Abstract
Future biodiversity loss threatens the integrity of complex ecological associations, including among hosts and parasites. Almost half of primate species are threatened with extinction, and the loss of threatened hosts could negatively impact parasite associations and ecosystem functions. If endangered hosts are highly connected in host–parasite networks, then future host extinctions will also drive parasite extinctions, destabilizing ecological networks. If threatened hosts are not highly connected, however, then network structure should not be greatly affected by the loss of threatened hosts. Networks with high connectance, modularity, nestedness and robustness are more resilient to perturbations such as the loss of interactions than sparse, nonmodular and non-nested networks. We analysed the interaction network involving 213 primates and 763 parasites and removed threatened primates (114 species) to simulate the effects of extinction. Our analyses revealed that connections to 23% of primate parasites (176 species) may be lost if threatened primates go extinct. In addition, measures of network structure were affected, but in varying ways because threatened hosts have fewer parasite interactions than non-threatened hosts. These results reveal that host extinctions will perturb the host–parasite network and potentially lead to secondary extinctions of parasites. The ecological consequences of these extinctions remain unclear.
This article is part of the theme issue ‘Infectious disease macroecology: parasite diversity and dynamics across the globe’.
Keywords: modularity, social network analysis, Anthropocene, coevolution, host–parasite networks, conservation
1. Introduction
The unprecedented rate of species extinctions is causing a biodiversity crisis [1]. When two organisms have coevolved to such an extent that one cannot live without the other (i.e. they have specialized on one another), extinction of one organism will frequently lead to the secondary extinction of the other [2]. For example, many butterfly and moth species are highly specific in the host plants they use at the larval stage, and the loss of those host plants causes local extirpations of the herbivore [3]. Generalists, however, can be maintained by a diverse pool of potential resources, and are thus less susceptible to extinction when one or more of their resources become scarce. In the case of parasites, a generalist that infects multiple host species is less likely to go extinct when one of its hosts is lost, as compared to a specialist parasite that infects only one or a few hosts [4–6]. The current loss of host species is also driving an unseen loss of parasites [7,8]. In this study, we use the ecological definition of parasites, which are organisms that live on or in another species (the host), obtaining a benefit from the host at some cost to the host [9]. These include macroparasites, such as helminths, as well as microparasites, including viruses, bacteria, protozoa and fungi. Parasites are potentially important to ecosystem functions, given the important role parasites play in regulating host populations [10].
Interactions among organisms can be viewed as networks in which the organisms are represented as nodes and interactions among them as edges [11,12]. The patterns of interactions between hosts and parasites can be represented as a two-mode, or bipartite, network, with edges connecting parasites to the hosts they infect [13,14]. Network structure is an emergent property of the interactions among organisms. Modularity is one structural property that measures the extent to which interactions are concentrated within subgroups compared to between groups [15]. Connectance is the fraction of realized interactions among organisms in the network [16]. In the sense of species interactions, nestedness is the pattern in which specialist species (those with few connections) interact with a proper subset of the species interacting with generalists [17]. In other words, host species with few interactions are connected to parasites that are also connected to hosts with more interactions. Each of these network attributes affects the tolerance of networks to the loss of species.
Variation in network structure influences the ability of communities to adapt to change [3]. Networks with high modularity restrict the effects of species losses to within subgroups rather than propagate the effects of those losses throughout the network, especially in trophic webs [12,18]. Networks with higher connectance experience fewer secondary extinctions when nodes are removed compared to networks in which interactions are sparse (i.e. low connectance [12,16]). Further, removing the most highly connected nodes in the network increases secondary extinctions of parasites compared to random removal [16]. Networks that are highly nested, especially mutualistic networks like plant–pollinator interactions, are tolerant to species loss because a core of generalist species maintain links to other species, while the loss of specialists has little effect on secondary extinctions [12,17]. These factors and more are important to understand how ecological networks may be affected in the future by host extinctions.
In host–parasite networks in which a subset of hosts is threatened with extinction, networks with high modularity, connectance and/or nestedness are predicted to exhibit little change in network structure and few secondary extinctions following the loss of hosts compared to networks with low modularity, connectance or nestedness [12,17]. If threatened hosts are highly connected in the network, however, then the loss of hosts will negatively impact the structure of the ecological networks. These alternative predictions have important consequences for understanding the future ecological interactions in the Anthropocene.
In addition to secondary extinctions of parasites, host extinctions can influence the structure of the overall host–parasite network [11,19]. Simulation approaches can be used to investigate the effects of species loss on network structure [11,16]. In this study, we first calculated the network structural statistics before and after removing threatened hosts from the network. The removal of hosts in the simulations can be adjusted according to the hypotheses to be tested to create realistic null models. For example, we simulated a trait that is random with respect to observed threat status but non-random with respect to phylogeny, such that phylogenetic effects of extinction risk can be considered [20]. To contrast the effects of threatened host loss with a plausible null model, we randomly removed from the full network the same proportion of hosts and calculated the change in network statistics. The connectivity, or centrality, of the host species is also important to the effects of host removal. Thus, we also simulated the removal of hosts in the network based on their connectivity, which is predicted to result in more secondary extinctions than random removal because highly connected nodes are responsible for the majority of interactions [11].
We investigated the structure of ecological networks in primates and their parasites. Primates, including humans, are known to be infected with over 1000 parasites [21,22]. Some parasites of primates are generalists, able to infect multiple host species including other animals. Other parasites are more specialized and only known to infect one or a few primate hosts. Almost 50% of primates are currently threatened with extinction due to human activities, including habitat loss, hunting and trafficking for the pet trade [23]. Threatened primates have fewer parasites than non-threatened primates [6]. This might reflect that the parasites of threatened hosts are specialists, and is consistent with coextinction theory because these threatened hosts may have already lost some of their parasite interactions [24]. These anthropogenic factors are, therefore, threatening not only diverse host species, including primates, but also the diverse parasites they host.
Given the high threat level of primates [23], and the unique parasites they are known to host [22], we investigated how host extinctions could affect the coextinction of their parasites and the structure of ecological networks. Our aim is to understand how species loss may affect the integrity of ecological networks. Using data on 213 primates and 763 parasites, representing 2319 interactions, we simulated host extinction to investigate the effects of coextinction on network characteristics. For parasites not known to infect other animals, we predict that the loss of threatened primate hosts will lead to the secondary extinctions of those parasites. Conversely, if parasites tend to be generalists that also infect non-primate hosts, relatively few secondary extinctions are expected when the primate host goes extinct. Further, if the network before the removal of threatened hosts has high modularity, connectance and/or robustness, and threatened hosts have few interactions in the network, then we predict that network structure will not be greatly affected after threatened hosts are removed, as compared to simulations in which host removal is random.
2. Methods
(a) . Data compilation
We extracted data from the Global Primate Parasite Database (parasites.nunn-lab.org), a subset of the Global Mammal Parasite Database [25], with data collection focused only on primates. The database version used in this study was downloaded on 25 February 2019. We obtained over 7900 lines of data, and culled this dataset to include only hosts identified to the species level and parasites identified at least to genus level. For most parasites, the species-level taxonomy was accurately recorded, but for cases in which species-level data were available for a genus, any records listed as ‘sp.’ were omitted. This resulted in data for 213 primate hosts and 763 parasites with 2319 primate–parasite interactions.
To determine whether primate parasites are also known to infect other animals, we compared our list of observed parasites from the primate compilation to the list of parasites infecting other animals based on available databases, including all parasite types for carnivores and ungulates in the Global Mammal Parasite Database v. 2.0 [25], viruses [26] and helminths across mammals [27], and other general compilations [28–30]. We note that there are differences in how data were collected and taxonomy among databases which made direct comparisons difficult. For example, while the primate dataset and Global Mammal Parasite Database focused exclusively on data from wild animals, and omitted any data that were from captive studies, other databases may include those studies. Further we modified parasite names in the other databases to match those in our primate data, but these discrepancies may still have affected the comparability of the datasets. This resulted in a dataset of 69 431 interactions between 9411 parasites and 9202 hosts. Parasites in our dataset could be grouped into those that are known to infect other animals in addition to primates, in contrast to those parasites only recorded in primates but not in other animals. Given the non-exhaustive nature of such databases, there may still be other hosts which the primate parasites infect which are not yet recorded. Detailed parasitological studies suggest that many of the parasites are true specialists, such as ectoparasites known to infect only certain lemur species [31], or pinworms which are frequently found to be host specialists [32].
Primate threat status was taken from the IUCN Red List (downloaded 18 February 2015). Species listed as Critically Endangered, Endangered, Near Threatened or Vulnerable were coded as Threatened, which included 108 of the 213 hosts in our dataset. Based on ongoing deforestation, habitat degradation, hunting, disease, illicit trade and other anthropogenic pressures, these species are highly likely to go extinct in the future without significant intervention. The geographical distribution of species was coded to the level of continent (or island, in the case of Madagascar), so that primate communities in different biogeographic regions could be compared separately and account for regional differences in host–parasite interactions.
(b) . Analyses
We transformed the host–parasite matrix into a bipartite network using the R package ‘bipartite’ [33] in the R statistical environment [34]. We calculated the following network structural statistics on the bipartite network in the ‘bipartite’ package using the function networklevel [17,35,36]. First, we measured connectance, which is the sum of realized edges divided by the maximum possible number of edges, or number of edges/(number of hosts * number of parasites), ranging from 0 to 1. Second, we calculated the number of compartments, which is a measure of modularity, where a compartment is a subset of the bipartite network in which nodes within the compartment do not have interactions with nodes in other compartments, based on Jordan block matrix algebra [36]. While there are many other measures of modularity, we chose this measure because it was specifically designed to assess the degree of cohesion within sub-communities given a bipartite network [36]. Third, we measure the nestedness temperature, which quantifies the degree of deviation from the isocline of perfect nestedness, in which interactions between specialist species are proper subsets of interactions among generalists. A temperature value of 0 indicates a perfectly nested set of interactions, while 100 indicates a perfectly non-nested matrix.
Lastly, we measured robustness, defined as a measure of the sensitivity of the network to node extinction. To do so, we used the functions second.extinct and robustness in the package ‘bipartite’. This approach first iteratively removes species one at a time (in this case, hosts), and calculates the number of secondary extinctions; i.e. the number of parasites with no other host interactions in the network. Robustness is the area under the curve of the number of hosts and parasites lost due to the removal of species. Values closer to one indicate that host removal has little effect on secondary extinctions, while values closer to zero indicate fragile networks in which the loss of hosts would quickly increase the loss of parasites. Species removal was based on random removal of hosts, and based on the degree distribution, with the sequence of removal iterated from the best- to least-connected hosts. The latter case is the most extreme scenario of extinction, in which the host with the most parasite connections is removed in the first iteration, then the second most connected host in the second iteration, and so on for each host.
To evaluate the effect of host extinction on network structure statistics, we removed the endangered hosts (51% of host nodes), removed isolates and re-calculated the statistics. We compared the observed change in network statistics to what would be expected based on two null distributions. Given the original host–parasite network (213 hosts, 763 parasites), in the first null distribution we removed the same percentage of nodes (51%) randomly. We created a second null distribution removing the same percentage of nodes based on a binary trait simulated to evolve under an equal rates model along the phylogeny, because closely related species tend to have a more similar threat status than distantly related species. Null distributions were based on 100 randomizations.
For the statistical significance of the difference between the values for observed current network properties and the null distributions, the proportion of observed values was considered significant if it was as extreme or more than the values in the simulated data in less than 0.05 or greater than 0.95 of the simulations.
3. Results
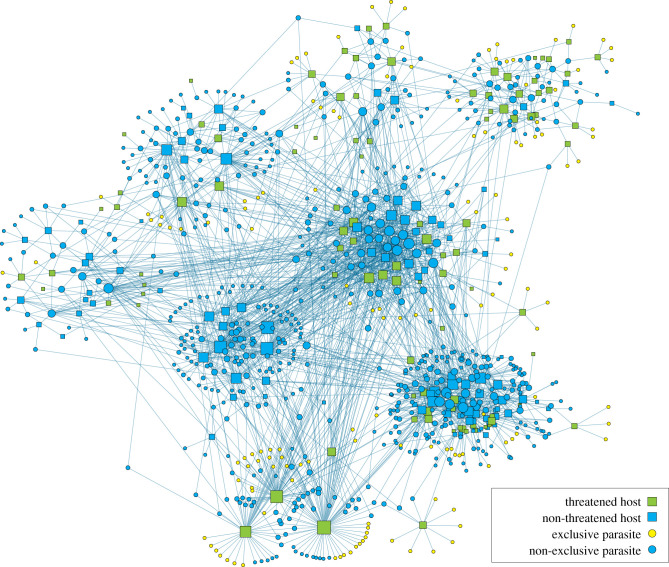
The overall host–parasite network was relatively sparse; with 2319 observed host–parasite interactions out of a possible 162 732 interactions, the connectance of 0.014 is low. Nine compartments were observed in the network. Of the 763 parasites in the primate database, 506 parasites were not recorded to infect other hosts in multiple databases across different host types. By contrast, 253 species were also recorded in non-primate hosts (figure 1).
Figure 1.
Host–parasite network for 213 primates and 763 parasites. Yellow circle nodes are parasites with threatened hosts and no other known hosts (exclusive), and green square nodes are primate hosts that are threatened with extinction. The blue squares are the non-threatened hosts and blue circles are parasites not known to infect threatened primates. Node size is scaled by degree. The plot layout used a community-weighted Fruchterman–Rheingold, which was then adjusted by hand to minimize node overlap. A digital version of this figure is provided as an electronic supplementary material file in which the vertex labels can be visualized.
(a) . Host extinction and network structure
The extinction of the 108 hosts that are currently listed as ‘threatened’ would result in lost interactions with 250 parasite species from the network. Of those 250 parasites that could be lost, 176 were not recorded in non-primate hosts, while 74 were known to infect other hosts. Thus, these 176 specialist parasites are likely to be lost due to primate host extinctions.
Host removal had varying effects on secondary extinction, depending on the simulation scenarios (figure 2). When hosts were removed based on the degree distribution, from least- to most-connected, the rate of secondary extinctions is low until almost all hosts are lost. By contrast, when hosts were removed from most- to least-connected, parasite species decayed quickly before plateauing. Random host removal had intermediate effects. Removal of hosts from most- to least-threatened overlapped the pattern of random removal until approximately half of hosts were removed, after which the rate of decay is slower and the shape of the curve more convex.
Figure 2.
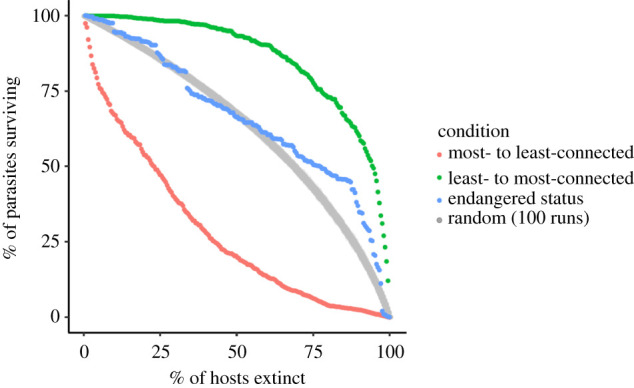
Patterns of secondary extinction under different scenarios of host loss. For the random extinction scenario, points for 100 runs are plotted together to illustrate the range of observed values, with an offset of 0.5 units on the horizontal axis to allow visualization of overlapping points.
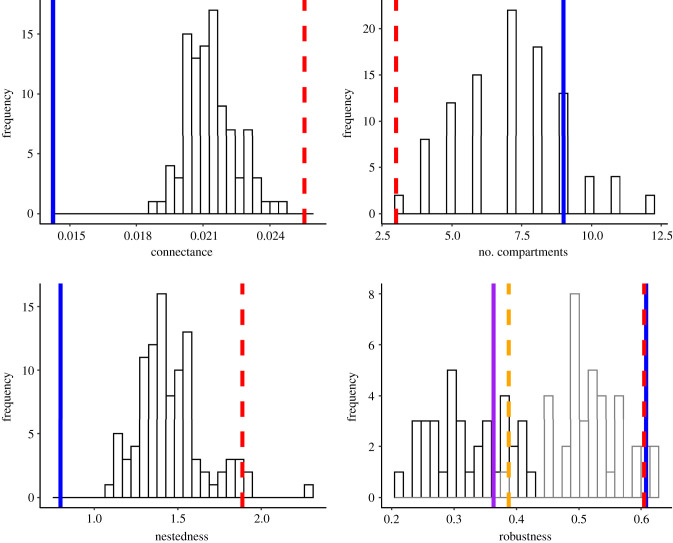
In addition to the potential for secondary extinctions, measures of network structure were perturbed by the removal of threatened hosts, though not universally. For connectance, the observed value, 0.014, reflects the sparse nature of the interaction network; a relatively small proportion of all possible edges are realized. When threatened hosts were removed, however, connectance almost doubled (0.025). Connectance was significantly lower before the removal of threatened hosts and were significantly higher after the removal of threatened hosts, compared to the null models (figure 3 and table 1). This reflects that threatened hosts contribute most to the sparsity of the network. There were nine compartments in the network before threatened host removal, while there were three after removing threatened hosts, significantly fewer than with random host removal (figure 3 and table 1), which ranged from 3 to 12 (figure 3).
Figure 3.
Global network structural statistics for the observed network and with the removal of threatened hosts, compared to a null model. The distribution represents statistics from 100 iterations of random removal of 51% of hosts from the network. For connectance, nestedness and the number of compartments, the observed statistics of the present network with 213 hosts are represented by the blue solid line, and the statistics when 51% of hosts that are threatened are removed are illustrated with red dashed line. For robustness, the two distributions represent if host removal in the robustness calculation was random (grey outline), or was based on the degree distribution (black outline). The observed robustness in the current network based on random host removal (blue solid line), and the observed robustness based on removing the best-connected hosts (purple solid line) are shown compared to the robustness expected with the loss of threatened hosts and random host removal (red dashed line), and the loss of threatened hosts and host removal based on degree distribution (orange dashed line). Results were similar based on a null distribution of removal of approximately 50% of hosts which were simulated to have a threat status with phylogenetic signal as in the observed network (see electronic supplementary material, figure SF1).
Table 1.
Results of statistical comparison between observed network statistics and the distribution of statistics expected based on the null model. For robustness, robustness1 represents the calculation in which hosts are removed at random, and robustness2 are the calculations in which hosts were removed based on the degree distribution, with hosts removed from the best- to worst-connected. Values given for hosts and parasites are the total number/those remaining after removing threatened hosts. * Value is significantly different from either of the null distributions; + indicates significantly higher than the null distributions, − indicates significantly lower; αtwo-tailed = 0.10. The probability was calculated as the proportion of null model values that were as extreme or more than the observed values.
| region | network metric | metric value pre-extinction | metric value post-extinction | range of values from null models (median) |
|---|---|---|---|---|
| global (hosts: 215/105; parasites: 763/513) | connectance | 0.014*− | 0.025*+ | 0.019–0.024 (0.021) |
| no. compartments | 9 | 3*− | 3–12 (7) | |
| nestedness | 0.80*− | 1.89*+ | 1.09–2.30 (1.42) | |
| robustness1 | 0.63*+ | 0.62*+ | 0.58–0.61 (0.60) | |
| robustness2 | 0.28 | 0.30*+ | 0.23–0.31 (0.27) | |
| Madagascar (hosts: 40/6; parasites: 122/38) | connectance | 0.052*− | 0.22 | 0.11–0.39 (0.20) |
| no. compartments | 6 | 3 | 1–6 (3) | |
| nestedness | 7.89*− | 39.06 | 21.71–64.12 (33.57) | |
| robustness1 | 0.61 | 0.60 | 0.38–0.63 (0.52) | |
| robustness2 | 0.36 | 0.39 | 0.22–0.49 (0.32) | |
| Africa (hosts: 64/20; parasites: 385/260) | connectance | 0.05*− | 0.06 | 0.05–0.08 (0.059) |
| no. compartments | 3 | 2 | 2–8 (5) | |
| nestedness | 2.43*− | 4.03 | 3.08–7.55 (4.25) | |
| robustness1 | 0.63*+ | 0.62*+ | 0.52–0.64 (0.61) | |
| robustness2 | 0.27*+ | 0.29*+ | 0.22–0.31 (0.26) | |
| Asia (hosts: 40/12; parasites: 179/121) | connectance | 0.05*− | 0.13*+ | 0.05–0.09 (0.067) |
| no. compartments | 5 | 2 | 2–8 (4) | |
| nestedness | 3.07*− | 14.89*+ | 3.56–10.53 (5.37) | |
| robustness1 | 0.59 | 0.56 | 0.51–0.60 (0.57) | |
| robustness2 | 0.23 | 0.29*+ | 0.19–0.30 (0.23) | |
| South America (hosts: 69/43; parasites: 234/188) | connectance | 0.04*− | 0.06 | 0.05–0.11 (0.07) |
| no. compartments | 2 | 2 | 1–4 (2) | |
| nestedness | 2.49*− | 3.69 | 3.37–12.31 (5.31) | |
| robustness1 | 0.61*+ | 0.61*+ | 0.52–0.61 (0.58) | |
| robustness2 | 0.28 | 0.27 | 0.20–0.36 (0.27) |
Nestedness temperature was significantly lower, reflecting a more nested interaction matrix, before the removal of threatened hosts, while after the temperature was significantly higher (less nested), compared to the null models (figure 3 and table 1). The robustness of the observed network to secondary extinctions was similar before and after the removal of threatened hosts (figure 3). The observed values of robustness before and after removing threatened hosts were significantly higher than the distribution of robustness in the null models (figure 3 and table 1). When hosts were removed in order from the best-connected to the least-connected, observed robustness was within the range of values expected based on random host removal, while it was significantly higher than the null after removing threatened hosts. When hosts were removed randomly, robustness was higher than the null before and after removing threatened hosts. Results were generally similar with the null model based on a binary trait simulated to evolve on the phylogeny according to the rate for the observed threat trait (electronic supplementary material, figure SF1).
(b) . Regional-level analyses
At the scale of different biogeographic regions, patterns were largely similar to those obtained for the global analysis (electronic supplementary material, SF2–6). In Madagascar, where the highest proportion of threatened hosts were found, only 6 out of the 40 lemur hosts in our database are not considered threatened (table 2, electronic supplementary material, SF2). Removal of those 34 threatened hosts would result in the loss of 84 parasites, 51 of which are not known to have other hosts. In other regions, 30–60% of the hosts were threatened, and 40–78 of their parasites that would be lost have no other known hosts (table 2).
Table 2.
Patterns of parasite secondary extinctions expected with the loss of threatened primate hosts.
| region (hosts) | parasites with other hosts | parasites with no other hosts | parasites lost | parasites lost with no other known hosts |
|---|---|---|---|---|
| global (213) | 253 | 511 | 250 | 176 |
| Madagascar (40) | 45 | 77 | 84 | 51 |
| Africa (64) | 165 | 220 | 124 | 78 |
| Asia (40) | 82 | 97 | 58 | 36 |
| South America (69) | 65 | 169 | 46 | 34 |
In terms of network structure across different regions, connectance and network temperature were significantly lower than the null models, and in Asia, the values after removing threatened hosts was significantly higher than the null models (table 2; electronic supplementary material, SF2–6). In Madagascar and Asia, robustness did not differ significantly from the null models, while it was significantly higher than the null models in Africa and South America (table 2; electronic supplementary material, SF2–6).
4. Discussion
Our findings revealed that if threatened primates go extinct, at least 176 parasites that have no other known hosts may also go extinct. The effects on network structure will vary, but the network exhibits properties that may buffer it from perturbations. We predicted that higher connectance values may limit the effects of host loss on changes in network structure. The connectance of the observed network was low because of the sparse nature of the interaction matrix; there are many unique host–parasite combinations, rather than a well-connected network. Removing threatened hosts would actually increase the connectance of the network by removing many of those unique host–parasite combinations, and the effect is stronger than if hosts are removed at random. We predicted that if the network were nested, it may be more tolerant of species losses than if it were non-nested. The observed network was more nested than if threatened hosts are lost, or if hosts were removed at random. Therefore, while the current network does have high nestedness, this structural feature may be eroded with host loss. Similarly, we predicted that if the network were highly modular, it may not be greatly affected by host loss. We found that the number of compartments in the network would decrease significantly with host loss. When hosts were removed iteratively at random, the robustness of the observed network to secondary extinctions was similar pre- and post-removal of threatened hosts, and higher than expected based on the random removal of half of hosts. This again reflects the unique host–parasite interactions of threatened hosts, with parasites that are not shared with non-threatened hosts. Conversely, if the best-connected hosts were removed first, the robustness of the network to secondary extinctions would be significantly lower, reflecting the strong effect of a small subset of well-connected host nodes to generating network structure.
Network properties such as high nestedness and modularity buffered networks of plants and their pollinators [37] and fish and their parasites [11] from secondary extinctions. This is because a relatively small subset of species makes up especially important ‘hubs’ in ecological networks, and the loss of weakly connected species does not greatly affect the overall structure [11,16]. Even when the best-connected nodes are removed first, networks can still exhibit tolerance to such perturbations when there are many generalist species that preserve links, despite the loss of other hosts [17,38,39]. While the current network structure of primate–parasite interactions may buffer the system, the loss of threatened hosts will degrade those properties. Our results showed that current networks are nested, but networks without threatened hosts and their parasites are significantly less nested than if hosts were removed at random. Similarly, the number of compartments significantly decreased in the global analysis. Therefore, the loss of threatened hosts may degrade the properties that confer tolerance to the primate–parasite interaction network.
Threatened hosts have fewer parasites than non-threatened hosts [6]. Threatened hosts are, therefore, not the ‘hubs’ structuring networks, and the loss of those hosts would not impact the structure of the primate–parasite network as much as if the generalist non-threatened hosts were lost. The lower parasite richness of threatened hosts may reflect that they have already experienced parasite losses. As host populations decline, the probability for parasite transmission can be impeded, causing parasites to go extinct before their hosts [24]. We may be observing the interaction network at a stage in the extinction trajectory during which these communities are already collapsing.
The patterns we observed are consistent with the hypothesis that the loss of threatened hosts will negatively impact network structure, but we also acknowledge that disentangling the effects of extinctions on networks is difficult because metrics of structure are strongly correlated to network size. Larger networks tend to be more nested [40] and more modular [41], and therefore removing approximately 50% of host nodes and their associated parasites will clearly affect these network structural properties. It is still unclear how local extinctions of hosts in sub-communities will affect global network structural properties. One way to address these questions is to examine multiple networks from the same regional communities of hosts at different localities which vary in the presence and abundance of hosts, especially the presence or absence of threatened hosts. It would then be possible to determine how those local communities of parasites are affected by host removal, with minimal changes to the overall size of the host network.
The results revealed the existence of threatened primates that host unique, potentially specialized parasites. The loss of the hosts may also result in the extinction of parasites, with an overall cascading erosion of biodiversity [17,42,43]. As shown for biodiversity loss more broadly [2,5], the loss of primate hosts could result in the loss of parasite species if those parasites do not have alternative hosts or cannot switch hosts. We found that 506 parasites are recorded to infect primates, but are not known from other hosts (66%). By contrast, 253 parasites are generalists that are known to infect a diversity of other hosts. Of the parasites known to infect only primates, 176 infect only threatened hosts, compared to 74 parasites known from threatened hosts that are also found in other hosts. Therefore, those 176 parasites are susceptible to secondary extinctions if their threatened hosts are lost. Changes to the host–parasite interactions have potentially negative consequences for wildlife health and ecosystem functions [2,19,44]. There is a growing appreciation for the need to conserve hosts and parasites, and the diversity of interactions among species more generally [4,43,45].
In conclusion, we showed how the loss of primate hosts would have varying impacts on the overall ecological network structure. In addition to affecting connectance, modularity, nestedness and robustness to secondary extinctions, there would also be important consequences for biodiversity more generally. Threatened primates host 176 parasites with no other recorded hosts, and the loss of these hosts would have unknown consequences for parasite extinction and ecosystem function. Future research should evaluate the potential for parasite species extinction, as well as possible spillover that may occur if parasites switch to new hosts. Parasites are an important component of ecosystems and thus understanding these ecological networks is crucial to conserving ecosystem functionality.
Acknowledgements
We appreciate the help of members of the Nunn lab and Duke Evolutionary Anthropology department for helpful feedback and discussions (especially C. Amoroso). Many thanks to Emily Sandberg and Marie Rogers for their assistance in data curation for the GPPD. This is Duke Lemur Center publication 1486.
Data accessibility
The data used and code used in this study are available as electronic supplementary material [46].
Authors' contributions
J.H. designed the study, conducted analyses and wrote the drafts of the manuscript. J.M. provided analytical expertize, created figures and contributed to the writing of the manuscript. C.L.N. led the data collection, provided analytical expertize and contributed to the manuscript. All authors approved the final version of the article.
Competing interests
We declare we have no competing interests.
Funding
We thank Duke University for support for J.H. during the creation of this manuscript, and a Duke University Network Analysis Center for a fellowship for him to attend the Social Networks and Health Workshop (NIH R25HD079352 ‘Focused Training in Social Networks and Health.’). Work on the Global Mammal Parasite Database was supported by the NSF (grant nos. DEB-0211908, EF-0723939/0904359 and BCS-1355902).
References
- 1.Pimm SL, Jenkins CN, Abell R, Brooks TM, Gittleman JL, Joppa LN, Raven PH, Roberts CM, Sexton JO. 2014The biodiversity of species and their rates of extinction, distribution, and protection. Science 344, 1246752. ( 10.1126/science.1246752) [DOI] [PubMed] [Google Scholar]
- 2.Colwell RK, Dunn RR, Harris NC. 2012Coextinction and persistence of dependent species in a changing world. Annu. Rev. Ecol. Evol. Syst. 43, 183-203. ( 10.1146/annurev-ecolsys-110411-160304) [DOI] [Google Scholar]
- 3.Robinson ML, Strauss SY. 2020Generalists are more specialized in low-resource habitats, increasing stability of ecological network structure. Proc. Natl Acad. Sci. USA 117, 2043-2048. ( 10.1073/pnas.1820143117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn RR, Harris NC, Colwell RK, Koh LP, Sodhi NS. 2009The sixth mass coextinction: are most endangered species parasites and mutualists? Proc. R. Soc. B 276, 3037-3045. ( 10.1098/rspb.2009.0413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koh LP, Dunn RR, Sodhi NS, Colwell RK, Proctor HC, Smith VS. 2004Species coextinctions and the biodiversity crisis. Science 305, 1632-1634. ( 10.1126/science.1101101) [DOI] [PubMed] [Google Scholar]
- 6.Altizer S, Nunn CL, Lindenfors P. 2007Do threatened hosts have fewer parasites? A comparative study in primates. J. Anim. Ecol. 76, 304-314. ( 10.1111/j.1365-2656.2007.01214.x) [DOI] [PubMed] [Google Scholar]
- 7.Hudson PJ, Dobson AP, Lafferty KD. 2006Is a healthy ecosystem one that is rich in parasites? Trends Ecol. Evol. 21, 381-385. ( 10.1016/j.tree.2006.04.007) [DOI] [PubMed] [Google Scholar]
- 8.Lafferty KD, et al. 2008Parasites in food webs: the ultimate missing links. Ecol. Lett. 11, 533-546. ( 10.1111/j.1461-0248.2008.01174.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nunn C, Altizer S, Altizer SM. 2006Infectious diseases in primates: behavior, ecology and evolution. New York: Oxford University Press. [Google Scholar]
- 10.Anderson RM, May RM. 1978Regulation and stability of host–parasite population interactions: I. Regulatory processes. J. Anim. Ecol. 47, 219-247. ( 10.2307/3933) [DOI] [Google Scholar]
- 11.Dallas T, Cornelius E. 2015Co-extinction in a host-parasite network: identifying key hosts for network stability. Sci. Rep. 5, 13185. ( 10.1038/srep13185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thébault E, Fontaine C. 2010Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329, 853-856. ( 10.1126/science.1188321%JScience) [DOI] [PubMed] [Google Scholar]
- 13.Bascompte J, Jordano P. 2014Mutualistic networks. Princeton, NJ: Princeton University Press. [Google Scholar]
- 14.Poulin R. 2010Network analysis shining light on parasite ecology and diversity. Trends Parasitol. 26, 492-498. ( 10.1016/j.pt.2010.05.008) [DOI] [PubMed] [Google Scholar]
- 15.Newman ME. 2006Modularity and community structure in networks. Proc. Natl Acad. Sci. USA 103, 8577-8582. ( 10.1073/pnas.0601602103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunne JA, Williams RJ, Martinez ND. 2002Network structure and biodiversity loss in food webs: robustness increases with connectance. Ecol. Lett. 5, 558-567. ( 10.1046/j.1461-0248.2002.00354.x) [DOI] [Google Scholar]
- 17.Memmott J, Waser NM, Price MV. 2004Tolerance of pollination networks to species extinctions. Proc. R. Soc. Lond. B 271, 2605-2611. ( 10.1098/rspb.2004.2909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krause AE, Frank KA, Mason DM, Ulanowicz RE, Taylor WW. 2003Compartments revealed in food-web structure. Nature 426, 282-285. ( 10.1038/nature02115) [DOI] [PubMed] [Google Scholar]
- 19.Strona G. 2015Past, present and future of host–parasite co-extinctions. Int. J. Parasitol. Parasites Wildl. 4, 431-441. ( 10.1016/j.ijppaw.2015.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jetz W, Freckleton RP. 2015Towards a general framework for predicting threat status of data-deficient species from phylogenetic, spatial and environmental information. Phil. Trans. R. Soc. B 370, 20140016. ( 10.1098/rstb.2014.0016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor LH, Latham SM, Mark E. 2001Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. B 356, 983-989. ( 10.1098/rstb.2001.0888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gómez JM, Nunn CL, Verdú M. 2013Centrality in primate–parasite networks reveals the potential for the transmission of emerging infectious diseases to humans. Proc. Natl Acad. Sci. USA 110, 7738-7741. ( 10.1073/pnas.1220716110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estrada A, et al. 2017Impending extinction crisis of the world's primates: why primates matter. Sci. Adv. 3, e1600946. ( 10.1126/sciadv.1600946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farrell MJ, Stephens PR, Berrang-Ford L, Gittleman JL, Davies TJ. 2015The path to host extinction can lead to loss of generalist parasites. J. Anim. Ecol. 84, 978-984. ( 10.1111/1365-2656.12342) [DOI] [PubMed] [Google Scholar]
- 25.Stephens PR, et al. 2017Global mammal parasite database version 2.0. Ecology 98, 1476. ( 10.1002/ecy.1799) [DOI] [PubMed] [Google Scholar]
- 26.Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. 2017Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646-650. ( 10.1038/nature22975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson CJ, Zipfel CM, Garnier R, Bansal S. 2019Global estimates of mammalian viral diversity accounting for host sharing. Nat. Ecol. Evol. 3, 1070-1075. ( 10.1038/s41559-019-0910-6) [DOI] [PubMed] [Google Scholar]
- 28.Wardeh M, Risley C, McIntyre MK, Setzkorn C, Baylis M. 2015Database of host–pathogen and related species interactions, and their global distribution. Sci. Data 2, 1-11. ( 10.1038/sdata.2015.49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw LP, Wang AD, Dylus D, Meier M, Pogacnik G, Dessimoz C, Balloux F. 2020The phylogenetic range of bacterial and viral pathogens of vertebrates. Mol. Ecol. 29, 3361-3379. ( 10.1111/mec.15463) [DOI] [PubMed] [Google Scholar]
- 30.Benesh DP, Lafferty KD, Kuris A. 2017A life cycle database for parasitic acanthocephalans, cestodes, and nematodes. Ecology 98, 882. ( 10.1002/ecy.1680) [DOI] [PubMed] [Google Scholar]
- 31.Klein A, Zimmermann E, Radespiel U, Schaarschmidt F, Springer A, Strube C. 2018Ectoparasite communities of small-bodied Malagasy primates: seasonal and socioecological influences on tick, mite and lice infestation of Microcebus murinus and M. ravelobensis in northwestern Madagascar. Parasites Vectors 11, 459. ( 10.1186/s13071-018-3034-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hugot J-P. 1999Primates and their pinworm parasites: the Cameron hypothesis revisited. Syst. Biol. 48, 523-546. ( 10.1080/106351599260120) [DOI] [PubMed] [Google Scholar]
- 33.Dormann CF, Gruber B, Fründ J. 2008Introducing the bipartite package: analysing ecological networks. R News 8/2, 1-20. [Google Scholar]
- 34.R Core Team. 2014R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See http://www.R-project.org/. [Google Scholar]
- 35.Burgos E, Ceva H, Perazzo RP, Devoto M, Medan D, Zimmermann M, Delbue AM. 2007Why nestedness in mutualistic networks? J. Theor. Biol. 249, 307-313. ( 10.1016/j.jtbi.2007.07.030) [DOI] [PubMed] [Google Scholar]
- 36.Dormann CF, Fründ J, Blüthgen N, Gruber B. 2009Indices, graphs and null models: analyzing bipartite ecological networks. Open Ecol. J. 2, 7-24. ( 10.2174/1874213000902010007) [DOI] [Google Scholar]
- 37.Olesen JM, Bascompte J, Dupont YL, Jordano P. 2007The modularity of pollination networks. Proc. Natl Acad. Sci. USA 104, 19 891-19 896. ( 10.1073/pnas.0706375104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verdú M, Valiente-Banuet A. 2008The nested assembly of plant facilitation networks prevents species extinctions. Am. Nat. 172, 751-760. ( 10.1086/593003) [DOI] [PubMed] [Google Scholar]
- 39.Saavedra S, Stouffer DB, Uzzi B, Bascompte J. 2011Strong contributors to network persistence are the most vulnerable to extinction. Nature 478, 233-235. ( 10.1038/nature10433) [DOI] [PubMed] [Google Scholar]
- 40.Nielsen A, Bascompte J. 2007Ecological networks, nestedness and sampling effort. J. Ecol. 95, 1134-1141. ( 10.1111/j.1365-2745.2007.01271.x) [DOI] [Google Scholar]
- 41.Sumner KM, McCabe CM, Nunn CL. 2018Network size, structure, and pathogen transmission: a simulation study comparing different community detection algorithms. Behaviour 155, 639-670. ( 10.1163/1568539X-00003508) [DOI] [Google Scholar]
- 42.Gómez A, Nichols E. 2013Neglected wild life: parasitic biodiversity as a conservation target. Int. J. Parasitol. Parasites Wildl. 2, 222-227. ( 10.1016/j.ijppaw.2013.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lafferty KD. 2012Biodiversity loss decreases parasite diversity: theory and patterns. Phil. Trans. R. Soc. B 367, 2814-2827. ( 10.1098/rstb.2012.0110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valiente-Banuet A, et al. 2015Beyond species loss: the extinction of ecological interactions in a changing world. Funct. Ecol. 29, 299-307. ( 10.1111/1365-2435.12356) [DOI] [Google Scholar]
- 45.Tylianakis JM, Laliberté E, Nielsen A, Bascompte J. 2010Conservation of species interaction networks. Biol. Conserv. 143, 2270-2279. ( 10.1016/j.biocon.2009.12.004) [DOI] [Google Scholar]
- 46.Herrera JP, Moody J, Nunn CL. 2021Predictions of primate–parasite coextinction. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Herrera JP, Moody J, Nunn CL. 2021Predictions of primate–parasite coextinction. Figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
The data used and code used in this study are available as electronic supplementary material [46].