Abstract
In Escherichia coli, elevated levels of free l-tryptophan (l-Trp) promote translational arrest of the TnaC peptide by inhibiting its termination. However, the mechanism by which translation-termination by the UGA-specific decoding release factor 2 (RF2) is inhibited at the UGA stop codon of stalled TnaC-ribosome-nascent chain complexes has so far been ambiguous. This study presents cryo-EM structures for ribosomes stalled by TnaC in the absence and presence of RF2 at average resolutions of 2.9 and 3.5 Å, respectively. Stalled TnaC assumes a distinct conformation composed of two small α-helices that act together with residues in the peptide exit tunnel (PET) to coordinate a single L-Trp molecule. In addition, while the peptidyl-transferase center (PTC) is locked in a conformation that allows RF2 to adopt its canonical position in the ribosome, it prevents the conserved and catalytically essential GGQ motif of RF2 from adopting its active conformation in the PTC. This explains how translation of the TnaC peptide effectively allows the ribosome to function as a L-Trp-specific small-molecule sensor that regulates the tnaCAB operon.
INTRODUCTION
Translational arrest peptides (APs) engage in specific interactions with the peptide exit tunnel (PET) of ribosomes to temporarily arrest translation (1,2). Translational arrest can facilitate certain regulatory mechanisms of gene expression; examples in bacteria include APs acting as antibiotic sensors (3–9), force sensors such as the secretion monitor proteins SecM (1) and VemP (10), small metabolite sensors such as SpeFL for L-ornithine (11) and TnaC for the amino acid L-Trp (12–17).
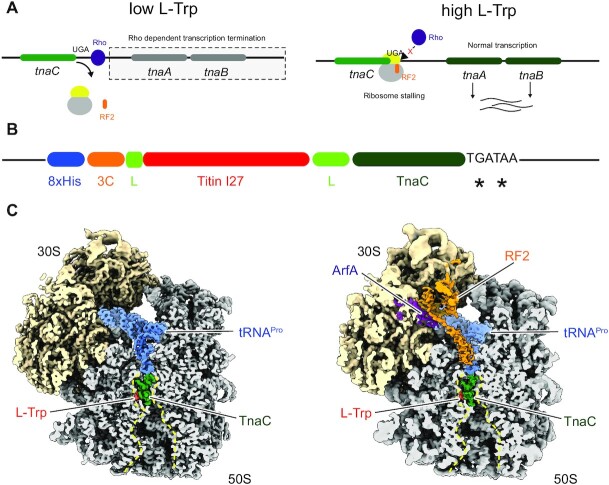
The TnaC system utilizes translational arrest to couple the expression of two tryptophan-processing genes (tnaA and tnaB) to the cellular level of free L-Trp. The tnaCAB operon comprises the tnaC uORF encoding a leader peptide, directly followed by a Rho-dependent transcription termination sequence and further downstream tnaA and tnaB. (14,15,18) (Figure 1A). As long as the Rho-dependent termination sequence on the nascent mRNA is accessible for Rho, transcription and thus expression of the downstream tnaA and tnaB is prevented (19). In this case, translation of the nascent mRNA occurs only at the TnaC leader peptide. Upon encountering the UGA stop codon of tnaC, translation-termination is mediated by the decoding release factor 2 (RF2) that specifically recognizes the UGA stop codon to effect release of the peptidyl-tRNAPro at the P-site of the peptidyl transferase centre (PTC) of the ribosome (13). Thereby, the ribosome is quickly released from the nascent mRNA and the Rho-factor binding site remains accessible. In contrast, when the cellular level of free L-Trp are high, RF2 action is inhibited and the TnaC-peptidyl-tRNAPro is trapped at the PTC. This results in an arrested ribosome at the UGA stop codon (20). The arrested ribosome at the end of the TnaC ORF masks the Rho-factor binding site and the termination of transcription is inhibited; the L-Trp processing genes tnaA and tnaB are then expressed (20).
Figure 1.
(A) Schematic illustration for L-Trp dependent regulation of the tnaCAB operon in E. coli. (B) Schematic illustration of the construct used to purify TnaC–RNCs. A reporter expressing the cleavable His8-3C-Titin-I27-TnaC stalling peptide from a pBAD plasmid was used in a E. coli KC6 ΔsmpB ΔssrA strain (25) and RNCs were purified as described in Materials and Methods. (C) Cryo-EM maps of the TnaC-stalled 70S-RNA, without (left panel) and with bound RF2 and ArfA. The peptide exit tunnel (PET) is outlined with a dashed line (yellow).
The molecular mechanism of L-Trp dependent translational arrest was previously shown to involve the interaction of free L-Trp with the TnaC AP in the PET of the 50S subunit of the Escherichia coli ribosome (16,21–24). This interaction between L-Trp and TnaC was suggested to result in a perturbation of the PTC that suppressed the ability of RF2 to effect release (13,21). Previous cryo-EM structures of TnaC-stalled E.coli ribosome-nascent chain complexes (RNCs) revealed the structure of the nascent TnaC peptide in the PET at resolutions of 5.5 Å (24), 3.8 Å (21) and 3.1 Å (25), but they were all obtained in the absence of RF2. A prior cryo-EM reconstruction of TnaC-stalled RNCs by our group (21) was interpreted to have two L-Trp molecules coordinated in the PET. However, in a recent higher resolution structure, it became clear that there is only one L-Trp molecule located between two very short α-helices formed by the nascent peptide (25). The molecular basis of the L-Trp binding to the nascent chain is an important part of the mechanism since it is causative for the allosteric inhibition of termination. All previous studies suggested that TnaC could potentially stabilize critical residues of the PTC in conformations that are incompatible with the accommodation of RF2 in the A-site. However, in the absence of RF2, none of the existing structures revealed the molecular basis of the release inhibition of the TnaC peptide from the ribosome, and thus of translational arrest.
We now report two cryo-EM structures of L-Trp-containing TnaC-stalled ribosome-nascent chain complexes (RNCs) in the absence and presence of RF2 (TnaC–RNC and RF2–TnaC–RNC) at average resolutions of 2.9 and 3.5 Å, respectively. These structures unambiguously display the key residues of TnaC and the PTC that were previously identified to be important for stalling (12,16,17,23). In contrast to the previous suggestions, the RF2-containing structure reveals that RF2 can be fully accommodated in the A-site, despite the presence of L-Trp in the PET. However, the PTC is locked in a state that prevents the catalytically essential GGQ motif of RF2 from adopting its productive conformation, thereby inhibiting peptide release. Moreover, we confirm that a single L-Trp molecule in the PET, as observed before (25), is sufficient to induce stalling.
MATERIALS AND METHODS
Materials
All chemical reagents were sourced from Sigma-Aldrich Merck Europe, except for n-dodecyl β-d-maltopyranoside (Anatrace), cOmplete™ EDTA-free protease inhibitor cocktail and E. coli tRNA (Roche), Talon® and Ni-NTA metal affinity resin (Clontech, Takara Biosciences). All chemicals used during the purification of RNCs were molecular biology grade.
The centrifuges and rotors used were manufactured by Beckman Coulter unless specified, the emulsifex by Avestin, and the microfluidizer by Microfluidics Corp. Centrifugal filter units for concentrating protein samples were procured from Merck-Millipore. Equipment for size exclusion chromatography (SEC) was manufactured by GE Healthcare.
Purification of TnaC stalled ribosome-nascent chain complexes (TnaC-RNCs) from E. coli
TnaC–RNCs were purified as previously described in a study investigating cotranslational folding of the I27 domain of titin (25). In brief, the TnaC stalling construct was expressed from a pBAD vector harboring the I27-TnaC insert (Figure 1B). It codes for a stalled nascent chain with an N-terminal His8-tag followed by an HRV-3C cleavage site, the 89 amino acid long titin I27 domain (flanked by linkers: GT on the N-terminal and SGSGSGSGGP on the C-terminal side) and the TnaC stalling sequence (NILHISVTSKWFNIDNKIVDHRP-stop).
An E. coli clone (in the KC6 ΔsmpB ΔssrA background) expressing the His8-tagged I27-TnaC construct was cultured in lysogeny broth (LB) to an A600 of 0.5. Expression of the I27-TnaC was induced by treating the cells with a final concentration of 0.3% arabinose and culturing them at 37°C for 1 h. For purification of TnaC–RNCs, all following buffers were supplemented with 2 mM l-tryptophan to induce translation pausing. Cells were cooled down on ice for 15 min before they were collected by centrifugation and resuspended in Buffer A at pH 7.5 (50 mM HEPES–KOH, 250 mM KOAc, 2 mM l-tryptophan, 0.1% (DDM) and cOmplete EDTA-free protease inhibitor cocktail (Roche). Cells were lysed in the Emulsifex at 8000 psi at 4°C. The resulting cell lysate was cleared by centrifugation at 30 000 × g for 30 min in the JA 25–50 rotor (Beckman-Coulter). The supernatant after this step was loaded on a 750 mM sucrose cushion (50 mM HEPES–KOH, 250 mM KOAc, 2 mM l-tryptophan, 0.1% DDM, 750 mM sucrose and cOmplete EDTA-free protease inhibitor), and centrifuged at 45 000 × g for 24 h in a Ti70 rotor (Beckman Coulter) to obtain a ribosome pellet, which was resuspended on ice in 200 μl Buffer A.
I27-TnaC RNCs were purified from the ribosome suspension via an N-terminal His8 tag. The suspension was bound to Talon® metal affinity resin (preincubated with 10 μg/ml E. coli tRNA) for 1 h at 4°C. The sample-bound resin was subsequently washed with 20–50 column volumes of wash buffer at pH 7.5 (50 mM HEPES–KOH, 10 mM Mg(OAc)2, cOmplete EDTA-free protease inhibitor, 250 mM sucrose and 2 mM l-tryptophan). RNCs were eluted by incubating the resin with 2 column volumes elution buffer (50 mM HEPES–KOH, 150 mM KOAc, 2 mM l-tryptophan 10 mM Mg(OAc)2, cOmplete EDTA-free protease inhibitor, 150 mM imidazole, 250 mM sucrose). The eluates were concentrated by centrifugation at 86 320 × g for 2.5 h in a TLA 100.3 rotor). The resulting RNC pellet was allowed to gently dissolve in a minimal volume of grid buffer at pH 7.5 (20 mM HEPES–KOH, 50 mM KOAc, 5 mM Mg(OAc)2, 125 mM sucrose, 2 mM l-tryptophan and 0.03% DDM).
The Lep50-RNCs for visualization of the nascent chain in absence of tryptophan were generated using a His8-3C-Lep50-TnaC construct, comprising the N-terminal 50 aa of the leader peptidase (Lep) (26) and the TnaC stalling sequence. E. coli BL21 (DE3) cells carrying this construct were cultured in lysogeny broth (LB) to an A600 of 0.6. After addition of 2 mM l-tryptophan, expression of the construct was induced by treating the cells with a final concentration of 0.2% arabinose and culturing them at 37°C for 1 h. The cells were collected by centrifugation and resuspended in buffer Ac (50 mM HEPES–KOH pH 7.5, 250 mM KOAc, 2 mM l-tryptophan, 0.1% DDM, 5 mM β-mercaptoethanol, 250 μg/ml chloramphenicol and cOmplete EDTA-free protease inhibitor cocktail (Roche)). Cells were lysed by sonication and the lysate was cleared by centrifugation at 30 000 × g for 30 min in the JA 25–50 rotor (Beckman-Coulter). The supernatant after this step was loaded on a 750 mM sucrose cushion in buffer Ac and centrifuged at 45 000 × g for 24 h in a Ti70 rotor (Beckman Coulter) to obtain a ribosome pellet, which was resuspended on ice in buffer Ac. Affinity purification of the Lep50-RNCs was performed analogously to I27-TnaC RNCs, except for wash buffer composition. Washing steps were performed with 10 column volumes each of buffers Ac, buffer Bc (50 mM HEPES–KOH pH 7.5, 500 mM KOAc, 2 mM l-tryptophan, 0.1% DDM, 5 mM β-mercaptoethanol, 250 μg/ml Chloramphenicol, 15 mM imidazole) and buffer Cc (50 mM HEPES–KOH pH 7.5, 150 mM KOAc, 0.06% DDM, 5 mM β-mercaptoethanol, 250 μg/ml chloramphenicol). Elution was performed in buffer Cc supplemented to 300 mM imidazole. The eluate was loaded on a linear 10–50% sucrose gradient in buffer Cc and centrifuged at 202 048 × g for 2.5 h using a SW 40 Ti rotor (Beckman Coulter). Gradients were fractionated using a Biocomp piston gradient fractionator and A260 was observed using a Biocomp Triax flow cell. The 70S fraction was collected and concentrated by centrifugation in a TLA 100.3 rotor at 86 320 × g. The resulting RNC pellet was allowed to gently dissolve in a minimal volume of grid buffer (20 mM HEPES–KOH pH 7.5, 150 mM KOAc, 5 mM Mg(OAc)2, 125 mM sucrose, 0.05% β-octylglucoside).
Purification of release factor 2 (RF2) from E. coli
E. coli RF2 was expressed from a pET11a vector and purified from BL21 (DE3) cells as previously described (27). Briefly, E. coli BL21 (DE3) cells harboring a His6-tagged RF2 in a pET11a vector were cultured at 37°C to an A600 of 0.5. Expression of RF2 was induced by treating the bacterial cultures with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and culturing them at 37°C for a further 1.5 h. Cells were subsequently collected by centrifugation (SLC-6000 from Sorvall), and the cell pellet was resuspended in lysis buffer at pH 7.5 (50 mM NaH2PO4, 200 mM NaCl, 5 mM imidazole), and lysed in a microfluidizer (M-110L) by passing the suspension through it three times at 18 000 psi. The obtained lysate was clarified by centrifugation at 30 000 × g for 15 min (SS34, Sorvall), and the supernatant was bound to Ni-NTA metal affinity resin for 1 h. The resin was washed with wash buffer at pH 7.5 (50 mM NaH2PO4, 200 mM NaCl, 10 mM imidazole), and RF2 was eluted by incubating the resin with elution buffer at pH 7.5 (50 mM NaH2PO4, 200 mM NaCl, 250 mM imidazole).
The eluates were subjected to size-exclusion chromatography (Superdex 200 10/300 GL) in a gel filtration buffer at pH 7.5 (50 mM HEPES–KOH, 50 mM KCl, 100 mM NaCl, 2% glycerol and 5 mM β-mercaptoethanol). Proteins were concentrated in centrifugal filters units (Amicon ultracel-30), the concentration determined using a nanodrop, and stored at –80°C for future use.
RNC-RF2 binding assay
2 pmol TnaC–RNCs were mixed with a tenfold molar excess of purified RF2 on ice and then incubated at 37°C for 15 min. Binding of RF2 to the TnaC–RNCs was confirmed by spinning the mixture through a 750 mM sucrose cushion followed by an analysis by SDS-PAGE.
Sample preparation for cryo-EM, data acquisition and processing
Data for the RF2-TnaC-RNC sample was collected at the LMU Munich, Germany on a 300 keV Titan Krios transmission electron microscope (Thermo Fisher Scientific). 2 pmol TnaC–RNCs with a tenfold molar excess of purified RF2 (described in the binding assay) were loaded on R3/3 copper grids pre-coated with 2 nm thick carbon (Quantifoil) and vitrified in liquid ethane (Vitrobot Mark IV from Thermo Fisher Scientific). Data was acquired at a nominal pixel size of 1.084 Å at the object scale under low-dose conditions (2.8 e–/Å2/frame) on a Falcon II direct electron detector at defocus ranges from –0.7 to –3.4 μm.
The image stacks of ten frames were aligned and corrected for beam-induced drift using MotionCor2 (28). The CTF for each micrograph was determined using GCTF (29). 15 719 good micrographs were selected from two individual data collections and 1 817 758 particles were picked using CrYOLO (30) with a general model for cryo-EM images. After 2D classification in RELION-3.1, 1 074 182 particles were subjected to several rounds of 3D classification. The major class (459,171 particles) contained a tRNA in the P-site and was chosen for high resolution refinement in CryoSPARC-2 (31) and RELION-3.1 (32). In CryoSPARC, after legacy homogenous refinement, tilt and higher order aberrations were corrected during the newly implemented CryoSPARC homogenous refinement using a mask for the 50S subunit. A similar procedure was performed in RELION-3.1 including correction of higher-order aberrations (33). The final resolution for both programs was 2.9 Å with local resolution ranging from 2.4 to 6.5 Å. RF2-containing classes were enriched by focused classification in RELION 3.1 with a soft mask around the translation factor binding site and the P-site. After several iterative rounds of classification (‘T’ value = 4), 27 026 particles with clear density for RF2 were isolated and refined in both CryoSPARC and RELION-3.1 to a resolution of 3.5 Å.
Structures of the E. coli 70S ribosome bound to ArfA and RF2 (34) (PDB: 5MDV) and the TnaC–RNC used to investigate co-translational folding of I27 titin (25) (PDB 6I0Y) were used as templates for molecular interpretation and refined model building. These structures were docked into the cryo-EM maps and used as a basis to refine the model of the nascent TnaC chain (residues 9–24) including the bound L-Trp and RF2.
Data collection, frame alignment and CTF determination for the Lep50-RNC control sample were performed as described for RNC-RF2. A total of 324 728 particles were picked from 1739 micrographs using RELION 3.1 (32). The particles were subjected to several rounds of 3D classification in RELION 3.1 (32) and a subset of 107,016 particles resembling 70S ribosomes with P-site tRNA were refined to a final resolution of 2.9 Å using CryoSPARC (v3.2) (31). The pixel size was refined to 1.092 Å during processing. The model of the TnaC–RNC was rigid body docked into the final local resolution filtered map, manually adjusted in Coot (35) and refined using PHENIX (36). Structures were visualized using ChimeraX (37).
RESULTS
Stalled ribosome-nascent chain complexes of TnaC in the absence and presence of RF2
RF2–TnaC–RNC complexes were assembled in vitro by reconstituting TnaC–RNCs and RF2 that were purified from E. coli cells via N-terminal histidine tags as previously described (25,27). These samples were subjected to cryo-EM and single-particle analysis. In silico sorting of the dataset yielded a majority of classes containing ribosomes with a P-site tRNA, and a subpopulation of particles representing TnaC–RNCs with bound RF2 (Supplementary Figure S1). Both maps were refined to molecular resolution (average resolution 2.9 Å for the TnaC–RNC and 3.5 Å for the TnaC–RF2–RNC) (Figure 1C and Supplementary Figure S2). As a control, we determined the structure of TnaC–RNCs purified in the absence of L-Trp supplemented with chloramphenicol to prevent hydrolysis of the peptidyl-tRNA (38–41) at 2.9 Å (Supplementary Figure S3).
For the RF2–TnaC–RNC complex, we also identified an additional density that corresponds to the ribosome rescue factor ArfA. ArfA was previously shown to sense translational stalling at truncated mRNAs, and to recruit RF2 in the absence of a stop codon in order to facilitate release of the truncated peptide (27,34,42–44).
Interactions between TnaC and the peptide exit tunnel
We refined our previous reconstruction of the TnaC–RNC (Table 1) including the TnaC nascent chain (25) from the CCA-end of tRNAPro to the constriction site within the PET (TnaC residues 9–24), and reached a local resolution for the nascent peptide and neighbouring regions of around 2.8 Å. The local resolution of the entire ribosome ranges from 2.4 to 6.5 Å (Supplementary Figure S2). For the control reconstruction, no density for L-Trp was present and the TnaC arrest peptide was mostly disordered (Supplementary Figure S3). However, the L-Trp containing structures revealed a clear density for a single free L-Trp that was found close to the constriction site formed by ribosomal proteins uL4 and uL22 (Figure 2). The well-resolved region of TnaC exhibits two single-turn α-helices: minihelix 1 (mH1) comprising residues 11–15, and minihelix 2 (mH2) comprising residues 17–21, in the upper region of the PET, spanning a distance of about 24 Å from the CCA-end to the constriction. The C-terminal proline P24 is connected to the CCA-end of tRNAPro and the preceding R23 sidechain stacks on the C2504:C2452 bases (Figure 2A).
Table 1.
Cryo-EM data collection, refinement and validation statistics.
| TnaC–RNC (EMDB- 12936) (PDB 7OIZ) | RF2–TnaC–RNC (EMDB- 12937) (PDB 7OJ0) | |
|---|---|---|
| Data collection and processing | ||
| Magnification | 129 151 | |
| Voltage (kV) | 300 | |
| Electron exposure (e–/Å2) | 28 | |
| Defocus range (μm) | 0.7–3.4 | |
| Pixel size (Å) | 1.084 | |
| Symmetry imposed | C1 | |
| Initial particle images (no.) | 1 074 182 | |
| Final particle images (no.) | 459 171 | 27 026 |
| Map resolution (Å) | 2.89 | 3.52 |
| FSC threshold | 0.143 | 0.143 |
| Refinement | ||
| Initial model used (PDB code) | 5MDV, 6I0Y | |
| Map sharpening B factor (Å2) | -20 | -20 |
| Model composition | ||
| Non-hydrogen atoms | 140 339 | 146 553 |
| Protein residues | 5602 | 6264 |
| RNA | 4474 | 4527 |
| Ligands | 313 | 312 |
| B factors (Å2) | 56.54 | 110.93 |
| Protein | 51.63 | 122.49 |
| RNA | 58.86 | 105.22 |
| Ligand | 38.10 | 73.37 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.006 |
| Bond angles (°) | 0.802 | 0.815 |
| Validation | ||
| MolProbity score | 1.39 | 1.70 |
| Clashscore | 3.72 | 7.12 |
| Poor rotamers (%) | 0.00 | 0.06 |
| Ramachandran plot | ||
| Favored (%) | 96.50 | 95.58 |
| Allowed (%) | 3.44 | 4.33 |
| Disallowed (%) | 0.05 | 0.08 |
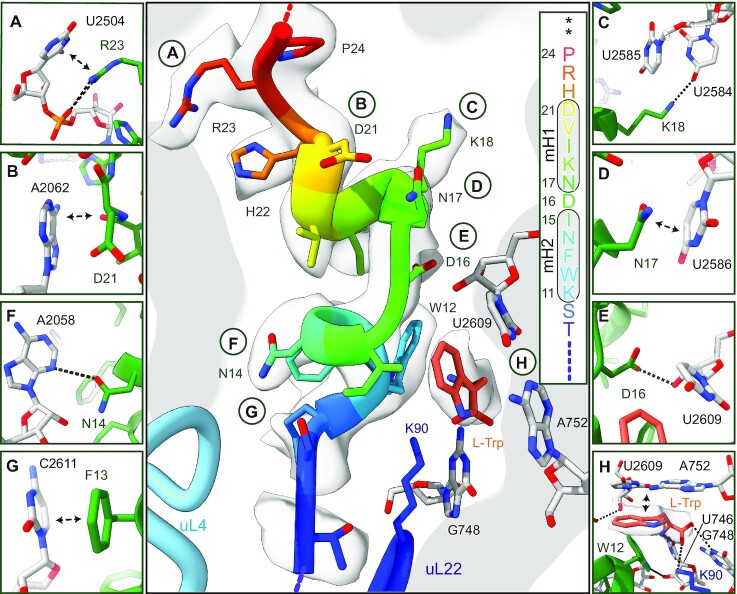
Figure 2.
Structure and molecular interactions of the TnaC-AP and L-Trp within the peptide exit tunnel. The central panel shows an overview and secondary structure of the TnaC-AP fitted into the electron potential map (transparent grey); panels (A–H) show zoomed views focusing on specific molecular interactions. See text for details. mH1 and mH2 = minihelix 1 and minihelix 2, asterisks indicate stop codons.
We also found that the amino acids essential for TnaC stalling (23) play a crucial role in forming the proximal compacted peptide structure. The peptide bond of D21 in mH1 stacks on A2062 (Figure 2B), and the essential D16 is located between the two minihelices and forms a hydrogen bond with K18. K18 forms an additional hydrogen bond with U2584 (Figure 2C). Within mH2, D16 and N17 are both involved in formation of the L-Trp binding pocket, and form a distinct network of interactions with the 23S ribosomal RNA (23S rRNA). These include stacking of the N17 side chain on the base of U2586 (Figure 2D), a hydrogen bond between D16 and U2609 (Figure 2E), hydrogen bonds between N14 and A2058 (Figure 2F), and stacking of F13 on C2611 (Figure 2G).
The Trp binding pocket accommodates a single free L-Trp
A single L-Trp molecule is bound in a pocket formed by the rRNA bases A752, U2609 and mH1 of TnaC (Figure 2H). These bases together with W12, were previously found to be essential for stalling (16,17,23). Substituting either or both of the bases likely disfavours an interaction between D16 in mH1 and U2609, thereby disrupting the geometry of the L-Trp binding pocket.
L-Trp itself is recognized by both the TnaC peptide and the ribosome. The indole ring is stacked on the A752:U2609 bases, and the adjacent W12 within the nascent TnaC may contribute to shaping the binding pocket to accommodate L-Trp. The configuration of the α-carbon of L-Trp finds a matching complementary environment formed by rRNA bases C746, G748 and amino acid K90 of uL22, the mutation of which was shown to abolish TnaC-mediated stalling (22,23). Together, these residues establish a hydrogen-bonding pattern that complements the stacking of the indole moiety, resulting in a high specificity for L-Trp. Notably, the indole amine does not appear to be involved in hydrogen bonding, explaining why N1-methylated L-Trp can still trigger stalling (45).
Rearrangements at the PTC impede translation termination by RF2
Previous structures of TnaC–RNCs (21,25) with bound L-Trp surmised that TnaC stabilizes critical residues of the PTC, such as U2585 and A2602, in a conformation that may be incompatible with the accommodation of RF2 and proper positioning of its GGQ-bearing domain 3 in the A-site. In the present study, we found a class of ribosomal particles associated with both RF2 and ArfA (Figure 1C). Notably, no density for mRNA is visible for its 3′ end in the mRNA channel of the 30S subunit indicating an endonucleolytic cleavage event during the in vivo generation or purification of the RNCs. Alternatively, the mRNA might be displaced as was previously observed in stalled ribosomes bound to ArfB (46). ArfA and RF2 appear to adopt identical positions when compared to previously visualized reconstituted ArfA-RF2 ribosome complexes (27,34,42–44). Despite the identical overall position of RF2, a critical difference between our TnaC-stalled and previous ArfA-RF2-70S complexes (27,34,42–44), is an almost complete disorder of the GGQ loop of RF2 (Figure 3A and B and Supplementary Figure S4). A comparison of the PTC between the RF2-bound and RF2-free maps shows that the densities for both, the PTC and the TnaC peptide, are essentially identical.
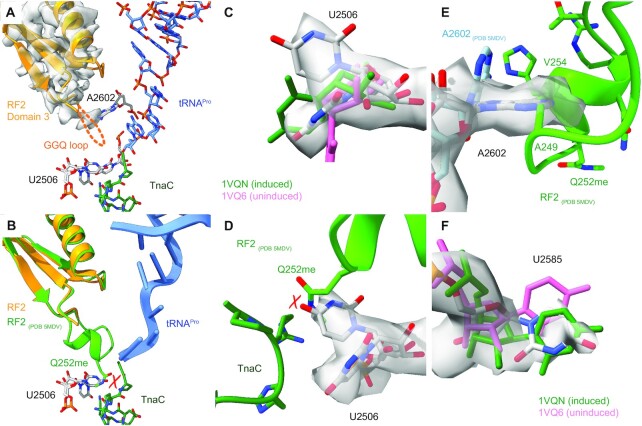
Figure 3.
(A) View focusing on the GGQ-harbouring RF2 domain 3 (density map in transparent grey) and the TnaC-AP-tRNAPro. No density is present for the GGQ-loop, indicating its flexibility. (B) Overlay of RF2 with a fully established GGQ-loop (green) as present in the ArfA-RF2-70S non-stop complex (34) with RF2 bound to the TnaC–RNC (yellow) showing a potential molecular clash of U2506 with methylated Q252 of RF2. (C) Density for U2506 (transparent grey), in comparison with its induced (1VQN) and uninduced (1VQ6) conformations (51). (D) Close-up of the clash between Q252me (5MDV) and U2506. (E) Position of U2585 compared with its induced and uninduced conformations. (F) The position of A2602 in the RF2–TnaC–RNC structure differs from the non-stop complex (5MDV) where the GGQ-loop is established.
To understand the silencing of the PTC, we inspected the conformation of three universally conserved PTC bases—U2506, U2585 and A2602— that have all been reported to be crucial for peptide-bond formation and release (47,48). During translation elongation and termination, these residues switch from an uninduced to an induced state. This rearrangement of the active site results from accommodation of the A-site—either by aminoacyl-tRNA or class I release factors—and allows for the attack of activated nucleophiles during catalysis, which in this case is a water molecule for termination (49–51). In both maps, we observe U2506 in two conformations, one where it pairs with G2583 and the other where it faces the uracil base towards the sidechain of R23 of TnaC (Figure 3C). While the first conformation resembles the induced conformation (1VQN), the latter position significantly differs from both the induced and uninduced states and would directly clash with methylated Q252 in the RF2 GGQ loop (Figure 3B, D and Supplementary Figure S4). We thus speculate that U2506 plays an active role in displacement of the GGQ loop, although the induced conformation of U2506 could still allow for accommodation of the GGQ loop.
As observed before in the absence of RF2 (21), we find A2602 to be in a stabilized conformation, which differs from the ArfA-RF2 accommodated structure (PDB 5MDV; Supplementary Figure S4) (34). As A2602 is also more disordered in our control structure, we believe that its stabilization is likely to be a result of the highly-structured TnaC in the tunnel. Yet, at the current resolution we cannot explain the detailed allosteric connection to the presence of the l-tryptophan-stabilized TnaC chain. Nevertheless, this stabilized conformation of A2602 would clearly clash with the GGQ loop of RF2 or with the residues immediately preceding (A249) or succeeding (V254) it (Figure 3E). Although the density of A2602 was less well-defined in the RF2-containing map, A2602 may still be rigid enough to affect the conformation of the GGQ loop. Finally, we did not find U2585 to be in a well-defined conformation, indicative of a high degree of flexibility. Nevertheless, weak density was visible for the induced conformation of U2585 (Figure 3F).
Taken together, our structures show that the binding of RF2 to TnaC–RNCs can in principle allow formation of the induced PTC state, at least for the base U2585. However, this base appears unusually flexible, thereby potentially being unfavourable for proper catalysis of the release reaction. Peptide release is primarily inhibited by preventing the formation of the rigid and active conformation of the RF2 GGQ-loop in the PTC. The observed disorder of the GGQ loop is likely a result of the synergistic steric interference of the critical PTC bases U2506 and A2602. This unusual conformation is either directly (U2506) or allosterically (A2602) stabilized by the TnaC AP. As a result, positioning of the nucleophilic water molecule for peptide release (49) is no longer possible.
DISCUSSION
When compared with other translational arrest peptides (1,2), the observed TnaC-mediated mechanism displays several of the typically observed features of arrest peptides. First, the nascent peptide engages in numerous defined contacts with the ribosomal tunnel between the PTC and the constriction, resulting in a distinct conformation of the peptide within the PET. Similar to the force-sensing APs of the cytomegalovirus (CMV-AP; encoded by the gp48 uORF2) (52) and VemP (10), TnaC adopts an α-helical secondary structure. However, in contrast to the other APs, its stabilization is dependent on the presence of L-Trp, for which it forms a binding pocket. Second, TnaC contains a C-terminal proline (P24), an amino acid with a rigid side-chain and slower peptidyl transfer kinetics. Terminal prolines are often observed in APs (2), for example in the CMV-AP, which also stalls at the stage of termination. Third, the TnaC AP arrests translation by perturbation of the PTC of the ribosome, resulting in inhibition of the RF2-mediated release reaction by preventing the formation of an ordered GGQ loop.
The suggested mechanism is consistent with previous observations that the peptidyl transfer reaction, in contrast to the release reaction, is not substantially impaired by the TnaC peptide (12,16,22,23). Interestingly, in the case of the eukaryotic CMV-AP, which prevents termination by eRF1, the GGQ loop can adopt its active conformation, but the PTC is still inhibited due to a complete repositioning of the base equivalent to E. coli U2585 (52). It is more difficult to understand however, how the release of the TnaC AP is efficiently catalysed by RF1, which also carries a GGQ loop (53). A possible explanation is that the GGQ motif and its RF1-specific flanking regions are capable of a stronger interaction with the ribosome, thereby overcoming the steric hindrance posed by the TnaC-stabilized PTC bases. This would be consistent with recent mutational analyses (53) and the known higher affinity of RF1 for ribosomes (54).
To our surprise, we also found the ribosome rescue factor ArfA associated with RF2 in all classes containing the release factor RF2. ArfA hasn’t been implicated with TnaC stalling so far, and we can only speculate about its involvement in this process. Several studies indicate that peptide-mediated stalling can lead to mRNA cleavage in or near the ribosomal A-site, especially in E. coli orB. subtilis cells with a non-functional trans-translation system (55–58). This cleavage is performed by a yet unknown endonuclease and was shown to be independent of RelE (55,56). It is possible that such a cleavage occurred during the course of a quality control event in our ΔsmpB ΔssrA E. coli strain when expressing the I27-TnaC construct, creating a substrate for ArfA-RF2. Another possible role of ArfA may be in displacement or disordering of mRNA in the mRNA channel as observed in cryo-EM structures of ArfB-bound stalled ribosomes with an mRNA present but not visible 3′ of the P-site (46). Yet, the functional role of such a displacement in the context of TnaC stalling is unclear and the role of ArfA needs to be elucidated in further studies.
In summary, our findings explain how the translation of the TnaC peptide effectively turns the ribosome into a small-molecule sensor that specifically recognizes a single L-Trp molecule, leading to the allosteric inhibition of RF2-dependent release and resulting in L-Trp dependent regulation of the tnaCAB operon.
DATA AVAILABILITY
Cryo-EM volumes and molecular models have been deposited at the Electron Microscopy Data Bank and Protein Data Bank with accession codes EMD-12936 (for the TnaC–RNC), EMD-12937 (for the RF2–TnaC–RNC), EMD-13180 (for the control TnaC–RNC) and PDB-7OIZ (for the TnaC–RNC), PDB-7OJ0 (for the RF2–TnaC–RNC), and PDB-7P3K (for the control TnaC–RNC). Materials are available from the authors on request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Ungewickell and S. Rieder for excellent technical assistance.
Notes
Present address: Ting Su, Pieris Pharmaceuticals GmbH, Hallbergmoos 85399, Germany.
Present address: Jingdong Cheng, Institutes of biomedical science, Shanghai Key Laboratory of Medical Epigenetics, International Co-laboratory of Medical Epigenetics and Metabolism (Ministry of Science and Technology), Fudan university, Dong’an Road 131, 200032 Shanghai, China.
Contributor Information
Ting Su, Gene Center Munich, Department of Biochemistry, University of Munich, Munich 81377, Germany.
Renuka Kudva, Department of Biochemistry and Biophysics, Stockholm University, Stockholm SE-10691, Sweden; Science for Life Laboratories, Solna 17165, Sweden.
Thomas Becker, Gene Center Munich, Department of Biochemistry, University of Munich, Munich 81377, Germany.
Robert Buschauer, Gene Center Munich, Department of Biochemistry, University of Munich, Munich 81377, Germany.
Tobias Komar, Gene Center Munich, Department of Biochemistry, University of Munich, Munich 81377, Germany.
Otto Berninghausen, Gene Center Munich, Department of Biochemistry, University of Munich, Munich 81377, Germany.
Gunnar von Heijne, Department of Biochemistry and Biophysics, Stockholm University, Stockholm SE-10691, Sweden; Science for Life Laboratories, Solna 17165, Sweden.
Jingdong Cheng, Gene Center Munich, Department of Biochemistry, University of Munich, Munich 81377, Germany.
Roland Beckmann, Gene Center Munich, Department of Biochemistry, University of Munich, Munich 81377, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
DFG fellowship through the Graduate School of Quantitative Biosciences Munich (QBM) (to T.S.); Ph.D. fellowship by Boehringer Ingelheim Fonds (to R.B.); DFG [SFB/TRR-174, BE1814/15-1, BE1814/1-1 to R.B.]; Knut and Alice Wallenberg Foundation [2017.0323]; Novo Nordisk Fund [NNF18OC0032828]; Swedish Research Council [621-2014-3713 to G.v.H.]. Funding for open access charge: DFG (to R.B.).
Conflict of interest statement. Ting Su, Renuka Ludva, Thomas Becker, Robert Buschauer, Tobias Komar, Otto Berninghausen, Gunnar von Heijne, Jingdong Cheng and Roland Beckmann declare that they have no conflicts of interest.
REFERENCES
- 1.Ito K., Chiba S.. Arrest peptides: cis-acting modulators of translation. Annu. Rev. Biochem. 2013; 82:171–202. [DOI] [PubMed] [Google Scholar]
- 2.Wilson D.N., Arenz S., Beckmann R.. Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr. Opin. Struct. Biol. 2016; 37:123–133. [DOI] [PubMed] [Google Scholar]
- 3.Arenz S., Juette M.F., Graf M., Nguyen F., Huter P., Polikanov Y.S., Blanchard S.C., Wilson D.N.. Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome. PNAS. 2016; 113:7527–7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arenz S., Meydan S., Starosta A.L., Berninghausen O., Beckmann R., Vazquez-Laslop N., Wilson D.N.. Drug sensing by the ribosome induces translational arrest via active site perturbation. Mol. Cell. 2014; 56:446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arenz S., Ramu H., Gupta P., Berninghausen O., Beckmann R., Vazquez-Laslop N., Mankin A.S., Wilson D.N.. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat. Commun. 2014; 5:3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta P., Liu B., Klepacki D., Gupta V., Schulten K., Mankin A.S., Vazquez-Laslop N.. Nascent peptide assists the ribosome in recognizing chemically distinct small molecules. Nat. Chem. Biol. 2016; 12:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koch M., Willi J., Pradere U., Hall J., Polacek N.. Critical 23S rRNA interactions for macrolide-dependent ribosome stalling on the ErmCL nascent peptide chain. Nucleic Acids Res. 2017; 45:6717–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen F., Starosta A.L., Arenz S., Sohmen D., Donhofer A., Wilson D.N.. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014; 395:559–575. [DOI] [PubMed] [Google Scholar]
- 9.Seip B., Innis C.A.. How widespread is metabolite sensing by ribosome-arresting nascent peptides. J. Mol. Biol. 2016; 428:2217–2227. [DOI] [PubMed] [Google Scholar]
- 10.Su T., Cheng J., Sohmen D., Hedman R., Berninghausen O., von Heijne G., Wilson D.N., Beckmann R.. The force-sensing peptide VemP employs extreme compaction and secondary structure formation to induce ribosomal stalling. eLife. 2017; 6:e25642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrero Del Valle A., Seip B., Cervera-Marzal I., Sacheau G., Seefeldt A.C., Innis C.A.. Ornithine capture by a translating ribosome controls bacterial polyamine synthesis. Nat Microbiol. 2020; 5:554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz-Vera L.R., Gong M., Yanofsky C.. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. PNAS. 2006; 103:3598–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong F., Ito K., Nakamura Y., Yanofsky C.. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro). PNAS. 2001; 98:8997–9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong F., Yanofsky C.. Instruction of translating ribosome by nascent peptide. Science. 2002; 297:1864–1867. [DOI] [PubMed] [Google Scholar]
- 15.Konan K.V., Yanofsky C.. Regulation of the Escherichia coli tna operon: nascent leader peptide control at the tnaC stop codon. J. Bacteriol. 1997; 179:1774–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez A.K., Gordon E., Sengupta A., Shirole N., Klepacki D., Martinez-Garriga B., Brown L.M., Benedik M.J., Yanofsky C., Mankin A.S.et al.. Interactions of the TnaC nascent peptide with rRNA in the exit tunnel enable the ribosome to respond to free tryptophan. Nucleic Acids Res. 2014; 42:1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez A.K., Shirole N.H., Murakami S., Benedik M.J., Sachs M.S., Cruz-Vera L.R.. Crucial elements that maintain the interactions between the regulatory TnaC peptide and the ribosome exit tunnel responsible for Trp inhibition of ribosome function. Nucleic Acids Res. 2012; 40:2247–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Botsford J.L., Demoss R.D.. Escherichia coli tryptophanase in the enteric environment. J. Bacteriol. 1972; 109:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gish K., Yanofsky C.. Evidence suggesting cis action by the TnaC leader peptide in regulating transcription attenuation in the tryptophanase operon of Escherichia coli. J. Bacteriol. 1995; 177:7245–7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong F., Yanofsky C.. Analysis of tryptophanase operon expression in vitro: accumulation of TnaC-peptidyl-tRNA in a release factor 2-depleted S-30 extract prevents Rho factor action, simulating induction. J. Biol. Chem. 2002; 277:17095–17100. [DOI] [PubMed] [Google Scholar]
- 21.Bischoff L., Berninghausen O., Beckmann R.. Molecular basis for the ribosome functioning as an L-tryptophan sensor. Cell Rep. 2014; 9:469–475. [DOI] [PubMed] [Google Scholar]
- 22.Cruz-Vera L.R., Rajagopal S., Squires C., Yanofsky C.. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell. 2005; 19:333–343. [DOI] [PubMed] [Google Scholar]
- 23.Cruz-Vera L.R., Yanofsky C.. Conserved residues Asp16 and Pro24 of TnaC-tRNAPro participate in tryptophan induction of Tna operon expression. J. Bacteriol. 2008; 190:4791–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seidelt B., Innis C.A., Wilson D.N., Gartmann M., Armache J.P., Villa E., Trabuco L.G., Becker T., Mielke T., Schulten K.et al.. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009; 326:1412–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian P., Steward A., Kudva R., Su T., Shilling P.J., Nickson A.A., Hollins J.J., Beckmann R., von Heijne G., Clarke J.et al.. Folding pathway of an Ig domain is conserved on and off the ribosome. PNAS. 2018; 115:E11284–E11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bornemann T., Jockel J., Rodnina M.V., Wintermeyer W.. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 2008; 15:494–499. [DOI] [PubMed] [Google Scholar]
- 27.Huter P., Muller C., Beckert B., Arenz S., Berninghausen O., Beckmann R., Wilson D.N.. Structural basis for ArfA-RF2-mediated translation termination on mRNAs lacking stop codons. Nature. 2017; 541:546–549. [DOI] [PubMed] [Google Scholar]
- 28.Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A.. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang K.Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016; 193:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D.et al.. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol. 2019; 2:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A.. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017; 14:290–296. [DOI] [PubMed] [Google Scholar]
- 32.Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H.. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018; 7:e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zivanov J., Nakane T., Scheres S.H.W.. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020; 7:253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James N.R., Brown A., Gordiyenko Y., Ramakrishnan V.. Translational termination without a stop codon. Science. 2016; 354:1437–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D, Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 36.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D, Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E.. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018; 27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen J.L., Moore P.B., Steitz T.A.. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 2003; 330:1061–1075. [DOI] [PubMed] [Google Scholar]
- 39.Bulkley D., Innis C.A., Blaha G., Steitz T.A.. Revisiting the structures of several antibiotics bound to the bacterial ribosome. 2010; 107:17158–17163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dunkle J.A., Xiong L., Mankin A.S., Cate J.H.. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. PNAS. 2010; 107:17152–17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svetlov M.S., Plessa E., Chen C.W., Bougas A., Krokidis M.G., Dinos G.P., Polikanov Y.S.. High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition. RNA. 2019; 25:600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demo G., Svidritskiy E., Madireddy R., Diaz-Avalos R., Grant T., Grigorieff N., Sousa D., Korostelev A.A.. Mechanism of ribosome rescue by ArfA and RF2. eLife. 2017; 6:e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma C., Kurita D., Li N., Chen Y., Himeno H., Gao N.. Mechanistic insights into the alternative translation termination by ArfA and RF2. Nature. 2017; 541:550–553. [DOI] [PubMed] [Google Scholar]
- 44.Zeng F., Chen Y., Remis J., Shekhar M., Phillips J.C., Tajkhorshid E., Jin H.. Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome. Nature. 2017; 541:554–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong F., Yanofsky C.. Reproducing tna operon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J. Biol. Chem. 2001; 276:1974–1983. [DOI] [PubMed] [Google Scholar]
- 46.Chan K.H., Petrychenko V., Mueller C., Maracci C., Holtkamp W., Wilson D.N., Fischer N., Rodnina M.V.. Mechanism of ribosome rescue by alternative ribosome-rescue factor B. Nat. Commun. 2020; 11:4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polacek N., Gomez M.J., Ito K., Xiong L., Nakamura Y., Mankin A.. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol. Cell. 2003; 11:103–112. [DOI] [PubMed] [Google Scholar]
- 48.Youngman E.M., Brunelle J.L., Kochaniak A.B., Green R.. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004; 117:589–599. [DOI] [PubMed] [Google Scholar]
- 49.Korostelev A.A.Structural aspects of translation termination on the ribosome. RNA. 2011; 17:1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmeing T.M., Huang K.S., Kitchen D.E., Strobel S.A., Steitz T.A.. Structural insights into the roles of water and the 2' hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell. 2005; 20:437–448. [DOI] [PubMed] [Google Scholar]
- 51.Schmeing T.M., Huang K.S., Strobel S.A., Steitz T.A.. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005; 438:520–524. [DOI] [PubMed] [Google Scholar]
- 52.Matheisl S., Berninghausen O., Becker T., Beckmann R.. Structure of a human translation termination complex. Nucleic Acids Res. 2015; 43:8615–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emmanuel J.S., Sengupta A., Gordon E.R., Noble J.T., Cruz-Vera L.R.. The regulatory TnaC nascent peptide preferentially inhibits release factor 2-mediated hydrolysis of peptidyl-tRNA. J. Biol. Chem. 2019; 294:19224–19235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adio S., Sharma H., Senyushkina T., Karki P., Maracci C., Wohlgemuth I., Holtkamp W., Peske F., Rodnina M.V.. Dynamics of ribosomes and release factors during translation termination in E. coli. eLife. 2018; 7:e34252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayes C.S., Sauer R.T.. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell. 2003; 12:903–911. [DOI] [PubMed] [Google Scholar]
- 56.Sunohara T., Jojima K., Yamamoto Y., Inada T., Aiba H.. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA. 2004; 10:378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loomis W.P., Koo J.T., Cheung T.P., Moseley S.L.. A tripeptide sequence within the nascent DaaP protein is required for mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 2001; 39:693–707. [DOI] [PubMed] [Google Scholar]
- 58.Drider D., DiChiara J.M., Wei J., Sharp J.S., Bechhofer D.H.. Endonuclease cleavage of messenger RNA in Bacillus subtilis. Mol. Microbiol. 2002; 43:1319–1329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM volumes and molecular models have been deposited at the Electron Microscopy Data Bank and Protein Data Bank with accession codes EMD-12936 (for the TnaC–RNC), EMD-12937 (for the RF2–TnaC–RNC), EMD-13180 (for the control TnaC–RNC) and PDB-7OIZ (for the TnaC–RNC), PDB-7OJ0 (for the RF2–TnaC–RNC), and PDB-7P3K (for the control TnaC–RNC). Materials are available from the authors on request.