Abstract
To reconstruct systematically hyperactive transcription factor (TF)-dependent transcription networks in squamous cell carcinomas (SCCs), a computational method (ELMER) was applied to 1293 pan-SCC patient samples, and 44 hyperactive SCC TFs were identified. As a top candidate, DLX5 exhibits a notable bifurcate re-configuration of its bivalent promoter in cancer. Specifically, DLX5 maintains a bivalent state in normal tissues; its promoter is hypermethylation, leading to DLX5 transcriptional silencing in esophageal adenocarcinoma (EAC). In stark contrast, DLX5 promoter gains active histone marks and becomes transcriptionally activated in ESCC, which is directly mediated by SOX2. Functionally, silencing of DLX5 substantially inhibits SCC viability both in vitro and in vivo. Mechanistically, DLX5 cooperates with TP63 in regulating ∼2000 enhancers and promoters, which converge on activating cancer-promoting pathways. Together, our data establish a novel and strong SCC-promoting factor and elucidate a new epigenomic mechanism - bifurcate chromatin re-configuration - during cancer development.
INTRODUCTION
Squamous cell carcinomas (SCCs) are aggressive malignancies derived from stratified epithelium of several organs, including esophagus (ESCC), head and neck (HNSCC), lung (LUSC), cervix (CESC), skin (SSCC), etc. SCCs are the most frequently occurring human cancers worldwide and represent a major cause of death. Notably, despite arising from different anatomical locations, SCCs share many unified genomic features, which are specific to squamous cell lineage (1,2). The most frequent genomic abnormalities converge on genes involved in various important biological processes, including squamous cell differentiation (e.g. TP63, SOX2, ZNF750 and NOTCH family), oxidative metabolism (e.g. NRF2), receptor tyrosine kinase signaling (e.g. FGFR1 and PIK3CA), as well as epigenetic regulation (e.g. EP300, MLL2, MLL3 and NSD1) (2–7). These genomic alterations occur much more frequently in SCCs relative to non-SCCs, highlighting their unique pathogenic significance in SCC biology. However, these shared genomic characteristics have not substantially improved clinical management of SCC patients, and effective targeted regimens are still unavailable for these cancers. Some forms of SCCs are highly lethal and have a very poor prognosis. For example, the 5-year survival rate of ESCC patients is ∼17% (8). Alarmingly, the incidence of SCCs, either individually or in aggregate, is increasing rapidly worldwide (9).
In contrast to genomic alterations, the epigenomic changes that are either shared or private in SCCs are less well-characterized. Most knowledge is derived from DNA methylation array data, which identified promoter hypermethylation in tumor suppressor genes, such as CDKN2A/B, LRP1B and RASSF1A (1). However, these changes are nonspecific to SCCs since they are commonly observed in most cancer types. Transcriptome profiling data from The Cancer Genome Atlas (TCGA) (4–7) and similar cell-of-origin (10) implies the presence of SCC-specific gene regulatory networks. However, such gene expression networks and their upstream TFs still await characterization in a pan-SCC manner.
Much of the DNA methylation work has focused on changes occurring at gene promoter regions, which are often hypermethylated in cancer cells (11,12). However, Whole Genome Bisulfite Sequencing (WGBS) has shown that reduced methylation in enhancer regions represents the most widespread change during both normal and cancer cell development (13–17). Importantly, our earlier work identified both cancer-specific enhancers and TF binding sites (TFBSs) based on this form of methylation alterations (17). To systematically characterize enhancer demethylation and associated mechanisms in cancer, we recently developed a computational method, named ELMER (Enhancer Linking by Methylation/Expression Relationships) (18,19). Specifically, ELMER uses methylation changes as key nodes and correlates them with gene expression to infer both the upstream TFs and downstream target genes for each TFBS. Instead of assuming that the nearest gene is the TFBS target, ELMER searches a local neighborhood of genes using correlation across samples. This is a crucially important aspect, as one of the major challenges in interpreting TFBSs and enhancers is that they do not typically regulate the nearest genes along the chromosome, but rather choose their target genes based on poorly understood processes that involve chromosome looping (19).
Here, we applied the ELMER program to 1293 TCGA pan-SCC samples to explore whether different types of SCC share SCC-specific gene regulatory networks and their associated upstream TFs.
MATERIALS AND METHODS
Patient samples
ESCC primary tumors and adjacent nonmalignant esophageal samples were collected from 235 ESCC patients following surgical resection between November 2007 and January 2011 at the Shantou Central Hospital. None of the patients had distant metastasis. The pathological stage was confirmed by pathologists according to the 8 th Edition of the American Joint Committee on Cancer (AJCC) Tumor-Node-Metastasis (TNM) Staging System (20). Ethical approval was obtained from the Ethics Committees of the Central Hospital of Shantou City and the Medical College of Shantou University.
Human cell lines
The source of cell lines was described in our previous studies (21–23). These cell lines were maintained at 37°C in 5% CO2 in either RPMI-1640 or DMEM medium supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 mg/ml).
Animal xenograft experiment
A total of forty-eight 4-week-old BALB/c-nude mice were obtained from the Experimental Animal Center of the Chinese Academy of Science (Guangzhou, China). Housing and all experimental procedures were approved by the Institutional Animal Care and Use Committee of Sun Yat-Sen University. After random allocation, 2 × 106 TE5-Scramble or TE5-DLX5-silencing cells, and 5 × 106 CAL27-Scramble or CAL27-DLX5-silencing cells were injected subcutaneously into the right flanks of nude mice. The tumors were measured every 4 days, and tumor volumes were calculated using the following formula: volume = length × width2/2. After 28 days, all mice were sacrificed, and tumor tissues were dissected for analyses.
RNA extraction, cDNA synthesis and qRT-PCR analysis
Total RNA was extracted from cell lines using Trizol reagent and used for cDNA synthesis and real-time PCR. The high-capacity cDNA reverse transcription kit (TaKaRa, Japan) was applied to generate complementary DNA. The SybrGreen-based real-time PCR was performed to measure gene expression. Relative expression levels were calculated using the ΔΔCt method and normalized to the level of GAPDH. Primer sequences are provided in Supplementary Table S1.
Western blotting and co-immunoprecipitation (Co-IP)
Cells were lysed with a buffer for protein extraction on ice. The Bradford method was used to measure the protein concentrations of the total lysates. Western blotting was performed using SDS-PAGE followed by conventional wet gel transfer. Nitrocellulose membrane (Bio-Rad) was incubated with primary antibodies overnight at 4°C and incubated with secondary antibodies for 1 h at room temperature. The nitrocellulose membrane was visualized using chemiluminescence kit (Thermo Fisher Scientific). 500 μg whole-cell lysate (for each experiment) were incubated with primary antibody or IgG, followed by IP on the rotary agitation overnight at 4°C. Upon incubation with Dynabeads Protein G (Thermo Fisher Scientific) for 5 h at 4°C, purification and Western blotting were performed using indicated antibodies. Antibodies used in this study are listed in Supplementary Table S2.
Plasmids, siRNAs and transfection
A myc-tagged full-length human DLX5 expression vector was purchased from BGI. Inc (China). The shRNAs were generated by inserting double-stranded oligonucleotides into pLKO.1-TRC (Addgene#10878). Retroviruses were generated through cotransfection with psPAX2 (Addgene #12260) and pMD2.G (Addgene#12259) into 293T cells using PEI transfection reagent. After 24 h, the supernatant was collected. Puromycin at 2 μg/ml was used for selecting cells expressing shRNAs. Bio-T transfection reagent (Bioland Scientific) and Lipofectamine RNAiMAX (Thermo Fisher Scientific) were used for transfection of DNA vectors and siRNAs, respectively. The sequences of shRNA and siRNA are listed in Supplementary Table S3.
Luciferase reporter assay
For the luciferase reporter assay, enhancer elements were cloned into firefly luciferase reporter vector pGL3-Promoter (Promega), and promoter regions were cloned into pGL3-Basic (Promega). A Renilla luciferase plasmid was used as a normalization control. Luciferase reporter activity was measured by the Dual-Luciferase Reporter Assay System (Promega) using a Luminometer (Promega). The primers used for cloning were listed in Supplementary Table S4.
Cell proliferation, colony formation and migration assays
Cells were seeded in 96-well plates (2000–5000 cells/well) and measured by CCK8 (Cell Counting Kit-8) staining. For colony formation assay, 500–1500 cells were seeded in 12-well plates and cultured for 2 weeks. After fixation and staining with 1% crystal violet, the number of colonies was counted. Transwell chambers were used to assess cell migration. 1 × 105–2 × 105 cells were plated on the top chamber with serum-free medium, and medium containing 10% FBS was used in the lower chamber. After 24 hours, migrated cells were fixed, stained with crystal violet, and photographed under a microscope. Five different fields of each well were counted and each sample was assayed in triplicate. The number of cells normalized with the control group, which presented the results in the form of relative multiples.
Immunohistochemistry (IHC)
IHC staining was performed using a polyclonal antibody against DLX5 (1:800, 10592-1-AP, Proteintech). The number of DLX5-positive cells was detected by a multispectral section automatic analysis system (Perkin Elmer, USA). Vectra 2.0.8 was performed for automatic image acquisition and generation. Nuance 3.0 software was used to build the spectral library and Inform 1.2 software was applied to score and analyze the image. DLX5 protein level was measured by an IHC score, which was calculated as the multiplication of the percent of positively stained cells and staining intensity using the formula: (% of 0)X0 + (% of 1+)X1 + (% of 2+)X2 + (% of 3+)X3.
Chromatin immunoprecipitation sequencing (ChIP-Seq) and ChIP-qPCR
ChIP was performed using a previously described protocol (22–24). Briefly, cells were cross-linked with 1% formaldehyde and then neutralized by 1.25M glycine. Cells were sonicated in a Bioruptor Sonicator (Diagenode, USA), followed by cell lysis. Sonicated lysates were incubated overnight at 4°C with indicated antibodies and then were incubated with magnetic beads for an additional 4 h. DNAs were eluted from precipitated immune-complex and sequenced in an Illumina Hiseq4000 (Illumina, USA) at BGI Inc. (China). Some of them was then validated by ChIP-qPCR. Primer sequences are provided in Supplementary Table S5.
RNA-Seq data analysis
The Kallisto pseudo aligner was used for the alignment of 50bp reads to reference the human hg19 Ensemble (V82) transcriptome. The Tximport Bioconductor package was used for gene-level analysis by measuring transcript abundances and counts summarization. DESeq/DESeq2 was used for differential gene expression analysis. Expressed genes (with average FPKM over 0.5) were used for GSEA analysis (25).
ChIP-Seq analysis
For ChIP-Seq analysis, sequencing reads were aligned to reference human genome (GRCh37/hg19) with BWA (0.7.12), with the removal of reads within the ENCODE blacklisted regions (26). Peaks were identified with MACS2, using parameters –bdg –SPMR –nomodel –extsize 200 -q 0.01 (27). Bigwig files were generated by bamCompare in DeepTools (v3.1.3) using parameters –binSize 10 –numberOfProcessors 5 –scaleFactorsMethod None –normalizeUsing CPM –ignoreDuplicates –extendReads 200 (28). Motifs were identified by Hypergeometric Optimization of Motif EnRichment (HOMER), using parameters -size 200 -len 8,10,12. The plotHeatmap and plotProfile functions were used for the co-binding analysis.
ELMER analyses
The ELMER program was performed as described previously (18). Briefly, HM450 array data from 4 TCGA cohorts (ESCC, HNSCC, LUSC and CESC) were downloaded and processed with SeSAMe (29), and their matched RNA-Seq data were downloaded from GDC (https://portal.gdc.cancer.gov/). Unsupervised analysis model was performed for the comparisons between tumor and matched normal samples from each cancer type. We used 160 944 probes that are >2 kbp from any TSS as annotated by GENCODE 28. ELMER version 2.9.5 was used with the following parameters: get.diff.meth(sig.diff) = 0.3, get.diff.meth(P_value) = 0.01, get.diff.meth (minSubgroupFrac) = 0.2, genome of reference = hg19, get.pair(Pe) = 10–3, get.pair(raw.pvalue) = 10–3 and get.pair(filter.probes) = TRUE, get.pair(permu.size) = 10 000, get.pair(minSubgroupFrac) = 0.4, get.enriched.motif(lower.OR) = 1.1, get.TFs(minSubgroupFrac) = 0.4. TF subfamilies were inferred from the TFs with the most significant anti-correlation scores. Using the classification from TFClass (30), candidate TFs were next identified within the TF subfamily (FDR q < 0.05).
Statistical analyses
Student's t-test and Mann–Whitney U test were used for the comparison of two groups. Categorical data were compared using chi-square test or Fisher's exact probability test. Correlation analysis was performed using Pearson coefficient. Univariate and multivariate Cox regression models were used to determine the hazard ratio (HR) of different variables. Statistical analyses were performed using GraphPad Prism software. For all tests, P values were two-sided; P < 0.05 was considered as statistically significant.
RESULTS
ELMER identifies novel hyperactive TFs across different types of SCC
As introduced earlier, to infer the transcriptional activity of TFs, ELMER calculates the methylation level of enhancer regions containing putative TFBSs of TFs and analyzes its correlation with the expression of corresponding TFs. We applied ELMER to four common forms of SCC (ESCC, HNSCC, LUSC and CESC) to identify cancer-specific TFs with higher transcriptional activity in tumors than adjacent normal samples. We analyzed paired RNA-Seq and DNA methylation array data from TCGA SCC samples, including ESCC (81 tumors versus 9 normal tissues), HNSCC (500 tumors versus 20 normal tissues), LUSC (370 tumors versus 7 normal tissues) and CESC (303 tumors versus 3 normal tissues).
Compared to adjacent normal tissues, a total of 44 hyperactive TFs were identified to be more active in different SCCs (FDR q value < 0.05, Figure 1A). Among them, many known master regulatory transcription factors (MRTFs) were found, such as TP63, SOX2, NFE2L2, GRHL2 (31), PITX1 (32) and FOSL1 (33), highlighting the capability of ELMER in discovering cancer-specific TFs. Showing TP63 as an example, its mRNA expression was significantly anti-correlated with the methylation level of enhancer regions containing its putative binding motif (Figure 1B). Specifically, SCC tumor samples had higher TP63 expression and lower methylation levels of its targeting enhancers relative to adjacent normal tissues (Supplementary Figure S1A, B).
Figure 1.
Identification of DLX5 as a hyperactive TF in SCCs. (A) Heatmap showing the q value of all enriched hyperactive TFs identified by the ELMER program (FDR q < 0.05). The novel candidates occurring in three SCC types are marked with red font. (B) Scatter plots comparing TP63 mRNA expression and DNA methylation at regions with predicted TP63-binding motif in CESC, ESCC and LUSC cohorts from TCGA. (C) The mRNA expression of DLX5 across 33 types of human cancers from TCGA. Each point represents one sample. SCC tumor samples are red, and EAC tumor samples are purple. (D) Relative cell viability of KYSE140, KYSE450 and KYSE70 cells upon knockdown of candidate TFs. SOX2 and TP63 were used as positive control. siMixture, siSOX2 + siTP63 + siDLX5.
On the other hand, nine TFs were shared by three different types of SCC, suggesting that different SCCs indeed exhibit similarity in the usage of TFs. Among them, three TFs (TP63, GRHL2, and FOSL1) have established functions in SCCs, while the biological functions of the remaining six (DLX5, ZFP64, HOXA11, HOXA10, POU6F2 and HOXD13) are hitherto unknown in SCCs. We next focused on these six candidates since this work was aimed to characterize hyperactive TFs shared by different SCCs.
Because cancer-specific TFs are often expressed in a cell-type-specific manner, we initially interrogated the TCGA RNA-Seq datasets containing 33 types of human cancers to determine the expression patterns of these six candidates. In general, except for HOXA10 and HOXA11, the other four candidates (DLX5, HOXD13, POU6F2 and ZFP64) showed higher SCC-specificity, with top 3 highest-expressed tumor types containing two different SCCs (Figure 1C; Supplementary Figure S1C-G). Next, as an initial loss-of-function screen, cell proliferation assay was performed focusing on DLX5, HOXD13, POU6F2 and ZFP64 with SOX2/TP63 as a positive control. Because all of these four genes were expressed at the highest levels in ESCC (Figure 1C; Supplementary Figure S1C-G), three different ESCC cell lines (KYSE140, KYSE450 and KYSE70) were used for screen (Supplementary Figure S1H). Compared with scramble control, silencing of either DLX5 or POU6F2 (but not HOXD13 or ZFP64) inhibited the proliferation of ESCC cells (Figure 1D). Interestingly, combined DLX5/SOX2/TP63 depletion had a greater effect than any single function of TF on the proliferation of ESCC cell lines (Figure 1D), which further illustrated the important role of DLX5 in ESCC growth. DLX5 was prioritized for further investigation, since (i) its knockdown produced the strongest anti-proliferative effect on all three ESCC cell lines (Figure 1D); (ii) it showed the most specific expression pattern in all four types of SCCs (Figure 1C; Supplementary Figure S1C–G). In the following biological and mechanistic studies, we focused primarily on ESCC as a representation of pan-SCC since DLX5 was expressed at the highest level in this type of SCCs, and performed validation experiments on other types of SCCs.
DLX5 is uniquely up-regulated, amplified and associated with poor survival in SCC patients
As expected, compared with adjacent normal tissues, the DLX5 mRNA level was significantly up-regulated in ESCC, HNSCC, and LUSC tumors (Figure 2A). To determine whether the up-regulation of DLX5 mRNA led to its protein over-expression, immunohistochemistry (IHC) was performed. We focused on ESCC because DLX5 was expressed at the highest level in this type of SCCs (Figure 1C). In a discovery cohort of 27 matched samples, DLX5 protein was significantly up-regulated in ESCC compared with adjacent normal esophageal epithelium (Figure 2B, C). The majority of DLX5 protein was observed in the nucleus, consistent with its reported function in transcriptional regulation. Notably, Kaplan-Meier analysis showed that higher DLX5 protein expression was significantly correlated with shorter overall survival (Figure 2D). To further verify the results, a larger sample cohort of 235 ESCC patients was used. Notably, the significant association was validated in the independent large sample cohort (Figure 2E, F). Using this much larger cohort, we performed multivariate Cox regression and identified the expression of DLX5 protein as an independent predictor of both overall survival (HR = 1.65; 95% confidence interval, 1.18–2.29; P < 0.01) and disease-free survival (HR = 1.53; 95% confidence interval, 1.10–2.12; P < 0.05, Supplementary Table S6).
Figure 2.
DLX5 is uniquely up-regulated, amplified and associated with poor survival in SCC patients. (A) Expression levels of DLX5 mRNA in TCGA normal and tumor samples in four types of SCCs. Each point represents one sample. (B, C) Representative IHC photos (B) and IHC scores (C) of DLX5 protein expression in ESCC. Scale bars: 50 μm. (D) Kaplan–Meier plots of the association between DLX5 protein expression and overall survival of 27 ESCC patients (Discovery cohort). (E, F) Kaplan–Meier plots of DLX5 protein expression with either overall (E) or disease-free survival (F) in 235 ESCC patients (validation cohort). (G) The segmented copy number profiles of chromosome 7 across four types of SCCs from TCGA. The DLX5 locus is marked with an arrow head. (H) Pearson correlation between DLX5 copy number and its mRNA expression. (I) Expression levels of DLX5 mRNA in TCGA SCC samples stratified based on gene dosage. ** P < 0.01; *** P < 0.001; ns, not significant.
To explore the mechanisms underlying DLX5 over-expression in SCCs, we first observed various degrees of copy-number gains (defined as log2 copy number (tumor/normal) > 0.5) (34) at DLX5 locus (Chr7q21.3) in SCCs (from 2.07% to 16.49% in different SCCs, Figure 2G). As anticipated, a significant positive correlation was found between DLX5 expression and its copy number in SCC samples (Figure 2H). Nevertheless, we reasoned that copy-number gains only accounted partially for the over-expression of DLX5 mRNA in SCCs. This is because relative to diploid samples, the mRNA abundance of DLX5 was increased by 1–2-fold in DLX5-amplified samples (Figure 2I); however, when compared with adjacent normal tissues, the DLX5 mRNA levels were up-regulated by 3–32-fold in SCCs (Figure 2A), suggesting that additional epigenomic mechanism(s) may further heighten the transcription of DLX5 in SCCs.
Bifurcate re-configuration of DLX5 bivalent promoter in cancer
In searching for potential epigenomic mechanism(s) regulating DLX5 transcription in SCCs, we unexpectedly noted that the DLX5 promoter and its flanking region were maintained in a ‘bivalent’ state across many different cell types profiled by the Roadmap consortium (35), including embryonic stem cells (ESCs) and esophagus epithelial cells (Figure 3A). In normal cells, bivalent chromatin has both repressive (H3K27me3) and active (H3K4me1/3) histone markers and is devoid of DNA methylation. Initially being discovered in ESCs, bivalent promoters are enriched in genes encoding developmental regulators (e.g. HOX genes) (36). The proposed physiological function of bivalent state is to allow gene rapid activation while maintaining repression in the absence of differentiation signals. Notably, it is well-established that promoters undergoing de novo DNA hypermethylation and transcriptional silencing in cancer cells are strongly enriched in bivalent genes (37–40). In other words, bivalent genes are highly prone to DNA hypermethylation and transcriptional repression during tumorigenesis. Therefore, it was intriguing that being accompanied by a bivalent promoter, the expression of DLX5 was up-regulated rather than repressed in SCCs.
Figure 3.
Chromatin bifurcate re-configuration of bivalent DLX5 promoter in cancer. (A) The chromatin states of ±1000 bp region of DLX5 TSS and its matched mRNA expression (bottom track) across 57 normal cell types from the Roadmap project. TssA, active TSS; TssAFlnk, flanking Active TSS; TxFlnk, transcription at gene 5' and 3'; Tx, strong transcription; TxWk, weak transcription; EnhG, genic enhancers; Enh, enhancers; ZNF/Rpts, ZNF genes & repeats; Het, heterochromatin; TssBiv, bivalent/poised TSS; BivFlnk; flanking bivalent TSS/Enhancer; EnhBiv, bivalent enhancer; ReprPC, repressed PolyComb; ReprPCWk, weak repressed PolyComb; Quies, quiescent/Low. (B) A pie chart of chromatin states for DLX5 promoter summarized from panel (A). (C) ChIP-Seq, WGBS and ATAC-Seq profiles across different cell types and tissues at DLX5 gene locus. ATAC-Seq data were from TCGA samples. ChIP-Seq profiles in embryonic stem cells and normal esophageal epithelium were from the Roadmap project. The rest of the data were either generated in this study (GSE142863) or from our internal samples we published recently (GSE106563 and GSE132686) (22,23). The scales of the top 10 tracks are shown; the scales of all WGBS tracks are 0–1 (β value); the scales of all ATAC-Seq tracks are 0–180 (CPM); the scales of all H3K27ac ChIP-Seq tracks of esophageal cancer cell lines are 0–7 (RPKM). (D) The ChIP-qPCR for H3K4me3 and H3K27me3 at the DLX5 promoter loci. Two different primer sets designed at the DLX5 gene promoter region, and NC was used as the negative control. Data are mean ± SD from three replicates per group. (E) A proposed model of chromatin configuration of DLX5 promoter from embryonic stem cells to normal esophageal epithelial cells, and then to either EAC or ESCC cells. (F) DNA methylation β values and (G) ATAC-Seq peak values of DLX5 promoter (–100, +1000 bp of TSS) across various TCGA cancer types with available WGBS data. CPM, counts per million.
In-depth examination showed that across all 57 cell types (with matched RNA-Seq and epigenomic data) profiled by the Roadmap consortium, 35 (60.8%) exhibited bivalent states at DLX5 TSS and its flanking regions (bivalent TSS, flanking bivalent TSS, and bivalent enhancer) and 14 (24.9%) displayed repressed states (Repressed PolyComb and Weak Repressed PolyComb), while only 6 (10.8%) showed active TSS (Figure 3B). This chromatin profile was consistent with the low expression of DLX5 mRNA in the majority of normal cell types (Figure 3A). Specifically, the RPKM value of DLX5 mRNA was <1 in 44 (77.2%) samples, with 33 (57.9%) having undetectable DLX5 expression (RPKM < 0.1). As a reference sample shown earlier (Figure 2A), the RPKM value of DLX5 was as low as 1.39 in esophageal epithelial cells. Moreover, independent large-scale transcriptome analyses of 61 normal samples (including projects of Human Protein Atlas, The Genotype-Tissue Expression, and Functional Annotation of Mammalian Genomes 5) also showed that 54 (88.5%) samples, including esophagus, had comparably low DLX5 mRNA (Supplementary Figure S2).
Using ESCs, normal esophageal epithelial cells, esophageal adenocarcinoma (EAC) and ESCC as representative cell types, we analyzed histone modification marks and further confirmed the bivalent chromatin re-configuration of DLX5 promoter (H3K4me3+/H3K27me3+) in these cell types (top 10 tracks, Figure 3C). We next analyzed our internal previously published WGBS and H3K27ac ChIP-Seq datasets generated from both normal and cancerous esophageal samples (21–23). Congruent with the bivalent state we observed in normal esophagus from the Roadmap consortium, DLX5 promoter was weakly methylated in our internal normal esophagus samples (11–15 tracks, Figure 3C). However, it became hypermethylated in EAC, while remained hypomethylated in ESCC (16–32 tracks, Figure 3C). Additionally, our internal ChIP-Seq/PCR of ESCC cells in this study showed that the H3K4me3 mark was highly enriched at the DLX5 promoter, while H3K27me3 was absent in ESCC (Figure 3C, D; Supplementary Figure S3A). In comparison, both H3K4me3 and H3K27me3 marks were nearly absent in EAC (Figure 3C, D). Consistently, H3K27ac signals at DLX5 promoter (indicative of active transcription) were undetectable in EAC cells but conspicuous in ESCC cells (bottom 18 tracks, Figure 3C). Indeed, ATAC-Seq results showed that the DLX5 promoter was completely inaccessible in TCGA EAC samples; by contrast, it had noticeable accessibility in ESCCs (Figure 3C). Further substantiating these epigenomic profiles, the DLX5 expression was 24-fold higher in ESCC than EAC samples, which we showed earlier (Figure 1C).
Together, these epigenomic characterizations reveal a notable bifurcate re-configuration of DLX5 promoter in cancer: DLX5 maintains a bivalent chromatin state in most normal tissue types, including both ESCs and normal esophagus epithelium; it becomes hypermethylated in EAC, which is consistent with the established feature of bivalent genes that are highly susceptible to DNA hypermethylation in cancer (38–40); in stark contrast, DLX5 promoter not only remains hypomethylated but also gains active histone markers and becomes transcriptionally activated in ESCC (Figure 3E).
To verify the bi-directional re-configuration of DLX5 promoter is not restricted to esophageal cancer, we next extended the epigenomic analyses of TCGA cancer types with available WGBS data. Notably, DLX5 promoter was hypermethylated in EAC, rectal, gastric, and colon cancers (β value > 0.25), and had the lowest methylation in bladder cancer, LUSC, and ESCC (Figure 3F; Supplementary Figure S3B). Consistent with these data, DLX5 was previously reported to be DNA hypermethylated in osteosarcoma (37). Further analysis of the ATAC-Seq of the above cancer types showed that bladder cancer, LUSC, and ESCC had the highest chromatic accessibility of DLX5 promoter (Figure 3G). Notably, a significant fraction (>35%) of bladder cancers are squamous cell type (41). Indeed, DLX5 mRNA was also highly expressed in bladder cancer (Figure 1C). Therefore, these data not only validate the bifurcate re-configuration of DLX5 bivalent promoter in other cancer types, but also suggest that its epigenetic conversion is lineage-specific since both of its epigenomic activation and overexpression are unique in SCCs.
SOX2 directly activates DLX5 in SCC by regulating its distal enhancer
Prompted by the finding of epigenomic bifurcate re-configuration of DLX5, we next sought to elucidate the mechanistic basis of its transcriptional activation in SCC cells. ChIP-Seq data showed that in addition to promoter, multiple distal regions of DLX5 had discernible H3K27ac signals, implying that they might function as putative enhancers (blue track, Figure 4A). To evaluate whether any of these distal regions have regulatory effect on DLX5 transcription, we re-analyzed a recent TCGA dataset which establishes ‘transcriptional links’ between distal regulatory elements and target genes, based on large-scale profiling of chromatin accessibility using ATAC-Seq with matched RNA-Seq across 23 cancer types, including ESCC (n = 11), HNSCC (n = 10), LUSC (n = 16) and CESC (n = 2) (42). Notably, within 500 kb of DLX5 TSS, a total of four distal transcriptional links for DLX5 expression were identified, all of which were located within the putative enhancer regions that had high H3K27ac signals (Figure 4A). As expected, being a positive control, the accessible degree of DLX5 promoter was positively correlated with its expression level (Figure 4A). We additionally performed ATAC-Seq on three different ESCC cell lines (KYSE140, TE5 and TT) and found that these putative DLX5 enhancers had comparable chromatin accessibility between our internal SCC cell lines and TCGA SCC patient samples (Figure 4A). These results suggested that these SCC cells could serve as a valid in vitro model for the epigenomic investigation of DLX5. To further determine whether these putative enhancers had functional impact on DLX5 transcription, we individually cloned each of them as well as the DLX5 promoter region (positive control) and performed luciferase reporter assay. Results showed that the reporter activity was significantly higher in all four enhancer-containing vectors compared with the control vector, with Enhancer-1 exhibiting the highest activity in both KYSE140 and TE5 cells (Figure 4B).
Figure 4.
SOX2 directly activates DLX5 in SCC by regulating its distal enhancer. (A) Various ATAC-Seq and ChIP-Seq profiles of DLX5 gene locus. Scatter plots showing the Pearson correlation of ATAC-Seq peaks versus DLX5 mRNA expression. ATAC-Seq data of SCC patient samples were from TCGA. SOX2 ChIP-Seq of HCC95 and SCC25 cells were from GSE104137 and GSE46837 (32,43). The rest of the data were either generated in this study (GSE142863) or from internal samples we published recently (GSE106563) (23). (B) The luciferase reporter activities of DLX5 enhancers and promoters measured in KYSE140 and TE5 cells in either the presence or absence of SOX2-silencing. Data are mean ± SD from three replicates per group. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Motif enrichment analysis of DLX5 enhancer and promoter regions as shown in panel (A). (D) Venn diagram of significant correlation (R > 0.3, P < 0.001) between the expression of SOX3, SOX10, SOX2, SOX6, SOX4 and DLX5 in four types of SCCs. (E) Relative expression of DLX5 and TP63 mRNA following silencing of SOX2 in KYSE140 and TE5 cells. Data are mean ± SD from three replicates per group. *P < 0.05; **P < 0.01; ***P < 0.001. (F) Western blotting of DLX5 and TP63 protein in KYSE140 and TE5 cells upon silencing of SOX2.
To identify upstream TF(s) regulating these distal enhancers and promoters, we performed motif enrichment analysis of these regulatory regions. Notably, the top-ranked consensus motifs were dominated by the SOX family, including SOX3, SOX10, SOX2, etc (Figure 4C). This result was encouraging given that SOX2 is one of the most well-defined oncogenic MRTFs promoting SCCs (43,44), which was also confirmed by our ELMER program (Figure 1). Since different members of a TF family can recognize highly similar or even identical motifs (e.g. SOX and GATA family), we speculated that other SOX family members were also identified likely due to the sequence similarity of their consensus motif. Importantly, the expression of DLX5 was mostly strongly correlated with that of SOX2 across 4 types of SCC samples (Figure 4D; Supplementary Figure S4A), suggesting that SOX2 is probably the truth hit of the motif analysis.
To validate this, we determined the regulation of SOX family members present in motif analysis on DLX5 transcription. Notably, the mRNA of DLX5 was unaffected upon individual knockdown of SOX3, SOX4, SOX6 and SOX10 in ESCC cell lines (Supplementary Figure S4B). However, both the mRNA and protein levels of DLX5 were significantly and specifically down-regulated when silencing SOX2 in SCC cell lines (Figure 4E, F; Supplementary Figure S5A, B). On the other hand, overexpression of SOX2 up-regulated the expression of DLX5 in SCC cell lines (KYSE150 and HSC3, Supplementary Figure S5B, C). To determine whether SOX2 is sufficient to drive the expression of DLX5 in other cell types, we enforced expression of SOX2 in esophageal epithelial cell lines (HET-1A) and esophageal adenocarcinoma cell lines (OE33). Notably, SOX2 only up-regulated the expression of DLX5 in esophageal epithelial cell lines (Supplementary Figure S5C). These results indicated that SOX2 activates DLX5 in esophageal epithelial cell lines and SCC cell lines, but not in adenocarcinoma cell lines. Indeed, there was no correlation observed between the expression of SOX2 and DLX5 in TCGA EAC and COAD samples (Supplementary Figure S5D). Moreover, the H3K27ac signals at both DLX5 enhancer and promoter regions were absent in OE33 and Lovo cell lines (Supplementary Figure S5E), congruent with their closed chromatin state in EAC and COAD (Figure 3G; Supplementary Figure S5E).
Furthermore, we analyzed our internal SOX2 ChIP-Seq data generated in three different ESCC cell lines (TE5, TT, and KYSE70) as well as publicly-available SOX2 ChIP-Seq data of an LUSC (HCC95) (43) and an HNSCC cell line (SCC25) (32). Indeed, SOX2 occupied two out of the four distal enhancers of DLX5 (Enhancer-1 and -2, Figure 4A). Moreover, the depletion of SOX2 markedly reduced the reporter activity of Enhancer-1 (Figure 4B). Consistently, the enrichment of H3K27ac at Enhancer-1 region was enhanced after overexpression of SOX2, while significantly reduced upon SOX2 knockdown in SCC cell lines (Supplementary Figure S5F-H). Considering TP63 is an important MRTF in SCCs (23,43), we further examined the regulatory effect of TP63 on DLX5 expression. We knockdown TP63 in ESCC cell lines (KYSE140 and TE5) by using two different shRNAs (one of them targets all TP63 transcripts). Unfortunately, the DLX5 expression was not down-regulated upon knocking down TP63 in ESCC cell lines (Supplementary Figure S6A). Subsequently, we overexpressed TP63 (ΔNp63, the predominantly expressed isoform in SCC) in SCC (HSC3 and KYSE150) cell lines and found that endogenous expression of TP63 did not significantly activate DLX5 gene expression (Supplementary Figure S6B). These results suggested that TP63 did not activate DLX5 in SCC. Indeed, all TP63 ChIP-seq of four SCC cell lines showed that there were no TP63 binding sites in the Enhancer-1 (an enhancer with regulatory effect confirmed above) and promoter region of DLX5 (Figure 4A; Supplementary Figure S6C). Also, knockdown DLX5 had no effect on SOX2 and TP63 expression in KYSE140 and TE5 cell lines (Supplementary Figure S6D), suggesting DLX5 did not form feedback regulation with SOX2/TP63.
These results demonstrate that SOX2 directly activates DLX5, a bivalent gene, in SCC cells by binding to its distal enhancer elements. This finding supports previous studies on the activation of bivalent factors during normal cell developmental process, which similarly entails the binding of lineage-specifying TFs to distal enhancers of target bivalent genes (such as MYOD1) (36,45).
DLX5 contributes to the viability and migration of SCC cells
Having established the mechanistic basis of DLX5 overexpression specifically in SCC, we next explored its functional significance in SCC biology. To this end, we accrued a total of 12 SCC cell lines from ESCC, HNSCC, and LUSC, and selected 10 cell lines exhibiting high DLX5 expression (relative to cell lines from EAC, STAD or COAD, Supplementary Figure S7A) for loss-of-function assays by using two independent shRNAs (Supplementary Figure S7B). Importantly, silencing of DLX5 by different shRNAs strongly impaired cell viability, migration and clonogenic capacity in all 10 SCC cells (Figure 5A–C; Supplementary Figure S8A-E). Some of the cell lines were randomly selected for validation using siRNA-mediated knockdown (Supplementary Figure S8F–H). Moreover, anchorage-independent growth assays showed that DLX5-knockdown markedly diminished both the size and number of SCC colonies in soft-agar (Figure 5D; Supplementary Figure S8I). Unfortunately, overexpression DLX5 did not promote cell proliferation and migration in SCC cell lines (Supplementary Figure S9A–C). In order to rule out off-target effects of RNA-interference, rescue assays were performed wherein an siRNA/shRNA against DLX5-3’UTR region and the open-reading-frame (ORF) of DLX5 were co-transfected into SCC cells (Supplementary Figure S7C). Indeed, DLX5 restoration significantly reversed siRNA/shRNA-mediated inhibition of cell proliferation and migration (Figure 5E, F; Supplementary Figure S9D, E). Besides, the function of DLX5 on cell proliferation and migration was further confirmed in primary cells from two HNSCC patients (Supplementary Figure S10). Furthermore, to confirm the functional contribution of DLX5 to SCC proliferation in vivo, ESCC and HNSCC xenograft tumor models were utilized. Importantly, silencing of DLX5 inhibited potently the growth of SCC xenografts in vivo (Figure 5G–I). These results together demonstrate that DLX5 contributes functionally to the proliferation, migration, and anchorage-independent growth of SCC cells.
Figure 5.
DLX5 is required for the proliferation, migration and anchorage-independent growth of SCC cells. (A) Short-term cell proliferation, (B) colony formation and (C) cell migration of various SCC cell lines upon DLX5 knockdown with two different shRNAs. Data are mean ± SD from three replicates per group. *P < 0.05; **P < 0.01; ***P < 0.001. (D) Soft-agar colony growth of KYSE140 and KYSE70 cells stably expressing either control shRNA (Scramble) or DLX5 shRNAs. Bars show the mean numbers of colonies (data are mean ± SD from three replicates per group). *P < 0.05; **P < 0.01. Rescue assays of (E) cell proliferation curves and (F) transwell migration of KYSE140 and TE5 cells from indicated groups. NC, Negative control; siUTR, siRNA against DLX5-UTR; VC, vector control; siUTR + DLX5, siRNA against DLX5-UTR region together with DLX5 expression vector. Data are mean ± SD from three replicates per group. *P < 0.05; **P < 0.01; ***P < 0.001. (G) Growth curves of xenograft tumors, (H) tumor images and (I) tumor weight measured at end point. n = 8. ***P < 0.001; ****P < 0.0001.
DLX5 interacts and cooperates with TP63 to control both distal and proximal regulatory elements
To elucidate molecular mechanisms underlying the functional contribution of DLX5 in SCCs, ChIP-Seq of DLX5 was performed to identify its genomic targets in both KYSE140 (ESCC) and CAL27 (HNSCC) cell lines. DLX5-binding peaks identified in these two cell lines were significantly and strongly overlapped and had a similar distribution (Supplementary Figure S11), consistent with our findings that DLX5 shared functions in different SCC cell types. Notably, in both cell lines, the sequence motifs most significantly enriched in DLX5-peaks were from the TP53/TP63 family (Figure 6A). Since TP63 is the best characterized SCC-specific MRTF (23,43), this result implies that DLX5 and TP63 may co-occupy genomic regions in SCC cells. Notably, co-immunoprecipitation (Co-IP) identified protein-protein interaction between endogenous DLX5 and TP63 using either TF as the ‘bait’ (Figure 6B), further suggesting the direct transcriptional cooperation between these two factors in SCCs.
Figure 6.
DLX5 interacts and cooperates with TP63 to regulate both distal and proximal regulatory elements. (A) Top 3 ranked motifs of DLX5 ChIP-Seq in KYSE140 and CAL27 cells. (B) Protein–protein interaction of DLX5 and TP63 as detected by Co-IP. (C) Heatmaps showing peaks of DLX5, TP63, H3K27ac and H3K4me3 ChIP-Seq in KYSE140 cells. Peaks were stratified into either TSS-proximal (<1.5 kb of TSS) or TSS-distal (>1.5 kb of TSS) groups and further grouped into either DLX5 solo-binding or DLX5/TP63 co-binding regions. Lines, peaks. (D–G) Line plots showing the average peak density of indicated factors in indicated genomic regions. (H) The percentage of down-regulated genes (Log2 fold change < –0.25) upon DLX5-knockdown in indicated groups. ***P < 0.001. (I) GSEA of differentially expressed genes upon DLX5-knockdown versus TP63-knockdown.
To explore the possible transcriptional cooperation between DLX5 and TP63, we analyzed their occupancy profiles generated identically from the same cell line (KYSE140). Validating the motif enrichment results, a total of 1939 (15.8%) DLX5-occupied genomic regions also had TP63-peaks in their vicinity (Figure 6C). To understand the impact of their cooperation on the epigenome, we compared chromatin markers of DLX5-occupied regions with either the presence or absence of TP63 co-binding. Among a total of 8473 TSS-distal regions occupied by DLX5, the intensity of H3K27ac was almost undetectable in DLX5 solo-binding regions (n = 7195, Figure 6C, D); in stark comparison, H3K27ac signals were markedly increased in regions with TP63 co-binding (n = 1278, Figure 6C, D). Similarly, in TSS-proximal regions, H3K27ac signals in DLX5/TP63 co-binding regions were also considerably higher than those in DLX5 solo-binding regions, albeit the magnitude of difference was less pronounced than that in distal elements (Figure 6C, E). There was not much of a difference in the signal of DLX5 itself with or without TP63 co-binding (Figure 6F), indicating that loading of DLX5 to the genome per se does not require assistance from TP63. As anticipated, the intensity of TP63 peaks in distal regions was significantly higher than that in TSS-proximal regions (Figure 6G).
To determine the effect of DLX5/TP63 co-binding on gene expression, we integrated RNA-Seq data generated upon DLX5 knockdown in the same KYSE140 cells. Notably, silencing of DLX5 led to downregulation of a significantly larger number of genes associated with co-binding than with solo-binding elements in both distal and proximal regions (Figure 6H). These data suggest that DLX5 cooperates with TP63 in the regulation of both TSS-distal and -proximal elements. To further substantiate this finding, we analyzed RNA-Seq data upon silencing of TP63 in KYSE140 cells. Indeed, gene set enrichment analysis (GSEA) showed that globally, genes downregulated upon DLX5-silencing were significantly enriched in those also reduced following TP63-depletion (Figure 6I). This result was reproduced in another ESCC cell line, TE5 (Figure 6I), reinforcing the notion that DLX5, in cooperation with TP63, positively regulates the activities of both enhancers and promoters in SCC cells.
DLX5/TP63 co-regulate cancer-promoting pathways in SCCs
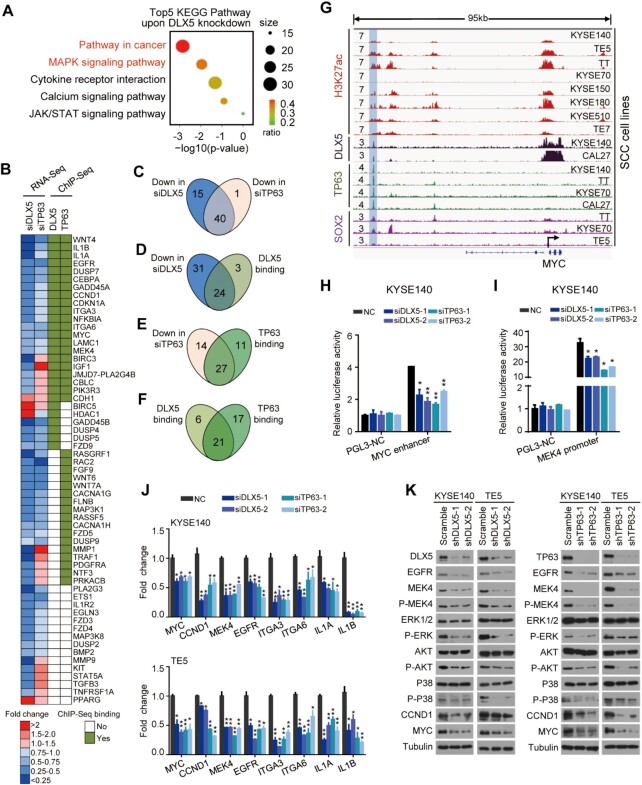
We next investigated target genes and signaling pathways downstream of DLX5 in SCC cells. KEGG pathway analyses of RNA-Seq data upon DLX5 knockdown showed that DLX5-regulated genes were most significantly enriched in ‘Pathway in cancer’ (ranking 1st) and ‘MAPK signaling pathway’ (ranking 2nd) (Figure 7A), congruent with our earlier results that DLX5 contributed to SCC cell viability, migration, and anchorage-independent growth (Figure 5).
Figure 7.
DLX5/TP63 co-regulate cancer-promoting pathways in SCC. (A) KEGG pathway analysis of the differentially expressed genes upon DLX5-knockdown (Log2 fold change > 1 or < –1). Dot size represents the number of genes enriched, and the color of circles denotes the enrichment ratio. (B) Heatmap of enriched genes in either ‘Pathway in cancer’ or ‘MAPK pathway’ upon DLX5-knockdown. (C–F) Venn diagrams showing the number of overlapped genes between indicated ChIP-Seq and RNA-Seq data. (G) IGV plots showing SOX2/TP63/DLX5 co-binding at a distal enhancer region of MYC. (H, I) The relative luciferase activity of MYC enhancer and MEK4 promoter following silencing of either DLX5 or TP63. Data are mean ± SD from three replicates per group. *P < 0.05; **P < 0.01. (J) mRNA expression of DLX5/TP63 co-regulated genes following silencing of either DLX5 or TP63. Data are mean ± SD from three replicates per group. *P < 0.05; **P < 0.01. (K) Western blotting of indicated proteins in KYSE140 and TE5 cells following silencing of either DLX5 or TP63.
Focusing on the components of these top two signaling pathways, we identified that a total of 55 and 41 genes were decreased upon DLX5- and TP63-knockdown, respectively (Figure 7B). Consistent with the regulatory cooperation between DLX5 and TP63, more than 70% of these genes exhibited similar change (that is, down-regulation) following the silencing of either DLX5 or TP63 (Figure 7B, C), suggesting that DLX5 and TP63 co-regulate these pathways in SCC cells. Moreover, ChIP-Seq data showed that 44% (24/55) and 59% (27/41) of the downregulated genes were directly occupied by DLX5 and TP63, respectively (Figure 7B, D, E), validating that these changes in gene expression were a result of the direct transcriptional regulation. Importantly, 77.8% (21/27) of DLX5 binding genes overlapped with those bound by TP63 (Figure 7B, F), reinforcing the notion of cooperative binding of these two TFs. These co-regulated target genes included many cancer-promoting factors with established functions in SCCs, such as CCDN1, MYC, EGFR, MEK4, ITGA6, etc (Figure 7B). An exemplary co-binding peak of DLX5/TP63 was shown in the enhancer region of MYC (Figure 7G), and the transcriptional activity of this element was validated by luciferase reporter assay (Figure 7H). Moreover, depletion of either DLX5 or TP63 significantly reduced the reporter activity of this element (Figure 7H). We also tested and verified an additional co-binding element in another target gene, MEK4 (Figure 7I; Supplementary Figure S12A).
To further confirm the co-regulation of DLX5/TP63, a total of 8 down-regulated genes as well as 4 not co-regulated genes (negative control) from Figure 7B were randomly selected, and their changes were readily verified by RT-qPCR using additional siRNAs targeting either DLX5 or TP63 in different cell lines (Figure 7J; Supplementary Figure S12B). Moreover, reduction in the protein levels of some of the selected genes (EGFR, MEK4, CCND1 and MYC) was confirmed by Western Blotting (Figure 7K). The measurement of the phosphorylation levels of key signaling molecules (ERK, AKT and P38) further verified that MAPK signaling pathway was indeed inhibited in either DLX5- or TP63-knockdown cells (Figure 7K). Since SOX2 is the upstream of DLX5 and TP63 (Figure 4 E, F), we also tested the regulation of SOX2 on DLX5/TP63 co-regulated genes. Obviously, silencing of SOX2 results in a similar change of these target genes as DLX5- or TP63-knockdown (Supplementary Figure S12C, D). Moreover, combinatorial knockdown of SOX2/TP63/DLX5 produced comparable effects with either single gene knockdown in every target gene tested (Supplementary Figure S12C, D). Notably, among the 15 DLX5/TP63 co-regulated genes, three of them were co-occupied by SOX2/DLX5/TP63, including MYC, EGFR, and ITGA6 (Figure 7G), which suggests that SOX2/TP63/DLX5 might co-regulate these genes. Indeed, the abundance of occupancy of H3K27ac at the enhancer of MYC substantially decreased after knocking down any of TP63, SOX2 or DLX5 (Supplementary Figure S12E). Moreover, the abundance of any one at the enhancer region of MYC decreased after knocking down the other two TFs (Supplementary Figure S12E), implying that DLX5, TP63 and SOX2 might form a complex at MYC enhancer. These data together strongly suggest that DLX5/TP63 co-regulate cancer-promoting pathways in SCCs.
DISCUSSION
Unbiased large-scale genomic studies have identified unified genetic lesions across different types of SCC. However, advances in the understanding of genomic alterations have not considerably improved clinical management of SCC patients. Thus, urgent needs still exist to further decipher the biological and pathological basis of SCCs. To this end, we utilized approaches complementary to genomic characterization, and performed epigenomic regulatory analyses across anatomically-distinct SCCs. Specifically, we inferred gene expression networks and their upstream hyper-activated TFs in SCCs using ELMER (18), a bioinformatic method that unbiasedly and systematically reconstructs gene regulatory networks by integrating matched DNA methylome and gene expression data. Particularly, ELMER measures the DNA methylation level of distal enhancers (instead of promoters), which are preferentially regulated by TFs in a cell-type-specific manner. Consistent with genetic changes, our epigenetic results showed that different types of SCC display notable similarity with respect to their hyperactive TFs. Indeed, multiple ELMER-identified TFs (such as TP63, SOX2, NFE2L2 and PITX1) have shared functions across different types of SCC. These results suggest that the ELMER program identifies bona fide SCC-specific TFs. Moreover, the list of candidate TFs discovered in the present study warrant further investigation.
Focusing on the top candidate TFs, we initially observed both genomic and epigenomic activation of DLX5 across different SCCs. Notably, further in-depth epigenomic analyses not only identified a bivalent state of DLX5 promoter in most normal cells, but also characterized complex chromatin re-configuration of this promoter in cancer. Being originally discovered in ESCs and later found in other cell types, bivalent promoters of many development-associated factors (such as the HOX genes) present a unique model of chromatin configuration and histone modifications (36). These bivalent promoters are commonly hypomethylated and marked by co-enrichment of H3K27me3 and H3K4me3, which are generated and maintained by histone ‘writers’ including SET1A/B, MLL and Polycomb Repressive Complex (PRC) (46). As an intermediate state, bivalent promoters are ‘poised’ for either timely activation in specific cell lineages or repression in other cell types. Functionally, bivalency is considered to fine-tune the expression of key factors during tissue development and to prevent unscheduled gene activation, thus contributing to the robustness and preciseness of the developmental process.
In cancer epigenome, independent findings established that bivalent promoters are highly susceptible to de novo DNA hypermethylation, such as those of CDKN2A/B, AXIN2, PAX5 and RUNX3(37–40). Nevertheless, despite that bivalent promoters contribute to over 75% of all hypermethylated promoters in cancers (37), reports have also sporadically shown an opposite alteration for them. For example, loss of the Polycomb mark H3K27me3 from bivalent promoters was found to result in the activation of genes associated with colon cancer progression (47). However, the activation of bivalent promoters in cancers has been seldom studied and poorly understood. The current work reveals that DLX5 represents another such bivalent gene that converts from a poised chromatin state in most normal tissues to an active state in SCCs. Importantly, we further established a bifurcate alteration pattern of DLX5 promoter in cancer, since its change in multiple other cancer types (such as EAC, gastric and colon cancers, and osteosarcoma (37)) is in the opposite direction, which fits the classic paradigm of cancer-associated DNA hypermethylation and transcriptional silencing (Figure 3). Therefore, our results suggest that depending on different cell lineages, one bivalent promoter can be converted to either active or repressed chromatin state during cancer development, highlighting the plasticity of this unique class of promoter. However, a possibility exists that the re-configuration of such bivalent promoters might already occur in the cell-of-origin of different cancer types, which warrants further investigation.
Mechanistically, how bivalent promoters become activated during normal development is incompletely understood. Distal enhancers appear to play an important role in this process by mediating the clearance of PcG proteins from bivalent promoters (45,48,49). For example, as a well-established enhancer/promoter paired gene, MYOD1 overexpression activates a self-regulatory mechanism by binding to a distal enhancer, leading to the conversion of the downstream bivalent promoter to an active state (45). Here in the context of cancer epigenetics, we showed that distal enhancers similarly contribute to the activation of DLX5 bivalent promoter, echoing the findings in normal cell developmental process. This resemblance suggests that the activation mechanism of bivalent promoters may be conserved across different cellular conditions, albeit under control of different upstream TFs. Indeed, our unbiased motif scan and validation experiments identified that SOX2 directly and specifically interacted with and activated DLX5 enhancer in SCC cells (Figure 4). SOX2 possibly evicts PRC complex at DLX5 promoter and recruits additional cofactors for ultimate DLX5 transcription activation. SOX2 does not upregulate DLX5 expression in adenocarcinoma cells might be due to a completely closed chromatin state, which leads to SOX2 fail binding of the regulatory elements of DLX5. In the previous studies, TP63 transcriptional regulation DLX5 has been well described in the development process (50–52), which mainly through the following mechanisms: tissue-specific enhancers, Mecp2-dependent chromatin looping, interaction between Evf2 ncRNA and DLX proteins as well as cis-acting regulation on the proximal promoter region (53–55). However, TP63 did not activate DLX5 expression in SCC cell lines. Indeed, at the DLX5 promoter and enhancer region that established ‘transcriptional links’, the TP63 binding sites were not found (Figure 4A; Supplementary Figure S6C). This may be due to different studies using different cell types that have variant enhancers and other different biological elements, such as the status of TP53 mutation. It is difficult to determine the exact mechanism of TP63 impact on DLX5 expression. After all, TP63 did not activate DLX5 expression in SCC cell lines, which warrants further investigation. Moreover, besides epigenetic activation, copy number gain of DLX5 locus was observed in TCGA SCC samples. The finding of complementary mechanisms converging to upregulate DLX5 expression specifically in SCCs underscores its functional significance in this cancer type, which was established and confirmed by our in vitro and in vivo experiments (Figure 5).
In normal conditions, DLX5 plays a role in osteogenesis by regulating osteocalcin gene transcription (56,57). DLX5 is also required for the development of the olfactory system and cranial neural crest (58,59). Moreover, DLX5 was identified as upstream regulators in NOTCH1-mediated squamous cell differentiation (60). In the context of cancer biology, the mechanistic basis for DLX5 overexpression is hitherto unknown, and the functional significance of DLX5 has only been implicated in ovarian and lung cancers (61–63). Moreover, the cistrome of DLX5 remained uncharacterized before. Here, we demonstrate that DLX5 is specifically upregulated in SCCs by SOX2 and identify DLX5 as an independent prognosis factor for ESCC patients. Functionally, DLX5 strongly promotes SCC cell proliferation, migration, anchorage-independent colony formation, as well as xenograft growth in vivo.
Transcriptional cooperation is a common working model of TFs in the regulation of gene expression. Well understood examples include SOX2/OCT4/NANOG in ESCs (64,65), and HNF1A/HNF4A/HNF6 in both liver and pancreas cells (66). In SCCs, we and others have identified the functional cooperation and interplay between TP63 and SOX2 (23,43). In the present study, our motif analysis unbiasedly identified TP63 as the most significant TF co-occupying DLX5-peaks in both ESCC and HNSCC cell lines. Besides, protein-protein interaction between DLX5 and TP63 was revealed by Co-IP experiments. Importantly, across the SCC cell epigenome, DLX5/TP63 co-occupied and co-regulated over 1900 cis-regulatory elements. Through integrating both ChIP-Seq and RNA-Seq data, we further showed that DLX5/TP63 co-regulated two critical signaling pathways for cancer: MAPK signaling and Pathway in cancer. Many important genes from these two pathways were directly co-occupied and co-regulated by DLX5/TP63, including CCND1, MYC, MEK4, EGFR, IL1A, IL1B, etc. Notably, combinatorial knockdown of DLX5/TP63 produced comparable effects with either single gene knockdown in every target gene tested (Supplementary Figure S12C), suggesting that DLX5 and TP63 co-dependent on each other in regulating fairly parted genes of cancer-promoting pathways. In lung cancer cells, DLX5 was found by ChIP-PCR to occupy the MYC promoter (63). However, our data additionally revealed that DLX5 also bound to a distal enhancer of MYC and it was the distal enhancer, but not the promoter, that was co-occupied and co-regulated by DLX5/TP63 (Figure 7G). Notably, three tumor-associated genes (MYC, EGFR and ITGA6) were co-occupied by DLX5/TP63/SOX2 implies that DLX5/TP63 might form a larger complex with other TFs (such as SOX2) at the regulatory elements of these genes (Figure 7G).
In summary, using a computational method, we systematically and unbiasedly identify a panel of hyperactive TFs shared among different types of SCC. As a novel hyperactive TFs, DLX5 shows a noteworthy bidirectional re-configuration of its bivalent promoter in cancer. Functionally, DLX5 is essential for SCC viability both in vitro and in vivo. Mechanistically, DLX5 interacts with TP63 and co-regulate thousands of cis-regulatory elements. These findings provide important mechanistic insights into transcriptional dysregulation in cancer biology.
DATA AVAILABILITY
The raw sequencing data generated in this study is available on NCBI GEO under the accession number GSE142863. The DLX5, TP63, H3K4me3, H3K27me3 ChIP-seq data of KYSE140 cells and DLX5, TP63 ChIP-seq data of CAL27 cells as well as H3K4me3, H3K27me3 ChIP-seq data of OE33 cells were stored in GEO under the accession number GSE142861; the RNA-seq data of KYSE140 cells with the absence and presence of DLX5 knockdown were stored in GEO under the accession number GSE142862. Our previously published ChIP-Seq and RNA-Seq data were stored in GEO under the accession number GSE106563, GSE132686 and GSE106564 (21–23).
Supplementary Material
Contributor Information
Yongsheng Huang, Guangdong Province Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, China.
Qian Yang, Institute of Oncologic Pathology, Medical College of Shantou University, Shantou, China.
Yueyuan Zheng, Department of Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Lehang Lin, Guangdong Province Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, China.
Xin Xu, Institute of Oncologic Pathology, Medical College of Shantou University, Shantou, China.
Xiu-E Xu, Institute of Oncologic Pathology, Medical College of Shantou University, Shantou, China.
Tiago C Silva, Department of Public Health Sciences, University of Miami, Miller School of Medicine, Miami, FL 33136, USA.
Masaharu Hazawa, Cell-Bionomics Research Unit, Innovative Integrated Bio-Research Core, Institute for Frontier Science Initiative, Kanazawa University, Kanazawa, 920-1192 Ishikawa, Japan.
Li Peng, Guangdong Province Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, China.
Haotian Cao, Guangdong Province Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, China.
Yanbing Ding, Department of Gastroenterology, Affiliated Hospital of Yangzhou University, Yangzhou University, Jiangsu, China.
Daning Lu, Guangdong Province Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, China.
Benjamin P Berman, Center for Bioinformatics and Functional Genomics, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Department of Developmental Biology and Cancer Research, Institute for Medical Research Israel-Canada, Hebrew University-Hadassah Medical School, Jerusalem, Israel.
Li-Yan Xu, Institute of Oncologic Pathology, Medical College of Shantou University, Shantou, China.
En-Min Li, Institute of Oncologic Pathology, Medical College of Shantou University, Shantou, China.
Dong Yin, Guangdong Province Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China [82073067, 81621004, 81872140 to D.Y., 81872306 to L.L., 81772532 to L.Y.X.]; Guangdong Science and Technology Department [2021A0505030084, 2019B020226003 to D.Y.]. Funding for open access charge: Natural Science Foundation of China; Guangdong Science and Technology Department.
Conflict of interest statement. None declared.
REFERENCES
- 1.Campbell J.D., Yau C., Bowlby R., Liu Y., Brennan K., Fan H., Taylor A.M., Wang C., Walter V., Akbani R.et al.. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 2018; 23:194–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dotto G.P., Rustgi A.K.. Squamous cell cancers: a unified perspective on biology and genetics. Cancer Cell. 2016; 29:622–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin D.C., Hao J.J., Nagata Y., Xu L., Shang L., Meng X., Sato Y., Okuno Y., Varela A.M., Ding L.W.et al.. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 2014; 46:467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012; 489:519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015; 517:576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network Integrated genomic and molecular characterization of cervical cancer. Nature. 2017; 543:378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network Integrated genomic characterization of oesophageal carcinoma. Nature. 2017; 541:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah M.A.Update on metastatic gastric and esophageal cancers. J. Clin. Oncol. 2015; 33:1760–1769. [DOI] [PubMed] [Google Scholar]
- 9.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A.. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018; 68:394–424. [DOI] [PubMed] [Google Scholar]
- 10.Sánchez-Danés A., Blanpain C.. Deciphering the cells of origin of squamous cell carcinomas. Nat. Rev. Cancer. 2018; 18:549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergman Y., Cedar H.. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013; 20:274–281. [DOI] [PubMed] [Google Scholar]
- 12.Baylin S.B., Herman J.G.. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000; 16:168–174. [DOI] [PubMed] [Google Scholar]
- 13.Hovestadt V., Jones D.T., Picelli S., Wang W., Kool M., Northcott P.A., Sultan M., Stachurski K., Ryzhova M., Warnatz H.J.et al.. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014; 510:537–541. [DOI] [PubMed] [Google Scholar]
- 14.Lin C.Y., Erkek S., Tong Y., Yin L., Federation A.J., Zapatka M., Haldipur P., Kawauchi D., Risch T., Warnatz H.J.et al.. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016; 530:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stadler M.B., Murr R., Burger L., Ivanek R., Lienert F., Schöler A., van Nimwegen E., Wirbelauer C., Oakeley E.J., Gaidatzis D.et al.. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011; 480:490–495. [DOI] [PubMed] [Google Scholar]
- 16.Ziller M.J., Gu H., Müller F., Donaghey J., Tsai L.T., Kohlbacher O., De Jager P.L., Rosen E.D., Bennett D.A., Bernstein B.E.et al.. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013; 500:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman B.P., Weisenberger D.J., Aman J.F., Hinoue T., Ramjan Z., Liu Y., Noushmehr H., Lange C.P., van Dijk C.M., Tollenaar R.A.et al.. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 2011; 44:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva T.C., Coetzee S.G., Gull N., Yao L., Hazelett D.J., Noushmehr H., Lin D.C., Berman B.P.. ELMER v.2: an R/Bioconductor package to reconstruct gene regulatory networks from DNA methylation and transcriptome profiles. Bioinformatics. 2019; 35:1974–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao L., Shen H., Laird P.W., Farnham P.J., Berman B.P.. Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biol. 2015; 16:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy R.A.WHO is in and WHO is out of the mouth, salivary glands, and jaws sections of the 4th edition of the WHO classification of head and neck tumours. Br. J. Oral Maxillofac. Surg. 2018; 56:90–95. [DOI] [PubMed] [Google Scholar]
- 21.Lin D.C., Dinh H.Q., Xie J.J., Mayakonda A., Silva T.C., Jiang Y.Y., Ding L.W., He J.Z., Xu X.E., Hao J.J.et al.. Identification of distinct mutational patterns and new driver genes in oesophageal squamous cell carcinomas and adenocarcinomas. Gut. 2018; 67:1769–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L., Huang M., Plummer J., Pan J., Jiang Y.Y., Yang Q., Silva T.C., Gull N., Chen S., Ding L.W.et al.. Master transcription factors form interconnected circuitry and orchestrate transcriptional networks in oesophageal adenocarcinoma. Gut. 2019; 69:630–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y., Jiang Y.Y., Xie J.J., Mayakonda A., Hazawa M., Chen L., Xiao J.F., Li C.Q., Huang M.L., Ding L.W.et al.. Co-activation of super-enhancer-driven CCAT1 by TP63 and SOX2 promotes squamous cancer progression. Nat. Commun. 2018; 9:3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin L., Huang M., Shi X., Mayakonda A., Hu K., Jiang Y.Y., Guo X., Chen L., Pang B., Doan N.et al.. Super-enhancer-associated MEIS1 promotes transcriptional dysregulation in Ewing sarcoma in co-operation with EWS-FLI1. Nucleic Acids Res. 2019; 47:1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou W., Triche T.J. Jr, Laird P.W., Shen H. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 2018; 46:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wingender E., Schoeps T., Haubrock M., Krull M., Dönitz J.. TFClass: expanding the classification of human transcription factors to their mammalian orthologs. Nucleic Acids Res. 2018; 46:D343–D347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang M.K., Chen W., Park N.H.. Regulation of epithelial cell proliferation, differentiation, and plasticity by grainyhead-like 2 during oral carcinogenesis. Crit. Rev. Oncog. 2018; 23:201–217. [DOI] [PubMed] [Google Scholar]
- 32.Sastre-Perona A., Hoang-Phou S., Leitner M.C., Okuniewska M., Meehan S., Schober M.. De novo PITX1 expression controls bi-stable transcriptional circuits to govern self-renewal and differentiation in squamous cell carcinoma. Cell Stem Cell. 2019; 24:390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Y., Wang C., Liu X., Mu W., Chen Z., Yu D., Wang A., Dai Y., Zhou X.. Molecular characterization of the microRNA-138-Fos-like antigen 1 (FOSL1) regulatory module in squamous cell carcinoma. J. Biol. Chem. 2011; 286:40104–40109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008; 455:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J.et al.. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K.et al.. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 37.Easwaran H., Johnstone S.E., Van Neste L., Ohm J., Mosbruger T., Wang Q., Aryee M.J., Joyce P., Ahuja N., Weisenberger D.et al.. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 2012; 22:837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohm J.E., McGarvey K.M., Yu X., Cheng L., Schuebel K.E., Cope L., Mohammad H.P., Chen W., Daniel V.C., Yu W.et al.. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007; 39:237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlesinger Y., Straussman R., Keshet I., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E.et al.. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007; 39:232–236. [DOI] [PubMed] [Google Scholar]
- 40.Widschwendter M., Fiegl H., Egle D., Mueller-Holzner E., Spizzo G., Marth C., Weisenberger D.J., Campan M., Young J., Jacobs I.et al.. Epigenetic stem cell signature in cancer. Nat. Genet. 2007; 39:157–158. [DOI] [PubMed] [Google Scholar]
- 41.Robertson A.G., Kim J., Al-Ahmadie H., Bellmunt J., Guo G., Cherniack A.D., Hinoue T., Laird P.W., Hoadley K.A., Akbani R.et al.. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017; 171:540–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corces M.R., Granja J.M., Shams S., Louie B.H., Seoane J.A., Zhou W., Silva T.C., Groeneveld C., Wong C.K., Cho S.W.et al.. The chromatin accessibility landscape of primary human cancers. Science. 2018; 362:eaav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe H., Ma Q., Peng S., Adelmant G., Swain D., Song W., Fox C., Francis J.M., Pedamallu C.S., DeLuca D.S.et al.. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J. Clin. Invest. 2014; 124:1636–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boumahdi S., Driessens G., Lapouge G., Rorive S., Nassar D., Le Mercier M., Delatte B., Caauwe A., Lenglez S., Nkusi E.et al.. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014; 511:246–250. [DOI] [PubMed] [Google Scholar]
- 45.Taberlay P.C., Kelly T.K., Liu C.C., You J.S., De Carvalho D.D., Miranda T.B., Zhou X.J., Liang G., Jones P.A.. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011; 147:1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voigt P., Tee W.W., Reinberg D.. A double take on bivalent promoters. Genes Dev. 2013; 27:1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hahn M.A., Li A.X., Wu X., Yang R., Drew D.A., Rosenberg D.W., Pfeifer G.P.. Loss of the polycomb mark from bivalent promoters leads to activation of cancer-promoting genes in colorectal tumors. Cancer Res. 2014; 74:3617–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seenundun S., Rampalli S., Liu Q.C., Aziz A., Palii C., Hong S., Blais A., Brand M., Ge K., Dilworth F.J.. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010; 29:1401–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vernimmen D., Lynch M.D., De Gobbi M., Garrick D., Sharpe J.A., Sloane-Stanley J.A., Smith A.J., Higgs D.R.. Polycomb eviction as a new distant enhancer function. Genes Dev. 2011; 25:1583–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo Iacono N., Mantero S., Chiarelli A., Garcia E., Mills A.A., Morasso M.I., Costanzo A., Levi G., Guerrini L., Merlo G.R.. Regulation of Dlx5 and Dlx6 gene expression by p63 is involved in EEC and SHFM congenital limb defects. Development. 2008; 135:1377–1388. [DOI] [PubMed] [Google Scholar]
- 51.Vera-Carbonell A., Moya-Quiles M.R., Ballesta-Martínez M., López-González V., Bafallíu J.A., Guillén-Navarro E., López-Expósito I.. Rapp-Hodgkin syndrome and SHFM1 patients: delineating the p63-Dlx5/Dlx6 pathway. Gene. 2012; 497:292–297. [DOI] [PubMed] [Google Scholar]
- 52.Kouwenhoven E.N., van Heeringen S.J., Tena J.J., Oti M., Dutilh B.E., Alonso M.E., de la Calle-Mustienes E., Smeenk L., Rinne T., Parsaulian L.et al.. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010; 6:e1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horike S., Cai S., Miyano M., Cheng J.F., Kohwi-Shigematsu T.. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005; 37:31–40. [DOI] [PubMed] [Google Scholar]
- 54.Feng J., Bi C., Clark B.S., Mady R., Shah P., Kohtz J.D.. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006; 20:1470–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zerucha T., Stühmer T., Hatch G., Park B.K., Long Q., Yu G., Gambarotta A., Schultz J.R., Rubenstein J.L., Ekker M.. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. 2000; 20:709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang H., Fan J., Cao Y., Gao R., Fan Z.. Distal-less homeobox 5 promotes the osteo-/dentinogenic differentiation potential of stem cells from apical papilla by activating histone demethylase KDM4B through a positive feedback mechanism. Exp. Cell Res. 2019; 374:221–230. [DOI] [PubMed] [Google Scholar]
- 57.Heo J.S., Lee S.G., Kim H.O.. Distal-less homeobox 5 is a master regulator of the osteogenesis of human mesenchymal stem cells. Int. J. Mol. Med. 2017; 40:1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sugii H., Grimaldi A., Li J., Parada C., Vu-Ho T., Feng J., Jing J., Yuan Y., Guo Y., Maeda H.et al.. The Dlx5-FGF10 signaling cascade controls cranial neural crest and myoblast interaction during oropharyngeal patterning and development. Development. 2017; 144:4037–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garaffo G., Conte D., Provero P., Tomaiuolo D., Luo Z., Pinciroli P., Peano C., D’Atri I., Gitton Y., Etzion T.et al.. The Dlx5 and Foxg1 transcription factors, linked via miRNA-9 and -200, are required for the development of the olfactory and GnRH system. Mol. Cell. Neurosci. 2015; 68:103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brooks Y.S., Ostano P., Jo S.H., Dai J., Getsios S., Dziunycz P., Hofbauer G.F., Cerveny K., Chiorino G., Lefort K.et al.. Multifactorial ERβ and NOTCH1 control of squamous differentiation and cancer. J. Clin. Invest. 2014; 124:2260–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan Y., Cheung M., Pei J., Menges C.W., Godwin A.K., Testa J.R.. Upregulation of DLX5 promotes ovarian cancer cell proliferation by enhancing IRS-2-AKT signaling. Cancer Res. 2010; 70:9197–9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kato T., Sato N., Takano A., Miyamoto M., Nishimura H., Tsuchiya E., Kondo S., Nakamura Y., Daigo Y.. Activation of placenta-specific transcription factor distal-less homeobox 5 predicts clinical outcome in primary lung cancer patients. Clin. Cancer Res. 2008; 14:263–2370. [DOI] [PubMed] [Google Scholar]
- 63.Xu J., Testa J.R.. DLX5 (distal-less homeobox 5) promotes tumor cell proliferation by transcriptionally regulating MYC. J. Biol. Chem. 2009; 284:20593–20601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G.et al.. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005; 122:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan J., Enge M., Whitington T., Dave K., Liu J., Sur I., Schmierer B., Jolma A., Kivioja T., Taipale M.et al.. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 2013; 154:801–813. [DOI] [PubMed] [Google Scholar]
- 66.Odom D.T., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K.et al.. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004; 303:1378–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data generated in this study is available on NCBI GEO under the accession number GSE142863. The DLX5, TP63, H3K4me3, H3K27me3 ChIP-seq data of KYSE140 cells and DLX5, TP63 ChIP-seq data of CAL27 cells as well as H3K4me3, H3K27me3 ChIP-seq data of OE33 cells were stored in GEO under the accession number GSE142861; the RNA-seq data of KYSE140 cells with the absence and presence of DLX5 knockdown were stored in GEO under the accession number GSE142862. Our previously published ChIP-Seq and RNA-Seq data were stored in GEO under the accession number GSE106563, GSE132686 and GSE106564 (21–23).