Abstract
Species are indisputable units for biodiversity conservation, yet their delimitation is fraught with both conceptual and methodological difficulties. A classic example is the taxonomic controversy surrounding the Gila robusta complex in the lower Colorado River of southwestern North America. Nominal species designations were originally defined according to weakly diagnostic morphological differences, but these conflicted with subsequent genetic analyses. Given this ambiguity, the complex was re-defined as a single polytypic unit, with the proposed “threatened” status under the U.S. Endangered Species Act of two elements being withdrawn. Here we re-evaluated the status of the complex by utilizing dense spatial and genomic sampling (n = 387 and >22 k loci), coupled with SNP-based coalescent and polymorphism-aware phylogenetic models. In doing so, we found that all three species were indeed supported as evolutionarily independent lineages, despite widespread phylogenetic discordance. To juxtapose this discrepancy with previous studies, we first categorized those evolutionary mechanisms driving discordance, then tested (and subsequently rejected) prior hypotheses which argued phylogenetic discord in the complex was driven by the hybrid origin of Gila nigra. The inconsistent patterns of diversity we found within G. robusta were instead associated with rapid Plio-Pleistocene drainage evolution, with subsequent divergence within the “anomaly zone” of tree space producing ambiguities that served to confound prior studies. Our results not only support the resurrection of the three species as distinct entities but also offer an empirical example of how phylogenetic discordance can be categorized within other recalcitrant taxa, particularly when variation is primarily partitioned at the species level.
Keywords: anomaly zone, conservation genomics, hybridization, phylogenomics, RADseq, taxonomic uncertainty
Significance
Conservation decisions are often taxon-centric, with conflicting evolutionary histories deconstructed via phylogenetic inference. Yet, evolutionary complexity in these situations is often a double-edged sword, with phylogenetic ambiguities and taxonomic uncertainties acting in concert to confuse coalescent histories. At best, this renders conservation efforts ineffective, while at worst it amplifies threats and compounds management. Herein, we demonstrate an effective approach that disentangles confusing phylogenetic signals, and does so within a region where biodiversity threats have been historically exacerbated by anthropogenic and geopolitical pressures–the American southwest. We employ in our test a unique species-complex of desert fish whose evolutionary context has not only been obscured by hybridization and rapid diversification, but also compounded by incomplete spatial and genomic sampling.
Introduction
Complex evolutionary histories remain consistently difficult to disentangle, despite a recent paradigm shift toward the development of increasingly comprehensive data sets (e.g., Edwards 2009; Giarla and Esselstyn 2015). Regardless of these efforts, phylogenetic uncertainty is still prevalent and has wide-ranging consequences, for example, on the study of macroevolutionary patterns (Stadler et al. 2016; Pereira and Schrago 2018), trait evolution (Hahn and Nakhleh 2016; Mendes et al. 2016; Wu et al. 2018), and ecological and biogeographic processes (Rangel et al. 2015; McVay et al. 2017).
Importantly, phylogenetic uncertainty also translates to taxonomic uncertainty. This is because modern systematic taxonomy fundamentally describes homology (i.e., Darwin’s [1859] “propinquity of descent” [Simpson 1961]), which by definition requires a phylogenetic context. Phylogenetic uncertainty in this sense can manifest itself as a soft polytomy (=“honest” uncertainty), the erroneous promotion of nonmonophyletic clades, or controversial “splitting” versus “lumping” of taxa. Incomplete or biased sampling is often a driver of this disparity (Ahrens et al. 2016; Reddy et al. 2017). Here, narrow taxon sampling may introduce substantial ascertainment bias (=systematic deviations due to sampling). On the other hand, a broader yet sparse sampling regime often fails to represent cryptic lineages (Heath et al. 2008)—with subsequent impacts on both the delimitation of species (Pante et al. 2015; Linck et al. 2019) and an understanding of their traits (Beaulieu and O’Meara 2018).
These sources of uncertainty culminate in topologies that often fluctuate with regard to sampling designs or methodologies, and this translates into taxonomic uncertainty (e.g., Pedraza-Marrón et al. 2019; Burbrink et al. 2020; Martin et al. 2021). Access to genome-scale data has alleviated some of these issues by offering a level of accuracy not possible with single-gene phylogenies (Philippe et al. 2005). However, their inherent complexity and heterogeneity introduce new problems, and consequently, additional sources of phylogenetic uncertainty.
Gene tree heterogeneity is a ubiquitous source of discordance in genomic data, and “noise” as a source of this variance must consequently be partitioned from “signal” (where “noise” is broadly categorized as systematic or stochastic error). Large genomic data sets can reduce stochastic error (Kumar et al. 2012), yet it still remains a prevalent issue when individual genes are examined (Springer and Gatesy 2016). On the other hand, systematic error in phylogenomics may represent a probabilistic bias toward incongruence that is inherent to the evolutionary process itself (Maddison 1997). This, in turn, exemplifies the complications introduced by genomic data: As genomic resolution increases, so also does the probability of sampling unmodeled processes (Rannala and Yang 2008; Lemmon and Lemmon 2013). This potential (i.e., simultaneously decreasing stochastic error as systematic error increases) yields a very real possibility of building a highly supported but ultimately incorrect tree.
Certain demographic histories are more predisposed to systematic error than others. For instance, when effective population sizes are large and speciation events exceptionally rapid, the time between divergence events may be insufficient to sort ancestral variation, such that the most probable gene topology will conflict with the underlying species branching pattern (Degnan and Rosenberg 2006). This results in what has been coined an “anomaly zone” of tree space (i.e., dominated by anomalous gene trees, or AGTs [Degnan and Rosenberg 2006]). Inferring species trees is demonstrably difficult in this region (Liu and Edwards 2009), and exceedingly so if additional sources of phylogenetic discordances, such as hybridization, are also apparent (Bangs et al. 2018). Here, historical or persistent gene flow both compresses apparent divergence in species-trees (Leache, Harris, et al. 2014) and similarly drives a predominance of AGTs which can supersede “correct” branching patterns in some regions of parameter space (Long and Kubatko 2018). The result is a confounding effect on the adequate delineation of phylogenetic groupings (e.g., a necessary step of biodiversity conservation), as well as a limitation in the downstream analysis of affected species trees (Bastide et al. 2018; Luo et al. 2018; Morales and Carstens 2018; Bangs et al. 2020).
In clades with such complex histories, it is often unclear where the source of poor support and/or topological conflict resides (Richards et al. 2018). To analytically account for gene tree conflict, it is necessary to categorize these sources and select approaches accordingly. Failure to do so promotes false confidence in an erroneous topology, as driven by model misspecification (Philippe et al. 2011). The overwhelmingly parametric nature of modern phylogenetics ensures that imperative issues will revolve around both the processes being modeled and what they actually allow us to ask from our data (Sullivan and Joyce 2005). However, the selection of methods that model processes of interest requires an a priori hypothesis so as to delimit which processes are involved. Diagnosing prominent processes is difficult in that a phylogenetic context is required from which to build such hypotheses. Fortunately, a wealth of information can be parsed from an otherwise “non-phylogenetic” signal (sensuPhilippe et al. 2005). For example, many statistical tests diagnose hybridization via its characteristic signature on the distribution of discordant topologies (e.g., Pease and Hahn 2015). Theoretical predictions regarding AGTs and the parameters under which they are generated are also well characterized (Degnan and Salter 2005; Degnan and Rosenberg 2009). Thus, by applying appropriate analytical approaches that sample many independently segregating regions of the genome, empiricists can still derive biologically meaningful phylogenies, despite the presence of complicated species histories (McCormack et al. 2009; Kumar et al. 2012).
Here, we demonstrate an empirical approach that infers species-histories and sources of subtree discordance when conflict originates not only from anomaly zone divergences but also hybridization. To do so, we used SNP-based coalescent and polymorphism-aware phylogenetic methods that bypass the necessity of fully resolved gene trees (Chifman and Kubatko 2014; Leache et al. 2014; De Maio et al. 2015). We combine coalescent predictions, phylogenetic network inference (Solís-Lemus and Ané 2016), and novel coalescent phylogeographic methods (Oaks 2019) to diagnose the sources of phylogenetic discordance and, by so doing, resolve a seemingly convoluted complex of study-species (the Gila robusta complex of the lower Colorado River basin). We then contextualize our results to demonstrate the downstream implications of “problematic” tree-space for threatened and endangered taxa, as represented by our study complex.
The Study Species
Few freshwater taxa have proven as problematic in recent years as the Gila robusta complex (Cyprinoidea: Leuciscidae) endemic to the Gila River basin of southwestern North America (fig. 1). The taxonomic debate surrounding this complex exemplifies an inherent conflict between the traditional rigidity of systematic taxonomy versus the urgency of decision-making for conservation and management (Forest et al. 2015). Our study system is the Gila River, a primary tributary of the lower basin Colorado River that drains the majority of Arizona and ∼11% of New Mexico. The critical shortage of water in this region is a major geopolitical driver for the taxonomic controversy surrounding the study species. As an example, the Lower Colorado River basin supplies approximately half of the total municipal and agricultural water requirements of the state of Arizona, and nearly two-thirds of its total gross state product (GSP) (Bureau of Reclamation 2012; James et al. 2014). This disproportionate regional reliance creates tension between the governance of a resource and its usage (e.g., Huckleberry and Potts 2019) which in turn magnifies the stakes involved in conservation policy (Minckley 1979; Carlson and Muth 1989; Minckley et al. 2003).
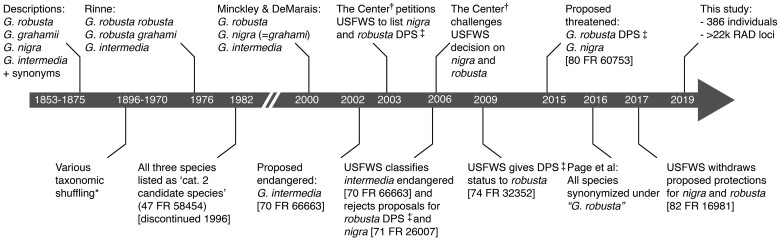
Fig. 1.
Timeline of the conservation status of Gila species endemic to the lower Colorado River basin [*See Copus et al. (2018) for a detailed overview of taxonomic synonymies; †“The Center” refers to the Center for Biological Diversity (501c3), Tuscon, AZ; ‡“DPS” = Distinct Population Segment as referenced in the United States Endangered Species Act (ESA 1973; 16 U.S.C. § 1531 et seq), here referring specifically to a lower basin sub-unit of Gila robusta]. Note that the timeline is not to scale.
We focused on three species (Roundtail chub, G. robusta; Gila chub, G. intermedia; and Headwater chub, G. nigra) that comprise a substantial proportion of the endemic Gila Basin ichthyofauna (=20% of 15 extant native species [excluding extirpated G. elegans, Ptychocheilus lucius, and Xyrauchen texanus]; Minckley and Marsh 2009). Historically, the focal taxa have been subjected to numerous taxonomic rearrangements (fig. 1). Until recently, the consensus was defined by Minckley and DeMarais (2000) on the basis of morphometric and meristic characters. These have since proven limited diagnostic capacity in the field, thus provoking numerous attempts at re-definition (Brandenburg et al. 2015; Moran et al. 2017; Carter et al. 2018). Genetic evaluations have been inconclusive to date (Schwemm 2006; Copus et al. 2018), leading to a contemporary recommendation that subsequently collapsed the complex into a single polytypic species (Page et al. 2016, 2017). Hybridization has also been implicated as an evolutionary mechanism in Gila and other codistributed Colorado River fishes (e.g., Bangs et al. 2018; Chafin et al. 2019; Corush et al. 2021), further complicating phylogenetic assessments to date.
Results
Phylogenetic Conflict in Gila
We formulated two hypotheses with regards to independent evolutionary sub-units. If populations represented a single polytypic species, then phylogenetic clustering should reflect intraspecific processes (e.g., structured according to stream hierarchy; Meffe and Vrijenhoek 1988). However, if a priori taxon assignments are evolutionarily independent, then they should be recapitulated in the phylogeny, irrespective of the drainage partition from which populations were sampled (see fig. 2). We also employed SNP-based methods that bypassed the derivation of gene trees (Leaché and Oaks 2017) given well-known issues associated with the application of supermatrix/concatenation approaches (Degnan and Rosenberg 2006; Edwards et al. 2016) and pervasive gene-tree uncertainty associated with short loci (Leaché and Oaks 2017). Of note, in order to accommodate both computational and methodological limits, as well as the population-centric nature of our a priori hypotheses, we focus on methods wherein phylogenetic tips comprise populations, not individuals.
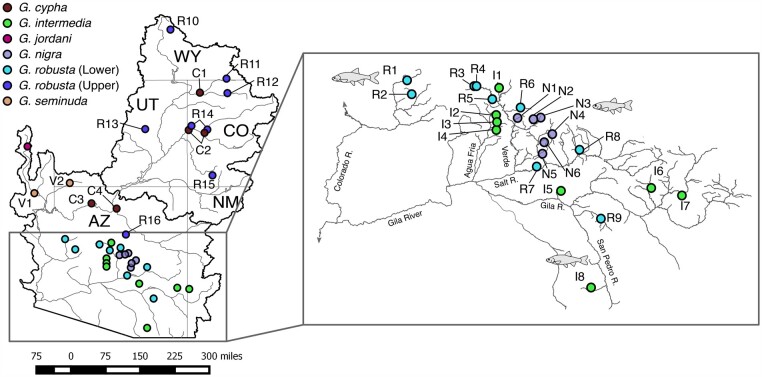
Fig. 2.
Sampling localities for Gila (n = 380 individuals) within the Colorado River Basin, southwestern North America. Locality codes are defined in Supplementary table S1, Supplementary Material online. Sympatric locations (R14 and C2) are slightly offset for visibility purposes. Map insert increases the viewing scale for sampling sites within the lower basin G. robusta “complex” (Bill Williams and Gila rivers).
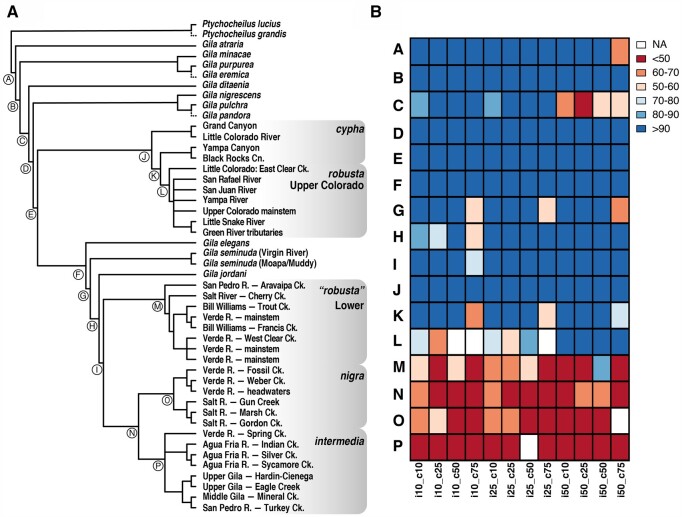
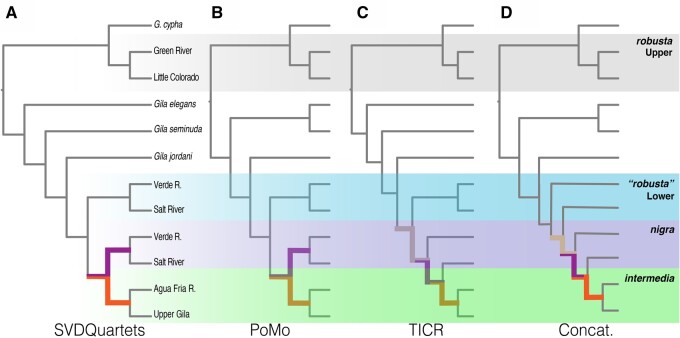
Tree reconstructions were relatively congruent across all three population methods (SVDquartets = fig. 3; PoMo, and TICR = fig. 4) . The concatenated individual-level supermatrix tree (supplementary fig. S1, Supplementary Material online) was also largely congruent with the population trees, but with two major disparities (discussed below). Bootstrap support was variable and declined with decreasing nodal depth in the SVDquartets analysis (fig. 3), whereas the vast majority of nodes in PoMo were supported at 100% (fig. 4A).
Fig. 3.
(A) Majority-rule consensus cladogram of SVDQuartets across 12 variably filtered SNP data sets varying from 7,357–21,007 SNPs and 256–347 individuals representing 16 Gila OTUs from the Colorado River Basin; Ptychocheilus spp. used as outgroups. (B) Binned bootstrap concordance values are reported for major nodes in the majority-rule consensus tree (A) labeled as (A–P). Supports are partitioned by data set, coded by the matrix occupancy threshold per individual (“i”) and per column (“c”; e.g., i50_c50 = 50% occupancy required per individual and per column). Dashed terminal branches indicate positions for taxa missing from >50% of data sets. For detailed locality information, refer to supplementary table S1, Supplementary Material online.
Fig. 4.
(A) PoMo phylogram across 12 Gila OTUs from the Colorado River basin. Branch lengths reflect the number of substitutions and inferred number of drift events per site. Branch supports (only shown for those <100%) represent concordance among 1,000 bootstrap replicates, inferred using a data set consisting of 281,613 nucleotides and 40 tips (i.e., populations); (B) Corresponding TICR phylogram reporting branch lengths in coalescent units, calculated from 31,465 quartets evaluated across 3,449 full alignments of ddRAD loci. For detailed locality information, refer to supplementary table S1, Supplementary Material online.
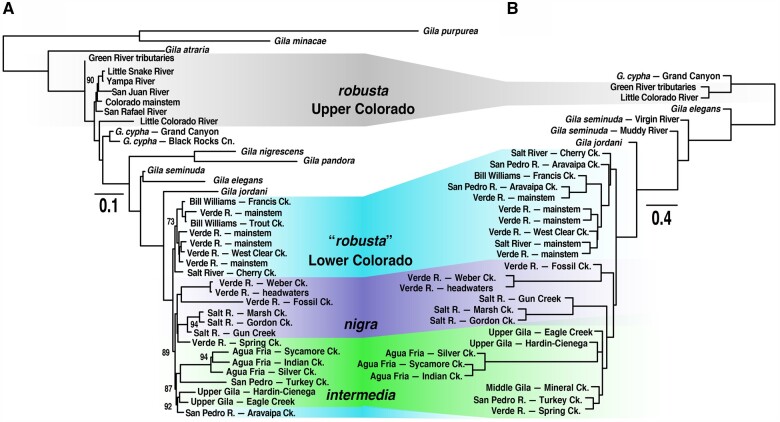
All analyses consistently supported the monophyly of a clade consisting of G. intermedia, G. nigra, and lower basin G. robusta (hereafter the “lower basin complex”). This clade had high bootstrap support in both SVDquartets and PoMo, and was universally placed as sister to G. jordani. Gila robusta was unequivocally polyphyletic in all analyses, forming two distinct groups geographically demarcated by the Grand Canyon. Lower basin G. robusta was monophyletic in all cases, save the concatenated tree, where it was paraphyletic (supplementary fig. S1, Supplementary Material online). It was also consistently recovered as sister to a monophyletic G. nigra + G. intermedia, with the exclusion of a single sample site (Aravaipa Creek) that nested within G. intermedia in the PoMo tree.
Topology was less consistent within the G. nigra + G. intermedia clade. Both were reciprocally monophyletic in the SVDquartets tree (albeit with low support; fig. 3), whereas PoMo yielded a monophyletic G. intermedia, with but one population (Spring Creek) contained within G. nigra (fig. 4A). The PoMo tree also conflicted with the other methods in the paraphyletic placement of upper basin G. robusta. We suspect this represents an artifact of well-known hybridization with sympatric G. cypha (Dowling and DeMarais 1993; Gerber et al. 2001; Douglas and Douglas 2007; Chafin et al. 2019).
Phylogenetic Conflict and Its Discrimination
The phylogenetic conflict was found to be variably attributable to either hybridization or rapid divergence. We found support for a single reticulation event connecting G. seminuda and G. elegans, an hypothesis consistent with prior interpretations (DeMarais et al. 1992). This model (i.e., with the number of reticulations [h] = 1) was selected as maximizing both first (L′[h] = L[h]—L[h−1]) and second-order (L′′[h] = L′[h + 1]—L′[h]) rate of change in phylogenetic network pseudolikelihood (supplementary figs. S2 and S3, Supplementary Material online; following Evanno et al. 2005), as computed using PhyloNetworks (Solís-Lemus and Ané 2016; Solís-Lemus et al. 2017). An alternative test using D-statistics (computed in Comp-D; Mussmann, Douglas, Bangs, et al. 2020) also supported introgression between G. elegans and G. seminuda ( = 0.302 across 86,400 tests; table1), as did analogous tests using distance-based networks (supplementary fig. S3, Supplementary Material online) and the H-statistic output by HyDe (Blischak et al. 2018). In the latter, P-values (from P = 7.8 × 10−9 to p = 5.6 × 10−8) supported a hybrid origin for G. seminuda from G. elegans and lower-basin progenitors. Introgression between upper basin G. robusta and G. cypha was also supported ( = −0.236 across 45,056 tests), corroborating other work (Chafin et al. 2019). No other introgressions were noted, thus rejecting the hypothesized hybrid origins of both G. jordani (Dowling and DeMarais 1993; Dowling and Secor 1997) and G. nigra (Demarais 1986; Minckley and DeMarais 2000).
Table 1.
Four-Taxon D-Statistic Tests of Admixture.
| P3 | P2 | P1 | Mean D | SD D | n | nSig/n (χ2) | nSig/n (Za) | nSig/n (Zb) |
|---|---|---|---|---|---|---|---|---|
| Cypha | jordani | Lower basin | 0.175 | 0.069 | 86,400 | 0.033 | 0.072 | 0.001 |
| Cypha | seminuda | Lower basin | 0.099 | 0.058 | 86,400 | 0.102 | 0.130 | 0.002 |
| elegans | jordani | Lower basin | −0.063 | 0.102 | 84,800 | 0.029 | 0.050 | 0.000 |
| elegans | robusta (lower) | nigra/int. | −0.026 | 0.109 | 413,600 | 0.014 | 0.047 | 0.001 |
| elegans | robusta (upper) | cypha | −0.236 | 0.064 | 45,056 | 0.380 | 0.415 | 0.045 |
| elegans | seminuda | Lower basin | 0.302 | 0.043 | 86,400 | 0.654 | 0.674 | 0.251 |
| jordani | robusta (lower) | nigra/int. | 0.087 | 0.055 | 601,600 | 0.042 | 0.072 | 0.001 |
| Nigra | int. (Salt) | int. (Verde) | 0.086 | 0.057 | 126,976 | 0.057 | 0.082 | 0.001 |
| robusta (lower) | intermedia | nigra | 0.041 | 0.074 | 793,600 | 0.001 | 0.002 | 0.000 |
| robusta (upper) | jordani | robusta (lower) | 0.165 | 0.085 | 168,000 | 0.050 | 0.081 | 0.001 |
| robusta (upper) | robusta (lower) | nigra/int. | −0.009 | 0.071 | 601,600 | 0.011 | 0.031 | 0.000 |
| robusta (upper) | seminuda | Lower basin | −0.017 | 0.081 | 180,800 | 0.030 | 0.053 | 0.004 |
| seminuda | jordani | Lower basin | −0.204 | 0.034 | 81,920 | 0.107 | 0.152 | 0.000 |
| seminuda | robusta (lower) | nigra/int. | 0.054 | 0.049 | 212,800 | 0.011 | 0.031 | 0.001 |
| atraria | robusta (upper) | cypha | 0.082 | 0.039 | 57,344 | 0.064 | 0.095 | 0.033 |
| nigrescens | robusta (lower) | nigra/int. | −0.075 | 0.127 | 485,472 | 0.023 | 0.079 | 0.002 |
| nigrescens | robusta (upper) | cypha | −0.039 | 0.032 | 53,248 | 0.040 | 0.066 | 0.005 |
| pandora | robusta (lower) | nigra/int. | −0.123 | 0.171 | 225,600 | 0.012 | 0.105 | 0.010 |
| pandora | robusta (upper) | cypha | −0.047 | 0.070 | 24,576 | 0.031 | 0.057 | 0.003 |
Notes.—Tests were performed for quartets sampled from n = 386 Gila individuals. Results are reported across n separate quartet samples per four-taxon test, randomly sampled without replacement, with site patterns calculated from 21,717 unlinked SNPs. Significance is reported as the proportion of tests at P < 0.05 (nSig/n) using chi-squared (χ2), Z-testa, and Z-test with Bonferroni correctionb. Positive and negative values of D suggest introgression of the P3 lineage with either P2 or P1, respectively. Results in bold were also supported by the phylogenetic network. See Supplementary table S1, Supplementary Material online for detailed locality information.
Multiple internode pairs were observed in the anomaly zone (fig. 5), as per tests developed by Linkem et al. (2016). In all cases, internode branches separating G. nigra and G. intermedia, and those separating their constituent lineages, reflected coalescent lengths that would yield anomalous gene trees. Not surprisingly, the internode separating G. jordani from the lower basin complex, and that of G. robusta from G. intermedia/G. nigra (fig. 5C; tan branches) also fell within the anomaly zone, per TICR and concatenated topology results.
Fig. 5.
Diagram comparing internode pairs within the anomaly zone, as determined using coalescent-unit transformed branch lengths mapped onto the (A) SVDQuartets, (B) PoMo, (C) TICR, and (D) concatenated trees (displayed here as cladograms). Paired internodes are color-coded: two successive internode branches of the same color = a pair of coalescent branch lengths falling within the anomaly zone; branches bicolored = branches involved in two separate significant anomalous divergences.
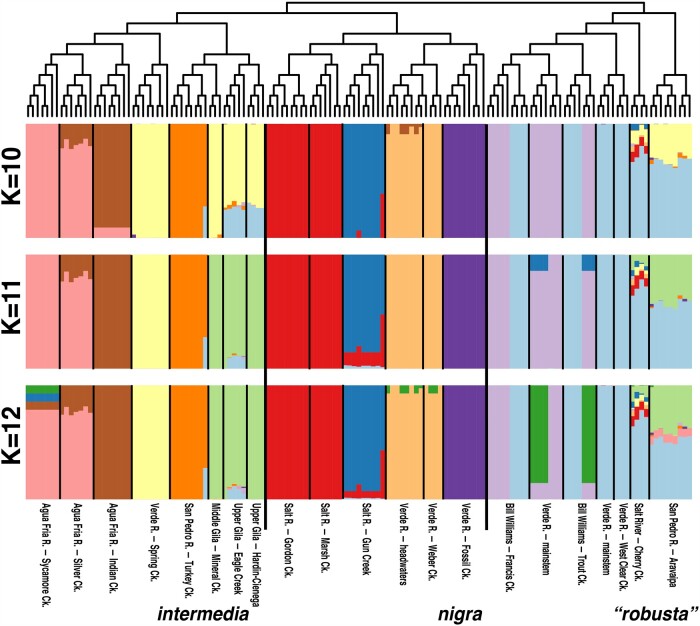
Population Assignment Tests and Contemporary Admixture
Assignment tests for the lower basin complex in the program Admixture (Alexander et al. 2009) revealed the optimal number of populations (K) as 11 (supplementary fig. S7, Supplementary Material online), thus we elected to display results for models of K = 10 through K = 12 (fig. 6). The result was fairly consistent across values of K, with all species generally displaying structure at the drainage or sub-drainage level. Gila nigra showed multiple clusters with little evidence for mixture within both Salt and Verde rivers (fig. 6), a pattern reflected in G. intermedia of the Agua Fria. The latter also showed a distinct grouping within each of the Verde and San Pedro River sites (each only represented by a single site; supplementary table S1, Supplementary Material online; fig. 6). A further distinction of Upper and Middle Gila River localities was seen at K > 11, whereas these were clustered with Verde River sites at K = 10. Gila robusta was relatively more homogenous, with little consistent drainage-level partitioning observed. Two anomalies were also seen in the results for G. robusta, in the formation of a phylogenetically inconsistent grouping involving Verde and Bill Williams samples, and an apparent admixture among G. robusta involving G. intermedia at Aravaipa Creek (fig. 6), though we note the latter was not corroborated by other tests of hybridization.
Fig. 6.
Admixture results for lower basin Gila robusta, G. intermedia, and G. nigra at K = 10–12. Individuals were obtained from 21 sites in the Gila River (fig. 2B). Results were generated for n = 140 individuals having <50% missing data for unlinked SNPs with a minor allele frequency ≥0.05 (=5,118 SNPs). Individuals are arranged according to the SVDQuartets individual-level phylogeny and are represented by stacked bar plots where colors are proportional to assignment probabilities as aggregated by Clumpak for 20 replicate runs of Admixture.
Biogeographic Hypotheses and Codivergence
The contemporary course of the Colorado River stemmed from the Pliocene erosion of the Grand Canyon and subsequent connection of the modern-day upper and lower basins, including stream capture of the Gila River (McKee et al. 1967; Minckley 1986). In the lower Colorado River basin, Gila then differentiated following one or more colonization events (e.g., Rinne 1976). Subsequent work (Douglas et al. 1999) supported this conclusion by examining contemporary phenotypic variation among all three species as a function of historical drainage connectivity, with the conclusion that body shape was most readily explained by Pliocene hydrography.
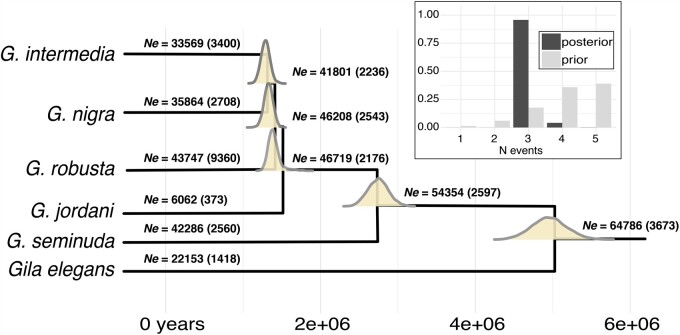
We tested if divergences were best explained by a model of in situ diversification following a single colonization event, or instead by multiple, successive colonizations. To do so, we compared divergence models using a Bayesian approach (Ecoevolity v0.3.2; Oaks 2019) that used a coalescent model (Bryant et al. 2012) to update a prior expectation for the number of evolutionary events across independent comparisons.
EcoEvolity model selection was found not to be impacted by alternative event priors (supplementary fig. S8, Supplementary Material online). The best-fitting model across all priors consistently demonstrated codivergence of G. jordani with the lower basin complex (G. robusta × G. intermedia and G. intermedia × G. nigra; fig. 7). The divergence of G. elegans and G. seminuda from a theoretical lower basin ancestor predates this putatively rapid radiation, suggesting a late-Miocene/early-Pliocene origin for G. elegans, although it is unclear if these estimates were impacted by the aforementioned introgression. Results for G. seminuda and the lower basin radiation indicated Pliocene and early Pleistocene divergences, respectively.
Fig. 7.
Posterior probability distributions for node ages and effective populations sizes (Ne) for six Gila OTUs from the Colorado River Basin. Estimates were derived from EcoEvolity and 2,000 randomly sampled full-length ddRAD locus alignments. Branches are annotated with a mean (std. dev.) Ne and posterior probabilities for divergence times are plotted on corresponding nodes. Units are in years, using a static mutation rate of 1.2 e−08 substitutions per year. The inset figure shows posterior probabilities for the total number of divergence events, as contrasted with a prior distribution weighted against codivergence (i.e., with all 5 nodes having different ages).
Discussion
Our objective was to determine if extensive geographic and genomic sampling could resolve the taxonomic recalcitrance found within the G. robusta complex. We applied diverse phylogenetic models, tests of hybridization, and predictions of parameter space within the anomaly zone to diagnose sources of discordance. In so doing, we also tested multiple hypothesized hybrid speciation events. We detected a single reticulation (G. seminuda), although other events with a lower component of genomic introgression may have also occurred. We documented rapid codivergence of lower basin taxa within the anomaly zone and were able to resolve these despite the prevalence of incomplete lineage sorting. This scenario (as outlined below) is consistent with the geomorphology of the region and, as such, seemingly represents adaptive radiation by our study complex, as facilitated by drainage evolution.
Methodological Artifacts and Conflicting Phylogenetic Hypotheses for Gila
Increased geographic and genomic sampling revealed the presence of diagnosable lineages within the G. robusta complex, with both rapid and reticulate divergences influencing inter-locus conflict. Phylogenetic hypotheses for our focal group had previously been generated using allozymes (Dowling and DeMarais 1993), Sanger sequencing (Schwemm 2006; Schönhuth et al. 2014), microsatellites (Dowling et al. 2015), and more recently RADseq (Copus et al. 2018). None could resolve relationships within the lower basin complex. To explain these contrasts, we argue that prior studies suffered from systematic artifacts and ascertainment biases that were overcome, at least in part, by our approach.
Incomplete or biased sampling is a familiar problem for biologists (e.g., Hillis 1998; Schwartz and McKelvey 2009; Ahrens et al. 2016), and we suggest it represented a major stumbling block for delineating the evolutionary history of Gila. Unfortunately, insufficient sampling is common in studies of threatened and endangered species, and its repercussions with regard to phylogenetic inference are severe (Hillis 1998). This fact is substantiated by the many examples in which increasingly comprehensive geographic sampling spurred a revision of phylogenetic hypotheses (e.g., Oakey et al. 2004; Linck et al. 2019). Likewise, incomplete sampling of genome-wide topological variation (e.g., Maddison 1997; Degnan and Rosenberg 2009) is an additional source of bias, especially when a very small number of markers are evaluated. These issues alone may explain the variation among prior studies. For example, Schwemm (2006) sampled extensively, including nearly all of the sites represented in our study, but assayed a far lower number of markers, reflecting technological constraints at the time (i.e., Sanger versus next-generation sequencing). Because anomalous gene trees are most probable under a scenario of rapid radiation (as documented herein), the reduced number of loci used by Schwemm (2006) could not recover a consistent species tree. In contrast, Copus et al. (2018) examined a data set containing 6,658 genomic SNPs (across 1,292 RAD contigs), but only did so across an extremely sparse sample (n = 19 individuals). A bioinformatic acquisition bias also may have impacted this study, in the form of strict filtering that disproportionately excluded loci with higher mutation rates, in turn diminishing the phylogenetic information content of the data set (Huang and Knowles 2016). This may explain the inability therein to discriminate G. robusta of the Little Colorado River from the remaining lower basin complex (Copus et al. 2018); a group which we have found to represent a different species entirely (e.g., Chafin et al. 2019; figs. 3 and 4).
A necessary consideration when validating phylogenetic hypotheses across methods (and data sets) is to gauge compatibility between the underlying evolutionary processes and those actually being modeled (Walker et al. 2018). In this sense, the consideration of statistical support metrics alone can be not only misleading but also promote false conclusions. For example, bootstrapping is by far the most prevalent method of evaluating support in phylogenetic data sets (Felsenstein 1985). While bootstrap concordances may be appropriate for moderately sized sequence alignments (e.g., Efron et al. 1996), they can be meaningless when applied to sufficiently large data sets (Gadagkar et al. 2005; Kumar et al. 2012). This is apparent in the high bootstrap support displayed for anomalous relationships in our study (supplementary fig. S1, Supplementary Material online). Phylogenetic signal also varies among loci, such that relatively few loci drive contentious relationships in many instances (Shen et al. 2017). This was indeed the case in Gila, where site-likelihood scores in all cases suggested that a minority of sites supported the recovered species trees (supplementary fig. S4, Supplementary Material online). Several discrepancies also reflected idiosyncrasies among the different approaches. For example, the PoMo topology has a paraphyletic upper basin G. robusta within which G. elegans, G. cypha, G. seminuda, G. jordani, and the lower basin complex were subsumed (fig. 4A). However, only ∼10% of SNPs supported this resolution (supplementary fig. S5, Supplementary Material online), a value far below the theoretical minimum SNP concordance factors (sCF) derived from completely random data (Minh et al. 2020). Of note, paraphyly is a well-known artifact when a bifurcating tree is inferred from reticulated species (Sosef 1997; Schmidt-Lebuhn 2012), with concatenation or binning approaches using genomic data being demonstrably vulnerable (Bangs et al. 2018). Thus, we tentatively attribute the observed paraphyly as an artifact of documented hybridization between G. cypha and G. robusta (Chafin et al. 2019), and the inability of PoMo to model hybridization. The lack of monophyly in G. seminuda is also potentially driven by hybridization, as indicated by TICR and the concatenation tree (supplementary fig. S1, Supplementary Material online).
In all cases, site-wide concordance was significantly predicted by subtending branch lengths, but not by node depths (supplementary fig. S6, Supplementary Material online). This suggests that site-wise concordance was unbiased in our analyses at either shallower or deeper timescales, but was instead affected by the length of time separating divergences. Some bioinformatic biases such as ortholog misidentification or lineage-specific locus dropout will disproportionally affect deeper nodes (Eaton 2017). We interpret the lack of correlation between node depth and site-wise concordance as an indication that these processes lack substantial bias. However, not all methods are equal with respect to their simplifying assumptions and the manner by which different sources of bias (e.g., bioinformatic versus biological) may drive the result. Given this, we deem it imperative to consider the biases and imperfections in both our data and the models we apply.
Complex Evolution and Biogeography of the Colorado River
The taxonomic instability of Gila is not uncommon for fishes in western North America, where puzzling patterns of diversity were generated by tectonism and vulcanism (Minckley 1986; Spencer et al. 2008). This issue is particularly emphasized when viewed through the lens of modern drainage connections (Douglas et al. 1999). Historic patterns of drainage isolation and intermittent fluvial connectivity not only support our genomic conclusions but also summarize the paleohistory of the Colorado River over temporal and spatial scales.
The earliest fossil record of Gila from the ancestral Colorado River dates back to mid-Miocene (Uyeno and Miller 1963), with subsequent Pliocene fossils representing typical “big river” morphologies now associated with G. elegans, G. cypha, and G. robusta (Uyeno and Miller 1965). The modern Grand Canyon region lacked any fluvial connection at the Miocene-Pliocene transition, due largely to regional tectonic uplifts that subsequently diverted the Colorado River (Spencer et al. 2001; House et al. 2005). Flows initiated in the early Pliocene (c.a. 4.9 mya; Sarna-Wojcicki et al. 2011) subsequently formed a chain of downstream lakes associated with the Bouse Formation (Lucchitta 1972; Spencer and Patchett 1997). The “spillover” from a successive string of Bouse Basin paleolakes was episodic, culminating in mid-Pliocene (House et al. 2008) with an eventual marine connection to the Gulf of California via the Salton Trough (Dorsey et al. 2007). Prior to this, the Gila River also drained into the Gulf (Eberly and Stanley 1978), and sedimentary evidence indicated isolation from the Colorado River until at least the northward mid-Pliocene extension of the Gulf (Helenes and Carreno 2014). This geomorphology is reflected in a broader phylogeographic pattern that underscores marked differences between resident fish communities in the upper and lower Colorado River basins (Hubbs and Miller 1948).
Intra-basin diversification also occurred as an addendum to hydrologic evolution. Although the course of the pluvial White River is now generally dry it seemingly represented a Pliocene/early Pleistocene tributary of a paleolake system that existed when the proto-Colorado River first extended into the modern-day lower basin (Dickinson 2013). This represented a potential colonization opportunity for upper basin fishes, a hypothesis that coincidentally aligns well with our rudimentary age estimate for Virgin River chub, G. seminuda (fig. 7). This early isolation, as well as the continued contrast between the spring-fed habitats and the high flows of the ancestral Colorado River, seemingly explain the unique assemblage of Gila and other fishes in the system (Hubbs and Miller 1948). Thus, as shown in other taxa (Burbrink and Gehara 2018), the biogeographic context (represented here as drainage evolution) is an important factor in the apparent reticulate evolution in Gila.
Phylogenetic signatures of the anomaly zone (fig. 5) coupled with codivergence modeling (fig. 7) suggest the diversification of lower basin Gila occurred rapidly postcolonization. Late Pliocene integration of the two basins provided an opportunity for dispersal into the lower basin tributaries. The Plio-Pleistocene regional climate was quite different, with a relatively mesic Pliocene as a precursor to a protracted monsoonal period extending through the early Pleistocene (Thompson 1991; Smith et al. 1993). The latter, in turn, may have yielded relatively unstable drainage connections (Huckleberry 1996). The potential for climate-driven instability, and the complex history of intra-drainage integration of Gila River tributaries during the Plio-Pleistocene (Dickinson 2015), lends support to the “cyclical-vicariance” model proposed by Douglas et al. (1999). Periods of isolation may have promoted an accumulation of ecological divergences that persisted postcontact and were sufficient to maintain species boundaries despite contemporary sympatric distributions and weak morphological differentiation. This hypothesis is also supported by the nonrandom mating found among G. robusta and G. nigra, despite anthropogenically induced contact (Marsh et al. 2017).
The relative timing of events inferred from our results is supported in the fossil record (Uyeno 1960; Uyeno and Miller 1963), but additional paleontological evaluations of Gila have been sparse. Thus, we hesitate to interpret these as absolute dates, given our fixed mutation rate for these analyses and uncertainty regarding the capacity of RADseq methods to yield an unbiased sampling of genome-wide mutation rate variation (e.g., Cariou et al. 2016). A noteworthy caveat was that less conservative summary metrics of the D-statistics also implicated some additional hybridization events (e.g., G. elegans × G. cypha; table 1) which were not corroborated by other methods. Recent studies have shown a vulnerability of the D-statistic to extreme demographic fluctuation (Amos 2020), which could explain these sporatic results (though we note hybridization between G. elegans × G. robusta [Corush et al. 2021] and G. robusta × G. cypha [Chafin et al. 2019] have been noted elsewhere). Thus, particularly given the contemporary decline of many species within the complex, hybridization remains difficult to disentangle using available methods, thereby emphasizing the importance of considering limitations of statistical approaches when interpreting results, especially if inferences will lead to far-reaching conclusions and management decisions.
Management Implications
A request by the Arizona Game and Fish Department to review the taxonomy of the Gila robusta complex prompted the American Fisheries Society (AFS) and the American Society of Ichthyology and Herpetology (ASIH) to recommend the synonymization of G. intermedia and G. nigra with G. robusta, owing in part to their morphological ambiguity and an imprecise taxonomic key (Carter et al. 2018). Given this, a proposal was subsequently withdrawn that would have extended protection to lower basin G. robusta and G. nigra at the federal level (USFWS 2017; fig. 1). As was the case prior to this withdrawal, G. intermedia alone is classified as endangered (USFWS 2005) under the United States Endangered Species Act (ESA 1973; 16 U.S.C. § 1531 et seq). Hence, the proposed synonymy within the complex has consequences that extend beyond taxonomy.
This study provides a much-needed resolution to this debate by defining several aspects: First, our study reinforced the recognition of G. robusta as demonstrably polyphyletic, with two discrete, allopatric clades corresponding to the upper and lower basins of the Colorado River (Dowling and DeMarais 1993; Schönhuth et al. 2014). Gila robusta was also monophyletic in both basins, with the exception of one population (Aravaipa Creek) that fell outside of the lower basin G. robusta in but one analysis (fig. 4). Of note, this population had been previously diagnosed as trending toward G. intermedia in terms of morphology (Rinne 1976; Demarais 1986). Although hybridization was not supported by D-statistics (table 1), this population did show mixed assignment in Admixture to a genotypic cluster otherwise comprised of Gila River G. intermedia (fig. 6). The fact that assignment proportions were consistent across individuals suggests that the admixture is historic (e.g., retention of introgressed alleles), rather than contemporary.
Of note, another anomaly in the Admixture results was the formation of phylogenetically spurious groupings in G. robusta from the Bill Williams drainage—here, we suspect one (or multiple) methodological artifacts. Firstly, missing information at critical sites (the presence of which is strongly implicated in our phylogenetic results; supplementary fig. S4, Supplementary Material online) can invoke spurious groupings in other clustering analyses (e.g., Martin et al. 2021). Secondly, given that our sampling regime herein was developed with a priority on phylogenetic hypotheses, per-site sample sizes are likely insufficient in number or representation to thoroughly evaluate subtle variation at the population level (e.g., Lawson et al. 2018). Our future research will investigate population structure in lower basin Gila, invoking a broader temporal and spatial depth in sampling, which will hopefully clarify this aspect.
These data, together with the geomorphic history of the region that promoted the diversification of endemic fishes (as above), clearly reject “G. robusta” as a descriptor of contemporary diversity. This underscores a major discrepancy in the taxonomic recommendations for the lower basin complex (Page et al. 2016). Given that the type locality of G. robusta is in the upper basin (i.e., the Little Colorado River), a pressing need is established to either determine taxonomic precedence for the lower basin “G. robusta,” or to provide a novel designation. A distinct possibility is the potential resurrection of a synonym, which would necessitate a detailed examination of type specimens prior to a formal recommendation. This may be appropriately adjudicated by the AFS-ASIH Names of Fishes Committee, as a follow-up to their earlier involvement.
The phylogenetic placement of G. intermedia and G. nigra is slightly more ambiguous. The short internodes and anomaly zone divergences identified herein explain previous patterns found in population-level studies, with elevated among-population divergence but scant signal uniting species (Dowling et al. 2015). This pattern was echoed (as in Dowling et al. 2015) in the analysis of population structure, where both species had relatively little exchange among drainages, with partitioning at the sub-drainage level also markedly present (fig. 6).
Tests of hybridization unequivocally rejected the previous hypothesis of hybrid speciation for G. nigra (Minckley and DeMarais 2000; Dowling et al. 2015), and instead demonstrated divergence in the “anomaly zone” as being explanatory. Thus, intermediacy in the body shape of G. nigra likely reflects differences accumulated during historic isolation (Douglas et al. 1999) and/or the retention of an adaptive ecomorphology (Douglas and Matthews 1992). These hypotheses warrant further exploration, with provisional results impinging upon future management decisions (Forest et al. 2015).
With regards to taxonomy, we confidently support G. intermedia as evolutionarily distinct from lower basin “robusta.” We likewise find phylogenetic support for G. nigra as an independent lineage, which together with prior evidence for assortative mating in a rare case of sympatry with lower basin “robusta” (Marsh et al. 2017) lends support to its continued recognition. Although, we note that genetic differentiation is but one component of an integrative species delimitation process. For management purposes, especially given morphological ambiguity within the complex, we echo a population-centric approach in practice (previously argued for by Dowling et al. 2015; Marsh et al. 2017). The emphasis is on fine-scale population structure (fig. 6) identified within broader phylogenetic groupings (e.g., figs. 3 and 4), congruent with the “3-species” hypotheses for lower basin Gila (Minckley and DeMarais 2000). We again note a formal taxonomic investigation is needed to clarify the appropriate designation of G. robusta.
Our case study of Gila emphasizes three primary components of a “Darwinian shortfall” in biodiversity conservation (Diniz-Filho et al. 2013): (i) The lack of comprehensive phylogenies; (ii) Uncertain branch lengths and divergence times; and (iii) insufficient models linking phylogenies with ecological and life-history traits. Taxonomic uncertainty in Gila is severely impacted by the first two, with taxonomic resolution prevented by the comingling of sparse phylogenetic coverage with temporal uncertainty. We must now address the relationships between ecology, life history, and phylogeny, so as to understand the manner by which phylogenetic groupings (identified herein) are appropriate as a surrogate for adaptive/functional diversity in Gila. For example: To what degree are Gila in the lower basin ecologically nonexchangeable (e.g., Crandall et al. 2000; Holycross and Douglas 2007)? How do they vary in their respective life histories? Is reproductive segregation maintained in sympatry (as in Marsh et al. 2017), and if so, by what mechanism?
Conclusions
The intractable phylogenetic relationships in Gila were resolved herein through improved spatial and genomic sampling. Our data, coupled with polymorphism-aware methods and contemporary approaches that infer trees, yielded a revised taxonomic hypothesis for Gila in the lower Colorado River. The geomorphic history of the Colorado River explains many anomalous patterns are seen in this and previous studies, wherein opportunities for contact and colonization were driven by the orogeny characteristic of the region. The signal of rapid diversification is quite clear in our data, as interpreted from patterns inherent to phylogenetic discord. We emphasize that discordance in this sense does not necessarily represent measurement error or uncertainty. Instead, it is an intrinsic component of phylogenetic variance that is not only expected within genomes (Maddison 1997), but also a necessary component from which to build hypotheses regarding the underlying evolutionary process (Hahn and Nakhleh 2016). Ignoring this variance in pursuit of a “resolved phylogeny” can lead to incorrect inferences driven by systematic error. Similarly, insufficient spatial or genomic sampling may also promote false confidence in anomalous relationships, particularly when character sampling is particularly dense whereas taxon sampling is sparse.
We reiterate that phylogenetic hypotheses, by their very nature, cannot exhaustively capture the underlying evolutionary process. One approach is to categorize phylogenetic (and “nonphylogenetic”) signals in those regions of the tree that are refractive to certain models (as done herein). We also acknowledge that attempts to reconstruct the past using contemporary observations represent a struggle against uncertainty and bias, with phylogenetic/taxonomic revisions expected as additional data are accrued. As such, we urge empiricists who engage in taxonomic controversies (such as herein) to interrogate their results for transparency. Sorting through conflicting recommendations invariably falls to natural resource managers that are mandated to implement conservation strategies based on “best available science,” but are rarely trained in phylogenetic inference or taxonomy. Unreported methodological or geopolitical biases only confound those efforts.
Materials and Methods
Taxonomic Sampling
A representative panel of n = 380 individuals (supplementary table S1, Supplementary Material online; fig. 2) was chosen primarily from field collections described in previous studies (Douglas et al. 2001; Douglas and Douglas 2007; Chafin et al. 2019), to include a broad geographic sampling of the complex as well as congeners. Several Gila species external to Colorado River drainage were also obtained from museum collections: Gila orcutti (Los Angeles County Museum of Natural History [LACM: 555990-1, 57271-1]), Gila atraria (Monte L. Bean Life Science Museum at Brigham Young University [BYU: 57580-4, 68470-4, 138751-2 61643-8]), and G. nigrescens (JJDE: 06-24 341:343, 06-16_259, and 06-16_267), G. minacae (JJDE: 06-20 302:306), and G. pulchra (Bell Museum at the University of Minnesota [JJDE: 06-15 238:239, 06-16 240, 06-17 241, and 06-18 242]). For the sake of clarity, we employed herein the nomenclature of Minckley and DeMarais (2000) and retained species-level nomenclature for all members of the Gila robusta complex. Additionally, we discriminate between G. robusta from the upper and lower basins of the Colorado River ecosystem (Chafin et al. 2019).
Given that no self-sustaining populations of wild Gila elegans exist, our samples were obtained from the Southwestern Native Aquatic Resources and Recovery Center (SNARRC; Dexter, NM). The genus Ptychocheilus served to root the Gila clade within the broader context of western leuciscids (Schönhuth et al. 2012, 2014, 2018).
Reduced-Representation Sequencing
Genomic DNA was extracted using either PureGene or DNeasy kits (Qiagen Inc.) and quantified via fluorometer (Qubit; Thermo-Fisher Scientific). Library preparations followed the published ddRAD protocol (Peterson et al. 2012), as in Chafin et al. (2019). Restriction enzyme and size-selection ranges were first screened using an in silico procedure, with various combinations of PstI, MspI, EcoRI, SbfI, and HpyCH4V being tested (Chafin et al. 2018). We excluded those enzyme combinations resulting in undesirable numbers of markers within a reasonable size selection range, or which suggested a prevalence of repetitive elements were excluded. Candidate enzyme pairings and size selection ranges were subsequently validated by quantifying digests for 15 representative samples on an Agilent 2200 TapeStation. Final library preparations were double-digested using a high-fidelity PstI (5′-CTGCAG-3′) and MspI (5′-CCGG-3′) following manufacturer’s protocols (New England Biosciences).
Digests were purified using bead purification at a 1.5X concentration (Ampure XP; Beckman-Coulter Inc.) and standardized prior to ligation at 100 ng per sample. Customized adapters (see Peterson et al. 2012) containing unique in-line barcodes were used to ligate samples with T4 DNA Ligase (New England Biosciences, Inc.) following manufacturer’s protocols. These were subsequently pooled in sets of 48, and size-selected at 250–350 bp (not including adapter length), using a Pippin Prep automated gel extraction instrument (Sage Sciences). Adapters were then extended in a 12-cycle PCR, using Phusion high-fidelity DNA polymerase (New England Biosciences Inc.), following the manufacturer’s protocol. This step completed adapters for Illumina sequencing and added an i7 index, which was unique to each library per lane. Libraries were pooled to n = 96 samples per lane (i.e., two sets of 48) at 10–20 nM concentration in 25 µl volumes for 100 bp single-end sequencing on an Illumina HiSeq 2500 at the University of Wisconsin Biotechnology Center (Madison, WI).
Data Processing and Assembly
Raw Illumina reads were demultiplexed and filtered using the pyRAD v3.0.6 pipeline (Eaton 2014). We removed reads containing >1 mismatch in the barcode sequence, or >5 low-quality base-calls (Phred Q < 20). Assembly of putative homologs was performed using de novo clustering in VSEARCH v2.15.0 (Rognes et al. 2016) using an 80% mismatch threshold. Loci were excluded according to the following criteria: >5 ambiguous nucleotides; >10 heterozygous sites in the alignment; >2 haplotypes per individual; <20X and >500X sequencing depth per individual; >70% heterozygosity per-site among individuals.
Our ddRAD approach generated 22,768 loci containing a total of 173,719 variable sites, of which one variable site per locus was sampled at random resulting in a data set of 21,717 single nucleotide polymorphisms (1,051 loci were monomorphic and thus were excluded). The mean per-individual depth of coverage across all retained loci was 79X. All relevant scripts for postassembly filtering and data conversion are available as open-source (github.com/tkchafin/scripts: concatenateNexus.py, filterLoci.py, makeHyde.py, nremover.pl, phylip2biNumNex.py, phylip2ecoevolity.pl, phylipFilterPops.pl).
Phylogenetic Inference
Using SVDquartets (Chifman and Kubatko 2014, 2015; as implemented in PAUP* v4.0, Swofford 2002), we first explored population trees across 12 variably filtered data sets using four differing occupancy thresholds (i.e., percentage of nonmissing data needed to maintain the locus) per SNP locus (i.e., 10, 25, 50, and 75%), along with three differing thresholds per individual (10, 25, and 50%). These filtered data sets ranged from 7,357–21,007 SNPs, with 8.48–43.65% missing data and 256–347 individuals. SVDquartets eases computation by inferring coalescent trees from randomly sampled quartets of species (i.e., optimizing among three possible unrooted topologies). It then generates a population tree via the implementation of a quartet-assembly algorithm (Snir and Rao 2012) that minimizes conflicts among quartet trees. Given run-time constraints (the longest was 180 days on 44 cores), all runs sampled quartets and were evaluated across 100 bootstrap pseudo-replicates.
We also used a polymorphism-aware method (PoMo; Schrempf et al. 2016) in IQ-Tree v1.6.7 (Nguyen et al. 2015) that considers allele frequencies rather than single nucleotides, thus allowing evaluation of change due to both substitution and drift. We used the entire alignment, to include nonvariable sequences so as to provide PoMo with empirical estimates of polymorphism. We filtered liberally using individual occupancy thresholds of 10% per-locus so as to maximize individual retention and per-population sample sizes. We then deleted populations that contained <2 individuals, and loci with ≥90% missing data in any single population (i.e., removing a locus even if highly prevalent in all other populations). This yielded a data set of 281,613 nucleotides and 40 groups. Nonfocal outgroups were excluded due to their disproportionate effect on missing data. Analyses were pseudo-replicated across 100 bootstraps.
We also calculated concordance factors (CFs) using a Bayesian concordance analysis in BUCKy v1.4.4 (Larget et al. 2010). The analysis was parallelized across all quartets via an adaptation of the TICR pipeline (Stenz et al. 2015), sampling every 1,000th iteration with four MCMC chains, each of length 10,000,000. The first 25% of sampled topologies for each quartet were discarded as burn-in. To prepare these data, we sampled all nonmonomorphic full gene alignments for which at least one diploid genotype at minimum could be sampled per population. We excluded outgroups and nonfocal Gila so as to maximize the number of loci retained. This yielded 3,449 genes across 31 sampled groups. Gene-tree priors were generated using MrBayes v.3.2.6 (Ronquist et al. 2012) with four independent chains, sampling each every 10,000 iterations, with a total chain length of 100,000,000 iterations and 50% discarded as burn-in. BUCKy was then run in parallel to generate quartet CFs across 31,465 quartets, using a chain length of 10,000,000, again with 50% burn-in. A population tree of quartet topologies was generated using QuartetMaxCut (Snir and Rao 2012) with the get-pop-tree.pl script from TICR (Stenz et al. 2015; https://github.com/nstenz/TICR).
Comparing Phylogenies and Estimating Site-Wise Conflict
To evaluate the performance of SVDquartets, TICR, and PoMo, we first computed site-wise log-likelihood scores (SLS) for each topology in IQ-Tree. Here, we used the population trees from SVDquartets, TICR, and PoMo as topological constraints in IQ-Tree (provided via -g), where population topologies served as a “skeletal” framework, with individual relationships within clades then optimized by IQ-Tree. This was done because we wished IQ-Tree to compute SLS scores only for each precalculated topology, which could then be compared to those computed from an unconstrained tree computed on concatenated data. All analyses employed a GTR model with empirical base frequencies and gamma-distributed rates and were assessed across 1,000 bootstrap pseudoreplicates. Analyses were also reduced to a subset of tips common across all variably filtered data sets. We quantified the phylogenetic signal supporting each resolution as the difference in site-wise log-likelihood scores (ΔSLS) between each population tree and the concatenation tree (Shen et al. 2017). We then calculated site-wise concordance factors (sCF) as an additional support metric (Minh et al. 2020).
Tests of Hybridization and Deep-Time Reticulation
D-statistics (Green et al. 2010; Eaton and Ree 2013) were calculated using Comp-D v2018-06-28 (Mussmann, Douglas, Bangs, et al. 2020). To further test hypotheses of reticulation, we used quartet concordance factors (CFs) as input for phylogenetic network inference using the SNaQ algorithm (PhyloNetworks v0.8.0; Solís-Lemus and Ané 2016; Solís-Lemus et al. 2017). The network was estimated under models of 0–5 hybrid nodes (h) that were evaluated using 48 independent replicates, with the model that maximized change in pseudolikelihood being judged best-fit.
Given the computational constraints of network inference, we reduced the data set to n = 2 populations per focal species (=12 total tips). We also explicitly tested for putative hybrid taxa (HyDe v0.4.2; Blischak et al. 2018), by using phylogenetic invariants to diagnose hybrid lineages, using all possible parent-descendant combinations. In this case, hybrids are detected by considering the ratio of two phylogenetic invariants which evaluate to zero for opposing topologies (Meng and Kubatko 2009; Chifman and Kubatko 2014, 2015). This ratio is incorporated into what Kubatko and Chifman (2019) refer to as the Hils statistic, H, which is compared with a normal distribution for hypothesis testing in hybrid taxa. Significance was assessed at a Bonferroni-corrected threshold = 5.7 × 10−8. We contrasted the resulting networks using a distance-based complement generated with the NeighborNet algorithm (Bryant and Moulton 2004), as implemented by SplitsTree4 (Huson 1998).
Finally, we visualized patterns of population differentiation and possible admixture using assignment tests (Admixture v1.3.0; Alexander et al. 2009). Here, we examined models with a priori population (K) varying from n = 1–16, with each K value evaluated with 20 replicates run in parallel (AdmixPipe v3.0; Mussmann, Douglas, Chafin, et al. 2020). Because Admixture and similar methods are strongly influenced by low-frequency variants (e.g., Linck and Battey 2019; Martin et al. 2021), we first filtered unlinked SNPs to only those exceeding a minor allele frequency (MAF) of 5%. Following the MAF filter, we additionally removed individuals missing >50% of genotypes, in those individual assignments with very large proportions of missing data can be dominated by uncertainty rather than biological signal (Martin et al. 2021). Finally, we chose the optimal K value as that which maximized classification in an Admixture cross-validation process. Results were then clustered and visualized (Clumpak web server; Kopelman et al. 2015).
Anomaly Zone Detection
Coalescent theory characterizes the boundaries of the anomaly zone in terms of branch lengths in coalescent units (Degnan and Rosenberg 2006). To test if contentious relationships in our tree fell within the anomaly zone, we first transformed branch lengths using quartet CFs (Stenz et al. 2015) , then tested if internode branch lengths fell within the theoretical boundary for the anomaly zone (Linkem et al. 2016). Code for these calculations are modified from Linkem et al. (2016) and are available as open-source (github.com/tkchafin/anomaly_zone).
Tests of Codivergence
Tests of codivergence were performed using the Bayesian method Ecoevolity v0.3.2 (Oaks 2019). Here, four independent MCMC chains were run with recommended settings (see documentation at phyletica.org/ecoevolity) and a burn-in that maximized effective sample sizes. Event models followed a Dirichlet process, with the concentration parameter exploring four alternative gamma-distributed priors (i.e., α = 2.0, β = 5.70; α = 0.5, β = 8.7; α = 1.0, β = 0.45; and α = 2.0, β = 2.18).
We randomly sampled 2,000 full-locus alignments, then examined potential codivergences in the lower-basin complex by selecting a series of pairwise comparisons: Gila elegans × G. robusta (lower); G. seminuda × G. robusta (lower); G. jordani × G. robusta (lower); G. intermedia × G. robusta (lower); and G. intermedia × G. nigra (lower). These targeted nodes are represented by “F,” “G,” “H,” “I,” and “N” in the SVDQuartets topology (fig. 3A).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This research was conducted, in partial fulfillment, by TKC of the Ph.D. degree in Biological Sciences at the University of Arkansas, as enabled by a Distinguished Doctoral Fellowship (DDF) award. Numerous agencies and organizations contributed field expertise, sampling, permit authorization, funding, and/or valuable comments: Arizona Game and Fish Department, Colorado Parks and Wildlife, Jicarilla Apache Game and Fish, Nevada Department of Wildlife, New Mexico Department of Game and Fish, United States Fish and Wildlife Service, United States Forest Service, National Park Service, United States Bureau of Reclamation, Utah Department of Natural Resources, Utah Division of Wildlife, and the Wyoming Game and Fish Department. Particular thanks are extended to J. Alves, M. Anderson, R. Anderson, P. Badame, K. Bestgen, M. Breen, M. Brouder, K. Breidinger, S. Bryan, P. Cavalli, B. DeMarais, T. Dowling, R. Fridell, K. Gelwicks, K. Guadalupe, J. Harter, B. Harvey, K. Hilwig, M. Hudson, D. Keller, J. Logan, C. McAda, S. Meiser, C. Minckley, T. Modde, K. Morgan, G. Munsen, F. Pfeifer, W. Radke, A. Rehm, S. Ross, M. Smith, R. Stevenson, R. Timmons, S. Tolentino, P. Unmack, A. Varelas, D. Weedman, K. Wilson, and E. Woodhouse (with apologies to anyone inadvertently overlooked). We also received tissues from the Bell Museum at the University of Minnesota, Los Angeles County Museum of Natural History, and the Monte L. Bean Life Science Museum at Brigham Young University (see accessions numbers in-text). We thank R. Feeney, D. Shiozawa, A. Simons, and the original collectors of accessioned materials. Additional sampling was completed by MED and MRD under permits provided by: Arizona Game and Fish Department, The Hualapai Tribe, The Navajo Nation, Nevada Department of Wildlife, U.S. National Park Service, and Wyoming Department of Game and Fish. Federal permit TE042961-0 (1434-HQ-97RU-01552 RWO #66) was provided by the U.S. Game and Fish Service. Sampling procedures were approved under Arizona State University Animal Care and Use Committee (ASU IACUC) permit 98–456R and Colorado State University Animal Care and Use Committee (CSU IACUC) permit 01–036A-01. Travel in the Grand Canyon was conducted under auspices of Grand Canyon National Park river use permits. We are also indebted to students, postdoctorals, and faculty who have promoted our research: A. Alverson, W. Anthonysamy, M. Davis, L. James, J. Pummill, A. Tucker. Funding was provided by several generous endowments from the University of Arkansas: The Bruker Professorship in Life Sciences (MRD), and the Twenty-First Century Chair in Global Change Biology (MED). This work was also in part facilitated by funding provided by the Arizona Game and Fish Heritage Fund, Colorado Parks and Wildlife, and U.S. Geological Survey Species at Risk Program. Additional analytical resources were provided by the Arkansas Economic Development Commission (Arkansas Settlement Proceeds Act of 2000) and the Arkansas High Performance Computing Center (AHPCC), and from an XSEDE Research Allocation [TG-BIO160065] to access the Jetstream cloud. The use of trade, product, industry or firm names is for informative purposes only and does not constitute an endorsement by the U.S. Government or the U.S. Fish and Wildlife Service. Links to non-Service Web sites do not imply any official U.S. Fish and Wildlife Service endorsement of the opinions or ideas expressed therein or guarantee the validity of the information provided. This work was also supported in part via a U.S. National Science Foundation (NSF) Postdoctoral Fellowship in Biology to TKC under Grant No. [DBI: 2010774]. The findings, conclusions, and opinions expressed in this article represent those of the authors, and do not necessarily represent the views of the U.S. Fish & Wildlife Service, funding agencies, nor other affiliated or contributing organizations.
Author Contributions
All authors contributed to research conceptualization and design. MED and MRD contributed to the sampling and coordination of agency efforts. TKC performed molecular work and bioinformatics. TKC, SMM, BTM, and MRB wrote relevant codes and contributed analyses. All authors contributed to writing the manuscript and approve the final submission.
Data Availability
Raw sequences are available via NCBI SRA associated with BioProject PRJNA558355. All additional relevant metadata, codes, assembled data sets, and unfiltered alignment files are available via the Open Science Framework (doi: 10.17605/OSF.IO/WABJT). Codes and custom scripts are also available as open-source under the GNU Public License via GitHub: github.com/tkchafin (doi: 10.5281/zenodo.5181290).
Literature cited
- Ahrens D, et al. 2016. Rarity and incomplete sampling in DNA-based species delimitation. Syst Biol. 65(3):478–494. [DOI] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos W.2020. Signals interpreted as archaic introgression appear to be driven primarily by faster evolution in Africa. R Soc Open Sci. 7(7):191900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs MR, Douglas MR, Mussmann SM, Douglas ME.. 2018. Unraveling historical introgression and resolving phylogenetic discord within Catostomus (Osteichthys: Catostomidae). BMC Evol Biol. 18(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs MR, Douglas MR, Chafin TK, Douglas ME.. 2020. Gene flow and species delimitation in fishes of Western North America: flannelmouth (Catostomus latipinnis) and Bluehead sucker (C. Pantosteus discobolus). Ecol Evol. 10(13):6477–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide P, Solís-Lemus C, Kriebel R, Sparks KW, Ané C.. 2018. Phylogenetic comparative methods on phylogenetic networks with reticulations. Syst Biol. 67(5):800–820. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, O’Meara BC.. 2018. Can we build it? Yes we can, but should we use it? Assessing the quality and value of a very large phylogeny of campanulid angiosperms. Am J Bot. 105(3):417–432. [DOI] [PubMed] [Google Scholar]
- Blischak PD, Chifman J, Wolfe AD, Kubatko LS.. 2018. HyDe: a python package for genome-scale hybridization detection. Syst Biol. 67(5):821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg WH, Kennedy JL, Farrington MA.. 2015. Determining the historical distribution of the Gila robusta complex (Gila chub, Gila intermedia, Headwater Chub, Gila nigra, and Roundtail Chub, Gila robusta) in the Gila River Basin, New Mexico, using morphological analysis. Roswell (NM: ): New Mexico Department of Game & Fish. [Google Scholar]
- Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A.. 2012. Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol Biol Evol. 29(8):1917–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Moulton V.. 2004. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 21(2):255–265. [DOI] [PubMed] [Google Scholar]
- Burbrink FT, et al. 2020. Interrogating genomic-scale data for Squamata (lizards, snakes, and amphisbaenians) shows no support for key traditional morphological relationships. Syst Biol. 69(3):502–520. [DOI] [PubMed] [Google Scholar]
- Burbrink FT, Gehara M.. 2018. The biogeography of deep time phylogenetic reticulation. Syst Biol. 67(5):743–755. [DOI] [PubMed] [Google Scholar]
- Bureau of Reclamation. 2012. Colorado River basin water supply and demand study. Washington (DC: ): USBR. [Google Scholar]
- Cariou M, Duret L, Charlat S.. 2016. How and how much does RAD-seq bias genetic diversity estimates? BMC Evol Biol. 16(1):240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CA, Muth RT.. 1989. The Colorado River: lifeline of the American Southwest. Can Spec Publ Fish Aquat Sci. 106:220–239. [Google Scholar]
- Carter JM, Clement MJ, Makinster AS, Crowder CD, Hickerson BT.. 2018. Classification success of species within the Gila robusta complex using morphometric and meristic characters—a re-examination. Copeia 106(2):279–291. [Google Scholar]
- Chafin TK, Douglas MR, Martin BT, Douglas ME.. 2019. Hybridization drives genetic erosion in sympatric desert fishes of western North America. Heredity 123(6):759–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafin TK, Martin BT, Mussmann SM, Douglas MR, Douglas ME.. 2018. FRAGMATIC: in silico locus prediction and its utility in optimizing ddRADseq projects. Conservation Genet Resour. 10(3):325–328. [Google Scholar]
- Chifman J, Kubatko L.. 2014. Quartet inference from SNP data under the coalescent model. Bioinformatics 30(23):3317–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chifman J, Kubatko L.. 2015. Identifiability of the unrooted species tree topology under the coalescent model with time-reversible substitution processes. J Theor Biol. 374:35–47. [DOI] [PubMed] [Google Scholar]
- Copus JM, Montgomery WL, Forsman ZH, Bowen BW, Toonen RJ.. 2018. Geopolitical species revisited: genomic and morphological data indicate that the roundtail chub Gila robusta species complex (Teleostei, Cyprinidae) is a single species. PeerJ 6:e5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corush JB, Fitzpatrick BM, Wolfe EL, Keck BP.. 2021. Breeding behavior predicts patterns of natural hybridization in North American minnows (Cyprinidae). J Evol Biol. 34(3):486–500. [DOI] [PubMed] [Google Scholar]
- Crandall KA, Bininda-Emonds OR, Mace GM, Wayne RK.. 2000. Considering evolutionary processes in conservation biology. Trends Ecol Evol. 15(7):290–295. [DOI] [PubMed] [Google Scholar]
- Darwin C.1859. On the origin of species by means of natural selection, or preservation of favoured races in the struggle for life. London (UK: ): John Murray. [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA.. 2006. Discordance of species trees with their most likely gene trees. PLoS Genet. 2(5):e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA.. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol. 24(6):332–340. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Salter LA.. 2005. Gene tree distributions under the coalescent process. Evolution 59(1):24–37. [PubMed] [Google Scholar]
- Demarais BD.1986. Morphological variation in Gila (Pisces: Cyprinidae) and geological history: lower Colorado River basin [Thesis]. [Tempe (AZ)]: Arizona State University.
- DeMarais BD, Dowling TE, Douglas ME, Minckley WL, Marsh PC.. 1992. Origin of Gila seminuda (Teleostei: Cyprinidae) through introgressive hybridization: implications for evolution and conservation. Proc Natl Acad Sci USA. 89(7):2747–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson WR.2013. Rejection of the lake spillover model for initial incision of the Grand Canyon, and discussion of alternatives. Geosphere 9(1):1–20. [Google Scholar]
- Dickinson WR.2015. Integration of the Gila River drainage system through the Basin and Range province of southern Arizona and southwestern New Mexico (USA). Geomorphology 236:1–24. [Google Scholar]
- Diniz-Filho JAF, Loyola RD, Raia P, Mooers AO, Bini LM.. 2013. Darwinian shortfalls in biodiversity conservation. Trends Ecol Evol. 28(12):689–695. [DOI] [PubMed] [Google Scholar]
- Dorsey RJ, et al. 2007. Chronology of Miocene-Pliocene deposits at Split Mountain Gorge, Southern California: a record of regional tectonics and Colorado River evolution. Geology 35(1):57–60. [Google Scholar]
- Douglas ME, Minckley WL, DeMarais BD.. 1999. Did vicariance mold phenotypes of western North American fishes? Evidence from Gila River cyprinids. Evolution 53(1):238–246. [DOI] [PubMed] [Google Scholar]
- Douglas ME, Douglas MR, Lynch JM, McElroy DM.. 2001. Use of geometric morphometrics to differentiate Gila (Cyprinidae) within the Upper Colorado River Basin. Copeia 2001(2):389–400. [Google Scholar]
- Douglas ME, Matthews WJ.. 1992. Does morphology predict ecology? Hypothesis testing within a freshwater stream fish assemblage. Oikos 65(2):213. [Google Scholar]
- Douglas MR, Douglas ME.. 2007. Genetic structure of humpback chub Gila cypha and roundtail chub G. robusta in the Colorado River ecosystem. Reston (VA: ): Grand Canyon Monitoring and Research Center. [Google Scholar]
- Dowling TE, Anderson CD, Marsh PC, Rosenberg MS.. 2015. Population structure in the Roundtail chub (Gila robusta complex) of the Gila River Basin as determined by microsatellites: evolutionary and conservation implications. PLoS One. 10(10):e0139832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling TE, DeMarais BD.. 1993. Evolutionary significance of introgressive hybridization in cyprinid fishes. Nature 362(6419):444–446. [Google Scholar]
- Dowling TE, Secor CL.. 1997. The role of hybridization and introgression in the diversification of animals. Annu Rev Ecol Syst. 28(1):593–619. [Google Scholar]
- Eaton DAR.2014. PyRAD: assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics 30(13):1844–1849. [DOI] [PubMed] [Google Scholar]
- Eaton DAR, Ree RH.. 2013. Inferring phylogeny and introgression using RADseq data: an example from glowering plants (Pedicularis: Orobanchaceae). Syst Biol. 62(5):689–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton DAR, Spriggs EL, Park B, Donoghue MJ.. 2017. Misconceptions on missing data in RAD-seq phylogenetics with a deep-scale example from flowering plants. Syst Biol. 66(3):399–412. [DOI] [PubMed] [Google Scholar]
- Eberly LD, Stanley TB.. 1978. Cenozoic stratigraphy and geologic history of southwestern Arizona. Geol Soc America Bull. 89(6):921–940. [Google Scholar]
- Edwards SV.2009. Is a new and general theory of molecular systematics emerging? Evolution. 63(1):1–19. [DOI] [PubMed] [Google Scholar]
- Edwards SV, et al. 2016. Implementing and testing the multispecies coalescent model: a valuable paradigm for phylogenomics. Mol Phylogenet Evol. 94(A):447–462. [DOI] [PubMed] [Google Scholar]
- Efron B, Halloran E, Holmes S.. 1996. Bootstrap confidence levels for phylogenetic trees. Proc Natl Acad Sci USA. 93(23):13429–13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J.. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 14(8):2611–2620. [DOI] [PubMed] [Google Scholar]
- Felsenstein J.1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791. [DOI] [PubMed] [Google Scholar]
- Forest F, Crandal KA, Chase MW, Faith DP.. 2015. Phylogeny, extinction and conservation: embracing uncertainties in a time of urgency. Philos Trans R Soc B Biol Sci. 370:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadagkar SR, Rosenberg MS, Kumar S.. 2005. Inferring species phylogenies from multiple genes: concatenated sequence tree versus consensus gene tree. J Exp Zool B Mol Dev Evol. 304(1):64–74. [DOI] [PubMed] [Google Scholar]
- Gerber AS, Tibbets CA, Dowling TE.. 2001. The role of introgressive hybridization in the evolution of the Gila robusta complex (Teleostei: Cyprinidae). Evolution 55(10):2028–2039. [DOI] [PubMed] [Google Scholar]
- Giarla TC, Esselstyn JA.. 2015. The challenges of resolving a rapid, recent radiation: empirical and simulated phylogenomics of Philippine shrews. Syst Biol. 64(5):727–740. [DOI] [PubMed] [Google Scholar]
- Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Nakhleh L.. 2016. Irrational exuberance for resolved species trees. Evolution. 70(1):7–17. [DOI] [PubMed] [Google Scholar]
- Heath TA, Hedtke SM, Hillis DM.. 2008. Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol. 46:239–257. [Google Scholar]
- Helenes J, Carreno AL.. 2014. Neogene sedimentary record of the Gulf of California: towards a highly biodiverse scenario. In: Wehncke EV, Lara-lAra JR, Alvarez-Borrego S, Ezcurra E, editors. Conservation Science in Mexico’s Northwest: ecosystem status and trends in the Gulf of Calfiornia. Riverside (CA: ): UC Riverside. [Google Scholar]
- Hillis DM.1998. Taxonomic sampling, phylogenetic accuracy, and investigator bias. Syst Biol. 47(1):3–8. [DOI] [PubMed] [Google Scholar]
- Holycross AT, Douglas ME.. 2007. Geographic isolation, genetic divergence, and ecological non-exchangeability define conservation units in a threatened sky-island rattlesnake. Biol Conserv. 134(1):142–154. [Google Scholar]
- House PK, et al. 2005. Birth of the lower Colorado River—stratigraphic and geomorphic evidence for its inception near the conjunction of Nevada, Arizona, and California. Boulder (CO: ): Geological Society of America. [Google Scholar]
- House PK, Pearthree PA, Perkins ME.. 2008. Stratigraphic evidence for the role of lake spillover in the inception of the lower Colorado River in southern Nevada and western Arizona. Boulder (CO: ): Geological Society of America. [Google Scholar]
- Huang H, Knowles LL.. 2016. Unforeseen consequences of excluding missing data from next-generation sequences: simulation study of RAD sequences. Syst Biol. 65(3):357–365. [DOI] [PubMed] [Google Scholar]
- Hubbs CL, Miller RR.. 1948. The zoological evidence: correlation between fish distribution and hydrogeographic history in the desert basin of western North America. Bull Univ Utah. 10:17–166. [Google Scholar]
- Huckleberry G.1996. Historical geomorphology of the Gila River. Tucson (AZ: ): Arizona Geological Survey. [Google Scholar]
- Huckleberry JK, Potts MD.. 2019. Constraints to implementing the food-energy-water nexus concept: governance in the Lower Colorado River Basin. Environ Sci Policy. 92:289–298. [Google Scholar]
- Huson DH.1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14(1):68–73. [DOI] [PubMed] [Google Scholar]
- James T, Evans A, Madly E, Kelly C.. 2014. The economic importance of the Colorado River to the Basin region. Tempe (AZ: ): Arizona State University. [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I.. 2015. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 15(5):1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubatko LS, Chifman J.. 2019. An invariants-based method for efficient identification of hybrid species from large-scale genomic data. BMC Ecol Evol. 19:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Filipski AJ, Battistuzzi FU, Kosakovsky Pond SL, Tamura K.. 2012. Statistics and truth in phylogenomics. Mol Biol Evol. 29(2):457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larget BR, Kotha SK, Dewey CN, Ané C.. 2010. BUCKy: gene tree reconciliation with concordance. Bioinformatics. 26(22):2910–2911. [DOI] [PubMed] [Google Scholar]
- Lawson DJ, van Drop L, Falush D.. 2018. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat Commun. 9(1):3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leache AD, et al. 2014. Species delimitation using genome-wide SNP data. Syst Biol. 63(4):534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leache AD, Harris RB, Rannala B, Yang Z.. 2014. The influence of gene flow on species tree estimation: a simulation study. Syst Biol. 63(1):17–30. [DOI] [PubMed] [Google Scholar]
- Leaché AD, Oaks JR.. 2017. The utility of single nucleotide polymorphism (SNP) data in phylogenetics. Annu Rev Ecol Evol Syst. 48(1):69–84. [Google Scholar]
- Lemmon EM, Lemmon AR.. 2013. High-throughput genomic data in systematics and phylogenetics. Annu Rev Ecol Evol Syst. 44(1):99–121. [Google Scholar]
- Linck E, Battey CJ.. 2019. Minor allele frequency thresholds strongly affect population structure inference with genomic data sets. Mol Ecol Resour. 19(3):639–647. [DOI] [PubMed] [Google Scholar]
- Linck E, et al. 2019. Dense geographic and genomic sampling reveals paraphyly and a cryptic lineage in a classic sibling species complex. Syst Biol. 68:956–966. [DOI] [PubMed] [Google Scholar]
- Linkem CW, Minin VNV, Leaché AD.. 2016. Detecting the anomaly zone in species trees and evidence for a misleading signal in higher-level skink phylogeny (Squamata: Scincidae). Syst Biol. 65(3):465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Edwards SV.. 2009. Phylogenetic analysis in the anomaly zone. Syst Biol. 58(4):452–460. [DOI] [PubMed] [Google Scholar]
- Long C, Kubatko L.. 2018. The effect of gene flow on coalescent-based species-tree inference. Syst Biol. 67(5):770–785. [DOI] [PubMed] [Google Scholar]
- Lucchitta I.1972. Early history of the Colorado River in the Basin and Range province. Geol Soc America Bull. 83(7):1933–1948. [Google Scholar]
- Luo A, Ling C, Ho SYW, Zhu CD.. 2018. Comparison of methods for molecular species delimitation across a range of speciation scenarios. Syst Biol. 67(5):830–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W.1997. Gene trees in species trees. Syst Biol. 46(3):523–536. [Google Scholar]
- De Maio N, Schrempf D, Kosiol C.. 2015. PoMo: an allele frequency-based approach for species tree estimation. Syst Biol. 64(6):1018–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh PC, Clarkson RW, Dowling TE.. 2017. Molecular genetics informs spatial segregation of two desert stream Gila species. Trans Am Fish Soc. 146(1):47–59. [Google Scholar]
- Martin BT, et al. 2021. The choices we make and the impacts they have: machine learning and species delimitation in North American box turtles (Terrapene spp.). Mol Ecol Resour. Advance Access published Feb 10, 2021, doi: 10.1111/1755-0998.13350. [DOI] [PubMed] [Google Scholar]
- McCormack JE, Huang H, Knowles LL.. 2009. Maximum likelihood estimates of species trees: how accuracy of phylogenetic inference depends upon the divergence history and sampling design. Syst Biol. 58(5):501–508. [DOI] [PubMed] [Google Scholar]
- McKee ED, Wislon RF, Breed WJ, Breed CS.. 1967. Evolution of the Colorado River in Arizona: an hypothesis developed at the Symposium on Cenozoic Geology of the Colorado Plateau in Arizona, August 1964. Flagstaff (AZ: ): Museum of Northern Arizona. [Google Scholar]
- McVay JD, Hipp AL, Manos PS.. 2017. A genetic legacy of introgression confounds phylogeny and biogeography in oaks. Proc R Soc B Biol Sci. 284:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffe GK, Vrijenhoek RC.. 1988. Conservation genetics in the management of desert fishes. Conserv Biol. 2(2):157–169. [Google Scholar]
- Mendes FK, Hahn Y, Hahn MW.. 2016. Gene tree discordance can generate patterns of diminishing convergence over time. Mol Biol Evol. 33(12):3299–3307. [DOI] [PubMed] [Google Scholar]
- Meng C, Kubatko LS.. 2009. Detecting hybrid speciation in the presence of incomplete lineage sorting using gene tree incongruence: a model. Theor Popul Biol. 75(1):35–45. [DOI] [PubMed] [Google Scholar]
- Minckley WL.1979. Aquatic habitats and fishes of the lower Colorado River, southwestern United States. Washington (DC: ): USBR. [Google Scholar]
- Minckley WL.1986. Geography of western North American freshwater fishes: description and relationships to intracontinental tectonism. In: Hocutt CH, Wiley EO, editors. Zoogeography of Western North American freshwater fishes. New York (NY: ): John Wiley and Sons. p. 519–613. [Google Scholar]
- Minckley WL, DeMarais BD.. 2000. Taxonomy of chubs (Telostei, Cyprinidae, genus Gila) in the American Southwest with comments on conservation. Copeia 2000(1):251–256. [Google Scholar]
- Minckley WL, Marsh PC.. 2009. Inland fishes of the greater Southwest: chronicle of a vanishing biota. Tucson (AZ: ): University of Arizona Press. [Google Scholar]
- Minckley WL, et al. 2003. A conservation plan for native fishes of the lower Colorado River. Bioscience 53(3):219. [Google Scholar]
- Minh BQ, Hahn MW, Lanfear R.. 2020. New methods to calculate concordance factors for phylogenomic datasets. Mol Biol Evol. 37(9):2727–2733.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales AE, Carstens BC.. 2018. Evidence that Myotis lucifugus “Subspecies” are five nonsister species, despite gene flow. Syst Biol. 67(5):756–769. [DOI] [PubMed] [Google Scholar]
- Moran CJ, O’Neill MW, Armbruster JW, Gibb AC.. 2017. Can members of the south-western Gila robusta species complex be distinguished by morphological features? J Fish Biol. 91(1):302–316. [DOI] [PubMed] [Google Scholar]
- Mussmann SM, Douglas MR, Bangs MR, Douglas ME.. 2020. Comp-D: a program for comprehensive computation of D-statistics and population summaries of reticulated evolution. Conservation Genet Resour. 12(2):263–267. [Google Scholar]
- Mussmann SM, Douglas MR, Chafin TK, Douglas ME.. 2020. AdmixPipe: population analyses in ADMIXTURE for non-model organisms. BMC Bioinfo. 21:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakey DD, Douglas ME, Douglas MR.. 2004. Small fish in a large landscape: diversification of Rhinichthys osculus (Cyprinidae) in Western North America. Copeia 2004(2):207–221. [Google Scholar]
- Oaks JR.2019. Full Bayesian comparative phylogeography from genomic data. Syst Biol. 68(3):371–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page LM, et al. 2017. Taxonomy of Gila in the Lower Colorado River Basin of Arizona and New Mexico. Fisheries 42(9):456–460. [Google Scholar]
- Page LM, et al. 2016. Final report of the AFS/ASIH Joint Committee on the names of fishes on the taxonomy of Gila in the Lower Colorado River basin of Arizona and New Mexico. Phoenix (AZ: ): Arizona Game and Fish Department. [Google Scholar]
- Pante E, et al. 2015. Species are hypotheses: avoid connectivity assessments based on pillars of sand. Mol Ecol. 24(3):525–544. [DOI] [PubMed] [Google Scholar]
- Pease JB, Hahn MW.. 2015. Detection and polarization of introgression in a five-taxon phylogeny. Syst Biol. 64(4):651–662. [DOI] [PubMed] [Google Scholar]
- Pedraza-Marrón CDR, et al. 2019. Genomics overrules mitochondrial DNA, siding with morphology on a controversial case of species delimitation. Proc R Soc B. 286(1900):20182924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AG, Schrago CG.. 2018. Incomplete lineage sorting impacts the inference of macroevolutionary regimes from molecular phylogenies when concatenation is employed: an analysis based on Cetacea. Ecol Evol. 8(14):6965–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE.. 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One. 7(5):e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, et al. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9(3):e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Delsuc F, Brinkmann H, Lartillot N.. 2005. Phylogenomics. Annu Rev Ecol Evol Syst. 36(1):541–562. [Google Scholar]
- Rangel TF, et al. 2015. Phylogenetic uncertainty revisited: implications for ecological analyses. Evolution 69(5):1301–1312. [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang Z.. 2008. Phylogenetic inference using whole genomes. Annu Rev Genomics Hum Genet. 9:217–231. [DOI] [PubMed] [Google Scholar]
- Reddy S, et al. 2017. Why do phylogenomic data sets yield conflicting trees? Data type influences the avian tree of life more than taxon sampling. Syst Biol. 66(5):857–879. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Brown JM, Barley AJ, Chong RA, Thomson RC.. 2018. Variation across mitochondrial gene trees provides evidence for systematic error: how much gene tree variation is biological? Syst Biol. 67(5):847–860. [DOI] [PubMed] [Google Scholar]
- Rinne JN.1976. Cyprinid fishes of the genus Gila from the lower Colorado River basin. Wasmann J Biol. 34: 65–107. [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F.. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 4:e2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarna-Wojcicki AM, et al. 2011. Age, composition, and areal distribution of the Pliocene Lawlor Tuff, and three younger Pliocene tuffs. Calif Nevada Geosph. 7(3):599–628. [Google Scholar]
- Schmidt-Lebuhn AN.2012. Fallacies and false premises-a critical assessment of the arguments for the recognition of paraphyletic taxa in botany. Cladistics 28(2):174–187. [DOI] [PubMed] [Google Scholar]
- Schönhuth S, et al. 2014. Phylogenetic relationships of North American western chubs of the genus Gila (Cyprinidae, Teleostei), with emphasis on southern species. Mol Phylogenet Evol. 70:210–230. [DOI] [PubMed] [Google Scholar]
- Schönhuth S, Shiozawa DK, Dowling TE, Mayden RL.. 2012. Molecular systematics of western North American cyprinids (Cypriniformes: Cyprinidae). Zootaxa 3586(1):281–303. [Google Scholar]
- Schönhuth S, Vukić J, Šanda R, Yang L, Mayden RL.. 2018. Phylogenetic relationships and classification of the Holarctic family Leuciscidae (Cypriniformes: Cyprinoidei). Mol Phylogenet Evol. 127:781–799. [DOI] [PubMed] [Google Scholar]
- Schrempf D, Minh BQ, De Maio N, von Haeseler A, Kosiol C.. 2016. Reversible polymorphism-aware phylogenetic models and their application to tree inference. J Theor Biol. 407:362–370. [DOI] [PubMed] [Google Scholar]
- Schwartz MK, McKelvey KS.. 2009. Why sampling scheme matters: the effect of sampling scheme on landscape genetic results. Conserv Genet. 10(2):441–452. [Google Scholar]
- Schwemm MR.2006. Genetic variation in the Gila robusta complex (Teleostei: Cyprinidae) in the lower Colorado River [Thesis]. [Tempe (AZ)]: Arizona State University.
- Shen XX, Hittinger CT, Rokas A.. 2017. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat Ecol Evol. 1:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG.1961. Principles of animal taxonomy. New York (NY: ): Columbia University Press. [DOI] [PubMed] [Google Scholar]
- Smith GA, Yang W, Cerling TE, Geissman JW.. 1993. Comparison of a paleosol-carbonate isotope record to other records of Pliocene-early Pleistocene climate in the western United States. Geology 21(8):691–694. [Google Scholar]
- Snir S, Rao S.. 2012. Quartet MaxCut: a fast algorithm for amalgamating quartet trees. Mol Phylogenet Evol. 62(1):1–8. [DOI] [PubMed] [Google Scholar]
- Solís-Lemus C, Ané C.. 2016. Inferring phylogenetic networks with Maximum Pseudolikelihood under incomplete lineage sorting. PLoS Genet. 12(3):e1005896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís-Lemus C, Bastide P, Ané C.. 2017. PhyloNetworks: a package for phylogenetic networks. Mol Biol Evol. 34(12):3292–3298. [DOI] [PubMed] [Google Scholar]
- Sosef MSM.1997. Hierarchical models, reticulate evolution and the inevitability of paraphyletic supraspecific taxa. Taxon 46(1):75–85. [Google Scholar]
- Spencer JE, Patchett PJ.. 1997. Sr isotope evidence for a lacustrine origin for the upper Miocene to Pliocene Bouse Formation, lower Colorado River trough, and implications for timing of Colorado Plateau uplift. Geol Soc Am Bull. 109(6):767–778. [Google Scholar]
- Spencer JE, et al. 2001. 40Ar/39Ar geochronology of the Hualapai Limestone and Bouse Formation and implications for the age of the lower Colorado River. Color River Orig Evol Gd Canyon, Arizona, Gd Canyon Assoc. 89–91. [Google Scholar]
- Spencer JE, Smith GR, Dowling TE.. 2008. Middle to late Cenozoic geology, hydrography, and fish evolution in the American Southwest. In: Reheis MC, Hershler R, Miller DM, editors Late cenozoic drainage history of the Southwestern Great Basin and Lower Colorado River region: geologic and biotic perspectives. Boulder (CO: ): Geological Society of America. [Google Scholar]
- Springer MS, Gatesy J.. 2016. The gene tree delusion. Mol Phylogenet Evol. 94(A):1–33. [DOI] [PubMed] [Google Scholar]
- Stadler T, Degnan JH, Rosenberg NA.. 2016. Does gene tree discordance explain the mismatch between macroevolutionary models and empirical patterns of tree shape and branching times? Syst Biol. 65(4):628–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenz NWM, Larget B, Baum DA, Ané C.. 2015. Exploring tree-like and non-tree-like patterns using genome sequences: an example using the inbreeding plant species Arabidopsis thaliana (L.) heynh. Syst Biol. 64(5):809–823. [DOI] [PubMed] [Google Scholar]
- Sullivan J, Joyce P.. 2005. Model selection in phylogenetics. Annu Rev Ecol Evol Syst. 36(1):445–466. [Google Scholar]
- Thompson RS.1991. Pliocene environments and climates in the western United States. Quat Sci Rev. 10(2–3):115–132. [Google Scholar]
- USFWS. 2005. Endangered and threatened wildlife and plants: listing Gila chub as endangered with critical habitat. Fed Regist. 70:66664–66721. [Google Scholar]
- USFWS. 2017. Endangered and threatened wildlife and plants: threatened species status for the Headwater chub and Roundtail chub distinct population segment. Fed Regist. 82:16981–16988. [Google Scholar]
- Uyeno T.1960. Osteology and phylogeny of the American cyprinid fishes allied to the genus Gila [Thesis]. [Ann Arbor (MI)]: University of Michigan.
- Uyeno T, Miller RR.. 1963. Summary of late Cenozoic freshwater fish records for North America. Occas Pap Museum Zool Univ Michigan. 631:1–34. [Google Scholar]
- Uyeno T, Miller RR.. 1965. Middle Pliocene cyprinid fishes from the Bidahochi Formation. Copeia. 1965(1):28–41. [Google Scholar]
- Walker JF, Brown JW, Smith SA.. 2018. Analyzing contentious relationships and outlier genes in phylogenomics. Syst Biol. 67(5):916–924. [DOI] [PubMed] [Google Scholar]
- Wu M, Kostyun JL, Hahn MW, Moyle LC.. 2018. Dissecting the basis of novel trait evolution in a radiation with widespread phylogenetic discordance. Mol Ecol. 27:3301–3316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences are available via NCBI SRA associated with BioProject PRJNA558355. All additional relevant metadata, codes, assembled data sets, and unfiltered alignment files are available via the Open Science Framework (doi: 10.17605/OSF.IO/WABJT). Codes and custom scripts are also available as open-source under the GNU Public License via GitHub: github.com/tkchafin (doi: 10.5281/zenodo.5181290).