Abstract
Recent studies have indicated that long noncoding RNA (lncRNA) and N6-methyladenosine (m6A) methylation modification play critical roles in human cancers; however, their regulation on cervical cancer is largely unclear. Here, our study tries to investigate the underlying mechanisms by which lncRNA FOXD2 adjacent opposite strand RNA 1 (FOXD2-AS1) modulates cervical cancer tumorigenesis. Results illuminated that FOXD2-AS1 expression was significantly upregulated in cervical cancer cells and tissue, which was closely correlated to the unfavorable prognosis. Functionally, gain and loss-of-function assays showed that FOXD2-AS1 promoted the migration and proliferation of cervical cancer cells. Besides, FOXD2-AS1 silencing repressed the tumor growth in vivo. Mechanistically, m6A methyltransferase methyltransferase-like 3 (METTL3) enhanced the stability of FOXD2-AS1 and maintained its expression. Moreover, FOXD2-AS1 recruited lysine-specific demethylase 1 (LSD1) to the promoter region of p21 to silence its transcription abundance. In conclusion, these findings support that METTL3/FOXD2-AS1 accelerates cervical cancer progression via a m6A-dependent modality, which may serve as a potential therapeutic target for cervical cancer.
Keywords: cervical cancer, FOXD2-AS1, METTL3, N6-methyladenosine, LSD1
Graphical abstract
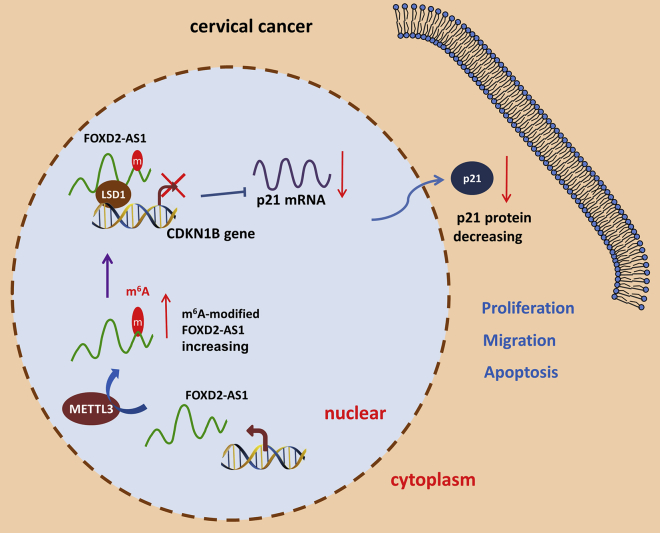
Long noncoding RNA (lncRNA) and N6-methyladenosine (m6A) methylation modification play crucial roles in human cancers. Ji et al. found that METTL3-induced FOXD2-AS1 accelerates cervical cancer progression via a m6A-dependent modality, which may serve as a potential therapeutic target for cervical cancer.
Introduction
Cervical cancer is one of the most common malignant tumors, accounting for a critical part of cancer-related deaths worldwide. Being limited by economic development and prevention, it is difficult to obtain effective diagnosis and treatment for a great number of sufferers, which gives rise to huge disease burden for families. Moreover, there are only a few established diagnostic markers, and a small amount of knowledge is available. The current understanding is that early diagnosis and timely intervention are more valuable for the clinical treatment. Therefore, it is imperative to identify the precise etiology and develop a novel therapeutic precept for cervical cancer.
Long noncoding RNA (lncRNA) is a group of functional noncoding RNA transcripts longer than 200 nt in length.1,2 More and more findings unveil that lncRNAs are engaged in multiple physiopathologic processes across the origin of neoplasia.3 Functionally, lncRNA could coordinate the pathogenesis, cell differentiation, proliferation, and apoptosis of human cancer cells. For instance, lncRNA XLOC_006390 has been found to facilitate the tumorigenesis of cervical cancer and metastasis through downregulating miR-331-3p and miR-338-3p expression.4 Moreover, lncRNA lncCCDST is downregulated in CC tissues and binds with pro-oncogenic DHX9 to promote cervical cancer motility and angiogenesis. The interaction of lncCCDST/DHX9 promotes the degradation of DHX9 by ubiquitin pathway.5 Therefore, the potential roles of lncRNAs for cervical cancer are still very charming and challenging.
N6-methyladenosine (m6A) methylation modification is the most common internal RNA modification in eukaryotes, acting as a critical upstream regulatory mechanism of RNA.6,7 In total RNA, nearly 0.1%–0.4% of adenosines are modified by m6A methylation.8 The m6A consensus motifs are primarily identified as RRACH (R for G/A/U, R for G/A, H for U/A/C).9 Increasing findings suggest that the level of mRNA and noncoding RNA may be correlated with the m6A methylation modification.10 For example, recent research indicates that m6A is highly enriched in lncRNA THOR transcripts, including UG(m6A)CU, GG(m6A)CU, and GA(m6A)CA motifs.11 In chronic kidney disease, m6A methyltransferase methyltransferase-like 3 (METTL3) acts as the main methyltransferase for MALAT1, and its m6A modification triggers the activation of the MALAT1/miR-145/FAK pathway.12
In cervical cancer, METTL3 has been identified to significantly upregulate and closely correlate with the poor prognosis.13 Moreover, METTL3 recruits a m6A reader YTHDF1 to enhance HK2 stability through targeting the 3′ UTR of HK2 mRNA.14 Regarding the regulatory manner, researchers found that METTL3 could bind with the lncRNAs to enhance these RNAs’ stability, thereby maintaining their expression or enrichment. For instance, METTL3 installs the m6A modification of lncRNA ABHD11-AS1 and enhances the stability of ABHD11-AS1 transcript to increase its expression.15 Together, these findings more or less indicate the critical roles of METTL3/lncRNAs in the cancer.
In the present research, our study tried to investigate the oncogenic role of lncRNA FOXD2-AS1 in the cervical cancer progression. Besides, we explored whether the genesis of lncRNA FOXD2-AS1 is associated with its m6A modification or not. Results demonstrated that METTL3/FOXD2-AS1 accelerates the cervical cancer progression via an m6A-dependent modality, which may provide a potential therapeutic target for cervical cancer.
Results
METTL3-induced lncRNA FOXD2-AS1 was upregulated in cervical cancer
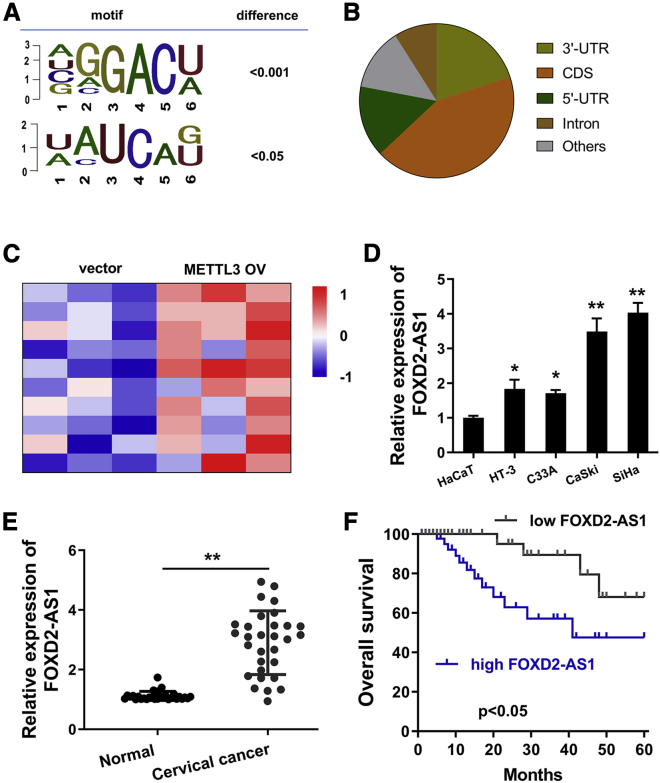
In the initial research, we performed the m6A methylated RNA immunoprecipitation sequencing (MeRIP-seq). Results demonstrated the m6A profile and motifs in the cervical cancer cells (SiHa) as compared with the normal cells (HaCat) (Figure 1A). The distribution situation of m6A modification included 3′ untranslated region (3′ UTR), CDS (coding sequence), 5′ UTR, and other regions (Figure 1B). In the MeRIP-seq, several candidate lncRNAs were found to carry m6A-modified sites. Then, to identify the candidate lncRNAs targeted by METTL3, we detected the candidate lncRNAs’ expression in the METTL3 overexpression cells. In the METTL3 overexpression transfection (SiHa cells), several candidate lncRNAs were detected using quantitative real-time RT-PCR, which was displayed using a heatmap (Figure 1C). In these lncRNAs, we noticed an oncogenic lncRNA FOXD2-AS1. The expression of FOXD2-AS1 was upregulated in the cervical cancer cells (Figure 1D). Moreover, the expression of FOXD2-AS1 was detected in cervical cancer tissue samples and normal tissue samples by quantitative real-time RT-PCR. Results showed that the FOXD2-AS1 level was significantly upregulated in the cervical cancer tissue as compared with the normal tissue (Figure 1E). Besides, survival analysis found that high expression of FOXD2-AS1 was correlated to the lower survival of cervical cancer patients (Figure 1F). These results suggested that METTL3-induced lncRNA FOXD2-AS1 was upregulated in cervical cancer.
Figure 1.
lncRNA FOXD2-AS1 was upregulated in cervical cancer
(A) m6A motifs were detected by m6A methylated RNA immunoprecipitation sequencing (MeRIP-seq). (B) The distribution situation of m6A modification included 3′ untranslated region (3′ UTR), CDS (coding sequence), 5′ UTR, and other regions. (C) Several candidate lncRNAs were detected using quantitative real-time RT-PCR and displayed using the heatmap. (D) FOXD2-AS1 expression was determined in cervical cancer cell lines (HT-3, C33A, CaSki, SiHa) and the normal cell line (HaCaT) using RT-PCR. (E) In cervical cancer tissue samples and normal tissue samples, RT-PCR detected the FOXD2-AS1 level. (F) Survival analysis using Kaplan-Meier test demonstrated the correlation within FOXD2-AS1 expression and survival of cervical cancer patients. All data were shown as mean ± SD. ∗∗p < 0.01.
METTL3 enhanced FOXD2-AS1 stability in cervical cancer cells
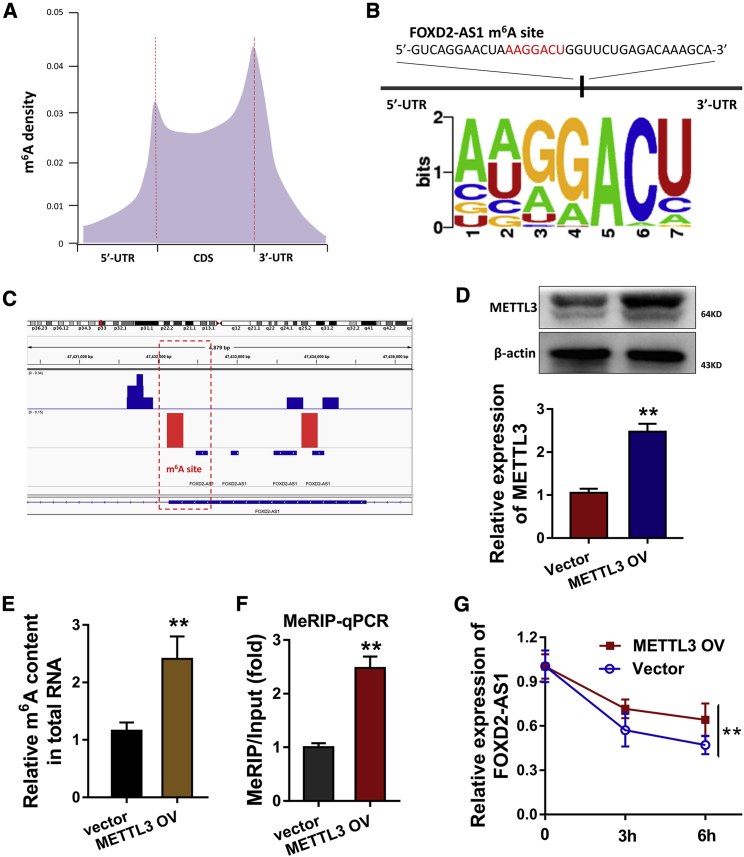
Regarding the upstream mechanism of FOXD2-AS1, we analyzed the potential interaction within FOXD2-AS1 and m6A modification using m6A methylated RNA immunoprecipitation sequencing (MeRIP-seq). MeRIP-seq unveiled the m6A profile in the cervical cancer cells, including 3′ UTRs, coding sites (CDSs), and 5′ UTR (Figure 2A). Moreover, MeRIP-seq displayed that there was a m6A site in the FOXD2-AS1 sequence (Figure 2B). Using the Integrative Genomics Viewer tool (IGV), we could discover the remarkable m6A site in the 3′ UTR of FOXD2-AS1 (Figure 2C). To identify the potential regulation of m6A toward FOXD2-AS1, we constructed the overexpression transfection of m6A methyltransferase METTL3 in SiHa cells (Figure 2D). Then m6A quantitative analysis found that METTL3 overexpression upregulated the m6A modification level of cervical cancer (Figure 2E). m6A methylated RNA immunoprecipitation quantitative real-time PCR (MeRIP-qPCR) analysis found that METTL3 overexpression upregulated the m6A enrichment of FOXD2-AS1 in SiHa cells (Figure 2F). For the FOXD2-AS1 stability, quantitative real-time RT-PCR assay demonstrated that METTL3 overexpression increased its stability when administrated with Act D (Figure 2G). In summary, FOXD2-AS1 was positively regulated by METTL3, and METTL3 maintained FOXD2-AS1 expression in cervical cancer.
Figure 2.
METTL3 enhanced FOXD2-AS1 stability in cervical cancer cells
(A) Metagene profiles showed the m6A profile in the cervical cancer cells, including 3′ UTR, coding sites (CDS), and 5′ UTR. (B) MeRIP-seq displayed the m6A site in the FOXD2-AS1 sequence. (C) Integrative Genomics Viewer tool (IGV) showed the m6A site in the 3′ UTR of FOXD2-AS1. (D) The METTL3 overexpression plasmids were transfected into SiHa cells. Western blot was performed to detect the METTL3 expression. (E) m6A quantitative analysis showed the m6A modification level of cervical cancer cells (SiHa) transfected with METTL3 overexpression. (F) MeRIP-qPCR analysis was performed to detect the m6A modification of FOXD2-AS1 using anti-IgG and anti-m6A antibodies in SiHa cells. Relative FOXD2-AS1 m6A enrichment of each group was normalized to Input group. (G) RNA stability assay was detected using quantitative real-time RT-PCR assay, reflecting the FOXD2-AS1 expression in SiHa cells when administrated with Act D. All data were shown as mean ± SD. ∗∗p < 0.01.
FOXD2-AS1 modulated the tumorigenesis of cervical cancer in vitro
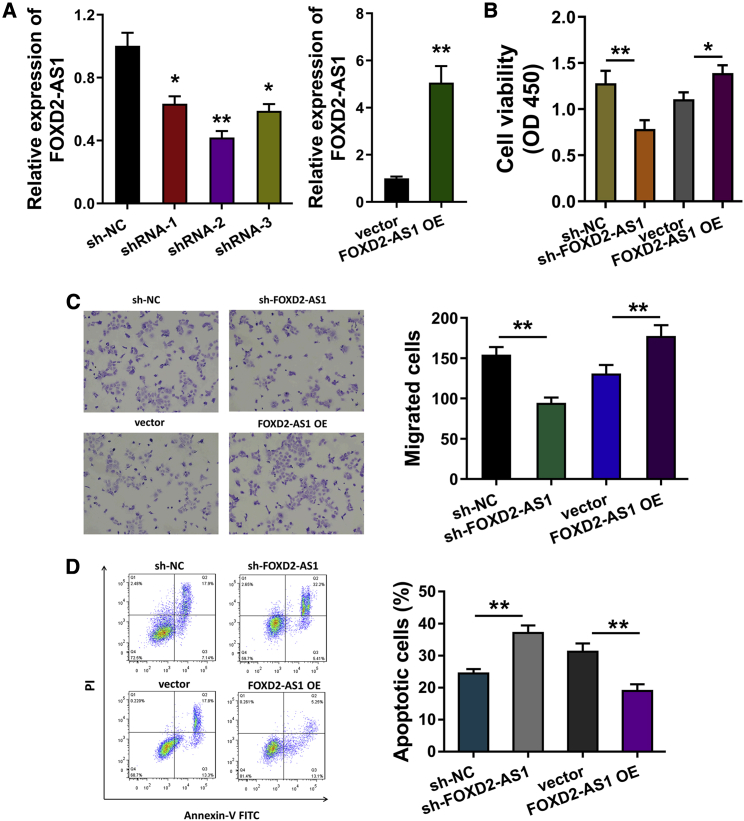
Given the ectopic high expression of FOXD2-AS1 in cervical cancer, its potential regulation on cervical cancer proliferation needed further investigation. First, gain-of-function and loss-of-function assays were constructed using pcDNA vector and short hairpin RNA (shRNA) stable transfection in SiHa cell line for FOXD2-AS1 knockdown and overexpression (Figure 3A). CCK-8 proliferative assay demonstrated that FOXD2-AS1 knockdown repressed the proliferation of SiHa cells, and FOXD2-AS1 overexpression promoted the proliferation (Figure 3B). Migration analysis found that FOXD2-AS1 knockdown repressed the migrative ability of SiHa cells, and FOXD2-AS1 overexpression promoted the migrative ability (Figure 3C). Flow cytometry apoptosis analysis found that FOXD2-AS1 knockdown facilitated the apoptosis of SiHa cells, and FOXD2-AS1 overexpression inhibited the apoptosis (Figure 3D). In summary, FOXD2-AS1 might modulate the proliferation and migration of cervical cancer in vitro.
Figure 3.
FOXD2-AS1 modulated the tumorigenesis of cervical cancer in vitro
(A) Gain-of-function and loss-of-function assays were constructed using pcDNA vector and short hairpin RNA (shRNA) stable transfection (SiHa cell line). Quantitative real-time RT-PCR detected the FOXD2-AS1 expression. (B) CCK-8 proliferative assay demonstrated the proliferation of cervical cancer cells with FOXD2-AS1 knockdown and overexpression in SiHa cells. (C) Migrative transwell analysis detected the migrative ability of cervical cancer cells transfected with FOXD2-AS1 shRNA or overexpression in SiHa cells. (D) Flow cytometry apoptosis analysis illustrated the apoptosis of cervical cancer cells with FOXD2-AS1 knockdown and overexpression in SiHa cells. All data were shown as mean ± SD. ∗∗p < 0.01.
FOXD2-AS1 recruited lysine-specific demethylase 1 (LSD1) to p21 promoter to reduce its expression
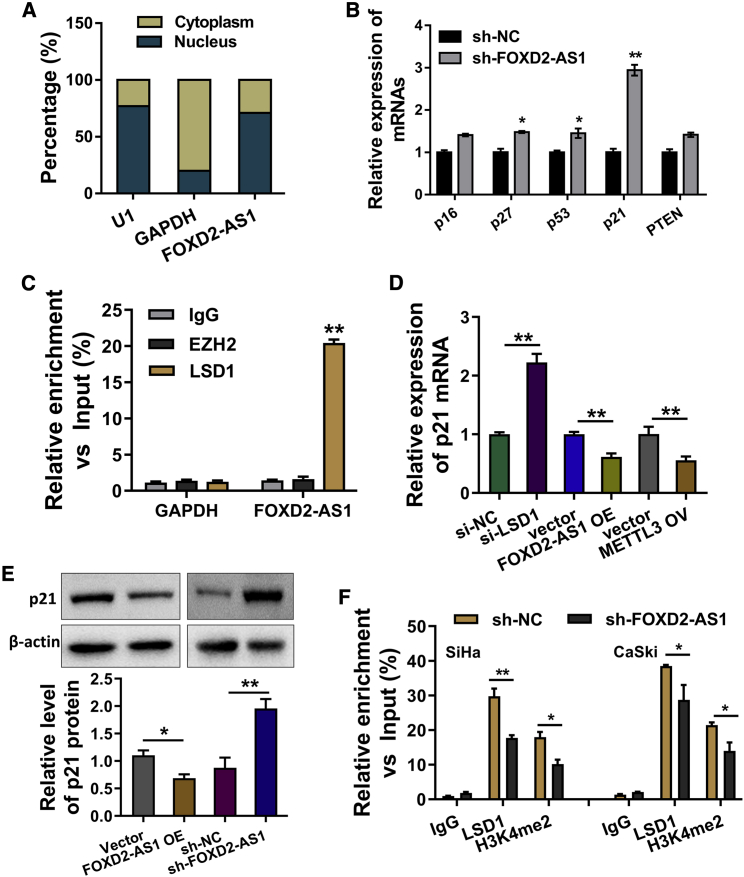
The subcellular location of FOXD2-AS1 was measured in cervical cancer cells, indicating that FOXD2-AS1 was principally distributed in the nucleus, rather than the cytoplasm (Figure 4A). In order to discover the potential target of FOXD2-AS1, we selected several candidate targets in the SiHa cells with FOXD2-AS1 knockdown (Figure 4B). RNA binding protein immunoprecipitation (RIP) showed that LSD1 could bind with FOXD2-AS1, rather than EZH2 (Figure 4C). In the SiHa cells transfected with LSD1 knockdown (si-LSD1), p21 mRNA was upregulated (Figure 4D). Besides, p21 mRNA levels were reduced in the FOXD2-AS1 overexpression or METTL3 overexpression transfection. Western blot analysis displayed that FOXD2-AS1 overexpression repressed the p21 protein level (Figure 4E). Chromatin immunoprecipitation (ChIP) demonstrated the occupancy of LSD1 and H3K4me2 of the promoter regions of p21 (Figure 4F). Taken together, FOXD2-AS1 recruited LSD1 to p21 promoter to reduce p21 expression.
Figure 4.
FOXD2-AS1 recruited LSD1 to p21 promoter to reduce p21 expression
(A) Subcellular location of FOXD2-AS1 was measured in SiHa cells using subcellular fractionation assay. (B) Several candidate targets were selected in the SiHa cells with FOXD2-AS1 knockdown, including p16, p27, p53, p21, and PTEN. (C) RNA binding protein immunoprecipitation (RIP) showed the binding of FOXD2-AS1 with LSD1 in SiHa cells, rather than EZH2. (D) p21 mRNA was detected in the SiHa cells transfected with LSD1 knockdown (si-LSD1). (E) Western blot analysis displayed the p21 protein level with FOXD2-AS1 knockdown or control. (F) Chromatin immunoprecipitation (ChIP) demonstrated the occupancy of LSD1 and H3K4me2 of the promoter regions of p21. All data were shown as mean ± SD. ∗∗p < 0.01; ∗p < 0.05.
FOXD2-AS1/LSD1/p21 axis regulated the tumorigenesis of cervical cancer
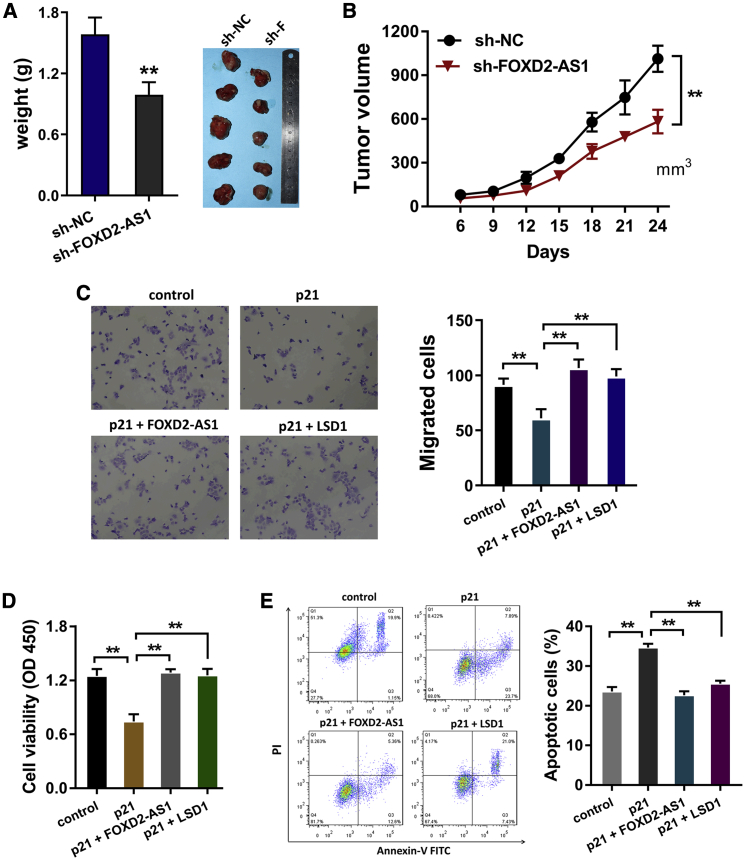
In vivo xenograft mice assay was performed to determine the role of FOXD2-AS1 on cervical cancer cells. Results showed that FOXD2-AS1 knockdown remarkably repressed the tumor weight (Figure 5A) and volume (Figure 5B). Migration analysis found that p21 overexpression repressed the migrative ability of cervical cancer cells, and the co-transfection of FOXD2-AS1 or LSD1 significantly recovered the migration phenotype (Figure 5C). CCK-8 proliferative assay demonstrated that p21 overexpression inhibited the proliferation of cervical cancer cells, and the co-transfection of FOXD2-AS1 or LSD1 significantly recovered the proliferative phenotype (Figure 5D). Flow cytometry apoptosis analysis found that p21 overexpression decreased the apoptotic rate, and the co-transfection of FOXD2-AS1 or LSD1 observably recovered the apoptotic phenotype (Figure 5E). In summary, these finding suggested that the FOXD2-AS1/LSD1/p21 axis regulated the proliferation and migration of cervical cancer (Figure 6).
Figure 5.
The FOXD2-AS1/LSD1/p21 axis regulated the tumorigenesis of cervical cancer
(A and B) In vivo xenograft mice assay was performed to determine the (A) tumor weight and (B) volume using cervical cancer cells (SiHa cells). (C) Migration analysis was performed using SiHa cells transfected with p21 overexpression plasmids and/or FOXD2-AS1 or LSD1 co-transfection. (D) CCK-8 proliferative assay was performed using SiHa cells transfected with p21 overexpression plasmids and/or FOXD2-AS1 or LSD1 co-transfection. (E) Flow cytometry apoptosis analysis was performed using SiHa cells transfected with p21 overexpression plasmids and/or FOXD2-AS1 or LSD1 co-transfection. All data were shown as mean ± SD. ∗∗p < 0.01.
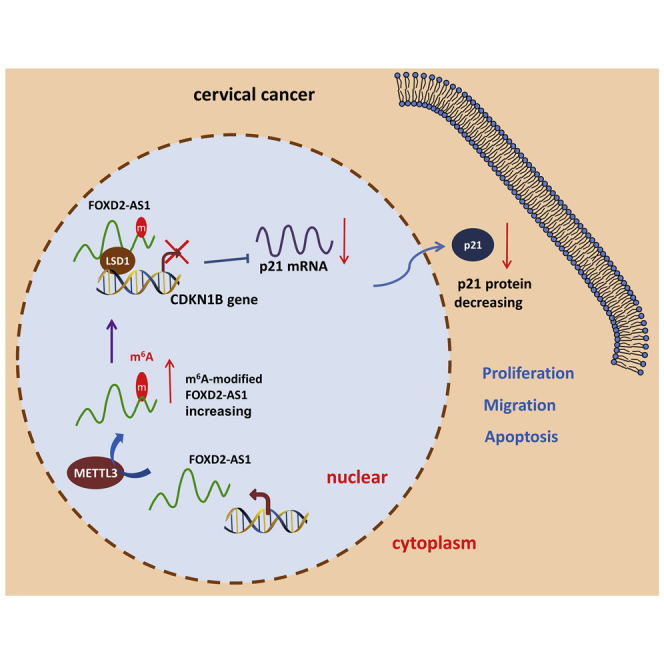
Figure 6.
METTL3/FOXD2-AS1 promotes the tumorigenesis of cervical cancer through the LSD1/p21 axis
Discussion
Increasing evidence has indicated that lncRNA is closely associated with the pathogenesis of cervical cancer.16,17 With the development of epigenetics and molecular biology, novel m6A methylation modification attracts increasingly more attention nowadays.18,19 Here, we find an interesting lightspot that m6A might participate in the cervical cancer pathogenesis together with lncRNA.
In the present research, we found that lncRNA FOXD2-AS1 was upregulated in the cervical cancer cells and tissue samples. The ectopic overexpression of lncRNA FOXD2-AS1 might imply its oncogenic role for cervical cancer pathogenesis. In numerous human cancers, FOXD2-AS1 has been identified to be a crucial oncogene. For example, lncRNA FOXD2-AS1 is significantly upregulated in hepatocellular carcinoma cells and aggravates the tumorigenesis via regulating the miR-206/MAP3K1 axis.20 lncRNA FOXD2-AS1 was significantly upregulated in esophagus cancer tissues compared with adjacent normal tissues and positively correlated with its clinicopathological parameters.21 In glioma, FOXD2-AS1 is closely linked to the cell-cycle modulation in low-grade glioma and glioblastoma through the FOXD2-AS1/miR-31/CDK1 axis.22
Given the ectopic overexpression of FOXD2-AS1 in cervical cancer, we set our eyes on potential upstream mechanisms of FOXD2-AS1 biogenesis. Using the MeRIP-seq, we found that there is a remarkable m6A peak in the 3′ end of the FOXD2-AS1 sequence. Further research found that m6A methyltransferase METTL3 could upregulate the RNA stability of FOXD2-AS1 in cervical cancer. In this type of maintenance, m6A key regulatory factor might enhance the stability of lncRNA. For example, m6A is highly enriched in lncRNA THOR transcripts, containing GAA(m6A)CA, GGA(m6A)CU, and UGA(m6A)CU motifs. The m6A-dependent RNA-protein interactions could maintain the oncogenic role of lncRNA THOR in cancers.11 In colon cancer, lncRNA NEAT1 is remarkably increased in colon cancer tissues and correlated with poor prognosis. m6A demethylase ALKBH5 could upregulate NEAT1 expression through demethylation.23 In non-small cell lung cancer, m6A methyltransferase-like 3 (METTL3) installs the m6A modification and enhances the stability of ABHD11-AS1 transcript to upregulate its expression,15 and our findings also clarify the potential m6A-lncRNA interaction in cervical cancer.
For the regulation of FOXD2-AS1, we found that FOXD2-AS1 could repress the expression of p21 in the cervical cancer cells. FOXD2-AS1 recruited the LSD1 on the promoter region of p21 gene. FOXD2-AS1 silencing could reduce the occupancy of LSD1 and H3K4me2 on the p21 promoter region. LSD1 is a critical transcriptional silence factor, and lncRNAs are found to interact with LSD1 to regulate gene expression. For example, lncRNA LINC00460 promotes the gastric cancer proliferation through downregulating tumor suppressor gene CCNG2, which is mediated by LSD1.24 lncRNA HIF1A-AS2 is ectopically overexpressed in trophoblasts cells and increases the trophoblasts cell migration and invasion function. HIF1A-AS2 could recruit LSD1 and epigenetically repress the expression of PHLDA1.25 Solid evidence has illustrated that p21 is a universal cell-cycle inhibitor, which is directly controlled by p53 and p53-independent manner. The discovery and achievements on p21 have illuminated its function on cellular growth control, stem cell phenotypes, cell stress response, and differentiation. In cervical cancer, p21 has similarly been identified to be a tumor repressor.26, 27, 28 Therefore, our data support that lncRNA FOXD2-AS1/LSD1/p21 might modulate the tumorigenesis of cervical cancer.
In summary, our work has identified the critical role of lncRNA FOXD2-AS1 in cervical cancer tumorigenesis. Moreover, m6A methyltransferase METTL3 enhances the stability of FOXD2-AS1 to maintain its expression. FOXD2-AS1 recruits LSD1 to the promoter of p21 to silence the expression, thereby accelerating the progression of cervical cancer. Overall, these findings provide a novel insight into exploiting therapeutic strategy for cervical cancer.
Materials and methods
Clinical specimens
A total of 30 fresh frozen cervical cancer tissues and normal tissues were collected from 2018 to 2019 during surgery at Shenzhen Baoan Women’s and Children’s Hospital (Table 1). None of the participants received any anti-cancer treatment before operation. This study has been approved by the Ethics Review Board of Shenzhen Baoan Women’s and Children’s Hospital. Written informed consents were received from all patients.
Table 1.
The correlations between FOXD2-AS1 and cervical cancer patients’ clinicopathological characteristics
| FOXD2-AS1 | p | |||
|---|---|---|---|---|
| Low (15) | High (15) | |||
| Age, years | 0.617 | |||
| <45 | 13 | 7 | 6 | |
| ≥45 | 17 | 8 | 9 | |
| Tumor size, cm | 0.008 | |||
| <4 | 11 | 8 | 3 | |
| ≥4 | 19 | 7 | 12 | |
| FIGO stages | 0.187 | |||
| I/II | 16 | 6 | 10 | |
| III/IV | 14 | 9 | 5 | |
| Tumor differentiation | 0.248 | |||
| Well/moderate | 18 | 10 | 8 | |
| Poor | 12 | 5 | 7 | |
| Lymph node metastasis | 0.173 | |||
| Positive | 10 | 4 | 6 | |
| Negative | 20 | 11 | 9 | |
Cell lines
The cervical cancer cell lines (CaSki, HT-3, SiHa, C33A) and human epidermal cells (HaCaT) were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China) and American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured with Dulbecco’s modified Eagle’s medium (DMEM; GIBCO, Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (GIBCO), 100 U/ml penicillin, and 100 mg/mL streptomycin (GIBCO). These cells were incubated in 5% CO2 incubator (Thermo Fisher, USA) with the humidified environment at 37°C.
Transfection
Transfection was conducted according to the manufacturer’s instructions. Overexpression and knockdown lentiviruses for FOXD2-AS1, as well as control lentivirus, were purchased from GeneChem Corporation (Shanghai, China). Stable clones were selected using puromycin (2 μg/ml; Sigma). Cells were harvested for mRNA analysis for transfection efficiency. The pcDNA3.1 plasmid carrying the full-length p21 and LSD1 sequences were synthesized by GeneCopoeia (Guangzhou, China). The plasmids were transfected into cervical cancer cells with Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific).
Quantitative real-time RT-PCR
Total RNA was isolated from cervical cancer cells and tissues using a miRNeasy Mini Kit (QIAGEN, Hilden, Germany). The concentration and purity were evaluated by ultraviolet spectrophotometry (NanoDrop ND2000; Thermo Fisher, Rockford, IL, USA). For cDNA, reverse transcription was performed using the SuperScript First-Stand Synthesis system (Invitrogen, USA). SYBR Green PCR Master Mix (Applied Biosystems) was subjected to qPCR on the ABI 7500 Detection System. GAPDH functioned as an internal reference. 2−ΔΔCt methods are conducted to calculate relative gene expression.
Western blotting analysis
The cells (SiHa) were lysed by radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich, St. Louis, MO, USA) containing the protease inhibitor phenylmethanesulphonyl fluoride (Beyotime Biotechnology, China) for 30 min at 4°C. Protein concentration was identified using BCA protein assay kit (Beyotime Biotechnology, China), which was separated using SDS-PAGE sample loading buffer (Beyotime Biotechnology, China) for electrophoresis. Then, the protein was transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA) and blocked with 5% non-fat milk at room temperature. After being washed by TBST twice, the PVDF membranes were incubated with primary antibodies (anti-METTL3 [Abcam], ab195352, 1:1,000; anti-p21 [Abcam], ab108349, 1:1,000) at 4°C overnight. After the PVDF membrane was washed by TBST, the membrane was incubated with secondary antibody (β-actin, 1:10,000; Sigma-Aldrich, USA) for 2 h at room temperature. Finally, proteins were detected by enhanced chemiluminescence (ECL) reagent (Millipore).
Cell proliferation, migration, and apoptosis assay
Cell viability was measured with CCK-8 assay. Cells (SiHa) were transfected and harvested with 0.25% trypsin and seeded in 96-well plates. A total of 1 × 103 cells/well were incubated with 10 μL of the CCK-8 reagent, with each well at 37°C for 2 h. The spectrophotometric absorbance was measured at 450 nm for each sample. Cell migration assay was performed using Matrigel chambers (BD Biosciences, USA). In brief, cells (5 × 104 cells/well, SiHa) suspended in DMEM/F12 culture medium without FBS were added into the upper chamber. Then 1 mL DMEM/F12 with 10% FBS was added to the lower compartment. After 24-h incubation, the cells residing on the upper surface were wiped off, and the cells that crossed the filter membrane were stained with 0.1% crystal violet. Cell number was quantified in five random fields under a microscope. For the apoptosis analysis, cells were washed in 100 μL of 1 × Binding Buffer and measured using fluorescein isothiocyanate (FITC) Annexin V Apoptosis Detection Kit I (BD Pharmagen, USA) following the manufacturer’s protocol.
m6A methylated RNA immunoprecipitation sequencing (MeRIP-seq)
Cervical cancer cells (SiHa) were washed by cold PBS twice and purified using Oligotex Direct mRNA Midi/Maxi Kit (Thermo Fisher) according to the manufacturer’s instructions. Then, mRNA concentration was quantified by a spectrophotometer. For immunoprecipitation (IP), mRNA (5 μg) was precipitated with Magna MeRIP m6A Kit (Millipore) according to the manufacturer’s instructions. The library was constructed using purified RNA fragments from IP and then sequenced with Illumina Nextseq500.
MeRIP-qPCR
MeRIP-qPCR was performed according to the published literature with slight modifications. In brief, total RNA (150 μg) was isolated from pretreated cells (SiHa) and randomly fragmented into 100-nt RNA fragments using ZnCl2. RNA samples were immunoprecipitated with antibodies (10 μg anti-m6A antibody or anti-mouse immunoglobulin G [IgG]; Millipore, Germany) pre-coated with magnetic beads to elute the m6A-modified RNA fragments from the beads with m6A in IP buffer (B5993; ApexBio Technology). MeRIP-qPCR was performed using the Magna MeRIP m6A Kit (Millipore, Germany) to detect the m6A modification quantitation. The RNA enrichment was analyzed by quantitative real-time RT-PCR.
m6A quantification
Total RNA was isolated from SiHa by TRIzol (Thermo Fisher) according to the manufacturer’s instruction. m6A content in total RNAs was detected using EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric) (Epigentek, USA). The m6A level was colorimetrically identified according to the absorbance at a wavelength of 450 nm.
RNA stability
Cells were treated with actinomycin D (1 μg/mL) and then collected at different time points. RNA was extracted from SiHa using TRIzol reagent (Invitrogen, Grand Island, NY, USA), and the RNA levels were measured using qRT-PCR.
RNA binding protein immunoprecipitation (RIP)
Cells (SiHa) were cultured and washed twice with ice-cold PBS and centrifuged and re-suspended in complete RIP lysis buffer (Merck Millipore) (140 mM NaCl, 20 mM Tris-HCl [pH 7.5], 0.05% Triton X-100) for 2 h. Antibody (5 μg) anti-Argonaute-2 (AGO2) (Abcam, MA, USA) or control rabbit IgG were pre-bound to protein A/G magnetic beads in IP buffer at 4°C overnight. Then, the expression of mRNAs was detected by quantitative real-time RT-PCR analysis.
ChIP assay
The ChIP assay was performed using the Magna ChIP Chromatin Immunoprecipitation Kit (Millipore) using SiHa cells according to the manual protocol. In brief, crosslinked chromatin was sonicated into fragments (200–1,000 bp) and then immunoprecipitated with anti-LSD1 (Millipore) and anti-H3K4me2 (Millipore). Then, the precipitated DNA-protein complex was eluted and quantified using quantitative real-time RT-PCR with SYBR-Green incorporation (Applied Biosystems, Foster City, CA, USA). The primers are listed in Table S1. Normal mouse IgG acted as a negative control.
Tumor formation assays
The tumor formation assays were approved by the Ethics Committee of Shenzhen Baoan Women’s and Children’s Hospital. All male BALB/c nude mice (5 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Company. A total 5 × 106 stably transfected SiHa cells were subcutaneously injected into the flank of nude mice. Every 3 days, tumor volume was detected using 0.5 × (L × W × W). At 3 weeks after injection, mice were sacrificed for tumor weight.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA, USA) and SPSS 20.0 (IBM Corporation, Armonk, NY, USA). Data were shown as mean ± standard deviation (SD). The differences between groups were analyzed using two-sided Student’s t test. The correlation within FOXD2-AS1 and clinicopathological features was calculated with chi-square test. Kaplan-Meier curve with log rank test was performed to analyze the survival outcome. Pearson’s correlation was performed to analyze the correlation. A p value less than 0.05 was considered as statistically significance.
Acknowledgments
This work was supported by the intra-hospital fund of Shenzhen Baoan Women’s and Children’s Hospital (Grant number BAFY 2020003), Shenzhen Key Medical Discipline Construction Fund (No. SZXK028), Shenzhen Science and Technology Innovation Committee Foundation (No. JCYJ20190809185001766). .
Author contributions
F.J. and Y.L. are the major performers for this study. S.C., X. Lin, and Y.Y. assisted with statistical analysis and literature novelty. Y.Z. and X. Luo are responsible for the design and funding.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2021.07.004.
Contributor Information
Yuanfang Zhu, Email: zhuyfdoctor@sina.com.
Xin Luo, Email: doctor_luoxin.edu@aliyun.com.
Supplemental information
References
- 1.Lu W., Cao F., Wang S., Sheng X., Ma J. LncRNAs: The Regulator of Glucose and Lipid Metabolism in Tumor Cells. Front. Oncol. 2019;9:1099. doi: 10.3389/fonc.2019.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan Y., Song D., Song X., Song C. The role of lncRNA MALAT1 in cardiovascular disease. IUBMB Life. 2020;72:334–342. doi: 10.1002/iub.2210. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y., Zhu Y., Xie Y., Ma X. The Role of Long Non-coding RNAs in Immunotherapy Resistance. Front. Oncol. 2019;9:1292. doi: 10.3389/fonc.2019.01292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luan X., Wang Y. LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p. J. Gynecol. Oncol. 2018;29:e95. doi: 10.3802/jgo.2018.29.e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding X., Jia X., Wang C., Xu J., Gao S.J., Lu C. A DHX9-lncRNA-MDM2 interaction regulates cell invasion and angiogenesis of cervical cancer. Cell Death Differ. 2019;26:1750–1765. doi: 10.1038/s41418-018-0242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu S., Zhang S., Wu X., Zhou X. m(6)A RNA Methylation in Cardiovascular Diseases. Mol. Ther. 2020;28:2111–2119. doi: 10.1016/j.ymthe.2020.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai F., Wu Y., Lu Y., An C., Zheng X., Dai L., Guo Y., Zhang L., Li H., Xu W., Gao W. Crosstalk between RNA m6A Modification and Non-coding RNA Contributes to Cancer Growth and Progression. Mol. Ther. Nucleic Acids. 2020;22:62–71. doi: 10.1016/j.omtn.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang M., Zhai Y., Zhang S., Dai X., Li Z. Roles of N6-Methyladenosine (m(6)A) in Stem Cell Fate Decisions and Early Embryonic Development in Mammals. J. Hematol. Oncol. 2020;8:782. doi: 10.3389/fcell.2020.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chokkalla A.K., Mehta S.L., Vemuganti R. Epitranscriptomic regulation by m6A RNA methylation in brain development and diseases. J. Cereb. Blood Flow Metab. 2020;40:2331–2349. doi: 10.1177/0271678X20960033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi Y.C., Chen X.Y., Zhang J., Zhu J.S. Novel insights into the interplay between m6A modification and noncoding RNAs in cancer. Mol. Cancer. 2020;19:121. doi: 10.1186/s12943-020-01233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H., Xu Y., Yao B., Sui T., Lai L., Li Z. A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death Dis. 2020;11:613. doi: 10.1038/s41419-020-02833-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu P., Zhang B., Chen Z., He Y., Du Y., Liu Y., Chen X. m6A-induced lncRNA MALAT1 aggravates renal fibrogenesis in obstructive nephropathy through the miR-145/FAK pathway. Aging (Albany NY) 2020;12:5280–5299. doi: 10.18632/aging.102950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z., Peng Y., Li J., Chen Z., Chen F., Tu J., Lin S., Wang H. N6-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat. Commun. 2020;11:2578. doi: 10.1038/s41467-020-16306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q., Guo X., Li L., Gao Z., Su X., Ji M., Liu J. N6-methyladenosine METTL3 promotes cervical cancer tumorigenesis and Warburg effect through YTHDF1/HK2 modification. Cell Death Dis. 2020;11:911. doi: 10.1038/s41419-020-03071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue L., Li J., Lin Y., Liu D., Yang Q., Jian J., Peng J. m6 A transferase METTL3-induced lncRNA ABHD11-AS1 promotes the Warburg effect of non-small-cell lung cancer. J. Cell. Physiol. 2021;236:2649–2658. doi: 10.1002/jcp.30023. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z., Sun Q., Guo J., Wang S., Song G., Liu W., Liu M., Tang H. GRSF1-mediated MIR-G-1 promotes malignant behavior and nuclear autophagy by directly upregulating TMED5 and LMNB1 in cervical cancer cells. Autophagy. 2019;15:668–685. doi: 10.1080/15548627.2018.1539590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Chang X., Zhang H., Yang Q., Pang L. LncRNA SOX2OT affects cervical cancer cell growth, migration and invasion by regulating SOX2. Cell Cycle. 2020;19:1391–1403. doi: 10.1080/15384101.2020.1750812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X., Peng W.X., Zhou H., Jiang J., Zhou X., Huang D., Mo Y.Y., Yang L. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27:1782–1794. doi: 10.1038/s41418-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y., Ouyang Z., Sui X., Qi M., Li M., He Y., Cao Y., Cao Q., Lu Q., Zhou S. Oocyte competence is maintained by m6A methyltransferase KIAA1429-mediated RNA metabolism during mouse follicular development. Cell Death Differ. 2020;27:2468–2483. doi: 10.1038/s41418-020-0516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu W., Feng H., Xu X., Huang X., Huang X., Chen W., Hao L., Xia W. Long noncoding RNA FOXD2-AS1 aggravates hepatocellular carcinoma tumorigenesis by regulating the miR-206/MAP3K1 axis. Cancer Med. 2020;9:5620–5631. doi: 10.1002/cam4.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi W., Gao Z., Song J., Wang W. Silence of FOXD2-AS1 inhibited the proliferation and invasion of esophagus cells by regulating miR-145-5p/CDK6 axis. Histol. Histopathol. 2020;35:1013–1021. doi: 10.14670/HH-18-232. [DOI] [PubMed] [Google Scholar]
- 22.Wang J., Li B., Wang C., Luo Y., Zhao M., Chen P. Long noncoding RNA FOXD2-AS1 promotes glioma cell cycle progression and proliferation through the FOXD2-AS1/miR-31/CDK1 pathway. J. Cell. Biochem. 2019;120:19784–19795. doi: 10.1002/jcb.29284. [DOI] [PubMed] [Google Scholar]
- 23.Guo T., Liu D.F., Peng S.H., Xu A.M. ALKBH5 promotes colon cancer progression by decreasing methylation of the lncRNA NEAT1. Am. J. Transl. Res. 2020;12:4542–4549. [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J., Lian Y., Yang R., Lian Y., Wu J., Liu J., Wang K., Xu H. Upregulation of lncRNA LINC00460 Facilitates GC Progression through Epigenetically Silencing CCNG2 by EZH2/LSD1 and Indicates Poor Outcomes. Mol. Ther. Nucleic Acids. 2020;19:1164–1175. doi: 10.1016/j.omtn.2019.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Wu D., Yang N., Xu Y., Wang S., Zhang Y., Sagnelli M., Hui B., Huang Z., Sun L. lncRNA HIF1A Antisense RNA 2 Modulates Trophoblast Cell Invasion and Proliferation through Upregulating PHLDA1 Expression. Mol. Ther. Nucleic Acids. 2019;16:605–615. doi: 10.1016/j.omtn.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang P., Liu J., Li W., Zeng X., Tang J. Role of p53 and p21 polymorphisms in the risk of cervical cancer among Chinese women. Acta Biochim. Biophys. Sin. (Shanghai) 2010;42:671–676. doi: 10.1093/abbs/gmq069. [DOI] [PubMed] [Google Scholar]
- 27.Wang N., Wang S., Zhang Q., Lu Y., Wei H., Li W., Zhang S., Yin D., Ou Y. Association of p21 SNPs and risk of cervical cancer among Chinese women. BMC Cancer. 2012;12:589. doi: 10.1186/1471-2407-12-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan Z.H., Zhang Y., Tian Y., Tan W., Li Y.H. IκB kinase b Mediating the Downregulation of p53 and p21 by Lipopolysaccharide in Human Papillomavirus 16+ Cervical Cancer Cells. Chin. Med. J. (Engl.) 2016;129:2703–2707. doi: 10.4103/0366-6999.193463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.