Abstract
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its rapid international spread has caused the coronavirus disease 2019 (COVID-19) pandemics, which is a global public health crisis. Thus, there is an urgent need to establish biological models to study the pathology of SARS-CoV-2 infection, which not only involves respiratory failure, but also includes dysregulation of other organs and systems, including the brain, heart, liver, intestines, pancreas, kidneys, eyes, and so on. Cellular and organoid models derived from human induced pluripotent stem cells (iPSCs) are ideal tools for in vitro simulation of viral life cycles and drug screening to prevent the reemergence of coronavirus. These iPSC-derived models could recapitulate the functions and physiology of various human cell types and assemble the complex microenvironments similar with those in the human organs; therefore, they can improve the study efficiency of viral infection mechanisms, mimic the natural host-virus interaction, and be suited for long-term experiments. In this review, we focus on the application of in vitro iPSC-derived cellular and organoid models in COVID-19 studies.
Keywords: induced pluripotent stem cell, organoid, cellular model, COVID-19, SARS-CoV-2
Introduction
Since its outbreak in 2019, the coronavirus disease (COVID-19) pandemics have infected more than 190 million people and caused more than 4 million deaths1. COVID-19 is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), an enveloped positive-sense single-stranded RNA virus. It enters the host cells using angiotensin-converting enzyme 2 (ACE2) as the cell surface receptor and transmembrane serine protease 2 (TMPRSS2) as the effector to cleave its spike protein (Hoffmann et al., 2020). SARS-CoV-2 spreads mainly through the respiratory tract (Lu et al., 2020). Respiratory failure is the most common cause of death in COVID-19 patients; meanwhile, severe fatal manifestations are also observed in other organs, such as the brain, heart, liver, intestines, and pancreas (Puelles et al., 2020). Therefore, it is of particular importance to find models that can imitate the natural host-virus interactions of SARS-CoV-2 in a variety of human cell types and organs, thus improving the study efficiency for identifying key molecular regulators and the underlying mechanisms of virus infection and disease progression.
There have been both animal models (e.g., transgenic mice expressing human ACE2 and non-human primates) and cell line models (e.g., African green monkey Vero E6 cells and human cancer cell lines) available for COVID-19 research (Takayama, 2020). However, animal models are quite costly and display very different physiological characteristics from human, and cell line models have limitations in reproducing the viral life cycle and the pathology of COVID-19 in different human organs and tissues that contain a variety of cell types (Liu et al., 2011). For example, the entry routes of SARS-CoV-2 vary between cell lines and human tissues, as do immune responses and host-virus interactions (Milewska et al., 2020). In addition, human cancer cells carry numerous tumor-associated mutations, such as P53 mutations, which could interfere the SARS-CoV-2 infection (Ma-Lauer et al., 2016). Therefore, there is an urgent need to establish more cost-efficient and human-relevant models for COVID-19 research.
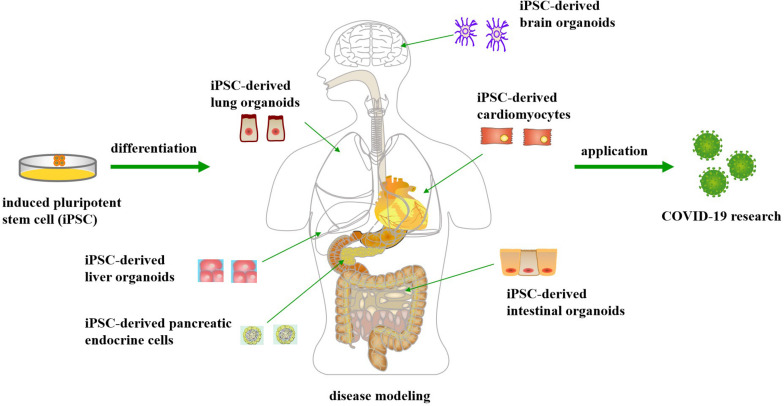
The emergence of human induced pluripotent stem cells (iPSCs) has enabled derivation of functional human cells or organoids to model human diseases, including infectious diseases, to develop new therapeutic approaches and to promote drug discovery (Yu et al., 2007; Luo et al., 2015a, 2018), without ethical issues like the human embryonic stem cells (Luo et al., 2014). For example, functional liver organoids generated by human iPSCs have been developed as personalized models of hepatitis B virus (HBV) infection, which is a powerful long-term platform for both research and drug screening of HBV (Nie et al., 2018). Recently, iPSC-derived cellular and organoid models have been utilized to simulate SARS-CoV-2 infections in multiple organs, not only the lung, but also the heart, brain, liver, intestines, and pancreas (Jacob et al., 2020; Yu et al., 2020) (Figure 1). These studies have demonstrated that SARS-CoV-2 can infect and propagate in a variety of cell types, leading to transcriptional alternations that indicate inflammatory responses and changes in cell function (Marshall, 2020). The purpose of this review is to describe the usefulness of these iPSC-derived cellular and organoid models in simulating human cellular physiology and tissue microenvironment, and enabling the study of host-virus interaction and drug screening for the COVID-19 disease, thus leading to a more comprehensive understanding of the SARS-CoV-2 pathogenesis.
FIGURE 1.
Application of human induced pluripotent stem cell-derived cellular and organoid models for COVID-19 research.
Lung Organoids
Human iPSC-derived airway and alveolar organoids have been developed and used for studying the processes of SARS-CoV-2 infection and transmission in the lungs (Pei et al., 2020). With these organoid models, the researchers have determined the cellular tropism of the virus. Ciliated cells, club cells, and alveolar type II cells (AT2) cells, which are arranged from the proximal to the distal airway and terminal alveoli in sequence, have been confirmed as SARS-CoV-2-targeted cells in the study (Mulay et al., 2021). Moreover, the viral infection downregulates metabolic processes, particularly the lipid metabolism, which, together with the already known upregulation of immune responses, is another molecular feature of SARS-CoV-2-infected cells (Pei et al., 2020). On the other hand, infected SARS-CoV-2 can decrease the level of its target ACE2 on the host cell surface through a variety of mechanisms. A comprehensive analysis of these information might enable better understanding about the viral pathogenesis and discovery therapeutic targets for the treatment of COVID-19 (Pei et al., 2020).
In addition, the early immune responses to viral infections were investigated using iPSC-derived AT2 (iAT2) cells (Huang J. et al., 2020). The results showed that AT2 cells are a central component of the inflammatory signaling that responds to SARS-CoV-2 infection within the first 24 h, with NF-κB signaling predominating this response (Huang J. et al., 2020). These findings are consistent with those in newly purified primary AT2 cells infected with SARS-CoV-2 (Mulay et al., 2021). They also observed cellular stress, toxicity, iAT cell death, and significant loss of surfactant genes expression in their model (Huang J. et al., 2020). These findings may be clinically relevant, as similar results were found in lung autopsies of multiple individuals who died of COVID-19 (Bradley et al., 2020; Hou et al., 2020). Other researchers have previously shown that the primary AT2 cells can be infected with SARS-CoV in the body (Qian et al., 2013); it has also been recently shown that AT2 cells may help promote lung regeneration in COVID-19 survivors (Chen J. et al., 2020). The correlation between AT2 cells and SARS-CoV-2 infection was further emphasized.
Furthermore, the researchers have demonstrated that human iPSC-derived lung cells and organoids could serve as powerful platforms for discovering and testing anti-SARS-CoV-2 drugs (Huang J. et al., 2020; Pei et al., 2020). They have demonstrated that Remdesivir, a predrug nucleotide analog that inhibits virus replication (Eastman et al., 2020), CB6, a human neutralizing antibody (Shi R. et al., 2020), as well as TMPRSS2 protease inhibition could effectively inhibit the replication of SARS-CoV-2 in the iPSC-derived models. These results are consistent with those in fundamental studies using primary cell models and in clinical trials (Beigel et al., 2020; Wang M. et al., 2020). Therefore, iPSC-derived in vitro human models could be employed to identify and test therapeutic entities for the treatment of COVID-19.
Taken together, the above results demonstrated that lung cells and organoids derived from human iPSCs can be utilized as pathophysiological models to study the potential mechanisms of SARS-CoV-2 transmission and to identify and test COVID-19 therapeutic agents (Pei et al., 2020).
Cardiomyocytes
There is growing evidence that patients with COVID-19 exhibit severe heart complications, elevated biomarkers of heart damage, and cardiac function deterioration, including cardiovascular complications such as cardiomyopathy, acute myocardial infarction, arrhythmia, and heart failure, greatly increasing the risk of death (Aggarwal et al., 2020a; Bansal, 2020; Long et al., 2020; Madjid et al., 2020; Shi S. et al., 2020). Several compounds and antibodies, such as Remdesivir, Olumiant + Remdesivir, Casirivimab + Imdevimab, Bamlanivimab + Etesevimab, Sotrovimab and Tocilizumab are currently licensed drug for treatment of COVID-19 patients under emergency use authorization2. However, there is limited safety information on these recommended drugs, especially since heart toxicity caused by the drug can lead to lethal complications, including myocardial ischemia, arrhythmias, and heart failure. Therefore, it is critical to evaluate any potential adverse effects on the cardiovascular system associated with current COVID-19 medications to avoid fatal side effects (Aggarwal et al., 2020b).
Human iPSC-derived cardiomyocytes (CMs) can be utilized to recapitulate cardiac pathophysiology and are considered as one of the most promising sources for cardic disease modeling, heart repair and cardiac toxicology screening (Mitcheson et al., 1998; Lan et al., 2013; Moreno and Pearson, 2013; Sharma et al., 2014, 2017; Burridge et al., 2016). In this context, iPSC-CMs are recommended as a reliable method of heart toxicity examination in the comprehensive in vitro proarrhythmia assay (CiPA), as a non-clinical safety pharmacological paradigm, to circumvent the limitations of existing methods used in preclinical safety assessment of drugs (Gintant et al., 2016; Goineau and Castagné, 2017; Sala et al., 2017). Choi et al. (2020) have shown that iPSC-CMs acutely treated with Remdesivir show a risk of arrhythmia and changes in the electrophysiological properties of myocardial cells in a dose-dependent manner, indicating that overdose or drug accumulation may lead to noteworthy adverse heart reactions, such as prolonged QT interstitial periods. In addition, they have demonstrated that iPSC-CMs not only allow SARS-CoV-2 infection, but also support the propagation of infectious viral particles (Choi et al., 2020).
Intestinal Organoids
Up to 50% of COVID-19 patients develop gastrointestinal symptoms associated with longer duration and increased severity of the disease (Luo et al., 2020; Wang F. et al., 2020; Wei et al., 2020; Xiao et al., 2020). However, it remains debatable whether the virus found in the intestines is contagious, as few studies have examined infectious viruses in feces (Zang et al., 2020). In cell culture, primary intestinal cells are highly susceptible to SARS-CoV-2 and can produce infectious viral particles. Intestinal organoids can quickly grow from adult stem cells derived from cells of large intestine and small intestine biopsy tissue (Sato et al., 2011). Stem cell-derived intestinal organoids have similar characteristics with primary intestinal cells and have been widely used to study viral infection (Sato et al., 2011; Ettayebi et al., 2016).
Studies have employed human iPSC-derived intestinal organoids to study SARS-CoV-2 tropisms in different intestinal cell types. In both in vivo and iPSC-derived organoid models, ACE2 is strongly expressed in the small intestine, as well as TMPRSS2. In contrast, colon organoids have lower ACE2 expression (Zang et al., 2020). In intestinal organs, TMPRSS4 performs the same functions as TMPRSS2 to support virus entry (Zang et al., 2020). Experiments have shown that two groups of small intestinal organoid models can be infected with SARS-CoV-2 (Lamers et al., 2020; Zang et al., 2020). In these organoid models, SARS-CoV-2, like its closest relative SARS-CoV, mainly infects mature enterocytes and dividing cells (Lamers et al., 2020; Zang et al., 2020). On the other hand, studies have shown that when the SARS-CoV-2 viruses are cultured in gastric fluid in the large intestine and small intestine, they quickly lose their infectious power (Simoneau and Ott, 2020; Zang et al., 2020). Therefore, even though viral particles were found in the feces occasionally, it might not be the primary pathway for virus transmission.
Furthermore, human iPSC-derived intestinal organoids generate valuable pathological models for studying the underlying mechanisms of intestinal SARS-CoV-2 infection. It is reported that SARS-CoV-2 actively infects both proximally and distally patterned intestinal organoids, thus resulting in production of infectious viral particles and significant transcriptional alterations, such as upregulation of the interferon-related genes, in multiple epithelial cell types (Mithal et al., 2021). Another organoid study shows that SARS-CoV-2 can infect all intestinal cell types investigated except goblet cells, and disrupt intestinal integrity, which might be the cause of diarrhea and other gastrointestinal symptoms associated with COVID-19 (Krüger et al., 2021).
More importantly, these organoids can serve as a potential platform for organ-specific drug testing and drug screening. For example, the researchers found that Remdesivir therapy inhibits SARS-CoV-2 viral replication in the intestinal organoids (Krüger et al., 2021). Therefore, clinical treatment with this drug may prevent intestinal damage caused by SARS-CoV-2 and relieve intestinal symptoms.
Brain Organoids
Approximately 36.4% of the COVID-19 patients develop a variety of neurological complications, ranging from loss of smell, nausea, dizziness, and headache to encephalopathy and stroke (Mao et al., 2020). RNA of SARS-COV-2 was found in the brains of some patients (Helms et al., 2020; Moriguchi et al., 2020). The mechanisms of SARS-COV-2 disrupting the brain-blood barrier and infecting the central nervous system (CNS) draw great concerns (Li et al., 2020). Studies have shown that CNS infections may lead to the pathophysiological and clinical manifestations associated with COVID-19 (Steardo et al., 2020). Therefore, it is necessary to establish a suitable in vitro model to study nerve infection by SARS-CoV-2.
IPSC-derived brain organoids are valuable tools for investigating the biological properties of SARS-CoV-2 in the CNS (Bullen et al., 2020; Jacob et al., 2020). Studies using iPSC-derive brain organoids find that choroid plexus epithelial cells are the main target of SARS-CoV-2 infection in the CNS; meanwhile, neurons, and astrocytes are sparsely infected (Jacob et al., 2020; Pellegrini et al., 2020). This finding is consistent with the discovery that the choroid plexus region is one of the hotspots of ACE2 expression in the CNS under inflammatory status, and it is more susceptible to SARS-CoV-2 infection than other regions (Chen R. et al., 2020). After SARS-CoV-2 infection, increased cellular remodeling and inflammatory responses were observed in choroid plexus epithelial cells (Chen R. et al., 2020). This finding provides an evidence that SARS-CoV-2 infection of the choroid plexus leads to disruptions in blood-cerebrospinal fluid barrier (BCB) integrity. Researchers have proposed that BCB decomposition can promote entry of the virus as well as immune cells expressing cytokines into the cerebrospinal fluid and brain tissue, potentially causing nerve inflammation (Pellegrini et al., 2020).
Whether SARS-CoV-2 propagates in the CNS remains controversial. Some studies have reported successful SARS-CoV-2 replication in brain organoids (Zhang et al., 2020), while the others suggest that the viral replication and proliferation are less efficient in the brain organoids (Ramani et al., 2020). These opposite results may be due to differences in the methods of establishing brain organoid models and the multiplicities of infection (MOI) used in these studies. In the former study, the neural progenitor cell (NPC) population is also found to be a target of SARS-CoV-2 (Ramani et al., 2020). This is an important finding as NPCs are responsible for repairing brain lesions caused by degenerative diseases or malignancies (Zhu et al., 2013, 2014; Luo and Zhu, 2014; Luo et al., 2015b). The impaired NPC population might be the reason for late or incomplete recovery of neurological manifestations in COVID-19 patients. On the other hand, it should be noted that although human brain organoids represent valuable models for in vitro research on SARS-CoV-2 infection, they merely have simplified structures (e.g., vein systems and blood-brain barriers) like the developing fetal brain, and lack mature cells, particularly asteroid cells and astrocytes (Jacob et al., 2020; Ramani et al., 2020).
Pancreatic Endocrine Cells
Single-cell RNA-seq analysis of primary human islets has indicated that both alpha cells and beta cells are positive for ACE2 and TMPRSS2 (Yang et al., 2020). Further validation experiments in humanized mouse model established by human iPSC-derived pancreatic endocrine cells confirm that both alpha cells and beta cells are susceptible to SARS-CoV-2 (Yang et al., 2020). Infected pancreatic endocrine cells display higher expression of pathways associated with apoptosis and viral infection, and lower expression of pathways associated with the normal functions of alpha cells and beta cells, thus leading to increased cell death and loss of cell identities (Yang et al., 2020). The infected cells are also expressing higher levels of chemokines, including CCL2, CXCL5, and CXCL6, and other degenerative factors and cytokines, which is similar with cells found in autopsies from COVID-19 patients (Blanco-Melo et al., 2020).
Liver Organoids
More than 50% of COVID-19 patients have symptoms of viral hepatitis (Ong et al., 2020). Particularly, the proportion of patients with liver damage in patients with severe symptoms is much higher than that in patients with mild symptoms (Huang C. et al., 2020). However, due to the lack of suitable research models, it was unclear whether the liver damages were caused by a direct viral infection or by systemic dysfunctions, such as cytokine storms.
Relevant studies have deployed human organoids as tools to study the correlations of SARS-CoV-2 infection and liver damage at both cellular and molecular levels (Huch et al., 2015; Dutta and Clevers, 2017). Some studies have established human liver organoid models which are capable of preserving the ACE2 + /TMPRSS2 + cholangiocyte population in long-term 3-dimensional (3D) cultures (Yang et al., 2020; Zhao et al., 2020). Further, the studies have confirmed that the cholangiocytes in the human liver organoids are permissive to SARS-CoV-2 infection and supporting strong viral propagation (Yang et al., 2020; Zhao et al., 2020). Moreover, SARS-CoV-2 infection induces cell death in the host cholangiocytes. Thus, these studies supports that liver damage in COVID-19 patients might be caused by gallbladder decomposition and subsequent accumulation of bile acid due to viral infection (Yang et al., 2020; Zhao et al., 2020).
Discussion and Future Perspectives
Most COVID-19 patients have mild respiratory symptoms; however, up to 20% of the patients develop severe pneumonia, leading to multi-organ failures and even death (Zhu N. et al., 2020). The development of iPSC technologies and the resulting differentiated cell models have dramatically accelerated studies of the pathogenesis of SARS-CoV-2 in various organs. Current studies have employed iPSC-derived cells and organoids, including iAT2 cells, cardiomyocytes, pancreatic endocrine cells, lung organoids, brain organoids, intestinal organoids, liver organoids, to investigate the underlying mechanisms of SARS-CoV-2 infection (Ardestani and Maedler, 2020; Bojkova et al., 2020; Huang J. et al., 2020; Jacob et al., 2020; Pei et al., 2020; Mithal et al., 2021). As COVID-19 could also cause kidney malfunctions (Chen N. et al., 2020), kidney organoids derived from iPSCs may be a potential research model as well (Phipson et al., 2019).
These iPSC-derived models are suited for leveraging the powers of the latest genetic tools, such as single-cell RNA-seq and CRISPR techniques, for COVID-19 research (Zhou et al., 2020). Single-cell RNA-seq techniques have been developed to investigate the viral tropisms and host transcriptional responses to viruses or external stimuli in complex organs and tissues (Zhu et al., 2018; Luo et al., 2019; Zhu D. et al., 2020). In the above studies, single-cell RNA-seq has been employed to screen for cell types that are positive of the SARS-CoV-2 receptor ACE2 and effector protease TMPRSS2, and to illustrate the transcriptional alternations after viral infection (Huang J. et al., 2020; Yang et al., 2020; Zang et al., 2020). Furthermore, the CRISPR system can be utilized to create genetically modified iPSC models for mechanism study of the candidate genetic factors (Luo et al., 2015c, 2016; Gkogkou et al., 2020; Kim et al., 2020; Zang et al., 2020; Yu, 2021). For example, with a CRISPR-engineered iPSC model, researchers have demonstrated that the single-nucleotide polymorphism rs4702, which is a common genetic variant located in the 3′ UTR of the protease FURIN, influences the SARS-CoV-2 permissiveness of alveolar and neuronal cells (Dobrindt et al., 2021).
On the other hand, there are a few limitations of iPSC-derived cellular and organoid platforms, such as inadequate complexity to reflect real tissue microenvironment and cell-cell interactions, and lack of real-time monitoring methods for 3D cultures. For examples, although the above studies have described the susceptibility of various cell types to SARS-CoV-2 infection with these iPSC derivatives, it is unclear whether these cell types are the primary targets for viral infection in COVID-19 without a more thorough analysis of samples from primary patients (Lamouroux et al., 2020). Besides, these iPSC-derived models are simplified ones compared to the fully functional and reacting human organs. In the future, these platforms should be exploited to produce more complex organoid models, including the immune system components that are missing from the current analysis (Yang et al., 2020).
Conclusion
In conclusion, human iPSC-derived cells and organoids can be used as ideal models for studying the mechanisms of viral infection and drug screening. Particularly, with the organoid models, the viral tropism and host responses of different cell types could be observed in a single system. These iPSC models help us better understand the pathogenesis of SARS-CoV-2 in different organs and systems, and provide powerful drug test and discovery platforms.
Author Contributions
YL and DZ conceived the study. YL, MZ, and DZ prepared the figure. YL, MZ, YpC, YoC, and DZ wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding. This work was supported by grants from the Guangzhou City Science, Technology and Innovation Commission (Nos. 201804010340 and 202002030077), the National Natural Science Foundation of China (Nos. 82002774 and 81401206), the Guangdong Province Natural Science Foundation (No. 2020A1515010065), the Guangdong Province Outstanding Youth Medical Talent Program (No. 110217110), the Liwan District Science and Technology Planning Project (No. 201804013), and the Third Affiliated Hospital of Guangzhou Medical University Elite Talent Fund Project (No. 110217103).
References
- Aggarwal G., Cheruiyot I., Aggarwal S., Wong J., Lippi G., Lavie C. J. (2020a). Association of Cardiovascular Disease With Coronavirus Disease 2019 (COVID-19) Severity: a Meta-Analysis. Curr. Probl. Cardiol. 45:100617. 10.1016/j.cpcardiol.2020.100617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal G., Henry B. M., Aggarwal S., Bangalore S. (2020b). Cardiovascular Safety of Potential Drugs for the Treatment of Coronavirus Disease 2019. Am. J. Cardiol. 128 147–150. 10.1016/j.amjcard.2020.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardestani A., Maedler K. (2020). Commentary: a Human Pluripotent Stem Cell-Based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Front. Endocrinol. 11:585922. 10.3389/fendo.2020.585922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M. (2020). Cardiovascular disease and COVID-19. Diabetes Metab. Syndr. 14 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J. H., Tomashek K. M., Dodd L. E., Mehta A. K., Zingman B. S., Kalil A. C. (2020). Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 383 1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B. E., Liu W. C., Uhl S., Hoagland D., Møller R., et al. (2020). Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181 1036–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Wagner J. U. G., Shumliakivska M., Aslan G. S., Saleem U., Hansen A. (2020). SARS-CoV-2 infects and induces cytotoxic effects in human cardiomyocytes. Cardiovasc. Res. 116 2207–2215. 10.1093/cvr/cvaa267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B. T., Maioli H., Johnston R., Chaudhry I., Fink S. L., Xu H., et al. (2020). Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet 396 320–332. 10.1016/s0140-6736(20)31305-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen C. K., Hogberg H. T., Bahadirli-Talbott A., Bishai W. R., Hartung T., Keuthan C. (2020). Infectability of human BrainSphere neurons suggests neurotropism of SARS-CoV-2. Altex 37 665–671. [DOI] [PubMed] [Google Scholar]
- Burridge P. W., Li Y. F., Matsa E., Wu H., Ong S. G., Sharma A. (2016). Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat. Med. 22 547–556. 10.1038/nm.4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wu H., Yu Y., Tang N. (2020). Pulmonary alveolar regeneration in adult COVID-19 patients. Cell Res. 30 708–710. 10.1038/s41422-020-0369-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 395 507–513. 10.1016/s0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Wang K., Yu J., Howard D., French L., Chen Z., et al. (2020). The Spatial and Cell-Type Distribution of SARS-CoV-2 Receptor ACE2 in the Human and Mouse Brains. Front. Neurol. 11:573095. 10.3389/fneur.2020.573095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. W., Shin J. S., Park S. J., Jung E., Park Y. G., Lee J., et al. (2020). Antiviral activity and safety of remdesivir against SARS-CoV-2 infection in human pluripotent stem cell-derived cardiomyocytes. Antiviral Res. 184:104955. 10.1016/j.antiviral.2020.104955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt K., Hoagland D. A., Seah C., Kassim B., O’Shea C. P., Murphy A., et al. (2021). Common Genetic Variation in Humans Impacts In Vitro Susceptibility to SARS-CoV-2 Infection. Stem Cell Rep. 16 505–518. 10.1016/j.stemcr.2021.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D., Clevers H. (2017). Organoid culture systems to study host-pathogen interactions. Curr. Opin. Immunol. 48 15–22. 10.1016/j.coi.2017.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman R. T., Roth J. S., Brimacombe K. R., Simeonov A., Shen M., Patnaik S., et al. (2020). Remdesivir: a Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 6 672–683. 10.1021/acscentsci.0c00489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettayebi K., Crawford S. E., Murakami K., Broughman J. R., Karandikar U., Tenge V. R. (2016). Replication of human noroviruses in stem cell-derived human enteroids. Science 353 1387–1393. 10.1126/science.aaf5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gintant G., Sager P. T., Stockbridge N. (2016). Evolution of strategies to improve preclinical cardiac safety testing. Nat. Rev. Drug Discov. 15 457–471. 10.1038/nrd.2015.34 [DOI] [PubMed] [Google Scholar]
- Gkogkou E., Barnasas G., Vougas K., Trougakos I. P. (2020). Expression profiling meta-analysis of ACE2 and TMPRSS2, the putative anti-inflammatory receptor and priming protease of SARS-CoV-2 in human cells, and identification of putative modulators. Redox Biol. 36:101615. 10.1016/j.redox.2020.101615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goineau S., Castagné V. (2017). Proarrhythmic risk assessment using conventional and new in vitro assays. Regul. Toxicol. Pharmacol. 88 1–11. 10.1016/j.yrtph.2017.05.012 [DOI] [PubMed] [Google Scholar]
- Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C., et al. (2020). Neurologic Features in Severe SARS-CoV-2 Infection. N. Engl. J. Med. 382 2268–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. (2020). Drosten and S. Pöhlmann, SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. J., Okuda K., Edwards C. E., Martinez D. R., Asakura T., Dinnon K. H., III (2020). SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 182 429–446.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Hume A. J., Abo K. M., Werder R. B., Villacorta-Martin C., Alysandratos K. D., et al. (2020). SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response. Cell Stem Cell 27 962–973.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Gehart H., van Boxtel R., Hamer K., Blokzijl F., Verstegen M. M. (2015). Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160 299–312. 10.1016/j.cell.2014.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Pather S. R., Huang W. K., Zhang F., Wong S. Z. H., Zhou H., et al. (2020). Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 27 937–950.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Koo B. K., Knoblich J. A. (2020). Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21 571–584. 10.1038/s41580-020-0259-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J., Groß R., Conzelmann C., Müller J. A., Koepke L., Sparrer K. M. J., et al. (2021). Drug Inhibition of SARS-CoV-2 Replication in Human Pluripotent Stem Cell-Derived Intestinal Organoids. Cell. Mol. Gastroenterol. Hepatol. 11 935–948. 10.1016/j.jcmgh.2020.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M. M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T. I., et al. (2020). SARS-CoV-2 productively infects human gut enterocytes. Science 369 50–54. 10.1126/science.abc1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouroux A., Attie-Bitach T., Martinovic J., Leruez-Ville M., Ville Y. (2020). Evidence for and against vertical transmission for severe acute respiratory syndrome coronavirus 2. Am. J. Obstet. Gynecol. 223 91.e1–91.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Lee A. S., Liang P., Sanchez-Freire V., Nguyen P. K., Wang L., et al. (2013). Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 12 101–113. 10.1016/j.stem.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. C., Bai W. Z., Hashikawa T. (2020). The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J. Med. Virol. 92 552–555. 10.1002/jmv.25728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q., Alvarez X., Wang H., Du Y., Zhu H., et al. (2011). Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J. Virol. 85 4025–4030. 10.1128/jvi.02292-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long B., Brady W. J., Koyfman A., Gottlieb M. (2020). Cardiovascular complications in COVID-19. Am. J. Emerg. Med. 38 1504–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395 565–574. 10.1016/s0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Zhang X., Xu H. (2020). Don’t Overlook Digestive Symptoms in Patients With 2019 Novel Coronavirus Disease (COVID-19). Clin. Gastroenterol. Hepatol. 18 1636–1637. 10.1016/j.cgh.2020.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Huang J., Tang Y., Luo X., Ge L., Sheng X., et al. (2019). Regional methylome profiling reveals dynamic epigenetic heterogeneity and convergent hypomethylation of stem cell quiescence-associated genes in breast cancer following neoadjuvant chemotherapy. Cell Biosci. 9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Li J., Zhu D., Fan Y., Li S., Sun X. (2014). High-resolution chromosomal microarray analysis of early-stage human embryonic stem cells reveals an association between X chromosome instability and skewed X inactivation. Cell Biosci 4:74. 10.1186/2045-3701-4-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Xu X., An X., Sun X., Wang S., Zhu D. (2016). Targeted Inhibition of the miR-199a/214 Cluster by CRISPR Interference Augments the Tumor Tropism of Human Induced Pluripotent Stem Cell-Derived Neural Stem Cells under Hypoxic Condition. Stem Cells Int. 2016:3598542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Zhu D. (2014). Combinatorial control of transgene expression by hypoxia-responsive promoter and microrna regulation for neural stem cell-based cancer therapy. Biomed. Res. Int. 2014:751397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Zhu D., Du R., Gong Y., Xie C., Xu X., et al. (2015a). Uniparental disomy of the entire X chromosome in Turner syndrome patient-specific induced pluripotent stem cells. Cell Discov. 1:15022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Zhu D., Lam D. H., Huang J., Tang Y., Luo X., et al. (2015b). A Double-Switch Cell Fusion-Inducible Transgene Expression System for Neural Stem Cell-Based Antiglioma Gene Therapy. Stem Cells Int. 2015:649080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Zhu D., Xu X., Ge L., Sun X., Chen G., et al. (2018). Generation of an induced pluripotent stem cell line from an adult male with 45,X/46,XY mosaicism. Stem Cell Res 27 42–45. 10.1016/j.scr.2018.01.003 [DOI] [PubMed] [Google Scholar]
- Luo Y., Zhu D., Zhang Z., Chen Y., Sun X. (2015c). Integrative Analysis of CRISPR/Cas9 Target Sites in the Human HBB Gene. Biomed. Res. Int. 2015:514709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madjid M., Safavi-Naeini P., Solomon S. D., Vardeny O. (2020). Potential Effects of Coronaviruses on the Cardiovascular System: a Review. JAMA Cardiol. 5 831–840. 10.1001/jamacardio.2020.1286 [DOI] [PubMed] [Google Scholar]
- Ma-Lauer Y., Carbajo-Lozoya J., Hein M. Y., Müller M. A., Deng W., Lei J., et al. (2016). p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl. Acad. Sci. U. S. A. 113 E5192–E5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., et al. (2020). Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 77 683–690. 10.1001/jamaneurol.2020.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. (2020). The lasting misery of coronavirus long-haulers. Nature 585 339–341. 10.1038/d41586-020-02598-6 [DOI] [PubMed] [Google Scholar]
- Milewska A., Kula-Pacurar A., Wadas J., Suder A., Szczepanski A., Dabrowska A., et al. (2020). Replication of Severe Acute Respiratory Syndrome Coronavirus 2 in Human Respiratory Epithelium. J. Virol. 94 e00957–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitcheson J. S., Hancox J. C., Levi A. J. (1998). Cultured adult cardiac myocytes: future applications, culture methods, morphological and electrophysiological properties. Cardiovasc. Res. 39 280–300. 10.1016/s0008-6363(98)00128-x [DOI] [PubMed] [Google Scholar]
- Mithal A., Hume A. J., Lindstrom-Vautrin J., Villacorta-Martin C., Olejnik J., Bullitt E., et al. (2021). Human Pluripotent Stem Cell-Derived Intestinal Organoids Model SARS-CoV-2 Infection Revealing a Common Epithelial Inflammatory Response. Stem Cell Rep. 16 940–953. 10.1016/j.stemcr.2021.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno L., Pearson A. D. (2013). How can attrition rates be reduced in cancer drug discovery? Expert. Opin. Drug Discov. 8 363–368. 10.1517/17460441.2013.768984 [DOI] [PubMed] [Google Scholar]
- Moriguchi T., Harii N., Goto J., Harada D., Sugawara H., Takamino J., et al. (2020). A first case of meningitis/encephalitis associated with SARS-Coronavirus-2. Int. J. Infect. Dis. 94 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay A., Konda B., Garcia G., Jr., Yao C., Beil S., Villalba J. M., et al. (2021). SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 35:109055. 10.1016/j.celrep.2021.109055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y. Z., Zheng Y. W., Miyakawa K., Murata S., Zhang R. R., Sekine K. (2018). Recapitulation of hepatitis B virus-host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine 35 114–123. 10.1016/j.ebiom.2018.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong J., Young B. E., Ong S. (2020). COVID-19 in gastroenterology: a clinical perspective. Gut 69 1144–1145. 10.1136/gutjnl-2020-321051 [DOI] [PubMed] [Google Scholar]
- Pei R., Feng J., Zhang Y., Sun H., Li L., Yang X., et al. (2020). Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS-CoV-2 infection. Protein Cell [Epub Online ahead of print]. 10.1007/s13238-020-00811-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Albecka A., Mallery D. L., Kellner M. J., Paul D., Carter A. P., et al. (2020). SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids. Cell Stem Cell 27 951–961.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B., Er P. X., Combes A. N., Forbes T. A., Howden S. E., Zappia L., et al. (2019). Evaluation of variability in human kidney organoids. Nat. Methods 16 79–87. 10.1038/s41592-018-0253-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles V. G., Lütgehetmann M., Lindenmeyer M. T., Sperhake J. P., Wong M. N., Allweiss L., et al. (2020). Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 383 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Travanty E. A., Oko L., Edeen K., Berglund A., Wang J., et al. (2013). Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am. J. Respir. Cell Mol. Biol. 48 742–748. 10.1165/rcmb.2012-0339oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A., Müller L., Ostermann P. N., Gabriel E., Abida-Islam P., Müller-Schiffmann A. (2020). SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 39:e106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala L., Bellin M., Mummery C. L. (2017). Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: has the time come? Br. J. Pharmacol. 174 3749–3765. 10.1111/bph.13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Stange D. E., Ferrante M., Vries R. G., Van Es J. H., Van den Brink S., et al. (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141 1762–1772. 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- Sharma A., Burridge P. W., McKeithan W. L., Serrano R., Shukla P., Sayed N., et al. (2017). High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci. Transl. Med. 9:eaaf2584. 10.1126/scitranslmed.aaf2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Marceau C., Hamaguchi R., Burridge P. W., Rajarajan K., Churko J. M., et al. (2014). Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circ. Res. 115 556–566. 10.1161/circresaha.115.303810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., et al. (2020). A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584 120–124. [DOI] [PubMed] [Google Scholar]
- Shi S., Qin M., Shen B., Cai Y., Liu T., Yang F., et al. (2020). Association of Cardiac Injury With Mortality in Hospitalized Patients With COVID-19 in Wuhan, China. JAMA Cardiol. 5 802–810. 10.1001/jamacardio.2020.0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau C. R., Ott M. (2020). Modeling Multi-organ Infection by SARS-CoV-2 Using Stem Cell Technology. Cell Stem Cell 27 859–868. 10.1016/j.stem.2020.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steardo L., Steardo L., Jr., Zorec R., Verkhratsky A. (2020). Neuroinfection may contribute to pathophysiology and clinical manifestations of COVID-19. Acta Physiol. 229:e13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K. (2020). In Vitro and Animal Models for SARS-CoV-2 research. Trends Pharmacol. Sci. 41 513–517. 10.1016/j.tips.2020.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wang H., Fan J., Zhang Y., Wang H., Zhao Q. (2020). Pancreatic Injury Patterns in Patients With Coronavirus Disease 19 Pneumonia. Gastroenterology 159 367–370. 10.1053/j.gastro.2020.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., et al. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X. S., Wang X., Niu Y. R., Ye L. L., Peng W. B., Wang Z. H., et al. (2020). Diarrhea Is Associated With Prolonged Symptoms and Viral Carriage in Corona Virus Disease 2019. Clin. Gastroenterol. Hepatol. 18 1753–1759.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. (2020). Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 158 1831–1833.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B. E., Gupta V., Wang P., Duan X. (2020). A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 27 125–136.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Jia R., Tang Y., Liu J., Wei B. (2020). SARS-CoV-2 infection and stem cells: interaction and intervention. Stem Cell Res. 46:101859. 10.1016/j.scr.2020.101859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. (2021). Organoids: a New Model for SARS-CoV-2 Translational Research. Int. J. Stem Cells 14 138–149. 10.15283/ijsc20169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318 1917–1920. [DOI] [PubMed] [Google Scholar]
- Zang R., Castro M.F. Gomez, McCune B. T., Zeng Q., Rothlauf P. W., Sonnek N. M., et al. (2020). TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 5:eabc3582. 10.1126/sciimmunol.abc3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. Z., Chu H., Han S., Shuai H., Deng J., Hu Y. F., et al. (2020). SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 30 928–931. 10.1038/s41422-020-0390-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ni C., Gao R., Wang Y., Yang L., Wei J., et al. (2020). Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell 11 771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Liu L. P., Fang M., Li Y. M., Zheng Y. W. (2020). A potential ex vivo infection model of human induced pluripotent stem cell-3D organoids beyond coronavirus disease 2019. Histol. Histopathol. 35 1077–1082. [DOI] [PubMed] [Google Scholar]
- Zhu D., Chen C., Purwanti Y. I., Du S., Lam D. H., Wu C., et al. (2014). Induced pluripotent stem cell-derived neural stem cells transduced with baculovirus encoding CD40 ligand for immunogene therapy in mouse models of breast cancer. Hum. Gene Ther. 25 747–758. 10.1089/hum.2013.160 [DOI] [PubMed] [Google Scholar]
- Zhu D., Lam D. H., Purwanti Y. I., Goh S. L., Wu C., Zeng J., et al. (2013). Systemic delivery of fusogenic membrane glycoprotein-expressing neural stem cells to selectively kill tumor cells. Mol. Ther. 21 1621–1630. 10.1038/mt.2013.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Rostami M. R., Zuo W. L., Leopold P. L., Crystal R. G. (2020). Single-Cell Transcriptome Analysis of Mouse Liver Cell-Specific Tropism and Transcriptional Dysregulation Following Intravenous Administration of AAVrh.10 Vectors. Hum. Gene Ther. 31 590–604. 10.1089/hum.2019.366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Zhao Z., Cui G., Chang S., Hu L., See Y. X., et al. (2018). Single-Cell Transcriptome Analysis Reveals Estrogen Signaling Coordinately Augments One-Carbon, Polyamine, and Purine Synthesis in Breast Cancer. Cell Rep. 25 2285–2298.e4. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. (2020). A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 382 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]