Abstract
The ribonucleoprotein (RNP) form of archaeal RNase P comprises one catalytic RNA and five protein cofactors. To catalyze Mg2+-dependent cleavage of the 5′ leader from pre-tRNAs, the catalytic (C) and specificity (S) domains of the RNase P RNA (RPR) cooperate to recognize different parts of the pre-tRNA. While ∼250–500 mM Mg2+ renders the archaeal RPR active without RNase P proteins (RPPs), addition of all RPPs lowers the Mg2+ requirement to ∼10–20 mM and improves the rate and fidelity of cleavage. To understand the Mg2+- and RPP-dependent structural changes that increase activity, we used pre-tRNA cleavage and ensemble FRET assays to characterize inter-domain interactions in Pyrococcus furiosus (Pfu) RPR, either alone or with RPPs ± pre-tRNA. Following splint ligation to doubly label the RPR (Cy3-RPRC domain and Cy5-RPRS domain), we used native mass spectrometry to verify the final product. We found that FRET correlates closely with activity, the Pfu RPR and RNase P holoenzyme (RPR + 5 RPPs) traverse different Mg2+-dependent paths to converge on similar functional states, and binding of the pre-tRNA by the holoenzyme influences Mg2+ cooperativity. Our findings highlight how Mg2+ and proteins in multi-subunit RNPs together favor RNA conformations in a dynamic ensemble for functional gains.
INTRODUCTION
Ribonucleoproteins (RNPs) are essential for fundamental cellular processes (e.g., mRNA splicing, RNA processing, translation). However, our understanding of how RNA structural changes induced by protein binding control the functional repertoire of complex RNPs is limited but growing. In some RNPs, this cooperation is required because proteins help RNAs overcome their weak affinity for sparse intracellular Mg2+, while in others proteins may either stabilize or nudge the RNA towards the native fold necessary for function (1–18). These advances, especially in the context of small RNPs, together with our increasing knowledge of RNA-protein interaction principles (19), provide a foundation to investigate the functional coupling between multiple components in larger RNPs. Nevertheless, because function often requires assembly of the entire RNP (20,21), establishing structure-function relationships for intermediate sub-complexes can be difficult even when detailed maps of the assembly pathway are available. In this regard, because archaeal and eukaryotic RNase P sub-assemblies have fractional activity (22–30), they are good models for studying large, multi-subunit RNPs as functional payoffs from RNA-protein cooperation can be dissected in a stepwise fashion.
The 5′-processing of precursor tRNAs (pre-tRNAs) mediated by the RNP form of RNase P is due to a single catalytic RNase P RNA (RPR) subunit that associates with a variable number of RNase P protein (RPP) subunits: one in Bacteria, up to five in Archaea, and up to 10 in Eukarya (nucleus) (31–36). This diversity in the overall make-up of the different RNase P RNP variants masks the striking conservation of the RPR’s active site, which remains constant even in the context of different auxiliary structural elements (32–35). In the absence of RPPs, many RPRs show weak activity in the presence of elevated concentrations of Mg2+, an essential cofactor (24,25,29,30,37). The crystal structure of bacterial RNase P (38) and the cryo-EM structures of archaeal and eukaryotic RNase P (39–41) provide high-resolution details about the arrangement of individual subunits and, importantly, highlight the uniformity amidst their diversity. They confirm findings from biochemical studies that all RPRs are modular in having a catalytic (C) domain that cleaves the 5′ leader of pre-tRNAs and a specificity (S) domain that binds the ‘elbow’ structure of the L-shaped tRNA (24,25,42,43). Conserved inter-domain interactions act as braces to forge RPR structures that can simultaneously recognize different parts of the pre-tRNA substrate (Supplementary Figure S1). For example, the long-range interaction between a GNRA tetraloop in the S domain and its dock site on the P1 (paired region 1) helix in the C domain is present in all RPRs (40,41,44,45).
The five archaeal RPPs (RPP21, RPP29, POP5, RPP30 and L7Ae; see (46) for an exception) are homologous to eukaryotic RPPs (47), allowing archaeal RNase P to serve as a surrogate for its more intractable eukaryotic cousin (25,48,49). Our in vitro reconstitutions of RNase P from different archaea (22,24–26) showed that RPP21•RPP29 and POP5•RPP30 work in pairs with the RPR to yield partial RNPs that are catalytically intermediate between the RPR alone and holoenzymes containing all RPPs (22,24,25). These binary complexes distinctly enhance the RPR’s apparent substrate affinity and rate of cleavage (22) while also synergistically rescuing mis-cleavage of non-canonical pre-tRNAs (23). Together, these four RPPs enable uniform cleavage of substrates that are otherwise processed at vastly different rates by the RPR alone. L7Ae, the fifth RPP, binds kink-turns in the RPR and potentiates archaeal RNase P function (7,26,27,50). To understand how the binding of different RPPs to specific regions of the RPR independently and collectively mediate structural changes that are essential for assembly and cleavage of diverse substrates, we focus on the interplay between Mg2+ and induced fit in Pyrococcus furiosus (Pfu) RNase P, an archaeal variant that we have previously characterized using biochemical, footprinting, and native mass spectrometry experiments (7,25,50,51).
Mg2+ plays two different roles in RNA function. First, diffuse, outer-sphere interactions as well as specific, inner-sphere interactions with Mg2+ help shape the tertiary structure of RNAs (52,53). Second, Mg2+ is essential for the activity of ribozymes including RNase P due to its involvement in a two metal-ion mechanism for phosphoryl transfer (37,54–56). While Pfu RPR alone in 500 mM Mg2+ and at 55°C is weakly active (25), addition of RPPs significantly decreases the Mg2+ requirement and enhances catalytic efficiency. POP5•RPP30 and RPP21•RPP29 independently decrease the Mg2+ requirement from 500 mM to 120 mM, and further decrease the requirement to 30 mM when working together (25). Additionally, inclusion of L7Ae decreases this amount to ∼10 mM for the 5-RPP combination (7). In most ribozymes, the roles of Mg2+ in structure and function are likely coupled.
Results from multiple kinetic studies point to a conformational change from the enzyme-substrate (ES) ground state to an activated ES* state before pre-tRNA cleavage. First, readout of the invariant tRNA ‘elbow’ by the S domain in bacterial and archaeal RPRs somehow rearranges the active site in the C domain. This inter-domain signaling, which modulates the location and affinity for catalytically relevant Mg2+, appears to be mediated by the substrate and constitutes an important determinant for rate and cleavage-site selection (24,57–60). Second, time-course analyses of the binding of a fluor-labeled pre-tRNA to Bacillus subtilis RNase P displayed biphasic kinetics, with an initial diffusion-limited encounter followed by a conformational change prior to cleavage (61). Third, kinetic studies with bacterial and archaeal RNase P suggest that the RPP(s) engender similar cleavage rates for different pre-tRNAs by shifting the equilibrium of the pre-cleavage isomerization from ES to ES* (22,24,62,63).
Despite the above findings, which suggest an induced-fit mechanism for RNase P, structural studies thus far indicate that the enzyme is largely pre-organized for catalysis. Superposition of the structures of Methanocaldococcus jannaschii (Mja) RNase P obtained in the absence or presence of tRNATyr did not show any substantive differences in the conformation of the RPR or RPPs (40); the same scenario was observed with bacterial RNase P (38), although subsequent SAXS studies of three phylogenetically distinct bacterial RPRs (64) suggested that conformational properties in solution may have been masked in the crystal due to specific inter-/intra-molecular interactions. Interestingly, cryo-EM structures of Saccharomyces cerevisiae RNase P in the absence or presence of pre-tRNAPhe revealed that a universally conserved uridine (U93) rotates drastically and points into the active site to coordinate and position a Mg2+ ion for pre-tRNA cleavage. Molecular dynamics (MD) simulations also showed that U93 has a dynamic conformation in the holoenzyme (RPR + 5 RPPs) but is constrained upon binding to the pre-tRNA (39).
Since Förster resonance energy transfer (FRET) measurements in solution have been employed to obtain dynamic information about RNA folding and RNP assembly (4,6,10,11,13,15,18,65), we sought to use FRET for identifying conformational changes during RNase P catalysis that may be inaccessible to cryo-EM- and X-ray crystallography-based snapshots. Specifically, our goal was to monitor Mg2+-induced, large-scale conformational changes in the Pfu RPR (± RPPs) and to investigate how these structural alterations correlate with the pre-tRNA cleavage activity of Pfu RNase P. Although our ensemble studies cannot shed light on either fast or slow movement(s) of specific residues/domains, we expected to gain insights into RPR conformational ensembles under equilibrium conditions. Indeed, our results highlight how Mg2+ promotes tertiary interactions between the Pfu RPR’s C and S domains that are important for dual anchoring of the pre-tRNA substrate and remodels the Pfu RNase P holoenzyme. Our findings have implications for understanding the assembly of multi-subunit, cellular RNPs.
MATERIALS AND METHODS
Cloning of Pfu RPR 5′3′ext, Pfu RPR ext mP1 and Thermus thermophilus (Tth) pre-tRNAGly
The protocols for cloning the Pfu RPR derivatives and pre-tRNA used in the studies reported here are described in the Supplementary Information (see Supplementary Table S1 for information on oligonucleotides used for cloning). All RNAs were generated by T7 RNA polymerase (T7 RNAP)-mediated run-off in vitro transcription (IVT) using as templates either linearized plasmid DNA or a PCR amplicon that included the T7 RNAP promoter (see Supplementary Information). As to nomenclature: Pfu RPR 5′3′ext refers to a Pfu RPR variant containing 23-nt extensions at both termini; Pfu RPR ext mP1 contains the extensions in Pfu RPR 5′3′ext and in addition lacks the two native terminal bp in P1.
Native MS analysis
All native MS experiments were conducted using a Q Exactive™ Ultra-High Mass Range (UHMR) Hybrid Quadrupole-Orbitrap™ mass spectrometer (Thermo Scientific) that was modified with a customized device for performing surface-induced dissociation (66,67). Samples were diluted to 1 μM in 800 mM NH4OAc and transferred to filament-containing borosilicate glass capillary tips (Sutter Instrument, Novato, CA) that were individually pulled in-house using a P-97 micropipette puller (Sutter Instrument). A platinum wire was then inserted into the sample solution, and a voltage of 0.7–1.5 kV was applied to directly infuse the sample into the UHMR mass spectrometer by nano-electrospray ionization. An in-source trapping voltage of -100 to -200 V and capillary temperature of 250°C were also applied to aid desolvation. Other instrument tune settings were as follows: scan range, 350–10,000 m/z; resolution (at 400 m/z), 3,125; microscans, 5; maximum injection time, 200 ms; S-lens radio frequency (RF) level, 200 V; source direct current (DC) offset, 21 V; injection flatapole DC, 9 V; inter flatapole DC, 8 V; bent flatapole DC, 6 V; transfer multipole DC, 0 V; C-trap entrance lens offset, 1.8 V; trapping gas pressure setting, 4. For all RNAs, except the synthetic internal fragment oligonucleotide, the mass spectrometer was set to high m/z ion transfer mode, with RF amplitudes of 700, 940 and 900 V applied to the injection flatapole, bent flatapole, and transfer multipole/higher-energy collisional dissociation cell, respectively; RF amplitudes of 150, 300 and 250 V, respectively, were used for the I-F oligonucleotide samples, which were analyzed in low m/z ion transfer mode.
Microscale thermophoresis (MST)
Equilibrium dissociation constants (KD) for binding of 5′-ext-Cy5–oligo (referred hereafter as Cy5–oligo; Supplementary Table S2, Millipore Sigma) to the 5′ extension of unlabeled Pfu RPR ext mP1 in the absence or presence of Pfu RPPs were determined using MST. All MST measurements were collected using a Monolith Pico instrument (NanoTemper Technologies, San Francisco, CA) and standard NanoTemper glass capillaries (MO-K022). Final 20-μl binding reactions containing 0.75 nM Cy5–oligo were incubated for 10 min at 37°C before initiating MST measurements.
Pfu RPR ext mP1 alone
Pfu RPR ext mP1 was folded in the presence of 10 mM MgCl2, serially diluted 1:1 in the appropriate 1× folding buffer (see below) and mixed with Cy5–oligo for a final Mg2+ concentration of either 10 or 500 mM.
Pfu RPR ext mP1 + 5 RPPs
Pfu RPR ext mP1 was folded in the presence of 2.5 mM MgCl2 and sequentially incubated for 5 min at 37°C with a 10-fold molar excess each of RPP21•RPP29, then L7Ae, and finally POP5•RPP30. Upon assembly of the holoenzyme, it was serially diluted 1:1 in Mg2+-free buffer and mixed with Cy5–oligo for a final Mg2+ concentration of either 0.033 or 30 mM.
During MST measurements, excitation power was set to 15% while MST power was set to medium. Initial fluorescence was measured for 5 s before the infrared laser was turned on for 30 s. Fluorescence was then monitored for an additional 5 s after the infrared laser was turned off. Data from MST measurements were plotted using Kaleidagraph (Synergy Software). KD values were calculated by plotting ΔFNorm versus the concentration of either Pfu RPR ext mP1 or Pfu RPR ext mP1 + 5 RPPs and by fitting the data to hyperbolic binding isotherms. Mean and standard deviation values for each condition were calculated from three technical replicates. Curve-fit errors did not exceed 25% in any trial.
RNase P activity assays
All activity assays and RNA folding were carried out in a thermal cycler (Bio-Rad, Formerly MJ Research Inc., Hercules, CA) and buffers sterilized using 0.22-μm syringe filters (VWR international, Radnor, PA). Separate assay buffers were prepared and pH-adjusted for each [Mg2+] tested. Prior to initiating cleavage assays, the RPR was incubated in ddH2O for 50 min at 50°C and 10 min at 37°C before an equal volume of appropriate 2X folding buffer was added. The RNA sample was then incubated for an additional 30 min at 37°C. Composition of the 2X folding buffer varied depending on whether RPR alone or RPR + 5 RPP complex was used. Refolded RPR was added to buffer containing appropriate [Mg2+] and, where appropriate, reconstituted with the RPPs. Cleavage was initiated by adding the appropriate substrate.
Pfu RPR ext mP1 alone
Activity of the RPR alone was tested at 50-mM intervals from 100 to 500 mM Mg2+. Prior to cleavage assays, RPR was refolded in a 0.5-ml PCR tube (Thermo Fisher or Axygen) by first incubating in ddH2O for 50 min at 50°C and for 10 min 37°C. Subsequently, the RPR was supplemented with an equal volume of 2X refolding buffer (100 mM HEPES–KOH [pH 8.4 at 25°C], 1.6 M NH4OAc, 20 mM MgCl2) and incubated for 30 min at 37°C. The final RPR concentration after refolding was 6 μM. First, 4.2 μl of 6 μM (for assays in 100–200 mM Mg2+) or 1.5 μM (for assays in 250–500 mM Mg2+) folded RPR were added to 6.3 μl 2× assay buffer and incubated for 2 min at 37°C. Each 2× buffer contained 50 mM HEPES–KOH (pH 8.4 at 25°C), 3.2 M NH4OAc, 5% (w/v) PEG 8000 and varying concentrations of MgCl2 (ranging from 190 to 990 mM). Tth pre-tRNAGly, a trace amount of which was labeled using 5′-γ-[32P]-ATP (PerkinElmer, Shelton, CT) and T4 PNK (NEB), was diluted in 1X refolding buffer to a final concentration of 720 μM. Substrate cleavage was initiated by addition of 2.1 μl of 720 μM Tth pre-tRNAGly, which had been pre-incubated for 2 min at 37°C. The final assay volume of 12.6 μl contained 2 μM (for assays in 100–200 mM Mg2+) or 0.5 μM (for assays in 250–500 mM Mg2+) RPR and 120 μM Tth pre-tRNAGly in 1X buffers (50 mM HEPES [pH 8.4 at 25°C], 2 M NH4OAc, 2.5% (w/v) PEG 8000 and 100–500 mM MgCl2). After addition of substrate, the reaction mixture was briefly vortexed and centrifuged for a few seconds before returning to the thermal cycler. After defined time intervals dictated by [Mg2+], 3-μl aliquots were removed from the reaction master mix and quenched with 17 μl loading dye. Cleavage products were separated using 10% (w/v) polyacrylamide/7 M urea gels. Tth pre-tRNAGly cleaved with Escherichia coli (Eco) RNase P was used as a positive control for RNase P cleavage.
During the first trial for each [Mg2+] tested, a sample containing only the substrate in assay buffer was also incubated for the entire duration of the time course to determine the extent of uncatalyzed breakdown. In the majority of cases, the extent of uncatalyzed breakdown was found to be negligible (<4%). For subsequent trials, one sample containing the substrate alone was incubated for only the longest time period in the time-course assay. These samples were used as negative controls. Initial velocities did not change whether or not the nominal uncatalyzed background cleavage was subtracted.
Pfu RPR ext mP1 + 5 RPPs
Activity assays using Pfu RPR ext mP1 + 5 RPPs were performed in a manner similar to the RPR alone assays, albeit with some differences. A 2X refolding buffer containing only 5 mM MgCl2 was used to accommodate the lower [Mg2+] tested in the assays with RPR + 5 RPPs. RPPs were purified as described elsewhere (25,50). First, 1.64 μl of 500 nM RPR was added to 8.2 μl assay buffer containing varying [Mg2+] and incubated for 5 min at 37°C. To reconstitute the RNP, RPR was mixed with 1.64 μl of 5 μM RPPs (in a specific order) and incubated for 5 min at 37°C following each addition. The order of addition was RPP21•RPP29, followed by L7Ae (C71V variant (50)), and finally POP5•RPP30. Due to the weak activity of the RPR + 5 RPPs with Tth pre-tRNAGly at 37°C, Eco pre-tRNATyr (68) was used as the substrate for assays using RPR + 5 RPPs. Unlabeled Eco pre-tRNATyr was mixed with a trace amount of 5′-[32P]-pre-tRNATyr and diluted to the desired final concentration in 100 mM HEPES–KOH (pH 8.4 at 25°C), 1.6 M NH4OAc. To initiate pre-tRNA cleavage, 1.64 μl of 10 μM Eco pre-tRNATyr, which had been pre-incubated at 37°C for 2 min, was added before the reaction was mixed by pipetting, briefly centrifuged, and returned to the thermal cycler. Each 16.4-μl assay contained 50 nM RPR, 500 nM RPPs and 1 μM Eco pre-tRNATyr, in 1X assay buffer [50 mM HEPES (pH 8.4 at 25°C), 800 M NH4OAc, 0.5–30 mM MgCl2, and 2.5% (v/v) PEG 8000]. After defined time intervals dictated by [Mg2+] used in each assay, 4-μl aliquots were withdrawn from the reaction master mix and quenched with 16 μl loading dye. Samples were analyzed by electrophoresis on 8% (w/v) polyacrylamide/7 M urea gels. Eco pre-tRNATyr cleaved by Eco RNase P was used as positive control for RNase P cleavage. Negative controls were prepared as described above for the RPR-alone reactions.
Data analysis
Polyacrylamide gels were scanned using a Typhoon RGB imager (Cytiva) and were quantitated using ImageQuant (Cytiva). Percent cleavage values were plotted against the time of incubation to obtain the slope (initial velocity), which in turn was used for subsequent turnover number calculations. Time courses were designed to ensure 30–40% cleavage and maintain linearity in product formation. While there were a few reactions that exceeded this threshold marginally, all curve fits yielded correlation coefficients (R2) > 0.96.
Ensemble FRET experiments
Buffers used for FRET experiments were identical to those used for activity assays. Cy5–Pfu RPR ext mP1 was mixed with a Cy3–labeled oligonucleotide (5′-ext-Cy3–oligo, referred hereafter as Cy3–oligo; Supplementary Table S2) complementary to the RPR’s 5′ extension, and folding was performed as described above. Folded RPR + Cy3–oligo samples were added to the appropriate buffers (± RPPs) and pre-incubated at 37°C prior to FRET measurements. RPR, RPP, and substrate concentrations were identical to those used in activity assays unless otherwise stated, and each FRET sample had a final volume of 55 μl.
Pfu RPR ext mP1 alone
Folded RPR + Cy3–oligo samples were diluted to a final concentration of 100 nM Cy5–RPR ext mP1 + 77 nM Cy3–oligo prior to preincubation at 37°C. Samples were pre-incubated for 10 min at 37°C prior to FRET measurements. Although we had intended to use 100 nM of Cy3–oligo, we discovered that the concentration used was sub-stochiometric (77 nM) due to an error in the stock concentration. Fortuitously, the lower concentration of Cy3–oligo, which we continued to use, minimized leakage of donor into acceptor channels.
Pfu RPR ext mP1 + 5 RPPs
Enzyme reconstitution conditions, including order of RPP addition and incubation time, were identical to those used for activity assays. Samples contained 50 nM Cy5–RPR ext mP1, 500 nM RPPs, and 100 nM Cy3–oligo. In addition to the Mg2+ concentrations tested in activity assays, FRET measurements were also collected at 0.033, 0.1 and 0.25 mM Mg2+. Samples were pre-incubated for 5 min at 37°C prior to FRET measurements.
Pfu RPR ext mP1 + 5 RPPs + pre-tRNATyr
For measurements conducted in the presence of Eco pre-tRNATyr, each sample contained 50 nM Cy5-RPR, 500 nM RPPs and 38.5 nM Cy3-oligo. We decreased the Cy3-oligo concentration to minimize potential non-specific binding of the Cy3–oligo to the excess pre-tRNA. However, two control experiments in which the Cy3–oligo concentration was varied revealed that Mg2+ dependent changes in EFRET were nearly identical when we added either 38.5 or 100 nM Cy3–oligo to Pfu RNase P in the absence or presence of substrate. Given the high affinity of the Cy3–oligo for the RPR (see below), we expect only a modest change in the concentration of bound species whether 38.5 or 100 nM Cy3–oligo was used. Following reconstitution, pre-incubated Eco pre-tRNATyr was added to a final concentration of 1 μM and FRET was measured immediately to minimize substrate turnover.
Fluorescence and FRET measurements
Pre-incubated samples were transferred to a 45-μl Starna Cells cuvette (16.45F-Q3/Z8.5), and fluorescence was measured at 37°C using a Horiba PTI Quantamaster fluorometer. Excitation and emission slits were set to 5 nm each, and a lamp intensity correction was applied as per the manufacturer's instructions. The excitation wavelength was first set to 510 nm (for Cy3) and emission was measured from 530 to 750 nm. Next, the excitation wavelength was set to 610 nm (for Cy5) and emission was measured from 630 to 750 nm. For each sample, a blank measurement of buffer alone was also taken. The Ratio A method (69) was used to calculate FRET efficiencies using an automated MATLAB script (70).
RESULTS
Site-specific fluor labeling of Pfu RPR ext mP1
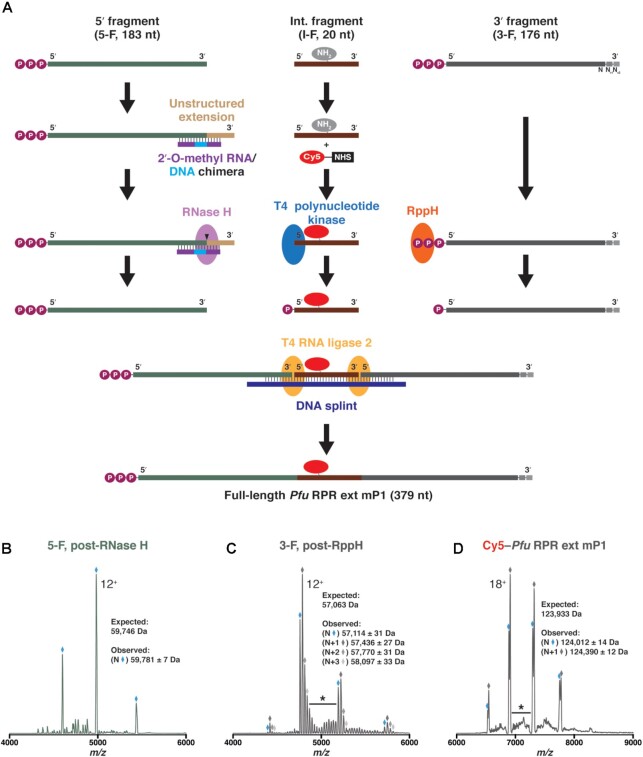
To conduct FRET measurements on the Pfu RPR, we sought to introduce the donor (Cy3) and the acceptor (Cy5) fluors in its C and S domains, respectively (Figure 1). While Cy3 was positioned on the C domain by virtue of a modified oligonucleotide that is complementary to an engineered extension, Cy5 was introduced in the S domain using a splint-ligation approach (Figures 1 and 2). For the latter, we synthesized three fragments of the Pfu RPR ext mP1: 5′-fragment (5-F, 1–183); internal fragment (I-F, 184–203); and 3′-fragment (3-F, 204–379) (Figure 2A). While 5-F and 3-F were made by IVT, I-F was chemically synthesized and obtained commercially. These three RNAs were then ligated using a DNA splint and T4 RNA ligase 2. Each fragment was processed individually and native MS was used to verify the mass of all components of the ligation reaction (Figure 2B–D; Supplementary Figure S2A-F; Supplementary Table S3; see Supplementary Information for additional experimental details).
Figure 1.
(A) A modified Pyrococcus furiosus RNase P RNA (Pfu RPR ext mP1) labeled with both Cy3 and Cy5 fluorophores. To generate Pfu RPR ext mP1, both the 5′- and 3′-termini were extended (inset, right) while the two terminal bp in the P1 helix were deleted. The cytidine at position 170 (wild-type numbering) was replaced with an amine-modified uridine (red) to facilitate internal Cy5 labeling via splint ligation (see Figure 2), and a Cy3-labeled DNA oligonucleotide was annealed to the 5′-end to obtain a dual-labeled RPR. (B) General scheme for Förster resonance energy transfer (FRET) studies. Increasing [Mg2+] is expected to promote interactions between the C (dark gray) and S (light gray) domains, thereby increasing the proximity of the Cy3 (donor) and Cy5 (acceptor) fluorophores and FRET efficiency.
Figure 2.
Use of splint ligation to internally label Pfu RPR ext mP1 with a Cy5 fluorophore. (A) The RPR was synthesized in three fragments: 5′-fragment (5-F, transcribed in vitro), internal fragment (I-F, commercially synthesized), and 3′-fragment (3-F, transcribed in vitro). Each fragment was processed individually prior to a two-step ligation using T4 RNA ligase 2 and a DNA splint. Mass spectrometry was used to verify the masses of the (B) 5′ fragment [5-F], post-RNase H cleavage; (C) 3′ fragment [3-F], post-RppH treatment; and (D) final Pfu RPR ext mP1 labeled with Cy5. Charge state distributions for the most intense species in each sample are indicated with diamonds, and the main charge state is labeled. In each spectrum, the species of intended length (N) is shown in blue while any prominent, 3′-extended species (N+#) are marked in varying shades of gray. Representative regions of low-intensity peaks that are indicative of additional heterogeneity have been denoted with an asterisk. In all spectra, the y-axis (not shown) represents relative intensity.
Establishing the utility of doubly fluor-labeled Pfu RPR ext mP1 for FRET
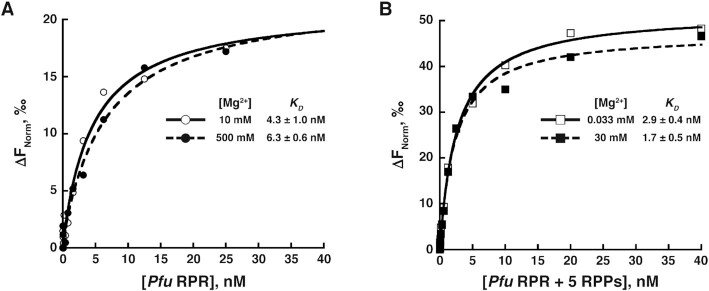
To prepare the RPR for ensemble FRET experiments, the RNA was refolded in the presence of the Cy3–oligo that is complementary to the 5′-extension of the RPR. To ensure that the observed FRET changes are due to structural reorganization caused by Mg2+ or RPP binding and are not from potentially altered binding of the Cy3–oligo to the 5′-extension, we performed microscale thermophoresis (MST) experiments to measure the binding affinity of Cy3–oligo to Pfu RPR at low and high Mg2+ concentrations and either in the absence or presence of RPPs. Due to MST instrument specifications, we used an oligonucleotide identical in sequence to that used in our FRET studies but labeled with Cy5 instead of Cy3 (Supplementary Table S2). In the absence of RPPs, we determined KD values of 4.3 ± 1.0 and 6.3 ± 0.6 nM at 10 mM and 500 mM Mg2+, respectively (Figure 3A). In the presence of RPPs, the KD value decreased modestly from 2.9 ± 0.4 nM at 0.033 mM Mg2+ to 1.7 ± 0.5 nM at 30 mM Mg2+ (Figure 3B). The mean and standard deviation values were calculated using data from three trials for each experimental condition (Supplementary Figure S3). Results from these MST experiments show that binding of the Cy5–oligo to the 5′-extension was unaffected by changes in either Mg2+ concentration or reconstitution with RPPs.
Figure 3.
Representative microscale thermophoresis (MST) measurements of Cy5-labeled oligonucleotide (Cy5–oligo) binding to (A) unlabeled Pfu RPR ext mP1 at 10 mM (⚬, solid line) and 500 mM (•, dashed line) Mg2+, and (B) Pfu RPR ext mP1 + 5 RPPs at 0.033 mM (□, solid line) and 30 mM (▪, dashed line) Mg2+. Plots were fit to a hyperbolic binding isotherm to yield KD values. In the inset tables, mean and standard deviation values were calculated from three technical replicates (see Supplementary Figure S3 for the primary data from individual trials).
We next determined the pre-tRNA cleavage activity of doubly fluor-labeled Pfu RPR ext mP1. At 450 mM Mg2+ and 37°C, cleavage of Tth pre-tRNAGly by Cy5–Pfu RPR ext mP1 + Cy3–oligo was ∼5-fold lower than that of the unlabeled Pfu RPR ext mP1; we observed a ∼3.3-fold decrease when we tested cleavage of Eco pre-tRNATyr by the holoenzyme assembled with Cy5-labeled RPR ext mP1 relative to the unlabeled counterpart (Supplementary Table S4; see Supplementary Information for additional details). Because the doubly fluor-labeled RPR was functional, albeit less active than the unmodified counterpart, we proceeded with the ensemble FRET experiments. The loss of activity that we observed with the fluor-labeled RPR is not surprising and has precedents. For example, two different Cy3– and Cy5–dual labeled guide RNAs that were individually assembled into a H/ACA snoRNP and used in single-molecule fluorescence studies displayed 4- to 6-fold lower pseudouridylation activity relative to the unlabeled counterpart (11).
Activity and ensemble FRET assays using Pfu RPR ext mP1
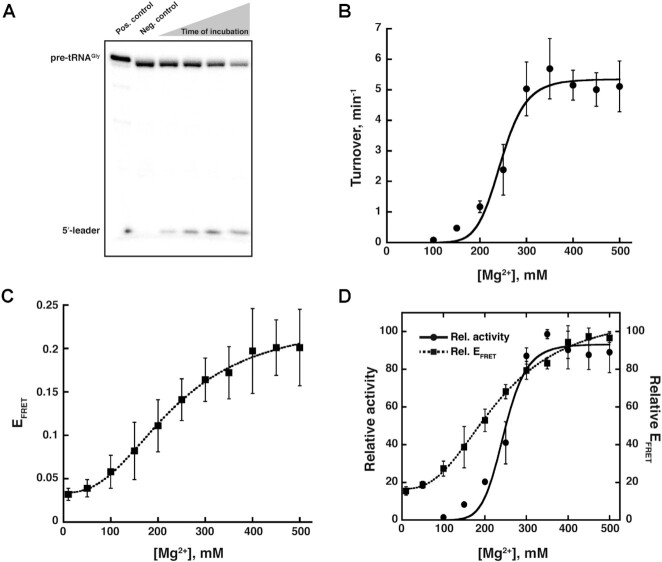
Cleavage of 5′-[32P]-pre-tRNAGly (120 μM) was assayed between 100 mM and 500 mM Mg2+, and monitored by denaturing PAGE (Figure 4A, Supplementary Figure S4); all experiments were performed at 37°C (see Supplementary Information for the rationale underlying choice of 37°C). RPR activity was weak at 100 mM Mg2+ but the activity increased nearly 70-fold when [Mg2+] was increased from 100 to 350 mM before plateauing (Figure 4B). A similar Mg2+-dependent activity profile was also observed with Eco pre-tRNATyr, a second substrate that we tested [Supplementary Figure S5 (25)].
Figure 4.
Effect of Mg2+ on the activity and inter-domain interactions of Pfu RPR ext mP1. (A) Representative data showing cleavage of γ-32P-ATP-labeled Thermus thermophilus (Tth) pre-tRNAGly by unlabeled Pfu RPR ext mP1 in 450 mM Mg2+ and at 37°C during a 20-min time-course (also, see Supplementary Figure S4 and S5). (B) Mg2+-dependent increase in the activity of unlabeled Pfu RPR ext mP1. Average turnover numbers calculated from four independent initial velocity measurements are plotted against [Mg2+]. (C) Mg2+-dependent increase in FRET efficiency for dual-fluor-labeled Pfu RPR ext mP1. Average FRET efficiencies obtained from three independent measurements are plotted against [Mg2+] (see Supplementary Figure S6 for additional data). (D) Overlay of turnover number (solid line) and FRET efficiency (dashed line), which shows that FRET efficiency correlates with the rate of catalysis as a function of [Mg2+]. Relative activity and EFRETvalues were obtained by normalization against the highest turnover number or FRET efficiency, respectively; since this reference value was not always at the same concentration of Mg2+ among the replicates, there is minor variability between panels B or C versus D but the trends are highly similar. In instances where error bars are not seen, the errors are smaller than the symbols used.
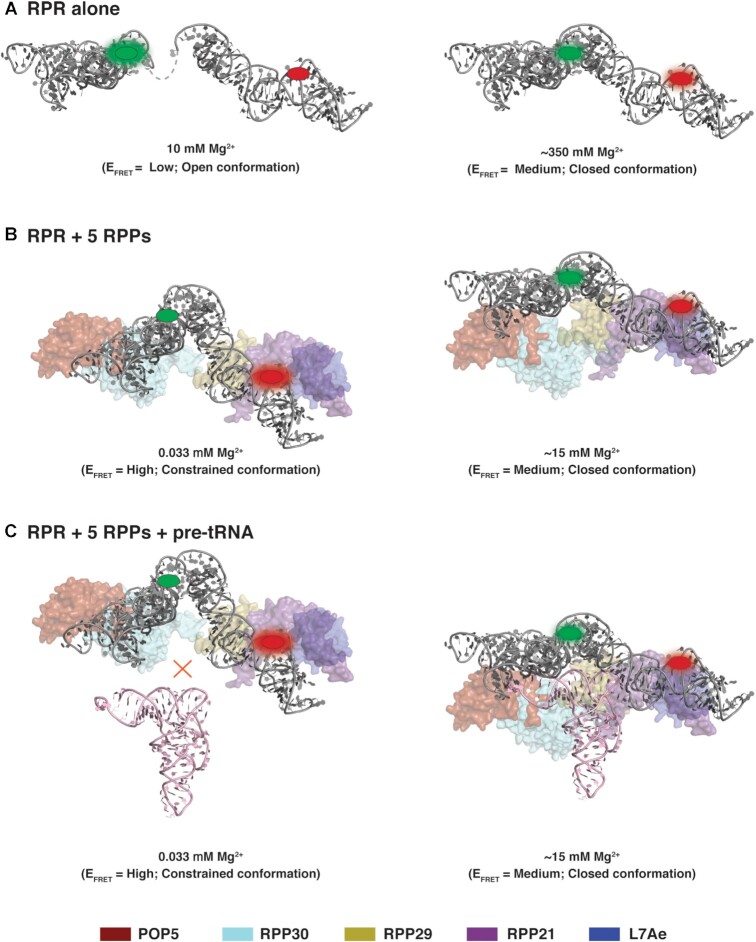
FRET was measured from 10 mM to 500 mM Mg2+ and at 37°C in the same buffers used for activity assays. Upon excitation of the Cy3 donor, Cy5 emission was observed with the doubly labeled Pfu RPR ext mP1 thus confirming FRET (Figure 4C and Supplementary Figure S6). Mirroring the [Mg2+]-dependent trend observed for turnover number, the FRET efficiency (EFRET) also showed a gradual increase with increasing [Mg2+], plateauing at 350–400 mM Mg2+ coinciding with the concentration where we observed maximal activity (see overlay in Figure 4D). These coincident trends highlight how FRET—a proxy for distance and contacts between the C and S domains—accurately tracks pre-tRNA cleavage. The increase in EFRET values from 0.03 ± 0.01 at 10 mM to 0.20 ± 0.03 at 450 mM Mg2+ indicates a shift from an ‘open’ to a ‘closed’ conformation (Figure 6A, Scheme I).
Figure 6.
Model of potential structural changes in archaeal RNase P (also, see Scheme I). Based on the ensemble FRET measurements, the RPR (gray) alone likely adopts an ‘open’ conformation (low FRET efficiency; A, left) at 10 mM Mg2+ while the complex of Pfu RPR + 5 RPPs exists in a ‘constrained’ conformation (high FRET efficiency; B, left) at 0.033 mM Mg2+. Neither state is competent for pre-tRNA cleavage (C, left). With increasing [Mg2+], both the RPR and the holoenzyme complex comprised of Pfu RPR + 5 RPPs undergo structural remodeling to converge on a common ‘closed’ active conformation (A–C, right). While there are caveats to using ensemble FRET measurements to establish a precise distance between two fluorophores, conservation of the active site and pre-tRNA anchors in different RNase P RNP variants provide some support for this qualitative model generated using the cryo-EM structure of Methanocaldococcus jannaschii (Mja) RNase P as template (40). Although we depict single conformations in this model for the sake of simplicity, each state is more accurately described by an ensemble. EFRET classified using 0.03 < low < 0.10 < medium < 0.30 < high < 0.60.
Our finding of moderate EFRET in 100–200 mM Mg2+, where the RPR is weakly active, suggests that less Mg2+ is required for formation of tertiary structure compared to function (Figure 4). It is possible that diffusely-bound Mg2+ ions promote RPR tertiary structure, which in turn is required for occupancy of site-specific Mg2+ in the active site.
Activity and ensemble FRET assays using the Pfu RNase P holoenzyme
To understand the effect of all five cognate RPPs on Mg2+-induced structural and functional changes in the RPR, we tested the activity of the RPR + 5 RPPs (POP5•RPP30, RPP21•RPP29, L7Ae) complex. Previous assays of Pfu RNase P at 55°C (7,50) informed the range of Mg2+ concentrations to be tested. The RPR + 5 RPP complex is active at 10 mM Mg2+ and 55°C. However, as our experiments were performed at 37°C, we explored a slightly wider range of Mg2+ concentrations (33 μM to 30 mM Mg2+) to account for a possible temperature-dependent shift in the [Mg2+] that is optimal for activity. As the POP5•RPP30 and RPP21•RPP29 complexes are stored in Mg2+ (50), the lowest possible [Mg2+] we could test was ∼33 μM.
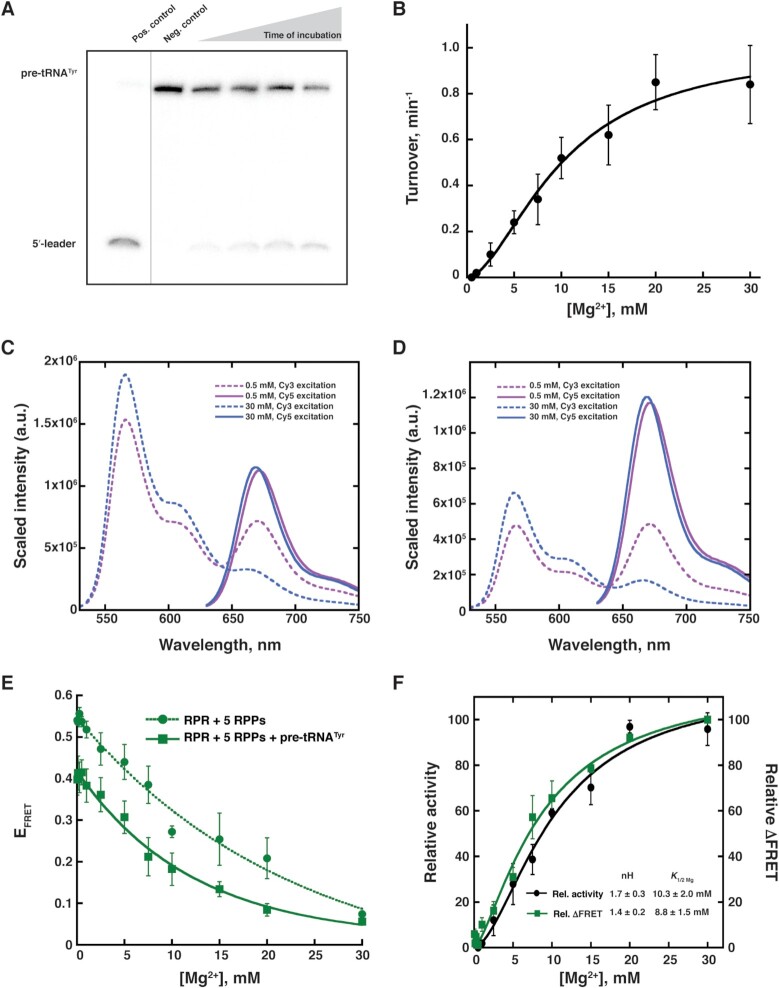
When cleavage of Eco pre-tRNATyr was analyzed by denaturing PAGE (Figure 5A), we observed activity starting at 1 mM Mg2+ (Figure 5B). As noticed with RPR alone, the holoenzyme showed a gradual increase in activity with increasing [Mg2+] and eventually plateaued at 20 mM (Figure 5B). The activity increased 42-fold as the [Mg2+] increased from 1 to 20 mM. While the actual turnover numbers with the RPR + 5 RPP complex were lower than that observed with the RPR alone (Figures 4 and 5), these assays entailed use of different substrates and assay conditions; also, note that the range of Mg2+ concentrations tested for the RPR and holoenzyme are quite different (Supplementary Figure S7).
Figure 5.
Effect of Mg2+ on the activity and inter-domain interactions of Pfu RPR ext mP1 reconstituted with all 5 RPPs. (A) Representative data showing cleavage of γ-32P-ATP-labeled Escherichia coli (Eco) pre-tRNATyr by unlabeled Pfu RPR ext mP1 + 5 RPPs in 30 mM Mg2+ and at 37°C during a 5-min time-course. For illustrative purposes, the gel was cropped to reposition non-adjacent lanes of interest; no other modifications were made to the image. (B) Mg2+-dependent increase in the activity of unlabeled Pfu RPR ext mP1. Average turnover numbers calculated from three independent initial velocity measurements are plotted against [Mg2+], and the data were fit to the Hill equation. Dual-fluor-labeled Pfu RPR ext mP1 + 5 RPPs (C) without and (D) with Eco pre-tRNATyr shows a decrease in FRET from low (0.5 mM; purple) to high (30 mM; red) Mg2+. A representative spectrum from one trial at each Mg2+ concentration is shown. Within each set, emission from direct Cy5 excitation was normalized against the sample with the highest signal. The same scaling factor was then applied to the corresponding emission spectrum obtained from Cy3 excitation (see Supplementary Figure S10 for additional data). (E) Mg2+-dependent increase in FRET efficiency for dual-fluor-labeled Pfu RPR ext mP1 + 5 RPPs without (dotted line) and with (solid line) Eco pre-tRNATyr. Average FRET efficiency from three independent measurements is plotted against [Mg2+]. (F) Comparison of relative turnover numbers (black) and ΔFRET values (green solid line). Data for Pfu RPR ext mP1 + 5 RPPs + Eco pre-tRNATyr were fit to the Hill equation to obtain the nH (Hill coefficient) and K1/2Mg2+ values in the inset (see Supplementary Figure S11 for additional data).
Although we used a stochiometric excess of RPPs to promote assembly of the Pfu RNase P holoenzyme and had some previous evidence from native MS for assembly of the RNP at low Mg2+ (7,51), we used two approaches to test the idea that the complete holoenzyme (RPR + all five RPPs) is assembled at very low and high [Mg2+]. First, to investigate if the activity observed with RPR + 5 RPPs could have resulted from partial assemblies, we performed pre-tRNATyr cleavage assays using RPR assembled with either RPP21•RPP29 + POP5•RPP30 or POP5•RPP30 + L7Ae. Of these two combinations, RPR + POP5•RPP30 + L7Ae was more active under the assay conditions tested but even this 3-RPP complex fared weaker than the 5-RPP complex between 1 and 15 mM Mg2+ (Supplementary Figure S8). Thus, activity at <15 mM Mg2+ is attributable to a complex containing 5 RPPs. Second, we assessed Pfu RNase P assembly using mass photometry (MP), a powerful tool to gain information on the oligomeric state of proteins either alone or in complexes (71,72). Despite being of lower mass accuracy than native MS, MP has the advantage of permitting measurements in non-volatile salts such as those typically used in assay buffers (e.g., Mg2+, HEPES). When we examined the Pfu RPR ± 5 RPPs at 33 μM and 10 mM Mg2+, we observed a mass range consistent with the RPR assembled into a complex with 5 RPPs (RPR + POP5•RPP30 + RPP21•RPP29 + L7Ae3; Supplementary Figure S9). However, the broad MP profiles also make it difficult to determine if the RPR is fully assembled into a stable RNP containing all five RPPs. The RPR and RPPs may be in dynamic equilibrium at low Mg2+, and increasing Mg2+ could enhance both the binding affinity of the RPPs (e.g. third copy of L7Ae; (7)) and the stability of the holoenzyme on account of gains in RPR and RPP co-folding. Nevertheless, it appears that the complete holoenzyme is assembled at very low and high [Mg2+].
The RPR was reconstituted with the 5 RPPs, and FRET was measured at 37°C. Surprisingly, unlike the RPR alone, the holoenzyme was associated with an EFRET = 0.54 ± 0.02 even at 33 μM Mg2+. Contrary to the trend observed with RPR alone, there was a decrease in the holoenzyme's EFRET with increasing [Mg2+], with a two-fold drop to 0.25 ± 0.06 at 15 mM Mg2+ (Figure 5C, E and Supplementary Figure S10). This finding leads to three inferences. First, the Mg2+-dependent self-organization in the holoenzyme is complete at much lower Mg2+ concentrations compared to the RPR alone (20 mM versus 400 mM; Figures 4D and 5E). Second, increasing [Mg2+] leads to an increase in distance between Cy3 and Cy5 in case of the holoenzyme, which contrasts sharply with the observed decrease in the case of RPR alone (Figures 4D and 5E). The gradual Mg2+-dependent decrease in EFRET observed with RPR + 5 RPPs might reflect Mg2+-induced remodeling of the RNP complex and suggests a shift from a ‘constrained’ to a ‘closed (not collapsed)’ conformation (Figure 6B, Scheme I). We recognize that other perspectives merit consideration since not all RNAs undergo protein-induced structural remodeling ((4); see Discussion). Lastly, similar EFRET values for the RPR without or with 5 RPPs at the Mg2+ concentration where their activities begin to plateau suggests that they each take a different path to reach a similar conformation that is optimal for catalysis.
The EFRET values for the RPR and holoenzyme, respectively, of 0.03 and 0.54 (at 33 μM Mg2+) prompted us to determine if this difference might arise from RPPs inducing changes in the local environment of the fluorophore and not by altering the RPR structure. When the RPR was assembled as part of sub-complexes (with POP5•RPP30 or RPP21•RPP29 + L7Ae), we obtained EFRET ∼0.4 to 0.45 in the presence of 33 μM Mg2+. Given the possibility of a ‘collapsed-state’ ensemble even in the sub-assemblies, it seems unlikely that the large FRET changes observed with the RPR + 5 RPPs would arise from perturbations to the local environment of the fluor especially with two suites of RPPs with different physicochemical properties. Also, protein-induced fluorescence enhancement typically results in ∼2- to 3-fold gains unlike what we observed.
Ensemble FRET measurements with Pfu RPR ext mP1 + 5 RPPs + pre-tRNATyr
We employed FRET to gain insights into the conformational changes in the holoenzyme in the presence of pre-tRNATyr (Figure 5D). We do not expect more than one percent of the substrate to be cleaved during the three-min period of each FRET measurement given the turnover number of the Cy5 Pfu RPR ext mP1 (Supplementary Table S4). As pre-tRNATyr is present in 20-fold excess over the holoenzyme, changes in FRET efficiency as a function of increasing [Mg2+] are therefore likely to reflect structural alterations in the enzyme-substrate (ES) complex. When FRET was measured in the presence of pre-tRNATyr, we observed a trend similar to that observed with the holoenzyme alone (Figure 5E, solid vs dotted lines). However, we extracted an important feature from these data by plotting the relative change in EFRET. We calculated ΔFRET = (MaxFRET – EFRET)/(MaxFRET – MinFRET) × 100 at each [Mg2+] tested. When either the turnover numbers or ΔFRET values were plotted as a function of Mg2+ (Figure 5F), they could be fit to the Hill equation and there was good agreement in the nH (Hill coefficient, 1.7 ± 0.3 or 1.4 ± 0.2, respectively) and K1/2Mg2+ (10.3 ± 2.0 and 8.8 ± 1.5 mM, respectively). These values indicate that at least two Mg2+ ions bind cooperatively, although the second ion might be bound weakly/transiently.
Since the abovementioned cooperativity is absent when we examined ΔFRET of the RPR + 5 RPP complex alone (Figure 5E, Supplementary Figure S11), substrate binding is associated with this cooperativity. However, we investigated if one assay variable might have contributed to the observed difference. We had used the Cy3–oligo at a two-fold excess over the RPR in case of the holoenzyme and a slightly sub-stoichiometric amount with the holoenzyme–substrate assays (see Materials and Methods) since we were concerned about potential non-specific binding of the Cy3–oligo to the excess pre-tRNA. Nevertheless, when we compared EFRET and the Hill equation curve-fits for the holoenzyme-substrate complex using either the two-fold excess or the sub-stoichiometric amount of the Cy3–oligo, the Hill coefficient and K1/2Mg2+ values were similar (nH = 1.4 or 1.5; K1/2Mg2+ 8.8 or 5.6 mM; Supplementary Figure S11). Thus, the difference that we observed between the holoenzyme and holoenzyme–substrate complex (Figure 5E and Supplementary Figure S11) with respect to Mg2+ cooperativity is due to substrate binding.
DISCUSSION
We elaborate below the implications of four findings from our study of Pfu RNase P: (i) FRET monitored using Cy3-RPRC domain and Cy5-RPRS domain correlates well with pre-tRNA cleavage activity; (ii) the RPR and the holoenzyme (RPR + 5 RPPs) traverse different Mg2+-dependent paths to converge on similar functional states (Figure 6); (iii) the Pfu RNase P holoenzyme (RPR + 5 RPPs) is remodeled by increasing Mg2+; and (iv) binding of the pre-tRNA substrate to the holoenzyme influences the cooperativity of Mg2+ binding and catalysis.
Interdomain interactions in Pfu RPR are essential for catalysis
The bacterial, archaeal, and eukaryotic RNase P holoenzyme-tRNA structures reveal the invariant use of a shallow pocket to bind the acceptor–T arm in the substrate (Supplementary Figure S1C and D) (38–41). In all three cases, cleavage between N-1 and N+1 in the pre-tRNA substrate depends on at least two specific contacts: the G19–C56 base pair (bp) of the tRNA elbow that forms π-π interactions with two interdigitated T loops in the RPR’s S domain, and the first bp of the acceptor stem (N+1 – N+72) that is recognized by the RPR’s C domain through either RNA or RNP contacts (38–41). If this dual anchor is used as a molecular ruler to trigger site-specific cleavage, optimal substrate recognition must require the C and S domains to interact and be oriented appropriately with respect to each other. Such a postulate, first inspired by biochemical and phylogenetic studies of bacterial RNase P (43,44,73), is applicable to Pfu RNase P. Pfu RPR displays optimal activity in 300–400 mM Mg2+, while the RPR + 5 RPP complex activity starts to plateau around 15–20 mM Mg2+ (Figures 4 and 5). The coincident EFRET values (0.15–0.25) at these optimal [Mg2+] suggests that a specific RPR conformation is required for pre-tRNA cleavage.
RPR and the holoenzyme achieve a common functional fold albeit through different paths
Even though the RPR conformation either alone or in the holoenzyme has a different ‘starting line’, it reaches a similar final destination albeit through different routes. The parallel Mg2+-dependent increase in EFRET and activity (Figure 4) for the Pfu RPR alone suggests that decreasing the distance between the C (Cy3) and S (Cy5) domains is associated with an increase in turnover number. The low EFRET at 10 mM Mg2+ is indicative of an RPROpen conformation; this state transitions to a medium EFRET conformation at 300–400 mM Mg2+ where the RPR is active (RPRClosed; Figure 6A; Scheme I). Clearly, both conformations are thermodynamically stable. Compared to the RPR, the high EFRET observed with the holoenzyme at <1 mM Mg2+ suggests the presence of an RNPConstrained conformation that is not functional. Increasing [Mg2+] from <1 to 15 mM results in the holoenzyme adopting a medium EFRET and active conformation (RNPClosed; Figure 6B and C, Scheme I). Thus, the RPR and the holoenzyme converge on similar final state(s) but after traversing different paths.

Scheme I
Mg2+-induced remodeling of the RNase P RNP
The EFRET values for the RPR and holoenzyme, respectively, are 0.03 and 0.54 (at 33 μM Mg2+) and 0.03 and 0.27 (at 10 mM Mg2+) (Figure 5; data not shown). These unexpected differences in EFRET values indicate that the RPR is present in two distinct conformational ensembles, one when alone and the other as part of the holoenzyme. Comparison of the bacterial RPR without and with its single RPP showed minimal changes in the RPR structure (38). Since there is no structure available for the archaeal or eukaryotic RPR alone, their remodeling (if any) upon binding to their cognate RPPs is unknown. However, MD simulations of the yeast RPR and holoenzyme (RPR + 9 RPPs) showed large fluctuations in the RPR backbone that increased the distance between the two structural elements needed for dual anchoring of the pre-tRNA substrate (39). Addition of the RPPs constrained the RPR’s dynamic behavior except in two regions that do not bind the RPPs (39). These MD simulations and our FRET studies suggest that the archaeal (and possibly eukaryotic) RPRs in the absence or presence of RPPs might sample a different conformational ensemble, perhaps allowing regulatory control of the holoenzyme not possible with the lone RPR.
The Mg2+-dependent decrease in EFRET and increased pre-tRNA cleavage observed with the holoenzyme (Figure 5) is consistent with a few not mutually exclusive conclusions. First, while the bacterial RPR forms three long-range, inter-domain interactions—L9-P1, L8-P4 and L18-P8 (38,45)—the archaeal and eukaryotic RPRs have only the L9-P1 contact and instead use RPPs as scaffolds to stabilize the final fold (39–41). The cryo-EM structure of Mja (archaeal) RNase P (40) illustrates how this scaffolding is made possible by stringing together five RPPs through protein-protein interactions. However, rather than a static structure, our data support the idea of Mg2+-induced distortion/wedging to rearrange the RPPs and convert the holoenzyme from a ‘constrained’ to a ‘closed (but less collapsed)’ conformation (Figure 6B, Scheme I). A similar strain-propagation model was proposed based on SHAPE studies of the bI5 group I intron with and without its two protein cofactors (2). Second, if Mg2+ and RPPs bind to the RPR in its folded state, their binding will be thermodynamically coupled and cause remodeling of the RNase P RNP. With FRET as an output, we plan to test this premise. Ongoing single-molecule FRET studies will also help determine the population distribution of different sub-assemblies and the effect of partial suites of RPPs on RPR structural conformations. We expect to map the hierarchy and cooperativity among RPPs during RNase P assembly and determine the contribution of such linkages to activity over a range of [Mg2+]. Such experiments have been instructive in revealing RNA structural dynamics and the temporal order of protein-binding events in telomerase, U4/U6 di-snRNP, and the H/ACA snoRNP (4,11,13).
We highlight some similarities and differences between our findings and previous studies that examined RNA folding and RNP assembly. First, ensemble and single-molecule FRET studies show that Na+ and Mg2+ are capable of promoting ‘closed’ RNA conformations by decreasing the rate of unfolding of the folded state (6,65). Mg2+ acts likewise in the case of Pfu RPR alone, although EFRET = 0.03 at 2 M NH4+ (in low Mg2+) suggests that offsetting the electrostatic repulsion by screening alone is insufficient (Figure 4C) and that Mg2+ is necessary to promote inter-domain interactions associated with a functional fold. Second, results from investigations of small RNPs or isolated modules of larger RNPs show how proteins can alter the equilibrium between an open and docked/closed state and outperform Mg2+ alone in influencing this conformational toggling. Steady-state FRET measurements revealed that L7Ae can bind an RNA containing a kink-turn even in the absence of metal ions and promote a kinked conformation that is observed with Mg2+ alone; a small increase in EFRET in the presence of L7Ae and Mg2+ (compared to either alone) suggests additional protein binding-mediated stabilization (15). Results from ensemble FRET and footprinting experiments with the Eco 16S rRNA 5′ domain showed that the native conformation (albeit not fully populated) is achieved by 10 mM Mg2+. At 4 mM Mg2+, however, S4 along with S16 and S20, which act cooperatively on the RNA either by stabilizing the native or destabilizing the non-native state, are necessary for rRNA folding (18). SMF studies with a yeast U2 snRNA model showed that either Mg2+ or the Cus2p protein stabilize the ‘open’ conformation required for catalysis rather than the ‘closed’ conformation needed for assembly (10). However, Mg2+ and Cus2p together also resulted in less frequent sampling of certain conformations promoted by Mg2+ alone. The decreased FRET with increased Mg2+ in the Cus2p-U2 snRNA complex mirrors the trend that we observed with Pfu RNase P (Figure 5E). These two cases show how Mg2+ and proteins engender a concerted distortion to transition the RNP from a compact assembly to an extended functional state. There is a key difference, however, between Pfu RNase P and other RNPs studied thus far. Typically, protein cofactors shift the equilibrium between pre-existing conformational ensembles of their respective RNA ligand. The high EFRET (∼0.5) of the Pfu RNase P holoenzyme that is not observed with the RPR alone suggests a new collapsed RPR conformation but the inability to parse heterogeneity in ensemble experiments precludes firm conclusions in this regard. Since ensemble averages mask the proportion of sub-populations, rare conformations (including those with high EFRET) in the RPR-alone sample will not be evident.
Coupling between induced fit and Mg2+ coordination
There appear to be no dramatic alterations in the structure of the holoenzyme upon binding to the substrate as judged by the similar FRET profiles (Figure 5E). This claim is consistent with the cryo-EM structures of Mja RNase P (40) that show little difference in the overall architecture of the RPR and RPPs in the absence or presence of tRNATyr. However, we found cooperativity in the Mg2+ response when the substrate is present. One interpretation of these findings is that the substrate participates in shaping the active site, perhaps even bringing with it a Mg2+ (74). Moreover, since bacterial RPR S-domain mutants that are incapable of reading the tRNA ‘elbow’ could be rescued by increasing Mg2+ (57,60), it appears that interdomain crosstalk leads to a rearrangement of Mg2+ in the active site. Our FRET studies confirm the interplay between the induced fit triggered by substrate binding and positioning of catalytically relevant metal ions. FRET studies with ‘elbow-lacking’ model substrates that cannot engender inter-domain signaling would provide further support for this notion of Mg2+ coordination that is associated with induced fit.
SUMMARY
The retention of RNA enzymes in the extant cellular pool of protein-based catalysts has been attributed to their ability to leverage substrate binding to effect long-range induced fit and efficiently sample conformational ensembles (75). Such gating mechanisms afford exquisite control of rate and fidelity, not unlike protein-only enzymes (76). Under low-Mg2+ cellular conditions, however, such regulation in catalytic RNAs may depend on proteins as exemplified by RPPs that fasten distal RPR structural elements to facilitate pre-tRNA recognition and local Mg2+-interaction networks essential for catalysis. Our finding that proteins may promote new RNA conformational ensembles that can be fine-tuned differently by cellular Mg2+ supports the notion that functional and regulatory gains shaped the evolutionary forces that favored a functional alliance between RNA and protein in essential cellular RNPs.
Supplementary Material
ACKNOWLEDGEMENTS
We are extremely grateful to Dr Lien Lai (Gopalan laboratory) for valuable feedback on the work and manuscript, and Hong Duc Phan (Gopalan laboratory) for assistance with illustrations; Dr Yi Luo, Dr Benjamin Donovan, and Khan Cox (Poirier laboratory) for their assistance with initial ensemble FRET experiments and data analyses; Drs Mark Foster (OSU), Julius Lucks (Northwestern University) and Ming Lei (Shanghai Institute of Precision Medicine) for useful suggestions; Xiao Ma (Foster laboratory) for providing Pfu L7Ae used in pilot studies; Dr Ruben Gonzalez (Columbia University) for guidance on fluor labeling; Dr. Dennis Bong (OSU) for supporting early studies using bPNAs; Dr Sergei Kazakov (Somagenics) for valuable advice on RNA ligation; Dr David Riddle (Wright Patterson Air Force Base) for generously facilitating initial MST studies through preliminary discussions and instrumentation access and support; Dr Dmitri Kudryashov (OSU) for kindly sharing the Monolith NT.115 (NanoTemper) instrument for MST studies; Dr Jennifer Ottesen (OSU) for access to her NanoDrop spectrophotometer; and Dr Richard Fishel and Dr. Ross Larue (OSU) for training and use of the mass photometer (MP) and Dr Dalton Snyder (OSU) for help with MP data acquisition.
Contributor Information
Ila A Marathe, Department of Microbiology, The Ohio State University, Columbus, OH 43210, USA; Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210, USA; Center for RNA Biology, The Ohio State University, Columbus, OH 43210, USA.
Stella M Lai, Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210, USA; Center for RNA Biology, The Ohio State University, Columbus, OH 43210, USA; Resource for Native Mass Spectrometry-Guided Structural Biology, The Ohio State University, Columbus, OH 43210, USA.
Walter J Zahurancik, Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210, USA; Center for RNA Biology, The Ohio State University, Columbus, OH 43210, USA.
Michael G Poirier, Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210, USA; Center for RNA Biology, The Ohio State University, Columbus, OH 43210, USA; Department of Physics, The Ohio State University, Columbus, OH 43210, USA.
Vicki H Wysocki, Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210, USA; Center for RNA Biology, The Ohio State University, Columbus, OH 43210, USA; Resource for Native Mass Spectrometry-Guided Structural Biology, The Ohio State University, Columbus, OH 43210, USA.
Venkat Gopalan, Department of Microbiology, The Ohio State University, Columbus, OH 43210, USA; Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210, USA; Center for RNA Biology, The Ohio State University, Columbus, OH 43210, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM120582 [M.G.P., V.H.W. to V.G., GM131626 [M.G.P.], P41 GM128577 to V.H.W.]; Ohio State University Comprehensive Cancer Center Pelotonia Fellowship Program (pre-doctoral to I.M. and post-doctoral to W.Z.]. Funding for instrumentation: NIH [GM114666-06S1] (to D.K.); NIH [S10OD023582 to Jane E. Jackman]; NIH [CA067007]; James Comprehensive Cancer Center (to R.F.). Funding for open access charge: Behrman Research Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bokinsky G., Nivon L.G., Liu S., Chai G., Hong M., Weeks K.M., Zhuang X.. Two distinct binding modes of a protein cofactor with its target RNA. J. Mol. Biol. 2006; 361:771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncan C.D., Weeks K.M.. Nonhierarchical ribonucleoprotein assembly suggests a strain-propagation model for protein-facilitated RNA folding. Biochemistry. 2010; 49:5418–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Froschauer E.M., Kolisek M., Dieterich F., Schweigel M., Schweyen R.J.. Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol. Lett. 2004; 237:49–55. [DOI] [PubMed] [Google Scholar]
- 4.Hardin J.W., Warnasooriya C., Kondo Y., Nagai K., Rueda D.. Assembly and dynamics of the U4/U6 di-snRNP by single-molecule FRET. Nucleic. Acids. Res. 2015; 43:10963–10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim H., Abeysirigunawardena S.C., Chen K., Mayerle M., Ragunathan K., Luthey-Schulten Z., Ha T., Woodson S.A.. Protein-guided RNA dynamics during early ribosome assembly. Nature. 2014; 506:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim H.D., Nienhaus G.U., Ha T., Orr J.W., Williamson J.R., Chu S.. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc. Natl Acad. Sci. U.S.A. 2002; 99:4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai L.B., Tanimoto A., Lai S.M., Chen W.Y., Marathe I.A., Westhof E., Wysocki V.H., Gopalan V.. A novel double kink-turn module in euryarchaeal RNase P RNAs. Nucleic Acids Res. 2017; 45:7432–7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayerle M., Bellur D.L., Woodson S.A.. Slow formation of stable complexes during coincubation of minimal rRNA and ribosomal protein S4. J. Mol. Biol. 2011; 412:453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pyle A.M.Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 2002; 7:679–690. [DOI] [PubMed] [Google Scholar]
- 10.Rodgers M.L., Tretbar U.S., Dehaven A., Alwan A.A., Luo G., Mast H.M., Hoskins A.A.. Conformational dynamics of stem II of the U2 snRNA. RNA. 2016; 22:225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt A., Hanspach G., Hengesbach M.. Structural dynamics govern substrate recruitment and catalytic turnover in H/ACA RNP pseudouridylation. RNA Biol. 2020; 10.1080/15476286.2020.1842984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroeder R., Barta A., Semrad K.. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 2004; 5:908–919. [DOI] [PubMed] [Google Scholar]
- 13.Stone M.D., Mihalusova M., O’Connor C.M., Prathapam R., Collins K., Zhuang X.. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007; 446:458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun L., Harris M.E.. Evidence that binding of C5 protein to P RNA enhances ribozyme catalysis by influencing active site metal ion affinity. RNA. 2007; 13:1505–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner B., Melcher S.E., Wilson T.J., Norman D.G., Lilley D.M.. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 2005; 11:1192–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weeks K.M., Cech T.R.. Protein facilitation of group I intron splicing by assembly of the catalytic core and the 5′ splice site domain. Cell. 1995; 82:221–230. [DOI] [PubMed] [Google Scholar]
- 17.Yu G., Zhao Y., Li H.. The multistructural forms of box C/D ribonucleoprotein particles. RNA. 2018; 24:1625–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abeysirigunawardena S.C., Woodson S.A.. Differential effects of ribosomal proteins and Mg2+ ions on a conformational switch during 30S ribosome 5′-domain assembly. RNA. 2015; 21:1859–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corley M., Burns M.C., Yeo G.W.. How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol. Cell. 2020; 78:9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen S.R., Rodgers M.L., Hoskins A.A.. Fluorescent labeling of proteins in whole cell extracts for single-molecule imaging. Methods Enzymol. 2016; 581:83–104. [DOI] [PubMed] [Google Scholar]
- 21.Tamaru D., Amikura K., Shimizu Y., Nierhaus K.H., Ueda T.. Reconstitution of 30S ribosomal subunits in vitro using ribosome biogenesis factors. RNA. 2018; 24:1512–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen W.Y., Pulukkunat D.K., Cho I.M., Tsai H.Y., Gopalan V.. Dissecting functional cooperation among protein subunits in archaeal RNase P, a catalytic ribonucleoprotein complex. Nucleic Acids Res. 2010; 38:8316–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen W.Y., Singh D., Lai L.B., Stiffler M.A., Lai H.D., Foster M.P., Gopalan V.. Fidelity of tRNA 5′-maturation: a possible basis for the functional dependence of archaeal and eukaryal RNase P on multiple protein cofactors. Nucleic Acids Res. 2012; 40:4666–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pulukkunat D.K., Gopalan V.. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res. 2008; 36:4172–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai H.Y., Pulukkunat D.K., Woznick W.K., Gopalan V.. Functional reconstitution and characterization of Pyrococcus furiosus RNase P. Proc. Natl Acad. Sci. U.S.A. 2006; 103:16147–16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho I.M., Lai L.B., Susanti D., Mukhopadhyay B., Gopalan V.. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc. Natl Acad. Sci. U.S.A. 2010; 107:14573–14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuhara H., Kifusa M., Watanabe M., Terada A., Honda T., Numata T., Kakuta Y., Kimura M.. A fifth protein subunit Ph1496p elevates the optimum temperature for the ribonuclease P activity from Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 2006; 343:956–964. [DOI] [PubMed] [Google Scholar]
- 28.Perederina A., Berezin I., Krasilnikov A.S.. In vitro reconstitution and analysis of eukaryotic RNase P RNPs. Nucleic Acids Res. 2018; 46:6857–6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikovska E., Svard S.G., Kirsebom L.A.. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl Acad. Sci. U.S.A. 2007; 104:2062–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pannucci J.A., Haas E.S., Hall T.A., Harris J.K., Brown J.W.. RNase P RNAs from some Archaea are catalytically active. Proc. Natl Acad. Sci. U.S.A. 1999; 96:7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altman S.A view of RNase P. Mol. Biosyst. 2007; 3:604–607. [DOI] [PubMed] [Google Scholar]
- 32.Ellis J.C., Brown J.W.. The RNase P family. RNA Biol. 2009; 6:362–369. [DOI] [PubMed] [Google Scholar]
- 33.Esakova O., Krasilnikov A.S.. Of proteins and RNA: the RNase P/MRP family. RNA. 2010; 16:1725–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans D., Marquez S.M., Pace N.R.. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006; 31:333–341. [DOI] [PubMed] [Google Scholar]
- 35.Lai L.B., Vioque A., Kirsebom L.A., Gopalan V.. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 2010; 584:287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schenking I., Hartmann R.K., Rossmanith W.. Evolutionary Biology—A Transdisciplinary Approach. 2020; Springer Nature Switzerland; 255–299. [Google Scholar]
- 37.Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S.. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983; 35:849–857. [DOI] [PubMed] [Google Scholar]
- 38.Reiter N.J., Osterman A., Torres-Larios A., Swinger K.K., Pan T., Mondragon A.. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010; 468:784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan P., Tan M., Zhang Y., Niu S., Chen J., Shi S., Qiu S., Wang X., Peng X., Cai G.et al.. Structural insight into precursor tRNA processing by yeast ribonuclease P. Science. 2018; 362:eaat6678. [DOI] [PubMed] [Google Scholar]
- 40.Wan F., Wang Q., Tan J., Tan M., Chen J., Shi S., Lan P., Wu J., Lei M.. Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme. Nat. Commun. 2019; 10:2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J., Niu S., Tan M., Huang C., Li M., Song Y., Wang Q., Chen J., Shi S., Lan P.et al.. Cryo-EM structure of the human ribonuclease P holoenzyme. Cell. 2018; 175:1393–1404. [DOI] [PubMed] [Google Scholar]
- 42.Guerrier-Takada C., Altman S.. Reconstitution of enzymatic activity from fragments of M1 RNA. Proc. Natl Acad. Sci. U.S.A. 1992; 89:1266–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loria A., Pan T.. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA. 1996; 2:551–563. [PMC free article] [PubMed] [Google Scholar]
- 44.Massire C., Jaeger L., Westhof E.. Phylogenetic evidence for a new tertiary interaction in bacterial RNase P RNAs. RNA. 1997; 3:553–556. [PMC free article] [PubMed] [Google Scholar]
- 45.Torres-Larios A., Swinger K.K., Krasilnikov A.S., Pan T., Mondragon A.. Cystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005; 437:584–587. [DOI] [PubMed] [Google Scholar]
- 46.Lai L.B., Chan P.P., Cozen A.E., Bernick D.L., Brown J.W., Gopalan V., Lowe T.M.. Discovery of a minimal form of RNase P in Pyrobaculum. Proc. Natl Acad. Sci. U.S.A. 2010; 107:22493–22498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall T.A., Brown J.W.. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA. 2002; 8:296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boomershine W.P., McElroy C.A., Tsai H.Y., Wilson R.C., Gopalan V., Foster M.P.. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc. Natl Acad. Sci. U.S.A. 2003; 100:15398–15403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kouzuma Y., Mizoguchi M., Takagi H., Fukuhara H., Tsukamoto M., Numata T., Kimura M.. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem. Biophys. Res. Commun. 2003; 306:666–673. [DOI] [PubMed] [Google Scholar]
- 50.Lai S.M., Lai L.B., Foster M.P., Gopalan V.. The L7Ae protein binds to two kink-turns in the Pyrococcus furiosus RNase P RNA. Nucleic Acids Res. 2014; 42:13328–13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma X., Lai L.B., Lai S.M., Tanimoto A., Foster M.P., Wysocki V.H., Gopalan V.. Uncovering the stoichiometry of Pyrococcus furiosus RNase P, a multi-subunit catalytic ribonucleoprotein complex, by surface-induced dissociation and ion mobility mass spectrometry. Angew. Chem. Int. Ed. Engl. 2014; 53:11483–11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Misra V.K., Draper D.E.. On the role of magnesium ions in RNA stability. Biopolymers. 1999; 48:113–135. [DOI] [PubMed] [Google Scholar]
- 53.Strulson C.A., Boyer J.A., Whitman E.E., Bevilacqua P.C.. Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions. RNA. 2014; 20:331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward W.L., Plakos K., DeRose V.J.. Nucleic acid catalysis: metals, nucleobases, and other cofactors. Chem. Rev. 2014; 114:4318–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warnecke J.M., Furste J.P., Hardt W.D., Erdmann V.A., Hartmann R.K.. Ribonuclease P (RNase P) RNA is converted to a Cd(2+)-ribozyme by a single Rp-phosphorothioate modification in the precursor tRNA at the RNase P cleavage site. Proc. Natl Acad. Sci. U.S.A. 1996; 93:8924–8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steitz T.A., Steitz J.A.. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. U.S.A. 1993; 90:6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brannvall M., Kikovska E., Wu S., Kirsebom L.A.. Evidence for induced fit in bacterial RNase P RNA-mediated cleavage. J. Mol. Biol. 2007; 372:1149–1164. [DOI] [PubMed] [Google Scholar]
- 58.Mao G., Srivastava A.S., Wu S., Kosek D., Lindell M., Kirsebom L.A.. Critical domain interactions for type A RNase P RNA catalysis with and without the specificity domain. PLoS One. 2018; 13:e0192873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinapah S., Wu S., Chen Y., Pettersson B.M., Gopalan V., Kirsebom L.A.. Cleavage of model substrates by archaeal RNase P: role of protein cofactors in cleavage-site selection. Nucleic Acids Res. 2011; 39:1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu S., Chen Y., Lindell M., Mao G., Kirsebom L.A.. Functional coupling between a distal interaction and the cleavage site in bacterial RNase-P-RNA-mediated cleavage. J. Mol. Biol. 2011; 411:384–396. [DOI] [PubMed] [Google Scholar]
- 61.Hsieh J., Fierke C.A.. Conformational change in the Bacillus subtilis RNase P holoenzyme–pre-tRNA complex enhances substrate affinity and limits cleavage rate. RNA. 2009; 15:1565–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niland C.N., Anderson D.R., Jankowsky E., Harris M.E.. The contribution of the C5 protein subunit of Escherichia coli ribonuclease P to specificity for precursor tRNA is modulated by proximal 5′ leader sequences. RNA. 2017; 23:1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun L., Campbell F.E., Zahler N.H., Harris M.E.. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. EMBO J. 2006; 25:3998–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kazantsev A.V., Rambo R.P., Karimpour S., Santalucia J. Jr, Tainer J.A., Pace N.R.. Solution structure of RNase P RNA. RNA. 2011; 17:1159–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schroeder K.T., Lilley D.M.. Ion-induced folding of a kink turn that departs from the conventional sequence. Nucleic Acids Res. 2009; 37:7281–7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.VanAernum Z.L., Gilbert J.D., Belov M.E., Makarov A.A., Horning S.R., Wysocki V.H.. Surface-Induced dissociation of noncovalent protein complexes in an extended mass range orbitrap mass spectrometer. Anal. Chem. 2019; 91:3611–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harvey S.R., VanAernum Z.L., Kostelic M.M., Marty M.T., Wysocki V.H.. Probing the structure of nanodiscs using surface-induced dissociation mass spectrometry. Chem. Commun. (Camb.). 2020; 56:15651–15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vioque A., Arnez J., Altman S.. Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli. J. Mol. Biol. 1988; 202:835–848. [DOI] [PubMed] [Google Scholar]
- 69.Clegg R.M.Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992; 211:353–388. [DOI] [PubMed] [Google Scholar]
- 70.Donovan B.T., Chen H., Jipa C., Bai L., Poirier M.G.. Dissociation rate compensation mechanism for budding yeast pioneer transcription factors. Elife. 2019; 8:e43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soltermann F., Foley E.D.B., Pagnoni V., Galpin M., Benesch J.L.P., Kukura P., Struwe W.B.. Quantifying protein-protein Interactions by molecular counting with mass photometry. Angew. Chem. Int. Ed. Engl. 2020; 59:10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sonn-Segev A., Belacic K., Bodrug T., Young G., VanderLinden R.T., Schulman B.A., Schimpf J., Friedrich T., Dip P.V., Schwartz T.U.et al.. Quantifying the heterogeneity of macromolecular machines by mass photometry. Nat. Commun. 2020; 11:1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pomeranz Krummel D.A., Altman S.. Verification of phylogenetic predictions in vivo and the importance of the tetraloop motif in a catalytic RNA. Proc. Natl Acad. Sci. U.S.A. 1999; 96:11200–11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kirsebom L.A., Trobro S.. RNase P RNA-mediated cleavage. IUBMB Life. 2009; 61:189–200. [DOI] [PubMed] [Google Scholar]
- 75.Cech T.R.Crawling out of the RNA world. Cell. 2009; 136:599–602. [DOI] [PubMed] [Google Scholar]
- 76.Johnson K.A.Role of induced fit in enzyme specificity: a molecular forward/reverse switch. J. Biol. Chem. 2008; 283:26297–26301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.