Abstract
Purpose
Artificial intelligence (AI) is playing an ever-increasing role in Neuroradiology.
Methods
When designing AI-based research in neuroradiology and appreciating the literature, it is important to understand the fundamental principles of AI. Training, validation, and test datasets must be defined and set apart as priorities. External validation and testing datasets are preferable, when feasible. The specific type of learning process (supervised vs. unsupervised) and the machine learning model also require definition. Deep learning (DL) is an AI-based approach that is modelled on the structure of neurons of the brain; convolutional neural networks (CNN) are a commonly used example in neuroradiology.
Results
Radiomics is a frequently used approach in which a multitude of imaging features are extracted from a region of interest and subsequently reduced and selected to convey diagnostic or prognostic information. Deep radiomics uses CNNs to directly extract features and obviate the need for predefined features.
Conclusion
Common limitations and pitfalls in AI-based research in neuroradiology are limited sample sizes (“small-n-large-p problem”), selection bias, as well as overfitting and underfitting.
Keywords: Artificial intelligence, Machine learning, Radiomics, Neuroradiology
Introduction
In recent years, artificial intelligence (AI) and machine learning (ML) have become widely used terms that evoke mixed feelings among neuroradiologists. Provocative statements by data scientists and politicians that radiology will become obsolete as a specialty within the next five years and that radiologists should no longer be trained sent shock waves through the field in the middle of the last decade. More than five years later, however, neuroradiologists are far from being replaced by algorithms and aside from temporary decreases due to the COVID pandemic, workloads have continued to rise. The overall attitude toward a mutually beneficial role of AI in radiology has become more favorable among both radiologists and data scientists in the last years.
A recent survey of chest radiologists and data scientists revealed that only a minority expect an obsolescence of radiology, while the majority predict that AI will have a positive impact on radiology in terms of job satisfaction, salaries, and the role of radiologists in society [1]. A survey by the European Society of Radiology of its members demonstrated that the biggest impact of AI was expected in the fields of breast, oncologic, thoracic, and neuroimaging [2]. This survey demonstrates an overall positive attitude of radiologists toward AI. Nevertheless, the current time frame of AI experience in neuroradiology is still limited, and further studies into the impact of AI on the field of neuroradiology will be needed in the future. These may also include analyses of the amounts and potential impact of capital investments and the respective financial projections on neuroradiology.
Accounting for this limited time frame, the impact of AI and ML on everyday clinical practice in neuroradiology has so far been modest. There are several reasons for this limited clinical translation. While AI and ML provide powerful tools for image analysis, most of these methods have initially been developed in the nonmedical field. The translation of these algorithms into the medical context has not been as straightforward as initially anticipated. Commonly, the infrastructure of radiological departments cannot accommodate the implementation of newly developed algorithms [3]. Nevertheless, there are endeavors to increasingly bridge this gap by developing platforms that allow for a more rapid integration of new AI algorithms into the clinical-neuroradiological workflow. In addition, data scientists and clinical neuroradiologists tend to speak a different professional language and misunderstandings often arise where different jargons are used.
Terminology
Artificial intelligence is a broad term denoting the branch of science that deals with machines performing tasks that otherwise require human intelligence. From the perspective of computer science, AI algorithms are not provided with direct instructions on how to perform a task. Using a segmentation task as an example, a conventional model would be explicitly instructed with a threshold to classify pixels of an input image. An AI model, on the other hand, can learn the threshold (or patterns) through examination of a large number of images using parts or the whole image and contextual information depending on the technique used.
Radiomics refers to a quantitative approach to medical imaging utilizing spatial distribution of signal intensities, such as entropy patterns, and other inherent imaging-based data that are not perceptible to the human eye. The underlying rationale for the use of radiomics is the assumption that electronic medical images contain information beyond visual perception that better reflect tissue properties and may improve diagnostic or prognostic accuracy [4, 5].
Machine learning is a field of AI in which algorithms are trained using known datasets, from which the machine “learns.” The developed algorithm then applies this knowledge to perform diagnostic tasks in unknown datasets [6].
Deep learning is a form of AI that is modelled on the structure of neurons of the brain. It utilizes artificial neural networks with multiple “hidden” layers to solve complex problems [7]. These “hidden layers” enable the machine to continually learn and incorporate newly acquired knowledge to improve its performance [7, 8]. Deep Learning can be unsupervised, semi-supervised, or supervised.
It is important to remember that radiomics, machine learning, and deep learning are not separate entities, but are often intricately intertwined. Methods of deep learning are, for example, commonly used in radiomics pipelines.
History of artificial intelligence in radiology
The field of AI in science and research can be traced as far back as 1950, when Alan Turing (considered by many to be the “father of AI”), questioned in his landmark paper: “Can machines think?” [9] This prompted researchers like John McCarthy to engage in a summer research project, to answer whether a machine can simulate human intelligence. McCarthy is credited with coining the term “artificial intelligence.” [9] In the same decade, pioneering work done by Frank Rosenblatt led to the construction of the “perceptron,” a machine intended for image recognition, built on principles of ML and neural networks [10]. The term “machine learning” was coined by Arthur Samuel in 1959 [11]. Several limitations of the perceptron were highlighted in the 1969 book (updated in 1988) by fellow AI scientists, Marvin Minsky and Seymour Papert, which purportedly led to a decrease in funding for AI research in the 1970s and 1980s—the so-called AI winter [12].
However, by this time, the potential for machine or computer-assisted diagnosis in radiology had already begun to be recognized. In their seminal paper highlighting the role that logic and probability play in physicians’ reasoning and approach to a patient’s case, Ledley et al. state that “errors in differential diagnosis result more frequently from errors of omission than from other sources” and proposed that computers may especially be suited to process information to aid the physician and avoid errors of omission [13]. In the early 1960s, the idea that computers could aid radiologists in image interpretation gained further traction when Lodwick et al. presented a method of digitizing plain radiographs as a basis for future exploration of computer-aided detection in lung cancer and also in prediction of disease activity [14]. The concepts of computer-aided detection (CAD) systems were introduced to radiology literature in the 1960s [15, 16]. The 1980s saw the introduction of image segmentation [17]. It was not until access to big data and an exponential increase in computational power in the 1990s and 2000s, however, that advanced artificial neural networks were able to “learn” and provide more diagnostic information beyond detection and segmentation alone. Over the last decade, publications in the field of AI in radiology has increased 7–eightfold when compared to the 2000s [18].
Study design for AI-based research in neuroradiology
When designing an AI-based study in neuroradiology, it is important to formulate a goal that is both relevant and feasible. The study team should be multi-disciplinary and include both neuroradiologists and data scientists [19, 20]. The aims of the study should be thoroughly discussed among the members of the study team. Generally, the neuroradiologist/clinician should provide input on the clinical relevance of the study, and the data scientist should comment on the feasibility of the project and the methods needed to reach the proposed goals. Members of the study team should reflect the specific needs of the project: While clinical expertise is required to select the patient cohort and provide relevant clinical information (e.g., date of diagnosis, type and duration of therapy, response to therapy), neuroradiologists are needed to select the appropriate imaging examinations and to define the regions of interest. Technical expertise is required for data de-identification and/or anonymization and other important steps throughout the project [21]. Data scientists are crucial to select the correct models and algorithms to ascertain that the developed algorithms will be robust and generalizable. From the study team, a project lead should be nominated who plans, coordinates, and supervises the project with the rest of the team.
Following the definition of the scope of the study, the study team must seek approval from the respective institutional review boards (IRB) or ethics committees according to local/national regulations. Regional, national, or transnational regulations such as the Health Insurance Portability and Accountability Act (HIPAA) in the USA or the General Data Protection Regulation in the European Union and European Economic Area must be adhered to [22]. Following IRB approval, the study cohort and (if needed) a control cohort must be defined using appropriate inclusion and exclusion criteria. The study cohort usually shares common features including demographic or disease characteristics and availability of imaging exams and the respective “ground truth.” This can be, for example, a tumor type, a molecular subtype, a laboratory value, response to therapy, progression-free survival, or overall survival. Following data de-identification, imaging data can often be made accessible from the local picture and archiving communication system (PACS). Imaging studies need to be explored, and a quality control needs to be performed to ensure sufficient image quality, and availability of the necessary images, e.g., CT contrast phases or MRI sequences needed for the particular study.
After verification of the available clinical and imaging data and ground truth, the study team must begin the labeling process (image annotation or delineation of lesion), which is a crucial step in the study. Depending on the scope of the study, neuroradiologists and other qualified members of the study team may need to:
Provide segmentation for a lesion
Annotate images with abnormal findings
Classify abnormalities according to radiological criteria [20, 23]
The type of label may vary depending on the scope of the study, but usually requires neuroradiological expertise. The labeled imaging data are then used as input for the machine learning model, while the ground truth serves as the reference that the model is expected to learn. The ground truth can be defined by pathologic, clinical, or imaging criteria [20]. Pathologic criteria may be derived from histopathology or molecular pathology. Clinical references may consist of follow-up exams, recurrent disease, or survival. Imaging ground truth may include alternative imaging modalities, follow-up imaging exams, response to therapy, or neuroradiologic reports. After data annotation, training, validation, and test datasets must be defined. The training dataset is used to optimize the machine learning model through minimizing loss (the error of predicted results compared to ground truth). Once the model is trained, a test dataset is used to test the performance of the model. The validation dataset is a separate cohort that is used to fine tune the model. In conventional machine learning algorithms such as SVM and RFs, the hyperparameters (e.g., number of trees in RF) must be fine-tuned using a validation dataset to maximize the performance on the test set. In deep learning, the validation cohort is used to determine the stoppage point for the training (optimization) of the model. If the optimization process is not stopped at a proper point, the model will overfit the training data meaning that it will memorize the training dataset and thus will be unable to perform well on the test dataset (poor generalizability). With large study cohorts, commonly used ratios are 80% of the dataset for the training cohort, 10% of the dataset for the validation cohort, and 10% of the dataset for the test cohort [20]. The validation and test datasets are set aside, while the algorithm is being trained. When there is only a limited sample size available, k-fold cross-validation is often applied [24]. When possible, external validation and testing datasets, i.e., datasets acquired at a different institution, are preferable as they allow for a more robust evaluation of the developed algorithm.
Together with data scientists, the specific type of learning process (supervised vs. unsupervised) and machine learning model (CNN, recurrent neural network (RNN), random forest (RF), among others) is then selected. Depending on the model type and available data, hyperparameter fine tuning can be conducted using the validation cohort to improve the model’s prediction. Eventually, several performance metrics assess the model’s performance using the prediction on the test dataset. These metrics commonly include sensitivity, specificity, accuracy, precision, recall, and F1 score. For classification tasks, the receiver operating characteristic (ROC) curve graphically illustrates the diagnostic performance at various thresholds, while the numeric value of the area under the ROC curve (AUC) can be used to compare the performance of different machine learning models [20, 25]. For segmentation tasks, Dice or Jaccard coefficients are calculated as a similarity metric at the pixel level [26].
Radiomics and deep radiomics
Radiomics is a frequently used analytic methodology in neuroradiological research. The general workflow in radiomics typically involves the following steps [4, 27]:
Image acquisition or retrieval
Identification of a region or volume of interest through manual, semi-automated, or automated delineation or segmentation
Extraction of quantitative image features from this region of interest, e.g., the size or shape or texture parameters within the region of interest
Statistical analyses of the image features
Assessment for a potential association between the radiomic features and a clinically relevant endpoint
A typical radiomics pipeline is shown in Fig. 1. As with any other research project, IRB/ethics committee approval must be obtained, and endpoints and inclusion and exclusion criteria need to be defined. Subsequently, the first step in a radiomics project in neuroradiology is to acquire images or, more frequently, to retrospectively identify image datasets that satisfy the inclusion and exclusion criteria. These images can, in principle, be any medical images. In neuroradiological radiomics, CT or MR images are most commonly used. It is important to avoid selection bias to ascertain generalizability of the results. To avoid selection bias, the included patients/datasets should not be “cherry-picked.” Clear inclusion and exclusion criteria need to be defined, and consecutive patients meeting these criteria should be included. It needs to be clear how the final sample was defined for the different datasets. The next step is to identify the region or volume of interest. In neuroradiology, this region of interest can, for example, be a brain tumor or even just a component of a brain tumor (e.g., the contrast-enhancing part of a brain tumor). As feature extraction yields data as continuous variables, radiomics provides a wider scope to capture detailed features of tumors when compared to subjective visual assessment alone [28]. The boundaries of the pre-specified region of interest must be defined [29]. Most commonly, this is done manually by a team member adept at interpreting these images. In neuroradiological projects, this is usually a neuroradiologist or neuroradiology fellow/trainee. When the radiomics research is published, the individuals who defined the regions of interest should be specified, including their levels of experience. This is also important when interpreting the results of a radiomics study, as the definition of boundaries can decisively influence the results and generalizability.
Fig. 1.
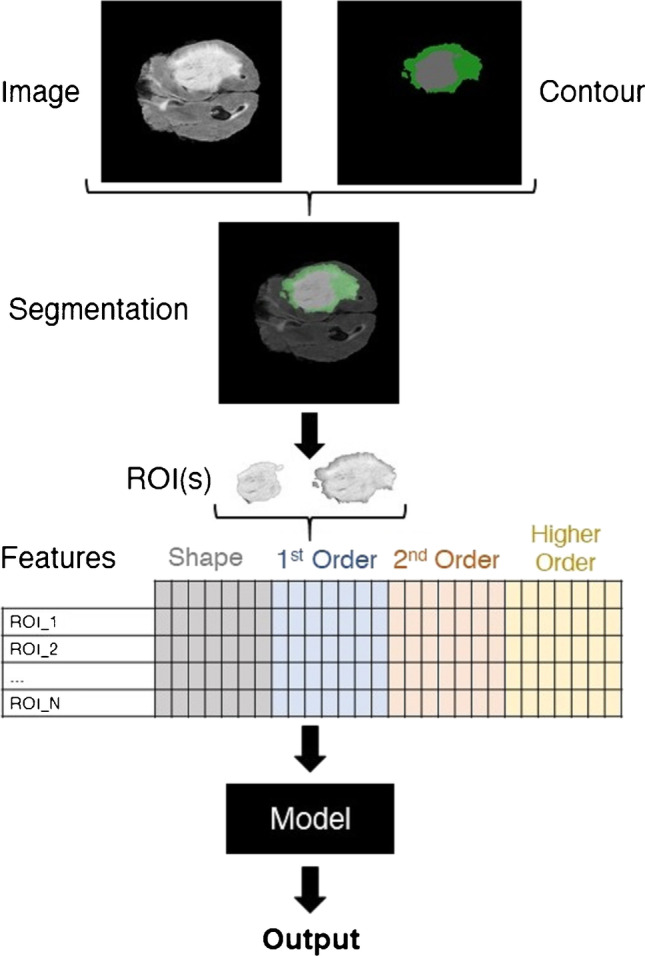
Example of a typical radiomics pipeline. Regions of interests (ROI) are created based on the neuroradiological images and binary masks are created. The corresponding radiomics features are extracted through applying predefined formulae to ROI numerical representations. A model is used to infer the output based on the input radiomics. The task for which the pipeline is implemented determines type of output. Classification, risk score assessment (regression), and survival analysis are the most common purposes of radiomics-based pipelines
Next, quantitative imaging features, i.e., radiomic features, are extracted and computed from the regions of interest. Traditionally, predefined radiomic features have been used to capture different traits of the regions of interest, such as texture, size, and shape, which may characterize the phenotype and potentially genotype of the tissue. Radiomic features usually include first and second-order statistical features. Standard libraries such as PyRadiomics are frequently used by the radiomics scientific community [30]. Once the features are calculated and prognostic features are selected, a predictive model such as binary classification (e.g., support vector machine (SVM) or a survival model (e.g., Cox regression) is developed for a given endpoint. Endpoints in neuroradiology are commonly the classification into different tumor types or molecular subtypes, or the prediction of response to therapy, progression-free survival, or overall survival. Recently, CNNs have been used to directly extract (deep radiomic) features from ROIs and, thus, forego the need for predefined features, a process which is also referred to as deep radiomics [31].
The main difference between radiomics and deep radiomics lies in how the features are calculated. In a radiomics pipeline, features are extracted based on predefined mathematical equations, designed by image analysis experts. Deep radiomics, on the other hand, are in fact weights of CNNs that are automatically set during the training phase and tailored to the specific task (e.g., molecular subtype of a brain tumor).
Preprocessing and feature selection
As specified by the Quantitative Imaging Biomarkers Alliance (QIBA), reproducibility of radiomic features is defined as repeat measurements in different settings, including at different locations, or with different operators or scanners [32, 33]. Imaging data irreproducibility is one of the main reasons for a significant variability in radiomic biomarkers, which may lead to unreliable and inaccurate predictive models based on radiomics [33]. Preprocessing is an important step to maximize the reproducibility of radiomic features. Most radiomic studies use conventional intensity normalization methods such as Gaussian, Z-score, and histogram matching in MR and CT images and bias field correction in MR images [34]. Normalization methods, such as Z-score, can be applied to the images, the extracted radiomic features, or both of these.
There are usually hundreds, if not thousands, of radiomic features that can be calculated for each region of interest. A large of number of these feature may be highly correlated with other features and thus be redundant. In addition, not all features carry predictive value with respect to the endpoint and may therefore lead to poor predictive performance. Having a large number of features for a relatively small number of regions of interest is referred to as the small-n-large-p problem, which occurs commonly in neuroradiological AI-based research and may lead to underfitting or overfitting.
Two main classes of feature reduction methods are used:
Unsupervised methods
Supervised methods
In unsupervised methods, features are removed based on their relationship (e.g., correlation) with other features independent of predictive value. Examples for unsupervised methods include principal component analysis (PCA), correlation matrix, and zero variance and near-zero variance.
In supervised methods, features are reduced based on the target variable. Least absolute shrinkage and selection operator (LASSO) and maximum relevance-minimum redundancy (mRMR) are popular supervised methods for feature reduction. LASSO employs a regression analysis on training data and removes features whose coefficients are zero. mRMR methods include a similar mechanism to add a feature into the selected set of features if it contributes to the outcome. However, the feature will not be appended to the set if it creates redundancy in conjunction with another feature in the set. In this context, linear correlation is the simplest form of redundancy.
Radiomics pipelines are used for a broad range of applications such as diagnosis, e.g., of brain tumor types or molecular subtypes, and prognosis, e.g., progression-free survival or overall survival. Consequently, the output might be a class label (e.g., tumor subtype), a risk score (e.g., treatment response), a time to event (in survival analysis pipelines), or even features which are aimed to be used in a hybrid pipeline (i.e., where other features such as clinical variables or deep features are combined).
A wide variety of models can be utilized in radiomic pipelines. For classification purposes, support vector machines (SVM), random forests, XGBoost, and feed-forward neural networks are the typical options. Cox regression has been widely used in survival analysis projects; however, other models such as survival trees do exist.
SVM models are large margin classifiers. They try to draw a border to classify data points into different classes, while the data points are in furthest possible distance from the border. Basic SVM models are linear, but if they are equipped with kernels, they become nonlinear. Kernels represent a means of mapping the data to a higher-dimensional space.
Decision trees are building blocks of random forests. Decisions trees are in fact nested if-else conditions. They repeatedly divide the data based on a threshold and value of one feature. Decision trees often suffer from overfitting. To tackle their overfitting problem, random forests ensemble multiple decision trees. As the naming implies, XGboost is a boosting-based algorithm. It iteratively adds decision trees in a way that each tree lowers the error of its previous trees. Tree-based models are popular options for classification of tabular data, such as radiomics.
A perceptron is a linear classifier which is capable of finding a line such that it separates different classes of data, assuming such a line exists. An activation function is a mapping which is applied to output of a perceptron and makes it nonlinear. Multiple perceptrons and their activation functions form a layer. Stack of layers makes a feed-forward neural network, sometimes called artificial neural network (ANN). Hence, ANNs are highly nonlinear classifiers.
Survival analysis is a distinct domain for machine learning models. It deals with censored data which can be imagined as a combination of an event’s occurrence (binary outcome) and its expected duration (time to event). Survival trees utilize a thresholding approach to fulfil the task, whereas cox regression establishes a baseline hazard and provides estimated hazard ratios at different times [35].
Machine learning
Machine learning algorithms can be divided into three areas:
Supervised learning
Unsupervised learning
Reinforcement learning
Supervised learning is the most prevalent approach in different application areas of AI including neuroradiology [36]. Supervised learning relies on labeled datasets. Depending on the task, the label might be a class, a continuous score, or even an entire image (e.g., pixel-wise labels or contours for segmentation pipelines). Unlike supervised techniques, unsupervised learning does not need labeled data [37, 38]. These techniques aim to uncover hidden structures of the dataset which makes them well suited for clustering, dimensionality reduction, and data visualization tasks. Examples of unsupervised techniques include superpixels, k-nearest neighbors (kNN), principal component analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE), and UMAP. Superpixels and kNN are utilized to cluster pixels of an image and have potential for segmentation tasks. PCA, t-SNE, and UMAP are dimensionality reduction techniques which are often used for feature fusion or data visualization goals.
Real-world machine learning has not necessarily conformed to the classical definitions. To battle existing limitations and to satiate application-specific demands, multiple machine learning branches have been formed. Semi-supervised learning is an established field where the model is trained on examples of a single class, i.e., to learn the distribution of healthy or abnormal cases [39]. Anomaly detection, normalization, and distribution mapping are typical use-cases of semi-supervised algorithms. Generative adversarial networks (GANs) and Gaussian mixture model (GMM) are two examples of semi-supervised methods. In comparison to supervised approaches, semi-supervised classifiers often land at a lower level in terms of performance. Nonetheless, when annotating the data is not feasible, semi-supervised models are often helpful. Active learning is another branch of ML where the model attempts to identify and exploit informative examples of the training dataset to reduce the size of the required training cohort and avoid being misled by outliers [40]. The informativeness of data in neuroradiology may be negatively influenced by factors such as high level of noise, scanner inconsistencies, and multi-institutional datasets. Therefore, active learning has great potential for neuroimaging applications. Dynamic learning refers to fine-tuning the model over time. Dynamic learning can help AI models to be continuously augmented based on expert’s feedback. Transfer learning is where a model trained with another dataset is fine-tuned over the target dataset [41, 42]. Transfer learning is an effective approach when sufficient data is not available to train the model from scratch.
Convolutional neural networks
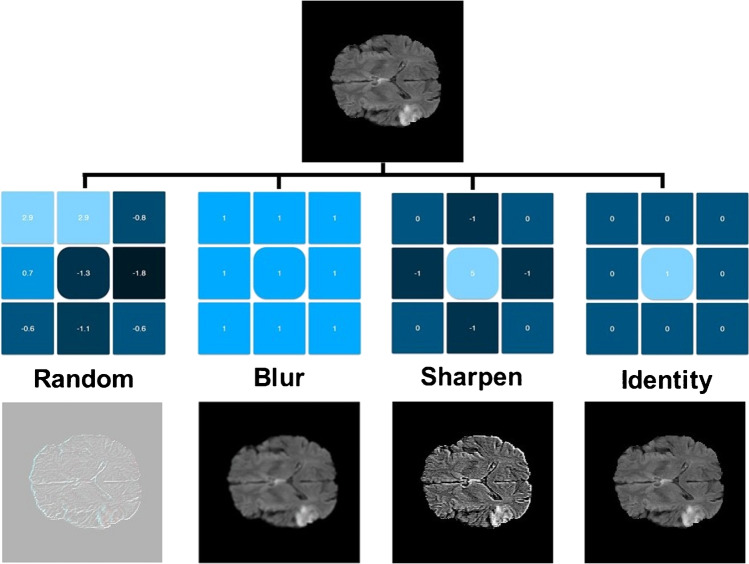
Convolutional neural networks (CNNs) are the basis of deep learning methods which excel at pattern recognition. They are useful for solving complex patterns from imaging data and are considered more robust when compared to conventional algorithms. Data-driven approaches based on CNNs do not require prior image interpretation or definition by human experts [43]. Figure 2 demonstrates a schematic drawing of a typical CNN. Kernel methods are frequently used for image processing. From a mathematical standpoint, kernels are matrices which are applied to images through convolution operation. The original image is separated and simplified into different feature representations by applying different kernels. In the training phase, the model learns how to weigh these feature representations in order to make the correct classification. Predefined kernels are available for different image processing operations such as blurring, sharpening, darkening, brightening, and edge detection. Examples of kernels are shown in Fig. 3. The core idea of CNNs is to learn advanced kernels from their training dataset. This has helped CNNs to surpass conventional neural networks in terms of performance in two ways:
CNNs have lower numbers of trainable parameters and thus are less prone to overfitting.
CNNs are not sensitive to location of the driving objects or patterns within the image.
Fig. 2.
Schematic of a convolutional neural network. Convolutional neural networks consist of convolution layers and fully connected layers, also known as dense layers. The convolution layers serve as feature extractors, and the fully connected layers are classifiers. Output of the network depends on the target task. For an N-class classification scenario, the network has N nodes in its output layer. Each of these will generate the probability of the input image belonging to their corresponding class
Fig. 3.
Utilizing kernels to manipulate images. Kernels are the essence of convolutional neural networks. These are predefined matrices customized for specific tasks such as sharpening and blurring images. In convolutional neural networks, the idea is to learn multiple kernels and utilize them to extract informative features from the input images (images were created using https://github.com/generic-github-user/Image-Convolution-Playground)
Multiple variations of CNNs have been devised to perform different tasks. AlexNet, ResNet, and DenseNet are three well-known architectures for classification ends. U-Net belongs to the family of models called variational auto-encoders (VAE) which is a popular option for segmentation. While a classifier such as AlexNet converts the input image into a set of probabilities that indicate the category (class or label) of the image (e.g., cancer vs. benign), U-Net generates a new image that highlights the ROI. The architecture of classifiers often includes down-sampling blocks whereas U-Net features both down-sampling and up-sampling elements to construct an image as output. Generative adversarial networks (GANs) comprise two main parts: generator and discriminator that are trained simultaneously to learn the distribution of the training dataset and generate never-before-seen images that lie within the same distribution as the training images. This enables them to be utilized for anomaly detection, image normalization, and distribution transformation.
It is an area of discussion whether CNNs or radiomics are preferable in the field of medical imaging and neuroradiology. A major incentive for favoring radiomics is the black-box nature of CNNs—the basis of the output of the CNN is not always readily explicable. However, not every radiomics pipeline is easily comprehensible either. The interpretation of second and higher-order radiomics features can be quite challenging. Some feature reduction methods such as PCA, on the other hand, may fuse different features in a non-reversible way. Finally, the model itself may not be transparent. If a feed-forward neural network is used, for example, it can be difficult to determine how much each feature contributed to the output.
Potential pitfalls in artificial intelligence–based studies in neuroradiology
There are multiple potential pitfalls that need to be considered not only when designing an AI-based study in neuroradiology, but also when interpreting the literature.
First and foremost, the research question must be important and address a relevant unsolved question and/or an unmet need. Sufficient data must be available to address the chosen question, and it must be kept in mind that AI-based research is generally “data-hungry.”
AI-based algorithms in neuroradiology must be repeatable and generalizable. There are multiple factors, however, that can limit the generalizability to another setting. For example, recently, Hoebel et al. found that normalization and intensity quantization have a significant effect on repeatability and redundancy of radiomic features [44]. Others found that certain radiomics feature classes were not equally robust across various acquired sequences and that classification accuracy is strongly influenced by image resolution [45, 46]. The lack of standardization of sequence acquisition, radiomic features extraction, and normalization is one of the main reasons why translation of this research into the real-world clinical and neuroradiological scenario is still hampered [46]. From the initial phases of devising the study design, it is important to avoid selection bias. Inclusion and exclusion criteria need to be clearly defined and adhered to, and it is imperative that these criteria lead to an unbiased study sample that can be generalized beyond one’s own institution. When feasible, external validation and test datasets are ideal to demonstrate the robustness and generalizability of the algorithm outside one’s own institution.
When placing the boundaries of the region or volume of interest, it needs to be ascertained that these boundaries are drawn correctly, consistently, and in a fashion that is appropriate for the research question that is to be addressed. Regions of interest should be placed by individuals well-versed in image interpretation.
The small-n-large-p problem inherent to most neuroradiological research projects and almost all AI-based neuroradiological research necessitates the need to employ strategies to mitigate the effects of limited sample sizes. It is important to avoid overfitting and underfitting of the model as much as feasible. In overfitting, the model has “overlearned and memorized” the specifics of the training dataset and does not perform well with previously unseen datasets, i.e., with the validation and test datasets. This means that it will likely not perform well in a real-world scenario. When underfitting occurs, the model has not learned enough with the training and is still in need to learn further with additional data. Figure 4 demonstrates a schematic for an optimum point in training, validation, and test cohorts in relation to the number of iterations. In retrospective studies, the ideal case is to have separate training, validation, and test cohorts representing the true distribution (in statistical language, representative independent samples of the population). A capable machine learning model will overfit to the training cohort as the number of iterations grows. It is translated to training error being converged to zero. During the training process, the validation cohort is monitored, and the training is stopped where the validation loss is minimized. Underfitting refers to the situation in which the model can still perform better on the validation set. Overfitting, on the other hand, is where the model is performing well on the training cohort and poor on the validation. The test cohort is kept unseen until the final evaluation of the model. If the cohorts are representative, minimum points of test and validation errors will correspond to each other. In the real world, especially in the domain of medical imaging, training, validation, and test errors are noisy. There might be a residual error in the training cohort, and validation error may fluctuate around its optimal point. Furthermore, optimum points of validation and test cohorts may not correspond to each other.
Fig. 4.

Schematic for an optimum point in training, validation, and test cohorts in relation to the number of iterations
Perhaps one of the best examples of pitfalls in AI-based studies in general are machine learning studies of chest CTs and radiographs to detect and prognosticate the coronavirus disease 2019 (COVID-19). In a recent meta-analysis of 2,212 studies published between January 1 and October 3, 2020, no machine learning model was found to be of potential clinical use due to methodological flaws and/or underlying biases [47]. A common shortcoming was failure to adhere to the mandatory criteria from the checklist for artificial intelligence in medical imaging (CLAIM) [48]. CLAIM serves as a guide to authors in presenting AI research in medical imaging and provides a framework addressing key concerns when reviewing manuscripts [48]. Further shortcomings were lack of external validation; missing sensitivity analysis of the reported model; failure to report demographics in data partitions; statistical tests used to assess significance of results or determine confidence intervals; and insufficient reporting of limitations, biases, or generalizability [48]. While this meta-analysis was targeted to evaluate AI-based studies of the chest for the prediction and prognosis of COVID-19, the issues and shortcomings reported apply in principle also to neuroimaging and underline the points made above. And last but not least, the ease of use and integration into the neuroradiologist’s workflow is an important prerequisite for a successful clinical translation. Algorithms that are not user friendly and require cumbersome offline processing usually fail to bridge the translational gap into the clinical realm.
Conclusion
Artificial intelligence has highlighted unprecedented opportunities in neuroradiological research. Imaging features beyond visual perception are used to generate diagnostic and prognostic information. There currently is a translational gap between AI-based research and clinical applications in the real-world neuroradiological setting, which will need be increasingly bridged in the future.
Abbreviations
- AI
Artificial intelligence
- AUC
Area under the ROC curve
- CNN
Convolutional neural networks
- CT
Computed tomography
- GAN
Generative adversarial networks
- LASSO
Least absolute shrinkage and selection operator
- ML
Machine learning
- MRI
Magnetic resonance imaging
- mRMR
Maximum relevance-minimum redundancy (mRMR)
- PCA
Principal component analysis
- ROC
Receiver operating characteristic
- ROI
Region of interest
- SVM
Support vector machine
- VAE
Variational auto-encoders
Author contribution
Matthias W. Wagner: literature search, critical review of the literature, writing of the manuscript, reviewing of the manuscript.
Khashayar Namdar: literature search, critical review of the literature, writing of the manuscript, reviewing of the manuscript.
Asthik Biswas: literature search, critical review of the literature, writing of the manuscript, reviewing of the manuscript.
Suranna Monah: literature search, critical review of the literature, writing of the manuscript, reviewing of the manuscript.
Farzad Khalvati: literature search, critical review of the literature, writing of the manuscript, reviewing of the manuscript.
Birgit B. Ertl-Wagner: literature search, critical review of the literature, writing of the manuscript, reviewing of the manuscript.
Matthias W Wagner and Khashayar Namdar contributed equally.
Farzad Khalvati and Birgit B. Ertl-Wagner contributed equally.
Funding
No funding was received for this review article.
Availability of data and material
Not applicable; this is a review article.
Code availability
Not applicable; this is a review article.
Declarations
Ethics approval.
No ethics approval was required for this review article. All ethical standards were adhered to.
Consent to participate.
Not applicable; this is a review article that did not involve enrolment of participants.
Consent for publication.
All authors consent to publication.
Conflict of interest
There is no conflict of interest for this review article for any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Eltorai AEM, Bratt AK, Guo HH. Thoracic radiologists' versus computer scientists' perspectives on the future of artificial intelligence in radiology. J Thorac Imaging. 2020;35:255–259. doi: 10.1097/RTI.0000000000000453. [DOI] [PubMed] [Google Scholar]
- 2.European Society of Radiology (ESR) Impact of artificial intelligence on radiology: a EuroAIM survey among members of the European Society of Radiology. Insights Imaging. 2019;10:105. doi: 10.1186/s13244-019-0798-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weikert T, Cyriac J, Yang S, Nesic I, Parmar V, Stieltjes B. A practical guide to artificial intelligence-based image analysis in radiology. Invest Radiol. 2020;55:1–7. doi: 10.1097/RLI.0000000000000600. [DOI] [PubMed] [Google Scholar]
- 4.Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, Bellomi M. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2:36. doi: 10.1186/s41747-018-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging. 2020;11:91. doi: 10.1186/s13244-020-00887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. Radiographics. 2017;37:505–515. doi: 10.1148/rg.2017160130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP. Deep learning in neuroradiology. AJNR Am J Neuroradiol. 2018;39:1776–1784. doi: 10.3174/ajnr.A5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaka H, Zhang E, Khan N. Artificial intelligence and deep learning in neuroradiology: Exploring the New Frontier. Can Assoc Radiol J. 2021;72:35–44. doi: 10.1177/0846537120954293. [DOI] [PubMed] [Google Scholar]
- 9.McCarthy J, Minsky ML, Rochester N, Shannon CE. A proposal for the Dartmouth Summer Research Project on Artificial Intelligence, August 31, 1955. AIMag. 2006;27(4):12. [Google Scholar]
- 10.Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev. 1958;65:386–408. doi: 10.1037/h0042519. [DOI] [PubMed] [Google Scholar]
- 11.Samuel AL. Some studies in machine learning using the game of checkers IBM. J Res Dev. 1959;3:210–229. [Google Scholar]
- 12.Eberhart RC, Dobbins RW. Early neural network development history: the age of Camelot. IEEE Eng Med Biol Mag. 1990;9:15–18. doi: 10.1109/51.59207. [DOI] [PubMed] [Google Scholar]
- 13.Ledley RS, Lusted LB. Reasoning foundations of medical diagnosis; symbolic logic, probability, and value theory aid our understanding of how physicians reason. Science. 1959;130:9–21. doi: 10.1126/science.130.3366.9. [DOI] [PubMed] [Google Scholar]
- 14.Lodwick GS, Keats TE, Dorst JP. The coding of roentgen images for computer analysis as applied to lung cancer. Radiology. 1963;81:185–200. doi: 10.1148/81.2.185. [DOI] [PubMed] [Google Scholar]
- 15.Myers PH, Nice CM, Becker HC, et al. Automated computer analysis of radiographic images. Radiology. 1964;83:1029–1033. doi: 10.1148/83.6.1029. [DOI] [PubMed] [Google Scholar]
- 16.Winsberg F, Elkin M, May J, et al. Detection of radiographic abnormalities in mammograms by means of optical scanning and computer analysis. Radiology. 1967;89:211–215. doi: 10.1148/89.2.211. [DOI] [Google Scholar]
- 17.Haralick RM, Shapiro LG. Image segmentation techniques. Computer Vision, Graphics, and Image Processing. 1985;29(1):100–132. doi: 10.1016/S0734-189X(85)90153-7. [DOI] [Google Scholar]
- 18.Pesapane F, Codari M, Sardanelli F. Artificial intelligence in medical imaging: threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur Radiol Exp. 2018;2:35. doi: 10.1186/s41747-018-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nancarrow SA, Booth A, Ariss S, Smith T, Enderby P, Roots A. Ten principles of good interdisciplinary team work. Hum Resour Health. 2013;11:19. doi: 10.1186/1478-4491-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montagnon E, Cerny M, Cadrin-Chênevert A, Hamilton V, Derennes T, Ilinca A, Vandenbroucke-Menu F, Turcotte S, Kadoury S, Tang A. Deep learning workflow in radiology: a primer. Insights Imaging. 2020;11:22. doi: 10.1186/s13244-019-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaremko JL, Azar M, Bromwich R, Lum A, Alicia Cheong LH, Gibert M, Laviolette F, Gray B, Reinhold C, Cicero M, Chong J, Shaw J, Rybicki FJ, Hurrell C, Lee E, Tang A, Canadian Association of Radiologists (CAR) Artificial Intelligence Working Group Canadian Association of Radiologists White Paper on ethical and legal issues related to artificial intelligence in radiology. Can Assoc Radiol J. 2019;70(2):107–118. doi: 10.1016/j.carj.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Custers B, Dechesne F, Sears AM, Tani T, van der Hof S. A comparison of data protection legislation and policies across the EU. Comput Law Secur Rev. 2018;34(2):234–243. doi: 10.1016/j.clsr.2017.09.001. [DOI] [Google Scholar]
- 23.Rios Velazquez E, Meier R, Dunn WD, Jr, Alexander B, Wiest R, Bauer S, Gutman DA, Reyes M, Aerts HJ. Fully automatic GBM segmentation in the TCGA-GBM dataset: Prognosis and correlation with VASARI features. Sci Rep. 2015;5:16822. doi: 10.1038/srep16822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez JD, Pérez A, Lozano JA. Sensitivity analysis of kappa-fold cross validation in prediction error estimation. IEEE Trans Pattern Anal Mach Intell. 2010;32:569–575. doi: 10.1109/TPAMI.2009.187. [DOI] [PubMed] [Google Scholar]
- 25.Walter SD. (2005) The partial area under the summary ROC curve. Stat Med. 2005;24(13):2025–2040. doi: 10.1002/sim.2103. [DOI] [PubMed] [Google Scholar]
- 26.Eelbode T, Bertels J, Berman M, Vandermeulen D, Maes F, Bisschops R, Blaschko MB. Optimization for medical image segmentation: theory and practice when evaluating with dice score or Jaccard index. IEEE Trans Med Imaging. 2020;39:3679–3690. doi: 10.1109/TMI.2020.3002417. [DOI] [PubMed] [Google Scholar]
- 27.Khalvati F, Zhang Y, Wong A, Haider MA. “Radiomics”, Encyclopedia of. Biomed Eng. 2019;2:597–603. [Google Scholar]
- 28.Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O. Deep learning with convolutional neural network in radiology. Jpn J Radiol. 2018;36(4):257–272. doi: 10.1007/s11604-018-0726-3. [DOI] [PubMed] [Google Scholar]
- 29.Li Q, Bai H, Chen Y, Sun Q, Liu L, Zhou S, Wang G, Liang C, Li ZC. A fully-automatic multiparametric radiomics model: towards reproducible and prognostic imaging signature for prediction of overall survival in glioblastoma multiforme. Sci Rep. 2017;7:14331. doi: 10.1038/s41598-017-14753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, Bussink J, Gillies RJ, Mak RH, Aerts HJWL. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711. doi: 10.1371/journal.pmed.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raunig DL, McShane LM, Pennello G, Gatsonis C, Carson PL, Voyvodic JT, Wahl RL, Kurland BF, Schwarz AJ, Gönen M, Zahlmann G, Kondratovich MV, O'Donnell K, Petrick N, Cole PE, Garra B, Sullivan DC, QIBA Technical Performance Working Group Quantitative imaging biomarkers: a review of statistical methods for technical performance assessment. Stat Methods Med Res. 2015;24:27–67. doi: 10.1177/0962280214537344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park JE, Park SY, Kim HJ, Kim HS. Reproducibility and generalizability in radiomics modeling: possible strategies in radiologic and statistical perspectives. Korean J Radiol. 2019;20:1124–1137. doi: 10.3348/kjr.2018.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carré A, Klausner G, Edjlali M, Lerousseau M, Briend-Diop J, Sun R, Ammari S, Reuzé S, Alvarez Andres E, Estienne T, Niyoteka S, Battistella E, Vakalopoulou M, Dhermain F, Paragios N, Deutsch E, Oppenheim C, Pallud J, Robert C. Standardization of brain MR images across machines and protocols: bridging the gap for MRI-based radiomics. Sci Rep. 2020;10:12340. doi: 10.1038/s41598-020-69298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh R, Mukhopadhyay K. Survival analysis in clinical trials: basics and must know areas. Perspect Clin Res. 2011;2:145–148. doi: 10.4103/2229-3485.86872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finck T, Schinz D, Grundl L, Eisawy R, Yigitsoy M, Moosbauer J, Pfister F, Wiestler B. Automated pathology detection and patient triage in routinely acquired head computed tomography scans. Invest Radiol. 2021;56(9):571–578. doi: 10.1097/RLI.0000000000000775. [DOI] [PubMed] [Google Scholar]
- 37.Baur C, Wiestler B, Muehlau M, Zimmer C, Navab N, Albarqouni S. Modeling healthy anatomy with artificial intelligence for unsupervised anomaly detection in brain MRI. Radiol Artif Intell. 2021;3:e190169. doi: 10.1148/ryai.2021190169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto A, Pereira S, Meier R, Wiest R, Alves V, Reyes M, Silva CA. Combining unsupervised and supervised learning for predicting the final stroke lesion. Med Image Anal. 2021;69:101888. doi: 10.1016/j.media.2020.101888. [DOI] [PubMed] [Google Scholar]
- 39.Han CH, Kim M, Kwak JT. Semi-supervised learning for an improved diagnosis of COVID-19 in CT images. PLoS One. 2021;16:e0249450. doi: 10.1371/journal.pone.0249450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao R, Namdar K, Liu L, Khalvati F. A transfer learning–based active learning framework for brain tumor classification. Front Artif Intell. 2021;2021:635766. doi: 10.3389/frai.2021.635766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schirmer MD, Venkataraman A, Rekik I, Kim M, Mostofsky SH, Nebel MB, Rosch K, Seymour K, Crocetti D, Irzan H, Hütel M, Ourselin S, Marlow N, Melbourne A, Levchenko E, Zhou S, Kunda M, Lu H, Dvornek NC, Zhuang J, Pinto G, Samal S, Zhang J, Bernal-Rusiel JL, Pienaar R, Chung AW. Neuropsychiatric disease classification using functional connectomics - results of the connectomics in neuroimaging transfer learning challenge. Med Image Anal. 2021;70:101972. doi: 10.1016/j.media.2021.101972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen KT, Schürer M, Ouyang J, Koran MEI, Davidzon G, Mormino E, Tiepolt S, Hoffmann KT, Sabri O, Zaharchuk G, Barthel H. Generalization of deep learning models for ultra-low-count amyloid PET/MRI using transfer learning. Eur J Nucl Med Mol Imaging. 2020;47:2998–3007. doi: 10.1007/s00259-020-04897-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park JE, Kickingereder P, Kim HS. Radiomics and deep learning from research to clinical workflow: neuro-oncologic imaging. Korean J Radiol. 2020;21:1126–1137. doi: 10.3348/kjr.2019.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoebel KV, Patel JB, Beers AL, Chang K, Singh P, Brown JM, Pinho MC, Batchelor TT, Gerstner ER, Rosen BR, Kalpathy-Cramer J. Radiomics repeatability pitfalls in a scan-rescan MRI study of glioblastoma. Radiol Artif Intell. 2020;3:e190199. doi: 10.1148/ryai.2020190199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baeßler B, Weiss K, Pinto Dos Santos D. Robustness and reproducibility of radiomics in magnetic resonance imaging: a phantom study. Invest Radiol. 2019;54:221–228. doi: 10.1097/RLI.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 46.Mayerhoefer ME, Szomolanyi P, Jirak D, Berg A, Materka A, Dirisamer A, Trattnig S. Effects of magnetic resonance image interpolation on the results of texture-based pattern classification: a phantom study. Invest Radiol. 2009;44:405–411. doi: 10.1097/RLI.0b013e3181a50a66. [DOI] [PubMed] [Google Scholar]
- 47.Roberts M, Driggs D, Thorpe M, Gilbey J, Yeung M, Ursprung S, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nature Machine Intelligence. 2021;3:199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- 48.Mongan J, Moy L, Kahn CE., Jr Checklist for artificial intelligence in medical imaging (CLAIM): a guide for authors and reviewers. Radiol Artif Intell. 2020;25:e200029. doi: 10.1148/ryai.2020200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable; this is a review article.
Not applicable; this is a review article.