Abstract
Antimicrobial peptides (AMPs) have recently been proposed as significant immunological factors involved in pathogenesis of coronavirus disease 19 (COVID-19). Human β-defensins (hBDs) are among these AMPs, but the evidence is not well detailed. Therefore, this case-control study analyzed levels of hBD1, hBD2, hBD3 and hBD4 in serum of 103 patients with severe COVID-19 and 105 healthy controls. Most patients were older than 45 years (80.6%), and more than 50% suffered from chronic diseases (cardiovascular and diabetes). Results revealed that median levels of hBD1 and hBD3 did not show significant differences between patients and controls. On the contrary, HBD2 levels were significantly decreased in patients compared to controls (1036 vs. 1289 ng/L; p < 0.001), while HBD4 levels were significantly increased (4.04 vs. 2.43 ng/L; p < 0.001). Receiver operating characteristic curve analysis demonstrated the predictive significance of hBD2 (area under the curve [AUC] = 0.795; 95% confidence interval [CI] = 0.729–0.861; p < 0.001) and hBD4 (AUC = 0.816; 95% CI = 0.756–0.876; p < 0.001) in discriminating between COVID-19 patients and controls. Logistic regression analysis (adjusted for age, gender and body mass index) confirmed the significance of hBD2 (odds ratio [OR] = 0.996; corrected p = 0.004) and hBD4 (OR = 4.948; corrected p < 0.001) in susceptibility to COVID-19. In conclusion, the study indicated that hBD2 showed low levels in serum of patients infected with severe COVID-19, while hBD4 showed elevated levels. These differences in HBDs were not influenced by age, gender, body mass index, or chronic disease.
Keywords: COVID-19, Human β-defensin, Age, Receiver operating characteristic, Logistic regression analysis
Abbreviations
- ALP
Alkaline phosphatase
- ALT
Alanine aminotransferase
- AMP
Antimicrobial peptide
- AST
Aspartate aminotransferase
- AUC
Area under curve
- BMI
Body mass index
- BUN
Blood urea nitrogen
- CI
Confidence interval
- COVID-19
Coronavirus disease 19
- CRP
C-reactive protein
- CT
Computed tomography
- ESR
Erythrocyte sedimentation rate
- hBD
human β-defensin
- IQR
Interquartile range
- LSD
Least significant difference
- MERS
Middle East respiratory syndrome-coronavirus
- OR
Odds ratio
- p:
Probability
- pc
Corrected probability
- RBG
Random blood glucose
- ROC:
Receiver operating characteristic
- rs
correlation coefficient
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SD
Standard deviation
- WBC
White blood cell count
1. Introduction
Since the outbreak of coronavirus disease 19 (COVID-19) in China in December 2019, the highly contagious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread globally to become a pandemic affecting millions of people in more than 200 countries with a mortality rate of approximately 2% [1]. An impressive number of studies have been conducted to understand the pathogenesis of this respiratory infection and the factors associated with its risk. These studies revealed that immunological factors play a key role in pathogenesis of COVID-19, virus persistence, and risk of severity and/or death [2], and dysregulated innate and adaptive immune responses have been well documented [3]. The dysregulation is primarily associated with increased production of pro-inflammatory cytokines, and accordingly the cytokine storm has been described as an important feature of COVID-19 pathogenesis that also correlates with disease severity [4]. In addition, antimicrobial peptides (AMPs) have recently been proposed as other immunological factors involved in pathogenesis of COVID-19 and have been considered as promising and potential candidates to combat SARS-CoV-2, but the evidence is not well detailed [[5], [6], [7]].
AMPs are multifunctional peptides that represent the first line of defense against various pathogens [8]. Defensins are a group of natural AMPs with powerful effects on viruses, bacteria and fungi [9]. Besides, defensins are viewed as multifunctional factors involved in the regulation of immune surveillance required to maintain homeostasis [10]. Protein structure studies have classified defensins into three groups; α-, β- and θ-defensins, which are synthesized in leukocytes and can also be produced by various epithelial cells and mucosal tissues [11]. Evolutionarily speaking, β-defensins are the common ancestor of all types of defensins in vertebrates [12]. In humans, β-defensins (hBDs) are the most abundant AMPs, and at present, more than 30 hBDs have been recognized, but only hBD1, hBD2, hBD3 and hBD4 have been well analyzed [13]. Their expression is constitutive, but infection-induced expression has been reported for hBD2 and hBD3 [14].
Although hBDs are primarily recognized as AMPs, they are also potent immunomodulators, and their role in regulating innate and adaptive immune responses is highly evolving [8]. In microbial infections, it has been indicated that hBDs levels change in response to viral, bacterial, and fungal infections, and thus their clinical relevance in these infections has been proposed [15]. In contrast to α-defensins, which have anti-viral effects, hBDs do not appear to affect virions, but rather inhibit viral replication by altering cellular functions [16]. In this context, it has been suggested that hBDs can modulate virus binding to host cell surface receptors and can also disrupt intracellular signaling, and through these mechanisms virus replication can be inhibited [17]. hBDs can also act as chemokines to augment and alter adaptive immune responses against viruses; for instance, hBD3 has been shown to be involved in the induction of chemokine release from monocyte-derived macrophages in HIV-infected patients and this can regulate cell migration into inflamed tissues [18]. Further, it has been shown that hBDs can regulate the gene expression of pro-inflammatory cytokines, and thus the level of these immunomodulators may be affected [19]. In addition, hBDs (for instance, hBD2 and hBD3) can also be induced by pro-inflammatory cytokines in response to invading pathogens [7]. In COVID-19, the pathogenesis has been characterized by pathological levels of pro-inflammatory cytokines particularly in severe cases [4], and thus hBDs may show dysregulated expression in COVID-19 patients. In line with these proposals, it is plausible to hypothesize that hBDs are altered in COVID-19 patients. To test this hypothesis, the levels of four hBDs (hBD1, hBD2, hBD3 and hBD4) were analyzed in serum of patients with severe COVID-19 infection.
2. Materials and methods
2.1. Cases and controls
During the period from September to December 2020, a case-control study was conducted on 103 patients with COVID-19 and 105 healthy volunteers (control group). Patients were admitted to COVID-19 care units in Baghdad hospitals, and were enrolled in study 4–5 days after admission. The RealLine SARS-CoV-2 kit (Bioron Diagnostics GmbH) was used to diagnose COVID-19 in nasopharyngeal swabs of patients on admission. The diagnosis was confirmed by a computed tomography (CT) scan of the chest. After 4–7 days of hospitalization, only patients who were willing to participate in the study and tested positive for molecular testing and CT scan indicating COVID-19 were included. In the initial protocol of our study, we took into account the identification of moderate, severe and critical patients, but were faced with the fact that the wards had mostly severe cases and, therefore, only cases with severe COVID-19 were enrolled in the study. The World Health Organization (WHO) Interim Guidance for determining disease severity were followed, and COVID-19 was deemed severe if the patient met one of the following conditions: severe respiratory distress, respiratory rate ≥30 breaths/minute and pulse oxygen saturation (SpO2) ≤ 93% on resting state [20]. Information regarding age, gender, body mass index (BMI) and chronic diseases (cardiovascular and diabetes) were recorded for patients and controls. In addition, patients were profiled for hemoglobin, platelet count, white blood cell count (WBC), erythrocyte sedimentation rate (ESR), random blood glucose (RBG), alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), blood urea nitrogen (BUN), creatinine, uric acid, total cholesterol and triglycerides. The control group included blood donors and health service personnel who had no respiratory infection in the past 12 months, and did not suffer from chronic diseases (respiratory allergy, diabetes and cardiovascular disease). Controls were initially examined for C-reactive protein (CRP) and erythrocyte sedimentation rate (ESR), and only subjects with CRP-seronegative and those with an ESR less than 20 mm/h were included. Besides, the results of serum tests for the anti-pathogen antibody panel in the Central Blood Bank (Baghdad) were also negative. All participants gave written consent after obtaining approval from the Ethics Committee of the Iraqi Ministry of Health and Environment.
2.2. Serum level of hBDs
Five milliliters of blood were drawn from each participant in a plain tube. The tube was centrifuged after blood clotting (3000 rpm for 15 min at 4 °C), and serum was collected and kept frozen at – 20 °C until assessment. Serum levels of hBDs (hBD1, hBD2, hBD3 and hBD4) were determined using enzyme-linked immunosorbent assay kits (Cat. No: E6426Hu, E6427Hu, E6428Hu and E3146Hu, respectively; Bioassay Technology Laboratory, China), and manufacturer's instructions were followed. The standard curve ranges of the kits were 0–400, 0–7000, 0–15000 and 0–15 ng/L, respectively.
2.3. Statistical analysis
Number and percentage were used to express categorical variables, and significant differences were assessed using Pearson Chi-square test. Parametric variables were given as mean ± standard deviation (SD) and significant differences were assessed using the least significant difference (LSD) test. Nonparametric variables were expressed as median and interquartile range (IQR), and Mann-Whitney U test was used to assess significant differences between medians. Receiver operating curve (ROC) analysis was performed to calculate area under the curve (AUC), 95% confidence interval (CI), cut-off value, sensitivity and specificity. The Youden index was used to optimize the cut-off value. Multinomial logistic regression analysis was applied to determine odds ratio (OR) and 95% CI. The analysis was adjusted for age (Model I), age and gender (Model II), or age, gender and body mass index (Model III). Correlations between variables were assessed by Spearman rank-order correlation analysis. A probability (p) value ≤ 0.05 was considered significant. The p-value was corrected using Bonferroni correction due to multiple comparisons. The statistical analysis was performed using IBM SPSS Statistics 25.0 (Armonk, NY: IBM Corp.) and GraphPad Prism version 8.0.0 (San Diego, California USA). G*Power software (version 3.1.9.7) was used to calculate power of sample size.
3. Results
3.1. Power of sample size
The sample sizes of patients (N = 103) and controls (N = 105) were computed in the G*Power software to calculate the power of sample size (1 – β error probability). It was found that at two-tailed α error probability of 0.05 and effect size convention of 0.5, the power of sample size was 0.95. Thus, statistical validation of the included samples was ensured.
3.2. Baseline characteristics of cases and controls
The mean age of COVID-19 cases was significantly higher than that of control subjects (56.9 ± 14.9 vs. 32.7 ± 8.7 years; p < 0.001). Most patients were older than 45 years (80.6%), while most healthy subjects were younger than 45 years old (87.6%). Patients and controls distributed by gender (males and females) or BMI (normal weight and overweight/obese) did not show significant differences. More than 50% of patients (57.3%) had chronic diseases (cardiovascular disease, diabetes or both) (Table 1 ).
Table 1.
Baseline characteristics of COVID-19 cases and controls.
| Characteristic | Cases (N = 103) | Controls (N = 105) | p-value | ||
|---|---|---|---|---|---|
| Mean age ± SD (years) | 56.9 ± 14.9 | 32.7 ± 8.7 | <0.001 | ||
| Age groups; years | <45 | 20 (19.4) | 92 (87.6) | <0.001 | |
| ≥45 | 83 (80.6) | 13 (12.4) | |||
| Gender; N (%) | Male | 76 (73.8) | 73 (69.5) | 0.495 | |
| Female | 27 (26.2) | 32 (30.5) | |||
| BMI; N (%) | Normal weight | 28 (27.2) | 39 (37.1) | 0.124 | |
| Overweight/obese | 75 (72.8) | 66 (62.9) | |||
| Chronic disease; N (%) | Yes | 59 (57.3) | 0 (0.0) | < 0.001 | |
| No | 44 (42.7) | 100 (100.0) | |||
SD: Standard deviation; BMI: Body mass index (kg/m2); p: Probability of least significant difference or Pearson Chi-square test. Significant p-value is bold-marked.
3.3. Baseline laboratory parameters of cases
COVID-19 cases were profiled for the laboratory parameters given in Table 2 . Some parameters were within the reference ranges while others were not. Means of WBC (12.3 ± 5.2 × 109/L), ESR (58.4 ± 27.0 mm/h), RBG (233.4 ± 128.9 mg/dL), BUN (61.0 ± 44.0 mg/dL) and triglycerides (219.6 ± 113.4 mg/dL) were above the reference range, while ALT occupied the upper limit of the reference range (58.3 ± 53.1 U/L) (Table 2).
Table 2.
Baseline laboratory parameters of COVID-19 cases.
| Parameter | Mean ± SD | Median (IQR: 5–95%) | Reference range† | Status |
|---|---|---|---|---|
| Hemoglobin (g/dL) | 14.3 ± 9.8 | 13.7 (10.0–17.4) | 11.6–16.6 | Normal |
| Platelets ( × 109/L) | 282.9 ± 135.9 | 251.0 (125.0–565.0) | 135–317 | Normal |
| WBC ( × 109/L) | 12.3 ± 5.2 | 11.5 (5.6–21.0) | 3.4–9.6 | Increased |
| ESR (mm/hour) | 58.4 ± 27.0 | 57.0 (14.0–98.0) | 0–29 | Increased |
| RBG (mg/dL) | 233.4 ± 128.9 | 185.0 (104.9–503.0) | 79–140 | Increased |
| ALT (U/L) | 58.3 ± 53.1 | 44.8 (14.5–162.0) | 7–55 | Upper limit |
| AST (U/L) | 42.8 ± 24.4 | 37.9 (15.2–87.0) | 8–48 | Normal |
| ALP (IU/L) | 91.3 ± 54.4 | 81.8 (45.8–146.3) | 40–129 | Normal |
| BUN (mg/dL) | 61.0 ± 44.0 | 47.0 (23.0–155.0) | 7–20 | Increased |
| Creatinine (mg/dL) | 1.1 ± 0.7 | 0.9 (0.5–2.8) | 0.74–1.35 | Normal |
| Uric acid (mg/dL) | 5.6 ± 2.1 | 5.8 (1.8–8.6) | 2.7–8.0 | Normal |
| Total cholesterol (mg/dL) | 184.6 ± 96.9 | 161.3 (93.0–396.0) | <200 | Normal |
| Triglycerides (mg/dL) | 219.6 ± 113.4 | 194.0 (80.8–428.0) | <150 | Increased |
WBC: White blood cell count; ESR: Erythrocyte sedimentation rate; RBG: Random blood glucose; ALT: Alanine aminotransferase; AST: Aspartate aminotransferase; ALP: Alkaline phosphatase; BUN: Blood urea nitrogen; SD: Standard deviation; IQR: Interquartile range; †: Data source from https://www.mayoclinic.org.
3.4. Serum levels of hBDs
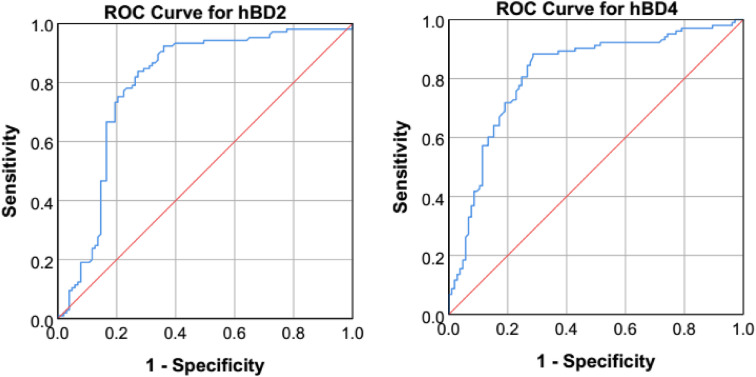
The median levels of hBD1 and hBD3 did not show significant differences between COVID-19 cases and controls. On the contrary, the levels of hBD2 were significantly decreased in cases compared to controls (1036 [IQR: 857–1187] vs. 1289 [IQR: 1214–1388) ng/L; p < 0.001), while the levels of hBD4 were significantly increased (4.04 [IQR: 3.32–5.07] vs. 2.43 [IQR: 2.09–3.06] ng/L; p < 0.001) (Fig. 1 ). ROC curve analysis revealed the predictive significance of hBD2 (AUC = 0.795; 95% CI = 0.729–0.861; p < 0.001; Youden index = 0.55; cut-off value = 1193 ng/L; sensitivity = 77.1%; specificity = 77.7%) and hBD4 (AUC = 0.816; 95% CI = 0.756–0.876; p < 0.001; Youden index = 0.56; cut-off value = 3.23 ng/L; sensitivity = 80.6%; specificity = 75.2%) in discriminating between COVID-19 cases and controls (Fig. 2 ). Logistic regression analysis (Models I, II and III) confirmed the significance of hBD2 (OR = 0.996, 0.996 and 0.996, respectively; pc = 0.004) and hBD4 (OR = 4.781, 4.926 and 4.948, respectively; pc < 0.001) in susceptibility to COVID-19 (Table 3 ).
Fig. 1.
Scatter dot plots of hBD1, hBD2, hBD3 and HBD4 levels in serum of COVID-19 cases and controls showing median (horizontal line) and interquartile range (vertical line). The significance of difference between medians was assessed by Mann-Whitney U test.
Fig. 2.
Receiver operating curve (ROC) analysis of hBD2 (AUC = 0.795; 95% CI = 0.729–0.861; p-value < 0.001; Youden index = 0.55; cut-off value = 1193 ng/L; sensitivity = 77.1%; specificity = 77.7%) and hBD4 (AUC = 0.816; 95% CI = 0.756–0.876; p-value < 0.001; Youden index = 0.56; cut-off value = 3.23 ng/L; sensitivity = 80.6%; specificity = 75.2%) in COVID-19 patients versus controls.
Table 3.
Logistic regression analysis of hBD1, hBD2, hBD3 and hBD4 in serum of COVID-19 cases and controls.
| hBD | Model I |
Model II |
Model III |
|||
|---|---|---|---|---|---|---|
| OR (95% CI)† | p (pc) | OR (95% CI)† | p (pc) | OR (95% CI)† | p (pc) | |
| hBD1 | 1.004 (0.996–1.011) | 0.343 (1.0) | 1.004 (0.996–1.012) | 0.351 (1.0) | 1.005 (0.997–1.012) | 0.232 (0.928) |
| hBD2 | 0.996 (0.993–.998) | 0.001 (0.004) | 0.996 (0.993–0.998) | 0.001 (0.004) | 0.996 (0.993–0.998) | 0.001 (0.004) |
| hBD3 | 0.999 (0.997–1.000) | 0.037 (0.148) | 0.998 (0.997–1.000) | 0.028 (0.112) | 0.998 (0.997–1.000) | 0.031 (0.124) |
| hBD4 | 4.781 (2.579–8.863) | < 0.001 (< 0.001) | 4.926 (2.627–9.239) | < 0.001 (< 0.001) | 4.948 (2.632–9.302) | < 0.001 (< 0.001) |
hBD: Human β-defensin; OR: Odds ratio; CI: Confidence interval; p: Probability; pc: Bonferroni-corrected probability; †: The reference category was controls, and the analysis was adjusted for age (Model I), age and gender (Model II), or age, gender and body mass index (Model III). Significant p-value is bold-marked.
3.5. Serum levels of hBDs stratified by characteristics of cases
COVID-19 cases stratified according to age group, gender, BMI and chronic diseases showed no significant differences between medians of hBD1, hBD2, hBD3 and hBD4 in each stratum (Table 4 ). A similar observation was made in controls (data not shown).
Table 4.
Median levels of hBD1, hBD2, hBD3 and hBD4 in serum of COVID-19 cases stratified according to characteristics of patients.
| Characteristic | Median (IQR: 25–75%); ng/L |
||||
|---|---|---|---|---|---|
| hBD1 | hBD2 | hBD3 | hBD4 | ||
| Age group | <45 years | 198.0 (151.5–307.3) | 1100 (955–1203) | 1580 (1470–2144) | 4.08 (3.29–5.13) |
| ≥45 years | 204.9 (158.5–238.1) | 1015 (852–1175) | 1532 (1220–1936) | 4.04 (3.32–5.03) | |
| p-value | 0.665 | 0.293 | 0.107 | 0.623 | |
| Gender | Male (N = 76) | 199.4 (157.6–234.8) | 1009 (853–1188) | 1583 (1332–1960) | 4.05 (3.32–4.93) |
| Female (N = 27) | 206.0 (162.8–307.4) | 1071 (954–1185) | 1453 (1142–2020) | 4.04 (3.19–5.79) | |
| p-value | 0.669 | 0.414 | 0.152 | 0.913 | |
| BMI | NOR (N = 28) | 227.4 (160.5–323.6) | 1106 (845–1339) | 1668 (1306–2160) | 4.43 (3.32–5.27) |
| O/O (N = 75) | 196.9 (157.6–229.8) | 1008 (866–1147) | 1532 (1220–1936) | 3.95 (3.26–5.03) | |
| p-value | 0.138 | 0.128 | 0.341 | 0.224 | |
| CHD | Yes (N = 59) | 197.2 (157.6–231.2) | 1053 (921–1189) | 1644 (1297–2050) | 4.18 (3.55–5.26) |
| No (N = 44) | 207.5 (156.8–258.3) | 1012 (844–1180) | 1491 (1152–1893) | 3.88 (3.15–4.81) | |
| p-value | 0.857 | 0.650 | 0.148 | 0.146 | |
BMI: Body mass index; NOR: Normal weight; O/O: Overweight/obese; CHD: Chronic disease; IQR: Interquartile range; hBD: Human β-defensin; p: Probability of Mann-Whitney U test.
3.6. Spearman's rank-order correlation analysis
Spearman rank-order correlation analysis was performed to estimate correlation coefficient (rs) between hBD1, hBD2, hBD3, hBD4, WBC, ESR and RBG. The analysis revealed that hBD1 was positively correlated with hBD2 (rs = 0.260; p < 0.001) and hBD3 (rs = 0.1765; p = 0.011); hBD2 was positively correlated with hBD3 (rs = 0.295; p < 0.001) and negatively correlated with hBD4 (rs = −0.166; p = 0.016), WBC (rs = −0.192; p = 0.005), ESR (rs = −0.459; p < 0.001) and RBG (rs = −0.436; p < 0.001); hBD3 was positively correlated with hBD4 (rs = 0.464; p < 0.001); hBD4 was positively correlated with WBC (rs = 0.269; p < 0.001), ESR (rs = −0.422; p < 0.001) and RBG (rs = 0.436; p < 0.001); WBC was positively correlated with ESR (rs = 0.370; p < 0.01) and RBG (rs = 0.180; p = 0.009); and ESR was positively correlated with RBG (rs = 0.665; p < 0.001) (Fig. 3 and Supplementary Table).
Fig. 3.
Scatter plot Spearman rank-order correlation coefficient (rs) for analysis between human β-defensins (hBD1, hBD2, hBD3 and hBD4), white blood cell count (WBC), erythrocyte sedimentation rate (ESR) and random blood glucose (RBG).
4. Discussion
The immune system plays an important role in the development of COVID-19, and understanding its functional components may pave the way for the development of control strategies. The current study focused on a group of these components, hBD1, hBD2, hBD3 and hBD4, due to their anti-viral and immunomodulatory properties [[5], [6], [7]]. The results showed that hBD2 was lower in serum of COVID-19 patients compared to healthy controls, while hBD4 showed higher levels. ROC curve analysis demonstrated the predictive significance of hBD2 and hBD4 in discriminating between COVID-19 patients and controls, and the recorded AUCs were 0.795 and 0.816, respectively. Besides, logistic regression confirmed the significance of both hBDs in susceptibility to COVID-19. hBD2 was proposed to be associated with a decreased risk of COVID-19 (OR = 0.996), while an increased risk was associated with hBD4 (OR = 4.948) after adjusting the analysis for age, gender and BMI. However, a point of concern must be addressed in this context, patients and controls were not age-matched, with most patients being >45 years old and most controls <45 years old. To investigate this issue, the levels of hBDs were analyzed in patients and controls after stratification by age groups (<45 and ≥ 45 years). The analysis revealed no age-related differences, and this may indicate that age had no effect on hBDs levels. In fact, other characteristics of patients or controls (gender, BMI and chronic disease) also did not influence the levels of hBDs (Table 4). These analyses indicate that hBD2 and hBD4 are involved in the pathogenesis of COVID-19 regardless of age, gender, BMI or chronic disease.
hBD2 is a cationic peptide that is expressed on the epithelial surfaces of various systems in the human body including the respiratory system. Viral, bacterial and fungal pathogens can also induce the expression of hBD2, and thus its potential in controlling respiratory infections caused by these pathogens has been proposed [19]. Indeed, hBD2 was initially identified as a peptide with anti-bacterial effects, but recent investigations have shown that this defensin is also effective against enveloped and non-enveloped viruses, and a number of studies have demonstrated dysregulated expression of hBD2 in respiratory infections caused by viral agents (influenza virus, respiratory syncytial virus, adenoviruses and rhinovirus) [[21], [22], [23], [24], [25]]. The anti-viral effects of hBD2 could be through direct interference with viral replication or indirectly by activating antiviral immune responses. In this context, it has been reported that hBD2 induces dendritic cell maturation and enhances their recruitment to the site of inflammation to promote adaptive immunity [26]. Besides, various pro-inflammatory cytokines have been described to participate in increasing the expression of hBD2 [27], and conversely it has been found that recombinant hBD2 was able to suppress dendritic cell-mediated secretion of pro-inflammatory cytokines [28]. Due to these biological properties, hBD2 has been proposed to have role in controlling SARS-CoV-2 infection [7]. In the current study, patients with severe COVID-19 showed a down-regulated expression of hBD2 and this may promote persistence of viral infection and contribute to disease severity. Unfortunately, there is no direct evidence to support or refute these preliminary findings. In the Middle East respiratory syndrome-coronavirus (MERS-CoV), the ability of hBD2 to enhance anti-viral immunity in vitro and in vivo was studied using the MERS-CoV protein receptor binding domain as a model antigen. The study concluded that HBD2 can activate the primary anti-viral innate immune response and may also potentiate the induction of an effective antigen-specific immunity [29]. However, in a recent investigation, the effects of hBDs (hBD2, hBD3 and hBD6) on SARS-CoV-2 infection were studied, but these defensins did not effectively inhibit viral replication [30]. No other study has been conducted in this regard; therefore, further studies are needed to unravel the role of hBD2 in controlling COVID-19 and/or disease severity.
In contrast to hBD2, hBD4 levels were significantly elevated in the serum of patients with severe COVID-19, and this was associated with an approximately 5-fold increased risk of SARS-CoV-2 infection. hBD4 is a new member of the hBD family of AMPs that shows expression in neutrophils and the lung tissue (bronchial and bronchiolar epithelium), but this expression and the pathological significance of hBD4 in respiratory infections have not been well elucidated [31]. This defensin is a cationic peptide with known anti-bacterial and anti-fungal properties [32,33], but the anti-viral effects of hBD4 have not been explored. This study is perhaps the first to examine the relationship between hBD4 and COVID-19 and the results may be promising as hBD4 is likely to be involved in the pathogenesis of SARS-CoV-2 infection. The effects of hBD4 on immune system functions have also not been investigated. However, and contrast to hBD1, hBD2 and hBD3, it has been shown that hBD4 cannot induce chemotaxis mediated by the chemokine receptor 6 (CCR6) [34]. Further, a recent study showed that HBD4 is highly expressed in human dental-derived stem cells after stimulation by pro-inflammatory cytokines. The study also documented the anti-inflammatory activity of hBD4 along with its anti-bacterial effects [35]. Besides, the present study illustrated that serum level of hBD4 was positively correlated with WBC and ESR (inflammatory markers), and this may suggest that hBD4 is linked to inflammation in COVID-19 patients. Taken together, these results suggest a role for hBD4 in immunity and inflammation and may be associated with susceptibility to COVID-19, but further studies in this context are warranted.
Regardless of these findings, the study faced some limitations. First, asymptomatic COVID-19 patients and patients with mild or critical illness were not studied. Second, although the levels of hBDs did not show any age-related differences, the sample size of patients in the <45-year-old group and controls in the ≥45-year-old group was small. Third, serum levels of some pro-inflammatory cytokines were not determined. Fourth, Data regarding viral load at the time of the study were not obtained.
In conclusion, the study indicated that hBD2 showed low levels in serum of patients infected with severe COVID-19, while hBD4 showed elevated levels. These differences in HBDs were not influenced by age, gender, BMI, or chronic disease.
Author contributions
Noor Al-Bayatee: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing – original draft, Visualization, Writing – review and editing.
Ali Ad'hiah: Conceptualization, Methodology, Investigation, Formal analysis, Data curation. Writing – review and editing, Supervision, Project administration.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We appreciate the kind assistance and cooperation of the medical staff at Dar Al-Salam Field Hospital, Al-Karkh General Hospital and Al-Furat General Hospital.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micpath.2021.105205.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wang C., Wang Z., Wang G., Lau J.Y.N., Zhang K., Li W. COVID-19 in early 2021: current status and looking forward. Signal Transduct. Target. Ther. 2021;6:1–14. doi: 10.1038/s41392-021-00527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabaan A.A., Al-Ahmed S.H., Garout M.A., Al-Qaaneh A.M., Sule A.A., Tirupathi R., Al Mutair A., Alhumaid S., Hasan A., Dhawan M., Tiwari R., Sharun K., Mohapatra R.K., Mitra S., Bin Emran T., Bilal M., Singh R., Alyami S.A., Moni M.A., Dhama K. Diverse immunological factors influencing pathogenesis in patients with COVID-19: a review on viral dissemination, immunotherapeutic options to counter cytokine storm and inflammatory responses. Pathogens. 2021;10:565. doi: 10.3390/pathogens10050565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tahaghoghi-Hajghorbani S., Zafari P., Masoumi E., Rajabinejad M., Jafari-Shakib R., Hasani B., Rafiei A. The role of dysregulated immune responses in COVID-19 pathogenesis. Virus Res. 2020;290:198197. doi: 10.1016/j.virusres.2020.198197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowaiye A.B., Okpalefe O.A., Adejoke O.O., Ogidigo J.O., Oladipo O.H., Ogu A.C., Oli A.N., Olofinase S., Onyekwere O., Abubakar A.R., Jahan D., Islam S., Dutta S., Haque M. Attenuating the effects of novel COVID-19 (SARS-CoV-2) infection-induced cytokine storm and the implications. J. Inflamm. Res. 2021;14:1487–1510. doi: 10.2147/JIR.S301784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mousavi Maleki M.S., Restamian M., Madanchi H. Antimicrobial peptides and other peptide-like therapeutics as promising candidates to combat SARS-CoV-2. Expert Rev. Anti Infect. Ther. 2021 doi: 10.1080/14787210.2021.1912593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang R., Jiang X., Qiao J., Wang Z., Tong A., Yang J., Yang S., Yang L. Antimicrobial peptide DP7 with potential activity against SARS coronavirus infections. Signal Transduct. Target. Ther. 2021;6:1–3. doi: 10.1038/s41392-021-00551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solanki S.S., Singh P., Kashyap P., Sansi M.S., Ali S.A. Promising role of defensins peptides as therapeutics to combat against viral infection. Microb. Pathog. 2021;155:104930. doi: 10.1016/j.micpath.2021.104930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond G., Beckloff N., Weinberg A., Kisich K. The roles of antimicrobial peptides in innate host defense. Curr. Pharmaceut. Des. 2009;15:2377–2392. doi: 10.2174/138161209788682325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L.J., Gallo R.L. Antimicrobial peptides. Curr. Biol. 2016;26:R14–R19. doi: 10.1016/j.cub.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Prasad S.V., Fiedoruk K., Daniluk T., Piktel E., Bucki R. Expression and function of host defense peptides at inflammation sites. Int. J. Mol. Sci. 2020;21:104. doi: 10.3390/ijms21010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huan Y., Kong Q., Mou H., Yi H. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front. Microbiol. 2020;11:582779. doi: 10.3389/fmicb.2020.582779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Machado L.R., Ottolini B. An evolutionary history of defensins: a role for copy number variation in maximizing host innate and adaptive immune responses. Front. Immunol. 2015;6:115. doi: 10.3389/fimmu.2015.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amerikova M., Pencheva El-Tibi I., Maslarska V., Bozhanov S., Tachkov K. Antimicrobial activity, mechanism of action, and methods for stabilisation of defensins as new therapeutic agents. Biotechnol. Biotechnol. Equip. 2019;33:671–682. doi: 10.1080/13102818.2019.1611385. [DOI] [Google Scholar]
- 14.Shelley J.R., Davidson D.J., Dorin J.R. The dichotomous responses driven by β-defensins. Front. Immunol. 2020;11:1176. doi: 10.3389/fimmu.2020.01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fruitwala S., El-Naccache D.W., Chang T.L. Multifaceted immune functions of human defensins and underlying mechanisms. Semin. Cell Dev. Biol. 2019;88:163. doi: 10.1016/J.SEMCDB.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klotman M.E., Chang T.L. Defensins in innate antiviral immunity. Nat. Rev. Immunol. 2006;6:447–456. doi: 10.1038/nri1860. [DOI] [PubMed] [Google Scholar]
- 17.Wilson S.S., Wiens M.E., Smith J.G. Antiviral mechanisms of human defensins. J. Mol. Biol. 2013;425:4965–4980. doi: 10.1016/j.jmb.2013.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrov V., Funderburg N., Weinberg A., Sieg S. Human β defensin-3 induces chemokines from monocytes and macrophages: diminished activity in cells from HIV-infected persons. Immunology. 2013;140:413–420. doi: 10.1111/imm.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meade K.G., O'Farrelly C. Β-Defensins: farming the microbiome for homeostasis and health. Front. Immunol. 2019;10:3072. doi: 10.3389/fimmu.2018.03072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization . Who; 2020. Clinical Management of Severe Acute Respiratory Infection when Novel Coronavirus (2019-nCoV) Infection Is Suspected: Interim Guidance 28 January 2020; p. 10.https://apps.who.int/iris/handle/10665/330893 [Google Scholar]
- 21.Chong K.T., Thangavel R.R., Tang X. Enhanced expression of murine β-defensins (MBD-1, -2,- 3, and -4) in upper and lower airway mucosa of influenza virus infected mice. Virology. 2008;380:136–143. doi: 10.1016/j.virol.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 22.Kota S., Sabbah A., Te H.C., Harnack R., Xiang Y., Meng X., Bose S. Role of human β-defensin-2 during tumor necrosis factor-α/NF- βB-mediated innate antiviral response against human respiratory syncytial virus. J. Biol. Chem. 2008;283:22417–22429. doi: 10.1074/jbc.M710415200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnason J.W., Murphy J.C., Kooi C., Wiehler S., Traves S.L., Shelfoon C., MacIejewski B., Dumonceaux C.J., Lewenza W.S., Proud D., Leigh R. Human β-defensin-2 production upon viral and bacterial coinfection is attenuated in COPD. PloS One. 2017;12 doi: 10.1371/journal.pone.0175963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J., Yang Y.L., Jang Y.S. Human β-defensin 2 is involved in CCR2-mediated Nod2 signal transduction, leading to activation of the innate immune response in macrophages. Immunobiology. 2019;224:502–510. doi: 10.1016/j.imbio.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding J., Chou Y.Y., Chang T.L. Defensins in viral infections. J. Innate Immun. 2009;1:413–420. doi: 10.1159/000226256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park M.S., Il Kim J., Lee I., Park S., Bae J.Y., Park M.S. Towards the application of human defensins as antivirals. Biomol. Ther. 2018;26:242–254. doi: 10.4062/biomolther.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patricia Rosete Olvera D., Cabello Gutiérrez C. Immune Response Act. Immunomodulation; IntechOpen: 2019. Multifunctional activity of the β-defensin-2 during respiratory infections. [DOI] [Google Scholar]
- 28.Koeninger L., Armbruster N.S., Brinch K.S., Kjaerulf S., Andersen B., Langnau C., Autenrieth S.E., Schneidawind D., Stange E.F., Malek N.P., Nordkild P., Jensen B.A.H., Wehkamp J. Human β-defensin 2 mediated immune modulation as treatment for experimental colitis. Front. Immunol. 2020;11:93. doi: 10.3389/fimmu.2020.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J., Yang Y.L., Jang S.-H., Jang Y.-S. Human β-defensin 2 plays a regulatory role in innate antiviral immunity and is capable of potentiating the induction of antigen-specific immunity. Virol. J. 2018;151(15):1–12. doi: 10.1186/S12985-018-1035-2. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu C., Wang A., Marin M., Honnen W., Ramasamy S., Porter E., Subbian S., Pinter A., Melikyan G.B., Lu W., Chang T.L. Human defensins inhibit SARS-CoV-2 infection by blocking viral entry. Viruses. 2021;13:1246. doi: 10.3390/v13071246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanagi S., Ashitani J.I., Ishimoto H., Date Y., Mukae H., Chino N.N., Nakazato M. Isolation of human β-defensin-4 in lung tissue and its increase in lower respiratory tract infection. Respir. Res. 2005;6:1–9. doi: 10.1186/1465-9921-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma H., Nagaraj R. Antimicrobial activity of human β-defensin 4 analogs: insights into the role of disulfide linkages in modulating activity. Peptides. 2012;38:255–265. doi: 10.1016/j.peptides.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 33.Sharma H., Nagaraj R. Human β-defensin 4 with non-native disulfide bridges exhibit antimicrobial activity. PloS One. 2015;10 doi: 10.1371/journal.pone.0119525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prahl A., Pazgier M., Alexandratos J., Lubkowski J. Human β-defensin 4 - defensin without the “twist,”. Postepy Biochem. 2016;62:349–361. https://pubmed.ncbi.nlm.nih.gov/28132490/ accessed. [PubMed] [Google Scholar]
- 35.Zhai Y., Wang Y., Rao N., Li J., Li X., Fang T., Zhao Y., Ge L. Activation and biological properties of human β defensin 4 in stem cells derived from human exfoliated deciduous teeth. Front. Physiol. 2019;10:1304. doi: 10.3389/fphys.2019.01304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.