the outbreak, 240 days starting from 20th January 2020. The model predicted that the outbreak is assumed to reach its peak in late May 2020 and will start to drop around early July 2020.
Panwar et al. [30] developed a rapid Covid-19 screeing tool name nCOVnet using CXRs. A total of 337 chest X-ray images were included in the dataset, with 192 of them being covid-19 positive individuals. They used the deep learning-based CNN model. In less than five seconds, the proposed model could detect a covid-19 positive patient with a claimed 97.62% accuracy. Hu et al. [31] used a model that relied primarily on Artificial Intelligence for real-time prediction of covid-19 to measure the size, duration, and end time of the outbreak in China. The authors collected data on confirmed covid-19 cases from January 11, 2020, to January 20, 2020, from the Surging News Network and from January 21, 2020, to February 27, 2020, from the WHO. A stacked encoder model was used for modeling of the transmission dynamics. Using clustering algorithms and auto-encoder, they grouped and evaluated the transmission. The model anticipated that the different time points at which the provinces would enter the plateau of the forecasted transmission dynamic curves varied from January 21 to April 19, 2020. Wang et al. [11] proposed the COVID-Net model, which was based on CNN. They also created a benchmark dataset for covid research: COVIDx, which was used to train and test the covid-Net model. The dataset consists of 13,975 CXR images taken from 13,870 patients. Their model achieved promising results on the said dataset.
Ghoshal and Tucker [6] collected data from two sources: (i) Dr. Joseph Cohen’s Github repository and (ii) Kaggle’s Chest X-Ray Images for the research. The dataset contained 5,941 PA chest radiography images in total. The dataset’s composition was, Normal: 1,583, covid-19: 68, Viral Pneumonia: 1,504, and Bacterial Pneumonia: 2,786, non-covid-19. To estimate the uncertainty in the model, they trained a Bayesian deep learning classifier by transfer learning on X-ray images. More reliable predictions were yielded by first estimating uncertainty in deep learning.
Narin et al. [32] developed a model using deep CNN to automatically detect the coronavirus infection. The dataset consisted of 100 chest X-ray images, out of which 50 were of covid-19 patients taken from the GitHub repository shared by Dr. Joseph Cohen, and 50 were of normal people. They used three different convolutional neural network-based models: (i) ResNet50, (ii) InceptionV3, and (iii) Inception- ResNetV2. The performance of the model was evaluated using five fold cross-validation techniques. The experimental outcomes confirmed that the pre-trained ResNet50 model gave the highest classification performance with 98% accuracy. InceptionV3 gave 97% accuracy and Inception-ResNetV2 gave 87% accuracy for the best case. Pandey et al. [13] proposed covid-19 outbreak prediction in India based on SEIR and regression model. The dataset was collected from Johns Hopkins University repository in the time period of January 30th, 2020 to March 30th, 2020, consisting of confirmed cases, death cases, and recovered cases in India. They used the SEIR model and regression model for evaluating the dataset. The performance of the models was evaluated using RMSLE. Their model achieved the RMSLE value of 1.75 for the regression model and 1.52 for the SEIR model. The RMSLE error rate between the regression model and SEIR model was found to be 2.01.
Even-though, researchers trying their level best, the coronavirus spreading rate is on peak. It is continuously increasing. The country-wise risk from this disease predicted using neural-network based model by [33]. The authors used a dataset that consisted of date, country, confirmed cases of covid-19, recovered cases, and total death. Further, the dataset was combined with weather data consisting of humidity, ozone, perception, dew, maximum temperature, minimum temperature, and UV. The complete dataset was taken from 22nd January 2020 to 10th March 2020. They selected five major features: active cases, Ozone, humidity, dew, and temperature. They used a Bayesian optimization guided shallow LSTM for predicting the country-specific risk of the covid-19. The results showed that the proposed model can outperform methods for 170 countries’ data and can be a useful tool for such risk categorization. The model can also predict the long-duration outbreak of such an epidemic.
Wang et al. [1] proposed a deep learning algorithm to screen for COVID- 19 disease using CT images. In addition to the previously diagnosed typical viral pneumonia, the authors collected 453 CT images of pathogen-confirmed covid-19 instances, out of which 217 images were used as the training set. The model consisted a Random selection of ROIs, feature extraction with CNN. Their model yielded an AUC of 0.90. Zheng et al. [3] used 499 CT images of covid-19 positive samples to train the model and 131 CT samples for testing. They developed a 3-dimensional CNN to detect covid-19. Their model used a 3D lung mask as input, and a pre-trained UNet generated the 3D lung mask and achieved 0.959 AUC value for the best case.
Apostolopoulos and Mpesiana [34] proposed an automatic detection technique of covid-19 from X-ray images using transfer learning and convolutional neural networks. Two datasets were used for the research, which were collected from available X-ray images on public medical repositories. Dataset_1 contained a total of 1428 X-ray images, out of which 224 images were of confirmed covid-19 disease,504 images of normal conditions, and 714 images with confirmed common bacterial pneumonia. Dataset_2 contained 224 images with confirmed covid-19 disease, 714 images with confirmed bacterial and viral pneumonia, and 504 images of normal conditions. They used transfer learn- ing with CNNs to evaluate both the dataset. The CNNs used for the study were VGG19, MobileNet v2, Inception, Xception, Inception ResNet v2. The results showed that MobileNet v2 obtained the best accuracy of 96.78%. The sensitivity of 98.66% and specificity of 96.46%.
Wangping et al. [35] proposed an extended susceptible-infected-removed prediction of the pandemic trend of covid-19 in Italy and compared it with Hunan, China. Dataset was taken from publicly available covid-19 data by Johns Hopkins University, having daily counts of confirmed cases, recovered cases, and deaths for the period of January 22, 2020, to April 02, 2020. The model calculated mean of basic reproductive number for covid-19 was 4.34 for Italy and 3.16 for Hunan, China. The model predicted that there would be 182,051 infected cases under the current situation, and the end of this would be August 05 in Italy. Salman et al. [36] proposed a model to detect covid-19 using artificial intelligence. The authors used a total of 260 chest X-ray images from the GitHub repository and Kaggle. Out of 260 images, 130 were of covid-19, and 130 were normal X-ray images. With Inceptionv3, their model obtained 100% accuracy and F1-score value for detecting covid-19.
Tiwari et al. [37] proposed a prediction method for the outbreak trend of coronavirus disease in India. The dataset, which included national information of reported cases, recovered cases, and death cases, was taken from Kaggle. To forecast India’s results over the next 22 days, the authors used data from China from January 22, 2020, to April 3, 2020. Using WEKA to forecast the day-wise number of reported cases, recovered cases, and death cases, they made a predictive model. The model predicted that the outbreak peak is expected to occur in India between the third and fourth weeks of April 2020. The total number of expected confirmed covid-19 cases could reach around 68,978, and by April 25, 2020, the deaths are predicted to be around 1557. By analysing data from Google trends using deep learning techniques, Ayyoubzadeh et al. [38] predicted trends in Iran’s covid-19 outbreak. From Feb 15, 2020, to Mar 18, 2020, the dataset consisted of daily new coronavirus cases in Iran and was taken from the WorldOmeter website. Google patterns were checked from Feb 10, 2020, to Mar 18, 2020 for covid-19-related terms. To estimate the number of positive covid-19 cases, linear regression and LSTM models were used. The models were evaluated using the RMSE. With an RMSE of 7.562, the linear regression model gave the best result.
Li et al. [39] used AI-based model to distinguish covid-19 from Community-acquired pneumonia on chest CT. The dataset consisted of 4,356 total 3D chest CT exams from 3,233 patients, which were collected from six medical centres. Out of 4,356 chest CT, 1296 (30%) CT were of confirmed covid-19 cases, 1735 (40%) were of community-acquired pneumonia (CAP), and 1325 (30%) were of non-pneumonia. They developed a model called COVNet which consisted of RestNet50 as the backbone. Their model achieved an AUC value of 0.96 in the independent test set. Ozturk et al. [40] used deep neural networks for covid-19 detection. They used 127 X-ray images diagnosed with covid-19, out of which 43 were female, and 82 were male cases. They also used the ChestX-ray8 database provided by Wang et al. [41]. The dataset consisted 500 normal and 500 pneumonia class samples. They developed DarkCovidNet; a deep learning model which was inspired by the DarkNet architecture [42]. Their model consisting seventeen convolution layers; where each convolutional layer followed by batch normalization and Leaky ReLU operations. The model achieved the accuracy value of 98.09% and 87.02% for binary and multi-class cases respectively.
For a correct diagnosis, quantitative measurements, and prognosis of covid-19 using CT, Zhang et al. [43] suggested the clinically relevant AI method. Using CC-CCII data containing 617,775 CT slices of 6752 CT scans from 4154 patients, the authors built a broad CT dataset. They used deep learning models and achieved 92.49% accuracy and the best case AUC value of 0.9813. A model called COVIDiagnosis-Net, a deep Bayes-squeezeNet-based diagnosis of coronavirus disease from X-ray images, was suggested by Ucar and Korkmaz [7]. They used a COVIDx dataset consisting of 5949 photographs of posteroanterior chest radiography from 2839 patients. 1583 standard, 4290 pneumonia, and 76 covid-19 cases of infection are included in the COVIDx dataset. They have developed a COVIDiagnosis-Net based on deep Bayes-SqueezeNet. Due to their inconsistent sample distributions, the offline augmentation process was used in the first stage for the raw X-ray input images. SqueezeNet is a pre-trained CNN model, and it is pre-trained on the ILSVRC-12 challenge ImageNet dataset. The overall accuracy of their model was 0.983.
Wang et al. [44] suggested a classifier of irregular respiratory patterns that may lead in an accurate way to a large-scale screening of people infected with covid-19. They first proposed a Respiratory Simulation Model (RSM) to address the data scarcity issues for training of the model. They used LSTM, GRU, BI-AT-LSTM, and BI-AT-GRU deep neural networks. BI-AT-GRU model achieved the highest accuracy of 94.5%. Chen et al. [45] proposed a time de- pendent SIR model for covid-19 with undetectable infected people. The authors collected the dataset from the National Health Commission of the People’s Republic of China (NHC). They used a time-dependent SIR model that is more flexible and more stable than direct methods of estimation than standard static SIR. The model’s one-day prediction errors were less than 3% for the number of confirmed cases prediction.
A lung infection quantification method of covid-19 in CT images with deep learning was suggested by Shan et al. [46]. For validation, the authors used 300 CT images and for training 249 CT images were used. They developed a segmentation method based on deep learning that used the “VB-Net” neural network in CT scan images to segment covid-19 infection regions. They used a human-in-the-loop (HITL) technique to help radiologists refine each case’s automated annotation to speed up the manual delineation of CT images for training. The proposed model yielded 91.6% Dice similarity coefficients between the automated and manual segmentation and a mean POI estimation error on the validation dataset of 0.3% for the entire lung. A large-scale screening approach for covid-19 from community-acquired pneumonia was suggested by Shi et al. [47] using infection size conscious classification. Dataset consists of 2,685 CT images, out of which 1658 were confirmed covid-19 cases, and 1,027 were CAP patients. The dataset was taken from three hospitals (i) Tongji Hospital of Huazhong University of Science and Technology, (ii) Shanghai Public Health Clinical Center of Fudan University, and (ii) China-Japan Union Hospital of Jilin University. All images were preprocessed to obtain the segmentation of both infections and lung fields. Their model achieved an accuracy value of 87.90% for the best case.
Hemdan et al. [48] proposed COVIDX-Net, a framework of deep learning to diagnose covid-19 using X-ray images. The dataset consisted of 50 chest X-ray images with 25 confirmed covid-19 cases. 80% of the dataset was used for training, and 20% was used for testing. The COVIDX-Net includes seven different architectures of deep convolutional neural network models i.e. i) VGG19, ii) DenseNet121, iii) InceptionV3, iv) ResNetV2, v) Inception-ResNet-V2, vi) Xception and vii) MobileNetV2. The model achieved the value of F1-scores as 0.89 and 0.91 for regular and covid-19 cases. Zhang et al. [49] suggested a method of screening for viral pneumonia on chest X-ray images using confidence-conscious anomaly detection. They used two X-ray image datasets: (i) X-viral and ii) X-covid. In the X-viral dataset, 5,977 cases of viral pneumonia, 18,619 cases of non-viral pneumonia, and 18,774 stable controls were collected from 390 township hospitals. The X-COVID dataset consisted 107 and 106 samples of confirmed and normal cases. They created a CAAD model focused on deep learning. EfficientNet was used as a function extractor for the CAAD model with the B0 architecture pre-trained on ImageNet. On the clinical X-viral dataset, the proposed model outperforms and for X-covid dataset, the model achieved an AUC of 83.61%.
Abbas et al. [50] used deep learning model for covid-19 detection. They used X-ray images of 80 samples of normal, 105 samples of covid-19, and 11 samples of SARS patient for the research. They developed a deep CNN called DeTraC which deals with the image dataset’s irregularities. Their model achieved a high accuracy of 95.12% with a sensitivity of 97.91% and a specificity of 91.8% in detecting covid-19 X-ray images from normal and severe acute respiratory syndrome cases. The limitation of their model is the limited sample used for the model development. The achieved accuracy may vary if the size of the dataset increases. Yang et al. [51] developed COVID- 19 datasets and used a multi-task learning technique to predict the covid-19 cases. The dataset contained 349 covid-19 CT images from 216 patients and 463 normal CT images. On the self-made dataset, they applied multi-task learning and contrastive self-supervised learning to achieve an accuracy of 89%.
Asnaoui et al. [52] proposed an automated method for detection and classification of Pneumonia based on X-ray images using deep learning. The authors presented a comparison of recent deep convolutional neural networks. The dataset contained 5,856 images, out of which 4,273 were of Pneumonia, and 1,583 were normal. The DCNNs used by them were VGG16 and VGG19, Inception_V3, Resnet 50, Inception_Resnet_V2, Densenet201, MobileNet_V2, Xception. The result showed that fine-tuned versions of Resnet50, MobileNet_V2, and Inception_Resnet_V2 showed highly satisfactory performance by achieving an accuracy value of more than 96%. Xception, VGG16, VGG19, Inception_V3, and DenseNet201 showed relatively low-performance with an accuracy of 84%. Jain et al. [53] proposed a deep learning-based method for detection and analysis of covid-19 using chest X-ray images. They used 6432 chest X-ray scans data for their research. The training dataset contained 5467 images, and the validation dataset contained 965 images. They used deep learning-based CNN models and compared the performance of ResNeXt, Xception, InceptionV3 models. Xception model performed the best as compared to other models with an accuracy of 97.97%.
Maghdid et al. [54] proposed a covid-19 diagnosis method from X-ray and CT images using deep learning and transfer learning algorithms. The dataset consisted of 170 X-ray images and 361 CT images of covid-19 disease collected from five different sources i) British Society of Thoracic Imaging (BSTI), Italian Society of Medical Radiology, iii) Radiopedia, iv) GitHub, and v) Kaggle. A simple CNN and modified pre-trained AlexNet model. The proposed model results showed an accuracy of up to 98% via a pre-trained network and 94.1% accuracy by using the modified CNN. Vaid et al. [55] developed a deep learning-based model to predict covid-19 through chest X-ray. They used a publicly available dataset that contained 181 frontal chest X-ray images of covid-19 positive patients. The dataset also contained 364 chest X-ray images of healthy people collected from NIH clinical centre. The overall ratio of covid-19 to healthy chest X-ray images in the dataset was 1:2. Authors implemented a transfer learning approach for the detection of covid-19 from the chest anterior-posterior radiographs of patients. The model achieved 96.3% accuracy.
Khalifa et al. [56] used GAN and transfer learning model for covid-19 detection. The dataset consisted of 5,863 X-ray images divided into two categories i) Normal and ii) Pneumonia. The authors used 10% of the dataset for training, and the remaining was used to test the model. They GAN and four deep transfer learning models, i.e., i) Alexnet, ii) Googlenet, iii) Squeezenet, and iv) Resnet18. The results showed that Resnet18 performed the best among all models and achieved an accuracy of 99% by using GAN as an image augmenter. Chaganti et al. [57] proposed an automated quantification method of abnormal CT patterns associated with covid-19 from chest CT. The dataset contained 9749 chest CT images. The model was trained with 901 samples whereas tested with 200 samples. The deep learning and deep models were used to measure and compute the severity of the disease.
Hall et al. [58] proposed a model to find covid-19 from chest X-ray using deep learning. 135 covid-19 chest X-ray images were collected from different sources such as GitHub, Radiopaedia, and SIRM. Additionally, 320 pneumonia chest X-ray images were taken from the Kaggle database for the research. They used a pre-trained Resnet50, VGG16, and their own developed CNN model. The models were trained on a balanced set of covid-19 and pneumonia chest X-rays. The model’s overall accuracy was 91.24%, with the true positive rate for covid-19 of 0.7879 and the AUC value of 0.94. Gozes et al. [59] proposed a coronavirus detection method and analysis on chest CT images using deep learning. The authors collected the data of 109 confirmed covid-19 patients from Zhejiang province, China, and used it as a testing dataset. They used ResNet50-2D convolutional neural network architecture and achieved a 0.948 AUC value for the best case.
A model called COVID-CAPS was introduced by Afshar et al. [60], which was a capsule network-based method for recognizing covid-19 cases from X-ray images. For typical thorax anomalies, the authors used a large dataset consisting of 94,323 frontal view chest X-ray images. The dataset, containing 112,120 X-ray images for 14 thorax anomalies, was taken from the NIH CXR dataset. They used 90% of the dataset for training and 10% for validation. They developed the COVID-CAPS model, which consists of four convolutional layers and three capsule layers. The first convolutional layer was followed by batch normalization, second convolutional layer was followed by average-age pooling, third layer was also the convolutional ones, fourth convolutional layers was reshaped to form the first capsule layer. COVID-CAPS achieved an accuracy of 95.7%, a sensitivity of 90%, a specificity of 95.8%, and an AUC value of 0.97. Using a pre-trained COVID-CAPS further improved the accuracy to 98.3% and specificity to 98.6%.
Gozes et al. [61] developed an automated CT image analysis tool for Coronavirus identification. They used a dataset containing 6,356 CT scans for their research. They used transfer deep learning models to develop an image analysis system and achieved a 0.996 AUC value for the best case. Dandekar and Barbastathis [62] proposed a neural network aided quarantine control model estimation of global covid-19 spread. The data was collected from the Chinese National Health Commission, which consisted of data for the infected and recovered case count in Wuhan, China. The authors used an augmented neural network model. Another deep learning model was proposed by [2] to detect and evaluate pneumonia cases during the covid-19. The dataset consisted of 5,863 chest X-ray images divided into two categories i) normal and ii) pneumonia. They used five CNN-based architectures i) ResNet34, ii) ResNet50, iii) DenseNet169, iv) VGG-19 and v) Inception ResNetV2 & RNN. Their model performed best for the pneumonia class.
Karim et al. [63] proposed the DeepCOVIDExplainer model, which gives an explainable covid-19 diagnosis based on chest X-ray. Authors used 15,959 CXR images of 15,854 patients which included normal, pneumonia and covid-19 cases and the dataset was collected from four different sources i) COVIDx dataset by Wang et al. [41], (ii) COVID chest X-ray dataset, iii) covid-19 patients lungs X-ray images and iv) CXR images. DenseNet-161 and VGG-19 performed the best on balanced and imbalanced datasets, while VGG-16 was the worst performer. DenseNet-161 achieved a precision value of 0.952, recall value of 0.945, and F1-score of 0.945, whereas VGG-19 obtained the precision, recall, and F1-score of 0.943, 0.935 0.939, respectively.
Luz et al. [64] proposed an efficient and effective deep learning model for covid-19 patterns detection through X-ray images. They collected 13,800 X-ray images divided into healthy, non-covid-19 pneumonia, and covid-19. Dataset was collected from three sources: i) RSNA Pneumonia detection challenge dataset, (ii) covid-19 image data collection, and iii) COVIDx dataset. Out of 13,800 X-ray images, 13,569 were used to train the proposed model, and other competing architectures and 231 were used for validation of the model. Their model achieved an overall accuracy of 93.9%. Hu et al. [65]proposed a deep learning model for covid-19 infection detection and classification through CT images. They collected 60 3D CT lung scans from the publicly available TCIA dataset. They used CNN, which was full convolutional, consisting of five convolutional blocks. This system could reliably differentiate covid-19 cases from patients with CAP and NP. The suggested model achieved high precision, accuracy, and AUC for the classification.
Wu et al. [66] proposed an explainable covid-19 diagnosis system by joint classification and segmentation. They created a dataset which contained 144,167 CT images, among which 3,855 images of 200 positive cases are pixel-level annotated, 64,771 images of the other 200 positive cases are patient-level annotated. The rest 75,541 images are from the 350 negative patients. They developed a Joint Classification and Segmentation (JCS) model. The classifier was based on the Res2Net network, and the architecture of the segmentation model used the VGG-16 backbone. The proposed model achieved an average sensitivity of 95.0%. Rajaraman et al. [10] proposed iteratively pruned deep learning ensembles for covid-19 detection in chest X-rays. Authors collected dataset from four sources: (i) Pediatric CXR dataset, (ii) RSNA CXR dataset, (iii) Twitter covid-19 CXR dataset, and (iv) Montreal covid-19 CXR dataset. They used 90% of the dataset for training and 10% for testing. They first evaluated the performance of customized CNN and a selection of ImageNet pre-trained CNN models, which were (i) VGG-16, (ii) VGG-19, (iii) InceptionV3, (iv) Xception, (v) InceptionResNet-V2, (vi) MobileNet-V2, (vii) DenseNet-201, and viii) NasNet-mobile. The best performing models were iteratively pruned to reduce complexity and improve memory efficiency. The iteratively pruned version of InceptionV3 performed the best, achieving an accuracy value of 99.01% and an AUC of 0.9972.
Minaee et al. [14] proposed a deep-COVID model for predicting covid-19 from chest X-ray images using deep transfer learning. The authors created a COVID-Xray-5k dataset, which contained 2,031 training images and 3,040 test images. The dataset was created from two publicly available sources (i) Chestxray-Dataset and (ii) ChexPert dataset. They used four pre-trained convolutional models (i) ResNet18, (ii) ResNet50, (iii) SqueezeNet, and iv) DenseNet-121. All the models achieved a sensitivity value of 97.5%, with SequeezeNet achieving the highest specificity of 97.8%.
Brunese et al. [67] used deep learning techniques to identify covid-19 from chest X-ray. They used a total of 6,523 chest X-rays, 250 of which were from covid-19 patients, 2,753 from patients with various respiratory illnesses, and 3,520 from healthy individuals. Using the VGG-16 transfer learning model, their model achieved an accuracy of 0.97 for the best case. Qiu et al. [68] proposed MiniSeg model, which is an extremely minimum network for efficient covid-19 segmentation model. The authors utilized an open access covid-19 CT segmentation dataset, which consisted of 100 axial CT images from 60 patients with covid-19. They developed MiniSeg model, which is a lightweight deep learning model for efficient covid-19 segmentation. MiniSeg adopted a novel multi-scale learning module, i.e., the attentive hierarchical spatial Pyramid (AHSP) module, to ensure its accuracy under the extremely minimum network size constraints. The results showed that the MiniSeg gave the best performance compared to the other 26 image segmentation methods and is also highly efficient.
Toǧaçar et al. [69] used deep learning models with the CXR dataset for covid-19 detection. A total of 458 chest X-ray images were collected from different sources, out of which 295 were of covid positive, 65 were of normal/healthy, and 98 were of pneumonia. For the best case, their model achieved 99.27% accuracy value. Punn and Agarwal [70] proposed an automated diagnosis of covid-19 using fine-tuned deep neural networks. They used two dataset: i) covid-19 Image dataset and ii) RSNA. covid-19 image dataset consisted of 108 posteroanterior CXR (PCXR) images and 45 PCXR images of other pneumonia. The RSNA dataset consisted of 470 PCXR images and 453 PCXR images of normal conditions. Their model achieved an accuracy of 96%. Cabras [71] proposed a Bayesian deep learning model for estimating the covid-19 trend in Spain. The dataset was collected from Instituto Carlos III de Madrid in Madrid2, Spain, and consisted of cumulative numbers of cases, deaths, and recovered. They used the LSTM network for analyzing sequences with the usual Bayesian Poisson-Gamma model for counts. Cohen et al. [72] proposed a covid-19 pneumonia severity predicting model based on analyzing chest X-ray using deep learning. The authors collected 94 posteroanterior CXR images from a public covid-19 data collection. The pre-training dataset consisted of 88,079 non-covid-19 images and was collected from seven different sources: i) RSNA Pneumonia Challenge, ii) CheXpert - Stanford University, iii) ChestX-ray8 - National Institutes of Health (NIH), iv) ChestX-ray8 - NIH with labels from Google, v) MIMIC-CXR - MIT, vi) PadChest - University of Alicante, and vii) OpenI. They used the DenseNet model from the TorchXRayVision library because DenseNet models have been shown to predict pneumonia well.
Ismael et al. [73] used deep feature extraction and pretrained CNN to classify covid-19 through chest X-ray images. The dataset contained a total of 380 chest X-ray images, out of which 180 were of covid-19 positive patients, and 200 were of normal/healthy individuals. For the deep feature extraction, they used pre-trained transfer learning models. They achieved 94.7% accuracy using ResNet50 model for deep feature extraction with SVM classifier. Ying et al. [74] suggested a deep learning model using CT images to reliably diagnose novel coronaviruses. The dataset contained chest CT scan images of 88 covid-19 patients, 101 bacterial pneumonia infected patients and 86 healthy individuals. The authors developed a CT diagnostic system focused on deep learning and named it DeepPneumonia to classify patients with covid-19. To extract the top-K information in the CT images and obtain the image-level predictions, they also designed a comprehensive relationship extraction neural network (DRE-Net). On pre-trained ResNet50, on which the feature pyramid network (FPN) was added, DRE-Net was built. The proposed model obtained an excellent AUC of 0.99.
An AI method for covid-19 diagnosis was proposed by Cheng Jin [75]. There were 10,250 CT images from three publicly accessible databases were collected: (i) LIDC-IDRI, (ii) Tianchi-Alibaba, and (iii) CC-CCII. They used deep learning-based AI system to clarify the attentional region characteristics. The model obtained an AUC value of 97.17% for the best case. He et al. [76] proposed a deep learning model for covid-19 diagnosis based on CT scans. They created a publicly available covid-19 CT dataset, which contained 349 positive and 397 negative CT scan images. To learn efficient and unbiased feature representations, they built a self-trans method that synergistically combines contrastive self-supervised learning with transfer learning. They used the self-trans approach with ResNet-50 and DenseNet-169. DenseNet-169 performed the best with an F1-score of 0.85 and an AUC value of 0.94.
Amyar et al. [77] proposed a Multi-Task deep learning-based CT imaging classification and segmentation for covid-19. The authors collected 1,044 CT images, which included 449 CT images of covid-19 patients, 98 with lung cancer, 397 of different kinds of pathology, and 100 of normal condition. The dataset was taken from two sources: (i) COVID-CT-dataset, (ii) http://medicalsegmentation.com/covid19/. They developed an MTL architecture based on three tasks: i) Covid vs non-covid classification, ii) covid lesion segmentation, and iii) Image reconstruction. An encoder and two decoders for reconstruction and segmentation with a multi-layer perceptron used to design the architecture. For the segmentation and ROC higher than 93% for the classification, the suggested model obtained a dice coefficient greater than 0.78. Fan et al. [78] proposed Inf-Net which was an automatic covid-19 lung infection segmentation model using CT images. Authors used publicly available covid-19 CT segmentation dataset which consisted of 100 axial CT images from different covid-19 patients. They developed a Lung Infection Segmentation Network (Inf-Net) which consisted of three reverse attention (RA) modules connected to the paralleled partial decoder (PPD). The parallel partial decoder contained five convolutional blocks of Res2Net. The model achieved a dice score of 0.579, sensitivity of 0.870, specificity of 0.974, precision of 0.500 and MAE of 0.047 for the best case. Wang et al. [79] used a deep learning system for covid-19 diagnostic and prognostic analysis. The dataset consisted of a total of 5,372 patients with CT images from seven cities or provinces. They used a deep learning system and achieved an AUC value of 0.90 on Wuhan city and Henan’s training dataset, 0.88 AUC value on the validation dataset of Heilongjiang, and 0.87 AUC value on the validation dataset of Anhui.
Farid et al. [80] proposed a model for analysis and prediction of covid-19. They used a dataset contained 51 images, each segregated into the severity of SARS and covid-19, and was taken from the publicly available Kaggle database. They used a combination of four image filters: (i) MPEG7 Histogram, (ii) Gabor, (iii) Pyramid of Rotation-Invariant Local Binary Pattern Histograms and iv) Fuzzy 64-bin Histogram along with a composed hybrid feature (CHFS) model for the diagnosis of covid-19. Instead of probability and a random selection, they also used the J48 decision tree classifier as a fitness function to speed up convergence and pick the best features. When used with Naive Bayes as a meta-classifier in a hybrid classification system, the proposed model gave a greater precision of 96.07%. Apostolopoulos [81] suggested a way to extract potentially representative bio-markers of coronavirus from X-ray images with a deep learning approach and pulmonary disease image data. The dataset contained 3,905 X-ray images corresponding to six diseases. They used MobileNet v2 convolutional neural network, which was trained from scratch. Their model achieved 99.18% accuracy, 97.36% sensitivity, and 99.42% specificity in detecting covid-19.
This section discussed the researches that use a deep learning framework for covid-19 detection and prediction. Many researchers used the CXR dataset, whereas many used datasets with multiple classes like-Confirmed covid-19 cases, Pneumonia, and Normal. One of the major limitations of the reported outcomes is the robustness of the model. The majority of the model trained and tested with a limited sample that does not validate the model’s outcome. Secondly, the models were developed with the dataset, which considered only local covid-19 patient information. The model developed with a particular demographic region dataset may not work well in another region. Hence, the robustness of such models is in doubt. Even though the prediction accuracies are high, a robust system is needed to detect this deadly virus in every part of the world.
Research Contributions with Machine Learning
This section describes the outcomes of the machine learning models with their limitations. The summary of key researches using the ML approaches are listed in Table 2. Ribeiro et al. [84] proposed a model for short-term forecasting of covid-19 in Brazil. The collected dataset contained cumulative confirmed cases of covid-19 from 10 states of Brazil that occurred until April 18 or 19 of 2020. In the task of predicting one, three, and six days ahead of the covid-19 cumulative verified cases, many machine learning models such as stacking, support vector regression and others were used. The experimental outcomes confirmed that the SVR and stacking ensemble learning performed the best. Tuli et al. [85] suggested a model using machine learning and cloud computing to forecast the growth and trend of the outbreak of covid-19. They used “our world in data” dataset for the research. The dataset is updated regularly based on WHO reports. The iterative weighting-based Weibull model performed better in terms of covid-19 predictions.
Table 2.
Research findings and limitations using Machine Learning frameworks
| Objective | Dataset | Time Frame | Source of Dataset | Model used | Performance | Limitation |
|---|---|---|---|---|---|---|
| Short-term forecasting of covid-19 in Brazil [84] | Dataset of confirmed cases of covid-19 from 10 states in Brazil | April 18, 2020 to April 19, 2020 | Not Available | ML based models | SVR and stacking performed the best | For one-day-ahead the applications are limited |
| Predicting the growth and trend of covid-19 outbreak [85] | Our World in Data by Hannah Ritchie | Gaussian model | Predictive model | May lead to worse of public health situation | ||
| Prediction of the spread of covid-19 in India [28] | covid-19 data of India | 30th January 2020 to 4th April 2020 | Not Available | LSTM and curve fitting method | High accuracy | Only accurate within a range |
| Prediction of covid-19 disease progression in India [86] | Not Available | Not Available | (i) Johns Hopkins University, (ii) Covid19India website, (iii) Kaggle-Covid-19 | Statistical machine learning model | R0 =2.75 | Prediction was not in sync with actual desease progression |
| Real time forecasting of covid-19 [9] | Not Available | Not Available | Not Available | Clustering techniques | Predictive model | More miss classification rate |
| Predicting the survival for severe covid-19 patients [87] | 375 patients data | Not Available | Tongi Hospital, Wuhan,China | XGBoost classifier | F1-score 0.92 | Sample size is relatively limited |
| Confirmation of covid-19 cases [15] | Not Available | 20th January 2020 to 12th March 2020 | Kagglea | LSTM-GRU based model | Accuracy of 87% | Accuracy for released case was only 40.5% |
| To predict mortality risk in patients [88] | 117,000 confirmed covid-19 patients data from 76 countries | Not Available | Not Available | ML based models | Accuracy: 93.75% | – |
| Analysis of covid-19 epidemic [4] | Daily reported cases | January 22, 2020 to April 1,2020 | Johns Hopkins University | ML and DL based models | Polynomial regression yielded minimum RMSE | Models were validated against the training dataset |
| Forecasting future of covid-19 [89] | Not Available | Not Available | Multiple sources | Regression model | Exponential smoothing (ES) performed best | Difficult to put an accurate hyperplane |
| To identify an intrinsic covid-19 virus [8] | 5000 unique viral genomic sequences | Jan 27, 2020 | Not Available | SVM and KNN | Accuracy:100% | Availability of coronavirus genomic signatures are limited |
| Pandemic prediction for Hungary [90] | Daily cases and number of deaths in Hungary | 4 March 2020 to 28 April 2020 | Not Available | Hybrid machine learning model | MLP-ICA and ANFIS gave promising results | Model is susceptible to major interruptions |
| covid-19 outbreak prediction [91] | Not Available | Not Available | WorldOmeter website | Machine learning algorithms | MLP and ANFIS achieved high generalization | Study provides only the initial benchmarking |
| Prediction of coronavirus clinical severity with AI [92] | 53 hospitalized patients with confirmed covid-19 | Not Available | Not Available | ML based classifiers | Accuracy: 80% | Limited size of the dataset with some incomplete data |
| Classification of covid-19 using CT images [96] | Not Available | Not Available | Moroccan Ministry to Healthb | ML based classifiers | Good predictive model | Localized dataset was taken to validate the model |
| Classification of covid-19 using CT images [93] | 150 CT abdominal images | Not Available | Societa Italina di Radiologia Medica e Interventistica | Mixed models | Accuracy : 99.68% | Performance was not validated with other CT image dataset |
| Forecasting of covid-19 outbreak in India [94] | Not Available | 22nd January 2020 to 10th April 2020 | Kaggle c | Multilayer perceptron | CI: 95.0% | Less number of attributes |
| Diagnosis of covid-19 using AI [95] | 1838 cough sounds and 3597 non-cough environmental sounds | Not Available | ESC-50 database | ML and DL based models | Accuracy: 95.60% | Limited quantity of the training and testing data |
Ndiaye et al. [82] developed a forecasting model for covid-19. They collected the data from website: https://www.tableau.com/covid-19- coronavirus-data-resources during January 21, 2020, to April 02, 2020. They used machine-learning approach for predicting time series data based on an additive model. Sourish Das [86] proposed a prediction model for covid-19 disease progression in India. The dataset used was taken from three sources: (i) John Hop- kins University repository, (ii) Covid19India website3, and iii)Kaggle-Covid-194. He used the popular epidemic SIR model to estimate the basic reproduction number at national and various state levels. The SIR model used to predict when the peak will be reached because it captures the epidemic’s inherent dynamism. The model calculated that India’s R0 =2.75 as of March 04, 2020, and is very much equal to Hubei/China at the early stage of disease progression and predicted Punjab’s situation and required immediate actions to curb the covid-19 spread. He concluded that if the nation-wide lock-down works, the cases are expected to be less than 66,224 by May 01, 2020.
Liu et al. [9] developed a forecasting model for real-time. Dataset used by them was collected by aggregating the data of covid-19 activity reports from China Centers for Disease Control and Prevention (CDC), Baidu Internet search frequencies, media activity reported by media cloud, covid-19 regular forecasts from an agent-based mechanistic model, and the Global Epidemic and Mobility Model (GLEAM). They used a clustering technique that allows covid-19 activity to be exploited in geo-spatial synchronicities across Chinese provinces. They called this method as ARGOnet augmented. In 27 out of 32 Chinese provinces, the model’s predictive power outperforms the collected baseline models. It could easily be applied to other geographies currently grappling with the covid-19 outbreak to help make decisions. Yan et al. [87] proposed a way of predicting the survival for severe COVID- 19 patients with a machine learning-based predictive model. They collected data from 375 patients, including 201 survivors from Tongji Hospital in Wuhan, China. They selected three key clinical features: i) lactic dehydrogenase (LDH), ii) lymphocyte, and iii) High-sensitivity C-reactive protein (hsCRP). They used a supervised XG- Boost classifier as the predictor. On the training dataset, the model achieved F1- score of 0.99 on Survival prediction and 0.98 on Death prediction, a precision score of 1.00 and 0.97 on Survival and Death prediction, respectively.
Dutta and Bandyopadhyay [15] proposed a machine learning approach for the confirmation of covid-19 cases. The dataset used for the research was taken from kaggle5 which contained cases from 20th January 2020 to 12th March 2020. The model achieved an accuracy of 87% for confirmed cases. Pourhomayoun and Shakibi [88] proposed a model to predict mortality risk in patients of covid-19 using artificial intelligence. They used a dataset having more than 117,000 confirmed covid-19 patients from 76 countries. They chose 42 features that contained demographic features such as age, sex, province, country, age, travel history, general medical information, and patient symptoms. To predict the mortality rate among covid-19 patients, machine learning based classifiers and neural network model was used. Using 10-fold cross-validation, the accuracy achieved by Neural Network was 93.75% for the best case. Punn et al. [4] proposed machine learning and deep learning algorithms to analyse the covid-19 epidemic. The data was collected from Johns Hopkins University’s official repository from January 22, 2020, to April 1, 2020. The dataset consisted of the daily reported case and daily time series summary tables. Authors used machine learning algorithms to achieve the better forecasting predictions.
Rustam et al. [89] proposed covid-19 future forecasting using supervised machine learning models. The dataset was obtained from the GitHub repository provided by the Center for Systems Science and Engineering, Johns Hopkins University. Four regression models, i) Linear regression, ii) LASSO regression, iii) SVM, and iv) exponential Smoothing were used for forecasting covid-19 future. The results showed that exponential smoothing performed the best given the dataset’s nature and size. LASSO and LR also performed well for forecasting to some extent to predict death rates and confirmed cases. SVM produced poor results in all the scenarios because of fluctuations in dataset values. Randhawa et al. [8] suggested a model that could recognize an intrinsic genomic signature of the covid-19 virus. Over 5,000 separate viral genomic sequences were included in the dataset, totalling 61.8 million bp, including 29 covid-19 virus sequences available as of 27 January 2020. The proposed method uses digital signal processing supervised machine learning for genome analyses. Total six machine learning models were used: i) Linear Discriminant, ii) Linear SVM, iii) Quadratic SVM, iv) Fine KNN, v) Subspace Discriminant, and vi) Subspace KNN. The method achieved 100% accurate covid-19 virus sequence classification and, within a few minutes using only raw DNA sequence data, discovered the most important relationship among over 5,000 viral genomes.
Covid-19 pandemic prediction for Hungary using a hybrid machine learning method was proposed by Pinter et al. [90]. Their model projected that the outbreak and overall morality would drop dramatically in Hungary by the end of May. Ardabili et al. [91] proposed prediction of covid-19 outbreak using machine learning approach. They collected data from WorldOmeter website6 for five countries: (i) Italy, (ii) Germany, (iii) Iran, (iv) the USA, and (v) China on total cases over 30 days. Jiang et al. [92] used a dataset of 53 hospitalized patients with confirmed covid-19. The dataset was evaluated with 11 features (i) lymphocyte count, (ii) white blood count, (iii) temperature, (iv) creatinine, (v) hemoglobin, (vi) gender, (vii) age, (viii) ALT, (ix) K+, (x) N+ and (xi) Myalgias. They used six machine learning algorithms i) Logistic regression, ii) KNN, iii) Decision tree (based on gain ratio), iv) Decision tree (based on Gini index), v) Random forests and vi) Support Vector Machine and the accuracy achieved was 50%, 80%, 70%, 70%, 70%, and 80%, respectively. Barstugan et al. [93] suggested using machine learning to identify covid-19 using CT images. From the Societa Italina di Radiologia Medica e Interventistica, the authors collected 150 CT abdominal photographs of 53 infected cases. Out of 150 CT images, they cut the patch regions. During the classifying process, they used 2-fold, 5-fold, and 10-fold cross-validation. With 10-fold validation, their model obtained the precision value of 99.68%.
Sujath et al. [94] proposed a machine learning forecasting model for covid-19 outbreak in India. They collected predominant information of covid-19 from 22nd January 2020 to 10th April 2020 from kaggle7. They used the Multilayer perceptron and Vector autoregression method for the predictions. Imran et al. [95] suggested an AI4covid-19 model that is an AI-enabled preliminary covid-19 diagnosis using cough sound samples through an app. The authors used the ESC-50 dataset, which is freely accessible and offers a large range of human and environmental sounds. The sound collection was divided into fifty grades, one of which was cough sounds. The suggested model used 1,838 cough sounds and 3,597 non-cough environmental sounds for training and testing. Data samples used for covid-19 diagnostic engine training included cough sounds from 70 patients with covid-19, 130 patients with pertussis, 96 patients with bronchitis, and 247 samples of usual cough from different individuals. The proposed AI4covid-19 model uses three distinct classifier systems in parallel, i.e., Deep transfer learning-based multi-class classifier (DTL-MC), a multi-class classifier based on classical machine learning (CML-MC), and binary classifier classification based on deep transfer learning (DTL-BC). The app reports a diagnosis only if all three classifiers return similar classification results. If the classifier does not approve,’ evaluation inconclusive’ will be returned by the app. The total precision obtained by the model was 95.60%, and the F1-Score achieved 95.61%.
This section discussed the researches that use a machine learning framework for covid-19 detection and prediction. One of the major limitations of the traditional machine learning-based model is manual feature extraction. The models need the features extracted manually from the input. This is entirely dependent on human expertise. Many times, features working for a problem do not fit in other similar problems; hence, again, a different set of features is needed to extract from the input. Researchers confirmed that the covid-19 virus is changing its structure continuously. Therefore, in such cases, a manual feature dependent model may not be a good choice. Here, automated feature extraction techniques are helpful to save human effort and get better contextual features from the input automatically.
Open Issues and Future Research Direction
Currently, the entire world putting their full effort to find the solution for the pandemic happened due to novel coronavirus. This article summarises the research works that were recently reported by the worldwide research community. The researchers widely used Deep learning based framework such as CNN, RNN, LSTM, and others for prediction and detection of covid-19 cases [1–4, 6, 7, 10, 27, 34, 36, 38–40, 43, 44, 46, 48, 49, 52, 54, 57–59, 64, 65, 68, 70–72, 74–77, 79, 81, 97]. Also, many of them used machine learning models such as random forest, Naive Bayes, logistic regression, and a similar one for the purpose of same [4, 8, 9, 15, 82, 84–96, 98]. The reported models claimed that the accuracy of covid-19 prediction is nearly 100% in many cases whereas in some cases it is 80-85%. Researchers mainly used the patient’s CT scan images [1, 3, 39, 43, 46, 93] and CXR images for their research [2, 6, 7, 10, 11, 14, 32, 34, 36, 40, 48–50, 52, 54, 56, 58, 60, 63, 64, 70, 72, 81]. It has been found that, compared to the other features such as symptoms of cold and cough, the features extracted from the CT-scan images is more effective for accurate predictions [47, 51, 54, 57, 59, 65, 66, 68, 74–80].
This article summarizes a total of 85 recent pieces of research out of which few research articles were reported using CT-scan images whereas few research articles were based on CXR. Many researches are reported either an analysis of the current pandemic or based on people survey’s report or used machine learning models with the features collected manually. Among all, the number of articles utilized the deep learning frameworks are 39 whereas machine learning-based models are used in 18 articles. This analysis confirmed that for covid-19 prediction, deep learning-based frameworks are more suitable.
The data sources used for predictions and detection of the covid-19 cases are listed in Tables 1, 2, 3. The majority of research work utilizes the source of data repository Kaggle, whereas some of them used Johns Hopkins University and WHO’s dataset for the research. Even though, many detection mechanisms are reported. The number of False-positive cases as well as False-negative predictions is not minimized which leads many people to remain un-traced. Due to this, the world is facing a second wave of this pandemic where the number of positive cases once again starts increasing day by day. Some of the Indian states like Delhi, Gujarat, Tamil Nadu also facing the second wave of the novel coronavirus infections.
Table 3.
List of datasets used for covid-19 research
The second wave of the covid-19 in a different portion of the world confirmed that it is still alive and needed a stronger solution to get rid of it. Countries like Russia, UK, and India reported that they are in the last phase of the trial for the covid-19 Vaccine whereas others are still in the process. Many research challenges exist in the current time to find the solution to this pandemic. One of the main challenges is identifying the key symptoms of covid-19, currently, the fever, cough, cold, and others are treated as main symptoms. However, these symptoms are common due to climate changes in the world, hence the model produces False-positive and False-negative predictions with such data. It is needed to find the key symptoms of the covid-19 disease which helps to reduce the miss-classification rate. Another issue is dynamic, researchers reported that the virus changes its structure with time, and hence it is being difficult to find a stable solution for it. Apart from this, the labeled CT scan images, CXR data are also not available at current time which is needed for fruitful research outcome.
Currently, researchers are actively working on getting the antidote for this deadly disease, and hence it is still an open issue for all. Researchers confirmed that the virus changing its patterns, and hence an antidote developed for a particular pattern, may not be ready to fight with their new variants. Future researchers may analyze the reported results and provide a better solution to minimize the false negative cases during the covid-19 predictions. Researchers may use the existing dataset listed in Table 3, or they may develop a dataset as per the model requirement, which provides better accuracy.
Challenges
This study provided a comprehensive summary of the reported covid-19 research with ML and DL techniques. Many existing models reported the highest accuracy and few of them achieved accuracy in the range of 70 to 80% also. Even though the reported accuracies are satisfactory, it has several limitations as well as challenges. As shown in Tables 1 and 2, many researchers uses a limited dataset for their research and hence, the developed model may not fit in general circumstances. The major issue for the researchers is the availability of confirmed covid-19 datasets. In the early phase of research, the annotated dataset was not available, and hence, with limited datasets, models are trained and tested. At the current time, the annotated dataset’s availability has a major challenge–the disease spreading at different rates in different parts of the world. Few places, the speed is very high, like in the UK, the average number of cases during the first wave is very high. In the second wave, India faces issues, and the average cases per day are very high compared to other countries. Hence, to prepare the antidote for this deadly virus, a demographic based annotated dataset might be helpful.
Conclusion
This article summarizes the reported works on novel coronavirus detection and prediction. The world, facing the second wave of the covid-19 pandemic, many countries like Italy, the USA, Russia, and India where the number of cases was decreased by the middle of October. However, again it was started increasing in the middle of November 2020. This paper focuses on summarizing the approaches that were used to predict that the people are infected or not from covid-19. Data like people’s CT scan and CXR are the main source for prediction. Many researchers used deep learning based models whereas comparatively less researchers prefer machine learning models for covid-19 disease predictions. Researchers confirmed that image-based data like CT scans and X-rays are more helpful for accurate predictions. This paper provided a brief summary of the reported works on both the deep learning and machine learning models and also highlight the limitations of the proposed work. Moreover, the source of the dataset used by the researchers was provided which might be helpful for the future researcher to start the work quickly. Many of the reported work, not cross-validated or not tested with the different dataset, and hence the reported accuracy may vary with the change of the dataset. In the future, researchers may develop a strong model that can easily differentiate the covid-19 infected people from others.
Abbreviations
- AHSP
Attentive hierarchical spatial pyramid
- AI
Artificial Intelligence
- ALT
Alanine amino transferase
- ANFIS
Adaptive network-based fuzzy inference system
- ARIMA
Autoregressive integrated moving average
- AUC
Area under ROC curve
- BI-AT-GRU
Bidirectional attention GRU
- BI-AT-LSTM
Bidirectional attention LSTM
- BSTI
British society of thoracic imaging
- CAAD
Confidence aware anomaly detection
- CAP
Community acquired pneumonia
- CDC
Centers for disease control and prevention
- CFR
Case fatality ratio
- CML-MC
Classical machine learning based multi class classifier
- CNN
Convolutional neural networks
- CT
Computed tomography
- CUBIST
Cubist regression
- CXR
Chest X-ray
- DCNN
Deep convolutional neural network
- DI2IN
Deep image to image network
- DNA
Deoxyribonucleic acid
- DNN
Deep neural network
- DRE-NET
Details relation extraction neural network
- DT
Decision tree
- DTL-BC
Deep transfer learning-based binary class classifier
- DTL-MC
Deep transfer learning-based multi class classifier
- DWT
Discrete wavelet transform
- ES
Exponential smoothing
- eSIR
Extended Susceptible-infected-removed
- FPN
Feature pyramid network
- GANS
Generative adversarial networks
- GLCM
Grey level co-occurrence matrix
- GLEAM
Global epidemic and mobility model
- GLRLM
Grey level run length matrix
- GLSZM
Grey-level size zone matrix
- GP
Gaussian process
- GRU
Gated recurrent unit
- HITL
Human-in-the-loop
- hsCRP
High-sensitivity C-reactive protein
- IP
Improvement percentage index
- JCS
Joint classification and segmentation
- KNN
K-nearest neighbour
- LASSO
Least absolute shrinkage and selection operator
- LDH
Lactic dehydrogenase
- LDP
Local Directional Pattern
- LR
Logistic regression
- LSTM
Long short-term memory
- LSTM-GRU
Long short-term memory GRU
- MAE
Mean absolute error
- MLDSP
Machine learning with digital signal processing
- MLP
Multi-layered perceptron
- ICA
Imperialist competitive algorithm
- MSE
Mean squared error
- NB
Naive Bayes
- NCP
Novel Coronavirus pneumonia
- NIH
National institutes of Health
- PA
Posterior-anterior
- PCXR
Posteroanterior CXR
- PPD
Paralleled partial decoder
- PR
Polynomial regression
- RF
Random forest
- RIDGE
Ridge regression
- RMLSE
Root mean squared logarithmic error
- RMSE
Root mean square error
- RNN
Recurrent neural network
- ROI
Regions of interest
- RSM
Respiratory simulation model
- RT
Regression tree
- SARS
Severe acute respiratory syndrome
- SD
Standard deviation
- SEIR
Susceptible-exposed-infectious-removed
- SIR
Susceptible, Infected, Recovered
- sMAPE
Symmetric mean absolute percentage error
- SVM
Support Vector Machine
- SVR
Support vector regression
- TL
Transfer learning
- UV
Ultraviolet
- VAR
Vector autoregression
- WHO
World Health Organization
- XGBoost
Extreme gradient boosting
Declarations
Conflict of Interest
There is no conflict of interest
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shivam Chahar, Email: shivam.chahar@gmail.com.
Pradeep Kumar Roy, Email: pkroynitp@gmail.com.
References
- 1.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X et al. (2020) “A deep learning algorithm using ct images to screen for corona virus disease (covid-19),” medRxiv [DOI] [PMC free article] [PubMed]
- 2.Hammoudi, K, Benhabiles H, Melkemi M, Dornaika F, Arganda-Carreras I, Collard D, Scherpereel A (2020) “Deep learning on chest x-ray images to detect and evaluate pneumonia cases at the era of covid-19,” arXiv:2004.03399 [DOI] [PMC free article] [PubMed]
- 3.Zheng C, Deng X, Fu Q, Zhou Q, Feng J, Ma H, Liu W, Wang X (2020) “Deep learning-based detection for covid-19 from chest ct using weak label,” medRxiv
- 4.Punn NS, Sonbhadra SK, Agarwal S (2020) “Covid-19 epidemic analysis using machine learning and deep learning algorithms,” medRxiv
- 5.Yang Z, Zeng Z, Wang K, Wong S-S, Liang W, Zanin M, Liu P, Cao X, Gao Z, Mai Z, et al. Modified seir and ai prediction of the epidemics trend of covid-19 in china under public health interventions. J Thorac Dis. 2020;12(3):165–174. doi: 10.21037/jtd.2020.02.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghoshal B, Tucker A (2020) “Estimating uncertainty and interpretability in deep learning for coronavirus (covid-19) detection,” arXiv:2003.10769
- 7.Ucar F, Korkmaz D (2020) “Covidiagnosis-net: deep bayes-squeezenet based diagnostic of the coronavirus disease 2019 (covid-19) from x-ray images,” Medical Hypotheses, p 109761 [DOI] [PMC free article] [PubMed]
- 8.Randhawa GS, Soltysiak MP, El Roz H, de Souza CP, Hill KA, Kari L. Machine learning using intrinsic genomic signatures for rapid classification of novel pathogens: Covid-19 case study. Plos one. 2020;15(4):e0232391. doi: 10.1371/journal.pone.0232391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu D, Clemente L, Poirier C, Ding X, Chinazzi M, Davis JT, Vespignani A, Santillana M (2020) “A machine learning methodology for real-time forecasting of the 2019-2020 covid-19 outbreak using internet searches, news alerts, and estimates from mechanistic models,” arXiv:2004.04019
- 10.Rajaraman S, Siegelman J, Alderson PO, Folio LS, Folio LR, Antani SK (2020) “Iteratively pruned deep learning ensembles for covid-19 detection in chest x-rays,” arXiv:2004.08379 [DOI] [PMC free article] [PubMed]
- 11.Wang L, Lin ZQ, Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao ASS, Vazquez JA. Identification of covid-19 can be quicker through artificial intelligence framework using a mobile phone-based survey when cities and towns are under quarantine. Infect Control Hospital Epidemiol. 2020;41(7):826–830. doi: 10.1017/ice.2020.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey G, Chaudhary P, Gupta R, Pal S (2020) “Seir and regression model based covid-19 outbreak predictions in india,” arXiv:2004.00958
- 14.Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ (2020) “Deep-covid: predicting covid-19 from chest x-ray images using deep transfer learning,” arXiv:2004.09363 [DOI] [PMC free article] [PubMed]
- 15.Bandyopadhyay SK, Dutta S (2020) “Machine learning approach for confirmation of covid-19 cases: positive, negative, death and release,” medRxiv
- 16.Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E, Bonten MM, Dahly DL, Damen JA, Debray TP et al. (2020) “Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal,” bmj, vol 369, pp 1–16 [DOI] [PMC free article] [PubMed]
- 17.Shoeibi A, Khodatars M, Alizadehsani R, Ghassemi N, Jafari M, Moridian P, Khadem A, Sadeghi D, Hussain S, Zare A et al. (2020) “Automated detection and forecasting of covid-19 using deep learning techniques: a review,” arXiv:2007.10785
- 18.Guo Y, Liu Y, Oerlemans A, Lao S, Wu S, Lew MS. Deep learning for visual understanding: a review. Neurocomputing. 2016;187:27–48. doi: 10.1016/j.neucom.2015.09.116. [DOI] [Google Scholar]
- 19.Kowsari K, Jafari Meimandi K, Heidarysafa M, Mendu S, Barnes L, Brown D. Text classification algorithms: a survey. Information. 2019;10(4):150. doi: 10.3390/info10040150. [DOI] [Google Scholar]
- 20.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Adv Neural Information Process Syst. 2012;25:1097–1105. [Google Scholar]
- 21.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 22.Roy PK, Singh JP (2020) Predicting closed questions on community question answering sites using convolutional neural network. Neural Comput Appl 32:10555–10572
- 23.Roy PK, Singh JP, Banerjee S. Deep learning to filter sms spam. Future Gener Computer Syst. 2020;102:524–533. doi: 10.1016/j.future.2019.09.001. [DOI] [Google Scholar]
- 24.Yan B, Tang X, Liu B, Wang J, Zhou Y, Zheng G, Zou Q, Lu Y, Tu W (2020) An improved method of covid-19 case fitting and prediction based on lstm arXiv:2005.03446
- 25.Roy PK (2020) Multilayer convolutional neural network to filter low quality content from quora. Neural Process Lett 52:805–821
- 26.Chakraborty T, Ghosh I. "Real-time forecasts and risk assessment of novel coronavirus (covid-19) cases: a data-driven analysis," Chaos. Solitons Fractals. 2020;135:109850. doi: 10.1016/j.chaos.2020.109850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chimmula VKR, Zhang L. Time series forecasting of covid-19 transmission in canada using lstm networks Chaos. Solitons Fractals. 2020;135:109864. doi: 10.1016/j.chaos.2020.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomar A, Gupta N. Prediction for the spread of covid-19 in india and effectiveness of preventive measures. Sci Total Environ. 2020;728:138762. doi: 10.1016/j.scitotenv.2020.138762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamzah FB, Lau C, Nazri H, Ligot DV, Lee G, Tan CL, Shaib M et al (2020) “Coronatracker: worldwide covid-19 outbreak data analysis and prediction. Bull World Health Organ 1(32). 10.2471/BLT.20.255695
- 30.Panwar H, Gupta P, Siddiqui MK, Morales-Menendez R, Singh V. Application of deep learning for fast detection of covid-19 in x-rays using ncovnet. Chaos, Solitons Fractals. 2020;138:109944. doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Z, Ge Q, Jin L, Xiong M (2020) “Artificial intelligence forecasting of covid-19 in china,” arXiv:2002.07112 [DOI] [PMC free article] [PubMed]
- 32.Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks arXiv:2003.10849 [DOI] [PMC free article] [PubMed]
- 33.Pal R, Sekh AA, Kar S, Prasad DK (2020) Neural network based country wise risk prediction of covid-19 arXiv:2004.00959
- 34.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wangping J, Ke H, Yang S, Wenzhe C, Shengshu W, Shanshan Y, Jianwei W, Fuyin K, Penggang T, Jing L, et al (2020) Extended sir prediction of the epidemics trend of covid-19 in italy and compared with hunan, china. Front Med 7:169. 10.3389/fmed.2020.00169 [DOI] [PMC free article] [PubMed]
- 36.Salman FM, Abu-Naser SS, Alajrami E, Abu-Nasser BS, Alashqar BA. Covid-19 detection using artificial intelligence. Int J Acad Eng Res (IJAER) 2020;4(3):18–25. [Google Scholar]
- 37.Tiwari S, Kumar S, Guleria K (2020) Outbreak trends of coronavirus disease–2019 in india: a prediction. Disaster Med Public Health Prep 14(5):e33–e38 [DOI] [PMC free article] [PubMed]
- 38.Ayyoubzadeh SM, Ayyoubzadeh SM, Zahedi H, Ahmadi M, Kalhori SRN. Predicting covid-19 incidence through analysis of google trends data in iran: data mining and deep learning pilot study. JMIR Public Health Surveill. 2020;6(2):e18828. doi: 10.2196/18828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q et al. (2020) Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct Radiology p In Press [DOI] [PMC free article] [PubMed]
- 40.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR (2020) Automated detection of covid-19 cases using deep neural networks with x-ray images Computers Biol Med p 103792 [DOI] [PMC free article] [PubMed]
- 41.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases in Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2097–2106
- 42.Redmon J, Farhadi A (2018) Yolov3: an incremental improvement CoRR, vol abs/1804.02767
- 43.Zhang K, Liu X, Shen J, Li Z, Sang Y, Wu X, Zha Y, Liang W, Wang C, Wang K, et al. Clinically applicable ai system for accurate diagnosis, quantitative measurements, and prognosis of covid-19 pneumonia using computed tomography. Cell. 2020;181(6):1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Hu M, Li Q, Zhang X-P, Zhai G, Yao N (2020) Abnormal respiratory patterns classifier may contribute to large-scale screening of people infected with covid-19 in an accurate and unobtrusive manner arXiv:2002.05534
- 45.Chen Y-C, Lu P-E, Chang C-S, Liu T-H. A time-dependent sir model for covid-19 with undetectable infected persons. IEEE Transaction Netw Sci Eng. 2020;7(4):3279–3294. doi: 10.1109/TNSE.2020.3024723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, Xue Z, Shi Y (2020) Lung infection quantification of covid-19 in ct images with deep learning arXiv:2003.04655
- 47.Shi F, Xia L, Shan F, Wu D, Wei Y, Yuan H, Jiang H, Gao Y, Sui H, Shen D (2020) Large-scale screening of covid-19 from community acquired pneumonia using infection size-aware classification arXiv:2003.09860 [DOI] [PubMed]
- 48.Hemdan EE-D, Shouman MA, Karar ME (2020) Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images arXiv:2003.11055
- 49.Zhang J, Xie Y, Liao Z, Pang G, Verjans J, Li W, Sun Z, He J, Li Y, Shen C et al. (2020) Viral pneumonia screening on chest x-ray images using confidence-aware anomaly detection arXiv:2003.12338 [DOI] [PMC free article] [PubMed]
- 50.Abbas A, Abdelsamea MM, Gaber MM (2020) Classification of covid-19 in chest x-ray images using detrac deep convolutional neural network arXiv:2003.13815 [DOI] [PMC free article] [PubMed]
- 51.Zhao J, Zhang Y, He X, Xie P (2020) Covid-ct-dataset: a ct scan dataset about covid-19 arXiv:2003.13865
- 52.Asnaoui KE, Chawki Y, Idri A (2020) Automated methods for detection and classification pneumonia based on x-ray images using deep learning arXiv:2003.14363
- 53.Jain R, Gupta M, Taneja S, Hemanth DJ. Deep learning based detection and analysis of covid-19 on chest x-ray images. Appl Intell. 2021;51(3):1690–1700. doi: 10.1007/s10489-020-01902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maghdid HS, Asaad AT, Ghafoor KZ, Sadiq AS, Khan MK (2020) Diagnosing covid-19 pneumonia from x-ray and ct images using deep learning and transfer learning algorithms arXiv:2004.00038
- 55.Vaid S, Kalantar R, Bhandari M. Deep learning covid-19 detection bias: accuracy through artificial intelligence. Int Orthop. 2020;44:1539–1542. doi: 10.1007/s00264-020-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khalifa NEM, Taha MHN, Hassanien AE, Elghamrawy S (2020) Detection of coronavirus (covid-19) associated pneumonia based on generative adversarial networks and a fine-tuned deep transfer learning model using chest x-ray dataset arXiv:2004.01184
- 57.Chaganti S, Grenier P, Balachandran A, Chabin G, Cohen S, Flohr T, Georgescu B, Grbic S, Liu S, Mellot F, et al. Automated quantification of ct patterns associated with covid from chest ct. Radiol Artif Intel. 2020;2(4):e200048. doi: 10.1148/ryai.2020200048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hall LO, Paul R, Goldgof DB, Goldgof GM (2020) Finding covid-19 from chest x-rays using deep learning on a small dataset arXiv:2004.02060
- 59.Gozes O, Frid-Adar M, Sagie N, Zhang H, Ji W, Greenspan H (2020) Coronavirus detection and analysis on chest ct with deep learning arXiv:2004.02640
- 60.Afshar P, Heidarian S, Naderkhani F, Oikonomou A, Plataniotis KN, Mohammadi A (2020) Covid-caps: A capsule network-based framework for identification of covid-19 cases from x-ray images arXiv:2004.02696 [DOI] [PMC free article] [PubMed]
- 61.Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, Bernheim A, Siegel E (2020) Rapid ai development cycle for the coronavirus (covid-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis arXiv:2003.05037
- 62.Dandekar R, Barbastathis G (2020) Neural network aided quarantine control model estimation of global covid-19 spread arXiv:2004.02752 [DOI] [PMC free article] [PubMed]
- 63.Karim M, Döhmen T, Rebholz-Schuhmann D, Decker S, Cochez M, Beyan O et al. (2020) Deepcovidexplainer: explainable covid-19 predictions based on chest x-ray images arXiv:2004.04582
- 64.Luz E, Silva P, Silva R, Silva L, Guimarães J, Miozzo G, Moreira G, Menotti D (2021) Towards an effective and efficient deep learning model for covid-19 patterns detection in x-ray images. Res Biomed Eng 1–14. 10.1007/s42600-021-00151-6
- 65.Hu S, Gao Y, Niu Z, Jiang Y, Li L, Xiao X, Wang M, Fang EF, Menpes-Smith W, Xia J, et al. Weakly supervised deep learning for covid-19 infection detection and classification from ct images. IEEE Access. 2020;8:118869–118883. doi: 10.1109/ACCESS.2020.3005510. [DOI] [Google Scholar]
- 66.Wu Y-H, Gao S-H, Mei J, Xu J, Fan D-P, Zhao C-W, Cheng M-M (2020) Jcs: an explainable covid-19 diagnosis system by joint classification and segmentation arXiv:2004.07054 [DOI] [PubMed]
- 67.Brunese L, Mercaldo F, Reginelli A, Santone A. Explainable deep learning for pulmonary disease and coronavirus covid-19 detection from x-rays. Computer Methods Programs Biomed. 2020;196:105608. doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qiu Y, Liu Y, Xu J (2020) Miniseg: an extremely minimum network for efficient covid-19 segmentation arXiv:2004.09750 [DOI] [PubMed]
- 69.Toğaçar M, Ergen B, Cömert Z. Covid-19 detection using deep learning models to exploit social mimic optimization and structured chest x-ray images using fuzzy color and stacking approaches. Computers Biol Med. 2020;121:103805. doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Punn NS, Agarwal S (2020) Automated diagnosis of covid-19 with limited posteroanterior chest x-ray images using fine-tuned deep neural networks arXiv:2004.11676 [DOI] [PMC free article] [PubMed]
- 71.Cabras S (2020) A bayesian-deep learning model for estimating covid-19 evolution in spain arXiv:2005.10335
- 72.Cohen JP, Dao L, Morrison P, Roth K, Bengio Y, Shen B, Abbasi A, Hoshmand-Kochi M, Ghassemi M, Li H et al. (2020) Predicting covid-19 pneumonia severity on chest x-ray with deep learning arXiv:2005.11856 [DOI] [PMC free article] [PubMed]
- 73.Ismael AM, Şengür A. Deep learning approaches for covid-19 detection based on chest x-ray images. Expert Syst Appl. 2021;164:114054. doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Zhao H, Jie Y, Wang R et al. (2020) Deep learning enables accurate diagnosis of novel coronavirus (covid-19) with ct images medRxiv [DOI] [PMC free article] [PubMed]
- 75.Jin C, Chen W, Cao Y, Xu Z, Zhang X, Deng L, Zheng C, Zhou J, Shi H, Feng J (2020) Development and evaluation of an ai system for covid-19 diagnosis medRxiv [DOI] [PMC free article] [PubMed]
- 76.He X, Yang X, Zhang S, Zhao J, Zhang Y, Xing E, Xie P (2020) Sample-efficient deep learning for covid-19 diagnosis based on ct scans medRxiv
- 77.Amyar A, Modzelewski R, Ruan S (2020) Multi-task deep learning based ct imaging analysis for covid-19: classification and segmentation medRxiv [DOI] [PMC free article] [PubMed]
- 78.Fan D-P, Zhou T, Ji G-P, Zhou Y, Chen G, Fu H, Shen J, Shao L. Inf-net: automatic covid-19 lung infection segmentation from ct images. IEEE Transactions Med Imag. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 79.Wang S, Zha Y, Li W, Wu Q, Li X, Niu M, Wang M, Qiu X, Li H, Yu H, et al. A fully automatic deep learning system for covid-19 diagnostic and prognostic analysis. Eur Respir J. 2020;56:1–11. doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Farid AA, Selim GI, Awad H, Khater A. A novel approach of ct images feature analysis and prediction to screen for corona virus disease (covid-19) Int J Sci Eng Res. 2020;11(3):1–9. [Google Scholar]
- 81.Apostolopoulos ID, Aznaouridis SI, Tzani MA. Extracting possibly representative covid-19 biomarkers from x-ray images with deep learning approach and image data related to pulmonary diseases. J Med Biol Eng. 2020;40:462–469. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ndiaye BM, Tendeng L, Seck D (2020) Analysis of the covid-19 pandemic by sir model and machine learning technics for forecasting arXiv:2004.01574
- 83.Pirouz B, Shaffiee Haghshenas S, Shaffiee Haghshenas S, Piro P. Investigating a serious challenge in the sustainable development process: analysis of confirmed cases of covid-19 (new type of coronavirus) through a binary classification using artificial intelligence and regression analysis. Sustainability. 2020;12(6):2427. doi: 10.3390/su12062427. [DOI] [Google Scholar]
- 84.Ribeiro MHDM, da Silva RG, Mariani VC, dos Santos Coelho L. Short-term forecasting covid-19 cumulative confirmed cases: perspectives for brazil Chaos. Solitons Fractals. 2020;135:109853. doi: 10.1016/j.chaos.2020.109853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tuli S, Tuli S, Tuli R, Gill SS. Predicting the growth and trend of covid-19 pandemic using machine learning and cloud computing. Internet Things. 2020;11:10022. doi: 10.1016/j.iot.2020.100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Das S (2020) Prediction of covid-19 disease progression in india: under the effect of national lockdown arXiv:2004.03147
- 87.Yan L, Zhang H-T, Xiao Y, Wang M, Sun C, Liang J, Li S, Zhang M, Guo Y, Xiao Y et al. (2020) Prediction of survival for severe covid-19 patients with three clinical features: development of a machine learning-based prognostic model with clinical data in wuhan medRxiv
- 88.Pourhomayoun M, Shakibi M (2020) Predicting mortality risk in patients with covid-19 using artificial intelligence to help medical decision-making medRxiv [DOI] [PMC free article] [PubMed]
- 89.Rustam F, Reshi AA, Mehmood A, Ullah S, On B-W, Aslam W, Choi GS. Covid-19 future forecasting using supervised machine learning models. IEEE access. 2020;8:101489–101499. doi: 10.1109/ACCESS.2020.2997311. [DOI] [Google Scholar]
- 90.Pinter G, Felde I, Mosavi A, Ghamisi P, Gloaguen R. Covid-19 pandemic prediction for hungary; a hybrid machine learning approach. Mathematics. 2020;8(6):890. doi: 10.3390/math8060890. [DOI] [Google Scholar]
- 91.Ardabili SF, Mosavi A, Ghamisi P, Ferdinand F, Varkonyi-Koczy AR, Reuter U, Rabczuk T, Atkinson PM. Covid-19 outbreak prediction with machine learning. Algorithms. 2020;13(10):249. doi: 10.3390/a13100249. [DOI] [Google Scholar]
- 92.Jiang X, Coffee M, Bari A, Wang J, Jiang X, Huang J, Shi J, Dai J, Cai J, Zhang T, et al. Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. CMC: Computers Mater Continua. 2020;63:537–51. [Google Scholar]
- 93.Barstugan M, Ozkaya U, Ozturk S (2020) Coronavirus (covid-19) classification using ct images by machine learning methods arXiv:2003.09424
- 94.Sujath R, Chatterjee JM, Hassanien AE. A machine learning forecasting model for covid-19 pandemic in India. Stoch Environ Res Risk Assess. 2020;34:959–972. doi: 10.1007/s00477-020-01827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Imran A, Posokhova I, Qureshi HN, Masood U, Riaz S, Ali K, John CN, Nabeel M (2020) Ai4covid-19: Ai enabled preliminary diagnosis for covid-19 from cough samples via an app arXiv:2004.01275 [DOI] [PMC free article] [PubMed]
- 96.Erraissi A, Banane M. Machine learning model to predict the number of cases contaminated by covid-19. Int J Comput Digital Syst. 2020;9:1–11. doi: 10.12785/ijcds/090101. [DOI] [Google Scholar]
- 97.Jamshidi M, Lalbakhsh A, Talla J, Peroutka Z, Hadjilooei F, Lalbakhsh P, Jamshidi M, La Spada L, Mirmozafari M, Dehghani M, et al. Artificial intelligence and covid-19: deep learning approaches for diagnosis and treatment. IEEE Access. 2020;8:109581–109595. doi: 10.1109/ACCESS.2020.3001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Metsky HC, Freije CA, Kosoko-Thoroddsen T-SF, Sabeti PC, Myhrvold C (2020) Crispr-based surveillance for covid-19 using genomically-comprehensive machine learning design BioRxiv