Abstract
We here describe sequencing and assembly of both the autosomes and the sex chromosome in Micropoecilia picta, the closest related species to the guppy, Poecilia reticulata. Poecilia (Micropoecilia) picta is a close outgroup for studying the guppy, an important organism for studies in evolutionary ecology and in sex chromosome evolution. The guppy XY pair (LG12) has long been studied as a test case for the importance of sexually antagonistic variants in selection for suppressed recombination between Y and X chromosomes. The guppy Y chromosome is not degenerated, but appears to carry functional copies of all genes that are present on its X counterpart. The X chromosomes of M. picta (and its relative Micropoecilia parae) are homologous to the guppy XY pair, but their Y chromosomes are highly degenerated, and no genes can be identified in the fully Y-linked region. A complete genome sequence of a M. picta male may therefore contribute to understanding how the guppy Y evolved. These fish species’ genomes are estimated to be about 750 Mb, with high densities of repetitive sequences, suggesting that long-read sequencing is needed. We evaluated several assembly approaches, and used our results to investigate the extent of Y chromosome degeneration in this species.
Keywords: genome sequence, genetic degeneration, sex chromosome
Significance
The sex chromosomes of the guppy (Poecilia reticulata) and its close relatives in the genus Micropoecilia species carry the same set of genes, but the Micropoecilia Y chromosomes has lost almost all genes present on the X, whereas the guppy Y is very similar to its X, raising a puzzle about how the guppy Y evolved. If the Micropoecilia Y carries a male-determining gene, it should be found in the genome sequence of a male, but our analyses did not detect any Y-linked genes in Micropoecilia picta. This result supports other evidence that the guppy Y does not share the same male-determining gene with M. picta, but has probably recently evolved a new gene with this function.
Introduction
The guppy, Poecilia reticulata, and the closely related Poecilid fish Micropoecilia picta and Micropoeciliaparae (also sometimes called Poecilia) are interesting for understanding the evolution of sex chromosome pairs. Here, we use the genus name Micropoecilia for the latter species, to avoid confusion about which lineage is being referred to. In the guppy, the chromosome that carries the male-determining locus is minimally differentiated from its X chromosome counterpart, with all genes apparently present on both members of the XY pair (Wright et al. 2017; Bergero et al. 2019a). This can be explained by rare recombination between most regions of this chromosome pair, in which no consistently completely male-specific variants have yet been detected (Bergero et al. 2019a; Darolti et al. 2020). In contrast, the M. picta and M. parae Y chromosomes are highly degenerated. In M. picta, few Y-linked genes were detected in either complete genome sequencing studies by an Illumina short-read approach (Darolti et al. 2019) or by a high-throughput sequencing approach based on PCR amplification and sequencing using primers designed from guppy coding regions, which estimated 99% gene loss (Bergero et al. 2019b). In both M. picta and M. parae, X-linked microsatellites were found to be hemizygous in males (Bergero et al. 2019b). These results indicate that recombination with the X is completely suppressed in this species, except in the pseudoautosomal region (PAR), where genetic mapping detects crossovers (as described in the accompanying manuscript—Charlesworth et al. 2021).
These species are closely related, with sequence divergence of only 2–3% (Pollux et al. 2014; see also Charlesworth et al. 2021), prompting the question of how and when the guppy Y chromosome evolved, and what can explain the difference between the Y chromosomes of the guppy and M. picta. One possibility is that the male-determining factor in these fish arose in a common ancestor before the split of Poecilia and Micropoecilia, and that (for unknown reasons) recombination became suppressed only in the latter. In this scenario, the guppy lineage maintained the same sex-determining locus, but did not evolve suppressed recombination, and remained physically small, accounting for the failure to detect any Y-specific region of its sex chromosome. If so, a complete M. picta genome sequence might detect a small Y-linked region that is also shared by the guppy, potentially allowing identification of candidates for the guppy male-determining gene. As will be seen below, our results show that no such Y-linked region is present in M. picta.
This result points to the alternative possibility that the guppy lineage underwent a turnover event in which the ancestral sex-determining locus was replaced by a new one on the same chromosome (Bergero et al. 2019b) and the accompanying manuscript (Charlesworth et al. 2021); the Y in the Micropoecilia lineage (and the common ancestor with the guppy) may even no longer carry a male-determiner, and sex determination may involve a balance between the X and autosomes, as in Drosophila (Bridges 1921). If so, this would support the view that the guppy Y evolved a new male-determining factor. The guppy male-determining factor remains to be discovered, but could be a single gene, like those of the pufferfish, Takifugu niphobles (Ieda et al. 2018) and Mugil cephalus (Curzon et al. 2021), where sex is controlled by variants within a single gene with copies on both members of the sex chromosome pair, and turnover events may also have occurred. Assemblies of both the X and Y in M. picta should therefore also help distinguish between these alternatives, and understand the origin of the guppy XY pair.
The highly degenerated state of the M. picta Y chromosome suggests that the sex chromosome pair could be simple to assemble if a single male is sequenced, The XY pair consists of two parts that should each assemble well: the fully sex-linked part is hemizygous in males, and has only haploid X chromosome sequences, whereas, in the pseudoautosomal part, the chromosome pair recombine, so that both members of the pair should have similar homologous sequences.
Here, we describe analyses that have yielded an assembly of a M. picta male genome sequence. Because these fish have genome sizes of around 750 Mb (Künstner et al. 2016; Fraser et al. 2020; Almeida et al. 2021), short-read sequencing is unlikely to produce a genome assembly suitable for studying the questions just outlined, due to several factors. Genomes of this size have high repetitive sequence and transposable element content (Tørresen et al. 2017) and are hard to physically sequence (Bustos et al. 2016) and to computationally assemble (Treangen and Salzberg 2011), in particular over extended stretches of repetitive content, which are expected in degenerated sex chromosomes. These repeats cause problems with contiguity as they greatly exceed the length of typical paired-end short reads, and may even have repeat intervals that occupy most or all of the length of a short read, such as 60- to 64-bp interval hypervariable mini-satellites, or multikilobase TEs. Long-read data offer the opportunity to resolve both simple and complex repetitive regions (Du and Liang 2019).
However, repeats still create problems, as seen in the examples of PacBio sequencing projects of the maize (Jiao et al. 2017) and human genomes (Shi et al. 2016). Here, we evaluate a genome sequence based on using Oxford Nanopore Technologies (ONT) sequencing data. These approaches are expected to generate longer sequences than PacBio sequencing, at the expense of a higher error rate, of up to 5–15% (Rang et al. 2018). Our aim is not to determine accurate sequences of genes, and an error rate of this magnitude could still allow us to achieve our first aim, to understand the organization of this species’ sex chromosome pair and compare it with that of the guppy. As described below, our data have an error rate of approximately 9%.
Furthermore, it is not yet excluded that the M. picta Y could carry sequences that are diverged from their X-linked counterparts, and have therefore remained undetected by previous studies. These might include potentially functional genic and other sequences, or degenerated ones, like those found in the partially degenerated neo-Y chromosomes of Drosophila miranda (Yi et al. 2003; Bartolomé and Charlesworth 2006) and D. busckii (Zhou and Bachtrog 2015). A second aim was therefore to assess the extent of the M. picta Y chromosome degeneration in more detail than in previous analyses, and to search for fully Y-linked alleles of X-linked genes. Such genes would allow us to estimate Y–X divergence, and estimate the time when the M. picta Y chromosome first stopped recombining with the X, and test whether this occurred after the split from the guppy lineage, or before the split. Given that the M. picta Y carries few genes, any XY genes, or Y-only genes identified in M. picta would also be interesting as candidates for male-determining factors, or male-essential genes, such as those found on the Drosophila Y (Carvalho et al. 2000, 2001; Vibranovski et al. 2008; Mahajan and Bachtrog 2017). If such genes are found in M. picta males, it would allow tests of whether they are also present in the guppy Y-linked region. Finally, a better assembly will be valuable for future work, including genome-wide association studies, genetic mapping, and assessment of chromosome rearrangements that could have contributed to suppression of Y chromosome recombination with its X counterpart.
Here, we describe our assembly of the M. picta genome sequences, and use of several reference genomes from close outgroup species, including the guppy (P. reticulata) female short-read and male PacBio assemblies (Künstner et al. 2016; Fraser et al. 2020) and the platyfish, X. maculatus (Schartl et al. 2013). Divergence between the guppy and M. picta for all site types is about 2–3%, and about 7% for synonymous sites (Pollux et al. 2014; Charlesworth et al. 2021), slightly less than synonymous site divergence from the platyfish, which is 8–10% (Charlesworth et al. 2021).
Results
Supplementary figure S1, Supplementary Material online, shows a summary of statistics for our raw sequence data, using NanoStat from the NanoPack package (De Coster et al. 2018). We evaluated several assembly approaches, considering the whole genome, and then the sex chromosome.
Evaluations of the Assembly as a Whole
The published assembly of a female P. reticulata assembly, based on Illumina short-read sequencing (Künstner et al. 2016), includes 40,143 contigs. Initial de novo assembly of the M. picta ONT data yielded a more contiguous assembly, with either Flye or Redbean software. Supplementary table S2, Supplementary Material online, shows that neither was superior in all assembly metrics, though, encouragingly, most of the genome size is covered in contigs longer than 50 kb in all assemblies. The initial assembly generated from Flye yielded a greater total assembly length, higher N50, maximum contig length, and percentage representation of expected genes, as assessed by the BUSCO software. Redbean yielded more contigs exceeding 50 kb, but fewer bases were covered in these longer contigs, and all three BUSCO completeness scores were low (supplementary table S2, Supplementary Material online). Further polishing with Racon and Medaka improved contiguity, at the expense of fragmenting larger contigs. N50 remained higher for the Flye than the Redbean assembly, but the number of longer contigs remained lower for Flye. Polishing greatly increased all three BUSCO completeness scores for the Redbean assembly, whereas those for the Flye assembly (whose initial scores were already high), were little changed (supplementary table S2, Supplementary Material online).
Analysis of Repetitive Sequences and Their Effects on Assembly
A/T-based simple repeats (particularly poly(A), poly(T) and tandem dinucleotide A/T, or trinucleotide poly(AT) sequences) are among the most difficult regions for assembly of genome sequences including those of the mouse, humans, Drosophila and C. elegans (Heydari et al. 2019), as reviewed in Chen et al. (2013). There is also evidence for AT-biased transposons (Tørresen et al. 2017) in cod genome sequence data, as well as AC and AG dinucleotide enrichment as both a genome-wide feature and an artefact of library preparation (Star et al. 2016; Tørresen et al. 2017). Resolution of high AT-repeat regions has been shown to specifically improve genome assembly in short-read and hybrid assemblies of a range of organisms (Heydari et al. 2019) and may be relevant for long-read sequences also. Problems assembling such regions may be exacerbated by commonly used short-read error correction (EC) tools, which differentially handle repetitive regions, and regions with relative coverage changes, leading to systematic introduction of breaks throughout these sites (Heydari et al. 2017). This is likely to have contributed to fragmentation in the published M. picta assembly (Darolti et al. 2019), where the EC tool Quake was used, and the female guppy reference genome assembly, which used EC modules of ALLPATHS-LG (Künstner et al. 2016), and is known to include assembly errors (Charlesworth et al. 2020; Darolti et al. 2020; Fraser et al. 2020). Polishing tools used for long-read assemblies often make similar assumptions about uniformity of coverage and base composition as short-read EC tools, and may therefore introduce problems in long-read sequences at such regions, which may be particularly abundant in the sex chromosomes, and specifically at the regions of most interest for our goals (see Introduction) identifying genome regions with suppressed recombination may particularly benefit from analysis that can deal with poly(A)/poly(T) sequences, as these are strongly correlated with low recombination rates in humans (Kong et al. 2002); similar problems are experienced in other sex chromosome assembly projects (Kim et al. 2013; Singhal et al. 2015). We therefore first investigated whether repetitive regions are impeding our assembly efforts, and whether a particular class of repeat is involved, in the hope that this information might improve the contiguity of the M. picta genome assembly, particularly the sex chromosomes. Consistent with this, lower coverage of genes in the M. picta sequence was associated with lower GC content (supplementary fig. S3, Supplementary Material online).
High numbers of poly(AT) repeats, compared with genome-wide expected values (see Materials and Methods), were found in the raw reads (supplementary table S3, Supplementary Material online), and specifically at the contig starts and ends in all four of the de novo assemblies described above, including those after polishing (supplementary tables S3–S6, Supplementary Material online); other dinucleotide repeats and poly(A) repeats were also found. The Redbean assembly had less repeat enrichment overall than the Flye assembly. 29-mer results for raw read data (not shown) were almost identical in terms of composition to the 15-mer results shown, confirming that extensive dinucleotide repeat regions are present. The 29-mer results also detected complex repeats. For comparison, supplementary table S7, Supplementary Material online, shows the results of the same analysis using the published P. reticulata female assembly. This suggested that neutral (AC and AG) repeats, AT-biased repeats, poly(AT), poly(A), and poly(T) repeats all contributed to preventing contig extension, similar to their effects in M. picta. The results for the X. maculatus assembly and guppy male PacBio assembly inferred effects of standard vertebrate telomeric sequences (GGGTTA) and neutral repeat sequences, and a slight effect of poly(G) sequences (supplementary tables S8 and S9, Supplementary Material online).
Hybrid Assembly with Repeat-Aware Correction of a Short-Read Library
Given the evidence just described that repetitive (especially AT-rich) regions are probably reducing the contiguity of our M. picta assemblies, we attempted to improve our assembly using software such as BrownieCorrector (Heydari et al. 2019) and KarecT (Allam et al. 2015), which are designed specifically to resolve repeat regions in short-read sequencing data. The analyses used a short-read library from a prior sequencing study of M. picta (Darolti et al. 2019) in a hybrid assembly approach, to see whether integrating an AT-resolved short-read library with our long-read data would assist assembly; the workflow is described in a supplementary methods file, Supplementary Material online. This did not improve our de novo assembly. The failure may be due to the short-read sequencing library that was available having a general lack of coverage at AT-rich regions, possibly due to difficulties with physically sequencing such regions. The supplementary methods file, Supplementary Material online, describes our analyses suggesting such a problem. Given this concern, this approach was not explored further. It could be valuable if a suitable sequence data set were available.
RaGOO Reference-Guided Assemblies
We therefore next employed a reference-guided assembly approach (Lischer and Shimizu 2017) to attempt to improve the genome assembly and generate pseudomolecules, potentially at the chromosome scale. Challenging cases such as bird sex chromosomes have been shown to benefit from this approach (Card et al. 2014; Wang et al. 2014). However, such approaches were originally designed for resequencing projects (Klein et al. 2011), and use for even closely related species can be problematic (Schneeberger et al. 2011), especially for sex chromosomes and other regions where the reference assembly is fragmented and complex (Heydari et al. 2017; Kolmogorov et al. 2018). Moreover, criteria for evaluating the quality of the reference-guided assemblies have not yet been widely adopted and generally accepted. We therefore evaluated whether our sequences yield good assemblies (see fig. 1 below).
Fig. 1.

Evaluations of RaGOO assemblies of Micropoecilia picta sequences using three different reference genome assemblies. The results for the sex chromosome are indicated by the blue box. (A) Number of contigs per RaGOO pseudomolecule for each chromosome. (B) Fraction of total pseudomolecule length covered by contigs with grouping scores >0.9.
As detailed in supplementary methods, Supplementary Material online, several reference genomes are available for use with M. picta, including the published guppy (P. reticulata) assembly of a female (Künstner et al. 2016), with known assembly errors, a PacBio assembly of a male guppy (Fraser et al. 2020). Synonymous site divergence between the guppy and M. picta is about 7%, and divergence for all site types is about 2–3% (Pollux et al. 2014; Charlesworth et al. 2021). We also used an X. maculatus assembly (Schartl et al. 2013), a species with slightly higher synonymous site divergence, of 8–10% divergence (Charlesworth et al. 2021). We did not use the X. helleri reference (Shen et al. 2016), with divergence similar to that of X. maculatus, because the length of its sex chromosome homolog is about 6 Mb shorter than in the X. maculatus reference.
The aim of these analyses was to use reference genomes to achieve an assembly. Before doing so, we needed to know that the extent of divergence from the outgroup species was not too high. As the tolerable divergence for successful reference-guided assembly is not yet clear, we first briefly assess the current evidence. Increased divergence clearly creates assembly difficulties (Card et al. 2014), depending on the sequencing approach and data generated (see supplementary methods, Supplementary Material online, for details). Exploratory comparisons of RaGOO with and without close reference genome sequences were described in the original methods paper (Alonge et al. 2019, based on assemblies of species in the plant genus Solanum). This study identified RaGOO metrics which can be used diagnostically to indicate improvements or reductions in assembly quality. We therefore monitored these metrics throughout our assembly work with different references. Evidence for good grouping and other scores is shown in supplementary table S2 and figures S4 and S5, Supplementary Material online. A grouping score >0.9 appears to be the most helpful single metric, and figure 1B shows results for all M. picta chromosomes.
The X axes order the chromosomes according to their linkage groups in the guppy, with the X. maculatus (platyfish, Xm) homologs indicated above their results in part A. In part A, the number of contigs for chromosome 2 is the total for both Xm7 and Xm24, as these are fused in the guppy but two distinct chromosomes in the platyfish. “Unpl. in ref.” indicates results of scores for contigs mapping to all unplaced contigs in the respective reference assemblies, whilst “Unpl. de novo” scores refer to contigs that remain unplaced after RaGOO scaffolding (ie. in “Chr0”). In part B, for the results using the X. maculatus reference genome, the value shown for the fused chromosome, chr2 is based on Xm7 (the value for Xm24 was similar [0.78] and is not shown separately).
Regardless of the reference assembly used, the M. picta sex chromosome, chromosome 12, assembled much less well than the autosomes. This is clear with both criteria shown in figure 1 (the number of contigs per RaGOO pseudomolecule for each chromosome, and the fraction of total pseudomolecule length covered by contigs with grouping scores >0.9), particularly for the X. maculatus assembly, with the sex chromosome being approximately two to three times more fragmented than the autosomes (fig. 1A). Only LG18 showed a level of fragmentation similar to that of the sex chromosome, though both these were less fragmented than the unplaced contigs. We conclude that the autosomes can be assembled fairly reliably, and that the sex chromosome fragmentation in these assemblies is clearly due to its special biological features. We also found higher grouping scores for the autosomes than the sex chromosome (fig. 1B), and a relatively much larger proportion of the sex chromosome had low values with all three reference genome employed (supplementary fig. S4, Supplementary Material online); for this chromosome, our M. picta assembly included 18 Mb (18,088,251 bp of the total 31,543,944 bp, or ∼60%) with grouping score >0.9 (for discussion of different assemblies, see supplementary methods file, Supplementary Material online). All RaGOO assemblies indicate similar numbers of broken contigs and possible misassemblies on the sex chromosome.
Overall, <1% of all our M. picta sequence data remained unlocalized after these analyses. Supplementary table S11, Supplementary Material online, summarizes key statistics, including numbers of contigs and introduced breaks (which may create misassemblies and should be treated with caution), with the guppy PacBio assembly estimated as approximately 1/3rd worse than the other assemblies, and the X. maculatus assembly marginally favored. Supplementary table S11, Supplementary Material online, shows statistics for the hybrid assembly based on this reference.
Attempts to Assemble the Sex Chromosome
Despite these problems, the sex chromosome assemblies may still be of value if they include stretches of high-confidence contigs. Although, as might be expected, the centromeric and terminal regions have lower scores than other regions, large parts of the sex chromosome have high location, orientation, and grouping scores (supplementary fig. S5, Supplementary Material online). Overall, approximately 40% of the sequences assigned to the sex chromosome can be localized and oriented. Although the X. maculatus assembly seems to perform well for most of the M. picta genome, the other references performed better for the sex chromosome. Problematic regions were particularly seen around the tip of this chromosome when the X. maculatus reference was used, and to a lesser extent in the guppy-based assemblies (see supplementary fig. S5, Supplementary Material online), suggesting that these regions may undergo more sequence evolution than other parts of the chromosome, or that this region’s highly variable base composition may create problems. Dot plots comparing the sex chromosome pseudomolecules and the reference chromosomes additionally identified the tip as highly repetitive in the X. maculatus-guided pseudomolecule (supplementary fig. S6, Supplementary Material online).
Manual inspection of broken contigs on Xm8/LG12 showed that the RaGOO process subjected a large proportion to undue fragmentation, as they were found assembled in their prebreaking sequence; 14 out of the 27 broken contigs using the Xm8-like pseudomolecule showed this effect, six of 19 using the female P. reticulata LG12, and at least 14 out of 30 using the PacBio male one. There were signs of probable direct mis-assembly from undue breakage, including a broken contig that was found with approximately the first and last thirds of the parent contig ordered in sequence but the middle absent. Likewise, there were several examples of a start or end of a parent contig being moved elsewhere on the same chromosome, particularly into repetitive regions. Such results are not unexpected, given that RaGOO validates introduced fragmentation based on comparison to the reference genome, which contain known mis-assemblies. Supplementary table S11, Supplementary Material online, includes lower bound estimates of the number of direct mis-assemblies related to broken contigs, based on the numbers of unique parent contigs with obviously erroneous fragmentation.
Examination of dot plots (see supplementary methods and fig. S6, Supplementary Material online) showed that RaGOO worked well outside repetitive regions, recovered the main patterns of synteny expected between pseudomolecules, and these plots support the view that handling repetitive regions can considerably improve the assembly.
Supplementary figure S2, Supplementary Material online, summarizes the depths of coverage of sequences on the sex chromosome; for genes in the first 21 Mb in guppy genome assembly, the proportion of reads covering the sequences averaged 100%, but the value declined steadily toward the chromosome terminus, and was only 60% for the region assembled distal to 26 Mb in the guppy. The terminal region also had extreme base composition for many sequences (supplementary fig. S3, Supplementary Material online).
Given these encouraging findings for much of LG12, we next attempted to use the M. picta contigs from the stage before implementing reference-guided analyses to search for long contigs that contain genes assembled in noncontiguous regions in the guppy assemblies (see Materials and Methods). If the sex chromosome is not syntenic in these two species, this analysis should reveal many such discrepancies. On the other hand, if the chromosomes are syntenic, such discrepancies can help to detect mis-assemblies in the guppy, particularly if they coincide with regions already identified as problematic or repetitive in the analyses already described.
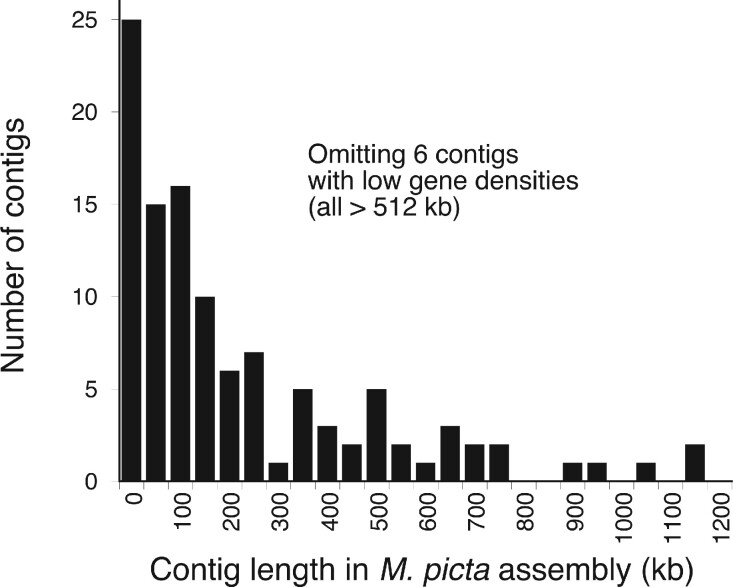
Using the guppy female assembly, almost all genes from the guppy LG12 are represented on 115 M. picta contigs, whose sizes are summarized in figure 2 and table 1, and the genes within them are in the same order as in the guppy, with very few exceptions (fig. 3), consistent with the results from the RaGOO analysis. The mean contig size is 414 kb (median 150), or 244 and 134 kb, respectively excluding six long contigs with very low gene densities, whose mean and median lengths are 1,371 and 1,265 kb, respectively (tables 1 and 2). For contigs other than these 6, the mean and median kilobases per gene are 37.20 and 30.05. The results based on the guppy male assembly are similar, with almost all genes from the guppy represented on 88 M. picta contigs, but with fewer guppy genes covered, mainly due to the lower representation of genes in the PacBio assembly, compared with the Illumina one (table 1).
Fig. 2.
Lengths of contigs containing sequences assembled on the guppy LG12. About 506 contigs were found using the guppy female assembly based on Illumina short-read sequencing (this excludes six contigs with low gene densities, all >512 kb; as described in the text, there were seven such contigs using the male guppy assembly).
Table 1.
Summary of Micropoecilia picta Contigs in Which Genes Assembled on the Guppy LG12 Were Detected Using Genes in the Female (Illumina) Assembly or the Male PacBio Assembly
| Female Assembly |
Male Assembly |
|||
|---|---|---|---|---|
| Total Length of Contigs (Mb) | Number of Contigs | Total Length of Contigs (Mb) | Number of Contigs | |
| Long contigs with normal gene density | 26.918 | 84 | 24.414 | 72 |
| Small contigs (<50 kb) | 0.671 | 25 | 0.238 | 9 |
| Contigs with low gene density | 20.852 | 6 | 26.075 | 7 |
| Total number | 115 | 88 | ||
Fig. 3.

Micropoecilia picta contigs to show that there are few regions of the guppy female assembly of LG12 that are not represented s. Alternating contigs are in black and gray, with large colored symbols indicating contigs that include genes assigned to more than one contiguous region in the guppy female assembly (see table 2 of the main text, which also shows those identified by the analysis of the guppy male assembly). Contigs denoted “LARGE” are the large, low-density ones mentioned in the text.
Table 2.
Micropoecilia picta Contigs That Include Genes Assigned to More Than One Contiguous Region in the Guppy LG12 Assembly from Either the Female Illumina Sequencing (Indicated by F in the Left-Hand Column) or the Male PacBio Sequencing (M in the Left-Hand Column)
| Contig and Assembly Used (F or M) | Region 1 | Region 2 | Numbers of Genes | ||
|---|---|---|---|---|---|
| From | To | From | To | ||
| Proximal to 10 Mb | |||||
| contig_1682 (M) | 3,480 | 36,891 | 3,295,789 | 3,614,517 | 3, 10 |
| contig_474 (M) | 4,694,901 | 4,793,198 | 8,134,430 | 8,243,264 | 4, 4 |
| Near 10 Mb and also terminal region | |||||
| contig_1832 (F) | 9,829,816 | 9,936,380 | 20,238,456 | 20,707,596 | 7 |
| 10,694,363 | 10,785,162 | 2, 14 | |||
| contig_1089 (F and M) | 11,019,033 | 12,013,746 | 24,505,075 | 24,640,561 | 18, 4 |
| 19,720,934 | 19,731,289 | 19,963,267 | 20,065,708 | 2, 3 | |
| Wholly within terminal region | |||||
| contig_305 (F and M) | 21,616,399 | 21,626,825 | 25,236,538 | 25,461,841 | 2, 5 |
| 25,802,356 | 25,935,960 | 7 | |||
| 20,402,707 | 20,637,445 | 25,858,867 | 25,886,114 | 4, 2 | |
| contig_42 (F) | 20,941,634 | 20,967,698 | 24,888,846 | 25,006,165 | 1, 7 |
| 19,720,934 | 19,731,289 | 19,963,267 | 20,065,708 | 2, 3 | |
| Scattered: contig_1818 (Female assembly only) | |||||
| 52,626 | 134,454 | ||||
| 10,457,608 | 10,532,228 | ||||
| 10,709,181 | 10,785,162 | ||||
| 10,784,208 | 10,932,419 | ||||
| 12,998,746 | 3,103,108 | ||||
| 13,462,512 | 13,622,358 | ||||
However, several large contigs had very low gene densities, often with only a single LG12 gene. The female and male guppy assemblies included six and seven such contigs, respectively (with total sizes of about 66 and 82 Mb, see supplementary table S13, Supplementary Material online). It seems likely that at least some of these are autosomal contigs that contain a gene that was incorrectly assembled on LG12 in the guppy. To investigate the other sequences in these M. picta contigs, we therefore did BLAT searches of the guppy genome assemblies using as queries all 1,500 bp nonoverlapping segments from each such contig or scaffold (see Materials and Methods and supplementary fig. S7B, Supplementary Material online). The results using unfiltered M. picta contigs or scaffolds confirm that these large M. picta contigs or scaffolds consist largely of autosomal sequences syntenic with regions in the guppy assemblies (supplementary fig. S8, Supplementary Material online). Most include small amounts of sequences that were assembled on LG12, probably representing short sequences shared with an autosome. A few larger regions could indicate chromosome rearrangements between the guppy and M. picta, or mis-assemblies of guppy sequences. Results described below suggest that rearrangements are few, at least for the sex chromosome pair. Genetic mapping in the guppy should help resolve such regions in the future. A large (almost 0.5 Mb) segment of contig_645 clearly belongs to LG7, but is assembled on LG12 in the guppy female genome, and is probably a mis-assembly (Charlesworth et al. 2021). Supplementary figure S8, Supplementary Material online, shows that the most likely positions for the mis-assembled sequences are at 6.3 Mb on LG7.
Because we filtered the BLAT results to include only unique hits in the guppy for the M. picta window queries, this analysis will not detect sequences with duplications in the guppy. As the guppy sex chromosome probably includes some duplicated regions (Charlesworth et al. 2020; Fraser et al. 2020), these could have been missed by our analysis. We therefore also manually searched the results from this second BLAT experiment for contiguous sets of M. picta query windows that find two or more distinct guppy target regions that could each be part of separate genes, suggesting nontandem duplication (supplementary fig. S7C, Supplementary Material online). We examined windows with multiple hits in the guppy, with a maximum of 20; among the 131 Mb included in these large contigs in total, only 129 such cases were found in the guppy male chr12 assembly results, and 142 in results using the female LG12. None of these showed the signal of gene duplications just described, but appear merely to reflect dispersed short repetitive sequences. We conclude that our M. picta contigs and scaffolds largely include single-copy guppy genes.
Only five M. picta contigs include sequences from two or more different regions of the guppy female LG12 assembly, and four when using the male assembly (table 2). These discrepancies also suggest possible errors in the guppy assembly. They are mainly confined to two regions, distal to 21 Mb in the female guppy assembly, and near 10 Mb, which should be tested by genetic mapping in the guppy (table 2 shows that they include several genes that could potentially be mapped). Encouragingly, where different contigs overlapped included parts of the same gene, they were in adjacent positions, based on the guppy female assembly (supplementary table S14, Supplementary Material online). In total, these contigs include 17.3 Mb of sequence, representing more than 2/3 of LG12. Overall, these results strongly suggest that this chromosome’s gene order in M. picta is generally very similar to that in the guppy, and that both assemblies are largely correctly ordered, although rearrangements cannot be excluded in the regions where no overlapping genes were found (eight of these were >1 Mb, including one 4.7 Mb region between 7,266,064 and 11,988,456 bp in the guppy female assembly that includes the problematic region already mentioned).
Discussion and Conclusions
The choice of the best assembly for reference-guided analyses in M. picta is not clear, and our results described above suggest that different reference assemblies may differ in their usefulness for different chromosomes. For the M. picta sex chromosome pair, the platyfish assembly appears to have produced better results than either guppy one, including the PacBio one (Fraser et al. 2020). This may simply reflect a better assembly of the platyfish genome.
Overall, the assembly appears better for the autosomes than for the sex chromosome pair. This is unexpected. If the XY pair consists of an X and a completely degenerated Y, we might expect the sequence from a single male individual, as studied here, to assemble better than the autosomes, since his recombining PAR sequences should be similar for both the Y and the X, and the Y-degenerated part will be haploid, whereas the autosomes will have heterozygous regions, including indels and transposable element insertions that will impede assembly. Such sequences might, however, cause sequencing and assembly problems, even in haploid genome regions.
Another possible explanation for the poorer assembly of the sex chromosome than the autosomes might be that the fully Y-linked region has retained sequences. These could be highly diverged from the homologous X-linked sequences, and differ by indels, impeding assembly. However, our results support previous evidence that the M. picta Y chromosome carries extremely few genes (Bergero et al. 2019b; Darolti et al. 2019). Our LG12 sequences therefore clearly largely reflect the X chromosome. Despite the assembly difficulties, our analyses suggest that genes in this chromosome are ordered very similarly to those in the guppy, though the results point to several regions where the guppy assembly of the sex chromosome may include errors, which should be tested by future genetic mapping.
In the Introduction, we mentioned the possibility that the M. picta Y chromosome might carry a small number of genes, potentially including a male-determining factor. As already explained, it is important to search for such genes, as they offer the best information about the time when the Y and X chromosomes stopped recombining and their sequences started diverging. Given the highly degenerated state of the Y, divergence from the X-linked homologues is expected to be high, but the Y-linked sequences should nevertheless be found among our reads. Although they would probably not assemble into the X chromosome, they might be assembled into the homologous chromosome of the outgroup, the platyfish. We found no evidence for the existence of any Y-linked sequences in any of our assemblies, including this outgroup one. We therefore conclude that the M. picta Y either has no remaining genes, or their sequences are highly diverged. Our BLAT minScore of 800 for 1,500 bp queries means that sequences should be detected unless they have <53% identity with the guppy query sequences (>47% divergence). If Y-linked sequences exist in M. picta, their divergence must greatly exceed that of guppy X-linked sequences from those of M. picta. The median divergence between the guppy and M. picta, estimated from a set of loci with long coding sequences from multiple chromosomes, is 2.7% for pooled synonymous and nonsynonymous sites (supplementary table S2, Supplementary Material online, of Charlesworth et al. 2021). The genes analyzed were selected for having long coding sequences, which are expected to be subject to selective constraints and therefore tend to have low divergence (supplementary table S2, Supplementary Material online, of Charlesworth et al. 2021). Our BLAT results, which include noncoding and nongenic regions, thus yield a higher divergence value, about 9.4% (the matches of our observed unique BLAT hits have a median identity 90.6%, based on a mean length of 1,313 bp, and 75% of such hits had identity >85.6%). The complete absence of any candidate Y-linked sequences among our BLAT targets suggests that M. picta has very few, or no, completely Y-linked sequences (rather than that these were not detected). These results therefore support the conclusion (Charlesworth et al. 2021) that, if completely Y-linked sequences do exist in M. picta, they must have been diverging from their X-linked alleles since before the split from the guppy lineage, in other words that recombination of the Y chromosome with the X was suppressed before the split between the Poecilia and Micropoecilia lineages.
Materials and Methods
Materials and Sequencing
The M. picta sample (RP09m) was a male collected by David Reznick in Trinidad (Cunipia River, part of the Caroni swamp, Trinidad, 10°36′18.89″N, 61°25′28.462″W, as described in supplementary table S5 of Bergero et al. 2019b). The sex was confirmed by genotyping several microsatellite markers on the XY pair (Bergero et al. 2019b). DNA for long-read sequencing was extracted using the Qiagen Genomic DNA Preparation kit with 100/G tips (Qiagen), and purified as described in supplementary methods file, Supplementary Material online. A library was made using the Oxford Nanopore ligation sequencing library protocol, and sequenced on a PromethION flowcell. Base-calling was performed using the Oxford Nanopore Technologies base-caller “guppy” (version 3.2.6+afc8e14). Supplementary figure S1, Supplementary Material online, summarizes the read lengths obtained.
Genome Assembly, Polishing, and Validation
The reads from all the flowcells were used to generate a genome assembly using the software Flye (version 2.7b) (Kolmogorov et al. 2019) with parameters “-x ont -p 0 -k 15 -AS 2 -s 0.05 -g 1 g -t 16” and Redbean (version 2.5) (Ruan and Li 2020) with parameters “-t 30 -p 19 -AS 2 -s 0.05 -L 2000 -g 1 g.” As no short read was available from the M. picta individual sequenced, the assemblies generated from Flye and Redbean were polished with four iterations of Racon version 1.3.3 (Vaser et al. 2017) followed by one iteration of medaka (version 0.10.0). QUAST version 5.0.2 (Gurevich et al. 2013) was then used to generate assembly validation metrics including the number of contigs and N50 BUSCO version 4.0.4 software (Simão et al. 2015) was used on to assess the assemblies’ quality in terms of gene completeness based on three different databases, Eukaryotes, Vertebrates, and Actinopterygii. The results are in supplementary table S2, Supplementary Material online. The Flye assembly has been deposited in the European Nucleotide Archive under accession number ERZ1744533 (https://www.ebi.ac.uk/ena/browser/text-search?query=PRJEB43222).
Evaluations of the Assembly
Analysis of Repetitive Sequences
To test whether repeats are responsible for contig breaking (and, if so, which class(es) of repeats), an analysis was done using the “count” command of Jellyfish v2.2.10 software (Marçais and Kingsford 2011) to quantify 15-mers and 29-mers in the raw M. picta sequencing data. Additionally, the first and last 100 bp of each contig was extracted from each of the M. picta de novo raw and polished assemblies described above. 15-mers were quantified for each genome, to identify the base composition at regions where contig extension fails. The same analysis was applied to the female P. reticulata short-read semiscaffolded assembly (Künstner et al. 2016), using the full set of 40,143 contigs to look for repeat profiles that may be found at nonextendable ends in both species. The male PacBio-based P. reticulata semiscaffolded assembly (Fraser et al. 2020) (n = 267 scaffolds) and male X. maculatus scaffolded assembly (Amores et al. 2014) (n = 101 scaffolds) were also profiled; we expected these to return telomeric and other “background” sequences exemplary of complete assemblies. To test for enrichment of particular 15-mers, we compared their representation with the base composition and tested the null hypothesis of equal representation of all 15-mers.
BLAT Searches
A final analysis of the unordered M. picta contigs the Flye assembly further evaluated the correspondence between gene orders in our reliable long M. picta contigs and in the guppy. These analyses used contigs polished with the “medaka” polisher (without filtering by length) as targets for queries in BLAT searches. A first BLAT experiment used as queries the 903 genes in the NCBI annotation of the guppy LG12, based on the female assembly of short-read Illumina sequences (Künstner et al. 2016); a similar analysis used the guppy male assembly from long-read (PacBio) sequencing (Fraser et al. 2020), with 596 genes annotated. For these first BLAT experiments, with guppy query sequences, custom Python3 scripts (in supplementary methods, Supplementary Material online) (scripts for BLAT searches) first collected 1,500 bp regions, or “windows,” from the guppy genes and formatted the products as a multi-FASTA file for use in the BLAT searches. The numbers of regions depended on the genes’ lengths, as follows: the first 1,500 bp was selected from genes <3 kb, the first and last from genes ≥3 and ≤10 kb, three regions of the same length, including one in the middle, from genes ≥10 and ≤30 kb, and four equally spaced regions for genes >30 kb. The minScore parameter for the BLAT searches was set at 800 bp (matches out of the 1,500 bp “windows”). Incorrect or unreliable hits were filtered out using the following conservative criteria for exclusion: genes with middle region(s) on a contig different from that with the start and end region, genes/regions with gaps exceeding 15 kb, or with hits across many contigs. Before filtering, 837 of the guppy LG12 genes used as queries (92.8%) were found in the M. picta contigs/scaffolds, and after filtering 788 (87.4%) using the female guppy assembly, and 408 (68.5%) with the male assembly.
As described in the Results section, we also did a second set of BLAT experiments using M. picta contigs as queries, to discover regions where our M. picta contigs or scaffolds suggested possible mis-assemblies in the guppy, and to test whether any sex-linked genes in M. picta have additional copies in the guppy (reflecting either duplications onto the sex chromosome, or of sex-linked genes to autosomal locations, or sequences present in two or more different regions of the sex chromosome). Supplementary figure S7, Supplementary Material online, includes diagrams of both BLAT experiments.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This project was supported by ERC (Grant No. 695225) (GUPPYSEX). We are very grateful to David Reznick and his assistants A. Van Alst and M. Charran, and R. Bassar, for samples of wild Micropoecilia picta from Trinidad, Dr Andres de la Filia for his DNA extraction protocol, and Edinburgh Genomics, University of Edinburgh, for sequencing. Edinburgh Genomics is party supported with core funding from NERC (UKSBS PR18037).
Data Availability
All raw and processed sequencing data generated in this study have been submitted to the European Nucleotide Archive (ENA: https://www.ebi.ac.uk/ena/browser/home) under study accession number and study unique name (PRJEB43222m and ena-STUDY-ED-22-02-2021-10:06:18:768-1014, respectively). The assembly accession number is ERZ1744533.
Literature Cited
- Allam A, Kalnis P, Solovyev V.. 2015. Karect: accurate correction of substitution, insertion and deletion errors for next-generation sequencing data. Bioinformatics 31(21):3421–3428. [DOI] [PubMed] [Google Scholar]
- Almeida P, et al. 2021. Divergence and remarkable diversity of the Y chromosome in guppies. Mol Biol Evol. 38(2):619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, et al. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 20(1):224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, et al. 2014. A RAD-Tag genetic map for the Platyfish (Xiphophorus maculatus) reveals mechanisms of karyotype evolution among Teleost fish. Genetics 197(2):625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé C, Charlesworth B.. 2006. Evolution of amino acid sequences and codon usage on the Drosophila miranda neo-sex chromosomes. Genetics 174(4):2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Gardner J, Bader B, Yong L, Charlesworth D.. 2019a. Exaggerated heterochiasmy in a fish with sex-linked male coloration polymorphisms. Proc Natl Acad Sci U S A. 116(14):6924–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Gardner J, Charlesworth D.. 2019b. Evolution of a Y chromosome from an X chromosome. Curr Biol. Advance Access published July 11, 2019, doi: 10.2139/ssrn.3417937.
- Bridges C.1921. Triploid intersexes in Drosophila melanogaster. Science 54(1394):252–254. [DOI] [PubMed] [Google Scholar]
- Bustos AD, Cuadrado A, Jouve N.. 2016. Sequencing of long stretches of repetitive DNA. Sci Rep. 6:36665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card D, et al. 2014. Two low coverage bird genomes and a comparison of reference-guided versus de novo genome assemblies. PLOS One 9(9):e106649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Dobo BA, Vibranovski MD, Clark AG.. 2001. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 98(23):13225–13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Lazzaro BP, Clark AG.. 2000. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci U S A. 97(24):13239–13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Bergero R, Graham C, Gardner J, Keegan K.. 2021. How did the guppy Y chromosome evolve? PLoS Genet. 17(8):e1009704. [DOI] [PMC free article] [PubMed]
- Charlesworth D, Bergero R, Graham C, Gardner J, Yong L.. 2020. Locating the sex determining region of linkage group 12 of guppy (Poecilia reticulata). G3 (Bethesda) 10(10):3639–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-C, Liu T, Yu C-H, Chiang T-Y, Hwang C-C.. 2013. Effects of GC bias in next-generation-sequencing data on de novo genome assembly. PLoS One 8(4):e62856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curzon A, et al. 2021. A novel c.1759T>G variant in follicle-stimulating hormone-receptor gene is concordant with male determination in the flathead grey mullet (Mugil cephalus). G3 (Bethesda) 11:3867–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darolti I, et al. 2019. Extreme heterogeneity in sex chromosome differentiation and dosage compensation in livebearers. Proc Natl Acad Sci U S A. 116(38):19031–19036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darolti I, Wright A, Mank J.. 2020. Guppy Y chromosome integrity maintained by incomplete recombination suppression. Genome Biol Evol. 12(6):965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C.. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34(15):2666–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Liang C.. 2019. Assembly of chromosome-scale contigs by efficiently resolving repetitive sequences with long reads. Nat Commun. 10(1):5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser B, et al. 2020. Improved reference genome uncovers novel sex-linked regions in the guppy (Poecilia reticulata). Genome Biol Evol. 12(10):1789–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. 2013. Quast: quality assessment tool for genome assemblies. Bioinformatics 29(8):1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydari M, Miclotte G, Demeester P, Van de Peer Y, Fostier J.. 2017. Evaluation of the impact of Illumina error correction tools on de novo genome assembly. BMC Bioinformatics 18(1):374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydari M, Miclotte G, YVd P, Fostier J.. 2019. Illumina error correction near highly repetitive DNA regions improves de novo genome assembly. BMC Bioinformatics 20(1):298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda R, et al. 2018. Identification of the sex-determining locus in grass puffer (Takifugu niphobles) provides evidence for sex-chromosome turnover in a subset of Takifugu species. PLoS One 13(1):e0190635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, et al. 2017. Improved maize reference genome with single-molecule technologies. Nature 546(7659):524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, et al. 2013. Reference-assisted chromosome assembly. Proc Natl Acad Sci U S A. 110:1785–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Ossowski S, Schneeberger K, Weigel D, Huson D.. 2011. A low coverage assembly tool for resequencing projects. PLOS One 6(8):e23455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, et al. 2018. Chromosome assembly of large and complex genomes using multiple references. Genome Res. 28(11):1720–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner P.. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37(5):540–546. [DOI] [PubMed] [Google Scholar]
- Kong A, et al. 2002. A high-resolution recombination map of the human genome. Nat Genet. 31(3):241–247. [DOI] [PubMed] [Google Scholar]
- Künstner A, et al. 2016. The genome of the Trinidadian guppy, Poecilia reticulata, and variation in the Guanapo population. PLoS One 11(12):e0169087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischer HEL, Shimizu K.. 2017. Reference-guided de novo assembly approach improves genome reconstruction for related species. BMC Bioinformatics 18(1):474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Bachtrog D.. 2017. Convergent evolution of Y chromosome gene content in flies. Nat Commun. 8(1):785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C.. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27(6):764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollux BJA, Meredith RW, Springer MS, Garland T, Reznick DN.. 2014. The evolution of the placenta drives a shift in sexual selection in livebearing fish. Nature 513(7517):233–236. [DOI] [PubMed] [Google Scholar]
- Rang F, Kloosterman W, Jd R.. 2018. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 19(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H.. 2020. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 17(2):155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M, et al. 2013. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat Genet. 45(5):567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, et al. 2011. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc Natl Acad Sci U S A. 108(25):10249–10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, et al. 2016. X. couchianus and X. hellerii genome models provide genomic variation insight among Xiphophorus species. BMC Genomics 17:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, et al. 2016. Long-read sequencing and de novo assembly of a Chinese genome. Nat Commun. 7:12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F, Waterhouse R, Ioannidis P, Kriventseva E, Zdobnov E.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Singhal S, et al. 2015. Stable recombination hotspots in birds. Science 350(6263):928–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B, et al. 2016. Preferential amplification of repetitive DNA during whole genome sequencing library creation from historic samples. Sci Technol Archaeol Res. 2(1):36–45. [Google Scholar]
- Tørresen O, et al. 2017. An improved genome assembly uncovers prolific tandem repeats in Atlantic cod. BMC Genomics 18(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T, Salzberg S.. 2011. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 13(1):36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R, Sović I, Nagarajan N, Šikić M.. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27(5):737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski M, Koerich L, Carvalho A.. 2008. Two new Y-linked genes in Drosophila melanogaster. Genetics 179(4):2325–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Ekblom R, Bunikis I, Siitari H, Höglund J.. 2014. Whole genome sequencing of the black grouse (Tetrao tetrix): reference guided assembly suggests faster-Z and MHC evolution. BMC Genomics 15:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A, et al. 2017. Convergent recombination suppression suggests a role of sexual conflict in guppy sex chromosome formation. Nat Commun. 8:14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Bachtrog D, Charlesworth B.. 2003. A survey of chromosomal and nucleotide sequence variation in Drosophila miranda. Genetics 164(4):1369–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D.. 2015. Ancestral chromatin configuration constrains chromatin evolution on differentiating sex chromosomes in Drosophila. PLoS Genet. 11(6):e1005331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed sequencing data generated in this study have been submitted to the European Nucleotide Archive (ENA: https://www.ebi.ac.uk/ena/browser/home) under study accession number and study unique name (PRJEB43222m and ena-STUDY-ED-22-02-2021-10:06:18:768-1014, respectively). The assembly accession number is ERZ1744533.