Abstract
As part of an ongoing surveillance program of antibiotic-resistant Streptococcus pneumoniae in Sofia, Bulgaria, 120 penicillin-resistant strains (PRSP) (most of them recovered from children hospitalized with pneumococcal disease) were analyzed by microbiological and molecular methods. Several unique features of this collection are of particular interest. (i) Most isolates (112 of 120) were also resistant to trimethoprim-sulfamethoxazole (SXT) (97 of 120 isolates, or 80%), and over 70% (86 of 120) of the isolates were resistant to at least three antibiotics in addition to penicillin. (ii) Close to 80% of all isolates were represented by large clusters of bacteria, each with a unique serotype, antibiotype, and chromosomal macrorestriction pattern (determined by pulsed-field gel electrophoresis), as well as unique restriction fragmentation length polymorphisms of the penicillin-binding protein genes pbp1a, pbp2x, and pbp2b. (iii) A large proportion (45 of 120, or 38%) of the strains belonged to two internationally spread epidemic clones of S. pneumoniae, the first expressing capsular type 23F and the second expressing serotype 9. (iv) A unique Bulgarian cluster composed of eight serotype 19F isolates was resistant to tetracycline, SXT, cefotaxime, and extremely high levels of penicillin and erythromycin. Nevertheless, this clone did not react with either the erm or the mef DNA probes, and thus the mechanism of macrolide resistance in this group of PRSP remains to be elucidated.
Several recent studies indicate that the frequency of penicillin-resistant and multidrug-resistant Streptococcus pneumoniae bacteria has continued to increase in the 1990s (22). Mechanisms involved include the acquisition of penicillin resistance genes by susceptible isolates, generating bacteria for which the penicillin MICs were moderately increased, and the geographic spread of genetic lineages (clones) for which penicillin MICs are most frequently elevated (1 μg/ml or higher) and that usually also carry resistance traits to other antimicrobial agents. One such epidemic penicillin-resistant clone is the “Spanish/USA clone,” usually expressing the 23F serotype and which has spread to several countries in Europe (6, 7, 14, 18, 21, 26, 30), North and South America (18, 28), the Far East (16, 25), and South Africa (11). A second, extensively spread clone, sometimes referred to as the “French/Spanish clone,” usually classified as serogroup 9 or serotype 14, and carrying resistance to penicillin and trimethoprim-sulfamethoxazole, has spread to several countries in Europe and South America (8, 9, 13, 17, 18, 28).
Methods of molecular epidemiology have begun to play an increasingly important role in identifying the nature of genetic traits carried by these strains and in tracing their routes of geographic dispersal.
A recent survey indicated that S. pneumoniae bacteria with reduced susceptibilities to penicillin (PRSP) constitute a high proportion (25%) of invasive and colonizing isolates in Bulgaria (22). In the follow-up study described here we used molecular fingerprinting techniques in order to better understand the nature and origin of the penicillin-resistant Bulgarian strains of S. pneumoniae. Our data indicate that a surprisingly large proportion of PRSP from Bulgaria for which penicillin MICs are 1 μg/ml or higher are members of the Spanish/USA or French/Spanish clone, suggesting the importation of these strains from abroad. The molecular fingerprinting techniques also identified three hitherto unseen PRSP lineages: one highly penicillin-resistant and multidrug-resistant cluster of serotype 19F and large clusters of serotypes 6B and 14, respectively, both of which were represented by isolates with low-level penicillin resistance accompanied by resistance to erythromycin and tetracycline. Each of these three clusters appears to be unique to this Bulgarian collection of PRSP.
MATERIALS AND METHODS
Bacterial strains.
We analyzed 120 clinical isolates of penicillin-resistant S. pneumoniae strains collected between 1991 to 1997 in Sofia, Bulgaria. Strains were isolated from inpatients at seven different hospitals.
Clinical sources, dates, hospitals, clinical diagnoses, and other relevant information related to the isolates are described in Table 1.
TABLE 1.
Penicillin-resistant clinical isolates of S. pneumoniae collected from 1991 to 1997 in Sofia, Bulgaria
| Isolate | Hospitala | Source of isolateb | Isolation datec | Patient
|
Serotype | MIC (μg/ml)d | Antibiotic resistance patterne | PFGE patternf | |
|---|---|---|---|---|---|---|---|---|---|
| Age | Sex | ||||||||
| Bul 52 | HID | Eye | 03/12/93 | 1 yr 5 mo | M | 23F | 2 | P, C, T, SXT, CTX | A1 |
| Bul 53 | HID | Throat | 03/12/93 | 28 yr | M | 23 | 2 | P, E, C, T, SXT, CTX | A1 |
| Sof 33 | I City Hospital | Nph | 08/11/93 | 1 yr | F | 23 | 2 | P, C, T, SXT | A1 |
| Sof 106 | HID | Nph | 11/12/93 | 5 mo | M | 23 | 2 | P, C, T, SXT | A1 |
| Sof 176 | HID | Nph | 24/02/94 | 2 yr 4 mo | M | 23 | 2 | P, C, T, SXT, CTX | A1 |
| Bul 117 | Pirogov | Sputum | 1996 | >18 yr | 23 | 2 | P, C, T, SXT | A2 | |
| Bul 118 | Pirogov | Sputum | 1996 | >18 yr | 23 | 2 | P, C, T, SXT, CTX | A2 | |
| Bul 121 | Pirogov | Sputum | 1996 | >18 yr | 23F | 2 | P, C, T, SXT | A2 | |
| Sof 282 | HID | Nph | 03/05/94 | 8 mo | M | 23 | 2 | P, C, T, SXT, CTX | A3 |
| Bul 22 | UH Queen Ioanna | Ear fluid | 12/04/92 | 3 yr | 23F | 4 | P, C, T, SXT, CTX | A4 | |
| Bul 116 | Pirogov | Pleural fluid | 1995 | >18 yr | 23 | 2 | P, C, T, SXT | A4 | |
| Bul 89 | HID | Throat | 07/03/95 | 7 yr | M | 23F | 2 | P, C, T, SXT | A5 |
| Bul 114 | Pirogov | Sputum | 1995 | >18 yr | 23 | 2 | P, C, T, SXT | A6 | |
| Bul 97 | HID | Throat | 12/06/95 | 8 yr | F | 23 | 0.12 | P, SXT | B1 |
| Bul 103 | HID | Throat | 04/11/95 | 2 yr 8 mo | F | 23 | 0.12 | P, SXT | B1 |
| Bul 119 | Pirogov | Sputum | 1996 | >18 yr | 23F | 0.25 | P, SXT | B1 | |
| Bul 120 | Pirogov | Sputum | 1996 | >18 yr | 23 | 0.25 | P, SXT | B2 | |
| Sof 57 | 28 Polyclinic | Nph | 16/11/93 | 2 yr | M | 23 | 0.12 | P, SXT | B3 |
| Bul 63 | HID | Throat | 04/02/94 | 3 yr | F | 23 | 0.5 | P | C1 |
| Sof 163 | HID | Nph | 10/02/94 | 2 yr | M | 23 | 0.25 | P | C1 |
| Sof 345 | HID | Nph | 06/07/94 | 8 mo | F | 23 | 0.5 | P | C2 |
| Bul 7 | HID | Eye | 24/10/91 | 7 mo | F | 23 | 0.5 | P, T, SXT | D1 |
| Bul 8 | HID | Throat | 05/11/91 | 4 yr | F | 23 | 0.25 | P, T, SXT | D1 |
| Sof 204 | HID | Nph | 17/03/94 | 9 mo | M | 23 | 0.5 | P, T, SXT | D2 |
| Bul 112 | HID | Nose | 23/05/96 | 5 yr | M | 23 | 0.5 | P | D3 |
| Bul 78 | Children’s Hospital | Ear fluid | 09/11/94 | 6 yr | 6 | 0.12 | P, E, T | E1 | |
| Bul 92 | HID | Nose | 05/04/95 | 1 yr | F | 6 | 0.12 | P, E, T | E1 |
| Sof 73 | 28 Polyclinic | Nph | 22/11/93 | 2 yr 3 mo | F | 6 | 0.12 | P, E, T | E1 |
| Sof 74 | 28 Polyclinic | Nph | 22/11/93 | 1 yr 2 mo | F | 6 | 0.12 | P, E, T | E1 |
| Sof 201 | HID | Nph | 14/03/94 | 2 yr 3 mo | M | 6 | 0.12 | P, E, T, SXT | E1 |
| Sof 228 | HID | Nph | 23/03/94 | 10 mo | M | 6 | 0.12 | P, E, T | E1 |
| Sof 229 | HID | Nph | 23/03/94 | 1 yr | M | 6 | 0.12 | P, E, T, SXT | E1 |
| Sof 245 | HID | Nph | 30/03/94 | 8 mo | M | 6 | 0.12 | P, E, T | E1 |
| Sof 320 | HID | Nph | 08/06/94 | 1 yr 4 mo | F | 6 | 0.12 | P, E, T | E1 |
| Sof 330 | HID | Nph | 22/06/94 | 1 yr 4 mo | F | 6 | 0.25 | P, E, T | E1 |
| Sof 361 | HID | Nph | 25/07/94 | 1 yr 2 mo | M | 6 | 0.12 | P, E, T, SXT | E1 |
| Sof 378 | HID | Nph | 03/08/94 | 9 mo | M | 6 | 0.12 | P, E, T, SXT | E2 |
| Bul 21 | HID | Ear fluid | 30/03/92 | 9 mo | M | 6 | 0.12 | P, E, T, SXT | E2 |
| Bul 108 | HID | Nph | 08/12/95 | 1 yr 1 mo | M | 6 | 0.12 | P, E, T, SXT | E2 |
| Sof 193 | HID | Nph | 09/03/94 | 2 yr | F | 6 | 0.25 | P, E, T | E2 |
| Sof 220 | HID | Nph | 21/03/94 | 1 yr 1 mo | F | 6 | 0.12 | P, E, T | E2 |
| Sof 222 | HID | Nph | 21/03/94 | 6 mo | F | 6 | 0.12 | P, E, T | E2 |
| Sof 240 | HID | Nph | 28/03/94 | 7 mo | F | 6 | 0.12 | P, E, T | E2 |
| Sof 248 | HID | Nph | 04/04/94 | 6 mo | M | 6 | 0.12 | P, E, T, SXT | E2 |
| Bul 35 | HID | Nose | 05/02/93 | 1 yr | M | 6B | 0.12 | P, E, T | E3 |
| Bul 107 | HID | Throat | 27/11/95 | 3 yr | F | 6 | 0.12 | P, E, T, SXT | E3 |
| Sof 200 | HID | Nph | 14/03/94 | 2 yr 7 mo | F | 6 | 0.12 | P, E, T | E3 |
| Bul 72 | HID | Throat | 10/06/94 | 4 yr | M | 6 | 0.12 | P, C, SXT | E4 |
| Sof 370 | HID | Nph | 01/08/94 | 6 mo | M | 6 | 4 | P, E, C, SXT, CTX | E5 |
| Sof 114 | HID | Nph | 17/12/93 | 5 mo | M | 6 | 0.12 | P, E, T, SXT | E6 |
| Bul 87 | HID | Throat | 01/03/95 | 2 yr 6 mo | M | 6A | 0.12 | P, E, C, T | F1 |
| Sof 386 | HID | Nph | 09/08/94 | 1 yr 5 mo | M | 6 | 0.12 | P, SXT | F2 |
| Bul 20 | HID | CSF | 28/03/92 | 36 yr | M | 6 | 0.12 | P | G |
| Bul 101 | HID | Nose | 17/10/95 | 5 yr | M | 6A | 0.12 | P, SXT | H |
| Bul 45 | HID | Nose | 08/06/93 | 1 mo | F | 9V | 2 | P, SXT | I1 |
| Bul 48 | HID | Throat | 04/11/93 | 19 yr | F | 9V | 2 | P, SXT | I1 |
| Bul 66 | HID | Nph | 21/03/94 | 1 mo | F | 9 | 1 | P, SXT | I1 |
| Bul 67 | HID | Nose | 13/04/94 | 5 yr | F | 9 | 1 | P, SXT | I1 |
| Bul 68 | HID | Nose | 17/05/94 | 1 yr 8 mo | M | 9 | 0.5 | P, SXT | I1 |
| Bul 69 | HID | Throat | 26/05/94 | 11 yr | F | 9V | 1 | P, SXT | I1 |
| Bul 70 | HID | Nose | 01/06/94 | 1 yr 6 mo | F | 9 | 2 | P, SXT | I1 |
| Bul 80 | Children’s Hospital | Ear fluid | 30/11/94 | 2 yr | 9 | 1 | P, SXT | I1 | |
| Bul 82 | HID | Nose | 25/12/94 | 1 yr 6 mo | M | 9 | 2 | P, SXT | I1 |
| Bul 85 | HID | Throat | 03/01/95 | 5 yr 6 mo | M | 9 | 1 | P, SXT | I1 |
| Bul 86 | HID | Nose | 01/03/95 | 7 yr | M | 9 | 1 | P, SXT | I1 |
| Bul 109 | HID | Throat | 11/12/95 | 2 yr 7 mo | F | 9V | 2 | P, SXT | I1 |
| Bul 113 | HID | Throat | 08/02/95 | 30 yr | M | 9A | 2 | P, SXT | I1 |
| Bul 132 | HID | CSF | 15/03/97 | 53 yr | F | 9 | 1 | P, SXT | I1 |
| Sof 41 | HID | Nph | 04/10/93 | 3 yr 7 mo | F | 9 | 2 | P, SXT | I1 |
| Sof 212 | HID | Nph | 18/03/94 | 2 yr 10 mo | F | 9 | 2 | P, SXT | I1 |
| Sof 309 | HID | Nph | 31/05/94 | 1 yr | F | 9 | 1 | P, SXT | I1 |
| Sof 313 | HID | Nph | 02/06/94 | 1 yr 6 mo | F | 9 | 1 | P, SXT | I1 |
| Bul 31 | HID | CSF | 05/12/92 | 59 yr | M | 9V | 2 | P, SXT | I1 |
| Bul 84 | HID | Ear fluid | 03/01/95 | 6 mo | M | 9A | 2 | P, SXT, CTX | I1 |
| Bul 111 | HID | Nose | 28/03/96 | 10 mo | F | 9 | 2 | P, SXT, CTX | I1 |
| Bul 125 | HID | Throat | 21/01/97 | 30 yr | F | 9 | 4 | P, SXT, CTX | I1 |
| Bul 76 | HID | Throat | 10/10/94 | 52 yr | F | 14 | 0.12 | P, E, C, SXT | J1 |
| Bul 79 | HID | Throat | 28/11/94 | 8 yr | M | 14 | 0.12 | P, E, C, SXT | J1 |
| Bul 123 | HID | Nose | 14/11/96 | 1 yr | M | 14 | 0.25 | P, E, C, SXT | J1 |
| Bul 124 | HID | CSF | 18/11/96 | 50 yr | F | 14 | 1 | P, E, C, SXT | J1 |
| Bul 127 | HID | Throat | 24/01/97 | 1 yr | M | 14 | 0.25 | P, E, C, SXT | J1 |
| Bul 128 | HID | Nose | 25/01/97 | 7 mo | F | 14 | 0.12 | P, E, C, SXT | J1 |
| Bul 129 | HID | Nose | 31/01/97 | 2 yr | M | 14 | 0.25 | P, E, C, SXT | J1 |
| Sof 24 | Children’s Hospital | Nph | 02/11/93 | 2 yr 1 mo | M | 14 | 0.25 | P, E, C, SXT | J1 |
| Bul 74 | HID | Nose | 06/07/94 | 1 yr 3 mo | F | 14 | 0.12 | P, E, C, SXT | J2 |
| Bul 126 | HID | Ear fluid | 16/01/97 | 5 mo | M | 14 | 0.12 | P, E, C, SXT | J2 |
| Sof 203a | HID | Nph | 11/03/94 | 2 yr | F | 14 | 0.5 | P, E, C, SXT | J2 |
| Sof 60 | Children’s Hospital | Nph | 17/11/93 | 2 yr 8 mo | F | 14 | 0.12 | P, E, C, SXT | J3 |
| Sof 183 | HID | Nph | 01/03/94 | 1 yr 5 mo | M | 14 | 0.12 | P, E, C, SXT | J3 |
| Sof 291 | HID | Nph | 05/05/94 | 7 mo | F | 14 | 0.25 | P, E, C, SXT | J4 |
| Sof 168 | HID | Nph | 11/02/94 | 2 yr 6 mo | F | 14 | 0.5 | P, SXT | J5 |
| Bul 23 | UH of Pulmonology | Nose | 21/04/92 | 4 yr | 19A | 8 | P, E, T, SXT, RIF, CTX | K1 | |
| Bul 29 | HID | Nose | 12/10/92 | 1 yr 11 mo | M | 19 | 8 | P, E, T, SXT, CTX | K1 |
| Bul 34 | HID | Eye | 05/02/93 | 1 yr | M | 19A | 8 | P, E, T, SXT, CTX | K1 |
| Bul 40 | UH Queen Ioanna | Ear fluid | 14/04/93 | 3 yr | F | 19A | 8 | P, E, T, SXT, CTX | K1 |
| Bul 91 | HID | Ear fluid | 29/03/95 | 4 mo | F | 19 | 8 | P, E, T, SXT, CTX | K1 |
| Bul 115 | Pirogov | Sputum | 1995 | >18 yr | 19F | 4 | P, E, T, SXT, RIF, CTX | K1 | |
| Sof 289 | HID | Nph | 05/05/94 | 1 yr 7 mo | F | 19 | 8 | P, E, T, SXT, CTX | K1 |
| Bul 16 | HID | Throat | 03/02/92 | 11 mo | F | 19 | 4 | P, E, T, SXT, RIF, CTX | K2 |
| Bul 105 | HID | Nose | 06/11/95 | 1 yr 9 mo | M | 19A | 0.25 | P, E, T, SXT | L |
| Bul 11 | HID | Eye | 21/01/92 | 5 yr | M | 15B | 0.25 | P, E, C, SXT | M1 |
| Bul 33 | HID | CSF | 21/12/92 | 54 yr | M | 15 | 0.5 | P, E, C, SXT | M1 |
| Bul 83 | HID | Nose | 29/12/94 | 2 yr | F | 15 | 0.25 | P, E, C, SXT | M1 |
| Bul 99 | HID | Nose | 04/07/95 | 52 yr | M | 15A | 0.5 | P, SXT | M2 |
| Sof 137 | HID | Nph | 22/01/94 | 2 yr | F | 15 | 0.5 | P, SXT | M2 |
| Sof 197 | HID | Nph | 12/03/94 | 1 yr 9 mo | M | 15 | 1 | P, SXT | M2 |
| Sof 179 | HID | Nph | 25/02/94 | 3 yr | M | 15 | 0.5 | P, E, C, SXT | M3 |
| Sof 382 | HID | Nph | 05/08/94 | 4 mo | M | 15 | 0.5 | P, E, C, SXT | M3 |
| Sof 383 | HID | Nph | 05/08/94 | 1 yr 3 mo | M | 15 | 0.5 | P, E, C, SXT | M3 |
| Bul 94 | HID | Nose | 18/05/95 | 1 yr 6 mo | M | 15 | 0.5 | P | M4 |
| Bul 73 | HID | Throat | 17/06/94 | 18 yr | M | 21 | 0.12 | P, SXT | N |
| Bul 100 | HID | Throat | 30/05/95 | 4 yr 6 mo | M | 21 | 0.12 | P, SXT | N |
| Bul 25 | HID | CSF | 16/02/92 | 72 yr | F | 4 | 0.12 | P | O |
| Sof 306 | HID | Nph | 27/05/94 | 3 mo | M | 33 | 0.25 | P | P |
| Bul 28 | HID | Eye | 06/06/92 | 2 yr | M | NTg | 2 | P, SXT, CTX | Q |
| Bul 42 | UH Queen Ioanna | Nose | 03/05/93 | 2 yr | M | NT | 0.5 | P, T, SXT | R1 |
| Sof 153 | HID | Nph | 03/02/94 | 11 mo | F | NT | 0.5 | P, SXT | R2 |
| Bul 98 | HID | Nose | 24/06/95 | 2 yr | M | NT | 2 | P, SXT, CTX | S |
| Sof 348 | HID | Nph | 11/07/94 | 4 mo | F | NT | 1 | P, SXT | T |
| Sof 353 | HID | Nph | 13/07/94 | 1 yr 5 mo | M | NT | 0.5 | P, SXT | T |
UH, abbreviation for the names of Bulgarian hospitals.
Nph, nasopharynx; CSF, cerebrospinal fluid.
Dates are given as day/month/year.
MIC of penicillin for the strain.
P, penicillin; C, chloramphenicol; T, tetracycline; SXT, trimethoprim-sulfamethoxazole; CTX, cefotaxime; E, erythromycin; RIF, rifampin.
Pattern subtypes (27) are indicated as subscript numerals.
NT, nontypeable.
Serotypes and susceptibility testing.
Isolates were identified as S. pneumoniae by their susceptibility to optochin and their bile solubility and by employing the API system (Biomerieux, Marcy l’Etoile, France). Serotypes were determined on the basis of capsular swelling after suspension in antisera (Dako Co., Carpinteria, Calif.). Susceptibilities of pneumococcal strains to antimicrobial agents were assessed by a microdilution assay in cation-adjusted Mueller-Hinton broth with 5% lysed horse blood or by agar dilution, as detailed in guidelines from the National Committee for Clinical Laboratory Standards (19). The E test was used for the determination of susceptibility to tetracycline and chloramphenicol. MICs were determined after 24 h of incubation at 37°C in 5% CO2. S. pneumoniae ATCC 49619 was included in each run as a control. MICs of penicillin and cefotaxime were also confirmed by the E test, following a procedure recommended by the manufacturer (AB Biodisk, Solna, Sweden).
The antimicrobial agents tested (penicillin G, erythromycin, chloramphenicol, trimethoprim-sulfamethoxazole [co-trimoxazole], and tetracycline) were obtained from commercial sources. Strains were defined as antibiotic susceptible, intermediate, or resistant according to guidelines from the National Committee for Clinical Laboratory Standards (19).
PFGE.
Preparation of chromosomal DNA, restriction by SmaI endonuclease, and pulsed-field gel electrophoresis (PFGE) were performed by methods described previously (23). A CHEF-DRII apparatus (Bio-Rad, Richmond, Calif.) was used for running the gels. Running conditions were as follows: 23 h at 11.3°C at 200 V, ramped with an initial forward time of 1 s and a final forward time of 30 s. Gels were stained with ethidium bromide and photographed.
Hybridization with DNA probes.
Gels to be hybridized were transferred to nylon membranes with the Vacuum Gene system (Pharmacia Biotech, Uppsala, Sweden). The resulting membranes were probed with the ECL kit (Amersham, Little Chalfont, United Kingdom) according to the manufacturer’s recommendations. The molecular weights of the hybridization signals and the corresponding SmaI fragments were determined by comparison with a molecular weight ladder.
Probes.
DNA probes for ermB and mefE genes were obtained through PCR using, as templates, S. pneumoniae 02J 1095 (for ermB) and 02J 1175 (for mefE) and, as primers, GAAAARGTACTAACCAAATA and AGTAAYGGTACTTAAATTGTTTAC (for ermB) and AGTATCATTAATCACTAGTGC and CGTAATAGATGCAATCACAGC (for mefE), generated on the basis of sequences published by Sutcliffe et al. (24).
It has been previously shown that some streptococcal isolates carry chloramphenicol acetyltransferase (CAT) genes belonging to both catpC194 and catpC221 classes (4, 29). In order to design primers, protein sequences of the CAT determinants belonging to both classes (GenBank accession no. V01277, J01754, K01998, X65462, S50737, X60827, S45036, X02529, and X02872) were aligned, and primers were designed to match the conserved regions. The pair of primers thus obtained, CATd (TTAGGYTATTGGGATAAGTTA) and CATr (CATGRTAACCATCACAWACAG), was used to amplify a 338-bp fragment internal to the cat gene (235 to 572 bp of the coding region of the cat gene of plasmid pC164). In order to generate the probe template DNA, the chloramphenicol-resistant strain 8249 was used in a PCR, with annealing at 47°C for 30 s and extension at 72°C for 1 min for 30 cycles. Primers used for generating the tetM probe were TETMd (TGGAATTGATTTATCAACGG) and TETMr (TTCCAACCATACAATCCTTG), designed to amplify a 1,080-bp region internal to tetM (15) and corresponding to nucleotides 529 to 1608 of GenBank accession no. X52632. Preparations of template and the strain used were the same as for the preparation of the CAT probe. PCR was performed with annealing at 50°C for 30 s and extension at 72°C for 1 min for 30 cycles.
Preparation of chromosomal DNA and fingerprinting of the pbp1a, pbp2b, and pbp2x genes.
Chromosomal DNA was prepared as described by Dowson et al. (5). The pbp2b and pbp2x genes were amplified as 1.5- and 2-kb fragments, respectively, by PCR as described by Dowson et al. (5) and Munoz et al. (18). The pbp1a gene was amplified as a 2.4-kb fragment by PCR using, as primers, the oligonucleotides PN1A up (dACTAGTTGCAACAACTTCTAG) (downstream of the start codon of pbp1a; GenBank accession no. M90527) and PN1A down (dGCTCATCATCAGATAGTTCA) (upstream of the stop codon of pbp1a) (18). The same PCR conditions were used in the cases of pbp2b and pbp2x. The amplification products (20 μl each) were purified with the Wizard PCR purification system, according to the manufacturer’s instructions, digested by restriction endonucleases HinfI (pbp1a), StyI (pbp2b), and MseI plus DdeI (pbp2x), and separated by electrophoresis in 6% nondenaturing polyacrylamide gels.
RESULTS
Clinical, demographic, and microbiological properties.
Table 1 summarizes the relevant properties of the clinical isolates examined, including their dates of isolation, infection sites, serotypes, penicillin resistance levels, antibiotic resistance patterns, and the assignment of PFGE patterns, as determined after SmaI digestion of chromosomal DNA preparations and separation by contour-clamped electrophoresis. Resistance to penicillin, chloramphenicol, tetracycline, trimethoprim-sulfamethoxazole, and cefotaxime were determined, as described in Materials and Methods. PFGE patterns and subtypes were defined according to the recommendations in reference 27.
While the collection period for the 120 Bulgarian isolates included the entire span of years between 1991 and 1997, the majority of the isolates (89 of 120) were collected between 1993 and 1995. Only two isolates originated in 1991, nine originated in 1996, and five originated in 1997.
The majority of PRSP (100 of 120) were from the Hospital of Infectious Diseases (HID) in Sofia, the capital of Bulgaria, from a variety of clinical sources from children under 5 years of age who had been hospitalized with pneumococcal disease.
The PRSP expressed 10 different capsular types with the majority (110 of 120) belonging to six pediatric serotypes: 29 isolates in serotype 6, 25 in serotype 23F, 22 in serotype 9, 15 in serotype 14, 10 in serotype 15, and 9 in serotype 19. The remaining 10 isolates were in serotypes 4, 21, and 33 or were nontypeable.
Penicillin resistance level and PFGE type.
For 48 (40%) of the 120 PRSP, penicillin MICs were 1 to 8 μg/ml, and for the other 72 (60%), they were between 0.1 and 0.5 μg/ml. We shall refer to these two groups of strains as high-level and low-level penicillin-resistant strains.
Most of the high-level PRSP (43 of 48, or 86%) belonged to three clusters, each with a distinct PFGE type. The first of these clusters (13 isolates) showed PFGE pattern A, characteristic of the internationally spread serotype 23F multiresistant Spanish/USA clone of S. pneumoniae (Fig. 1). PFGE pattern A was present in six subtypes, defined according to the recommendations in reference 27. The second most frequent PFGE pattern (arbitrarily referred to as pattern I and represented by 22 isolates) was that of another internationally spread (French/Spanish) S. pneumoniae clone, which was also resistant to trimethoprim-sulfamethoxazole and expressed serotype 9A or 9V. Chromosomal macrorestriction patterns of the French/Spanish clone generated by two different restriction endonucleases are shown in Fig. 2A and B. Isolates with a PFGE pattern similar to that of the serotype 9 strains but expressing capsular type 14 are shown in Fig. 2C, and the hybridization of the same strain with the mef DNA probe is shown in Fig. 2D.
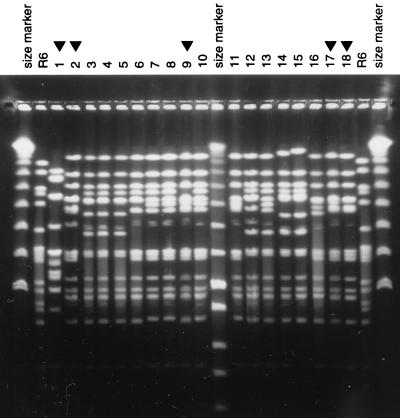
FIG. 1.
PFGE patterns of penicillin-resistant serotype 23F isolates from Bulgaria. SmaI digests of chromosomal DNAs from Bulgarian serotype 23 isolates were analyzed by PFGE. Lanes 3 to 6, strains Sof 33, 106, 176, and 282; lanes 7 and 8, Bul 121 and 118; lanes 10 to 16, Bul 117, 116, 114, 89, 52, 53, and 22. Similar PFGE patterns from penicillin-resistant isolates originating in other countries are also shown: lanes 2, 9, 17, and 18, serotype 23F isolates from Cleveland, Ohio (Clev 2), South Korea (KS), Mexico (HIM 176), and New York, N.Y. (SVM C35), respectively. Molecular size markers (λ) as well as digests of strain R6 were also included as molecular size references. Lane 1 contains a serotype 23F isolate with a completely different PFGE pattern unrelated to the PFGE pattern variants shown in the rest of the lanes.
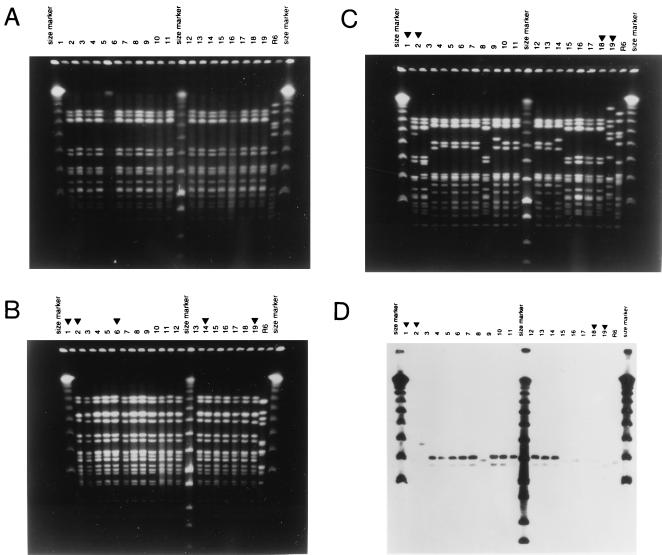
FIG. 2.
PFGE patterns of penicillin-resistant serotype 9 isolates from Bulgaria. (A) Lanes 1 through 15 show PFGE patterns of the following isolates: Bul 31, 45, 48, 66, 67, 68, 69, 70, 80, 82, 84, 85, 86, 109, and 113. Lanes 16 through 19 show PFGE patterns of isolates Sof 41, 212, 309, and 313. The outer lanes show λ molecular size markers. Additional molecular size markers include a low-molecular-weight λ ladder (between lanes 11 and 12) and the pattern of the standard laboratory strain R6. (B) ApaI was used for the fragmentation of chromosomal DNA from Bulgarian isolates which occupy lanes 3 to 5 (Bul 31, 45, and 66), lanes 7 to 13 (Bul 68, 70, 80, 82, 85, 109, and 111), and lanes 15 to 18 (Bul 113 and 125 and Sof 41 and 212). Penicillin-resistant S. pneumoniae strains from other countries, showing similar properties including PFGE pattern, are in lanes 1 and 2 (French isolates FR 2 and 12), lane 6 (Ireland 3), and lanes 14 and 19 (URU 135 [Uruguay] and AR 631 [Argentina]). The serotype of FR 2, URU 135, and AR 631 was 14; all other isolates expressed capsular type 9. (C and D) PFGE patterns of penicillin-resistant serotype 9 and serotype 14 isolates of S. pneumoniae from Bulgaria. (C) SmaI digests of chromosomal DNAs were analyzed by PFGE. Lanes 3 to 11, Bul 74, 76, 79, 123, 124, 125, 126, 127, and 128; lanes 12 to 14, Sof 24, 60, and 203a; lanes 15 to 17, Bul 111, 45, and 113. Isolates showing comparable PFGE patterns and/or serotypes from different countries are also shown in lane 1 (France), lane 2 (COL 51 [Colombia]), lane 18 (URU 135 [Uruguay]), and lane 19 (12337 TO [Slovakia]). Strains COL 5 and Bul 125, 111, 113, and 45 belong to serogroup 9. All other isolates belong to serotype 14. Molecular size markers are described in the legend to Fig. 1. (D) DNA fragments separated in panel C were transferred to membranes and tested with a DNA probe specific for the mefE gene. Strains in lanes 1, 2, 8, and 15 to 19 were susceptible to erythromycin.
A third highly penicillin-resistant cluster with a novel PFGE pattern (pattern K; eight isolates) appears to represent a hitherto undescribed genetic lineage for which penicillin MICs are high (4 to 8 μg/ml) and which expresses serotype 19F or 19A (Fig. 3A). Hybridization patterns with the tetM DNA probe are shown in Fig. 3B. The remaining five high-level PRSP each had six different PFGE types (see lanes 10 to 14 in Fig. 3A) and four different serotypes.
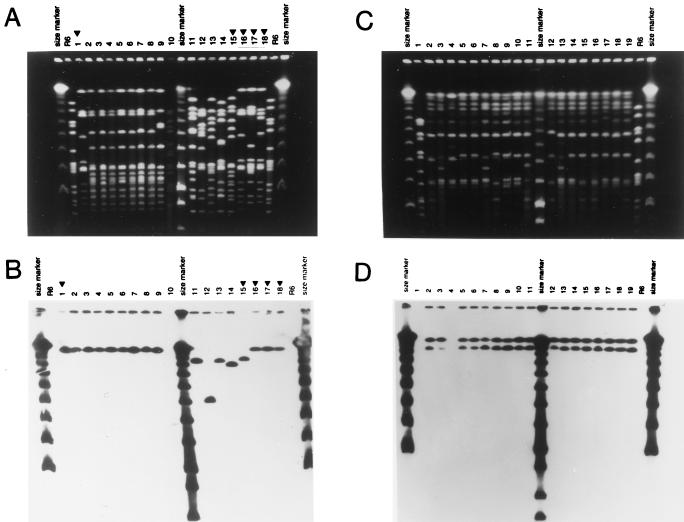
FIG. 3.
PFGE patterns of penicillin-resistant serotype 19 isolates of S. pneumoniae from Bulgaria. (A) SmaI digests of chromosomal DNAs were analyzed by PFGE. Lanes 2 to 14, Bul 23, 29, 34, 40, 91, 115, 289, 16, 105, 62, 75, 130, and 253. Lanes 1 and 16 to 18, Hun 524, 524, 27, and 48 from Hungary. Lane 15, Cs 22 (Slovakia). Molecular size markers are described in the legend to Fig. 1. (B) SmaI digests shown in panel A were tested with a DNA probe for tetM, after being transferred to nylon membranes. (C and D) PFGE patterns of penicillin-resistant serotype 6 isolates from Bulgaria (“Bulgarian clone”). (C) SmaI digests of chromosomal DNAs were analyzed by PFGE. Lanes 1 to 8 contain isolates Bul 20, 21, 35, 72, 78, 92, 107, and 108. Lanes 9 to 19 contain isolates Sof 73, 74, 114, 193, 200, 201, 220, 222, 228, 229, and 240. Molecular size markers are described in the legend to Fig. 1. (D) SmaI digests were tested by Southern hybridization with a DNA probe for ermB.
Most of the low-level PRSP (52 of 70, or over 70%) were also represented by clusters (which were arbitrarily defined as PFGE types shared by at least five independent isolates). The most prominent of these clusters consisted of 24 isolates belonging to serogroup 6 with PFGE type E (Fig. 3C) and showing a doublet band of hybridization with ermB (Fig. 3D). A second cluster of 14 isolates with serotype 14 and PFGE type J was also identified. Nine additional isolates with serotype 15 had PFGE type M, and a group of five strains with serogroup 23 had PFGE type B. The rest of the 18 low-level PRSP were represented by bacteria with ten different PFGE types and six distinct serotypes.
While the PRSP in this collection of 120 bacteria exhibited a total of 20 different PFGE types and 10 different serotypes, more than 75% of all these pneumococcal isolates (93 of 120), both among the low- and high-level resistant strains, could be accounted for by as few as six distinct PFGE types and six associated serotypes.
Resistance to antibiotics other than penicillin.
Perhaps the most unique feature of this collection of PRSP was that the overwhelming majority of the isolates (112 of 120, or 93%) showed resistance to at least one and usually more than two (86 of 120, or 71%) generically different antibacterial agents in addition to penicillin. Only 8 of the 120 isolates showed resistance to penicillin alone, and this small group of bacteria were represented by six different PFGE types. The single most frequent resistance trait accompanying penicillin resistance was resistance to trimethoprim-sulfamethoxazole (97 of 120 strains, or 80%).
The nature of antibiotic resistance mechanisms.
Table 2 summarizes the results of experiments in which the Bulgarian PRSP showing resistance to a variety of antimicrobial agents other than penicillin were tested with DNA probes for the appropriate genetic determinants. All isolates showing tetracycline resistance gave a positive hybridization with the tetM DNA probe, and most isolates exhibiting resistance to chloramphenicol were probe positive with the cat16 DNA probe. However, a group of chloramphenicol-resistant isolates gave no signal with this particular DNA probe (Table 2). We have no explanation for this discrepancy at the present time. Erythromycin-resistant isolates showed three kinds of results. Most, but not all, of the highly erythromycin-resistant isolates reacted with the ermB DNA probe, while strains for which erythromycin MICs were low (4 to 8 μg/ml) reacted with the mefE probe. The third group of isolates, all with high-level erythromycin resistance (MICs of 500 to over 1,000 μg/ml), gave negative results with both the ermB and mefE probes. These latter isolates belonged to a single clone of serogroup 19 and had a common PFGE pattern referred to as pattern K. The table also shows the antibiotic resistance phenotypes, serotypes, and PFGE patterns as well.
TABLE 2.
Southern hybridization with DNA probes of Bulgarian penicillin- and multidrug-resistant isolates of S. pneumoniaea
| Isolate | Serotype | Antibiotic resistance pattern | MIC (μg/ml)b
|
DNA probe hybridizationc (kb)
|
PFGE patternd | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P-AD | E-AD | T-ET | C-ET | ermB | mefE | tetM | cat | ||||
| Bul 52 | 23F | P, C, T, SXT, CTX | 2 | ≤0.06 | 32 | 24 | – | – | >400 | >400 | A1 |
| Bul 53 | 23 | P, E, C, T, SXT, CTX | 2 | 8 | 32 | 64 | – | Positive | >400 | >400 | A1 |
| Sof 33 | 23 | P, C, T, SXT | 2 | ND | 8 | 32 | – | – | >400 | >400 | A1 |
| Sof 106 | 23 | P, C, T, SXT | 2 | ND | 6 | 16 | – | – | >400 | >400 | A1 |
| Sof 176 | 23 | P, C, T, SXT, CTX | 2 | ND | 16 | 32 | – | – | >400 | >400 | A1 |
| Bul 117 | 23 | P, C, T, SXT | 2 | ND | 16 | 16 | – | – | >400 | >400 | A2 |
| Bul 118 | 23 | P, C, T, SXT, CTX | 2 | ND | 8 | 8 | – | – | >400 | >400 | A2 |
| Bul 121 | 23F | P, C, T, SXT | 2 | ND | 16 | 16 | – | – | >400 | >400 | A2 |
| Sof 282 | 23 | P, C, T, SXT, CTX | 2 | ND | 16 | 24 | – | – | >400 | >400 | A3 |
| Bul 22 | 23F | P, C, T, SXT, CTX | 4 | ND | 32 | 32 | – | – | >400 | >400 | A4 |
| Bul 116 | 23 | P, C, T, SXT | 2 | ND | 12 | 32 | – | – | >400 | >400 | A4 |
| Bul 89 | 23F | P, C, T, SXT | 2 | ND | 8 | 16 | – | – | >400 | >400 | A5 |
| Bul 114 | 23 | P, C, T, SXT | 2 | ND | 8 | 16 | – | – | >400 | >400 | A6 |
| Bul 78 | 6 | P, E, T | 0.12 | 2,048 | 96 | ND | 400/>450 | ND | 400/>450 | – | E1 |
| Bul 92 | 6 | P, E, T | 0.12 | 2,048 | 96 | ND | 400/>450 | ND | 400/>450 | – | E1 |
| Sof 73 | 6 | P, E, T | 0.12 | 2,048 | 48 | ND | 400/>450 | ND | 400/>450 | – | E1 |
| Sof 74 | 6 | P, E, T | 0.12 | 2,048 | 16 | ND | 400/>450 | ND | 400/>450 | – | E1 |
| Sof 201 | 6 | P, E, T, SXT | 0.12 | 2,048 | 128 | ND | 400/>450 | ND | 400/>450 | – | E1 |
| Sof 228 | 6 | P, E, T | 0.12 | 2,048 | 64 | ND | 400/>450 | ND | 400/>450 | – | E1 |
| Sof 229 | 6 | P, E, T, SXT | 0.12 | 2,048 | 48 | ND | 400/>450 | ND | 400/>450 | – | E1 |
| Bul 21 | 6 | P, E, T, SXT | 0.12 | 2,048 | 32 | ND | 400/>450 | ND | 400/>450 | – | E2 |
| Bul 108 | 6 | P, E, T, SXT | 0.12 | 2,048 | 32 | ND | 400/>450 | ND | 400/>450 | – | E2 |
| Sof 193 | 6 | P, E, T | 0.25 | 2,048 | 48 | ND | 400/>450 | ND | 400/>450 | – | E2 |
| Sof 220 | 6 | P, E, T | 0.12 | 2,048 | 64 | ND | 400/>450 | ND | 400/>450 | – | E2 |
| Sof 222 | 6 | P, E, T | 0.12 | 1,024 | 96 | ND | 400/>450 | ND | 400/>450 | – | E2 |
| Sof 240 | 6 | P, E, T | 0.12 | 2,048 | 32 | ND | 400/>450 | ND | 400/>450 | – | E2 |
| Bul 35 | 6B | P, E, T | 0.12 | 2,048 | 64 | ND | 400/>450 | ND | 400/>450 | – | E3 |
| Bul 107 | 6 | P, E, T, SXT | 0.12 | 2,048 | 128 | ND | 400/>450 | ND | 400/>450 | – | E3 |
| Sof 200 | 6 | P, E, T | 0.12 | 1,024 | 64 | ND | 400/>450 | ND | 400/>450 | – | E3 |
| Sof 114 | 6 | P, E, T, SXT | 0.12 | 1,024 | 32 | ND | 400/>450 | ND | 400/>450 | – | E6 |
| Bul 72 | 6 | P, C, SXT | 0.12 | ≤0.06 | 0.25 | 16 | Negative | ND | Negative | ND | E4 |
| Bul 76 | 14 | P, E, C, SXT | 0.12 | 4 | ND | 16 | ND | 100 | – | Negative | J1 |
| Bul 79 | 14 | P, E, C, SXT | 0.12 | 8 | ND | 32 | ND | 100 | – | Negative | J1 |
| Bul 123 | 14 | P, E, C, SXT | 0.25 | 8 | ND | 32 | ND | 100 | – | Negative | J1 |
| Bul 124 | 14 | P, E, C, SXT | 1 | 8 | ND | 32 | ND | 100 | – | Negative | J1 |
| Bul 127 | 14 | P, E, C, SXT | 0.25 | 8 | ND | 32 | ND | 100 | – | Negative | J1 |
| Bul 128 | 14 | P, E, C, SXT | 0.12 | 4 | ND | 32 | ND | 100 | – | Negative | J1 |
| Sof 24 | 14 | P, E, C, SXT | 0.25 | 8 | ND | 64 | ND | 100 | – | Negative | J1 |
| Bul 74 | 14 | P, E, C, SXT | 0.12 | 8 | ND | 32 | ND | 100 | – | Negative | J2 |
| Bul 126 | 14 | P, E, C, SXT | 0.12 | 8 | ND | 32 | ND | 100 | – | Negative | J2 |
| Sof 203a | 14 | P, E, C, SXT | 0.5 | 8 | ND | 32 | ND | 100 | – | Negative | J2 |
| Sof 60 | 14 | P, E, C, SXT | 0.12 | 4 | ND | 32 | ND | 100 | – | Negative | J3 |
| Bul 23 | 19A | P, E, T, SXT, RIF, CTX | 8 | 1,024 | 32 | ND | Negative | Negative | >400 | – | K1 |
| Bul 29 | 19 | P, E, T, SXT, CTX | 8 | 512 | 64 | ND | Negative | Negative | >400 | – | K1 |
| Bul 34 | 19A | P, E, T, SXT, CTX | 8 | 1,024 | 48 | ND | Negative | Negative | >400 | – | K1 |
| Bul 40 | 19A | P, E, T, SXT, CTX | 8 | 1,024 | 48 | 1.5 | Negative | Negative | >400 | – | K1 |
| Bul 91 | 19 | P, E, T, SXT, CTX | 8 | 1,024 | 24 | 2 | Negative | Negative | >400 | – | K1 |
| Bul 115 | 19F | P, E, T, SXT, RIF, CTX | 4 | 512 | 24 | 1 | Negative | Negative | >400 | – | K1 |
| Sof 289 | 19 | P, E, T, SXT, CTX | 8 | 512 | 16 | ND | Negative | Negative | >400 | – | K1 |
| Bul 16 | 19 | P, E, T, SXT, RIF, CTX | 4 | 512 | 32 | ND | Negative | Negative | >400 | – | K2 |
| Bul 105 | 19A | P, E, T, SXT | 0.25 | 2,048 | 64 | ND | 130 | Negative | 130 | – | L |
| Bul 11 | 15B | P, E, C, SXT | 0.25 | 8 | ND | 32 | ND | >450 | – | Negative | M1 |
| Bul 33 | 15 | P, E, C, SXT | 0.5 | 8 | ND | 32 | ND | >450 | – | Negative | M1 |
| Bul 83 | 15 | P, E, C, SXT | 0.25 | 16 | ND | 32 | ND | >450 | – | Negative | M1 |
| Sof 179 | 15 | P, E, C, SXT | 0.5 | 32 | ND | 32 | ND | >450 | – | Negative | M3 |
| Sof 382 | 15 | P, E, C, SXT | 0.5 | 32 | ND | 32 | ND | >450 | – | Negative | M3 |
| Sof 383 | 15 | P, E, C, SXT | 0.5 | 16 | ND | 48 | ND | >450 | – | Negative | M3 |
P, penicillin; E, erythromycin; T, tetracycline; C, chloramphenicol; SXT, trimethoprim-sulfamethoxazole; CTX, cefotaxime; RIF, rifampin. ND or –, not done.
MICs of antibacterial agents were obtained either by agar dilution (AD) or by the E test (ET).
Molecular size (in kilobases) of hybridizing fragment(s).
Pattern subtypes (27) are indicated as subscript numerals.
Polymorphism of the penicillin-binding protein genes pbp1a, pbp2x, and pbp2b.
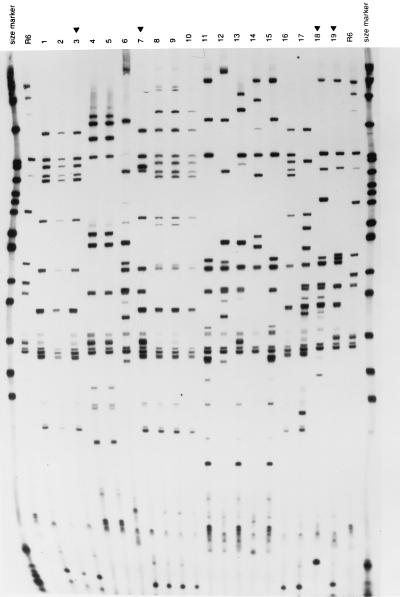
Appropriate primers were used to amplify the penicillin resistance genes pbp1a, pbp2x, and pbp2b from representative isolates of the PRSP belonging to the different PFGE types, and restriction fragment length polymorphism (RFLP) fingerprints were compared after restriction with a combination of endonucleases (Fig. 4A to C). The number assignments used to differentiate pbp gene fingerprints (Table 3) were arbitrary and were used simply to indicate their identities. Pneumococcal isolates with PFGE patterns A, E, I, and K each exhibited unique RFLP fingerprint patterns for the pbp1a and pbp2b genes. In the case of isolates with PFGE patterns A, E, and I, fingerprint patterns generated from the pbp2x gene were also unique and characteristic of these three PFGE types. However, in the case of the bacteria with PFGE pattern K, large isolate-to-isolate variation was observed in the pbp2x fingerprint pattern (six distinct RFLPs in eight different isolates). It is interesting to note that this particular group of isolates represents the unique serogroup 19 clone with a high level of penicillin resistance and multidrug resistance (Table 3).
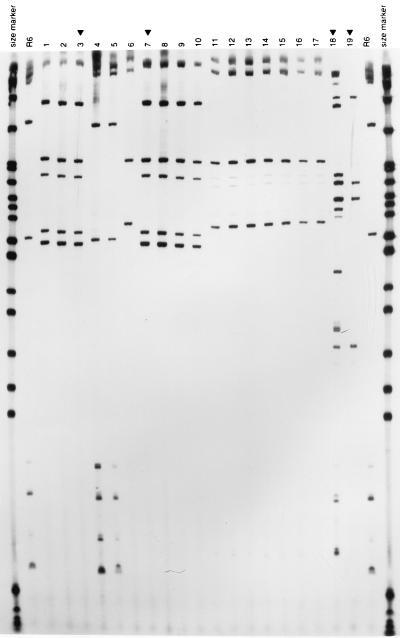
FIG. 4.
RFLP patterns of the pbp genes from penicillin-resistant isolates of S. pneumoniae from Bulgaria. (left panel) Fingerprints of pbp1a genes generated by HinfI; (middle panel) fingerprints of pbp2b genes generated by StyI; (right panel) fingerprints of pbp2x genes generated by MseI-DdeI. The outer lanes contain molecular size markers (pBR322 digested with HpaII). Lanes marked with R6 contain the penicillin-susceptible laboratory strain. Lanes marked with a black triangle contain the following additional control strains: lanes 3, Clev 2 (control for the serogroup 23F international clone A); lanes 7, IC 2 (control for another multiresistant internationally spread strain isolated in Iceland); lanes 18, Hun 524 (a serotype 19A multiresistant strain from Hungary); lanes 19, Cs 22 (a serotype 19A multiresistant strain from the Czech Republic). The rest of the lanes contained the following strains: lanes 1 and 2, Bul 89 and 22 (PFGE pattern A); lanes 4 to 6, Bul 72 and 78 and Sof 370 (PFGE pattern E; for the first two strains penicillin MICs were low and for the third they were high); lanes 8 to 10, Bul 45, 125, and 85 (PFGE pattern I); lanes 11 to 17, Bul 23, 29, 40, 91, and 115, Sof 289, and Bul 16 (all serogroup 19 and PFGE pattern K).
TABLE 3.
Fingerprint patterns of pbp1a, pbp2b, and pbp2x genes of Bulgarian penicillin-resistant isolates of S. pneumoniae
| Isolate | Source of isolatea | Serotype | MIC (μg/ml)b | Resistance patternc | Fingerprint pattern
|
PFGE patternd | ||
|---|---|---|---|---|---|---|---|---|
| pbp1a | pbp2b | pbp2x | ||||||
| Bul 52 | Eye | 23F | 2 | P, C, T, SXT, CTX | 1 | 1 | 1 | A1 |
| Bul 53 | Throat | 23 | 2 | P, E, C, T, SXT, CTX | 1 | 1 | 1 | A1 |
| Sof 33 | Nasopharynx | 23 | 2 | P, C, T, SXT | 1 | 1 | 1 | A1 |
| Sof 106 | Nasopharynx | 23 | 2 | P, C, T, SXT | 1 | 1 | 1 | A1 |
| Sof 176 | Nasopharynx | 23 | 2 | P, C, T, SXT, CTX | 1 | 1 | 1 | A1 |
| Bul 117 | Sputum | 23 | 2 | P, C, T, SXT | 1 | 1 | 1 | A2 |
| Bul 118 | Sputum | 23 | 2 | P, C, T, SXT, CTX | 1 | 1 | 1 | A2 |
| Bul 121 | Sputum | 23F | 2 | P, C, T, SXT | 1 | 1 | 1 | A2 |
| Sof 282 | Nasopharynx | 23 | 2 | P, C, T, SXT, CTX | 1 | 1 | 1 | A3 |
| Bul 22 | Ear fluid | 23F | 4 | P, C, T, SXT, CTX | 1 | 1 | 1 | A4 |
| Bul 116 | Pleural fluid | 23 | 2 | P, C, T, SXT | 1 | 1 | 1 | A4 |
| Bul 89 | Throat | 23F | 2 | P, C, T, SXT | 1 | 1 | 1 | A5 |
| Bul 114 | Sputum | 23 | 2 | P, C, T, SXT | 1 | 1 | 1 | A6 |
| Clev 2 | Ear fluid | 23F | 2 | P, E, C, T | 1 | 1 | 1 | A3 |
| Bul 78 | Ear fluid | 6 | 0.12 | P, E, T | 2 | 2 | 2 | E1 |
| Bul 92 | Nose | 6 | 0.12 | P, E, T | 2 | 2 | 2 | E1 |
| Sof 73 | Nasopharynx | 6 | 0.12 | P, E, T | 2 | 2 | 2 | E1 |
| Sof 74 | Nasopharynx | 6 | 0.12 | P, E, T | 2 | 2 | 2 | E1 |
| Sof 201 | Nasopharynx | 6 | 0.12 | P, E, T, SXT | 2 | 2 | 2 | E1 |
| Sof 228 | Nasopharynx | 6 | 0.12 | P, E, T | 2 | 2 | 2 | E1 |
| Sof 229 | Nasopharynx | 6 | 0.12 | P, E, T, SXT | 2 | 2 | 2 | E1 |
| Sof 245 | Nasopharynx | 6 | 0.12 | P, E, T | 2 | 2 | 2 | E1 |
| Sof 320 | Nasopharynx | 6 | 0.12 | P, E, T | 2 | 2 | 2 | E1 |
| Sof 330 | Nasopharynx | 6 | 0.25 | P, E, T | 2 | 2 | 2 | E1 |
| Sof 361 | Nasopharynx | 6 | 0.12 | P, E, T, SXT | 2 | 2 | 2 | E1 |
| Sof 378 | Nasopharynx | 6 | 0.12 | P, E, T, SXT | 2 | 2 | 2 | E2 |
| Bul 21 | Ear fluid | 6 | 0.12 | P, E, T, SXT | 2 | 2 | 2 | E2 |
| Bul 108 | Nasopharynx | 6 | 0.12 | P, E, T, SXT | 2 | 2 | 2 | E2 |
| Sof 193 | Nasopharynx | 6 | 0.25 | P, E, T | 2 | 2 | 2 | E2 |
| Sof 220 | Nasopharynx | 6 | 0.12 | P, E, T | 2 | 2 | 2 | E2 |
| Sof 222 | Nasopharynx | 6 | 0.12 | P, E, T | 2 | 2 | 2 | E2 |
| Sof 240 | Nasopharynx | 6 | 0.12 | P, E, T | 2 | 2 | 2 | E2 |
| Sof 248 | Nasopharynx | 6 | 0.12 | P, E, T, SXT | 2 | 2 | 2 | E2 |
| Bul 35 | Nose | 6B | 0.12 | P, E, T | 2 | 2 | 2 | E3 |
| Bul 107 | Throat | 6 | 0.12 | P, E, T, SXT | 2 | 2 | 2 | E3 |
| Sof 200 | Nasopharynx | 6 | 0.12 | P, E, T | 2 | 2 | 2 | E3 |
| Bul 72 | Throat | 6 | 0.12 | P, C, SXT | 2 | 2 | 2 | E4 |
| Sof 114 | Nasopharynx | 6 | 0.12 | P, E, T, SXT | 2 | 2 | 2 | E6 |
| Sof 370 | Nasopharynx | 6 | 4 | P, E, C, SXT, CTX | 3 | 6 | 3 | E5 |
| IC 2 | Ear fluid | 6B | 1 | P, E, C, T | 1 | 3 | 4 | B |
| Bul 45 | Nose | 9V | 2 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 48 | Throat | 9V | 2 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 66 | Nasopharynx | 9 | 1 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 67 | Nose | 9 | 1 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 69 | Throat | 9V | 1 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 70 | Nose | 9 | 2 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 80 | Ear fluid | 9 | 1 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 82 | Nose | 9 | 2 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 85 | Throat | 9 | 1 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 86 | Nose | 9 | 1 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 109 | Throat | 9V | 2 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 113 | Throat | 9A | 2 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 132 | CSF | 9 | 1 | P, SXT | 1 | 1 | 5 | I1 |
| Sof 212 | Nasopharynx | 9 | 2 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 31 | CSF | 9V | 2 | P, SXT | 1 | 1 | 5 | I1 |
| Bul 84 | Ear fluid | 9A | 2 | P, SXT, CTX | 1 | 1 | 5 | I1 |
| Bul 125 | Throat | 9 | 4 | P, SXT, CTX | 1 | 1 | 5 | I1 |
| Bul 23 | Nose | 19A | 8 | P, E, T, SXT, RIF, CTX | 3 | 4 | 6 | K1 |
| Bul 29 | Nose | 19 | 8 | P, E, T, SXT, CTX | 3 | 4 | 3 | K1 |
| Bul 34 | Eye | 19A | 8 | P, E, T, SXT, CTX | 3 | 4 | 3 | K1 |
| Bul 40 | Ear fluid | 19A | 8 | P, E, T, SXT, CTX | 3 | 4 | 7 | K1 |
| Bul 91 | Ear fluid | 19 | 8 | P, E, T, SXT, CTX | 3 | 4 | 8 | K1 |
| Bul 115 | Sputum | 19F | 4 | P, E, T, SXT, RIF, CTX | 3 | 4 | 6 | K1 |
| Sof 289 | Nasopharynx | 19 | 8 | P, E, T, SXT, CTX | 3 | 4 | 1 | K1 |
| Bul 16 | Throat | 19 | 4 | P, E, T, SXT, RIF, CTX | 3 | 4 | 9 | K2 |
| Hun 524 | Ear fluid | 19A | 2 | P, E, C, T | 4 | 2 | 10 | D5 |
| Cs 22 | URT | 19A | 2 | P, C, T | 5 | 5 | 11 | C1 |
| R6 | 0.03 | 2 | 6 | 12 | ||||
CSF, cerebrospinal fluid. URT, upper respiratory tract.
MIC of penicillin for the strain.
P, penicillin; C, chloramphenicol; T, tetracycline; SXT, trimethoprim-sulfamethoxazole; CTX, cefotaxime; E, erythromycin; RIF, rifampin.
Pattern subtypes (27) are indicated as subscript numerals.
DISCUSSION
The examination of the PRSP from Bulgaria with microbiological and molecular techniques has revealed several interesting features of this collection.
A surprisingly large fraction of the isolates (93 of 120, or close to 80%) were represented by clusters of strains, each exhibiting unique antibiotypes, serotypes, and PFGE patterns and unique RFLPs of their pbp genes as well. Also clonal was the distribution of other antibiotic resistance genes identified by a variety of DNA probes. Most interesting was the distribution of the macrolide resistance genes. For instance, all isolates of the unique serogroup 6 cluster, with the arbitrarily assigned PFGE pattern E and for which penicillin MICs were 0.1 μg/ml, reacted with the ermB DNA probe (a doublet of hybridizing bands) and showed an extremely high level of erythromycin resistance (MIC range, 1 to 2 mg/ml). The two identical SmaI fragments also gave a strong hybridization signal with their tetM probes.
A second unique cluster of serotype 14 pneumococci, for which penicillin MICs were between 0.1 and 1 μg/ml and with a common PFGE pattern called pattern J, reacted with the mefE DNA probe and erythromycin MICs for it were in the range of 4 to 8 μg/ml.
A third unique clone belonging to serogroup 19, with extremely high levels of penicillin and erythromycin resistance (assigned to PFGE pattern K), was phenotypically erythromycin and streptogramin-B resistant but remained clindamycin susceptible (unpublished data). This group of isolates did not react with either the ermB or mefE probes, suggesting that the macrolide resistance of these bacteria is accomplished by a mechanism yet to be determined.
The reason for the large numbers of clonal clusters in this collection is not fully clear. It is conceivable that part of the clustering may be related to nosocomial spread. For instance, PRSP with the clonal assignment I and M were all from the HID. On the other hand, isolates with the PFGE assignment of A, J, or K were recovered in four of the seven hospitals participating in the survey.
Special comments are appropriate for the two clusters to which we assigned PFGE patterns A and I. The first of these, represented by 13 isolates, had serotype 23F and was resistant to penicillin, tetracycline, chloramphenicol, and trimethoprim-sulfamethoxazole. In all these properties this cluster was indistinguishable from one of the internationally spread (Spanish/USA) clones of pneumococci. Bulgarian isolates belonging to this clone also generated RFLP patterns from their pbp1a, pbp2x, and pbp2b genes which were identical to the corresponding DNA fingerprints obtained from the authentic members of the Spanish/USA clone. A single isolate (Bul 53) was also resistant to erythromycin (erythromycin MIC, 8 μg/ml) and gave a positive signal with the mefE DNA probe.
The second cluster, composed of 22 isolates and belonging to serogroup 9, was assigned the PFGE pattern I. The penicillin resistance of this cluster (MIC, 1 to 2 μg/ml) was accompanied only by a resistance to trimethoprim-sulfamethoxazole. All these properties, as well as the RFLP patterns generated from the pbp1a, pbp2x, and pbp2b genes of this cluster, indicated that these bacteria were representatives of a second widely spread (French/Spanish) S. pneumoniae clone with high frequencies in several Western European and South American countries (3, 28). Our observations suggest that the two clones with PFGE patterns A and I, respectively, were introduced into Bulgaria from abroad.
While all members of the PRSP clone with PFGE pattern I from the Bulgarian collection expressed capsular polysaccharide 9 (9A, 9V, and unfactored), pneumococcal isolates with exactly the same PFGE pattern, RFLPs of pbp genes, and antibiotype have been reported to express the capsular polysaccharide 14 in isolates from several other countries, for instance, in France, Uruguay, Argentina, and Colombia (2, 8, 28). The PFGE patterns in this particular cluster were identified by using two different restriction enzymes (SmaI and ApaI) for the PFGE separation of DNA fragments (see Fig. 2A and B). This observation already noted by others (9) suggests that this clone of PRSP may be the product of capsular transformation events, which must have occurred in vivo (1, 3, 20).
The comparison of the fingerprint patterns in Fig. 4 allows several interesting conclusions. The two Bulgarian serotype 23 isolates with PFGE pattern A (Bul 89 and 22; Fig. 4, lanes 1 and 2) showed pbp1a, pbp2x, and pbp2b RFLPs identical to those of an authentic representative of the international clone A (strain Clev 2, an isolate from Cleveland in 1989; Fig. 4, lane 3), confirming that these three strains not only had common chromosomal backgrounds but identical pbp genes as well. Lanes 4 and 5 in Fig. 4 show pbp gene fingerprints of two representatives of the low-level penicillin-resistant (MIC, 0.1 μg/ml) PFGE type E cluster from Bulgaria. The identities of all the RFLP patterns are apparent. As expected for these isolates for which penicillin MICs are low, they also shared gene profiles for pbp1a that are virtually identical with those of the penicillin-susceptible control strain R6, while they differed from R6 in their pbp2x and pbp2b patterns. A third Bulgarian strain, belonging to the same clonal background (PFGE pattern E) but with high-level penicillin resistance (Sof 370; MIC, 4.0 μg/ml), was run in lane 6 (Fig. 4). As expected, the pbp1a, pbp2b, and pbp2x gene fingerprints of this isolate differed from those of strains Bul 72 and Bul 78, two strains sharing the same PFGE pattern E background. Interestingly, the pbp1a and pbp2x fingerprints of Sof 370 differed from those of strain R6, while the pbp2b fingerprints of Sof 370 were indistinguishable from the corresponding pbp fingerprint of the penicillin-susceptible R6 strain. It has been suggested that the mosaicism of the pbp1a, pbp2x, and pbp2b genes is a common feature of pneumococci with high-level resistance to penicillin. Sof 370 appears to be an exception, since the pbp2b gene of this isolate had an RFLP pattern similar to that of the penicillin-susceptible control strain. It should be noted that the MIC of cefotaxime for strain Sof 370 was also high (4 μg/ml), a resistance that is supposed to involve altered forms of pbp1a and pbp2x but not pbp2b (12).
RFLPs of the pbp genes from three representatives of serogroup 9 (Bul 45, Bul 125, and Bul 85; Fig. 4, lanes 8 to 10), sharing the PFGE pattern I typical of the second international clone, and seven representatives of the highly penicillin-resistant PFGE pattern K clone (lanes 11 to 17) showed that members of each of these two clones shared a set of identical pbp1a and pbp2b fingerprints, each typical of their particular clone. The serogroup 9 isolates had a common pbp2x pattern as well. On the other hand, this was not the case for representatives of the clone with PFGE pattern K, for which each of the seven isolates had a distinct pbp2x fingerprint, one of which was actually indistinguishable from the pbp2x fingerprint of the international clone A.
Additional examples of the existence of similar pbp genes in diverse genetic backgrounds are shown in Fig. 4. For instance, the pbp2x fingerprint of strain Bul 29 (PFGE pattern K) is identical to that of strain Sof 370 (PFGE pattern E). Close similarities exist between the pbp1a gene fingerprint profiles of strains with clonal type A (Fig. 4, lanes 1 to 3) and those of the multiresistant clone from Iceland (lane 7), as well as the pbp1a fingerprints of the serogroup 9 (PFGE pattern I) isolates. Clones of type A and type I also had very similar pbp2b fingerprints. Together these findings provide further examples of the extensive genetic exchanges that occur among genetic lineages of S. pneumoniae in vivo.
ACKNOWLEDGMENTS
These investigations received partial support from grant RO1 AI 37275 from the National Institutes of Health, the Fogarty Foundation, and the Bodman/Achelis Fund.
The expert advice and assistance of Shangwei Wu in the analysis of pbp fingerprints are gratefully acknowledged.
REFERENCES
- 1.Barnes D M, Whittier S, Gilligan P H, Soares S, Tomasz A, Henderson F W. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis. 1995;171:890–896. doi: 10.1093/infdis/171.4.890. [DOI] [PubMed] [Google Scholar]
- 2.Coffey T J, Berrón S, Daniels M, Garcia-Leoni M E, Cercenado E, Bouza E, Fenoll A, Spratt B G. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988–1994): novel major clones of serotypes 14, 19F and 15F. Microbiology. 1996;142:2747–2757. doi: 10.1099/13500872-142-10-2747. [DOI] [PubMed] [Google Scholar]
- 3.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 4.David F, de Cespedes G, Delbos F, Horaud T. Diversity of chromosomal genetic elements and gene identification in antibiotic-resistant strains of Streptococcus pneumoniae and Streptococcus bovis. Plasmid. 1993;29:147–153. doi: 10.1006/plas.1993.1017. [DOI] [PubMed] [Google Scholar]
- 5.Dowson C G, Hutchison A, Spratt B G. Extensive remodelling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989;3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 6.Ferroni A, Nguyen L, Gehanno P, Boucot I, Berche P. Clonal distribution of penicillin-resistant Streptococcus pneumoniae 23F in France. J Clin Microbiol. 1996;34:2707–2712. doi: 10.1128/jcm.34.11.2707-2712.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figueiredo A M S, Austrian R, Urbaskova P, Teixeira L A, Tomasz A. Novel penicillin-resistant clones of Streptococcus pneumoniae in the Czech Republic and in Slovakia. Microb Drug Resist. 1995;1:71–78. doi: 10.1089/mdr.1995.1.71. [DOI] [PubMed] [Google Scholar]
- 8.Gasc A M, Geslin P, Sicard A M. Relatedness of penicillin-resistant Streptococcus pneumoniae serogroup 9 strains from France and Spain. Microbiology. 1995;141:623–627. doi: 10.1099/13500872-141-3-623. [DOI] [PubMed] [Google Scholar]
- 9.Hall L M C, Whiley R A, Duke B, George R C, Efstratiou A. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J Clin Microbiol. 1996;34:853–859. doi: 10.1128/jcm.34.4.853-859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanavaki S, Karabela S, Marinis E, Legakis N J. Antibiotic resistance of clinical isolates of Streptococcus pneumoniae in Greece. J Clin Microbiol. 1994;32:3056–3058. doi: 10.1128/jcm.32.12.3056-3058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klugman K P, Coffey T J, Smith A, Wasas A, Meyers M, Spratt B G. Cluster of an erythromycin-resistant variant of the Spanish multiply resistant 23F clone of Streptococcus pneumoniae in South Africa. Eur J Clin Microbiol Infect Dis. 1994;13:171–174. doi: 10.1007/BF01982193. [DOI] [PubMed] [Google Scholar]
- 12.Laible G, Spratt B G, Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP2X genes in penicillin-resistant clinical isolates of S. pneumoniae. Mol Microbiol. 1991;5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 13.Lefevre J C, Bertrand M A, Fauçon G. Molecular analysis by pulsed-field gel electrophoresis of penicillin-resistant Streptococcus pneumoniae from Toulouse, France. Eur J Clin Microbiol Infect Dis. 1995;14:491–497. doi: 10.1007/BF02113426. [DOI] [PubMed] [Google Scholar]
- 14.Marchese A, Schito G C, Tomasz A. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Preponderance of multidrug-resistant clones among penicillin-resistant isolates of Streptococcus pneumoniae from Italy, abstr. C-75; p. 59. [Google Scholar]
- 15.Martin P, Trieu-Cuot P, Courvalin P. Nucleotide sequence of the tetM tetracycline resistance determinant of the streptococcal conjugative transposon Tn1545. Nucleic Acids Res. 1986;14:7047–7058. doi: 10.1093/nar/14.17.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGee L, Klugman K P, Friedland D, Lee H-J. Spread of the Spanish multiresistant serotype 23F clone of Streptococcus pneumoniae to Seoul, Korea. Microb Drug Resist. 1997;3:253–262. doi: 10.1089/mdr.1997.3.253. [DOI] [PubMed] [Google Scholar]
- 17.Moissenet D, Valcin M, Marchand V, Garabedian E N, Geslin P, Garbard-Chenon A, Vu-Thien H. Molecular epidemiology of Streptococcus pneumoniae with decreased susceptibility to penicillin in a Paris children’s hospital. J Clin Microbiol. 1997;35:298–301. doi: 10.1128/jcm.35.1.298-301.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munoz R, Coffey T J, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, Tomasz A. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. 6th ed. Approved standard M2-A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 20.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;177:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 21.Reichmann P, Varon E, Gunther E, Reinert R R, Luttikens R, Marton A, Geslin P, Wagner J, Hakenbeck R. Penicillin-resistant Streptococcus pneumoniae in Germany: genetic relationship to clones from other European countries. J Med Microbiol. 1995;43:377–385. doi: 10.1099/00222615-43-5-377. [DOI] [PubMed] [Google Scholar]
- 22.Setchanova L. Clinical isolates and nasopharyngeal carriage of antibiotic-resistant Streptococcus pneumoniae in Hospital for Infectious Diseases, Sofia, Bulgaria, 1991–1993. Microb Drug Resist. 1995;1:79–84. doi: 10.1089/mdr.1995.1.79. [DOI] [PubMed] [Google Scholar]
- 23.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 24.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarasi A, Chong Y, Lee K, Tomasz A. Spread of the serotype 23F multidrug-resistant Streptococcus pneumoniae clone to South Korea. Microb Drug Resist. 1997;3:105–109. doi: 10.1089/mdr.1997.3.105. [DOI] [PubMed] [Google Scholar]
- 26.Tarasi A, Sterk-Kuzmanovic N, Sieradzki K, Schoenwald S, Austrian R, Tomasz A. Penicillin-resistant and multidrug-resistant Streptococcus pneumoniae in a pediatric hospital in Zagreb, Croatia. Microb Drug Resist. 1995;1:169–176. doi: 10.1089/mdr.1995.1.169. [DOI] [PubMed] [Google Scholar]
- 27.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomasz A, Corso A Members of the PAHO/Rockefeller University Workshop: G. Echaniz-Aviles, M. C. de C. Brandileone, T. Camou, E. Castaneda, O. Figueroa, A. Rossi, and E. P. Severina. Molecular epidemiological characterization of penicillin-resistant Streptococcus pneumoniae invasive pediatric isolates recovered in six South American countries. Microb Drug Resist. 1998;4:195–207. doi: 10.1089/mdr.1998.4.195. [DOI] [PubMed] [Google Scholar]
- 29.Trieu-Cuot P, de Cespédès G, Bentorcha F, Delbos F, Gaspar E, Horaud T. Study of heterogeneity of chloramphenicol acetyltransferase (CAT) genes in streptococci and enterococci by polymerase chain reaction: characterization of a new CAT determinant. Antimicrob Agents Chemother. 1993;37:2593–2598. doi: 10.1128/aac.37.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaz Pato M V, de Carvalho C B, Tomasz A The Multicenter Study Group. Antibiotic susceptibility of Streptococcus pneumoniae isolates in Portugal. A multicenter study between 1989 and 1993. Microb Drug Resist. 1995;1:59–69. doi: 10.1089/mdr.1995.1.59. [DOI] [PubMed] [Google Scholar]