Abstract
Gametocytogenesis, the process by which malaria parasites produce sexual forms that can infect mosquitoes, is essential for the transmission of malaria. A transcriptional switch of the pfap2-g gene triggers sexual commitment, but how the complex multi-step process is precisely programed remains largely unknown. Here, by systematic functional screening of a panel of ApiAP2 transcription factors, we identify six new ApiAP2 members associated with gametocytogenesis in Plasmodium falciparum. Among these, PfAP2-G5 (PF3D7_1139300) was found to be indispensable for gametocytogenesis. This factor suppresses the transcriptional activity of the pfap2-g gene via binding to both the upstream region and exonic gene body, the latter is linked to the maintenance of local heterochromatin structure, thereby preventing initiation of sexual commitment. Removal of this repressive effect through pfap2-g5 knockout disrupts the asexual replication cycle and promotes sexual commitment accompanied by upregulation of pfap2-g expression. However, the gametocytes produced fail to mature fully. Further analyses show that PfAP2-G5 is essential for gametocyte maturation, and causes the down-regulation of pfap2-g and a set of early gametocyte genes activated by PfAP2-G prior to gametocyte development. Collectively, our findings reveal a regulation cascade of gametocyte production in malaria parasites, and provide a new target for transmission blocking interventions.
INTRODUCTION
Gametocytogenesis in malaria parasites is composed of two sequential stages; sexual commitment, in which a trophozoite commits to differentiate into a gametocyte rather than a schizont, and development, the biological process by which the merozoite transforms into a gametocyte (1,2). Identification of the regulators controlling the behavior of parasites pre- and post-sexual commitment can inform the design of transmission-blocking interventions for the control of malaria. Growing evidence indicates that one member of the apicomplexan apetala (AP2) transcription factor (ApiAP2) family, AP2-G, is a master regulator of sexual commitment in both human and rodent malaria parasites (3–10). In addition, studies with rodent and human malaria parasites have shown that additional ApiAP2 proteins, such as AP2-G2 and AP2-G3, co-regulate gametocytogenesis in conjunction with AP2-G, suggesting that a regulatory network involving ApiAP2 proteins regulates sexual commitment and gametocyte development (4,6,11–13). However, the molecular basis for the entire process is not well understood.
In the human malaria parasite Plasmodium falciparum, epigenetic regulators such as PfHP1 and PfHda2 repress sexual commitment via silencing of the pfap2-g gene in a heterochromatic environment, and GDV1 positively regulates this process by antagonizing HP1 (14–17). In addition, a metabolic pathway involving lysophosphatidylcholine also regulates sexual differentiation during the earliest known events upstream of PfAP2-G-mediated commitment (17,18). However, direct regulators of the pfap2-g gene remain largely uncharacterized. It was recently demonstrated that pfap2-g is able to positively autoregulate itself through the binding of PfAP2-G to its upstream regulatory region (URR) (9). Such a positive feedback loop likely contributes to a rapid response to environmental cues. However, how this feedback is regulated is not known. Moreover, the molecular mechanism(s) regulating the entire multi-step process of gametocytogenesis remain unresolved. To explore potential mutual regulation between PfAP2-G and other ApiAP2 factors, we utilized CRISPR–Cas9-mediated gene disruption to screen ApiAP2 transcriptional factors that have unknown functions using a gametocyte-producing parasite strain, P. falciparum NF54 (19). In addition to PfAP2-G, other ApiAP2 factors such as PfSIP2, PfAP2-Tel, PfAP2-I and PfAP2-exp are known to regulate telomere biology or gene expression in P. falciparum (9,20–24). Excluding these, the functions of the remaining twenty-two ApiAP2 genes remain uncharacterised.
Here we report that the ApiAP2 PF3D7_1139300, herein named PfAP2-G5, is crucial for both sexual commitment and development via regulation of the transcription of pfap2-g and/or its downstream target genes. We find that PfAP2-G5 suppresses the expression levels of pfap2-g via binding to the upstream regulatory region directly or through recruitment to the exonic gene body involved in the maintenance of local heterochromatin structure, thereby preventing the initiation of sexual commitment. Removal of this repressive effect by knockout of pfap2-g5 induces higher pfap2-g expression levels and disrupts asexual replication, but parasites fail to develop into mature gametocytes. We show that PfAP2-G5 is essential for gametocyte development through down-regulation of pfap2-g and a set of early gametocyte genes which are activated by PfAP2-G during commitment to gametocytogenesis.
MATERIALS AND METHODS
Parasite culture, plasmid construction and transfection
Plasmodium falciparum NF54 and 3D7 (G7 clone) parasites were cultured in vitro as described previously (25). Briefly, parasites were cultured in medium containing 0.5% Albumax II with O type fresh red blood cells, and a gas phase with 5% O2, 5% CO2 at 37°C. Ring stage synchronization was achieved by repeated 5% sorbitol treatments. To disrupt the opening reading frame (ORF) of the ApiAP2 TFs coding sequence in NF54 and 3D7 wild-types, we generated CRSIPR/Cas9 knockout vectors (26). First, complementary oligonucleotides encoding the guide RNA were inserted into pL6 plasmid between Xho I and Avr II sites. A ∼1-kb homologue sequence of the target genes which contained a several bases deletion was then generated using Asc I and Afl II sites by the In-Fusion PCR Cloning System. To create the GFP-expressing or 3× HA-tag parasite lines, we prepared transfection vectors carrying the sequence of gfp or 3× HA-tag, and fused them to the 3′end of pfap2-g5, pfap2-g or pfap2-g4, respectively. To generate promoter region mutation in pfap2-g and pfap2-g5_trun_rc parasites, the same strategy was adopted. Sequences coding different drug resistance genes were inserted into a pL6 plasmid between the Afl II and Hind III sites (see related supplementary Figures for more details of each construct).
These vectors were transfected to synchronized ring-stage parasites at ∼5% parasitemia with 100 μg of plasmid sgRNA and 80 μg pUF-Cas9 by electroporation as previously described (26). Transfected parasites were selected by DSM1, WR99210, G418 or Blasticidin S deaminase (BSD) according to the selective marker gene used in each plasmid. After selection under drug pressure for continuous 2–3 weeks, the transgenic parasites appeared. To confirm successful editing of different genes, parasites were subjected to PCR diagnosis or PCR-sequencing of integration site with genomic DNA (gDNA), or Western-blots with total protein extract lysed in SDS-PAGE loading buffer. The established lines were sub-cloned by limiting dilution cloning. All primers and sgRNA sequences used and the editing positions are listed in Supplementary Table S1.
Gametocytogenesis assay
Gametocyte induction was performed as previously described with minor modifications (27,28). Briefly, parasites were maintained in culture at < 5% parasitemia at 4% hematocrit. When cultures reached 3–5% rings, parasites were synchronized twice in two continuous intraerythrocytic cycles by 5% sorbitol solution. Old RBCs were replaced by fresh RBCs in the culture following multilayer Percoll gradient centrifugation at a late stage in the cycle prior to the gametocytogenesis assay. At day 0, the tightly synchronized ring-stage culture was adjusted to a 2% parasitemia in six-well plates without the addition of fresh RBCs during gametocytogenesis. The medium of the culture was changed every day by the addition of 5 ml of complete medium (ICM with 0.5% AlbuMax II) without disturbing the RBC layer at the bottom of the well. From Day 5 (G0), the cultures were treated with 50 mM N-acetyl-d-glucosamine (NAG) for 5 consecutive days to kill the asexual replicating forms after sexual commitment. Gametocyte production rate (%) was determined with reference to the gametocytemia at day 8 by dividing the parasitemia at day 3 determined by examination of Giemsa's solution-stained thin blood smears. Each experiment had three biological replicates and the experiment was repeated three times.
Quantitative reverse transcription PCR (qRT-PCR)
For qRT-PCR, total RNA was isolated from highly synchronous parasites with a combination of TRIzol (Invitrogen) treatment and using a Direct-zol RNA Kit (Zymo Research) as per the manufacturer's instructions. 1 μg total RNA was used to reverse transcribe and dye couple cDNA as described previously (Takara, No. 2641A). The RT-qPCR was performed in instrument (Takara, No. RR820A) on QuantStudio 7 Flex with an initial denaturing at 95°C for 30 s, followed by 40 cycles of 5 s at 95°C, 20 s at 54°C, 7 s at 56°C, 7 s at 59°C and 27 s at 62°C. The gene Seryl-tRNA synthetase (PF3D7_0717700) was used as endogenous control. Data were analysed using the ΔΔCt method (29). All qPCR experiments were performed for three biological replicates. All primers used for qPCR were listed in Supplementary Table S1.
Fluorescence assay
Live-cell fluorescence microscopy, immunofluorescence assays (IFAs) and flow cytometry were performed as described (29). For IFA, synchronized schizont-stage parasites were lysed with 0.15% saponin and fixed with 4% paraformaldehyde (Sigma) for 10 min, then blocked with 1% BSA (Sigma) in PBS (Gibco) and subsequently incubated with antibodies on microscope slides. pfap2-g-gfp::3D7 and pfap2-g-gfp:: pfap2-g5_trun were detected with primary antibody rabbit anti-GFP (Abcam, ab290), 1:1000. The secondary antibody (Alexa-Fluor-568-conjugated anti-mouse, 2 μg/ml, ThermoFisher Scientific) was diluted by 1:250. For live-cell fluorescence microscopy, parasites were Percoll-purified and centrifuged at 2000 rpm for 3 min. The pellet was resuspended in 1 ml pre-warmed 1% BSA (Sigma) in PBS (Gibco). The nuclei of parasites were stained by Hoechst (Invitrogen, H3570) (1:1000) and incubated for 15 min at 37°C in the dark, after which the cells were pelleted, washed and resuspended in 100 μl PBS. 5 μl sample was deposited on a microscope slide and covered with a coverslip. Images were taken using a Nikon A1R microscope at 100× magnification, acquired via NIS Elements software and processed using Adobe Photoshop CS5.
For flow cytometry, schizont-stage parasites of the 3D7 or PfAP2-G-GFP lines enriched by Percoll treatment were stained with Hoechst as for live-cell fluorescence microscopy, and subjected to flow cytometry analysis. About 20 000 cells were acquired (counting all events except debris) by gating on size (FSC and SSC) to draw gate 1. All cells in gate 1 were plotted on FSC-A versus FSC-H and gate 2 was drawn to exclude potential doublets and clumps. Cells in gate 2 were plotted on SSC versus V450 (Hoechst) and gate 3 was drawn to select infected cells. Infected cells were plotted on SSC versus GFP (FITC) and the gate was drawn on the basis of WT control (3D7 or NF54) and uninfected RBC population stained in the same way and analysed at the same time. Flow cytometry data were collected using the BD FACSVerseTM Flow Cytometer (Cat. No. 651155) and analysed using FlowJo 10.
Western blots
Synchronized schizont-stage parasites were isolated by treatment of infected RBC (iRBCs) with 0.15% saponin in PBS on ice for 10 min. Parasite pellets were washed by 1× PBS twice, then dissolved in 100 μl 1× SDS-loading buffer (Bio-Rad) and boiled at 100°C for 5 min. Total proteins were loaded onto SDS-PAGE gels with suitable amount according to the theoretical molecular weight of target proteins. Proteins were transferred to a PVDF membrane by semi-dry (Trans-Blot® SD Semi-Dry Electrophoretic Transfer Cell) for 2 h at room temperature or wet transfer (Trans-Blot® Cell) overnight at 4°C, and visualized by exposing with the Bio-Rad ChemiDoc XRS system. The antibody used in this study was mouse anti-GFP (Sigma), anti-HA (Roche) and rabbit anti-aldolase (Abcam). The experiment was done with ECL western blotting kit (GE healthcare).
RNA-seq library preparation and high throughput sequencing
RNA-seq analysis were performed as described (30). Briefly, parasites were synchronized twice to obtain asexual ring (10–15 hpi), trophozoite (25–30 hpi), schizont (40–45 hpi) at continuous timepoints during the process of gametocyte induction. Total RNA was isolated using TRIzol according to the manufacturer's protocol. Further library preparation for strand-specific RNA-seq was carried out by poly(A) selection with the KAPA mRNA Capture Beads (KAPA), and fragmented to about 300–400 nucleotides (nt) in length, then all subsequent steps were performed according to a KAPA Stranded mRNA-Seq Kit Illumina platform (KK8421) and sequenced on an Illumina HiSeq Xten system to generate 150 bp pair-end reads.
ChIP-seq and ChIP-qPCR
ChIP-seq assay was carried out as previously described with minor modifications (9,31). Synchronized parasites at different stages were harvested and cross-linked immediately with 1% paraformaldehyde (Sigma) by rotating for 10 min at 37°C, then quenched with 0.125 M glycine for 5 min. The culture was resuspended with 1× PBS and parasites were released from iRBC with 0.15% saponin for 10–15 min on ice. The washed nuclei were isolated by incubation with 2 ml of Lysis Buffer (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA pH8.0, 0.1 mM EGTA pH 8.0, 1 mM DTT, 0.25% NP40, 1× protease inhibitors cocktail) for 30 min on ice, and followed by dounce homogenization for 100 strokes. Afterwards the nuclei were resuspended in 200 μl of SDS Lysis Buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH8.0) for sonication to generate 200–500 bp fragments in length for 20 min at 5% duty factor, 200 cycles per burst, 75 W of peak incident power with Covaris M220 sonicator. The sonicated chromatin was diluted ten-fold with dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl pH8.0, 150 mM NaCl, 1× protease inhibitors cocktail) and precleared with Protein A/G magnetic beads (Pierce) for 2 h. The precleared chromatin supernatant was incubated with 1 μg rabbit anti-GFP (Abcam, ab290) or rat monoclonal anti-HA (Roche, 11867431001) and rabbit IgG (Abcam) as control or 3 μg anti-H3K9ac (Millipore, 07-352) and anti-H3K9me3 (Abcam, ab8898) and Protein A/G magnetic beads at 4°C overnight with a small volume of non-immunoprecipitated input material kept separately. The protein A/G agarose-antibody-chromatin complex was washed once for 5 min at 4°C under agitation with Low Salt Immune Complex Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.0, 150 mM NaCl), High Salt Immune Complex Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.0, 500 mM NaCl) and LiCl Immune Complex Wash Buffer (0.25 M LiCl, 1% NP-40, 1% deoxycolate, 1 mM EDTA, 10 mM Tris–HCl pH 8.0), then washed twice at room temperature with TE Buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA pH 8.0). Protein–DNA complex was eluted with 200 μl fresh Elution Buffer (1% SDS, 0.1 M NaHCO3). Crosslinking was reversed by incubating overnight at 45°C and DNA was treated with RNase A at 37°C for 30 min and Proteinase K at 45°C for 2 h.
ChIP-DNA was extracted using MinElute PCR purification kit (Qiagen, 28006), then used to prepare libraries or analyzed by qPCR. To prepare sequencing libraries, ChIP-DNA was end-repaired (Epicentre No. ER81050), adding protruding 3′ A base (NEB No. M0212L), adapter ligation (NEB No. M2200L), size selection and amplification of libraries (KAPA Biosystems, KB2500) with a PCR program: 1 min at 98°C, 12 cycles of 10 s at 98°C, 1 min at 65°C; finally extended 5 min at 65°C. Amplified libraries were sequenced on an Illumina HiSeq Xten system to generate 150 bp pair-end reads. For ChIP-qPCR, values of samples and control samples were presented as fold enrichment in the immunoprecipitated sample versus the negative control (a mock IP performed with non-immune IgG). Two biological replicates were used for each ChIP-seq experiment and three replicates for ChIP-qPCR. Those qPCR primers used were listed in Supplementary Table S1.
Strand-specific RNA-seq data analysis
Low-quality and adaptor sequences were trimmed from the reads using cutadapt (v1.16) with parameters: -a AGATCGGAAGAGC -AAGATCGGAAGAGC –trim-n -m 75 -q 20,20. Then, the reads were mapped to the P. falciparum 3D7 genome (Pf 3D7 v32, obtained from PlasmoDB) using Hisat2 strand-specific mode (v2.1.0) (32) with parameters: –rna-strandness RF –dta –no-discordant –no-mixed –no-unal. Samtools (v1.9) was used to filter low mapping quality reads and transfer the mapping results from sam format to position sorted bam format. After that, mapped reads were subsequently assembled into transcripts guided by the PlasmoDB gff annotation files (Pf 3D7 v32) using featureCounts (v1.6.1) with parameters: -M -p -B -C for all; -s 2 for sense transcripts; -s 1 for antisense transcripts. Finally, the expression levels of each gene were quantified and normalized as FPKM (fragments per kilobase of transcript sequence per million read pairs mapped) based on featureCounts raw outcome in R. Both sense and antisense read counts were merged for library normalization.
ChIP-seq data analysis
Low-quality and the adaptor sequences were trimmed from the reads using cutadapt (v1.16) with parameters: -a AGATCGGAAGAGC -A AGATCGGAAGAGC –trim-n -m 50 -q 20,20. Then, the reads were mapped to the P. falciparum 3D7 genome (Pf 3D7 v32, obtained from PlasmoDB) using Bowtie2 (v2.3.4.3) (33) with parameters: -N 0 –no-discordant –no-mixed –no-unal. Samtools (v1.9) was used to filter low mapping quality reads and transfer the mapping results from sam format to position sorted bam format. Next, the duplicated reads were removed by markdup from sambamba (v0.6.8). Then, the bam files were converted to bigwig files using bamCoverage from the deeptools suite (v3.1.3) (34) with parameters: –normalizeUsing RPKM –binSize 10 –smoothLength 30 -ignore Pf3D7_API_v3 Pf_M76611. The Integrative Genomics Viewer (IGV) was used to show the signal of ChIP-seq in certain genomic region in a track view.
Peak calling
MACS2 (v2.1.1) (35) was used to perform peak calling with the p-value cutoff of 0.01 and threshold cutoff 5-fold. IgG was used as control. The summit of peaks in intergenic region were used in further analysis using the BEDTools suite. The called summits of peaks were annotated to genes using annotatePeaks.pl from Homer (v4.10.1). Two biological replicates were used for each ChIP-seq experiment and subsequent analyses focused on genes bound by certain protein within 5 kb upstream in both replicates. For visualization, bam files were converted to bigwig files by MACS2 (v2.1.1) with the parameter –bdg. The bigwig files from ChIP sample were normalized to IgG sample by bigwigCompare from the deeptools suite (v3.1.3) and viewed using IGV.
Motif searching
Within 5 kb upstream to each gene, the summits with highest score were retained and the ±200 bp sequences surrounding these summits were trimmed for motif searching. Then MEME-ChIP suite (v5.1.1) (36) was used in motif enrichment analysis. In brief, DREME was used to identify enriched motifs between 5 and 10 bp in the trimmed sequences as compared to random P. falciparum genomic intervals of similar length. CentriMo was used to detect enrichment of previously characterized functional motifs for ApiAP2 family in the trimmed sequences. Finally, TOMTOM was used to compare the de novo discovered motifs to previously in silico discovered motifs; a match required a minimum overlap of four positions. Motif sequence logos were generated by MEME-ChIP suite.
RESULTS
CRISPR–Cas9-mediated gene disruption screen identifies the upstream regulators of pfap2-g
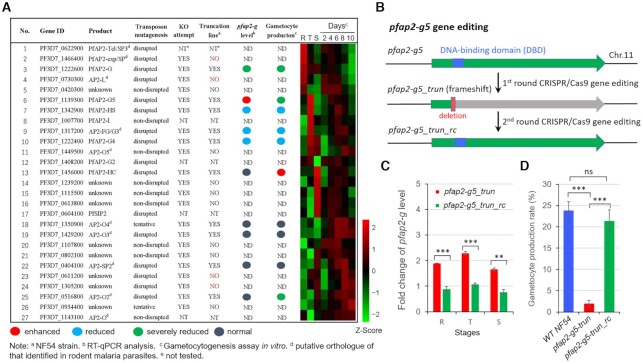
A total of twenty-two ApiAP2 genes were targeted for functional disruption via the CRISPR–Cas9 gene editing system in the P. falciparum NF54 strain. This approach enables a rapid preliminary screening of the effects of ApiAP2 proteins on the regulation of pfap2-g expression, though truncated products may be produced in some cases. After two to three rounds of independent transfection, we successfully obtained knockout lines for ten ApiAP2 genes (Figure 1A). Among these, four members were successfully disrupted by transposon mutagenesis previously (37). These conflicting results may be due to the failure of complete loss-of-function of target genes via piggyBac insertion in the 5′UTR or CDS, i.e. truncated proteins will be also produced as in the frameshift approach.
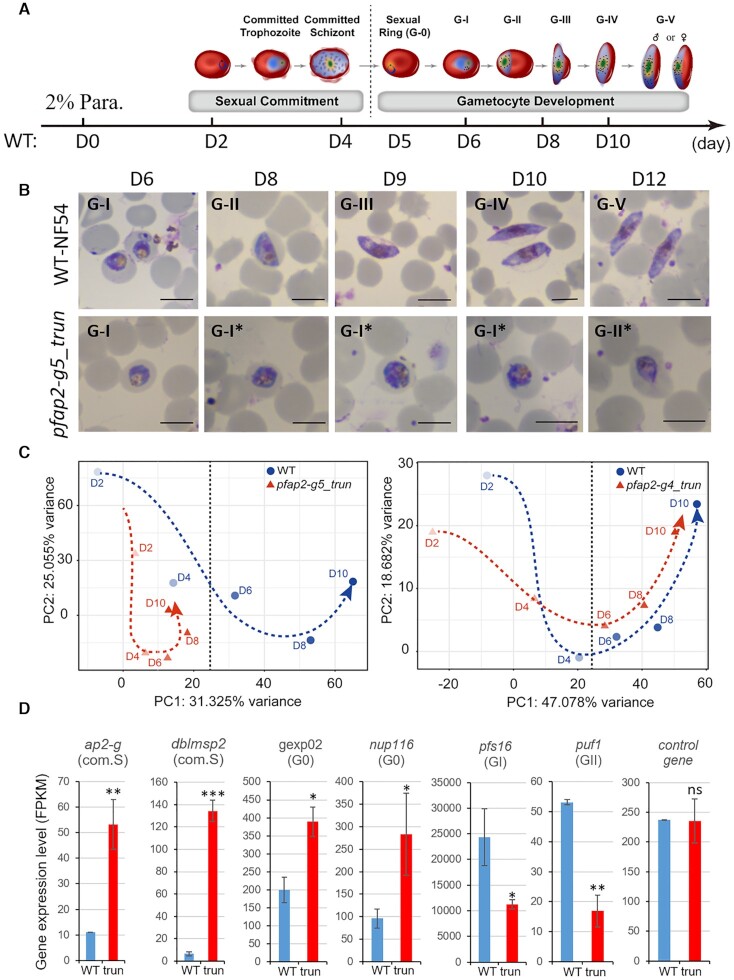
Figure 1.
Functional screening of ApiAP2 family for gametocyte production. (A) Phenotype summary of a CRISPR–Cas9 knockout screen of ApiAP2 genes in NF54 strains. The transcriptional abundancies of the pfap2-g gene were measured by RT-qPCR analysis for three biological replicates for each transgenic parasite line at 30–40 hpi. Gametocyte production rates were quantified by measuring the proportion of all gametocytes among infected RBCs, when mature forms were observed. Expression data of individual stages including asexual rings (R), trophozoites (T) and schizonts (S), and continuous timepoints over the course of gametocytogenesis (see Figure 5A) were measured by RNA-seq. RNA-seq data was normalized as Z-Scores. Conflicting results between transposon mutagenesis and frameshift-based KO approaches are indicated in red font, and the days on the top of the heatmap indicate the stages during gametocytogenesis. For detailed information, see Supplementary Table S2. (B) Schematic representation of the constructs of pfap2-g5_trun and pfap2-g5_trun_rc lines. The pfap2-g5_trun line was used a parental line for the 2nd transfection. (C) Fold-change in pfap2-g transcription of pfap2-g5_trun::NF54 or pfap2-g5_trun_rc::NF54 lines over WT NF54 line in ring (R), trophozoite (T), schizont (S). The data are from two replicates of RNA-seq analysis. (D) Gametocyte formation rate in NF54, pfap2-g5_trun::NF54 and pfap2-g5_trun_rc::NF54 line. The statistical analysis: ***P< 0.001, **P< 0.01, *P< 0.05, ‘ns’: not significant (unpaired two-tailed Student's t-test). The error bars represent s.e.m. for two replicates (C) or three replicates (D).
We next examined the influence of each ApiAP2 gene knockout on the mRNA abundance of pfap2-g and on gametocyte production in vitro. Quantitative RT-PCR (RT-qPCR) analysis showed that three ApiAP2 genes [PF3D7_1342900, PF3D7_1317200 (putative AP2-G3), PF3D7_1222400] positively correlated with pfap2-g transcription and gametocytogenesis, i.e. the knockout lines exhibited significantly lower levels of pfap2-g transcripts as well as reduced gametocyte production (Figure 1A and Supplementary Table S2).
Previously, single-cell RNA sequencing (scRNA-seq) has been used to investigate the dynamic transcriptome during sexual commitment in P. falciparum. Notably, two ApiAP2 genes, PF3D7_1139300 and Pf3D7_1222400 were shown to be transcribed sequentially following the transcription of pfap2-g, suggesting their involvement in sexual commitment (5). Consistent with this, our functional screen of the ApiAP2 family validated the positive regulatory role of Pf3D7_1222400 in gametocytogenesis (Figure 1A and Supplementary Figure S1). As two AP2-G-associated ApiAP2 factors (AP2-G2, AP2-G3) have been named in malaria parasites previously (4,12,13), we herein name this factor PfAP2-G4. Together with PfAP2-I, which is a regulatory partner of PfAP2-G (9), these PfAP2-G-associated ApiAP2 factors were predominantly transcribed in trophozoites, the stage at which cell fate is determined for differentiation into asexually replicating parasites or gametocytes (2). This suggests a potential ApiAP2-mediated regulatory network initiated when asexual forms commit to gametocytogenesis following environmental cues. In addition, the putative pfap2-o2 gene knockout displayed decreased gametocyte production, but with no change in pfap2-g gene expression, suggesting an unknown down-stream function of this factor on gametocytogenesis.
PfAP2-G5 is essential for asexual replication by preventing sexual commitment
Strikingly, one knockout line, PF3D7_1139300, which we herein name pfap2-g5, displayed a significant increase in pfap2-g mRNA abundance compared to the parental NF54 strain, but had a severe defect in gametocyte production (Figure 1A). Moreover, complementation of the pfap2-g5 gene by a second-round of gene editing, reversing the mutation, rescued the phenotype (Figure 1B–D and Supplementary Figure S2A and B). This unexpected phenotype in the pfap2-g5_trun line suggests that the function of PfAP2-G5 is not limited to the commitment stage, and also challenges the idea that the expression level of pfap2-g is positively associated with gametocyte production in malaria parasites (2,10).
We then generated an additional pfap2-g5 knockout line in the 3D7-G7 strain with a different genetic background to further validate this finding. This clone undergoes a very low-frequency of gametocytogenesis due to transcriptional silencing of the pfap2-g gene (25). We found that although the mRNA abundance of pfap2-g also increased by 2–5-fold in the resulting pfap2-g5_trun::3D7 transgenic parasite line asexual parasites (as observed in pfap2-g5_trun::NF54 line), the gametocyte production rate was significantly reduced (Supplementary Figure S2C and D). Western blot analysis validated the loss and regain of full-length PfAP2-G5 protein by generation of a GFP-tagging line with the WT NF54, KO and RC lines as parental parasites (Figure 3B and Supplementary Figure S3). These results for both pfap2-g5_trun lines indicate that PfAP2-G5 is required for not only gametocyte conversion but also gametocyte development.
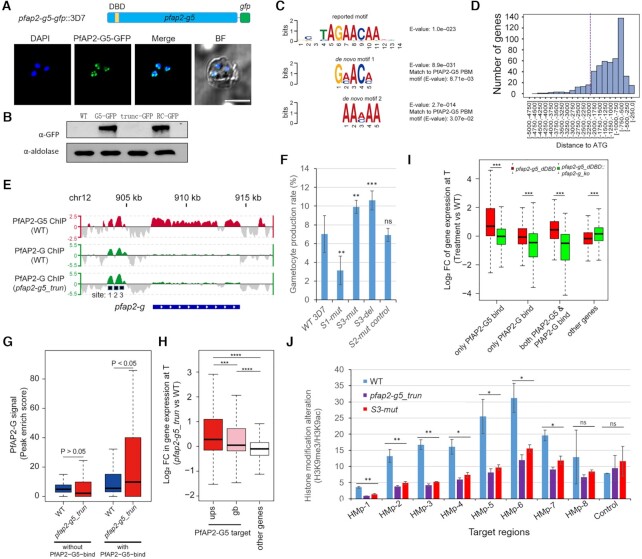
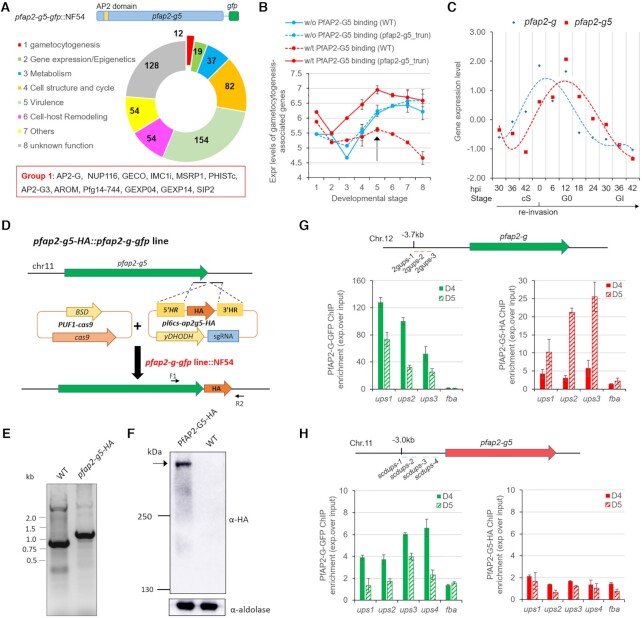
Figure 3.
PfAP2-G5 represses PfAP2-G expression via targeting the URR for maintenance of the integrity of local heterochromatin structure surrounding pfap2-g gene. (A) Upper: Schematic representation of the construct of pfap2-g5-gfp::3D7. Bottom: Live-cell fluorescence assay of PfAP2-G5-GFP (green). Nuclei are stained with DAPI (blue). Scale bar: 5 μm. (B) Western-blot analysis of pfap2-g5-gfp, pfap2-g5_trun-gfp, pfap2-g5_trun-rc-gfp lines at schizont stage. The Pfaldolase was used as an internal control. (C) DREME logos for the de novo motif enriched within trophozoite-stage PfAP2-G5 binding sites upstream of its target genes in vivo and the matching reported ApiAP2 binding motif (the top one) in vitro identified by Tomtom (39). (D) The distribution of PfAP2-G5-binding sites relative to the start codon of target genes. (E) Track view of the normalized signals (Log2-transformed ChIP/input ratio) at the pfap2-g gene locus for PfAP2-G5 in WT parasites (upper; red) and PfAP2-G in WT and pfap2-g5_trun parasites (bottom; green) in asexual trophozoites. One of the two biological replicates is shown. Three sites with mutations are indicated by black squares at the bottom. (F) Gametocyte production in parasites lines shown in the motif mutagenesis analysis (Supplementary Figure S10). (G) Box plots showing PfAP2-G binding signal (peak enrich score calculated from MACS2; higher score means higher enrichment) on the PfAP2-G5 target genes or non-target genes in WT and pfap2-g5_trun parasites in asexual trophozoites. The average values from two independent replicates are shown. P value: unpaired Wilcoxon rank test. (H) Box plots showing fold change (log2) in transcriptional levels of pfap2-g5_trun over WT parasites of target genes bound by PfAP2-G5 at upstream (ups) regions or gene body (gb) regions and non-target genes in asexual trophozoites by RNA-seq (the detailed transcriptomes are shown in Supplementary Table S5). The average values from two independent replicates are shown. P value: unpaired Wilcoxon rank test. (I) Box plots showing fold change (log2) in transcriptional levels of pfap2-g5_dDBD over WT parasites (red) or pfap2-g5_dDBD::pfap2-g_ko: over WT parasites of different groups of target genes, i.e. only PfAP2-G5-binding at upstream (ups) regions, only PfAP2-G-binding at upstream (ups) regions, both PfAP2-G5 and PfAP2-G-binding at upstream (ups) regions, and other non-binding genes for the two factors in asexual trophozoites by RNA-seq (the detailed transcriptomes are provided in Supplementary Table S9). The average values from two independent replicates were shown. P value: paired Wilcoxon rank test. (J) ChIP-qPCR analysis of histone modification changes (H3K9me3 over H3K9ac) at the pfap2-g gene locus in WT, pfap2-g5_trun, and S3-mut lines. The target regions for amplification are shown in Supplementary Figure S10G. Control gene: fba. In the (G-I): Boxes indicate the 25th to 75th percentiles. The horizontal lines within the boxes are the median values. The statistical analysis: ****P< 0.0001, ***P< 0.001, **P< 0.01, *P< 0.05, ns: not significant (unpaired two-tailed Student's t-test). The error bars represent s.e.m. for three replicates (F, J).
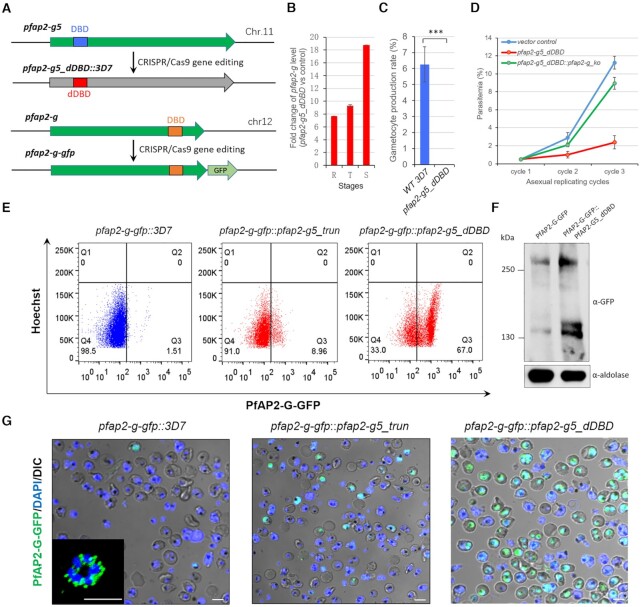
However, the fact that gametocyte production was not completely abolished in either line indicates that the frameshift-based knockout employed here may not completely disrupt the function of PfAP2-G5 due to the possible presence of truncated products. To investigate this further, we generated another loss-of-function line via deletion of the single AP2 DNA-binding domain (dDBD) within the coding region of PfAP2-G5 in 3D7 (here named pfap2-g5_dDBD) (Figure 2A and Supplementary Figures S4A and B). Surprisingly, while we observed a much higher expression of pfap2-g in this line, gametocyte production was completely abolished (Figure 2B and C). This finding was also confirmed in the NF54 line (Supplementary Figure S5).
Figure 2.
Loss of PfAP2-G5 promotes sexual commitment by activating pfap2-g gene but abolishes gametocyte production. (A) Schematic representation of pfap2-g5_dDBD line (upper) and pfap2-g-gfp line (bottom). DBD: DNA-binding domain. (B) Fold-change in pfap2-g transcription of pfap2-g5_dDBD over 3D7 line in asexual stages. The data are from RNA-seq analysis with two independent replicates. (C) Gametocyte formation rate in pfap2-g5_dDBD and 3D7 lines. (D) Growth rate analysis of pfap2-g5_dDBD, pfap2-g5_dDBD::pfap2-g_ko and vector control lines. (E) FACS assays of PfAP2-G-positive sub-population signal with late-stage parasites (30–40 hpi) from 3D7, pfap2-g5_trun, pfap2-g5_dDBD lines carrying PfAP2-G-GFP fusion protein. The detailed gating strategy is shown in Supplementary Figure S6. The nuclei are stained with Hoechst. (F) Western-blot analysis of PfAP2-G-GFP in pfap2-g-gfp and pfap2-g-gfp::pfap2-g5_dDBD lines by using anti-GFP antibody. The Pfaldolase was used as an internal control. (G) Live-cell fluorescence assays of PfAP2-G signal (green) with late stage parasites (30–40 hpi) from 3D7, pfap2-g5_trun, pfap2-g5_dDBD lines carrying PfAP2-G-GFP fusion protein. The nuclei are stained with DAPI (blue). Scale bar: 5 μm. The statistical analysis: ***P< 0.001, **P< 0.01, *P< 0.05, ns: not significant (unpaired two-tailed Student's t-test). The error bars represent s.e.m. for two biological replicates (B), or three replicates (C, D).
Moreover, an in vitro growth-rate assay revealed a serious growth defect in this line during the intraerythrocytic stages, whereas only moderate growth phenotypes were observed in the frameshift-based KO lines (Figure 2D and Supplementary Figures S2E and S2F). Meanwhile, whole genome sequencing analysis showed no nonsynonymous mutations in the CDS of known gametocytogenesis-associated genes (Supplementary Tables S3 and S4). These data confirm the essential role of PfAP2-G5 in maintenance of the asexual replication cycle via prevention of sexual commitment for human malaria parasites. In addition, we note that the frameshift KO approach may only produce a disrupted phenotype for ApiAP2 genes screened in this study (Figure 1A).
To further characterize the gametocyte null phenotype of pfap2-g5_dDBD parasites, we first generated a C-terminal GFP-tagged PfAP2-G protein in the WT 3D7 strain (pfap2-g-gfp::3D7), and performed a second round of CRISPR–Cas9 transfection to generate two lines of pfap2-g-gfp:: pfap2-g5_trun and pfap2-g-gfp:: pfap2-g5_dDBD) (Figure 2A and Supplementary Figure S4A–D). These GFP-tagged lines enabled us to evaluate the expression of PfAP2-G at the protein level upon the loss of PfAP2-G5. A flow cytometry (FACS) quantification assay showed that the proportion of PfAP2-G positive cells was dramatically enhanced in pfap2-g5_dDBD parasites (∼67%) compared to WT (∼1.5%) or pfap2-g5_trun line (∼9%) (Figure 2E and Supplementary Figure S6A). This result was confirmed by Western Blot and live-cell fluorescence analysis (Figure 2F and G).
PfAP2-G5 suppresses pfap2-g expression through binding to the upstream region
To investigate the underlying mechanism of PfAP2-G5-dependent suppression of pfap2-g gene expression in asexual replicating parasites, we profiled the binding sites of the two ApiAP2 transcription factors (PfAP2-G and PfAP2-G5) in the 3D7 genome of asexual stage parasites with PfAP2-G-GFP and PfAP2-G5-GFP transgenic lines (Figures 2F and 3A and B). Chromatin immunoprecipitation-sequencing (ChIP-seq) analysis with two highly reproducible replicates for PfAP2-G5 with trophozoite-stage PfAP2-G5-GFP parasites using the ChIP-grade commercial anti-GFP antibody (9) revealed that PfAP2-G5 binds to the upstream regions of 517 genes involved in different physiological processes, such as gametocytogenesis, pathogenesis, cell-host remodeling, etc (2,5,9,23,38) (Supplementary Figures S7A, C, S8A and Supplementary Table S5). De novo motif calling in PfAP2-G5-binding regions and motif distribution within the URR of pfap2-g gene suggested two putative motifs (GAACA or AACAA) matching the previously reported motif (AGAACAA) predicted by utilizing protein binding microarrays (PBMs) in vitro (39) (E-value = 8.71e–03 and 3.07e–02, respectively) (Figure 3C and Supplementary Figure S10B). Further examination revealed PfAP2-G5 mainly binds within –0.5 to –2.0 kb upstream the start codon (proximal upstream regulatory region (URR)) of the target genes (Figure 3D and Supplementary Figures S9A and B).
The overall mRNA abundancies of the 517 target genes increased significantly upon pfap2-g5 knockout (Supplementary Figures S7B, D, S8B and Supplementary Table S5). Intriguingly, the gametocytogenesis-associated group 1 genes exhibited an apparent upregulation compared to other target genes, suggesting a crucial role for PfAP2-G5 in regulating gametocyte production (Supplementary Table S4). In addition, the mRNA abundancies of some invasion and virulence genes also increased (Supplementary Figure S8B and Supplementary Table S5). This, together with the phenotypes of PfAP2-G5 in different parasite strains, strongly suggests that PfAP2-G5 is involved in transcriptional repression during gametocytogenesis.
Surprisingly, we observed that both PfAP2-G5 and PfAP2-G bind to the URR region (–2.0 to –3.5 kb) of pfap2-g and share a similar profile (Figure 3E). Three sub-regions (R1, R2, R3) were partitioned within the URR based on the ChIP-signal profiles and motif distribution of PfAP2-G and PfAP2-G5 (Supplementary Figure S10A). A motif search showed that multiple copies of either the PfAP2-G-binding motif (GTAC) or PfAP2-G5-binding motif 2 (AACAA or TTGTT) were present in the URR sequence of the pfap2-g gene, whereas only two PfAP2-G5-binding motif 1 copies were distributed within R1 or in the internal site between the R1 and R2 regions (Supplementary Figure S10A). Comparative ChIP-seq assays showed an altered pattern of the PfAP2-G binding in target genes including pfap2-g upon PfAP2-G5 KO (Figure 3E and G). These findings reveal a reciprocal co-regulation of PfAP2-G and PfAP2-G5 on the expression level of pfap2-g.
To explore how PfAP2-G5 and PfAP2-G co-ordinate in regulating the expression of pfap2-g, we conducted motif (AACAA or TTGTT) mutation in R1 (S1-mut) or R3 (S3-mut or S3-del) for PfAP2-G5 in the PfAP2-G5-GFP::3D7 line using the CRISPR–Cas9 system. A random mutation was introduced in R2 as a control (S2-mut-control) (Supplementary Figures S10A and S11). RT-qPCR analysis showed that pfap2-g gene expression is significantly increased upon either mutation or deletion of the R3 PfAP2-G5-binding motif, whereas mutation of the R1 PfAP2-G5-binding motif significantly reduced pfap2-g mRNA abundance (Supplementary Figure S10C). This indicates that the mutation of the PfAP2-G5-binding motif in R1 may have interfered with the localization of PfAP2-G at this site. Meanwhile, ChIP-qPCR showed PfAP2-G5 binding is almost completely ablated following mutation or deletion of R3 (Supplementary Figure S10D). As a control, we also conducted motif (GTAC) mutation in R1 (mut1) or R2 (mut2) for PfAP2-G (Supplementary Figure S10A an S11). Only the mut1 line produced clear phenotypes as described previously (9) (Supplementary Figures S10E and F). Together, these results confirm that PfAP2-G5 represses the pfap2-g gene through binding to the URR. We then investigated whether parasites bearing mutations or deletions in R3, which leads to pfap2-g activation, were unable to produce gametocytes as observed in the pfap2-g5_dDBD line. Strikingly, gametocytogenesis was enhanced in both the S3-mut or S3-del lines (Figure 3F). The seemingly opposite phenotypes observed following PfAP2-G5 knockout or binding disruption demonstrate that PfAP2-G5 is required for both sexual commitment and downstream gametocyte development.
Activation of pfap2-g accounts for the phenotypes of PfAP2-G5 KO parasites
Considering the potential interaction of PfAP2-G5 and PfAP2-G, we systematically assessed the overlapping ChIP target genes between the two transcriptional factors. A total of 126 genes were bound by both factors at the upstream regions in trophozoites, and most gametocytogenesis-associated genes were targeted (Supplementary Figure S12A and Supplementary Tables S6–S8). However, no direct physical interaction was observed in the Co-IP/WB analysis, though the fluorescent foci were closely associated in the nucleus as detected by IFA assay (Supplementary Figures S12D and E).
We next examined the triple overlapping ChIP target genes among PfAP2-G5, PfAP2-G and PfAP2-I since the coordination of gene regulation between PfAP2-G and PfAP2-I has been described recently (9). There are 28 genes co-regulated by the three ApiAp2 factors, including gametocytogenesis- and merozoite invasion-associated genes (Supplementary Figures S12B and C and Supplementary Table S8). This further supports the hypothesis that these merozoite invasion genes are involved in gametocytogenesis via unknown mechanisms (1,9).
We next evaluated the global effect of PfAP2-G5 KO on PfAP2-G enrichment at target gene loci. As observed in the case of pfap2-g, global PfAP2-G binding signals significantly increased in the pfap2-g5_trun line (Figure 3G). The mRNA abundances of PfAP2-G5-binding genes were also significantly elevated (Figure 3H and Supplementary Figure S9F and Supplementary Tables S5 and S11). These results raise the concern that the increased expression levels of PfAP2-G5 binding genes may be a result of either the removal of PfAP2-G5 on the target gene loci, or alternatively, the activation of pfap2-g. To explore this issue, we generated a dual gene KO line (pfap2-g5_dDBD::pfap2-g_ko) (Supplementary Figure S4E), and examined the differential gene expression pattern with RNA-seq data.
Interestingly, while the overall expression level of those genes only targeted by PfAP2-G5 were unchanged, other genes bound by PfAP2-G only or both PfAP2-G and PfAP2-G5 showed decreased expression levels compared with that of the WT line (Figure 3I and Supplementary Table S9). These data not only confirm that PfAP2-G is a transcriptional activator of its target genes, but also suggests that the de-repression of PfAP2-G5-binding genes is mainly a consequence of overexpression of pfap2-g upon PfAP2-G5 KO.
We then examined the growth rate of this dual gene KO line. Surprisingly, to a great extent, the severe growth defect of pfap2-g5_dDBD parasites was rescued when the pfap2-g gene was also disrupted (Figure 2D). Finally, the PfAP2-G-GFP-positive subpopulation of the pfap2-g5-dDBD line isolated by FACS-sorting failed into asexual replicating cycles (Supplementary Figure S6B and C). These data confirm that the activation of PfAP2-G promotes sexual commitment in either NCC or SCC pathways (7), thereby interfering with asexual replication.
PfAP2-G5 represses gene expression in heterochromatic genes via chromatin alteration
It is known that heterochromatin levels control the transcriptional activity of pfap2-g (14–16). ChIP-seq data revealed that PfAP2-G5 binds to the exonic gene body of target genes including pfap2-g (Figure 3E and Supplementary Figures S9C and D). This raises the possibility that PfAP2-G5 may affect gene expression, including that of pfap2-g, through interaction with the local heterochromatin structure. To assess this possibility, we then investigated whether activation of pfap2-g expression following removal of PfAP2-G5 from the URR is also linked to local chromatin alteration at this locus. We performed ChIP-qPCR analysis for two representative histone markers, H3K9me3 and H3K9ac, with trophozoite-stage parasites from pfap2-g5_trun, S3-mut or WT line, respectively. Intriguingly, we observed an alteration of heterochromatin modification (H3K9me3) surrounding the upstream and coding regions of pfap2-g gene (Figure 3J and Supplementary S10G).
Previous studies have uncovered several upstream positive (HP1 (14) or Hda2) and negative (GDV1) regulators of heterochromatin structure (14–16). Here, no change in HP1 or Hda2 expression level was observed in the pfap2-g5_trun line, whereas GDV1 expression levels were dramatically reduced (Supplementary Figure S13A). This suggests that PfAP2-G5 is critical for the maintenance of the heterochromatin microenvironment surrounding the pfap2-g locus via a mechanism independent of HP1 and Hda2. More importantly, this factor is likely able to antagonize the effect of decreased level of GDV1. We note, however, that the observed local chromatin remodeling in the two pfap2-g-deficent lines may be compromised due to the death of sexually committed parasites. An opposing effect of chromatin alteration (heterochromatinization) on the pfap2-g gene locus was observed in the pfap2-g4_trun line (Supplementary Figure S13B), which is consistent with the reduced expression of PfAP2-G in this line (Supplementary Figure S1G).
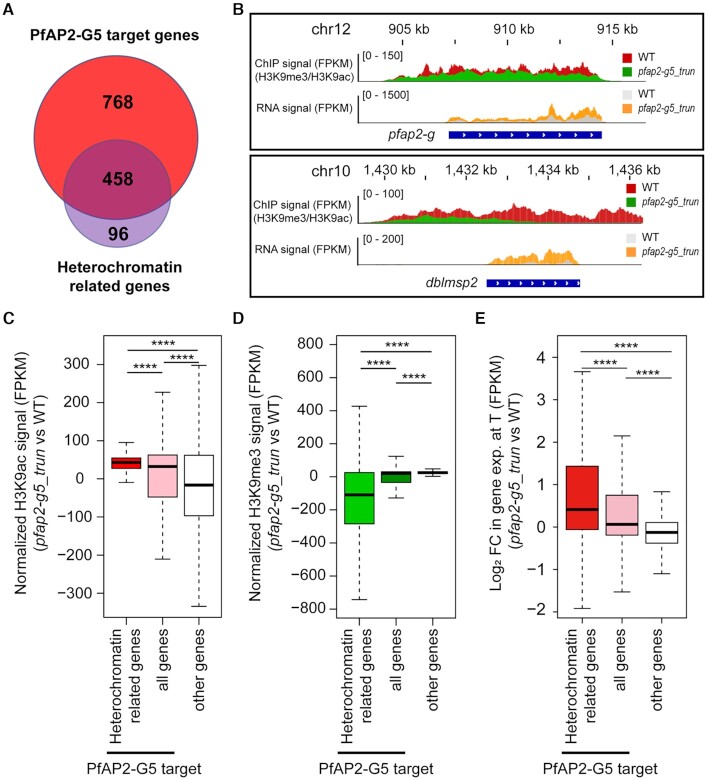
The above result suggests that PfAP2-G5 may regulate heterochromatic gene expression in genes other than pfap2-g (Supplementary Table S10). In fact, 458 out of 554 heterochromatic genes are also bound by PfAP2-G5 mostly at the exonic gene bodies (Figure 4A and Supplementary Figure S9E and Supplementary Table S11). The loss of PfAP2-G5 induced chromatin alteration toward euchromatic structure, i.e. switching from H3K9me3 to H3K9ac, which is favorable for heterochromatic gene expression (Figure 4B–E). Of note, the effect of gene dysregulation for those exonic gene body-binding (gb) targets was relatively weaker than that of upstream-binding (ups) targets (Figure 3H and Supplementary Figure S9F). Together, these finding suggest a complementary mechanism of PfAP2-G5-mediated gametocytogenesis control.
Figure 4.
PfAP2-G5 regulates heterochromatic genes via chromatin alteration. (A) Venn diagram showing overlap between target genes bound by PfAP2-G5 in asexual trophozoites and heterochromatic genes (Supplementary Tables S5, S10 and S11). (B) Track view of the histone modification signal (H3K9me3/H3K9ac ratio) and in transcriptional levels (FPKM) in WT or pfap2-g5_trun parasites at the pfap2-g gene locus (upper) and dblmsp2 gene locus (bottom) in asexual trophozoites. (C, D) Box plots showing the change of H3K9ac (C) and H3K9me3 (D) of pfap2-g5_trun subtract WT parasites of heterochromatin related genes bound by PfAP2-G5, all target genes bound by PfAP2-G5 and other non-target genes in asexual trophozoites by ChIP-seq. The average values from two independent replicates were shown. P value: unpaired Wilcoxon rank test. (E) Box plots showing fold change (log2) in transcriptional levels of pfap2-g5_trun over WT parasites of heterochromatin related genes bound by PfAP2-G5, all target genes bound by PfAP2-G5 and other non-target genes in asexual trophozoites by RNA-seq. The average values from two independent replicates are shown. P value: unpaired Wilcoxon rank test. In the (C–E) Boxes indicate the 25th to 75th percentiles. The horizontal lines within the boxes are the median values. The statistical analysis: ****P< 0.0001, ***P< 0.001, **P< 0.01, *P< 0.05, ns: not significant.
PfAP2-G5 is essential for gametocyte maturation through transcriptional repression of early gametocyte genes
It is important to know at which developmental stage the PfAP2-G5 KO parasites have been arrested during gametocyte production (Figure 5A). Examination of parasite morphology via Giemsa's solution-stained thin-smear microscopy suggests that knockout of PfAP2-G5 leads to developmental defects in early gametocytes likely at the GI stages, though a few GII-like parasites were also observed (Figure 5B and Supplementary Figure S14). In order to understand the mechanism underlying the defective development of gametocytes in PfAP2-G5 KO parasites, we profiled the dynamic transcriptome of the pfap2-g5_trun::NF54 line over the course of gametocytogenesis with WT NF54 control as described previously (8) (Supplementary Figure S15A). In P. falciparum, gametocytogenesis comprises two sequential processes: sexual commitment (D2-D4) and gametocyte development (D4-D12) as shown in Figure 5A. The second process is composed of two stages: early gametocyte development (D4-D6) and gametocyte maturation (D6–D12). Specifically, the developmental stage at D4 corresponds to committed schizonts, and that of D5 approximately corresponds to sexual rings (G0). Principle component analysis (PCA) of gene expression revealed the trajectory from sexual commitment to gametocyte maturation in WT parasites. In contrast, knockout of the pfap2-g5 gene led to arrest at a time point between sexual commitment (D4) and early gametocyte development (D6). For the pfap2-g4_trun line, however, a similar pattern was observed to that observed in the WT NF54 line during gametocyte development in spite of considerable differences in the putative ‘commitment cycle’ (Figure 5C).
Figure 5.
PfAP2-G5 is essential for the development of early-stage gametocytes. (A) A schematic model of the continuous developmental stages of parasites from sexual commitment to mature gametocytes. The transition from sexual commitment (committed schizont) to developmental stage (sexual ring) is indicated by a dashed vertical line. The sampling time points (day) for transcriptome analysis of WT line are indicated at the bottom. Abbreviation indicate parasitemia (Para.). Parasite drawings were plotted with Adobe Illustrator software commercially licensed to Tongji University. (B) Representative images of Giemsa's solution stained thin smears of WT NF54 and pfap2-g5_trun parasites during gametocytogenesis in vitro (D6–D12). The asexual forms were killed by NAG treatment for 5 consecutive days from G0 stage (D5). Scale bar: 5 μm. *: dying parasites with abnormal morphology. More images are also shown in Supplementary Figure S12. (C) PCA analysis showing transcriptomic profiles of pfap2-g5_trun::NF54 versus NF54 and pfap2-g4_trun::NF54 versus NF54 parasite lines over the course of gametocytogenesis measured by RNA-seq. The KO lines are shown with dashed red lines and WT with blue lines. Dashed black vertical lines indicate the transition from committed schizonts and sexual rings. (D) Gene expression levels of some representative gametocytogenesis-associated marker genes in WT and pfap2-g5_trun lines detected by RNA-seq. Control: seryl-tRNA synthetase (PF3D7_0717700). ***P< 0.001, **P< 0.01, *P< 0.05, ns: not significant (unpaired two-tailed Student's t-test). The error bars represent s.e.m. for two biological replicates of RNA-seq anlaysis.
We then attempted to interrogate this phenomenon at the gene level by categorizing putative gametocytogenesis-associated genes into five classes based on the peaks of their expression over the course of sexual commitment to gametocyte development (see the full gene list in Supplementary Table S4), i.e., early commitment, committed schizont (e.g. pfap2-g and dblmsp2), sexual ring (e.g. gexp02 and nup116), GI (e.g. Pfs16) and GII (e.g. Puf1) (2,5,9,10,16). In general, these classes of genes match to the continuous developmental stages of gametocytes during gametocytogenesis, i.e. cS (early or middle commitment stages, class 1), G0 (class 2), GI (class 3), GII (class 4) and GIII-V (class 5). Dynamic transcriptome analysis of the pfap2-g5_trun line revealed a disrupted pattern of gene expression for GIII-V genes, corresponding to the stages after sexual rings (Figure 5D and Supplementary Figures S15B and S15C and Supplementary Table S4). Therefore, we suggest that PfAP2-G5-mediated rapid suppression of these early gametocyte genes at sexual ring stages is critical for the completion of gametocyte development. Taken together, we demonstrate that PfAP2-G5 is not only critical for prevention of sexual commitment in asexual replicating cycles, but is also essential for the development of early gametocytes.
PfAP2-G5 and PfAP2-G reciprocally co-regulate early gametocyte-associated genes
In order to investigate the mechanistic role of PfAP2-G5 in regulating early gametocyte development following sexual commitment, we attempted to identify PfAP2-G5 target genes using ChIP-seq analysis with PfAP2-G5-GFP parasites harvested at D5 (corresponding approximately to G0) (Figure 5A). There are 540 potential PfAP2-G5 target genes with binding peaks in their upstream regions at this stage, and 314 target genes with signals at the exonic gene bodies (Figure 6A and Supplementary Figure S16A, Supplementary Tables S12 and S13). Strikingly, of the eight groups of PfAP2-G5 target genes, only Group 1, which are involved in gametocytogenesis, exhibited much higher mRNA abundancies during the stages from sexual commitment and early gametocyte development in pfap2-g5_trun parasites (Supplementary Figure S16B). Recent studies have revealed that AP2-G activates a group of genes during sexual commitment, and it has been hypothesized that they are associated with early gametocyte development (5,8–10). Importantly, the pfap2-g gene and its target genes are rapidly down-regulated following sexual commitment by an unknown mechanism.
Figure 6.
Reciprocal regulation between PfAP2-G5 and PfAP2-G is involved in the dynamic expression of EG genes at the early gametocyte stages. (A) Upper: schematic representation of the construct of pfap2-g5-gfp::NF54. Middle: functional categorization of the 540 PfAP2-G5-bound genes at the time point D5 approximately corresponding to G0. Gene numbers are shown in the circular graph. Bottom: gene list of the gametocytogenesis group in the red rectangle. The full list of PfAP2-G5 target genes is shown in Supplementary Table S12. (B) Transcriptional dynamics of all reported gametocytogenesis-associated genes in pfap2-g5_trun and WT NF54 lines measured by RNA-seq. These genes are classified into two groups bound by PfAP2-G5 (red lines) or not (blue lines) in their upstream regions. The arrow indicates the timepoint when PfAP2-G5-bound gametocytogenesis genes are precipitously down-regulated. ***P < 0.001, unpaired Wilcoxon rank test. ns: not significant. (C) Transcriptional profiles of pfap2-g (blue line) and pfap2-g5 (red line) genes across the sexual commitment and early gametocyte development from the microarray data of Josling et al. (9). The timepoints corresponding to the stage of committed schizonts (cS), sexual rings (G0) and gametocyte I (GI) are indicated below the axis x. (D) Schematic representation of the generation of pfap2-g5-HA::pfap2-g-gfp line in the NF54 line. The location of individual PCR primer for gDNA PCR validation are indicated in this gene locus (primer sequences are listed in Supplementary Table S1). (E) PCR validation of the HA-fusion genes in pfap2-g-gfp and WT lines. (F) Western-blot analysis of PfAP2-G5-HA in pfap2-g-gfp line by using anti-HA antibody. The Pfaldolase was used as an internal control. The full-length PfAP2-G5-HA protein is indicated by an arrow. (G, H) ChIP-qPCR analysis of PfAP2-G (anti-GFP) and PfAP2-G5 (anti-HA) for the binding levels within the upstream region of pfap2-g or pfap2-g5 gene. For pfap2-g gene, only the URR region was measured as shown in Supplementary Figure S10. The samples were harvested in D4 or D5 (see Figure 5A) for PfAP2-G5-HA::PfAP2-G-GFP::NF54 line. The fba (PF3D7_1444800) gene was used as negative control. The error bars represent s.e.m. for two biological replicates of RNA-seq analysis (B) or three replicates of ChIP-qPCR (G, H).
We then profiled the transcription dynamics of all reported gametocytogenesis-associated genes to which PfAP2-G5 is predicted to bind or not, in both the WT and pfap2-g5_trun parasite lines. Strikingly, gametocytogenesis-associated genes with PfAP2-G5 binding, most of which are early gametocyte (EG) genes, were significantly down-regulated in WT parasites from D4 onwards and these genes were released from suppression in the pfap2-g5_trun line. This suggests that repression of early gametocyte genes is required for gametocyte development. In contrast, those genes to which PfAP2-G5 does not bind remained highly expressed following D4 in both WT and pfap2-g5_trun lines (Figure 6B). Furthermore, 6-h interval transcription profiles of the development of sexual-stage-committed schizonts to sexual rings (9) revealed that both pfap2-g and pfap2-g5 share a similar expression pattern. Of note, pfap2-g reached maximal transcription on D4 (the endpoint of sexual commitment) whereas pfap2-g5 was maximally transcribed at the G0 stage (Figure 6C). Taken together, these results show that low expression of pfap2-g5 at the precommitment stage activates pfap2-g gene expression and this facilitates sexual commitment. Increasing expression of pfap2-g5 after sexual commitment is required for the maturation of sexually committed forms to gametocytes (Supplementary Figure S2G).
In addition, the sequential expression peaks of PfAP2-G and PfAP2-G5 shown in Figure 5C are consistent with the possibility that PfAP2-G activates pfap2-g5 to repress pfap2-g itself after EG genes are activated. To address the potential reciprocal regulation of these genes during sexual commitment and early gametocyte development, we tagged PfAP2-G5 endogenously with HA in the pfap2-g-gfp line (Figure 6D–F), and then performed ChIP-qPCR analyses for both proteins in parallel from the same parasite extracts. This enabled profiling of the dynamic binding of the two regulators on the upstream regions of pfap2-g and pfap2-g5 in committed schizonts (D4) or sexual rings (D5), respectively. Surprisingly, PfAP2-G binds to the upstream region of pfap2-g5 and pfap2-g at the committed schizont stage, but binding of PfAP2-G5 to pfap2-g was observed in sexual rings. No significant autoregulation of the pfap2-g5 gene was observed, though there was a minor enrichment of PfAP2-G5 on its own promoter (Figure 6G and H). Consistent with this finding, motif mapping analysis revealed that a cluster of PfAP2-G-binding motifs were present in the promoter region of pfap2-g5, in addition to the presence of a small number of scattered PfAP2-G5-binding motifs (Supplementary Figure S16C). These data confirm the co-regulation of PfAP2-G and PfAP2-G5 for early gametocyte development. Moreover, based on the ChIP-seq data, we speculate that PfAP2-G5 is also involved in the direct repression of early gametocyte genes activated by PfAP2-G. The overall ChIP signals of PfAP2-G5 on early gametocyte genes were significantly stronger than that of those genes involved in gametocyte maturation (Supplementary Figure S16D).
DISCUSSION
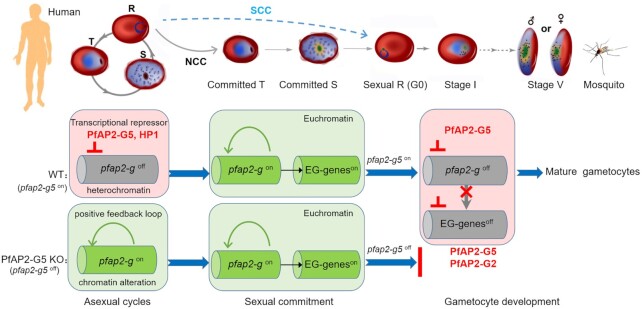
The present study uncovers a cascade of multiple ApiAP2 transcription factors involved in gametocytogenesis in malaria parasites. While many studies have reported that translational repression is a key mechanism underlying sexual differentiation and development in eukaryotes including protozoan Plasmodium (40–42), here we describe an entirely novel repression pathway at the transcriptional level that controls both sexual commitment and development in human malaria parasites. As a repressive transcription factor, PfAP2-G5 suppresses the expression of PfAP2-G via direct binding to its promoter or occupies the exonic gene body for maintenance of the integrity of the local heterochromatin environment surrounding the pfap2-g locus in asexual replicating parasites, thereby preventing the initiation of sexual commitment. When this repressive effect is removed, local chromatin alteration occurs at the pfap2-g gene locus, which activates pfap2-g gene expression and recruits PfAP2-G to the URR region. This positive autoregulation mechanism then promotes PfAP2-G-positive cells to develop to sexual commitment. At the late stage of the sexual commitment cycle, PfAP2-G activates a set of EG genes involved in early gametocyte development, and upregulates pfap2-g5. Subsequently, the expression of pfap2-g and this set of EG genes appear to be down-regulated by PfAP2-G5 in sexual rings allowing the maturation of gametocytes (Figure 7). Such reciprocal regulation along with the sequential developmental stages between the two ApiAp2 factors (and possibly additional factors including AP2-I and AP2-G2 (43) (44) represents a precisely programmed cascade for the regulation of gametocyte production in P. falciparum (Supplementary Table S8). Systematic comparative ChIP-seq and RNA-seq analyses confirm the co-regulation of some genes by the two physically interacting ApiAp2 factors (Supplementary Figure S17 and Table S14).
Figure 7.
A model of how PfAP2-G5 programs the regulation cascade for gametocytogenesis in P. falciparum parasites. In the asexual cycle, PfAP2-G5 suppresses the pfap2-g gene to prevent the initiation of sexual commitment (left) via maintenance of the HP1-dependent heterochromatin structure at the pfap2-g gene locus. When it is removed from the upstream regulatory region of pfap2-g, sexual commitment is triggered by either NCC or SCC pathway upon pfap2-g gene activation, which is accompanied with local chromatin alteration. Subsequently, pfap2-g activates a set of early gametocyte (EG) genes (middle), but their transcription activities including pfap2-g itself will be suppressed by PfAP2-G5 during early gametocyte development, or parasite development is arrested (right).
The mechanism underlying the mediation of the dynamic distribution of PfAP2-G5 in the URR of pfap2-g in response to environmental cues remains elusive. In rodent malaria parasites, the ortholog of PfAP2-G5 has not been functionally investigated due to failure of a CRISPR–Cas9-mediated knockout attempt (11). Interestingly, another rodent Plasmodium ApiAp2 protein, PbAP2-G2 (4), is also involved in early gametocyte development via a distinct mechanism; it represses genes associated with asexual replication via direct binding to their promoter regions, thus facilitating sexual differentiation (11). A recent report shows that the PfAP2-G2 is also involved in gametocyte maturation though repression of early gametocyte gene transcription. This finding indicates that PfAP2-G2 is another downstream regulator of PfAP2-G. Co-IP/MS analysis revealed a potential interaction between PfAP2-G5 and PfAP2-G2, suggesting an essential role for the transcriptional repression complex of early gametocyte genes for gametocyte maturation (43). In the future, more experimental evidence is required to confirm the necessity of EG gene repression for gametocyte development.
It is of note that there are two pathways underlying the conversion from asexual replicating forms to sexual rings (G0), i.e. next cycle conversion (NCC) and same cycle conversion (SCC). The latter has been relatively neglected following the identification of PfAP2-G as a ‘master regulator’ of gametocytogenesis (4,9), but we now recognize it as an important complementary mechanism to the classic NCC pathway (7). PfAP2-G-positive late-stage parasites will directly develop into sexually committed rings without the sexual commitment cycle. Though it is unclear as to the exact contribution of the SCC pathway to gametocytogenesis, both pathways are dependent on the presence of PfAP2-G5. The accumulation of PfAP2-G upon the removal of PfAP2-G5 from pfap2-g gene locus may trigger either NCC or SCC for the production of sexual rings. It is likely that both will be arrested at the early gametocyte stage due to the absence of the PfAP2-G5/G2-mediated transcriptional repression of early gametocyte genes (Figure 7). However, more experiments are needed to validate the involvement of PfAP2-G5 in the SCC pathway.
In other eukaryotes, this mechanism is sometimes utilized to regulate non-coding (45) or protein-coding gene expression, e.g., direct promoter repression by BCL11A controls the developmental transition between fetal and adult hemoglobin in humans (46). These findings reveal complex roles of transcription factors in regulating biological processes. In conclusion, our study provides a simplified control mechanism governing the entire process of gametocytogenesis which expands our understanding of the complex mechanism of transmission in malaria parasites (2,47).
DATA AVAILABILITY
All data is available in the main text or the supplementary materials. The raw and processed high-throughput sequencing data of this study have been deposited in Gene Expression Omnibus (GEO) database under accession number GSE149774. The raw data of Flow Cytometry experiments of this study have been deposited in FlowRepository with Repository ID, FR-FCM-Z47H.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Scherf (Institut Pasteur, Paris) for technical and material support; A. Cowman (The University of Melbourne) and M. Jacobs-Lorena (Johns Hopkins Malaria Research Institute) for critical comments of this manuscript.
Author contributions: Q.Z., J.C., L.G. and S.S. conceived and designed the experiments. X.S., C.W., Y.Z., X.H. and M.Z. generated all gene knock-out and knock-in transgenic parasite lines and performed ChIP-seq, RNA-seq, fluorescence assay. J.T., X.H. and G.G performed gametocytogenesis assay and prepared parasite samples for RNA-seq and ChIP-seq. S.S., M.L., L.W., Q.Z., G.Y., J.H. and C.J. performed all the bioinformatics analysis. Q.Z. and R.C wrote the manuscript with contribution from L.J., J.C., L.G., X.S., S.S. and X.Y.
Contributor Information
Xiaomin Shang, Laboratory of Molecular Parasitology, Research Center for Translational Medicine, Key Laboratory of Arrhythmias of the Ministry of Education of China, East Hospital, School of Medicine, Tongji University, Shanghai 200120, China.
Shijun Shen, Key Laboratory of Spine and Spinal Cord Injury Repair and Regeneration of Ministry of Education, Orthopaedic Department of Tongji Hospital, Shanghai Key Laboratory of Signaling and Disease Research, the School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Jianxia Tang, National Health Commission Key Laboratory of Parasitic Disease Control and Prevention, Jiangsu Provincial Key Laboratory on Parasite and Vector Control Technology, Jiangsu Institute of Parasitic Diseases, Wuxi 214064, China.
Xiaoqin He, National Health Commission Key Laboratory of Parasitic Disease Control and Prevention, Jiangsu Provincial Key Laboratory on Parasite and Vector Control Technology, Jiangsu Institute of Parasitic Diseases, Wuxi 214064, China.
Yuemeng Zhao, Laboratory of Molecular Parasitology, Research Center for Translational Medicine, Key Laboratory of Arrhythmias of the Ministry of Education of China, East Hospital, School of Medicine, Tongji University, Shanghai 200120, China; Unit of Human Parasite Molecular and Cell Biology, Key Laboratory of Molecular Virology and Immunology, Institut Pasteur of Shanghai, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Changhong Wang, Laboratory of Molecular Parasitology, Research Center for Translational Medicine, Key Laboratory of Arrhythmias of the Ministry of Education of China, East Hospital, School of Medicine, Tongji University, Shanghai 200120, China.
Xiaohui He, Laboratory of Molecular Parasitology, Research Center for Translational Medicine, Key Laboratory of Arrhythmias of the Ministry of Education of China, East Hospital, School of Medicine, Tongji University, Shanghai 200120, China.
Gangqiang Guo, Laboratory of Molecular Parasitology, Research Center for Translational Medicine, Key Laboratory of Arrhythmias of the Ministry of Education of China, East Hospital, School of Medicine, Tongji University, Shanghai 200120, China; Key Laboratory of Spine and Spinal Cord Injury Repair and Regeneration of Ministry of Education, Orthopaedic Department of Tongji Hospital, Shanghai Key Laboratory of Signaling and Disease Research, the School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Meng Liu, Key Laboratory of Spine and Spinal Cord Injury Repair and Regeneration of Ministry of Education, Orthopaedic Department of Tongji Hospital, Shanghai Key Laboratory of Signaling and Disease Research, the School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Liping Wang, Key Laboratory of Spine and Spinal Cord Injury Repair and Regeneration of Ministry of Education, Orthopaedic Department of Tongji Hospital, Shanghai Key Laboratory of Signaling and Disease Research, the School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Qianshu Zhu, Key Laboratory of Spine and Spinal Cord Injury Repair and Regeneration of Ministry of Education, Orthopaedic Department of Tongji Hospital, Shanghai Key Laboratory of Signaling and Disease Research, the School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Guang Yang, Key Laboratory of Spine and Spinal Cord Injury Repair and Regeneration of Ministry of Education, Orthopaedic Department of Tongji Hospital, Shanghai Key Laboratory of Signaling and Disease Research, the School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Cizhong Jiang, Key Laboratory of Spine and Spinal Cord Injury Repair and Regeneration of Ministry of Education, Orthopaedic Department of Tongji Hospital, Shanghai Key Laboratory of Signaling and Disease Research, the School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Meihua Zhang, National Health Commission Key Laboratory of Parasitic Disease Control and Prevention, Jiangsu Provincial Key Laboratory on Parasite and Vector Control Technology, Jiangsu Institute of Parasitic Diseases, Wuxi 214064, China.
Xinyu Yu, National Health Commission Key Laboratory of Parasitic Disease Control and Prevention, Jiangsu Provincial Key Laboratory on Parasite and Vector Control Technology, Jiangsu Institute of Parasitic Diseases, Wuxi 214064, China.
Jiping Han, Unit of Human Parasite Molecular and Cell Biology, Key Laboratory of Molecular Virology and Immunology, Institut Pasteur of Shanghai, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Richard Culleton, Division of Molecular Parasitology, Proteo-Science Centre, Ehime University, Matsuyama, Ehime 790-8577, Japan; Department of Protozoology, Institute of Tropical Medicine, Nagasaki University, Nagasaki, Japan.
Lubin Jiang, Unit of Human Parasite Molecular and Cell Biology, Key Laboratory of Molecular Virology and Immunology, Institut Pasteur of Shanghai, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
Jun Cao, National Health Commission Key Laboratory of Parasitic Disease Control and Prevention, Jiangsu Provincial Key Laboratory on Parasite and Vector Control Technology, Jiangsu Institute of Parasitic Diseases, Wuxi 214064, China; Center for Global Health, School of Public Health, Nanjing Medical University, Nanjing 211166, China; Public Health Research Center, Jiangnan University, Wuxi 214122, China.
Liang Gu, Key Laboratory of Spine and Spinal Cord Injury Repair and Regeneration of Ministry of Education, Orthopaedic Department of Tongji Hospital, Shanghai Key Laboratory of Signaling and Disease Research, the School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Qingfeng Zhang, Laboratory of Molecular Parasitology, Research Center for Translational Medicine, Key Laboratory of Arrhythmias of the Ministry of Education of China, East Hospital, School of Medicine, Tongji University, Shanghai 200120, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2018YFA0507300]; National Natural Science Foundation of China (NSFC) [81630063, 81971959, 31771419, 31721003, 81971967]; National Key R&D Program of China [2020YFC1200105]; Jiangsu Provincial Project of Invigorating Health Care through Science, Technology and Education and the Jiangsu Provincial Commission of Health. Funding for open access charge: National Key R&D Program of China [2020YFC1200105].
Conflict of interest statement. None declared.
REFERENCES
- 1.Josling G.A., Llinás M.. Sexual development in Plasmodium parasites: knowing when it's time to commit. Nat. Rev. Microbiol. 2015; 13:573–587. [DOI] [PubMed] [Google Scholar]
- 2.Josling G.A., Williamson K.C., Llinás M.. Regulation of sexual commitment and gametocytogenesis in malaria parasites. Annu. Rev. Microbiol. 2018; 72:501–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kafsack B.F.C., Rovira-Graells N., Clark T.G., Bancells C., Crowley V.M., Campino S.G., Williams A.E., Drought L.G., Kwiatkowski D.P., Baker D.A.et al.. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature. 2014; 507:248–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinha A., Hughes K.R., Modrzynska K.K., Otto T.D., Pfander C., Dickens N.J., Religa A.A., Bushell E., Graham A.L., Cameron R.et al.. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature. 2014; 507:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poran A., Notzel C., Aly O., Mencia-Trinchant N., Harris C.T., Guzman M.L., Hassane D.C., Lemento O.E., Kafsack B.F.C.. Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature. 2017; 551:95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kent R.S., Modrzynska K.K., Cameron R., Philip N., Billker O., Waters A.P.. Inducible developmental reprogramming redefines commitment to sexual development in the malaria parasite Plasmodium berghei. Nat. Microbiol. 2018; 3:1206–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bancells C., Llorà-Batlle O., Poran A., Nötzel C., Rovira-Graells N., Elemento O., Kafsack B.F.C., Cortés A.. Revisiting the initial steps of sexual development in the malaria parasite Plasmodium falciparum. Nat. Microbiol. 2019; 4:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Biljon R., van Wyk R., Painter H.J., Orchard L., Reader J., Niemand J., Llinás M., Birkholtz L.-M.. Hierarchical transcriptional control regulates Plasmodium falciparum sexual differentiation. BMC Genomics. 2019; 20:920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Josling G.A., Russell T.J., Venezia J., Orchard L., van Biljon R., Painter H.J., Llinás M.. Dissecting the role of PfAP2-G in malaria gametocytogenesis. Nat. Commun. 2020; 11:1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llorà-Batlle O., Michel-Todó L., Witmer K., Toda H., Fernández-Becerra C., Baum J., Cortés A.. Conditional expression of PfAP2-G for controlled massive sexual conversion in. Sci. Adv. 2020; 6:eaaz5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuda M., Iwanaga S., Kaneko I., Kato T.. Global transcriptional repression: An initial and essential step for Plasmodium sexual development. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:12824–12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modrzynska K., Pfander C., Chappell L., Yu L., Suarez C., Dundas K., Gomes A.R., Goulding D., Rayner J.C., Choudhary J.et al.. A knockout screen of ApiAP2 genes reveals networks of interacting transcriptional regulators controlling the Plasmodium life cycle. Cell Host Microbe. 2017; 21:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C., Li Z., Cui H., Jiang Y., Yang Z., Wang X., Gao H., Liu C., Zhang S., Su X.Z.et al.. Systematic CRISPR–Cas9-mediated modifications of Plasmodium yoelii ApiAP2 genes reveal functional insights into parasite development. mBio. 2017; 8:e01986-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brancucci N.M.B., Bertschi N.L., Zhu L., Niederwieser I., Chin W.H., Wampfler R., Freymond C., Rottmann M., Felger I., Bozdech Z.et al.. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe. 2014; 16:165–176. [DOI] [PubMed] [Google Scholar]
- 15.Coleman B.I., Skillman K.M., Jiang R.H.Y., Childs L.M., Altenhofen L.M., Ganter M., Leung Y., Goldowitz I., Kafsack B.F.C., Marti M.et al.. A Plasmodium falciparum histone deacetylase regulates antigenic variation and gametocyte conversion. Cell Host Microbe. 2014; 16:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filarsky M., Fraschka S.A., Niederwieser I., Brancucci N.M.B., Carrington E., Carrio E., Moes S., Jenoe P., Bartfai R., Voss T.S.. GDV1 induces sexual commitment of malaria parasites by antagonizing HP1-dependent gene silencing. Science. 2018; 359:1259–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Usui M., Prajapati S.K., Ayanful-Torgby R., Acquah F.K., Cudjoe E., Kakaney C., Amponsah J.A., Obboh E.K., Reddy D.K., Barbeau M.C.et al.. Plasmodium falciparum sexual differentiation in malaria patients is associated with host factors and GDV1-dependent genes. Nat. Commun. 2019; 10:2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brancucci N.M.B., Gerdt J.P., Wang C., De Niz M., Philip N., Adapa S.R., Zhang M., Hitz E., Niederwieser I., Boltryk S.D.et al.. Lysophosphatidylcholine regulates sexual stage differentiation in the human malaria parasite Plasmodium falciparum. Cell. 2017; 171:1532–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zanghi G., Vembar S.S., Baumgarten S., Ding S., Guizetti J., Bryant J.M., Mattei D., Jensen A.T.R., Renia L., Goh Y.S.et al.. A specific PfEMP1 is expressed in P. falciparum sporozoites and plays a role in hepatocyte infection. Cell Rep. 2018; 22:2951–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeninga M.D., Quinn J.E., Petter M.. ApiAP2 transcription factors in apicomplexan parasites. Pathogens. 2019; 8:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flueck C., Bartfai R., Niederwieser I., Witmer K., Alako B.T., Moes S., Bozdech Z., Jenoe P., Stunnenberg H.G., Voss T.S.. A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog. 2010; 6:e1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sierra-Miranda M., Vembar S.S., Delgadillo D.M., Avila-Lopez P.A., Herrera-Solorio A.M., Amado D.L., Vargas M., Hernandez-Rivas R.. PfAP2Tel, harbouring a non-canonical DNA-binding AP2 domain, binds to Plasmodium falciparum telomeres. Cell. Microbiol. 2017; 19:e12742. [DOI] [PubMed] [Google Scholar]
- 23.Santos J.M., Josling G., Ross P., Joshi P., Orchard L., Campbell T., Schieler A., Cristea I.M., Llinas M.. Red blood cell Invasion by the malaria parasite is coordinated by the PfAP2-I transcription factor. Cell Host Microbe. 2017; 21:731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martins R.M., Macpherson C.R., Claes A., Scheidig-Benatar C., Sakamoto H., Yam X.Y., Preiser P., Goel S., Wahlgren M., Sismeiro O.et al.. An ApiAP2 member regulates expression of clonally variant genes of the human malaria parasite Plasmodium falciparum. Sci. Rep. 2017; 7:14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan Y., Shen S., Wei G., Tang J., Zhao Y., Wang F., He X., Guo G., Shang X., Yu X.et al.. Rrp6 regulates heterochromatic gene silencing via ncRNA RUF6 decay in malaria parasites. mBio. 2020; 11:e01110-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghorbal M., Gorman M., Macpherson C.R., Martins R.M., Scherf A., Lopez-Rubio J.J.. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR–Cas9 system. Nat. Biotechnol. 2014; 32:819–821. [DOI] [PubMed] [Google Scholar]
- 27.Saliba K.S., Jacobs-Lorena M.. Production of Plasmodium falciparum gametocytes in vitro. Methods Mol. Biol. 2013; 923:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delves M.J., Straschil U., Ruecker A., Miguel-Blanco C., Marques S., Dufour A.C., Baum J., Sinden R.E.. Routine in vitro culture of P. falciparum gametocytes to evaluate novel transmission-blocking interventions. Nat. Protoc. 2016; 11:1668–1680. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q., Siegel T.N., Martins R.M., Wang F., Cao J., Gao Q., Cheng X., Jiang L., Hon C.-C., Scheidig-Benatar C.et al.. Exonuclease-mediated degradation of nascent RNA silences genes linked to severe malaria. Nature. 2014; 513:431–435. [DOI] [PubMed] [Google Scholar]
- 30.Siegel T.N., Hon C.C., Zhang Q., Lopez-Rubio J.J., Scheidig-Benatar C., Martins R.M., Sismeiro O., Coppée J.Y., Scherf A.. Strand-specific RNA-Seq reveals widespread and developmentally regulated transcription of natural antisense transcripts in Plasmodium falciparum. BMC Genomics. 2014; 15:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Rubio J.J., Mancio-Silva L., Scherf A.. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in Malaria parasites. Cell Host and Microbe. 2009; 5:179–190. [DOI] [PubMed] [Google Scholar]
- 32.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016; 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang M., Wang C., Otto T.D., Oberstaller J., Liao X., Adapa S.R., Udenze K., Bronner I.F., Casandra D., Mayho M.et al.. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science. 2018; 360:eaap7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahlgren M., Goel S., Akhouri R.R.. Variant surface antigens of Plasmodium falciparum and their roles in severe malaria. Nat. Rev. Microbiol. 2017; 15:479–491. [DOI] [PubMed] [Google Scholar]
- 39.Campbell T.L., De Silva E.K., Olszewski K.L., Elemento O., Llinas M.. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. Plos Pathogens. 2010; 6:e1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mair G.R., Braks J.A., Garver L.S., Wiegant J.C., Hall N., Dirks R.W., Khan S.M., Dimopoulos G., Janse C.J., Waters A.P.. Regulation of sexual development of Plasmodium by translational repression. Science. 2006; 313:667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lasonder E., Rijpma S.R., van Schaijk B.C., Hoeijmakers W.A., Kensche P.R., Gresnigt M.S., Italiaander A., Vos M.W., Woestenenk R., Bousema T.et al.. Integrated transcriptomic and proteomic analyses of P. falciparum gametocytes: molecular insight into sex-specific processes and translational repression. Nucleic Acids Res. 2016; 44:6087–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindner S.E., Swearingen K.E., Shears M.J., Walker M.P., Vrana E.N., Hart K.J., Minns A.M., Sinnis P., Moritz R.L., Kappe S.H.I.. Transcriptomics and proteomics reveal two waves of translational repression during the maturation of malaria parasite sporozoites. Nat. Commun. 2019; 10:4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh S., Santos J.M., Orchard L.M., Yamada N., van Biljon R., Painter H.J., Mahony S., Llinas M.. The PfAP2-G2 transcription factor is a critical regulator of gametocyte maturation. Mol. Microbiol. 2020; 115:1005–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Y., Qiao D., Wen Y., Bi Y., Chen Y., Huang Z., Cui L., Guo J., Cao Y.. PfAP2-G2 is associated to production and maturation of gametocytes in Plasmodium falciparum via regulating the expression of PfMDV-1. Front. Microbiol. 2020; 11:631444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu A.C.K., Patel H., Chia M., Moretto F., Frith D., Snijders A.P., van Werven F.J.. Repression of divergent noncoding transcription by a sequence-specific transcription factor. Mol. Cell. 2018; 72:942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu N., Hargreaves V.V., Zhu Q., Kurland J.V., Hong J., Kim W., Sher F., Macias-Trevino C., Rogers J.M., Kurita R.et al.. Direct promoter repression by BCL11A controls the fetal to adult hemoglobin switch. Cell. 2018; 173:430–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venugopal K., Hentzschel F., Valkiūnas G., Marti M.. Plasmodium asexual growth and sexual development in the haematopoietic niche of the host. Nat. Rev. Microbiol. 2020; 18:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the main text or the supplementary materials. The raw and processed high-throughput sequencing data of this study have been deposited in Gene Expression Omnibus (GEO) database under accession number GSE149774. The raw data of Flow Cytometry experiments of this study have been deposited in FlowRepository with Repository ID, FR-FCM-Z47H.