Abstract
Abstract
The emergence of SARS-CoV-2 has brought the world to a standstill, and till date, effective treatments and diagnostics against this idiosyncratic pathogen are lacking. As compared to the standard WHO/CDC qPCR detection method, which consumes several hours for detection, CRISPR-based SHERLOCK, DETECTR, and FELUDA have emerged as rapid diagnostic tools for the detection of the RNA genome of SARS-CoV-2 within an hour with 100% accuracy, specificity, and sensitivity. These attributes of CRISPR-based detection technologies have taken themselves one step ahead of available detection systems and are emerging as an inevitable tool for quick detection of the virus. Further, the discovery of Cas13s nucleases and their orthologs has opened a new corridor for exploitation of Cas13s as an antiviral therapy against SARS-CoV-2 and other viral diseases. One such approach is Prophylactic Antiviral CRISPR in huMAN cells (PACMAN), which needs a long haul to bring into therapy. The approval of SHERLOCK as the first CRISPR-based SARS-CoV-2 test kit by the FDA, for emergency diagnosis of COVID-19 patients, has given positive hope to scientists that sooner human trials of CRISPR-based therapy will be ratified. In this review, we have extensively reviewed the present CRISPR-based approaches, challenges, and future prospects in the light of diagnostics and therapeutics against SARS-CoV-2.
Key points
• The discovery of Cas12 and Cas13 siblings allowed scientists to detect the viral genes.
• Cas13d’s identification aided scientists in precisely cleaving the SARS-CoV-2 ssRNA.
• CRISPR-Cas system acts as “molecular detector and antiviral proctor.”
Keywords: CRISPR/Cas, SARS-CoV-2, SHERLOCK, PACMAN, FELUDA, DETECTR, COVID-19
Introduction
The emergence of severe acute respiratory syndrome–related coronavirus 2 (SARS-CoV-2), a single-stranded RNA beta coronavirus, in late 2019 from the Wuhan district of China, has become one of the major threats to mankind throughout the globe. The outbreak of coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 virus infection has led to an ongoing public health emergency in almost every country of the world (Ksiazek et al. 2003; Kazi et al. 2020; Singhal 2020). The prevalent symptoms of this disease are difficulty in breathing, chest and muscle pain, dry cough, fever, headache, diarrhea, cytokine storm, and even death in severe conditions. Upon infection, it takes an average of 5–14 days to show the viral symptoms, or it can be asymptomatic as well. The SARS-CoV-2 was found to be highly transmissible and human–human transmission, which primarily occurs in the form of droplets. Hence, it can spread through direct contact with the infected person and indirect contact with surfaces of the objects used by the infected person (Yuki et al. 2020; Esakandari et al. 2020). The transmission potential of the virus has resulted in rapid inflation of COVID-19 cases across the world and has become one of the major concerns globally. Despite the growing menace, so far, there are no clinically approved antiviral drugs, against the virus. Recently, researchers have formulated few drugs in repurposed form which are in the different stages of clinical development and attempting to design new therapeutic strategies which are available commercially (Chakravarti et al. 2021). Since the virus is highly transmissible, in addition to general social distancing, identification of infected individuals and screening their contacts for possible quarantine measures is one of the major steps to curb the community transmission of this virus (Huang et al. 2020). It is also suggested that the development of rapid and accurate diagnostic tools might help to change the current scenario by controlling the rapid spread of SARS-CoV-2. Currently, the available approach to diagnose the novel coronavirus (2019-nCoV) clinically is predominantly relying upon real-time PCR-based methods which requires 4–6 h as well as expensive resources to detect the pathogen. Parallelly, there are serological approaches are in place to detect the contagion, but the process is time-consuming, cost-effective, and unreliable. However, considering the growing need for even more extensive testing, the fact that RT-PCR requires advanced infrastructure and up to 24 h to deliver the results, warrants a better alternative (Lübke et al. 2020; Wei et al. 2021). Therefore, scientists have developed CRISPR (clustered regularly interspaced palindromic repeats)-based easy-to-use kits for detecting SARS-CoV-2 from the contagion samples in 30–60 min, just like a pregnancy test which does not require any sophisticated instruments or well-established lab and can be used in the home, airports, and even in railway stations for faster and efficient detection of contagion compared to WHO (World Health Organization)/CDC (Centers for Disease Control and Prevention) approved RT-PCR method (Surjit et al. 2006; Hou et al. 2020).
CRISPR is an adaptive immune system derived from archaea and bacteria and is consisted of a CRISPR array, an RNA-guided endonuclease (Cas), a single guide RNA (sgRNA), and a protospacer-adjacent motif (PAM) sequence at the 3′ end of the targeted DNA sequence that has been optimized for genome modification in human cells in vitro and in vivo. The CRISPR and its associated Cas mainly work by interacting with the guide RNA, complementary to the target sequence, which leads to the cleavage at a precise site in the target, generating double-stranded DNA breaks (DSBs). These DSBs are then repaired by an endogenous cellular repair pathway, i.e., either by non-homologous end joining (NHEJ) or homology-directed repair (HDR) pathways (Jinek et al. 2012; Amitai and Sorek 2016; Adli 2018; Gupta et al. 2019). The discovery of CRISPR has revolutionized the scientific world by showing its impact in a wide variety of applications ranging from drug development and the discovery of biomarkers, cancer therapy, agriculture, and HIV treatment, and the list is growing (Singh et al. 2021). Since the emergence of CRISPR-based technology, tremendous efforts are being reinforced to broaden its potential in medicinal therapeutics and use this technology in disease diagnosis and treatment (Gupta et al. 2021). In 2016, the CRISPR-Cas9-based diagnostics was utilized for the first time to detect the Zika virus (Pardee et al. 2016), and in 2017, it was used for the detection of Staphylococcus aureus (Guk et al. 2017). There are different types of CRISPR-Cas systems, being discovered progressively, and recently, scientists have identified two potent Cas nucleases: Cas12a (Cpf1) and Cas13a (C2c2) that have broadened the utilization of CRISPR in the area of diagnostics and therapeutic treatments of several diseases such as cancer, anemia, viral diseases, and neurogenerative diseases (Gao et al. 2016; Abudayyeh and Gootenberg 2020).
There are three different CRISPR-based methods, discovered by three distinct groups, for the detection and identification of viral RNA from the given viral samples. One of these is introduced by Mammoth Biosciences in 2018 named DETECTR (DNA Endonuclease-Targeted CRISPR Trans Reporter) that detects targeted DNA sequences using Cas12a. This DETECTR method was adapted to identify SARS-CoV-2 RNA in about 30 min (Chen et al. 2018). The second one was introduced by Feng Zhang’s group, named SHERLOCK (Specific High Sensitivity Enzymatic Reporter unLocking) in 2017, a CRISPR-based nucleic acid detection method that detects targeted RNA sequence instead of DNA using Cas13 nucleases in about 60 min (https://sherlock.bio/better-faster-affordable-diagnostic-testing/.), and the third group is led by Debjyoti Chakraborty, senior scientist at Institute of Genomics and Interactive Biology (CSIR-IGIB), who discovered FELUDA (FNCAS9 Editor Linked Uniform Detection Assay) to identify SARS-CoV-2 virus by targeting the genetic material of the virus using Cas9 nucleases in about 45 min (Azhar et al. 2020; Table 1). In this review article, an attempt has been undertaken to catalogue the ongoing endeavors that how CRISPR technology is being used for the detection and diagnosis of SARS-CoV-2 and how it can be used as an antiviral therapeutic platform for the treatment of such viral diseases.
Table 1.
Divergence between the best known SARS-CoV-2 CRISPR-based detection methods
| Method name | CRISPR/Cas platform | Detection based on | Detection time | Founder | Reference |
|---|---|---|---|---|---|
| DETECTR (DNA Endonuclease Targeted CRISPR Trans Reporter) | CRISPR/Cas 12a | dsDNA or ssDNA | 30 min | Mammoth Biosciences | Chen et al. (2018) |
| SHERLOCK (Specific High Sensitivity Enzymatic Reporter unLocking) | CRISPR/Cas 13 | RNA | 60 min | Sherlock Biosciences | Abbott et al. (2020) |
| FELUDA (FNCAS9 Editor Linked Uniform Detection Assay) | CRISPR/Cas 9 | cDNA | 45 min | CSIR-TIFR | Azhar et al. (2020) |
Utilization of Cas12a (Cpf1) in SARS-CoV-2 detection
Cas12a nuclease belongs to class II, type V CRISPR-Cas system and was first characterized by Feng Zhang’s Lab at MIT in 2015 (Zetsche et al. 2015; Xiao et al. 2020). The discovery of Cas12a has brought a new outbreak in genome editing, empowering researchers to enable a whole new level of precision because of the following advantages over standard SpCas9: (i) Cas12a generates 5′ overhangs instead of blunt ends created by SpCas9, rendering in efficient DNA repair of the cleaved ends; (ii) Cas12a recognizes different PAM sequence, i.e., 5′TTTN3′ instead of 5′NGG3′ SpCas9 PAM sequence, which allows to edit A-T-rich sequences as well as provide flexibility in choosing target sites; and (iii) Cas12a requires only one 42 nucleotides long sgRNA, i.e., crRNA as compared to Cas9 which require a tracrRNA along with crRNA; further, the smaller size of Cas12a facilitates easy and efficient delivery into cells via low capacity vectors such as AAV (Zetsche et al. 2015; Li et al. 2018; Yan et al. 2019).
In 2018, Mammoth Biosciences has exploited the potentiality and precision of Cas12a and introduced an approach to detect sensitive DNA using LbCas12a isolated from Lachnospiraceae bacterium ND2006 and named this technique as DETECTR (DNA Endonuclease-Targeted CRISPR Trans Reporter). Recently DETECTR accompanied with RT-LAMP (Reverse-transcription-Loop Mediated Isothermal Amplification) was implemented to detect the SARS-CoV-2, and it was found that the virus can be detected with high precision within 30 min (Chen et al. 2018; Ali et al. 2020). RT-LAMP is the modified version of LAMP (Loop-Mediated Isothermal Amplification) and was first developed by Tsugunori Notomi et al. in the year 2000. LAMP is a novel, robust, affordable method to amplify DNA rapidly with high specificity and efficiency under isothermal reaction conditions. This method employs Bst DNA polymerase with high strand displacement activity and a set of specially designed specific primers that recognize 6 or more distinct sequences on the target DNA. In the case of RT-LAMP, Bst DNA/RNA polymerase is used which is a mixture of Bst polymerase and extremely thermostable reverse transcriptase (60 to 65 °C tolerant), which is used for isothermal amplification reaction of RNA via cDNA and detection in a single step. RT-LAMP is an ideal amplification technology for point of care/point of need assays against SARS-CoV-2 detection. Further, RT-LAMP allows multiplexing with different techniques such as Mg precipitation, colorimetric, agarose gel electrophoresis, and fluorescence, which provides more vantage in terms of rapidity, sensitivity, and specificity to detect the SARS-CoV-2 or like viruses or any infectious pathogen in 10–20 min even with low level of sample concentrations (Notomi et al. 2000; Ali et al. 2020; Subali and Wiyono 2021).
During the study, they used contrived patient samples of SARS-CoV-2 and detected two genes N-gene and E-gene using LbCas12a and synthetically designed Cas12a guide-RNAs (gRNAs). To detect the E-gene, sgRNAs from three SARS-like coronaviruses [SARS-CoV-2 (NC_045512), SARS-CoV (NC_004718), and bat-SL-CoV2C45 (MG772933)] were designed, whereas, for the N-gene, they only designed Cas12a gRNAs from the SARS-CoV-2 genome. They also demonstrated that this method can detect SARS-CoV-2 with no cross-reactivity for N-gene gRNA, compared to the E-gene detection, which showed cross-reactivity for E-gene gRNA with related SARS-like coronavirus strains (Chen et al. 2018; Ali et al. 2020). The approach is almost equivalent to the methods adopted by the WHO and CDC qRT-PCR assays for identifying specific SARS-CoV-2 or related SARS-like coronavirus (Lübke et al. 2020; Ali et al. 2020).
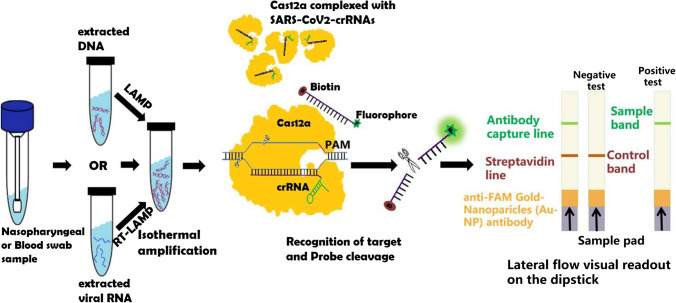
The DETECTR assay can be broadly described in three important steps: (i) pre-amplification of viral RNA or control RNA by RT-LAMP (Box 1) using LAMP primers against N-gene, E-gene, and RNaseP, since the target detection is challenging for the CRISPR when the target DNA concentration is extremely low (lower than 10 nM). The RT-LAMP reaction was optimized and carried out at 62 °C for 20–30 min independently for N-gene, E-gene, and RNase P; (ii) trans-cleavage assay, where LbCas12a is pre-incubated with compatible gRNA to form LbCas12a-gRNA complex for 10 min at 37 °C followed by the addition of cleavage reporter (FAM-biotin) to the LbCas12a-gRNA complexes. After which, LbCas12a-gRNA complexes are mixed with the amplicon in the ratio of 1:10, and the trans-cleavage assay is allowed for 10 min at 37 °C (Gootenberg et al. 2018; Bai et al. 2019; Javalkote et al. 2020; Broughton et al. 2020; Ali et al. 2020). The pre-amplification process and the trans-cleavage assay can be separated into two-pot reactions or be merged as a one-pot reaction. The operation of the assay in two-step largely complicates the process as it requires multiple manual operations and also upturns the potential risk of contamination. However, the addition of all the components (amplification and CRISPR reagents) in one pot would decrease the efficiency due to the possible cross-reactivity and digestion of the initial amplification products by the Cas/crRNA complex. Therefore, to run the one-pot reaction, the Cas12 can be isolated from the rest of the components by the addition of the Cas12 protein in a droplet on the tube wall, followed by the mixing of the Cas12 protein with the reaction mixture after the completion of the amplification process, to start the CRISPR assay (Ding et al. 2020); (iii) finally, a commercial lateral flow dip strip is added to the reaction tube, and result was interpreted in ≤ 2 min, visually. The first single band near the sample pad (control line) indicates a negative result, while a single band or two bands close to the top of the strip (test line) indicates a positive result. Further, bands were quantified using the ImageJ gel analyzer tool, and the intensity of the band signal was normalized to the maximum. The detection sensitivity of the DETECTR system was found to be in the range of femtomolar (fM) to attomolar (aM) of DNA/ssDNA with 70–300 copies/μl of input sample, and the detection time is recorded as 20–30 min which is much faster than the conventional RT-PCR detection method (Yuan et al. 2020; Broughton et al. 2020; Ding et al. 2020; Fig. 1). Though the lateral flow assay (LFA) platform is the most common colorimetric readout system for CRISPR sensing methods, as it is user-friendly, cost-effective, and accessible, various readout mechanisms such as fluorescence (Bai et al. 2019; Yuan et al. 2020; Ding et al. 2020), electrochemical (Dai et al. 2019), and electronic (Shao et al. 2019) have also been introduced for CRISPR-based assays. Primarily, Cas12- or Cas13-based sensing systems relied upon the fluorescence-based approaches for the detection of target sequence recognition. However, the involvement of bulky and expensive fluorescence readers has limited the applicability and accessibility of this platform. Moreover, the electronic readout using nanopore sensors has been extremely promising for dCas9- and Cas12a-based assays, while its implementation for CRISPR-based SARS-CoV-2 detection is yet to be developed (Yang et al. 2018; Shao et al. 2019).
Fig. 1.
Schematic representation of SARS-CoV-2-DETECTR experimental workflow. In DETECTR, nucleic acid is amplified using reverse transcriptase-recombinase polymerase amplification (RT-RPA) or reverse transcriptase-loop-mediated isothermal amplification (RT-LAMP) technology followed by recognition of target dsDNA by Cas12a-crRNA complex, which triggers the Cas12a enzyme which possesses collateral/trans-cleavage activity and thus cleaves fluorescence tagged reporter ssDNA which produces visual readout analyzed on dip strip paper by lateral flow assay
Utilization of Cas13a (C2c2) in SARS-CoV-2 detection
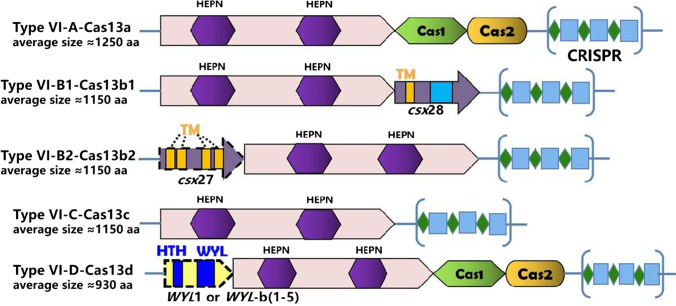
The identification of a novel class-2, type-VI effector molecule called Cas13a (C2c2), in 2016 by Feng Zhang, has broadened the application spectrum of the CRISPR/Cas system beyond anything, with its ability to target ssRNA without neutering the genome. Based on the presence of extra ORF region, adaptation genes (Cas1/Cas2) and presence of WYL-domain-associated accessory proteins, type VI CRISPR Cas system are further classified into Cas13a (C2c2), Cas13b, Cas13c, and Cas13d (CasRx), as shown in Fig. 2 (Abudayyeh et al. 2017; Yan et al. 2019; Abudayyeh and Gootenberg 2020). Further, all the four types VI-Cas 13 system have two HEPN domains in distinct positions, which demonstrates the RNA-guided RNA-targeting RNase activity. Type-VI CRISPR-Cas13s system was composed of RNA-guided RNase Cas13, 64–66 nt crRNA/gRNA (consisting of 24–30 nt spacer region which is complementary to the target site), and protospacer-flanking site (required in case of bacteria but not for mammalian and plant cell). The Cas13 protein recognizes the short hairpin structures in the crRNA and forms Cas13-crRNA complexes followed by the binding of the target ssRNA to the complementary 24–30 nt spacer region of the crRNA resulting in Cas13-mediated cleavage of the target ssRNA (Abudayyeh et al. 2017; Abudayyeh and Gootenberg 2020). However, collateral activity (non-specific cleavage of any nearby ssRNA regardless of complementary to the spacer region) persists in the case of bacteria but is found to be undetectable in human cells and even in plants (Abudayyeh et al. 2017). Further studies also revealed that Cas13s can be used in multiplexed targeting applications as it can process its CRISPR array and produces individual crRNAs for targeting multiple RNAs (Gootenberg et al. 2018; O’Connell 2019). Moreover, Cas13s treatment does not induce mutation in the crRNA target site and produces minimal off-target effects as compared to the Cas9 and Cas12 treatments (O’Connell 2019; https://www.synthego.com/blog/crispr-coronavirus-detection). This robust ability of the Cas13s allows researchers to use it in several applications such as transcript knockdown, imaging of live-cell transcript, and RNA editing and repair. Not only that, the collateral activity of Cas13s provides scientists to use this approach for diagnostic purposes and led to the discovery of SHERLOCK (Specific High Sensitivity Enzymatic Reporter unLocking) nucleic acid detection system. In this crRNA-Cas13a complex cleaves, the target ssRNA along with the collateral RNase activity is leveraged to cleave fluorescent reporters tagged RNA once it binds to the target, and this allowed Gootenberg et al. to design a rapid, sensitive, and specific nucleic acid detection system (Abudayyeh and Gootenberg 2021).
Fig. 2.
Structural representation of different type-VI CRISPR-Cas13 orthologs with their approximated amino acid (aa) size of each Cas13 sub-type. Abbreviations: HEPN higher eukaryotes and prokaryotes nucleotide-binding domain, HTH helix-turn-helix domain, WYL WYL domain, TM transmembrane spanning the region. Within each CRISPR array, green diamonds symbolize the CRISPR repeats, while blue rectangles symbolize the spacer sequences
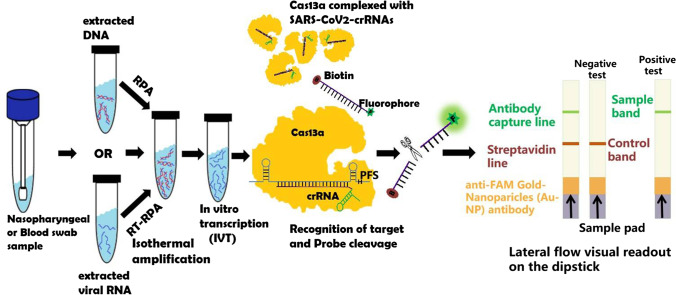
After refining over the years, Zhang et al. used this SHERLOCK system to detect SARS-CoV-2, in which they recognize two signature genes—the S gene and the ORF1ab gene with the help of two crRNAs. Once the virus-specific genes are recognized, it cleaves the target RNA along with the collateral RNA tagged with fluorophore or biotin and provides a visual readout using a commercially available dipstick test paper, within an hour and with setup time of < 25 min, similar to a pregnancy test kit (Abudayyeh and Gootenberg 2021). The SHERLOCK detection system involves mainly three basic steps to detect the COVID-19 virus: (i) amplification of the pathogenic RNA through reverse transcriptase-recombinase polymerase amplification (RT-RPA) followed by T7 RNA polymerase mediated in-vitro transcription of amplified DNA to generate the corresponding ssRNA. Compared to RT-LAMP, RT-RPA is faster and can be performed at room temperature, making it suitable for point of care applications; (ii) detection of pre-amplified viral RNA sequence with the help of target-specific crRNA and Cas13 nucleases; and (iii) visual readout using a dipstick test paper which detects cleaved collateral reporter RNA with labeled ends on specific antibody band, and the result was interpreted in ≤ 2 min, through the naked eye. The first single band near the sample pad (control line) indicates a negative result, while a single band or two bands close to the top of the strip (test line) indicates a positive result. Further, bands were quantified using the ImageJ gel analyzer tool, and the intensity of the band signal was normalized to the maximum (Gootenberg et al. 2017; Kellner et al. 2019; Abudayyeh and Gootenberg 2021; https://www.synthego.com/blog/crispr-coronavirus-detection; Fig. 3).
Fig. 3.
Schematic representation of SARS-CoV-2-SHERLOCK experimental workflow. In SHERLOCK, nucleic acid is amplified using reverse transcriptase-recombinase polymerase amplification (RPA or RT-RPA) technology followed by in vitro transcription process (IVT). The ssRNA is recognized by Cas13a-crRNA complex resulting in the activation of Cas13a enzyme, which possesses collateral/trans-cleavage activity and thus cleaves fluorescence tagged reporter ssRNA probe producing visual readout analyzed on dip strip paper by lateral flow assay
The SHERLOCK reaction can be carried out either in a two-step reaction or in a one-pot reaction. In two-step reactions, the nucleic acid is amplified using RPA technology followed by in vitro transcription and then Cas13-based detection, while in a one-pot reaction, all the components and enzymes are run simultaneously. However, one-pot reaction has several advantages over two-step reaction, as the lesser risk of contamination and faster detection time, and can be used for quantification and high-throughput application, but difficulty in one-pot reaction protocol optimization and less sensitivity make it disadvantageous over two-step reaction (Kellner et al. 2019; Lall 2020). Recently, due to increasing COVID-19 cases worldwide, the Food and Drug Administration (FDA) granted emergency use authorization to the first CRISPR-based test kit (SHERLOCK™-CRISPR-SAR-CoV-2) developed by Sherlock Biosciences which can yield results within an hour at 100% accuracy, sensitivity, specificity, and less expensive than RT-PCR test (https://sherlock.bio/sherlock-biosciences-receives-fda-emergency-use-authorization-for-crispr-sars-cov-2-rapid-diagnostic/).
SHERLOCK vs DETECTR
The development of several approaches for CRISPR-based diagnostics has revolutionized the detection of SARS-CoV-2 and has opened up a new window for further improvement and applicability of these techniques in the field of diagnostics and therapeutics. Among the two popular detection protocols, the detection sensitivity of the SHERLOCK system was found to be around 2 attomolar (2 aM) of RNA with 10–100 copies/μl of sample input, while the sensitivity of the DETECTR system was found to be in the range of femtomolar (fM) to attomolar (aM) of DNA with 70–300 copies/μl of input sample, because of the weak collateral activity of Cas12a as compared to the Cas13a. Moreover, Cas13a does not require PAM/PFS preferences at the target site, but Cas12a requires the PAM for cleavage at the target site. Thus, Cas13a-based SHERLOCK system stands out to be the most promising approach for CRISPR-based diagnostics. Further, SHERLOCK can regulate the expression profile of genes without altering the genome, but DETECTR has to modify the genome sequence to alter gene profile (Gootenberg et al. 2017; Lau et al. 2020; https://www.synthego.com/blog/crispr-coronavirus-detection). Hence, this robustness, flexibility, and specificity provide a broader target range to SHERLOCK as compared to the DETECTR. However, both SHERLOCK and DETECTR have shown advantages over the qPCR detection method because of the low cost of its components, does not require sophisticated lab and bulky instruments, and less time-consuming, but the problem of absolute quantification of nucleic acid using SHERLOCK and DETECTR persists, and thus standard PCR methods are more appropriate for such application (Lau et al. 2020).
Utilization of Cas9 in SARS-CoV-2 detection
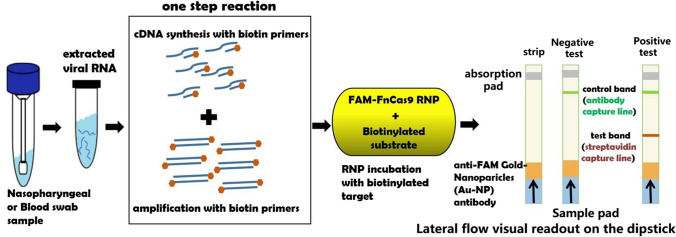
To date, the CRISPR/Cas9 system has rendered itself an easy-to-use, effective, and accessible genome editing platform with a broad range of applications in numerous fields. In addition, it has also found its place in the CRISPR-based diagnostics of SARS-CoV-2. A Cas9 ortholog from Francisella novicida with very high specificity and minimal off-target activity both under in vitro and in vivo conditions was used to develop FELUDA (FNCAS9 Editor Linked Uniform Detection Assay), to identify the SARS-CoV-2 virus by targeting the genetic material of the virus (Acharya et al. 2019). Like the gold-standard detection approach by RT-PCR, the swab sample for RNA extraction is collected from the nasopharyngeal area for the FELUDA test, followed by the conversion of the viral RNA to cDNA and its amplification to multiple copies in a single step using biotinylated primers. The amplification part is important for increasing the probability of detection. The sgRNAs were designed against the PAM containing conserved regions (NSP8 and nucleocapsid phosphoprotein) of SARS-CoV-2. Then, a FELUDA mixture comprised of amplified viral DNA, the guide RNA, and the Cas9 protein is prepared. The guide RNA directs the Cas9 protein to bind only with the viral DNA. The Cas9 reads the SARS-CoV-2 sequence like a bar code and binds with the matching sequence of the viral DNA. When a paper strip is immersed in the mixture, the complex goes towards it due to lateral flow. Remarkably, FELUDA-based lateral flow assay could distinguish SARS-CoV-2 synthetic DNA on a paper strip only when SARS-CoV-2-specific RNP was used to interrogate the substrate, where a single line indicates a negative result, while the double line, a positive result—much like a home pregnancy test. Moreover, FELUDA, by virtue of high specificity of FnCAS9-RNP, was also able to differentiate SARS-CoV-2 and SARS-CoV-1 sequences that only differed by a single nucleotide. Moreover, FELUDA is the world’s first COVID-19 detection test that employs Cas9 and has been established as a machine-independent and cost-effective alternative to current testing protocols by successfully detecting the presence of viral signature from low amounts of total RNA obtained from COVID-19 patients within 1 h (Acharya et al. 2019; Azhar et al. 2020; Fig. 4).
Fig. 4.
Schematic representation of FELUDA experimental workflow to detect SARS-CoV-2. FELUDA-based lateral flow assay could distinguish SARS-CoV-2 synthetic DNA on a paper strip only when SARS-CoV-2-specific RNP was used to interrogate the substrate, where a single line indicates a negative result, while the double line, a positive result
Identification and quantification of SARS-CoV-2 and their emerging variant strains using CRISPR-based methods
Among several tools developed so far, CRISPR-based methods SHERLOCK, DETECTR, and FELUDA techniques have shown rapid, 100% accuracy, sensitivity, and high specificity detection of SARS-CoV-2 but are not quantitative as compared to the standard PCR method. Recently, researchers have slightly modified the above methods which can not only detect the different variants of the SARS-CoV-2 efficaciously but also can detect the viral load and generate absolute quantification within 1 h. (Kaminski et al. 2021) These CRISPR-methods are named as follows: first, RAY (Rapid variant AssaY), an advanced version of FELUDA which uses multiplexing of crRNAs of a different mutated version of SARS-CoV-2 simultaneously accompanied with FnCAS9 (Kumar et al. 2021); second, miSHERLOCK (Minimally Instrumented) which uses multiplexing of crRNAs of a mutated version of SARS-CoV-2 simultaneously accompanied with Cas12a, and addition to that, it requires diagnostic chamber which includes sample concentrator, incubator, and fluorescence reporter and also supported with a mobile application that automatically interprets and quantifies the mutated variants (de Puig et al. 2021); third, RADICA (RApid DIgital Crispr Approach) which uses multiplexing of crRNAs of SARS-CoV-2, SARS-CoV, and MERS-CoV simultaneously accompanied with Cas12a followed by isothermal amplification of the extracted DNA and partitioning of the extracted DNA into a commercially available integrated chip. As the amplification begins, Cas12a cleaves the + ssDNA strand, and the fluorescence signal is detected by the detector and gives the absolute quantification of the virus particle (Wu et al. 2021).
CRISPR system as an antiviral platform
The discovery of CRISPR as a genome editor has revolutionized the scientific world with its broad range of applicability ranging from drug development, cancer therapy, agriculture, HIV treatment, sickle-cell anemia, and many more. Since then, scientists are putting tremendous efforts to use this technology in the detection/diagnosis and treatment of various diseases. Recently, researchers are recreating this technology by exploring more Cas protein orthologs such as Cas12 and Cas13 family nucleases, to detect and fight against the emerging deadly viruses SARS-CoV-2 by targeting the house-keeping genes of SARS-CoV-2, thus acting as an antiviral agent against COVID-19 (Yan et al. 2019; Straiton 2020). Further discovery of Cas13d has helped the scientists to target and cleave ssRNA of SARS-CoV-2. The uniqueness and the potentiality of Cas13d make it a promising target for the development of antiviral agents because it can be programmed to target and inhibit any ssRNA viruses with high precision in human cells, without genomic loss of the targeted gene, and also it does not require any stringent PAM/PFS sequence to cleave the target unlike Cas9 and Cas12 enzyme. Further, the delivery of Cas13d (967 aa) along with CRISPR array containing two or more crRNA via adeno-associated virus (AAV) into the mammalian cells is facilitated by the small size of the construct. By virtue of Cas13’s high catalytic activity in mammalian cells, it can efficiently clear the lung tissue and facilitate targeted inhibition of the SARS-CoV-2 genome. Moreover, Cas13d which targets ssRNA does not any report of off-target effects but in the case of Cas9 and Cas12 endonucleases which target DNA were found to have off-target effects (Konermann et al. 2018; Bayat et al. 2018; Freije et al. 2019). Thus, making the CRISPR-Cas13 system a prominent antiviral therapeutic tool to combat viral infections such as SARS-CoV-2.
Abbott et al. discovered CRISPR-Cas13-based therapy treatment called Prophylactic Antiviral CRISPR in huMAN cells (PACMAN) against SARS-CoV-2 using Cas13d derived from Ruminococcus flavefaciens. They demonstrated that PACMAN effectively degrades the genomic sequences of SARS-CoV-2 and influenza A virus (IAV) in human lung epithelial cells and stops its replication efficiently. Therefore, PACMAN can be a potential pan-coronavirus inhibition strategy to control the emerging variants of disease-causing coronavirus like SARS-CoV-2, SARS, and MERS that reside in animals and humans (Abbott et al. 2020). Recently, Nguyen et al. used the CRISPR-Cas13d system to degrade the SARS-CoV-2 RNA genome by designing specific crRNAs against ORF1ab (replicase) and S (spike) genes of it. Researchers have also shown that targeting the highly conserved RdRp gene of SARS-CoV-2 also inhibits viral replication (Nguyen et al. 2020; Lotfi and Rezaei 2020).
Since, PACMAN shows full potential to be used as a molecular tool for the detection and treatment of various diseases such as viral, anemia, cancer, and many more at the transcriptome level without altering the genome but still faces basic challenges when it comes to its delivery (Amirkhanov and Stepanov 2019). The large size of the CRISPR components often makes the in-cell delivery tedious. However, due to the small size of Cas13d, viral vectors are opted to deliver such CRISPR-based antivirals into the cells in vitro, but limitations like immunogenicity and duration of Cas protein expression still persist (Lino et al. 2018; Freije et al. 2019; Straiton 2020). Moreover, in vivo delivery of PACMAN using AAV vectors to the target tissues is still to be optimized to gain better insight into its mechanism, efficiency, fidelity, and toxicity. However, non-viral vectors such as inorganic nanoparticles, lipid nanoparticles, exosome encapsulation, cell-penetrating peptides, lipoplexes, and polyplexes can be used as an alternative delivery system of PACMAN into the cells or target organs/tissues to overcome the limitation of toxicity and immunogenicity, off-target effects, and scale-up the duration of Cas enzyme expression in the in vitro and in vivo (Lino et al. 2018; Abbott et al. 2020). Recently, researchers at Stanford University have synthesized a synthetic peptide termed lipitoids which is non-toxic and expeditiously carries nucleotides into human cells by encapsulating them into nanoparticles. They demonstrated that when PACMAN combined with lipitoid is added in a sample of SARS-CoV-2-affected human epithelial lung cells, it was able to reduce 90% of the SARS-CoV-2 from the solution. Therefore, lipitoid a non-viral delivery system, which can be used efficiently to deliver PACMAN into human cells. However, to achieve the best results, the next step will be to deliver PACMAN/lipitoid in vivo against a live SARS-CoV-2 virus and its related coronavirus variants (Lino et al. 2018; Nguyen et al. 2020; Abbott et al. 2020).
Implications of CRISPR in vaccine development
In response to control the outbreak of the SARS-CoV-2 pandemic, there is an urgent need to develop vaccines and specific antiviral drugs against the SARS-CoV-2 virus. However, the development of the vaccine is a long and complex process and takes 5 to 10 years to get commercialized. But because of the rapid inflation of COVID-19 cases globally, WHO permitted the use of seventeen COVID-19 vaccine candidates under the act of Emergency Use Authorization (EUA). These seventeen COVID-19 vaccine candidates were developed under 1 year with valid experimental (pre-clinical and clinical) data but still need long-term extensive research and should meet all the regulatory norms which are under process. Further, recent WHO’s landscape of COVID-19 vaccine development worldwide states that there are 294 vaccine candidates which are in the clinical (110 vaccine candidates) and pre-clinical (184 vaccine candidates) stage (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines). Traditionally, most of these vaccines work by injecting the viral proteins or weakened viruses or RNA into the human body and thus boosting the immune system to generate antibodies against it and thus neutralizing the virus. However, the evolution of mutant variants of SARS-CoV-2 makes the scenario worst as these vaccines might fail or become less effective against it. Therefore, there is an urgency for the acceleration of vaccine production or the finding of alternative approaches to detect and neutralize such mutant SARS-CoV-2 variants. To meet this, CRISPR approaches are being utilized by the researchers which shows great potential in producing safer, multivalent, recombinant viral vaccines; engineered B cells; and antiviral defense systems in humans to fight against such mutagenic pathogens and thus overcome the limitation of a traditional vaccine. For instance, Atasoy et al. implemented NHEJ-CRISPR/Cas9 (Non-Homologous End Joining) combined with CRE-LOX system and successfully deleted the virulence genes [thymidine kinase (TK) and unique short 4 (US4)] of infectious laryngotracheitis virus (ILTV) and simultaneously inserted foreign gene of Newcastle virus [inserted fusion (F) gene] without affecting replication of ILTV and expression of F protein and thus successfully generated multivalent recombinant vaccine vector which can be used for the protection against viral pathogens (Atasoy et al. 2019). Similarly, Chang et al. successfully generated recombinant HVT-H7N9-HA bivalent vaccine using HDR-CRISPR/Cas9 (homology-directed repair) accompanied with erythrocyte binding assay. They inserted the H7N9-HA expression cassette inside the UL45 and UL46 (Integrin region) of HVT (turkey herpes virus) using CRISPR/Cas9. This H7N9-HA cassette upon transfection expresses the avian influenza virus (AVI) antigen, hemagglutinin (HA) glycoprotein (Chang et al. 2019). Johnson et al. have shown that they can efficiently engineer B cells with CRISPR/Cas9 to express mAbs against different antigens. Hence, this strategy may be adapted as an alternative method to vaccination by inserting precisely engineered nucleic acid stretch for antibodies against SARS-CoV-2 in the host B cell genome to express the antibody that is specific for SARS-CoV-2 without introducing a version of the virus. This may further potentially eliminate the need of repeated injections and may allow fighting such viral outbreaks within a month instead of years to make traditional vaccines for the same (Johnson et al. 2018; Faiq 2020). Further, Abbott et al. have shown that PACMAN can be utilized as an alternative genetic vaccine approach to kill or disable the SARS-CoV-2 by targeting and destroying the intracellular viral genome and its mRNAs, hence halting the viral replication in humans without the requisite to introduce the virus itself. Furthermore, they have stated that Cas13d not only can neutralize SARS-CoV-2 but also has the potential to neutralize mutant SARS-CoV-2 variants and also the viruses that are found in animal reservoirs which may infect humans, by delivering multiple crRNAs simultaneously that target different regions of the virus and thus not allowing the virus to escape through mutation (Abott et al. 2020; Patchsung et al. 2020). In one of the most interesting recent studies, Blanchard et al. delivered polymer formulated mRNA encoded Cas13a and crRNAs for PB1 and PB2 which are highly conserved regions of influenza virus A (IVA) and for replicase and nucleocapsid genes of SARS-CoV-2 to the respiratory tract of IVA-infected mice and SARS-CoV-2-infected hamsters using nebulizer, respectively. They found that Cas13a debauched the IVA-RNA in lung tissue of mice efficaciously. On the other hand, Cas13a reduces the replication of SARS-CoV-2 in hamsters, thus palliating both the viral infections expeditiously (Blanchard et al. 2021). However, these studies are just a base-born and require more extensive pre-clinical studies to look into limitations like immunogenicity, delivery of CRISPR components, the fate of Cas protein expression, and off-target effects before it goes to the clinical stage.
Limitations and enhancement of CRISPR-Cas-based diagnostic and therapeutic tools
CRISPR-based sensing has exhibited its great potential in specific, sensitive, and affordable detection of nucleic acid in the last couple of years, and presently SARS-CoV-2, but still this arena needs much more attention for overcoming the challenges associated with this technology for the development of “sample-in-answer-out” CRISPR-based point of care devices. According to the WHO guidelines, the key features of a point of care diagnostic device are affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable (ASSURED) (Nouri et al. 2020). Abiding by these guidelines, the assay runtime of the CRISPR-based methods is one of the most crucial drawback, since sample preparation involves the most part of the CRISPR-based diagnostic methods, as well as separate sample preparations also increases the complexity of the process and in turn reduces the acceptability and applicability (Lino et al. 2018; Abbott et al. 2020; Nouri et al. 2020). This necessitates the development of a single-step diagnostic test for efficient and rapid point of care application. Recently, few studies have been carried out to accelerate and simplify the process of SARS-CoV-2 sample preparation, and several studies have also inspected sample preparation-free assay, using RT-PCR and RT-LAMP (Crannell et al. 2014; Rabe and Cepko 2020). Hence, further research should be concentrated on the development of a fully rationalized process by impeccably combining the sample preparation with the CRISPR assay for efficient and user-friendly operation, i.e., raw sample-in-answer-out. Apart from the lacking of the sample preparation process for CRISPR-based detection, performance enhancement of the CRISPR sensing at the final step also requires distinguished attention. Currently, to improve the performance of CRISPR sensing with respect to signal enhancement, nanomaterials have been introduced (Yang et al. 2018; Shao et al. 2019; Rabiee et al. 2020). A simple visual detection system has been recently developed using quantum dots, as an ultra-brightness indicator, for CRISPR-Cas12a assay to sense viral DNA targets (Ding et al. 2020; Yuan et al.; Rabiee et al. 2020). Hence, this sensing system has also omitted the use of bulky and complicated sensing instruments and employed a handheld flashlight to distinguish between positive and negative samples. Additionally, artificial intelligence (AI) can also be combined with this sensing technology for improved and smoother data storage, classification, and sharing. For instance, Ibrahim et al. have introduced the internet and machine learning with CRISPR sensing for wireless transmission of signals over the cloud to support and accelerate the decision-making processes. This opens up new corridors for further research, focusing on the enhancement of CRISPR sensing performances in the near future (Ibrahim et al. 2020).
Conclusion and future perspectives
The emergence of highly infectious SARS-CoV-2 and their natural variants resulting from rapid evolution and diversification has drastically affected human health. To address these ongoing health issues, emphasis has been given to the development of reliable and scalable diagnostic tools. Among several tools developed so far, CRISPR-based SHERLOCK, DETECTR, and FELUDA techniques hold considerable promise for practical application in the treatment and the diagnostics of COVID-19 caused by SARS-CoV-2. Such ready-to-use portable diagnostic kits are available at low cost and as such can be used at the hospital and resource-limited areas such as airports, schools, small clinics, and even at home as well. These CRISPR-based diagnostics have the potential to detect SARS-CoV-2 and the emerging variants and can provide results within an hour with 100% accuracy, sensitivity, and high specificity as compared to the standard PCR method. Thus, screening and early detection of the COVID-19 affected populations at an individual level will impact society in preventing the spread of transmission of SARS-CoV-2 viral infection by imposing quarantine measures immediately. Although the CRISPR-based detection tool has proven to be promising, it is still limited to the lab. Therefore, further translational research is very much required for lab to land transition through large-scale production of such rapid diagnostic kits and commercialization. The development of low-cost and portable microfluidic CRISPR-based cartridges and lyophilized reagents can aid to carry out the COVID-19 test at any location. However, the application of the kit will require well-trained personnel experienced in the technique.
The discovery of CRISPR-Cas13 further extended the practical applications such as RNA editing and repair, RNA knockdown, live-cell transcript imaging, and most currently being used as a novel therapeutic approach to cure COVID-19. Further, research on Cas13d nucleases has also led to the foundation for the development of new technology termed PACMAN which has intelligibly shown that CRISPR-Cas13d with specific crRNA can cleave the target viral ssRNA inside the SARS-CoV-2 viral infected cells by preventing the expression of protein-coding genes prudent for the survival and replication of the viruses. Thus, the CRISPR-Cas system acts as a “molecular detector and antiviral proctor.” PACMAN technology emerges as a potential antiviral strategy to combat not only SARS-CoV-2 but also several variants of viral strains with pandemic potency. Even so, the current delivery system of PACMAN uses an AAV vector as a vehicle, but it might be inapplicable for in vivo delivery, because of toxicity and immunogenicity. Therefore, the assessment of other delivery systems such as non-viral vectors may add benefits to the researchers to explore and refine the current technology to gain more cognition about the fate of delivery approach, duration of Cas13d expression, and toxicity of each component for future medicinal therapeutic advancements. Thus, CRISPR-Cas13 demands more attention to fully exploit its potential and overcome the associated challenges, for better application with salutary results for today and even for decades to come. Finally, we conclude that CRISPR assays are rapid, scalable, and customizable without losing specificity and sensitivity to detect SARS-CoV-2 and its variant strains with limited resources at home or any point of care. However, it still needs extensive studies to overcome the complexity of designing crRNAs by developing more effective bioinformatic tools to predict crRNAs specificity and activity. Further, modification of Cas enzyme or reporter molecule, incorporation of non-primer signal amplification strategies, ruling out separate sample preparation steps, and multiplexing with different Cas proteins such as Cas13b-d, Cas12b, and Cas14 for detection are some of the potential areas to meliorate and reshape the technology and extend its sensing strategies and thus open a new gateway for the researcher to carry out more extended research in the field and leverage this new technology for the betterment of human health.
Acknowledgements
All the authors acknowledge and thank their respective institutes. RG also acknowledges the UGC and GoI for National Fellowship. Mr. Kazi sincerely acknowledges Council of Scientific & Industrial Research (CSIR), India, for senior research fellowship (File No.: 09/202(0070)/2017-EMR-I). Authors also like to acknowledge the DBT, Govt of India [grant number BT/PR26301/GET/119/258/2017], and Govt of West Bengal Department of Biotechnology WBDBT [grant number – BT/P/Budget/RD-74/2017] for their support, and Indian Council of Medical Research for sponsoring Emeritus Professorship to Dr Syamal Roy.
Declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, Chemparathy A, Chmura S, Heaton NS, Debs R, Pande T, Endy D, La Russa MF, Lewis DB, Qi LS. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181:865–876.e12. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh, OO, Gootenberg J (2020) Cas13 — Zhang Lab. https://zlab.bio/cas13. Accessed 8 Jun 2021
- Abudayyeh OO, Gootenberg JS. CRISPR diagnostics. Science (80-) 2021;372:914–915. doi: 10.1126/science.abi9335. [DOI] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya S, Mishra A, Paul D, Ansari AH, Azhar M, Kumar M, Rauthan R, Sharma N, Aich M, Sinha D, Sharma S, Jain S, Ray A, Jain S, Ramalingam S, Maiti S, Chakraborty D. Francisella novicida Cas9 interrogates genomic DNA with very high specificity and can be used for mammalian genome editing. Proc Natl Acad Sci U S A. 2019;116:20959–20968. doi: 10.1073/pnas.1818461116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adli M. (2018). The CRISPR tool kit for genome editing and beyond. Nature Communications, 9(1). 10.1038/s41467-018-04252-2 [DOI] [PMC free article] [PubMed]
- Ali Z, Aman R, Mahas A, Rao GS, Tehseen M, Marsic T, Salunke R, Subudhi AK, Hala SM, Hamdan SM, Pain A, Alofi FS, Alsomali A, Hashem AM, Khogeer A, Almontashiri NAM, Abedalthagafi M, Hassan N, Mahfouz MM. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288:198129. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirkhanov RN, Stepanov GA. Systems of delivery of CRISPR/Cas9 ribonucleoprotein complexes for genome editing. Russ J Bioorganic Chem. 2019;45:431–437. doi: 10.1134/S1068162019060025. [DOI] [Google Scholar]
- Amitai G, Sorek R. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol. 2016;14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- Atasoy MO, Rohaim MA, Munir M. Simultaneous deletion of virulence factors and insertion of antigens into the infectious laryngotracheitis virus using NHEJ-CRISPR/Cas9 and Cre-Lox system for construction of a stable vaccine vector. Vaccines. 2019;7(4):207. doi: 10.3390/vaccines7040207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Lin H, Li H, Zhou Y, Liu J, Zhong G, Wu L, Jiang W, Du H, Yang J, Xie Q, Huang L. Cas12a-based on-site and rapid nucleic acid detection of african swine fever. Front Microbiol. 2019;10:2830. doi: 10.3389/fmicb.2019.02830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayat H, Naderi F, Khan AH, Memarnejadian A, Rahimpour A (2018). The Impact of CRISPR-Cas System on Antiviral Therapy. Advanced Pharmaceutical Bulletin, 8(4):591–597. 10.15171/apb.2018.067 [DOI] [PMC free article] [PubMed]
- Blanchard EL, Vanover D, Bawage SS, Tiwari PM, Rotolo L, Beyersdorf J, Peck HE, Bruno NC, Hincapie R, Michel F, Murray J, Sadhwani H, Vanderheyden B, Finn MG, Brinton MA, Lafontaine ER, Hogan RJ, Zurla C, Santangelo PJ. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat Biotechnol. 2021;39(6):717–726. doi: 10.1038/s41587-021-00822-w. [DOI] [PubMed] [Google Scholar]
- Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, Zorn K, Gopez A, Hsu E, Gu W, Miller S, Pan CY, Guevara H, Wadford DA, Chen JS, Chiu CY. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti R, Singh R, Ghosh A, Dey D, Sharma P, Velayutham R, Roy S, Ghosh D. A review on potential of natural products in the management of COVID-19. RSC Adv. 2021;11:16711–16735. doi: 10.1039/D1RA00644D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Ameen F, Sealy JE, Sadeyen JR, Bhat S, Li Y, Iqbal M. Application of HDR-CRISPR/Cas9 and erythrocyte binding for rapid generation of recombinant turkey herpesvirus-vectored avian influenza virus vaccines. Vaccines. 2019;7(4):192. doi: 10.3390/vaccines7040192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar M, Phutela R, Ansari AH, Sinha D, Sharma N, Kumar M, Aich M, Sharma S, Rauthan R, Singhal K, Lad H, Patra PK, Makharia G, Chandak GR, Chakraborty D, Maiti S (2020) Rapid, field-deployable nucleobase detection and identification using FnCas9. bioRxiv. 10.1101/2020.04.07.028167
- Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science (80-) 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crannell ZA, Rohrman B, Richards-Kortum R. Equipment-free incubation of recombinase polymerase amplification reactions using body heat. PLoS One. 2014;9:e112146. doi: 10.1371/journal.pone.0112146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Somoza RA, Wang L, Welter JF, Li Y, Caplan AI, Liu CC. Exploring the trans-cleavage activity of CRISPR-Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew Chemie - Int Ed. 2019;58:17399–17405. doi: 10.1002/anie.201910772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Puig H, Lee RA, Najjar D, Tan X, Soekensen LR, Angenent-Mari NM, Donghia NM, Weckman NE, Ory A, Ng CF, Nguyen PQ, Mao AS, Ferrante TC, Lansberry G, Sallum H, Niemi J, Collins JJ. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci Adv. 2021;7(32):eabh2944. doi: 10.1126/sciadv.abh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Yin K, Li Z, Lalla RV, Ballesteros E, Sfeir MM, Liu C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun. 2020;11:4711–4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esakandari H, Nabi-Afjadi M, Fakkari-Afjadi J, Farahmandian N, Miresmaeili SM, Bahreini E (2020) A comprehensive review of COVID-19 characteristics. Biological Procedures Online, 22(1). 10.1186/s12575-020-00128-2 [DOI] [PMC free article] [PubMed]
- Faiq MA. B-cell engineering: a promising approach towards vaccine development for COVID-19. Med Hypotheses. 2020;144:109948. doi: 10.1016/j.mehy.2020.109948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, Metsky HC, Luo CY, Abudayyeh OO, Gootenberg JS, Yozwiak NL, Zhang F, Sabeti PC. Programmable inhibition and detection of RNA viruses Using Cas13. Mol Cell. 2019;76:826–837.e11. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Yang H, Rajashankar KR, Huang Z, Patel DJ. Type v CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 2016;26:901–913. doi: 10.1038/cr.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science (80-) 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a and Csm6. Science (80-) 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guk K, Keem JO, Hwang SG, Kim H, Kang T, Lim EK, Jung J. A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosens Bioelectron. 2017;95:67–71. doi: 10.1016/j.bios.2017.04.016. [DOI] [PubMed] [Google Scholar]
- Gupta D, Bhattacharjee O, Mandal D, Sen MK, Dey D, Dasgupta A, Kazi TA, Gupta R, Sinharoy S, Acharya K, Chattopadhyay D, Ravichandiran V, Roy S, Ghosh D. CRISPR-Cas9 system: a new-fangled dawn in gene editing. Life Sci. 2019;232:116636. doi: 10.1016/j.lfs.2019.116636. [DOI] [PubMed] [Google Scholar]
- Gupta R, Gupta D, Ahmed KT, Dey D, Singh R, Swarnakar S, Ravichandiran V, Roy S, Ghosh D (2021) Modification of Cas9, gRNA and PAM: key to further regulate genome editing and its applications. In: Progress in Molecular Biology and Translational Science. Elsevier B.V., pp 85–98 [DOI] [PubMed]
- Hou T, Zeng W, Yang M, Chen W, Ren L, Ai J, Wu J, Liao Y, Gou X, Li Y, Wang X, Su H, Gu B, Wang J, Xu T (2020) Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog 16. 10.1371/journal.ppat.1008705 [DOI] [PMC free article] [PubMed]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim AU, Al-Turjman F, Sa’id Z, Ozsoz M (2020) Futuristic CRISPR-based biosensing in the cloud and internet of things era: an overview. Multimed Tools Appl 1–29. 10.1007/s11042-020-09010-5 [DOI] [PMC free article] [PubMed]
- Javalkote VS, Kancharla N, Bhadra B, Shukla M, Soni B, Sapre A, Goodin M, Bandyopadhyay A, Dasgupta S (2020) CRISPR-based assays for rapid detection of SARS-CoV-2. Methods 9:S1046-2023(20)30217-6. 10.1016/j.ymeth.2020.10.003 [DOI] [PMC free article] [PubMed]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (80-) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MJ, Laoharawee K, Lahr WS, Webber BR, Moriarity BS (2018) Engineering of primary human B cells with CRISPR/Cas9 targeted nuclease. Sci Rep 8(1). 10.1038/s41598-018-30358-0 [DOI] [PMC free article] [PubMed]
- Kaminski MM, Abudayyeh OO, Gootenberg JS, Zhang F, Collins JJ. CRISPR-based diagnostics. Nature. Biomed Eng. 2021;5(7):643–656. doi: 10.1038/s41551-021-00760-7. [DOI] [PubMed] [Google Scholar]
- Kazi TA, Bhattacharya R, Mukhopadhyay BC, Mitra S (2020) Animal hosts of coronavirus : unsung reason behind recurring outbreaks animal hosts of coronavirus : unsung reason behind recurring outbreaks. Int J Cell Sci Mol Biol. 10.19080/IJCSMB.2020.06.55569
- Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD (2018). Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 173(3):665–676.e14. 10.1016/j.cell.2018.02.033 [DOI] [PMC free article] [PubMed]
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling A-E, Humphrey CD, Shieh W-J, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang J-Y, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/nejmoa030781. [DOI] [PubMed] [Google Scholar]
- Kumar M, Gulati S, Ansari AH, Phutela R, Acharya S, Azhar M, Murthy J, Kathpalia P, Kanakan A, Maurya R, Vasudevan JS, S A, Pandey R, Maiti S, Chakraborty D (2021) FnCas9-based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip. ELife 10. 10.7554/elife.67130 [DOI] [PMC free article] [PubMed]
- Lall S (2020) SHERLOCK-based one-step test provides rapid and sensitive Covid-19 detection | MIT News | Massachusetts Institute of Technology. https://news.mit.edu/2020/sherlock-based-one-step-test-provides-rapid-sensitive-covid-19-detection-0505. Accessed 8 Jun 2021
- Lau A, Ren C, Lee LP. Critical review on where CRISPR meets molecular diagnostics. Prog Biomed Eng. 2020;3:012001. doi: 10.1088/2516-1091/abbf5e. [DOI] [Google Scholar]
- Li SY, Cheng QX, Liu JK, Nie XQ, Zhao GP, Wang J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018;28:491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino CA, Harper JC, Carney JP, Timlin JA. Delivering crispr: a review of the challenges and approaches. Drug Deliv. 2018;25:1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfi M, Rezaei N. (2020). CRISPR/Cas13: A potential therapeutic option of COVID-19. Biomedicine & Pharmacotherapy, 131, 110738. 10.1016/j.biopha.2020.110738 [DOI] [PMC free article] [PubMed]
- Lübke N, Senff T, Scherger S, Hauka S, Andrée M, Adams O, Timm J, Walker A (2020) Extraction-free SARS-CoV-2 detection by rapid RT-qPCR universal for all primary respiratory materials. J Clin Virol 130. 10.1016/j.jcv.2020.104579 [DOI] [PMC free article] [PubMed]
- Nguyen TM, Zhang Y, Pandolfi PP. Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020;30:189–190. doi: 10.1038/s41422-020-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri R, Jiang Y, Lian XL, Guan W. Sequence-specific recognition of HIV-1 DNA with solid-state CRISPR-Cas12a-assisted nanopores (SCAN) ACS Sensors. 2020;5:1273–1280. doi: 10.1021/acssensors.0c00497. [DOI] [PubMed] [Google Scholar]
- O’Connell MR. Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR–Cas systems. J Mol Biol. 2019;431:66–87. doi: 10.1016/j.jmb.2018.06.029. [DOI] [PubMed] [Google Scholar]
- Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O’Connor DH, Gehrke L, Collins JJ. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Patchsung M, Jantarug K, Pattama A, Aphicho K, Suraritdechachai S, Meesawat P, Sappakhaw K, Leelahakorn N, Ruenkam T, Wongsatit T, Athipanyasilp N, Eiamthong B, Lakkanasirorat B, Phoodokmai T, Niljianskul N, Pakotiprapha D, Chanarat S, Homchan A, Tinikul R, Uttamapinant C. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4(12):1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Prabhune M (2020) CRISPR NEWS. Synthego. https://www.synthego.com/blog/crispr-coronavirus-detection. Accessed 5 Jun 2020
- Rabe BA, Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc Natl Acad Sci U S A. 2020;117:24450–24458. doi: 10.1073/pnas.2011221117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiee N, Bagherzadeh M, Ghadiri AM, Salehi G, Fatahi Y, Dinarvand R. ZnAl nano layered double hydroxides for dual functional CRISPR/Cas9 delivery and enhanced green fluorescence protein biosensor. Sci Rep. 2020;10:1–15. doi: 10.1038/s41598-020-77809-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao N, Han X, Song Y, Zhang P, Qin L. CRISPR-Cas12a coupled with platinum nanoreporter for visual quantification of SNVs on a volumetric bar-chart chip. Anal Chem. 2019;91:12384–12391. doi: 10.1021/acs.analchem.9b02925. [DOI] [PubMed] [Google Scholar]
- Singh R, Chandel S, Ghosh A, Dey D, Chakravarti R, Roy S, Ravichandiran V, Ghosh D. Application of CRISPR/Cas system in the metabolic engineering of small molecules. Mol Biotechnol. 2021;63:459–476. doi: 10.1007/s12033-021-00310-1. [DOI] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straiton J. CRISPR vs COVID-19: how can gene editing help beat a virus? Biotechniques. 2020;69:327–329. doi: 10.2144/btn-2020-0145. [DOI] [PubMed] [Google Scholar]
- Subali AD, Wiyono L (2021) Reverse transcriptase loop mediated isothermal amplification (RT-LAMP) for COVID-19 diagnosis: a systematic review and meta-analysis. Pathogens Glob Health: 1–11. 10.1080/20477724.2021.1933335 [DOI] [PMC free article] [PubMed]
- Surjit M, Liu B, Chow VTK, Lal SK. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Suryawanshi H, Djandji A, Kohl E, Morgan S, Hod EA, Whittier S, Roth K, Yeh R, Alejaldre JC, Fleck E, Ferrara S, Hercz D, Andrews D, Lee L, Hendershot KA, Goldstein J, Suh Y, Mansukhani M, Williams Z. Field-deployable, rapid diagnostic testing of saliva for SARS-CoV-2. Sci Rep. 2021;11:5448. doi: 10.1038/s41598-021-84792-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tay JK, Goh CK, Chan C, Lee YH, Springs SL, Wang DY, Loh KS, Lu TK, Yu H. Digital CRISPR-based method for the rapid detection and absolute quantification of nucleic acids. Biomaterials. 2021;274:120876. doi: 10.1016/j.biomaterials.2021.120876. [DOI] [PubMed] [Google Scholar]
- Xiao G, He X, Zhang S, Liu Y, Liang Z, Liu H, Zhang J, Ou M, Cai S, Lai W, Zhang T, Ren L, Zhang G (2020) Cas12a/Guide RNA-based platform for rapid and accurate identification of major mycobacterium species. J Clin Microbiol 58. 10.1128/JCM.01368-19 [DOI] [PMC free article] [PubMed]
- Yan F, Wang W, Zhang J. CRISPR-Cas12 and Cas13: the lesser known siblings of CRISPR-Cas9. Cell Biol Toxicol. 2019;35:489–492. doi: 10.1007/s10565-019-09489-1. [DOI] [PubMed] [Google Scholar]
- Yang W, Restrepo-Pérez L, Bengtson M, Heerema SJ, Birnie A, Van Der Torre J, Dekker C. Detection of CRISPR-dCas9 on DNA with solid-state nanopores. Nano Lett. 2018;18:6469–6474. doi: 10.1021/acs.nanolett.8b02968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C, Tian T, Sun J, Hu M, Wang X, Xiong E, Cheng M, Bao Y, Lin W, Jiang J, Yang C, Chen Q, Zhang H, Wang H, Wang X, Deng X, Liao X, Liu Y, Wang Z, Zhang G, Zhou X. Universal and naked-eye gene detection platform based on the clustered regularly interspaced short palindromic repeats/Cas12a/13a system. Anal Chem. 2020;92:4029–4037. doi: 10.1021/acs.analchem.9b05597. [DOI] [PubMed] [Google Scholar]
- Yuki K, Fujiogi M, Koutsogiannaki S (2020) COVID-19 pathophysiology: A review. Clinical Immunology 215:108427. 10.1016/j.clim.2020.108427 [DOI] [PMC free article] [PubMed]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, Van Der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]