Abstract
Lysozyme enzymes provide classic examples of molecular adaptation and parallel evolution, however, nearly all insights to date come from chicken-type (c-type) lysozymes. Goose-type (g-type) lysozymes occur in diverse vertebrates, with multiple independent duplications reported. Most mammals possess two g-type lysozyme genes (Lyg1 and Lyg2), the result of an early duplication, although some lineages are known to have subsequently lost one copy. Here we examine g-type lysozyme evolution across >250 mammals and reveal widespread losses of either Lyg1 or Lyg2 in several divergent taxa across the mammal tree of life. At the same time, we report strong evidence of extensive losses of both gene copies in cetaceans and sirenians, with an additional putative case of parallel loss in the tarsier. To validate these findings, we inspected published short-read data and confirmed the presence of loss of function mutations. Despite these losses, comparisons of selection pressures between intact g- and c-type lysozyme genes showed stronger purifying selection in the former, indicative of conserved function. Although the reasons for the evolutionary loss of g-type lysozymes in fully aquatic mammals are not known, we suggest that this is likely to at least partially relate to their hairlessness. Indeed, although Lyg1 does not show tissue-specific expression, recent studies have linked Lyg2 expression to anagen hair follicle development and hair loss. Such a role for g-type lysozyme would explain why the Lyg2 gene became obsolete when these taxa lost their body hair.
Keywords: Lysozyme g, Cetacea, Sirenia, Chiroptera, parallelism, pseudogenization
Significance
We conduct the most comprehensive study to date of the evolutionary history of g-type lysozyme genes in mammals. By incorporating newly published mammalian genomes, we compared g-type lysozyme gene sequences from >250 species and found independent gene losses of both genes across divergent groups. Intriguingly, we report the first losses of Lyg2 in bats, and we show that a small number of taxa—the cetaceans and manatees as well as the tarsier—have lost both genes. The selective drivers for the inactivation of the Lyg1 and Lyg2 genes in mammals are not known, however, we suggest that the distribution of losses points to a link with hairlessness.
Introduction
Studies on the evolution of the lysozyme gene family in vertebrates have shed important insights into our understanding of molecular adaptation and parallelism (Stewart et al. 1987; Messier and Stewart 1997). For example, c-type lysozymes from divergent lineages of herbivorous mammals—in which lysozyme breaks down commensal bacteria involved in foregut fermentation—show identical parallel amino acid replacements (Zhang and Kumar 1997). In bats and some other mammals, lineage-specific duplication of the c-type lysozyme has also been documented and linked to functional diversification across different tissues (Hammer et al. 1987; Pacheco et al. 2007; Liu et al. 2014).
In contrast to c-type lysozyme, the evolution of g-type lysozyme has been less well-studied, and its function is relatively poorly characterized. G-type lysozyme was first isolated from goose egg whites, and its gene (Lyg) was subsequently characterized from chicken tissue (Nakano and Graf 1991). Since then, it has been found in diverse taxa, including vertebrates and invertebrates (Callewaert and Michiels 2010). Structurally similar to c-type lysozyme (six α-helixes and three β-sheets) (Moreno-Cordova et al. 2020), g-type lysozyme has been implicated in antimicrobial activity based on its ability to hydrolyze the β-1,4 glycosidic bonds in peptidoglycan, a constituent of bacterial cell walls. The active enzymatic sites in the g-type lysozymes include glutamic acid 73 (Glu-73)—which is located in the α4 helix and appears to be an acid catalyst—as well as two aspartate residues at positions 86 and 97 (Asp-86 and Asp-97) in β2 and β3 sheets, which might act as basic catalysts (Kawamura et al. 2006; Helland et al. 2009; Moreno-Cordova et al. 2020). Support for a role in immunity comes from reports of high expression levels in immune tissues from many fishes (Gao et al. 2016; Liu et al. 2016; Zhang et al. 2018), together with its upregulation in response to the infection of fish with pathogens (Mohapatra et al. 2019).
The genome of the ancestral amniotes was reported to have possessed three Lyg genes, which are retained in genomes of some extant reptile and bird lineages (Irwin 2014). Although only one of these three ancestral genes was inferred to have been retained in mammals, an ancient gene duplication event early in mammalian evolution resulted in two g-type lysozymes genes (Lyg1 and Lyg2) being present in many mammalian genomes (Irwin and Gong 2003; Irwin 2014). At the time, some lineages are known to have subsequently lost one copy, although inferences to date have been based on limited taxon coverage and the causes of such losses remain unclear (Irwin 2014). The two mammalian g-type lysozymes have been shown different expression patterns, with the Lyg1 widely expressed across different organs, and associated with tumor-related immune responses in humans (Liu et al. 2017). On the other hand, Lyg2 is highly expressed in the skin (Irwin 2014) and appears related to hair follicle development (Wang et al. 2019; Wiener et al. 2020) and antibacterial immunity (Huang et al. 2011).
The proliferation of available mammalian genomes provides new opportunities to determine the drivers of molecular evolution. Thus, to gain a more complete understanding of potential ecological and physiological factors underlying the pseudogenization of g-type lysozymes, here we examine the evolution of Lyg1 and Lyg2 genes across >250 mammals, covering 24 orders and all major taxonomic clades. We predict that if g-type lysozyme plays a critical function in immune defense, then, like chicken-type lysozyme gene (Lyz), at least one copy will be retained. In addition, if losses are related to changes in ecology, then we might expect associations with habitat use and/or diet.
Results
Lyg Genes Acquisitions
We retrieved gene sequences for 247 Lyg1 and 232 Lyg2 orthologs from 255 mammalian genomes across all major classes, including 196 species not previously examined, and identified hitherto unreported losses of both genes in multiple lineages across mammals (supplementary table S1, Supplementary Material online).
Identification of Lyg Pseudogenes Based on Published Data
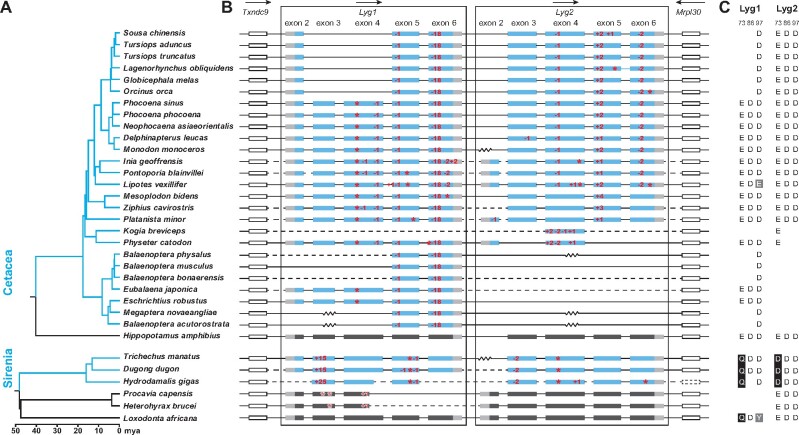
For Lyg1, pseudogenes were identified in 56 species, including all cetaceans and sirenians, and most even-toed ungulates, except some cervids, musk deer (Moschus moschiferus), giraffe (Giraffa tippelskirchi), okapi (Okapia johnstoni), and hippopotamus (Hippopotamus amphibius) (supplementary fig. S1, Supplementary Material online). In the case of the cetaceans, a 1-bp frame-shifting deletion in exon 5 was shared by all 26 species examined, implying that this mutation occurred early in the evolution of modern whales and likely was the pseudogene generating mutation. In sirenians, a premature stop mutation as well as a 1-bp deletion in exon 5, were shared by all species examined, indicating a loss in the ancestor of these species (fig. 1). We also found different loss-of-function mutations in some bats (Chiroptera) and primates, as well as in Hyracoidea (hyraxes), wallaby (Macropus eugenii), and the edible dormouse (Glis glis). In addition, we were unable to detect this gene, or could only find a gene fragment, in several species of the orders Diprotodontia, Primates, Carnivora, and Rodentia (see supplementary table S1, Supplementary Material online for detailed species information).
Fig. 1.
Loss of Lyg genes in aquatic mammals based on published sequences. (A) Species trees for Cetacea and Sirenia (blue clades), and their close relatives (black clades), with divergence time shown (Upham et al. 2019). (B) Coding regions of the Lyg1 and Lyg2 genes from each species is shown, which are located between the Txndc9 and Mrpl30 genes. For aquatic mammals, exons for the coding regions of Lyg1 and Lyg2 are shown in blue, with untranslated regions in gray. The two flanking genes are represented by hollow rectangles, with the dotted one showing a missing gene. Only exons are drawn to scale, with introns indicated by horizontal lines. The zig-zag line represents a gap in the genomic sequence within a scaffold, whereas the dashed line indicates a gap as the sequences are from different scaffolds. Arrows above the genes indicate the direction of gene transcription. Frameshift indels and premature stop codons are indicated in red. Indels with lengths that are multiples of 3, but not more than nine bases long (three amino acids), are not marked in the exons. (C) Numbering of the key catalytic amino acid residues 73, 86, and 97 are based on the goose Lyg positions, with the substitutions shaded in either black (for site 73) or gray (for both 86 and 97).
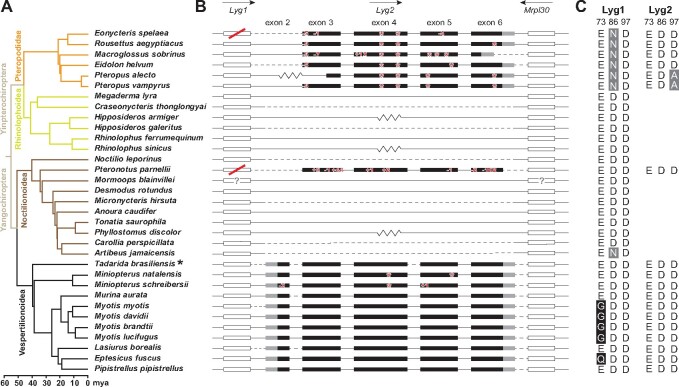
Intact Lyg2 genes were not found in the genomes of all cetaceans and sirenians examined, as well as many bat species, three primate species, the northern tree shrew (Tupaia belangeri), and the nine-banded armadillo (Dasypus novemcinctus) (supplementary table S1, Supplementary Material online). In the Sirenia, an indel (exon 3) and a stop codon (exon 4) were shared across all species. In contrast, we observed no shared mutation across all of the cetaceans examined (fig. 1) and thus no clear signature of ancestral losses was seen based on maximum parsimony reconstruction (supplementary fig. S2, Supplementary Material online). For both bats and primates, the distributions of losses were not monophyletic. For example, in bats, while all members of the family Pteropodidae (suborder Yinpterochiroptera) shared the same inactivating nonsense mutation (in exon 4), members of the genus Miniopterus (suborder Yangochiroptera) shared a different inactivating mutation (in exon 4), while no Lyg2 gene sequence was found in the genomes of species from the superfamilies Rhinolophoidea and Noctilionoidea (except for Parnell’s mustached bat [Pteronotus parnellii], which possesses an indel mutation not seen in any other bat). In contrast to these findings, most species from the superfamily Vespertilionoidea, including vespertilionid bats and possibly a molossid species (Tadarida brasiliensis), have a Lyg2 gene with an intact coding region (fig. 2). Like the Lyg1, based on partial sequences, we cannot confirm whether or not the Lyg2 genes from several species of eulipotyphlan, rodent, primate, and monotreme are pseudogenes (supplementary table S1, Supplementary Material online).
Fig. 2.
Loss of Lyg2 genes in bats based on published sequences. (A) Clades of different colors represent the different superfamilies of bats (Upham et al. 2019). For species marked with an asterisk, putative duplicated exons may exist. (B) Exons with coding regions for Lyg2 and its two flanking genes are displayed. The coding and untranslated regions are indicated in black and gray rectangles, respectively. Flanking genes (Lyg1 and Mrpl30) are indicated by hollow rectangles. Only exons are shown to scale, with introns indicated by horizontal lines. Arrows above the genes indicate the direction of gene transcription. Inactivating mutations in Lyg2 are marked with red numbers and asterisks. Lyg1 genes that do not have a complete open reading frame are marked with a red strike. Flanking genes that are not on the same scaffold and those that are in the same scaffold but whose sequence is not continuous with Lyg2 are indicated by the dotted and zig-zag lines, respectively. Flanking genes marked by question marks indicate that their positions are not the expected pattern. (C) Display of the key catalytic amino acid residue sites in Lyg1 and Lyg2. Numbering is based on goose Lyg. Black shading indicates that the amino acid change might have a large impact on the function of g-type lysozyme (site 73), whereas amino acid changes in gray shading might not affect the function (sites 86 and 97).
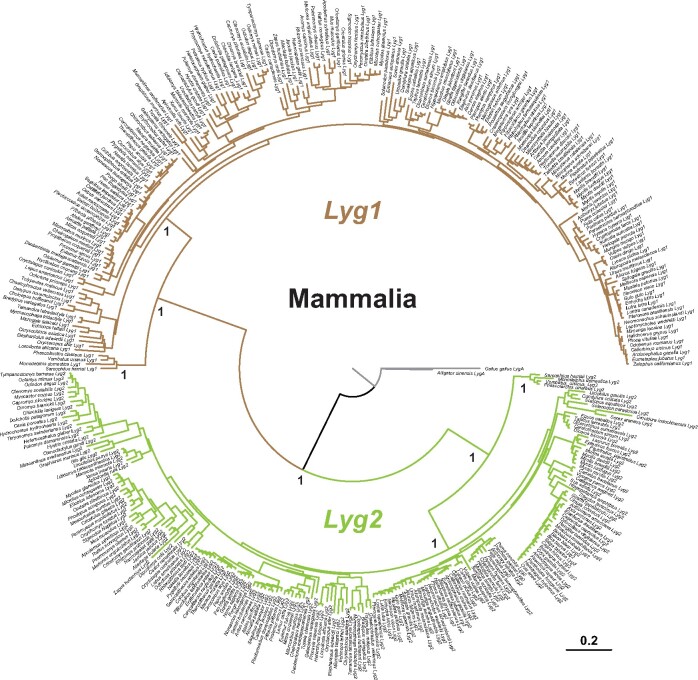
For intact g-type lysozyme genes, a phylogenetic tree was reconstructed based on either Bayesian or neighbor-joining (N-J) methods. Two clades represent mammalian Lyg1 and Lyg2 genes are highly supported in both trees, and within each clade, genes from marsupials and eutherians group together respectively (fig. 3 and supplementary fig. S3, Supplementary Material online).
Fig. 3.
The Bayesian phylogenetic tree reconstructed based on intact Lyg coding sequences. The Lyg1 (brown) and Lyg2 (green) clades are highly supported by Bayesian posterior probabilities. The amniote LygA genes from Alligator sinensis and Gallus gallus are used as outgroups.
Verification of Pseudogene Mutations Using Short-Read Data
Given that inferred pseudogenes based on genome assemblies should be treated with caution due to potential assembly errors, we validated each observed inactivating mutation by checking the short-read genomic and/or transcriptomic data set wherever these were available. Note that for one taxon, Piliocolobus tephrosceles, short-read data were not available for checking. Of 14 unique pseudogenes checked (9 Lyg1 and 5 Lyg2), we found that five were actually assembly errors and nine were real pseudogenes, although, in two of these latter cases, functional and nonfunctional copies were found together, implying possible allelic diversity (supplementary table S2, Supplementary Material online).
In the case of Lyg1, all of the mutations in the cetaceans and sirenians were supported by short-read genomic data, whereas discrepancies were seen for some gene sequences in other lineages. For example, short-read data from the roe deer (Capreolus capreolus) did not show evidence for the premature stop codon found in the genomic sequence (supplementary fig. S4A, Supplementary Material online), whereas short-read data supported the presence of a premature stop codon in exon 2 in the pronghorn (Antilocapra americana). In the Saiga antelope (Saiga tatarica), we found evidence for a premature stop codon in exon 3 in some but not all of the short-reads, which might indicate the presence of two alleles, only one of which is nonfunctional (supplementary fig. S4A, Supplementary Material online).
We also used the short-read data to examine putative loss-of-function mutations in other mammalian orders and found additional conflicts with the predicted gene sequences from the genome data sets. For example, within bats, short-read data suggested that exon 6 sequence of Lyg1 from Parnell’s mustached bat (suborder Yangochiroptera) was intact, in contrast to the 21-bp mismatch present in the genomic sequence (supplementary fig. S4A, Supplementary Material online). Similarly, we identified Lyg1 transcripts from the lung transcriptome of the lesser dawn bat (Eonycteris spelaea) (suborder Yinpterochiroptera) that did not have the 1-bp inactivating deletion seen in the genomic sequence (supplementary fig. S4B, Supplementary Material online).
In contrast to the bats, short-read data for primate Lyg1 pseudogenes supported the predicted gene sequences found in the genomic sequences. For example, the ring-tailed lemur (Lemur catta) showed stop mutations in exons 4. Additionally, mutations that would prevent the translation of a complete Lyg1 in the Philippine tarsier (Carlito syrichta) were also recovered by raw short-read data. In Rodentia, raw data supported the presence of a mutation in the start codon of Lyg1 in the dormouse, as well as the existence of a possible alternative out-of-frame ATG start codon outside. However, the putative indel mutations in exons 3 and 6 seen in the wallaby genomic sequence could not be confirmed with raw sequence data, although the coding region of this gene was still not complete (supplementary fig. S4B, Supplementary Material online).
We also examined short-read data of Lyg2 and confirmed all of the putative disrupting mutations in cetaceans, as well as the presence of a shared mutation in sirenians. Short-read data from Coquerel’s sifaka (Propithecus coquereli) recovered a premature stop codon in the last exon, although two different bases indicated the presence of two alleles, of which one is a pseudogene (supplementary fig. S4C, Supplementary Material online). Raw data also confirmed the presence of a large deletion in exon 6 in the small-eared galago (Otolemur garnettii) and deletion at the 5′-terminus of exon 4 in the Philippine tarsier (Carlito syrichta), respectively. Short-read data from the northern tree shrew failed to support the observed premature stop codon five amino acids upstream of the typical C-terminus, although the gene sequence was not complete (supplementary fig. S4C, Supplementary Material online). Finally, short-read transcriptome data recovered an alternative start codon in the nine-banded armadillo.
Verification of Lyg2 Pseudogene Mutations Using PCR
Finally, for three cetaceans (killer whale, Orcinus orca; harbor porpoise, Phocoena phocoena; and Sowerby’s beaked whale, Mesoplodon bidens) and one bat species (P. parnellii), to verify their pseudogene generating mutations for which tissue was available, we performed PCR and re-sequenced sections (exon 4, 5, or 6) of the Lyg2 genes. In each case, the mutations were identical to those observed in the genomic sequences, supporting our interpretations of wider patterns of pseudogenization (supplementary fig. S5, Supplementary Material online).
Loss of Lyg Genes in Mammals
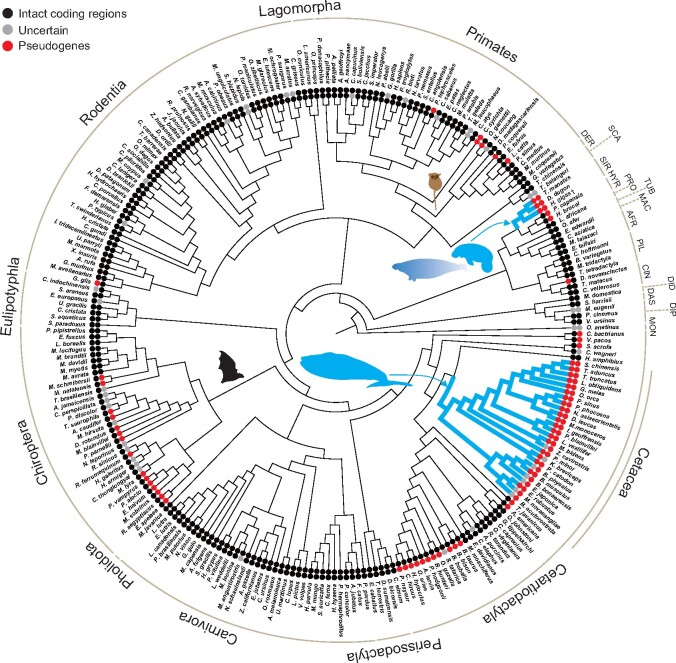
Taken together, using the verified pseudogene sequences, our data show that the losses of the Lyg1 and Lyg2 genes have occurred multiple times, and that cetaceans, sirenians, and also a tarsier have inactivated copies of both genes (fig. 4). All Lyg genes analyzed and re-examined in this study are summarized in supplementary table S3, Supplementary Material online.
Fig. 4.
The Lyg1 (outer circles) and Lyg2 (inner circles) genes of mammals, with pseudogenes validated. Color of circles represent different states of the genes, with black indicating complete open reading frames, gray being uncertain, and red being pseudogenes. Silhouettes are used to highlight species that have lost both of their Lyg1 and Lyg2 genes, with the extinct Steller’s sea cow indicated by a fading pattern, and also bats, showing extensive losses of Lyg2. Fully aquatic species are indicated by the bold blue branches. Abbreviations for the orders are: DER, Dermoptera; SCA, Scandentia; PIL, Pilosa; CIN, Cingulata; SIR, Sirenia; HYR, Hyracoidea; PRO, Proboscidea; TUB, Tubulidentata; MAC, Macroscelidea; AFR, Afrosoricida; DID, Didelphimorphia; DAS, Dasyuromorphia; DIP, Diprotodontia; and Mon, Monotremata.
Molecular Evolution of Lysozyme Genes
We compared the intensity of selection on Lyg pseudogenes from whales, sirenians, or bats to functional genes from other mammals, respectively. Pseudogenes of Lyg1 and Lyg2 from the three groups showed significant signals of relaxed selection compared with functional genes (table 1).
Table 1.
Selection Intensity for Pseudogenes in Whales, Sirenians and Bats
| Gene | Focal Clades | Backgrounds | Selection Intensity | P-Value |
|---|---|---|---|---|
| Lyg1 | Cetartiodactylan pseudogenes | Intact Lyg1 | 0 | <0.001 |
| Sirenian pseudogenes | 0.31 | 0.011 | ||
| Lyg2 | Cetartiodactylan pseudogenes | Intact Lyg2 | 0.04 | <0.001 |
| Sirenian pseudogenes | 0.11 | 0.008 | ||
| Chiropteran pseudogenes | 0.13 | <0.001 |
We also estimated the selection regimes acting on the mature protein region of g-type lysozyme across mammals, with a view to determining whether the observed gene losses (e.g., in whales and sirenians) might stem from the relaxed selection that occurs across mammals. We compared these rates to those for Lyz. Site-specific models were applied separately to multiple sequence alignments Lyg1, Lyg2, and Lyz coding sequences from 74 mammals that had single copies of each of these genes in their genomes (supplementary table S4, Supplementary Material online). These analyses showed that both g-type lysozyme genes (mature protein region) evolve more conservatively than c-type lysozyme, with Lyg2 experiencing the strongest purifying selection, and no sites were found to display evidence for positive selection (table 2).
Table 2.
Selective Pressure on Mature Protein Region of Mammalian Lysozyme Genes
| Gene | Model | ℓ | P-Value | Parameters | Site(s) under Positive Selectiona |
|---|---|---|---|---|---|
| Lyg1 | M1a | −6139.11 |
|
||
| M2a | −6138.76 | 0.702 |
|
106, 118 | |
| M8a | −6115.27 |
|
|||
| M8 | −6113.02 | 0.034* |
|
40, 71, 81, 99, 106, 116, 118, 133, 139 | |
| Lyg2 | M1a | −6311 |
|
||
| M2a | −6311 | 1 |
|
190 | |
| M8a | −6286.36 |
|
|||
| M8 | −6286.16 | 0.531 |
|
52, 113, 187, 190 | |
| Lyz | M1a | −4878.91 |
|
||
| M2a | −4847.18 | 0** |
|
33, 55, 68, 90, 96, 112, 137, 144 | |
| M8a | −4866.97 |
|
|||
| M8 | −4840.66 | 0** |
|
33, 55, 68, 90, 96, 112, 137, 144 |
P < 0.05,
P < 0.01.
Positively selected sites (probability > 0.99) shown in bold with underlines.
Discussion
In this study, we undertook the most detailed comparison of g-type lysozymes across mammals to date and uncovered extensive losses of both loci in multiple divergent taxa. For Lyg1, we corroborated a previous report of its loss in cetaceans and several lineages of even-toed ungulates (Irwin 2014). In addition, we identified several previously undescribed cases of losses in all sirenians and hyraxes, some primates, and also the dormouse. We also uncovered evidence of functional genes in some cervids as well as the giraffe, okapi, and hippopotamus, implying independent losses of this gene within the Cetartiodactyla clade. Like Lyg1, Lyg2 also experienced degradation in cetaceans and sirenians, as well as in most lineages of bats, the armadillo, and in some primates. Although most bats lack Lyg2, members of the bat family Vespertilionidae, and possibly also one member of the Molossidae, were found to possess a functional Lyg2 gene, suggesting that, like Lyg1 in artiodactyls, this locus has undergone multiple inactivation events within a single order of mammals. For both Lyg1 in artiodactyls and Lyg2 in Chiroptera, no shared inactivating mutations were identified across all species, supporting the inference of multiple parallel losses.
Despite the rampant losses we observed for both genes in cetaceans, an inspection of the key catalytic sites at positions 73, 86 and 97 (based on g-type lysozyme numbering) showed that no substitutions occurred at these sites prior to the inferred inactivating mutation. In contrast, both the Lyg1 and Lyg2 genes in sirenians show a replacement change at amino acid site 73 (E73Q and E73D, respectively). Substitutions at site 73 have previously been shown to impact enzymatic function (Kawamura et al. 2006), and, thus it is plausible that either a change in activity occurred to these genes in this clade prior to the pseudogenization of g-type lysozymes or the residues changed after the inactivation events.
Interestingly, we also observed several amino acid substitutions at site 73 in the Lyg1 of vespertilionid bats, again suggesting potential changes in enzymatic activity. In contrast, nonvespertilionid bats showed amino acid substitutions at either site 86 or 97, but not both sites. Previous work has indicated that these latter two residues can significantly affect function when they occur together, however, replacements at individual sites appear to have a less functional consequence (Helland et al. 2009). The pattern of Lyg1 sequence conservation alongside Lyg2 loss in nonvespertilionid bats could point to some form of compensation between these enzymes. If this is the case, then the inferred functional changes in the Lyg1 of vespertilionid bats could help to explain why these taxa have retained their intact Lyg2.
Taken together, our results indicate that both g-type lysozyme genes have only been lost in two groups of fully aquatic mammals, and in the tarsier, although additional genome sequence data are necessary to determine whether all tarsier species have lost both g-type lysozyme genes. The phylogenetic trees reconstructed based on functional Lyg1 and Lyg2 genes generally recovered the major mammalian groups, as previously reported (Irwin 2014), suggesting a conserved lysozyme function during the diversification of mammals. Although pseudogenes from whales, sirenians, and bats showed relaxed selection, site-model estimates of selection pressures for functional g-type lysozyme genes and the immune-related c-type lysozyme revealed stronger purifying selection acting on the mature g-type lysozyme. This implies that where g-type lysozymes have been retained in mammals then they are likely to be functionally important. Consistent with this, we found that the residues at the three critical sites (73, 86, and 97) were relatively conserved in functional copies of Lyg2 in study taxa, although much more variations were observed at these sites in functional copies of Lyg1.
In general, the functions of Lyg1 and Lyg2 in mammals are not well-defined and could be diverse (Irwin 2014). Lyg1 contains a derived amino residue substitution at the critical enzymatic site 73 and has been implicated in tumor suppression in humans (Liu et al. 2017). In contrast, protein sequences in other mammals that have retained the ancestral residue at site 73 (same state as in Lyg2) might continue to have antimicrobial activity, although this needs to be confirmed by experimental assays. In some cases, detected pseudogenes showed a loss of critical sites, however, it is unlikely that these critical sites would have had any functional consequences in these orthologs. Thus the intriguing patterns of gene loss of g-type lysozymes across mammals raise questions about the underlying triggers that led to relaxed selection in some lineages.
Some insights into the possible roles of Lyg1 and Lyg2 come from expression data. The former gene does not appear to show strong tissue-specific expression patterns, with expression reported across several organs and a peak in the kidney (Liu et al. 2017). In contrast, Lyg2 is highly expressed in specific tissues, notably the skin (Irwin 2014), eyes, and testes (Huang et al. 2011). Given that the encoded gene product of Lyg2 is predicted to retain antibacterial activity (Irwin 2014)—and thus might play a role in immunity (Huang et al. 2011)—it is plausible that expression in the skin might serve as the first line of defense. If so, then the fact that whale skin has been reported to regenerate much faster than human skin might lead to a reduced necessity for antimicrobial activity in these species (Hicks et al. 1985). If both g-type lysozyme enzymes have roles in immunity then it is also possible that they act to complement each other, such that the loss of one is compensated for by the retention of the other form. Such a scenario could help to account for why most taxonomic groups show the loss of only one copy. Further experimental work is thus needed to elucidate the functional relationship between the g-type lysozymes, as well as between these and c-type lysozyme, which also shows antimicrobial properties.
Aside from a potential function in immunity, there is an emerging specific link between Lyg2 activity and hair development. Indeed, Lyg2 expression has been recorded in anagen hair follicles (Wiener et al. 2020) and may act via the Wnt signaling pathway (Wang et al. 2019). Such a role for g-type lysozyme would explain why Lyg2 gene became obsolete in taxa that have lost their body hair. Recent results from pathological hairlessness in humans support this hypothesis; Wang et al. (2021) screened transcriptome data sets and found that Lyg2 was one of only 107 genes downregulated in patients across different alopecia phenotypes, including those with patchy to complete hairlessness (Wang et al. 2021). In this respect, it is interesting that the armadillo and bats, both of which have also lost their Lyg2 genes, are also characterized by partial body hairlessness, on the carapace and wings, respectively. That said, hairlessness does not explain gene loss in primates, implying that additional drivers might be important.
Previous studies of cetaceans and sirenians have reported multiple cases of pseudogenization, including parallel losses, that have been attributed to an aquatic niche (McGowen et al. 2014; Sun et al. 2017; Huelsmann et al. 2019; Lopes-Marques et al. 2019). Intriguingly, some such genes encode proteins that function in either immunity or hairlessness. For example, the KLK8 gene showing antibacterial activity in the skin has been deactivated in aquatic mammals (Hecker et al. 2017). Similarly, some hair-related genes have become degraded in cetaceans, including the Hairless (Hr) gene (Chen et al. 2013; Sharma et al. 2018). We thus hypothesize that relaxed selection resulting in the loss of functional Lyg1 and Lyg2 genes might relate to two separate aspects of cetacean skin morphology covering, respectively, immunity and hairlessness. Under this scenario, the evolutionary loss of both g-type lysozymes in fully aquatic mammals is a coincidence.
Materials and Methods
Searching G-Type Lysozyme Gene Sequences
To identify Lyg1 and Lyg2 genes in mammalian species, we conducted BLAST searches (12/2020) of genomes in the NCBI database (www.ncbi.nlm.nih.gov) using the human nucleotide sequences as queries (GenBank accession numbers BC029126 and BC100882 for LYG1 and LYG2, respectively). We then manually searched the identified genomic sequences to annotate the coding regions for both genes using the query sequences, and human annotation, as guides. Species and identified genes are listed in supplementary table S1, Supplementary Material online. For genes in which one or more taxa showed either missing/incomplete (a string of Ns) exons or putative duplicated exons, then we classified these as “uncertain” for the purposes of this study. For genes that contained premature stop codons, frameshift indel(s) leading to a premature stop codon(s), or lack of exon(s), and when the gene is located in a long scaffold possessing the two flanking genes, then we considered them to be pseudogenes. When the Lyg gene had all of the expected exons and predicted an intact open reading frame, then the genes were classified as functional. Putative pseudogene sequences were confirmed by searching the raw sequence data in the SRA database.
Verification of Lyg Gene Sequences
Part of the coding regions of Lyg2 genes from representative whale and bat species were verified by re-sequencing. Genomic DNA for three species of cetaceans (killer whale, harbor porpoise, and Sowerby’s beaked whale) and one bat (Parnell’s mustached bat) was used for PCR amplification. For the cetaceans, three species-specific pairs of PCR primers were designed to amplify the Lyg2 exons 4–6 genomic sequence. For Parnell’s mustached bat Lyg2 gene, a pair of primers that amplify part of exon 4 were designed. Primers are listed in supplementary table S5, Supplementary Material online. PCR products of the expected size were generated and then purified using a TIANquick Midi Purification Kit (Tiangen) and ligated into pGEM-T Easy vector (Promega) for cloning. At least three clones from each amplification were sequenced on an ABI 3730 (Applied Biosystems).
Phylogenetic Reconstruction for Intact Lyg Genes
The complete Lyg coding sequences (187 Lyg1 and 185 Lyg2) were aligned using ClustalW implemented in MEGA X, and then the phylogenetic tree was reconstructed by the N-J method in this software (Kumar et al. 2018). The maximum composite likelihood model was used and 2,000 bootstrap replications were performed. A Bayesian phylogenetic tree was also reconstructed by MrBayes 3 (Ronquist et al. 2012). K80+Γ model was selected according to the corrected Akaike information criterion by jModeltest 2 (Darriba et al. 2012). Ten million Markov chain generations were performed, with the first 4 million discarded before tree summarization. The LygA genes from a reptile (Alligator sinensis) and an avian (Gallus gallus) were used as outgroups, with the GenBank accession numbers are XM_006026334.2 and XM_416898.7.
Molecular Evolutionary Analysis of Lysozyme Gene Sequences
To infer the ancestral state for a frame-shifting insertion in exon 5 of the cetacean Lyg2, we first aligned the cetacean Lyg2 sequences using MEGA X (Kumar et al. 2018) and then reconstructed the ancestral state using the parsimony method with Mesquite 3 software (Maddison and Maddison 2019). To test for differences in selection intensity between Lyg pseudogenes (order Cetartiodactyla, Sirenia, or Chiroptera, respectively) and the other intact genes, we used the software RELAX (Wertheim et al. 2015).
Mammalian Lyz coding sequences were downloaded from NCBI (supplementary table S4, Supplementary Material online) to allow a comparison of the selective constraints acting upon these two types of lysozyme genes. For the analysis of selective constraints, we only used species that had single copies of Lyg1, Lyg2, and Lyz. The ω values (ratio of the rates of nonsynonymous to synonymous substitutions) were also estimated in Codeml in the PAML 4 package (Yang 2007).
To estimate the selection pressure acting on each of the three genes (all based on mature protein region) we ran and compared site models. The models tested were: 1) M1a (Null hypothesis: nearly neutral evolution) and M2a (Alternative hypothesis: positive selection); 2) M8a (Null hypothesis: β distribution with ω = 1) and M8 (Alternative hypothesis: β distribution with ω > 1). A species tree, based on a published topology (Upham et al. 2019), was used for ω estimation. Likelihood ratio tests were used to compare each pair of models and determine if the differences were statistically significant (Wong et al. 2004).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities [GK202102006] to Y.L. and [2020TS050] to H.C., the Natural Science Basic Research Program of Shaanxi to Y.L. [2021JM-197], the Ministry of Science and Technology of the People’s Republic of China [2016YFD0500300] to Y.L. and S.Z., the National Natural Science Foundation of China [32070407] to S.Z., and the European Research Council Starting grant [310482] to S.J.R.
Author Contributions
Y.L. designed the project. Y.L., S.Z., and S.J.R. contributed experimental materials and samples; X.Z. did the experiments; X.Z., C.H., G.L., and Y.L. analyzed the data; X.Z., D.M.I., S.J.R., and Y.L. wrote the paper.
Data Availability
New Lyg2 sequences from this study have been submitted to GenBank, with the accession numbers MW988587-MW988590. All other gene sequences used in the analyses are listed either in main text or in supplementary tables S1 and S4, Supplementary Material online.
Literature Cited
- Callewaert L, Michiels CW.. 2010. Lysozymes in the animal kingdom. J Biosci. 35(1):127–160. [DOI] [PubMed] [Google Scholar]
- Chen Z, Wang ZF, Xu SX, Zhou KY, Yang G.. 2013. Characterization of hairless (Hr) and FGF5 genes provides insights into the molecular basis of hair loss in cetaceans. BMC Evol Biol. 13:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, et al. 2016. The mucosal expression signatures of g-type lysozyme in turbot (Scophthalmus maximus) following bacterial challenge. Fish Shellfish Immunol. 54:612–619. [DOI] [PubMed] [Google Scholar]
- Hammer MF, Schilling JW, Prager EM, Wilson AC.. 1987. Recruitment of lysozyme as a major enzyme in the mouse gut: duplication, divergence, and regulatory evolution. J Mol Evol. 24(3):272–279. [DOI] [PubMed] [Google Scholar]
- Hecker N, Sharma V, Hiller M.. 2017. Transition to an aquatic habitat permitted the repeated loss of the pleiotropic KLK8 gene in mammals. Genome Biol Evol. 9:3179–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helland R, Larsen RL, Finstad S, Kyomuhendo P, Larsen AN.. 2009. Crystal structures of g-type lysozyme from Atlantic cod shed new light on substrate binding and the catalytic mechanism. Cell Mol Life Sci. 66(15):2585–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks BD, St Aubin DJ, Geraci JR, Brown WR.. 1985. Epidermal growth in the bottlenose dolphin, Tursiops truncatus. J Invest Dermatol. 85(1):60–63. [DOI] [PubMed] [Google Scholar]
- Huang P, et al. 2011. Characterization and expression of HLysG2, a basic goose-type lysozyme from the human eye and testis. Mol Immunol. 48(4):524–531. [DOI] [PubMed] [Google Scholar]
- Huelsmann M, et al. 2019. Genes lost during the transition from land to water in cetaceans highlight genomic changes associated with aquatic adaptations. Sci Adv. 5(9):eaaw6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin DM.2014. Evolution of the vertebrate goose-type lysozyme gene family. BMC Evol Biol. 14:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin DM, Gong Z.. 2003. Molecular evolution of vertebrate goose-type lysozyme genes. J Mol Evol. 56(2):234–242. [DOI] [PubMed] [Google Scholar]
- Kawamura S, Ohno K, Ohkuma M, Chijiiwa Y, Torikata T.. 2006. Experimental verification of the crucial roles of Glu73 in the catalytic activity and structural stability of goose type lysozyme. J Biochem. 140(1):75–85. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, et al. 2017. LYG1 exerts antitumor function through promoting the activation, proliferation, and function of CD4+ T cells. Oncoimmunology 6(4):e1292195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QN, et al. 2016. Molecular identification and expression analysis of a goose-type lysozyme (LysG) gene in yellow catfish Pelteobagrus fulvidraco. Fish Shellfish Immunol. 58:423–428. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. 2014. Adaptive functional diversification of lysozyme in insectivorous bats. Mol Biol Evol. 31(11):2829–2835. [DOI] [PubMed] [Google Scholar]
- Lopes-Marques M, et al. 2019. The singularity of Cetacea behavior parallels the complete inactivation of melatonin gene modules. Genes 10:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR.. 2019. Mesquite: a modular system for evolutionary analysis. Version 3.61. Available from: http://www.mesquiteproject.org.
- McGowen MR, Gatesy J, Wildman DE.. 2014. Molecular evolution tracks macroevolutionary transitions in Cetacea. Trends Ecol Evol. 29(6):336–346. [DOI] [PubMed] [Google Scholar]
- Messier W, Stewart CB.. 1997. Episodic adaptive evolution of primate lysozymes. Nature 385(6612):151–154. [DOI] [PubMed] [Google Scholar]
- Mohapatra A, Parida S, Mohanty J, Sahoo PK.. 2019. Identification and functional characterization of a g-type lysozyme gene of Labeo rohita, an Indian major carp species. Dev Comp Immunol. 92:87–98. [DOI] [PubMed] [Google Scholar]
- Moreno-Cordova EN, et al. 2020. Molecular characterization and expression analysis of the chicken-type and goose-type lysozymes from totoaba (Totoaba macdonaldi). Dev Comp Immunol. 113:103807. [DOI] [PubMed] [Google Scholar]
- Nakano T, Graf T.. 1991. Goose-type lysozyme gene of the chicken: sequence, genomic organization and expression reveals major differences to chicken-type lysozyme gene. Biochim Biophys Acta. 1090(2):273–276. [DOI] [PubMed] [Google Scholar]
- Pacheco MA, et al. 2007. Stomach lysozymes of the three-toed sloth (Bradypus variegatus), an arboreal folivore from the Neotropics. Comp Biochem Physiol A. 147(3):808–819. [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, et al. 2018. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat Commun. 9(1):1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CB, Schilling JW, Wilson AC.. 1987. Adaptive evolution in the stomach lysozymes of foregut fermenters. Nature 330(6146):401–404. [DOI] [PubMed] [Google Scholar]
- Sun XH, et al. 2017. Comparative genomics analyses of alpha-keratins reveal insights into evolutionary adaptation of marine mammals. Front Zool. 14:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upham NS, Esselstyn JA, Jetz W.. 2019. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 17(12):e3000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, et al. 2021. CCL13 is upregulated in alopecia areata lesions and is correlated with disease severity. Exp Dermatol. 30(5):723–732. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. 2019. m6A methylation analysis of differentially expressed genes in skin tissues of coarse and fine type Liaoning cashmere goats. Front Genet. 10:1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K.. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 32(3):820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener DJ, et al. 2020. Transcriptome profiling and differential gene expression in canine microdissected anagen and telogen hair follicles and interfollicular epidermis. Genes 11:884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WS, Yang Z, Goldman N, Nielsen R.. 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168(2):1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kumar S.. 1997. Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol. 14(5):527–536. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang H, Song W, Cui D, Wang L.. 2018. Identification and characterization of a novel goose-type and chicken-type lysozyme genes in Chinese rare minnow (Gobiocypris rarus) with potent antimicrobial activity. Genes Genomics. 40(6):569–577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New Lyg2 sequences from this study have been submitted to GenBank, with the accession numbers MW988587-MW988590. All other gene sequences used in the analyses are listed either in main text or in supplementary tables S1 and S4, Supplementary Material online.