Abstract
Background
APOE4 is the strongest genetic risk factor for Alzheimer’s disease (AD), and obesity is a strong environmental risk factor for AD. These factors result in multiple central nervous system (CNS) disturbances and significantly increase chances of AD. Since over 20% of the US population carry the APOE4 allele and over 40% are obese, it is important to understand how these risk factors interact to affect neurons and glia in the CNS.
Methods
We fed male and female APOE3 and APOE4 knock-in mice a high-fat diet (HFD-45% kcal fat) or a "control" diet (CD-10% kcal fat) for 12 weeks beginning at 6 months of age. At the end of the 12 weeks, brains were collected and analyzed for gliosis, neuroinflammatory genes, and neuronal integrity.
Results
APOE3 mice on HFD, but not APOE4 mice, experienced increases in gliosis as measured by GFAP and Iba1 immunostaining. APOE4 mice on HFD showed a stronger increase in the expression of Adora2a than APOE3 mice. Finally, APOE3 mice on HFD, but not APOE4 mice, also showed increased neuronal expression of immediate early genes cFos and Arc.
Conclusions
These findings demonstrate that APOE genotype and obesity interact in their effects on important processes particularly related to inflammation and neuronal plasticity in the CNS. During the early stages of obesity, the APOE3 genotype modulates a response to HFD while the APOE4 genotype does not. This supports a model where early dysregulation of inflammation in APOE4 brains could predispose to CNS damages from various insults and later result in the increased CNS damage normally associated with the APOE4 genotype.
Keywords: Obesity, High-fat diet, Apolipoprotein E, APOE, Inflammation, Gliosis
Background
Apolipoprotein E (APOE) 4 is the strongest genetic risk factor for Alzheimer’s disease (AD) [47]. It is present in nearly 25% of the US population and over 50% of AD patients [51]. There are three different APOE alleles: APOE2, APOE3, and APOE4. These alleles encode single amino acid differences at position 112 or 158 [28]. APOE3 is the most common allele and thus APOE3 homozygotes are defined as having an average risk of AD. Meanwhile, APOE4 increases risk of AD: Heterozygous APOE4 carriers are 3-4 times more likely to get AD, while APOE4 homozygous individuals are 15 times more likely [28]. APOE4 can increase the risk of cognitive deficits in healthy individuals and in APOE4 knock-in mouse models [10]. Compared to homozygous APOE3 mice, homozygous APOE4 mice have increased cognitive deficits and decreased neuronal integrity without developing AD pathology [41]. APOE4 mice crossed with mice that exhibit AD pathology have increased plaque accumulation and increased inflammation when compared to homozygous APOE3 mice [42, 49].
Obesity is a strong environmental risk factor for AD [5, 35] and has been reported to affect over 40% of the US population [13]. Obesity is often accompanied by metabolic disturbances such as increases in adipose tissue, hyperglycemia, hypercholesterolemia, insulin resistance, and has increasingly been associated with cognitive deficits [13, 24]. Obesity in rodents can be modeled through high-fat diets (HFD) [25, 27]. Like in humans, rodents on HFD develop increased adipose tissue, glucose intolerance, and insulin resistance [21, 26]. In the central nervous system (CNS), HFD has been associated with increased inflammation (gliosis, cytokine expression) [40, 53] and behavioral deficits (Morris Water Maze), delayed matching, and non-matching task [7, 32].
Here, we investigate how diet induced obesity affects inflammation in the CNS in APOE mice. Since APOE4 is the strongest genetic risk factor for AD and the prevalence of obesity is increasing, it is imperative to understand how the two risks interact and contribute to the eventual inflammatory state of AD. We previously found that HFD induced metabolic syndrome in both APOE3 and APOE4 mice with a stronger effect in APOE4 mice [21]. Here, we have identified the effects of HFD on brain inflammation.
Methods
Animals/diet
Male and female APOE3 and APOE4 knock-in mice, expressing the human APOE allele in the mouse APOE gene locus, on a C57BL/6J background (the gift of Patrick Sullivan) were used. These mice model AD risk without pathological changes. They exhibit normal CNS expression of the APOE gene, but differ in gene expression profiles and behavior across the APOE genotypes [41, 48, 56]. They were fed either a HFD (45% kcal fat, Research Diets-D12451) or ingredient-matched "control" diet (CD) (10% kcal fat, Research Diets-D12450H) for 12 weeks beginning at 6 months of age [21]. HFD fat content was predominantly lard-based, making it high in omega-6 fatty acids (ingredients: 177 g lard, 25 g soybean oil in 858 g of diet), while CD fat content was predominantly soybean oil based making it higher in omega-3 fatty acids (ingredients: 20 g lard, 25 g soybean oil in 1055 g of diet). Food and water were provided ad libitum. At the end of the 12 weeks, mice were euthanized and brains collected for analysis. Mice were euthanized by CO2 inhalation and brains perfused with 1x PBS. Brains of APOE3 and APOE4 mice on CD and HFD (n = 13-15 per genotype and diet, 5-9 per sex) were extracted, and hemi-sections were divided for immunofluorescence (IF) assays, Golgi staining, and biochemical assays. All experiments followed the guidelines of the Georgetown University Institutional Animal Care and Use Committee.
Immunofluorescence
For IF (N = 3-4 per genotype and diet, 1-2 per sex), one brain hemisphere was fixed in 4% PFA and sucrose before freezing in cold 2-methylbutene and slicing at 30 μm. IF stains were performed for Iba1 (Wako, Cat #: 019-19741), GFAP (Cell Signaling, Cat #: 3670S), Doublecortin (DCX, ThermoFisher, Cat #: 481200), NeuN (Chemicon, Cat #: MAB377), and cFOS (Abcam, Cat # ab190289). Stains were imaged using a Zeiss AxioSkop at either 10× (Iba1, GFAP, and cFOS) or 20× (co-stains and DCX) magnification. Staining was analyzed using Image J, with quantification from 4 brain slices/mouse. Hippocampal images were taken from the stratum radiatum and stratum oriens of CA1 and CA3, and the CA4 and the molecular layers (MO) of the dentate gyrus (DG). They were also taken from the arcuate nucleus, the dorsal medial, and lateral hypothalamus, and from multiple layers of the cerebral cortex (Fig. 1). Iba1 and GFAP were analyzed as the percent area covered by IF staining in the image. DCX and cFos expression were analyzed as the counts of antibody positive cells. All quantification was conducted blinded.
Fig. 1.
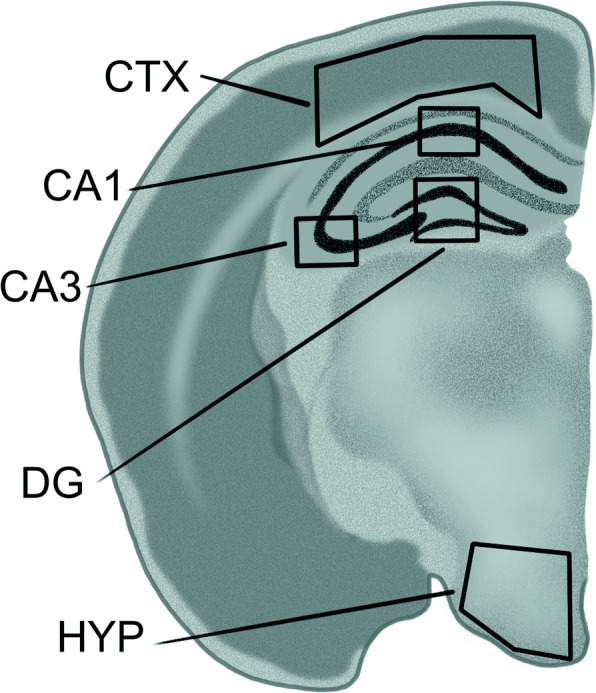
Illustration of mouse brain hemisphere with areas analyzed for IF outlined
Golgi staining
Brain hemisections were placed in Golgi staining solutions according to recommendations (FD Rapid GolgiStain Kit (PK401)) and sliced at 150 μm. Images of the basal shaft (BS) and apical oblique (AO) dendrites of neurons in the entorhinal cortex were acquired with an Olympus XB51 microscope at 60× magnification with oil immersion. A minimum of 10 neurons were imaged for each mouse (N = 4-5 mice per genotype and diet, 2-3 per sex). Dendritic lengths were measured, and spines counted using Image J. All quantification was conducted in a blinded manner.
RNA analyses
Cortex from one brain hemisphere (N = 5-7 per genotype and diet, 2-4 per sex) was used for RNA isolation. RNA was isolated using the Direct-zol RNA Miniprep kit (Zymo Research R2051) and analyzed by Georgetown University’s Genomic and Epigenomics Shared Resource to determine concentration and purity. For preliminary studies of brain inflammation, equal amounts of four samples for each condition were pooled and analyzed on the NanoString nCounter Mouse Neuroinflammation Panel (Cat #XT-CSO-MNROI1, N = 4 pooled per genotype and diet), which measures transcripts of 770 neuroinflammatory genes. For studies of brain metabolism, three independent samples from each condition were analyzed on the NanoString nCounter Mouse Metabolic Pathways Panel (Cat #XT-CSO-MMP1-12, N = 3 per genotype and diet), which detects transcripts of 768 metabolic genes. All data were analyzed using nSolver 4.0. Background was subtracted from negative controls and samples were normalized to the positive controls and housekeeping genes. Data were further analyzed by normalized count and fold difference, and differences were compared across diet and genotype.
mRNA species that demonstrated at least a twofold difference were subsequently analyzed using qRT-PCR. cDNA (N = 5-7 per genotype and diet, 2-4 per sex) was produced from extracted RNA using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Cat #: 4368814). qRT-PCR for Adora2a, Arc, cFOS, C3, Egr1, IL-3, IL-6, and TNFα were run with the sequences and temperatures described in Table 1. All qRT-PCR were performed in triplicates using the Power SybrGreen Mix (Applied Biosystems) and a 7900HT fast qRT-PCR system. Results were normalized to housekeeping gene GAPDH. Data were analyzed as fold difference compared to CD APOE3 samples as described [29].
Table 1.
Primer sequences for qRT-PCR
| Primer | Sequence | Temperature |
|---|---|---|
| Adora2a |
Fwd: AGCAACCTGCAGAACGTCACAAAC Rev: TGGCAATAGCCAAGAGGCTGAAGA |
60 |
| Arc |
Fwd: GGAGGGAGGTCTTCTACCGTC Rev: CCCCCACACCTACAGAGACA |
57.5 |
| C3 |
Fwd: GACGCCACTATGTCCATCCT Rev: CCAGCAGTTCCAGGTCCTTTG |
57.5 |
| cFos |
Fwd: CTCTGGGAAGCCAAGGTC Rev: CGAAGGGAACGGAATAAG |
55 |
| Egr1 |
Fwd: GAGGAGTTATCCCAGCCAA Rev: GGCAGAGGAAGACGATGAAG |
57.5 |
| GAPDH |
Fwd: GTGTTTCCTCGTCCCGTAGA Rev: ATTCCGTTCACACCGACCTT |
55, 57.5, 60 |
| IL3 |
Fwd: CCTGGGACTCCAAGCTTCAA Rev: GACAATAGAGCTGCAATTCAACGT |
57.5 |
| Il6 |
Fwd: ACGGCCTTCCCTACTTCACA Rev: CATTTCCACGATTTCCCAGA |
57.5 |
| TNF-a |
Fwd: GGTGCCTATGTCTCAGCCTCTT Rev: GCCATAGAACTGATGAGAGGGAG |
60 |
Statistical analyses
All data are expressed as mean ± standard deviation. Comparisons among APOE genotypes and diets were analyzed by two-way ANOVAs with Sidak’s multiple comparison test. Statistical significance was determined by a probability error of p < 0.05. Sample sizes were included throughout the “Methods” section for each assay. All analyses and graph plotting were done using GraphPad Prism 8.
Results
HFD increases Iba1 immunoreactivity in APOE3 and APOE4 mice
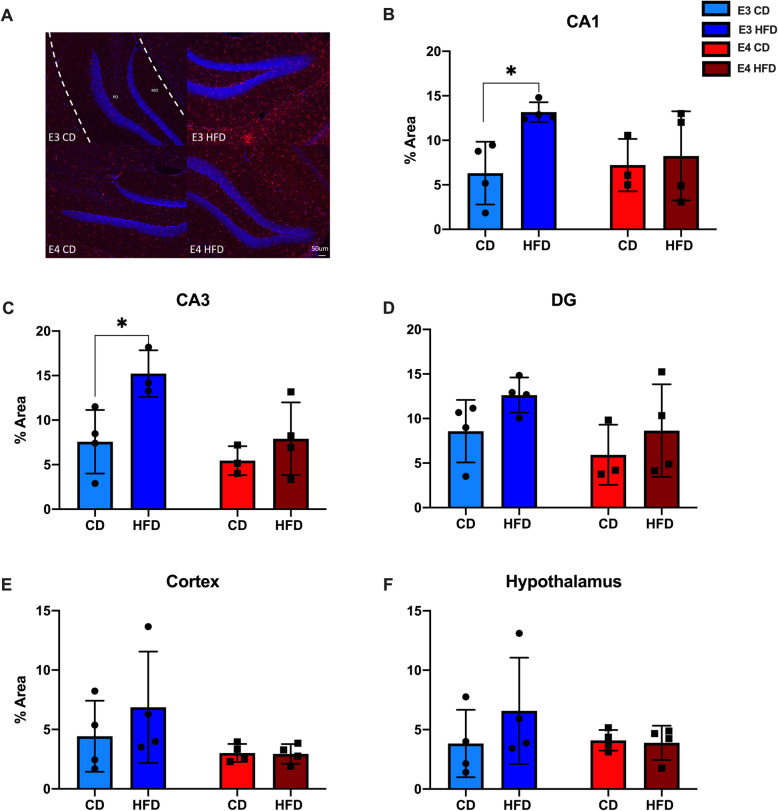
To examine whether HFD increased microglial activation, we measured Iba1 immunoreactivity in the hippocampus (HPC), cortex (CTX), and hypothalamus (HYP) of APOE3 and APOE4 mice either fed HFD or CD (Fig. 2A). In CA1 of the HPC, HFD APOE3 mice had significantly more Iba1 immunoreactivity when compared to CD APOE3 mice (Fig. 2B, p = 0.035). There were no significant differences between HFD and CD APOE4 mice. Similarly, in CA3 of the HPC, HFD APOE3 mice had significantly more Iba1 immunoreactivity when compared to CD APOE3 mice (Fig. 2C, p = 0.024) and there were no significant differences between HFD and CD APOE4 mice. In the dentate gyrus (DG) of the HPC, there were no significant differences among the groups, although the pattern mirrored CA1 and CA3 results (Fig. 2D). In the CTX and HYP, neither HFD APOE3 nor HFD APOE4 mice had significant differences in Iba1 immunoreactivity when compared to CD mice (Fig. 2E-F). For all areas, there were no statistically significant differences between HFD APOE3 and HFD APOE4 mice or between CD APOE3 and CD APOE4 mice.
Fig. 2.
HFD APOE3 mice, but not APOE4 mice show increased Iba1 immunoreactivity. A Images of Iba1 immunoreactivity in DG of the HPC. B-F Comparison of HFD effects on Iba1 immunoreactivity in CA1, CA3, and DG of the HPC, the CTX, and HYP. E3 CD: light blue, APOE3 mice on a CD, E3 HFD: dark blue, APOE3 mice on a HFD, E4 CD: light red, APOE4 mice on a CD, E4 HFD: dark red. N = 3-4, two-way ANOVA, *p < 0.05
HFD increases GFAP immunoreactivity in APOE3 and APOE4 mice
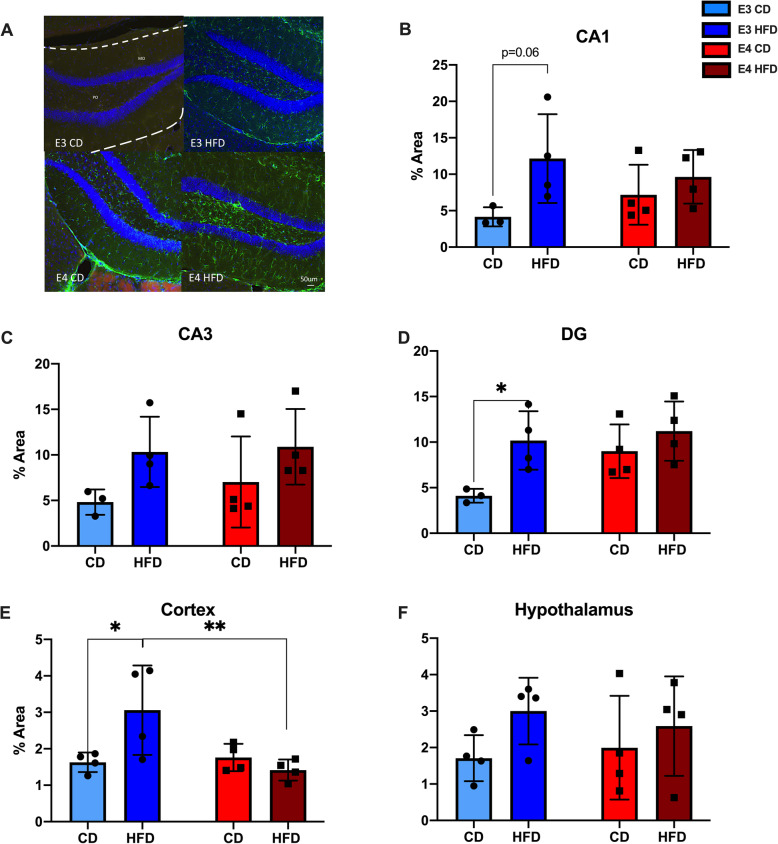
To examine whether HFD increased astrocytic activation, we analyzed GFAP immunoreactivity in the same brain regions (Fig. 3A). In CA1 of the HPC, HFD APOE3 mice trended toward significantly more GFAP immunoreactivity than CD APOE3 mice (Fig. 3B, p = 0.067); there was no difference between HFD APOE4 and CD APOE4 mice. In CA3 of the HPC, neither HFD APOE3 nor HFD APOE4 mice had significantly more GFAP immunoreactivity when compared to CD mice (Fig. 3C). In the DG of the HPC, HFD APOE3 mice had significantly more GFAP immunoreactivity when compared to CD APOE3 mice (Fig. 3D, p = 0.035), and there was no significant difference between HFD APOE4 and CD APOE4 mice. In the CTX, HFD APOE3 mice also had significantly more GFAP immunoreactivity than CD APOE3 mice (Fig. 3E, p = 0.02); again, there was no difference between HFD APOE4 and CD APOE4 mice. In the HYP, there were no significant differences by diet or genotype (Fig. 3F). When compared across genotypes, there were no differences except for in the CTX where HFD APOE3 mice had significantly more GFAP immunoreactivity than HFD APOE4 mice (Fig. 3E, p = 0.009). Thus, HFD was associated with higher measures of gliosis (Iba1 and GFAP) across brain regions in APOE3 mice, but not in APOE4 mice.
Fig. 3.
HFD APOE3 mice, but not APOE4 mice show increased GFAP immunoreactivity. A Images of GFAP immunoreactivity in DG of the HPC. B-F Comparison of HFD effects on GFAP immunoreactivity in CA1, CA3, and DG of the HPC, the CTX, and HYP. N = 3-4, two-way ANOVA, *p < 0.05, **p < 0.005
HFD increases immediate early gene expression in APOE3, but not APOE4 mice
To identify genes related to neuroinflammation or metabolism that may be related to HFD and APOE, we used a NanoString neuroinflammatory panel and a NanoString metabolic panel, and assessed over 1400 genes involved in neuroinflammation and metabolism. The panels included categories such as adaptive immune response, apoptosis, astrocyte function, inflammatory signaling, innate immune response, lipid metabolism, glucose transport, glycolysis, and fatty acid oxidation and synthesis. The panels also included APOE expression, which did not significantly differ by diet or genotype. From these categories, the two genes that had the highest expression, indicated by the neuroinflammatory panel, and that showed more than a twofold change after HFD were two immediate early gene (IEGs), cFos and Arc. IEGs can be altered by chronic activation of the innate immune system; under inflammatory conditions, increased IEG expression has also been associated with glia-induced increases in neuronal activity [43].
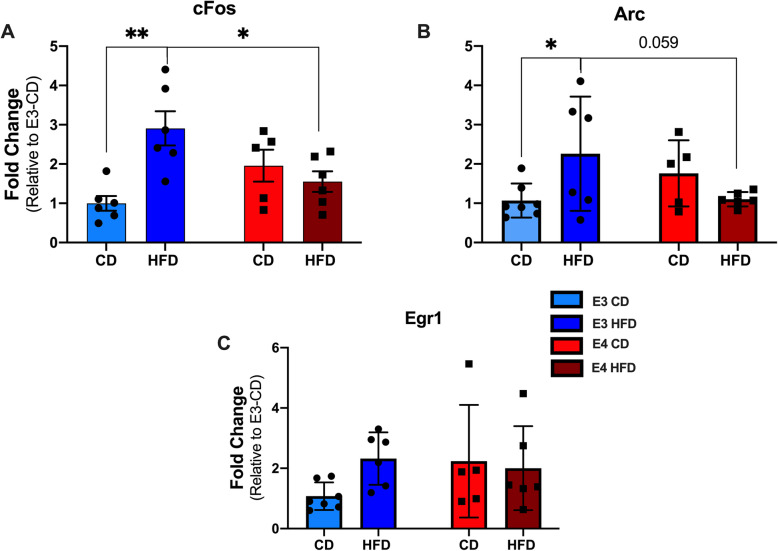
To examine IEG expression across individual brain samples, we used qRT-PCR. Consistent with the pilot data, HFD APOE3 mice had significantly higher cFos gene expression than CD APOE3 mice (Fig. 4A, 66%, p = 0.001), while there was no significant difference between HFD APOE4 and CD APOE4 mice. When compared across APOE genotypes, there were no differences in expression between CD APOE3 mice and CD APOE4 mice, but HFD APOE3 mice had significantly higher gene expression than HFD APOE4 mice (Fig. 4A, p = 0.013).
Fig. 4.
HFD APOE3 mice, but not APOE4 mice have increased IEG expression. A Comparison on HFD effects on fold changes in cFOS gene expression. B Comparison on HFD effects on fold changes in Arc gene expression. C Comparison on HFD effects on fold changes in Egr1 gene expression. N = 5-6, two-way ANOVA, *p < 0.05, **p < 0.005
Similar to the pattern of expression of cFos, Arc levels were significantly higher in HFD APOE3 mice compared to CD APOE3 mice (Fig. 4B, 55%, p = 0.04); again, there was no significant difference between HFD APOE4 and CD APOE4 mice. When compared across APOE genotypes, HFD APOE3 mice trended toward higher gene expression than HFD APOE4 mice (Fig. 4B, p = 0.059), and there were no differences in expression between CD APOE3 mice and CD APOE4 mice. To further test the effects of diet on IEG, we analyzed Erg1, another IEG involved in learning and memory that was not represented on the neuroinflammation panel. Erg1 did not show significant differences between diet or genotype, although its pattern of expression did mirror cFOS and Arc (Fig. 4C).
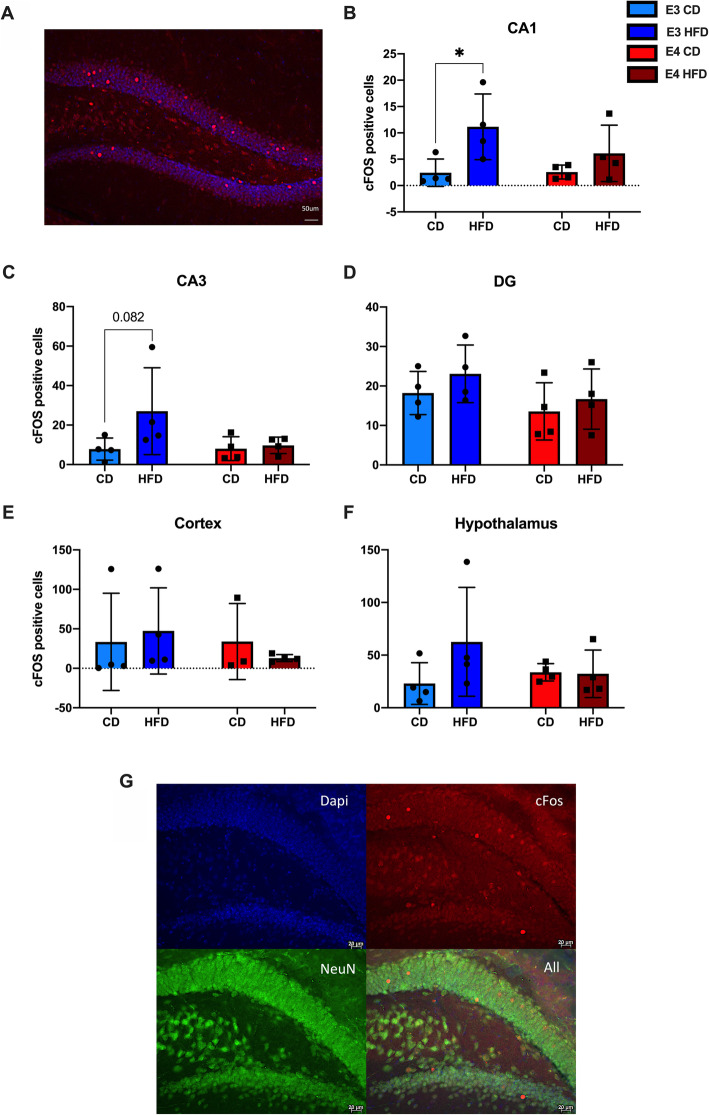
To analyze IEG protein in the brain across APOE genotypes and diet, we used IF for cFOS and quantified cFOS positive cells in the HPC, CTX, and HYP (Fig. 5A). In CA1 of the HPC, HFD APOE3 mice showed increased cFOS-positive cells when compared to CD APOE3 mice (Fig. 5B, p = 0.03), consistent with the mRNA data. There was no difference between HFD APOE4 and CD APOE4 mice and no differences when compared across genotypes. In CA3, HFD APOE3 mice trended toward more cFOS positive cells when compared to CD APOE3 mice (Fig. 5C, p = 0.082). In the DG, there were no significant differences, although the expression mirrored the findings in CA1 and CA3 (Fig. 5D). There were no significant differences in the CTX or HYP (Fig. 5E-F). Since cFOS can be present in either glia or neurons [2], we co-stained for cFOS and NeuN to test whether the cFOS was colocalized to neurons. All cFOS-positive cells co-stained for NeuN, across genotypes and diets (Fig. 5G).
Fig. 5.
HFD APOE3 mice, but not APOE4 mice have increased cFOS immunoreactivity. A Image of cFOS positive cells in the DG. B-F Quantification of cFOS positive cells in CA1, CA3, and DG of the HPC, the CTX, and HYP. G Representative image of cFOS positive cells colocalized with neurons. N = 3-4, two-way ANOVA, *p < 0.05
HFD differentially affects other neuroinflammatory genes in APOE3 and APOE4 mice
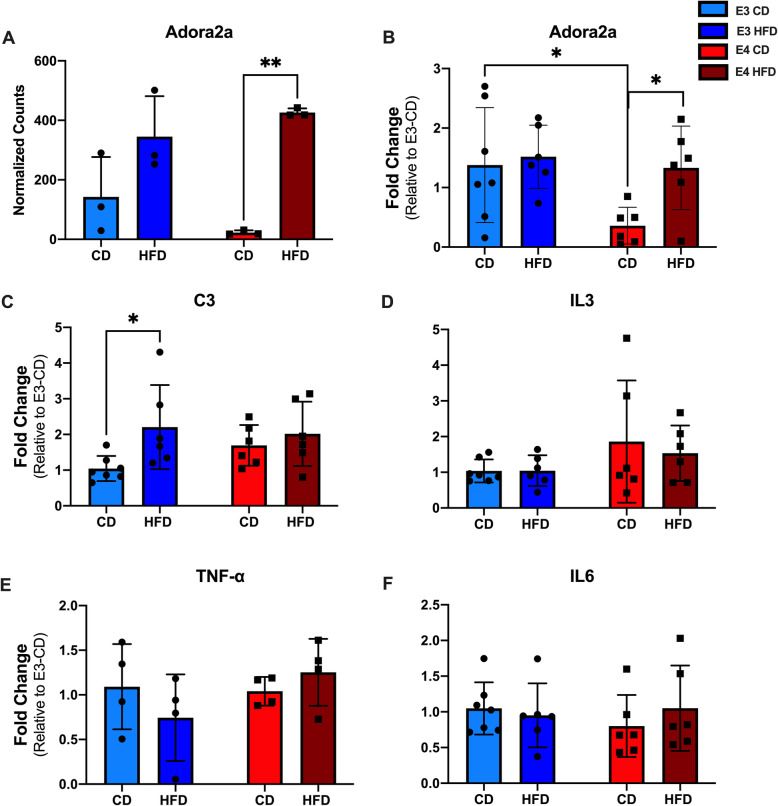
Along with IEGs, the NanoString nCounter mRNA analysis panel indicated changes in several other genes related to neuroinflammation: Adora2a, C3, and IL-3. Adora2a is an anti-inflammatory gene with properties protective against tissue damage [37, 50]. HFD APOE4 mice exhibited significantly higher levels of Adora2a gene expression than CD APOE4 mice both by NanoString nCounter analysis (Fig. 6A, p = 0.002) and qRT-PCR analysis (Fig. 6B, p = 0.046). There were no differences between HFD and CD APOE3 mice. When compared across APOE genotypes, there were no significant differences in Adora2a expression between mice with the NanoString nCounter analysis, but with qRT-PCR, CD APOE3 mice had significantly higher gene expression than CD APOE4 mice (Fig. 6B, p = 0.029). There were no differences between HFD APOE3 and HFD APOE4 mice. Thus, HFD increased Adora2a expression, particularly in the APOE4 mice.
Fig. 6.
HFD alters specific neuroinflammatory genes. A Comparison on HFD effects on NanoString nCount of Adora2a gene expression. B Comparison on HFD effects on fold changes in Adora2a gene expression. C Comparison on HFD effects on fold changes in C3 gene expression. D Comparison on HFD effects on fold changes in IL-3 gene expression. E Comparison on HFD effects on fold changes in TNF-a gene expression. F Comparison on HFD effects on fold changes in IL-6 gene expression. A N = 3, two-way ANOVA, *p < 0.003. B-F N = 4-6, two-way ANOVA, *p < 0.05
The remaining genes indicated by the neuroinflammatory panel were only analyzed by qRT-PCR because of the lower mRNA expression levels. The complement factor C3 plays an important role in innate immunity. HFD APOE3 mice exhibited significantly higher levels of C3 gene expression than CD APOE3 mice (Fig. 6C, p = 0.032). There were no differences between HFD and CD APOE4 mice and no differences by APOE genotype. To examine C3 activity, we ran a western blot and examined C3 and cleaved C3; however, we did not find any difference in C3 or cleaved C3 levels (data not shown). For the other neuroinflammatory gene identified (IL-3), as well as the common markers of general inflammation TNF-α and IL-6, there were no differences by diet or genotype (Fig. 6D-F).
HFD does not alter neurogenesis or spine density in APOE3 and APOE4 mice
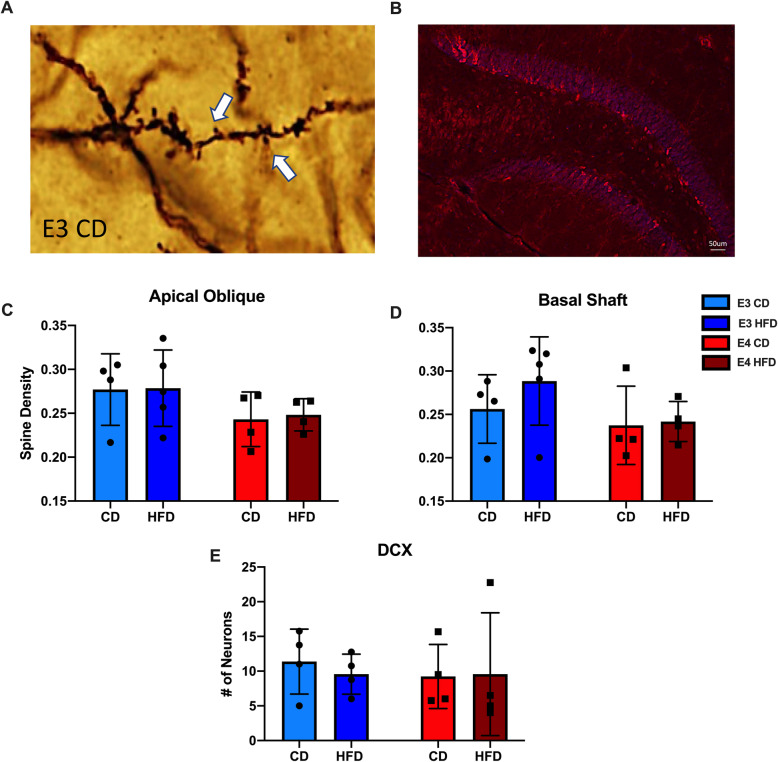
Neuronal degeneration has been associated with obesity and inflammation [3]. To test whether HFD here affected neuronal integrity, we examined spine density and hippocampal neurogenesis (Fig. 7A-B). For spine density, Golgi-stained pyramidal neurons in the entorhinal cortex were analyzed for dendritic spine density, quantified on the apical oblique and basal shaft dendrites. There were no significant differences in spine density across genotypes or diets (Fig. 7C-D). To examine whether HFD affects neurogenesis, we analyzed neurons in the DG of the HPC by DCX immunostaining. There were no significant differences in neurogenesis across genotypes or diets (Fig. 7E).
Fig. 7.
HFD does not alter neurogenesis or spine density in APOE3 and APOE4 mice. A Image of spines quantified from Golgi staining. B Image of DCX positive cells. C-D Quantification of spine density on the Apical Oblique and Basal Shaft. E Quantification of DCX positive cells
Discussion
It is important to understand how environmental factors combine with genetic factors to affect inflammation and alter the risk of AD. Here, we focused on the rapidly growing environmental risk factor, obesity, and the strongest genetic risk factor, APOE4, and sought to understand how the combination affects inflammation. Using a mouse model of the human APOE alleles, we found that HFD induced increased microglia and astrocyte expression in APOE3, but not APOE4 brains. This same pattern continued with increases in the IEGs cFOS and Arc in APOE3, but not APOE4 brains. These APOE-related CNS effects occur along with multiple metabolic disturbances: weight gain, adipose tissue accumulation, and glucose intolerance. These peripheral changes occurred predominantly in APOE4 mice; no behavioral changes were seen as a result of the HFD [21].
Previously HFD has been associated with increases in both microglial and astrocytic activity in wild-type mice [40, 53]. The APOE mice in our study are similar to control mice in that they do not have accumulations of Aβ or phospho-tau. Furthermore, the APOE3 mice act as controls and perform similarly to wild-type mice. Consistent with the study of wild-type mice, APOE3 mice in our study showed an increase in microgliosis and astrocytosis, although APOE4 mice did not. Another study showed HFD caused a decrease in CD68 (a marker of monocyte-derived cells) in APOE4 mice [17]. While data support that APOE4 mice have an increased response to short-term noxious stimuli such as LPS treatment or injury [23, 52, 57], there is evidence of a decreased response to the chronic inflammation caused by obesity [8, 17]. In one study, APOE3 and APOE4 mice were fed a HFD for approximately 7 months, and there were APOE3-specific increases in expression of TNFα, IL-6, and CD36 when compared to APOE4 mice [8]. APOE4 mice also exhibited lower expression of LPS immune sensors indicating a decreased ability for innate immune detection [8]. We propose that APOE3 mice have wild-type responses to chronic inflammatory conditions, while APOE4 mice may more readily respond to specific acute stimuli but have a more muted response to chronic stimuli.
Several studies have addressed the effects of HFD in AD mouse models related to APOE. HFD increases both Aß and phospho-tau pathology across AD mouse models [22, 27]; similar effects are seen in AD-APOE mouse models, but the responses differ by APOE genotype. In EFAD mice (mice that possess both a human APOE genotype and 5xFAD transgenes) on a HFD, there was an increase in gliosis and AD pathology in E4FAD mice, but not in E3FAD mice [34]. In APP/APOE mice (APOE mice crossed with APP/PS1ΔE9), there was an increase in AD pathology in APOE4, but not APOE3 mice on HFD [36]. The HFD may be affecting an already compromised inflammatory system responding to the increased amyloid found in the APOE4 mice.
We propose that, while chronic inflammation is damaging, inflammation at an early stage of obesity may be neuroprotective, and generally not experienced in APOE4 mice. Inflammation is associated with neuroprotection in early disease stages [15, 45] and the response in the APOE3 mice could be indicative of a similar mechanism. In AD mouse models, APOE3 has been associated with increases in microglial-plaque interactions when compared to APOE4 mice [46], an example of a positive inflammatory response in the APOE3 CNS (although there might be differences based on plaque morphology [42]). The inflammatory response from APOE3 mice can be beneficial for Aß clearance and, without it, the APOE4 mice may suffer.
In addition to inflammatory processes, glia are also heavily involved in lipid metabolism. Increased gliosis in the APOE3 mice may be a response to increased lipids and processing of lipids. Fatty acids can cross the blood-brain barrier and be taken up and stored by astrocytes; this ability prevents lipid induced neuronal damage [4]. Microglia also respond to increases in lipids and, when healthy, assist in the clearance of excess lipids [30]. The measures of astrogliosis and microgliosis in APOE3 brains could reflect changes to increased CNS lipids, and the APOE4 mice may have a deficit in responding to lipid-related stressors.
One gene significantly altered by diet and APOE genotype in the NanoString metabolic panel was Adora2a (also known as the adenosine A2A receptor), which is associated with inflammation [11]. While the panel had multiple metabolic categories, none of the genes specifically associated with metabolism were altered. Adora2a is expressed in the cortex and hippocampus; after CNS damage, there is an increase of Adora2a expression on glia, which is associated with increased activity and inflammation [6]. In multiple neurodegenerative conditions, there is an increase in Adora2a resulting in neuronal excitotoxicity and cell death [6]. Here, we see an increase in Adora2a in APOE4 mice on HFD, indicating potential enhancement in glutamatergic activity [11], which could lead to neuronal damage if prolonged. While the APOE4 mice do not demonstrate increased gliosis at this stage of developing obesity, there could be stronger inflammation and neuronal damage if increased Adora2a expression persisted.
An APOE3-specific response to HFD was further emphasized through expression of IEGs indicated by the NanoString neuroinflammatory panel and confirmed by qRT-PCR. We found that HFD increased cFOS and Arc expression twofold in APOE3 mice, with no effects in APOE4 mice. This expression was observed in neurons, with the counts of cFOS-positive neurons reproducing the effects of HFD on mRNA measures. IEGs are genes that are often rapidly and transiently expressed as direct responses to stimuli such as novel environments or injury. IEG expression is also associated with alterations in synaptic plasticity, with either up- or downregulation of IEGs being associated with increased and decreased LTP and memory retention, respectively [1, 12, 33]. Changes in IEG activity are also associated with behavioral tasks such as novel object recognition, and fear conditioning, indicative of their role in learning [33].
IEG expression also changes from neuronal stress or damage. IEG expression is increased in AD brain [31]. These increases have been linked to more GFAP positive astrocytes and thioflavin-stained plaques [2] and in Aβ-mediated apoptosis [31]. There are also alterations in neuronal IEG expression after traumatic brain injuries [14, 38] and after chronic LPS-induced inflammation [43, 44]. After chronic LPS infusion, mice were tested on a memory paradigm and those treated with LPS had increased IEG expression. This increased IEG expression was directly correlated with increased microglia activity [44]. Overall, the chronic inflammation led to gliosis and that resulted in increased IEG activity and cognitive deficits [44]. We propose a similar process in our study, with HFD, in the APOE3 but not APOE4 mice, increasing gliosis and that gliosis is leading to altered IEG expression.
In many of the published studies, IEG expression was measured after behavioral stimulations such as learning and memory challenges. Here, we have not directly induced IEG expression, suggesting another mechanism. There may be APOE3 specific neuronal adaptations to HFD expressed through IEG activity. IEG increases have been noted without additional stimuli in the HYP after an extended period on HFD, resulting in adipose tissue storage [54]. IEG activity also acts as an indicator of neuronal activity resulting in long-term adaptations within neural circuits [55]. The increased IEG expression we are seeing could affect the processing and storage of specific lipid components of the HFD, and that could lead to increased protection in APOE3 mice. Lack of HFD-induced IEG activity, as in the APOE4 mice, could lead to the more detrimental outcomes. The NanoString neuroinflammatory panel only indicated robust changes in the IEGs in APOE3 mice on HFD. There was no indication of more severe neuroinflammation, further emphasizing that the mice are not currently suffering from detrimental neuroinflammation. Rather, the APOE3 mice may be exhibiting compensational neuronal adaptations in response to HFD.
The APOE3 mice exhibited several CNS alterations associated with HFD that the APOE4 mice did not. These APOE3 mice also showed decreased metabolic disturbances compared to APOE4 mice [21]. We propose that during HFD, APOE3 mice are able to induce protective inflammatory pathways that have positive peripheral and CNS effects. These mice were 10 months old at the end of all experimentation and were on HFD for a total of 4 months; therefore, we may be seeing the preliminary effects of HFD as they continue to gain weight. Prolonged obesity could result in more detrimental effects such as increased noxious inflammation, neuronal damage, and behavioral deficits. APOE4 mice on HFD for extended periods or on diets with higher fat contents exhibit increased inflammation and behavioral deficits [18–20].
The APOE4 allele is the ancestorial version with the APOE3 allele potentially evolving as a result in shifts in diet availability [9, 16, 39]. Evolutionarily, APOE3 may allow humans to compensate for increased diet availability more easily and to process dietary fats, while APOE4 remains better suited for environments where diet availability is scarce.
Conclusion
Overall, we observed increases in HFD-induced gliosis and IEG expression specific to the mice expressing APOE3 and an increase in Adora2a expression in mice expressing APOE4. These changes were apparent without robust changes in an array of other inflammatory genes. This work suggests that initial gliosis and IEGs may perform a novel protective role in response to early stages of obesity. More broadly, this approach of combining strong genetic and environmental factors is necessary for providing insight into personal risks of important CNS impairment and potential for AD development.
Limitations
While these data identify specific CNS alterations in APOE3 mice that do not occur in APOE4, the study has important limitations. It does not identify any specific mechanisms that lead to the CNS alterations caused by HFD, or the downstream consequences of those alterations. In addition, the conclusions about HFD inducing gliosis in APOE3 mice are based on small sample numbers, with only three to four animals (of mixed sex) per group. For qRT-PCR analyses of the identified genes, we examined two to three animals per sex per group, and we did not observe any trends suggesting differences by sex. Given the importance of sex in the risk of AD, further studies could be powered to examine the combined effects of APOE genotype, and diet, and sex. Finally, while we observed strong differences in gliosis in the HPC, a larger sample size may have allowed us to better test for significant differences in the other areas.
Acknowledgements
Christi Anne Ng for artistic contribution to Fig. 1.
Abbreviations
- AD
Alzheimer’s disease
- AO
Apical oblique
- APOE
Apolipoprotein E
- BS
Basal shaft
- CNS
Central nervous system
- CTX
Cortex
- DCX
Doublecortin
- HFD
High-fat diet
- HPC
Hippocampus
- IEG
Immediate early gene
- LTP
Long-term potentiation
Authors’ contributions
NJ designed, analyzed, and interpreted the data. NJ also drafted and revised the work. KW analyzed the data. GWR supervised the work. All authors read and approved the final manuscript.
Funding
This work was supported by National Institutes of Health Grants R01 NS100704, R21 AG074139, and F99 NS118746-01.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All experiments followed the guidelines of the Institutional Animal Care and Use Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nahdia S. Jones, Email: nsj10@georgetown.edu
Katarina Q. Watson, Email: kqw2@georgetown.edu
G. William Rebeck, Email: gwr2@georgetown.edu.
References
- 1.Abraham WC, Dragunow M, Tate WP. The role of immediate early genes in the stabilization of long-term potentiation. Mol Neurobiol. 1991;5(2-4):297–314. doi: 10.1007/BF02935553. [DOI] [PubMed] [Google Scholar]
- 2.Anderson AJ, Cummings BJ, Cotman CW. Increased immunoreactivity for Jun- and Fos-related proteins in Alzheimer’s disease: association with pathology. Exp Neurol. 1994;125(2):286–295. doi: 10.1006/exnr.1994.1031. [DOI] [PubMed] [Google Scholar]
- 3.Arnold SE, Lucki I, Brookshire BR, Carlson GC, Browne CA, Kazi H, Bang S, Choi BR, Chen Y, McMullen MF, Kim SF. High fat diet produces brain insulin resistance, synaptodendritic abnormalities and altered behavior in mice. Neurobiol Dis. 2014;67:79–87. doi: 10.1016/j.nbd.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber CN, Raben DM. Lipid metabolism crosstalk in the brain: glia and neurons. Front Cell Neurosci. 2019;13:212. doi: 10.3389/fncel.2019.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besser LM, Gill DP, Monsell SE, Brenowitz W, Meranus DH, Kukull W, Gustafson DR. Body mass index, weight change, and clinical progression in mild cognitive impairment and Alzheimer disease. Alzheimer Dis Assoc Disord. 2014;28(1):36–43. doi: 10.1097/WAD.0000000000000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boison D, Chen JF, Fredholm BB. Adenosine signaling and function in glial cells. Cell Death Differ. 2010;17(7):1071–1082. doi: 10.1038/cdd.2009.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinel AL, Andre C, Aubert A, Ferreira G, Laye S, Castanon N. Cognitive and emotional alterations are related to hippocampal inflammation in a mouse model of metabolic syndrome. PLoS One. 2011;6(9):e24325. doi: 10.1371/journal.pone.0024325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dose J, Schloesser A, Torres GG, Venkatesh G, Hasler R, Flachsbart F, Lieb W, Nebel A, Rimbach G, Huebbe P. On a Western diet, APOEvarepsilon4 is associated with low innate immune sensing, but not APOEvarepsilon3. J Allergy Clin Immunol. 2018;142(1346-1349):e1349. doi: 10.1016/j.jaci.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 9.Finch CE, Stanford CB. Meat-adaptive genes and the evolution of slower aging in humans. Q Rev Biol. 2004;79(1):3–50. doi: 10.1086/381662. [DOI] [PubMed] [Google Scholar]
- 10.Flowers SA, Rebeck GW. APOE in the normal brain. Neurobiol Dis. 2020;136:104724. doi: 10.1016/j.nbd.2019.104724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomes CV, Kaster MP, Tome AR, Agostinho PM, Cunha RA. Adenosine receptors and brain diseases: neuroprotection and neurodegeneration. Biochim Biophys Acta. 2011;1808(5):1380–1399. doi: 10.1016/j.bbamem.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20(11):3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hales CM, Carroll MD, Fryar CD, Ogden CL (2017) Prevalence of obesity among adults and youth: United States, 2015-2016. NCHS Data Brief:1-8. [PubMed]
- 14.Hayes RL, Yang K, Raghupathi R, McIntosh TK. Changes in gene expression following traumatic brain injury in the rat. J Neurotrauma. 1995;12(5):779–790. doi: 10.1089/neu.1995.12.779. [DOI] [PubMed] [Google Scholar]
- 15.Hooten KG, Beers DR, Zhao W, Appel SH. Protective and toxic neuroinflammation in amyotrophic lateral sclerosis. Neurotherapeutics. 2015;12(2):364–375. doi: 10.1007/s13311-014-0329-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huebbe P, Dose J, Schloesser A, Campbell G, Gluer CC, Gupta Y, Ibrahim S, Minihane AM, Baines JF, Nebel A, Rimbach G. Apolipoprotein E (APOE) genotype regulates body weight and fatty acid utilization-Studies in gene-targeted replacement mice. Mol Nutr Food Res. 2015;59(2):334–343. doi: 10.1002/mnfr.201400636. [DOI] [PubMed] [Google Scholar]
- 17.Janssen CI, Jansen D, Mutsaers MP, Dederen PJ, Geenen B, Mulder MT, Kiliaan AJ. The effect of a high-fat diet on brain plasticity, inflammation and cognition in female ApoE4-knockin and ApoE-knockout mice. PLoS One. 2016;11(5):e0155307. doi: 10.1371/journal.pone.0155307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson LA, Torres ER, Impey S, Stevens JF, Raber J. Apolipoprotein E4 and insulin resistance interact to impair cognition and alter the epigenome and metabolome. Sci Rep. 2017;7(1):43701. doi: 10.1038/srep43701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson LA, Torres ER, Weber Boutros S, Patel E, Akinyeke T, Alkayed NJ, Raber J. Apolipoprotein E4 mediates insulin resistance-associated cerebrovascular dysfunction and the post-prandial response. J Cereb Blood Flow Metab. 2019;39(5):770–781. doi: 10.1177/0271678X17746186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones NS, Rebeck GW. The synergistic effects of APOE genotype and obesity on Alzheimer’s disease risk. Int J Mol Sci. 2018;20(1). 10.3390/ijms20010063. [DOI] [PMC free article] [PubMed]
- 21.Jones NS, Watson KQ, Rebeck GW (2019) Metabolic disturbances of a high fat diet are dependent on APOE genotype and sex. eneuro:ENEURO.0267-0219.2019. [DOI] [PMC free article] [PubMed]
- 22.Julien C, Tremblay C, Phivilay A, Berthiaume L, Emond V, Julien P, Calon F. High-fat diet aggravates amyloid-beta and tau pathologies in the 3xTg-AD mouse model. Neurobiol Aging. 2010;31(9):1516–1531. doi: 10.1016/j.neurobiolaging.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 23.Keene CD, Cudaback E, Li X, Montine KS, Montine TJ. Apolipoprotein E isoforms and regulation of the innate immune response in brain of patients with Alzheimer’s disease. Curr Opin Neurobiol. 2011;21(6):920–928. doi: 10.1016/j.conb.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller KB, Lemberg L. Obesity and the metabolic syndrome. Am J Crit Care. 2003;12(2):167–170. doi: 10.4037/ajcc2003.12.2.167. [DOI] [PubMed] [Google Scholar]
- 25.Kesby JP, Kim JJ, Scadeng M, Woods G, Kado DM, Olefsky JM, Jeste DV, Achim CL, Semenova S. Spatial cognition in adult and aged mice exposed to high-fat diet. PLoS One. 2015;10(10):e0140034. doi: 10.1371/journal.pone.0140034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohjima M, Sun Y, Chan L. Increased food intake leads to obesity and insulin resistance in the tg2576 Alzheimer’s disease mouse model. Endocrinology. 2010;151(4):1532–1540. doi: 10.1210/en.2009-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin B, Hasegawa Y, Takane K, Koibuchi N, Cao C, Kim-Mitsuyama S. High-fat-diet intake enhances cerebral amyloid angiopathy and cognitive impairment in a mouse model of Alzheimer’s disease, independently of metabolic disorders. J Am Heart Assoc. 2016;5(6). 10.1161/JAHA.115.003154. [DOI] [PMC free article] [PubMed]
- 28.Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9(2):106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Loving BA, Bruce KD. Lipid and lipoprotein metabolism in microglia. Front Physiol. 2020;11:393. doi: 10.3389/fphys.2020.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcus DL, Strafaci JA, Miller DC, Masia S, Thomas CG, Rosman J, Hussain S, Freedman ML. Quantitative neuronal c-fos and c-jun expression in Alzheimer’s disease. Neurobiol Aging. 1998;19(5):393–400. doi: 10.1016/S0197-4580(98)00077-3. [DOI] [PubMed] [Google Scholar]
- 32.McNeilly AD, Williamson R, Sutherland C, Balfour DJ, Stewart CA. High fat feeding promotes simultaneous decline in insulin sensitivity and cognitive performance in a delayed matching and non-matching to position task. Behav Brain Res. 2011;217(1):134–141. doi: 10.1016/j.bbr.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Minatohara K, Akiyoshi M, Okuno H. Role of immediate-early genes in synaptic plasticity and neuronal ensembles underlying the memory trace. Front Mol Neurosci. 2015;8:78. doi: 10.3389/fnmol.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moser VA, Pike CJ (2017) Obesity accelerates Alzheimer-related pathology in APOE4 but not APOE3 mice. eNeuro 4. [DOI] [PMC free article] [PubMed]
- 35.Naderali EK, Ratcliffe SH, Dale MC. Obesity and Alzheimer’s disease: a link between body weight and cognitive function in old age. Am J Alzheimers Dis Other Demen. 2009;24(6):445–449. doi: 10.1177/1533317509348208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nam KN, Wolfe CM, Fitz NF, Letronne F, Castranio EL, Mounier A, Schug J, Lefterov I, Koldamova R. Integrated approach reveals diet, APOE genotype and sex affect immune response in APP mice. Biochim Biophys Acta Mol Basis Dis. 2018;1864(1):152–161. doi: 10.1016/j.bbadis.2017.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohta A, Sitkovsky M. Role of G-protein-coupled adenosine receptors in downregulation of inflammation and protection from tissue damage. Nature. 2001;414(6866):916–920. doi: 10.1038/414916a. [DOI] [PubMed] [Google Scholar]
- 38.Phillips LL, Belardo ET. Expression of c-fos in the hippocampus following mild and moderate fluid percussion brain injury. J Neurotrauma. 1992;9(4):323–333. doi: 10.1089/neu.1992.9.323. [DOI] [PubMed] [Google Scholar]
- 39.Raichlen DA, Alexander GE. Exercise, APOE genotype, and the evolution of the human lifespan. Trends Neurosci. 2014;37(5):247–255. doi: 10.1016/j.tins.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivera P, Perez-Martin M, Pavon FJ, Serrano A, Crespillo A, Cifuentes M, Lopez-Avalos MD, Grondona JM, Vida M, Fernandez-Llebrez P, de Fonseca FR, Suarez J. Pharmacological administration of the isoflavone daidzein enhances cell proliferation and reduces high fat diet-induced apoptosis and gliosis in the rat hippocampus. PLoS One. 2013;8(5):e64750. doi: 10.1371/journal.pone.0064750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez GA, Burns MP, Weeber EJ, Rebeck GW. Young APOE4 targeted replacement mice exhibit poor spatial learning and memory, with reduced dendritic spine density in the medial entorhinal cortex. Learn Mem. 2013;20(5):256–266. doi: 10.1101/lm.030031.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez GA, Tai LM, LaDu MJ, Rebeck GW. Human APOE4 increases microglia reactivity at Abeta plaques in a mouse model of Abeta deposition. J Neuroinflammation. 2014;11(1):111. doi: 10.1186/1742-2094-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosi S. Neuroinflammation and the plasticity-related immediate-early gene Arc. Brain Behav Immun. 2011;25(Suppl 1):S39–S49. doi: 10.1016/j.bbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosi S, Ramirez-Amaya V, Vazdarjanova A, Esparza EE, Larkin PB, Fike JR, Wenk GL, Barnes CA. Accuracy of hippocampal network activity is disrupted by neuroinflammation: rescue by memantine. Brain. 2009;132(9):2464–2477. doi: 10.1093/brain/awp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russo MV, McGavern DB. Inflammatory neuroprotection following traumatic brain injury. Science. 2016;353(6301):783–785. doi: 10.1126/science.aaf6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephen TL, Cacciottolo M, Balu D, Morgan TE, LaDu MJ, Finch CE, Pike CJ. APOE genotype and sex affect microglial interactions with plaques in Alzheimer’s disease mice. Acta Neuropathol Commun. 2019;7(1):82. doi: 10.1186/s40478-019-0729-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stocker H, Mollers T, Perna L, Brenner H. The genetic risk of Alzheimer’s disease beyond APOE epsilon4: systematic review of Alzheimer’s genetic risk scores. Transl Psychiatry. 2018;8(1):166. doi: 10.1038/s41398-018-0221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan PM, Mace BE, Maeda N, Schmechel DE. Marked regional differences of brain human apolipoprotein E expression in targeted replacement mice. Neuroscience. 2004;124(4):725–733. doi: 10.1016/j.neuroscience.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 49.Tai LM, Balu D, Avila-Munoz E, Abdullah L, Thomas R, Collins N, Valencia-Olvera AC, LaDu MJ. EFAD transgenic mice as a human APOE relevant preclinical model of Alzheimer’s disease. J Lipid Res. 2017;58(9):1733–1755. doi: 10.1194/jlr.R076315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thiel M, Chouker A, Ohta A, Jackson E, Caldwell C, Smith P, Lukashev D, Bittmann I, Sitkovsky MV. Oxygenation inhibits the physiological tissue-protecting mechanism and thereby exacerbates acute inflammatory lung injury. PLoS Biol. 2005;3(6):e174. doi: 10.1371/journal.pbio.0030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ungar L, Altmann A, Greicius MD. Apolipoprotein E, gender, and Alzheimer’s disease: an overlooked, but potent and promising interaction. Brain Imaging Behav. 2014;8(2):262–273. doi: 10.1007/s11682-013-9272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vitek MP, Brown CM, Colton CA. APOE genotype-specific differences in the innate immune response. Neurobiol Aging. 2009;30(9):1350–1360. doi: 10.1016/j.neurobiolaging.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wanrooy BJ, Kumar KP, Wen SW, Qin CX, Ritchie RH, Wong CHY. Distinct contributions of hyperglycemia and high-fat feeding in metabolic syndrome-induced neuroinflammation. J Neuroinflammation. 2018;15(1):293. doi: 10.1186/s12974-018-1329-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xin X, Storlien LH, Huang XF. Hypothalamic c-fos-like immunoreactivity in high-fat diet-induced obese and resistant mice. Brain Res Bull. 2000;52(4):235–242. doi: 10.1016/S0361-9230(00)00228-8. [DOI] [PubMed] [Google Scholar]
- 55.Yap EL, Greenberg ME. Activity-regulated transcription: bridging the gap between neural activity and behavior. Neuron. 2018;100(2):330–348. doi: 10.1016/j.neuron.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao N, Ren Y, Yamazaki Y, Qiao W, Li F, Felton LM, Mahmoudiandehkordi S, Kueider-Paisley A, Sonoustoun B, Arnold M, Shue F, Zheng J, Attrebi ON, Martens YA, Li Z, Bastea L, Meneses AD, Chen K, Thompson JW, St John-Williams L, Tachibana M, Aikawa T, Oue H, Job L, Yamazaki A, Liu CC, Storz P, Asmann YW, Ertekin-Taner N, Kanekiyo T, Kaddurah-Daouk R, Bu G. Alzheimer’s risk factors age, APOE genotype, and sex drive distinct molecular pathways. Neuron. 2020;106(727-742):e726. doi: 10.1016/j.neuron.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu Y, Nwabuisi-Heath E, Dumanis SB, Tai LM, Yu C, Rebeck GW, LaDu MJ. APOE genotype alters glial activation and loss of synaptic markers in mice. Glia. 2012;60(4):559–569. doi: 10.1002/glia.22289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.