Abstract
Wound biofilms must be identified to target disruption and bacterial eradication but are challenging to detect with standard clinical assessment. This study tested whether bacterial fluorescence imaging could detect porphyrin‐producing bacteria within a biofilm using well‐established in vivo models. Mouse wounds were inoculated on Day 0 with planktonic bacteria (n = 39, porphyrin‐producing and non‐porphyrin‐producing species, 107 colony forming units (CFU)/wound) or with polymicrobial biofilms (n = 16, 3 biofilms per mouse, each with 1:1:1 parts Staphylococcus aureus/Escherichia coli/Enterobacter cloacae, 107 CFU/biofilm) that were grown in vitro. Mouse wounds inoculated with biofilm underwent fluorescence imaging up to Day 4 or 5. Wounds were then excised and sent for microbiological analysis. Bacteria‐matrix interaction was assessed with scanning electron microscopy (SEM) and histopathology. A total of 48 hours after inoculation with planktonic bacteria or biofilm, red fluorescence was readily detected in wounds; red fluorescence intensified up to Day 4. Red fluorescence from biofilms persisted in excised wound tissue post‐wash. SEM and histopathology confirmed bacteria‐matrix interaction. This pre‐clinical study is the first to demonstrate the fluorescence detection of bacterial biofilm in vivo using a point‐of‐care wound imaging device. These findings have implications for clinicians targeting biofilm and may facilitate improved visualisation and removal of biofilms.
Keywords: biofilms, fluorescence, optical imaging, porphyrins, wounds
1. INTRODUCTION
Bacteria in wounds are a major clinical challenge that burdens patients and health care systems worldwide.1, 2 The presence of bacteria in acute and chronic wounds can delay or prevent wound healing3, 4 and is the primary cause of escalating infection‐related complications.5, 6 Confirmation of bacterial presence is achieved through wound sampling and microbiological analysis, which is costly, takes several days, and can produce erroneous results; thus, many clinicians may decide not to sample chronic wounds at all.7
The development of bacterial biofilms represents a further challenge to conventional diagnosis, treatment, and wound healing.8 Biofilms contain sessile, polymicrobial communities of microorganisms encased in an exo‐polysaccharide (EPS) layer.9, 10 This EPS matrix promotes strong adherence and protection from environmental factors.9, 10 Bacteria encased in biofilms delay wound healing and can be up to 1000 times more resistant to antimicrobials and antibiotics than planktonic (free‐floating) bacteria.11, 12 Guidelines mandate that biofilms be mechanically disrupted (eg, debridement) to allow antibacterial strategies to have an effect.3, 13 However, there is currently no point‐of‐care, real‐time method for clinicians to locate and target regions of biofilm.
Non‐contact fluorescence imaging has emerged as a method to visualise wound tissue and bacterial fluorescence at the point of care,14, 15, 16, 17, 18 without any need for contrast agents. Clinical studies using the handheld MolecuLight i:X (MolecuLight Inc., Toronto, Canada) fluorescence imaging device have demonstrated its ability to visualise red or cyan fluorescence from bacteria.19 This endogenous bacterial fluorescence is attributed to the production of porphyrins or pyoverdines.20, 21, 22, 23 Porphyrins are natural intermediates in the heme pathway in the vast majority of bacterial species; δ‐aminolevulinic acid (ALA), readily available in in vivo tissues,24, 25 is an essential precursor to the production of these porphyrins.23 In contrast, pyoverdines are cyan‐fluorescing siderophores produced uniquely by Pseudomonas spp.26 Clinically, bacterial fluorescent signatures from wounds have been shown to correlate with moderate to heavy bacterial loads.14, 15, 16, 17, 18 Multisite clinical trials assessing red bacterial porphyrin fluorescence in wounds reported a positive predicted value of >95% for detecting bacterial loads of ≥104 CFU/g.15, 17, 27
In addition to clinical studies, fluorescence detection of bacteria has been evaluated in several pre‐clinical studies.28, 29, 30 In vitro studies performed on agar supplemented with ALA identified 28 common wound pathogens (Gram positive, Gram negative, aerobes, and anaerobes) that fluoresce red when illuminated by the MolecuLight i:X device (eg, Staphylococcus, Proteus, Klebsiella, Bacteroides).28 These studies also report that non‐porphyrin‐producing bacteria (eg, Enterococcus, Streptococcus) do not emit detectable red fluorescence, nor do most yeasts.28 Additional in vivo studies on wounds inoculated with planktonic Staphylococcus aureus have demonstrated that no exogenous additives or contrast agents are required to elicit readily detectable red fluorescence, indicating that bacteria in wounds can take up sufficient ALA from host tissue to produce porphyrins.30 When supplemented with ALA, red fluorescence from bacteria in biofilms have been detected in vitro; bacterial‐EPS matrix interactions were confirmed by scanning electron microscopy (SEM) and histopathology.28 This red fluorescence signal from porphyrins does not distinguish between planktonic and biofilm‐encased bacteria, but it can provide point‐of‐care information in real time for clinicians to visualise bacteria and target their treatments and provides immediate feedback on treatment effectiveness.18, 27, 30, 31, 32
To our knowledge, in vivo assessments of detectable fluorescence from bacteria in wound biofilms have not previously been reported. The current study had three aims:
Establish that detected red fluorescence is from bacteria and not from a host immune cell response or other factors by comparing murine models inoculated with porphyrin‐producing bacteria (Staphylococcus aureus, Escherichia coli), non‐porphyrin‐producing bacteria (Enterococcus faecalis, Streptococcus agalactiae), and phosphate‐buffered saline (PBS) control.
Determine in vitro whether bacteria in biofilms can take up ALA from surrounding culture media within a relatively short timeframe (24 hours).
Determine in vivo whether bacteria within biofilms emit red fluorescence from endogenous porphyrins at a level detectable by point‐of‐care fluorescence imaging.
2. MATERIALS AND METHODS
2.1. Murine chronic wound model inoculated with monomicrobial planktonic bacteria
Bacteria were freshly cultured and resuspended at a concentration of 4 MacFarland Standard (12.0 x 108 CFU/mL), suspended in PBS. A total of 39 adult female NCr (athymic) mice (Charles River Laboratories) were surgically wounded and inoculated with 10 μL of bacterial inoculate (12.0 x 108 CFU/mL) or sterile PBS as a negative control. Two bilateral dorsal skin wounds were surgically incised over the scapular regions on the mouse posterior. The wound to the left of the spine was inoculated with bacteria, while the wound to the right of the spine served as a negative control. The wounds were covered with Tegaderm to minimise cross‐contamination. The experimental groups were divided based on their monomicrobial bacterial inoculate and included: S. aureus (ATCC: 29213, n = 14 mice), E. faecalis (ATCC 49533, n = 5), Pseudomonas aeruginosa (ATCC: 27853, n = 6), E. coli (ATCC: 25922, n = 3), S. agalactiae (ATCC: 12386, n = 3), and PBS control (n = 8). Mice were then imaged every other day under anaesthesia up to 11 days. A subset of mice was euthanised on Day 5 to evaluate bacterial load and immune infiltration in the wound. This animal study was performed at the University of Toronto Animal Facility at the Donnelly Centre for Cellular and Biomolecular Research following the Animal Use Protocol 20012241.
2.2. In vitro polymicrobial biofilm model
The biofilm media was made up of 50% bovine plasma and 50% Bolton's broth, modified from previously described in vitro polymicrobial biofilm models.33, 34, 35, 36, 37 Media were inoculated with 1:1:1 ratio of wound pathogens S. aureus (ATCC 25923), Enterobacter cloacae (ATCC 13047), and E.coli (ATCC 25922) at a 107 CFU inoculating dose of bacteria per biofilm in 7 mL of biofilm media, with a sterile, scratched pipette tip acting as a scaffold. Biofilms were incubated at 37°C with shaking at 220 rpm for 4 days. Where indicated, positive controls were exposed to 5 mM of ALA for the last 24 hours of incubation to induce red fluorescence. Post‐incubation, in vitro polymicrobial biofilms appear as growing semi‐solid masses attached to the pipette tip submerged in biofilm media. To prepare for in vivo transplantation, in vitro biofilms were lifted out of the biofilm media, washed with 1 mL 1X PBS to remove planktonic cells, and removed from the pipette tip with tweezers. These semi‐solid biofilms were then applied to the mouse wound as described below and seen in Figure 3.
FIGURE 3.
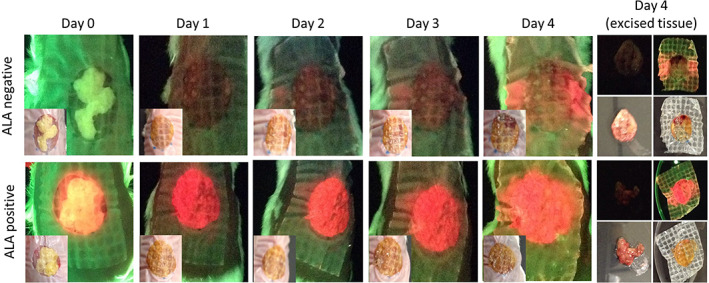
In vivo detection of fluorescence from bacteria within biofilms. Wounds were inoculated with polymicrobial biofilms grown in the absence of δ‐aminolevulinic acid (ALA) (ALA negative, n = 12). As a positive control, a subset of wounds were inoculated with biofilm grown in the presence of supraphysiological levels of ALA (ALA positive, n = 3). Standard (inset) and fluorescence images were captured daily from Days 0 to 4. Red fluorescence in the ALA negative group was evident on fluorescence images by Day 1, and fluorescent signal intensified up to Day 4 when wounds were excised, washed, and reimaged. Compared with Day 4 in vivo fluorescent signals, red fluorescent signal from excised wound bed material was decreased, much of which was because of a portion of the biofilm biomass being retained on the bandage
2.3. In vitro assessment of ALA uptake capability
Experiments were performed to determine the behaviour and timeline of ALA uptake and subsequent biofilm fluorescence. Three biofilms that had been exposed to ALA for the last 24 hours of incubation (“ALA endogenous”) were transferred to a chocolate agar (Thermo Scientific, does not contain ALA) to observe relative decreasing fluorescent signal over time. Three biofilms grown in the absence of ALA were transferred to Porphyrin Test Agar (Remel, Thermo Scientific), which contains ALA, the essential precursor for porphyrin production, to observe the potential uptake of ALA over time (“ALA exogenous”). As a negative control, three ALA negative biofilms were transferred onto chocolate agar to confirm no fluorescence induction. All were imaged with the MolecuLight i:X device (standard and fluorescent modes) at 24, 48, 72, and 96 hours.
2.4. Murine chronic wound model inoculated with biofilm
The non‐lethal surgical excision mouse model was used to study chronic infection, as previously described.33, 34, 35, 38, 39, 40, 41, 42, 43, 44 Adult, female, Swiss Webster mice were anaesthetised with an intraperitoneal injection of sodium pentobarbital. After a surgical plane of anaesthesia was reached, the backs were shaved, and the mice were administered a full‐thickness, dorsal, 1.0 x 1.0 cm excisional skin wound to the level of panniculus muscle with surgical scissors. In vitro polymicrobial biofilms grown either in the presence (“ALA positive”) or absence of ALA (“ALA negative”) were transplanted into the wound bed,33 and wounds were covered with a semipermeable polyurethane dressing (OPSITE dressing) to prevent contractile wound healing and contamination.35 Mice were given subcutaneous saline as fluid replacement and allowed to recover under warming lights. This non‐lethal, surgical excision mouse model has been used by us and others33, 34, 35, 38, 39, 40, 41, 42, 43, 44 to study chronic wound infections, and it has been shown that the infections formed in these wounds are biofilm‐associated, display increased tolerance to antimicrobials, delay wound healing, and can be easily monitored and analysed.33, 34, 35, 38, 39, 40, 41, 42, 43, 44 Mice were imaged with the MolecuLight i:X device (standard and fluorescent modes) on Days 0 to 4; on Day 4, the mice were euthanised with an intraperitoneal injection of Fatal‐Plus (Vortech Pharmaceuticals, Ltd.), and material was extracted from the wound bed. This extracted material was composed of material from the wound bed that likely contained skin tissue, bacteria and biofilm, and wound exudate and can be seen in Figure 3. Fluorescence images were taken before and after washing the excised wound bed material with 1X PBS to remove planktonic cells. This mouse model was reviewed and approved by the Institutional Animal Care and Use Committee of Texas Tech University Health Sciences Center (protocol number 07044). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
2.5. Microbial analysis
Culture‐based microbial analysis was conducted both in house and through a College of American Pathologists (CAP)‐certified clinical diagnostic laboratory. In‐house microbial analysis was conducted by homogenising excised mouse wound bed material in 1X PBS, which was then serially diluted and plated on selective and differential media to determine CFUs for each bacterial species. Mannitol Salt Agar (Fisher Scientific) was used to evaluate S. aureus, and EMB (Levine) Agar (Fisher Scientific) was used to evaluate E. coli and E. cloacae. Wound bed samples were also transported to the Clinical Laboratory Science Department at Covenant Medical Center (Lubbock, Texas) in enriched thioglycolate medium (Fisher Scientific) and evaluated on the automated system BD Phoenix (Becton Dickinson, M50). The polymicrobial nature of the ex vivo wound bed samples was confirmed using both methods of evaluation.
2.6. Histopathology
Histopathology was conducted at the Department of Pathology, Texas Tech University Health Sciences Center (Lubbock, Texas). Wound bed material (which may include host tissue and biofilm) from biofilm‐infected mice were embedded in paraffin and sectioned to a thickness of 5 μm, and slides were prepared. Slides were then stained with Gram stain, and Periodic acid‐Schiff (PAS) matrix stains. Haematoxylin and Eosin (H&E) staining was performed at the Pathology Research Project at University Health Network (Toronto, ON). Histopathological analysis of wounds inoculated with planktonic bacterial species was completed at the Pathology Research Project (Toronto, ON). Wound bed material (which may include host tissue and bacteria) was excised; embedded in paraffin; sectioned to a thickness of 5 μm; and stained for H&E, Gram, and macrophage content (F4/80+ antibody [clone Cl:A3‐1, MCA497, BioRad]). Stained slides were scanned at 40x using an Aperio slide scanner (ScanScope AT2) and imaged using Aperio ScanScope software.
2.7. Scanning electron microscopy
SEM was conducted at the Texas Tech University College of Arts and Sciences Microscopy Imaging Core Facility (Lubbock, Texas). Excised wound bed material samples were fixed in a solution of 2.5% glutaraldehyde and 2.0% paraformaldehyde in 0.05 M sodium cacodylate buffer at a pH of 7.4. After fixation, samples were rinsed in three changes of 0.05 M sodium cacodylate buffer, postfixed in 1.0% osmium tetroxide in 0.05 M sodium cacodylate buffer, rinsed in buffer, dehydrated through a graded series of alcohol, and dried at the critical point using a Tousimis Autosamdri‐814 dryer. The samples were then mounted, sputter coated with a thin layer of Au/Pd Alloy for conductivity, and imaged on the Hitachi S/N 4300 field emission scanning electron microscope. Four different imaging magnifications—400, 3.5 K, 6 K, and 10 K—were obtained. To obtain a systematic, unbiased set of SEM images from each sample, regions to be imaged were selected using the clock method, whereby images are captured at the 12, 3, 6, and 9 o'clock positions relative to a random starting location.
3. RESULTS
3.1. In vivo detection of fluorescence from bacteria
To evaluate if bacteria produce detectable fluorescence in vivo, adult female NCr mice were wounded and inoculated with various planktonic bacterial species (S. aureus, E. coli, P. aeruginosa, E. faecalis, and S. agalactiae). Mouse wounds then underwent fluorescence imaging every 2 to 3 days up to a total of 11 days (Figure 1A). The wound located on the dorsal to the left of the spine was inoculated with a 10‐μL bacterial suspension of 4 McFarland Standard of the specific bacterial species, while the wound to the right of the spine acted as an internal, non‐inoculated control. Cross‐contamination of these wounds occurred in 84% (26/31) of the mice; therefore, a complete negative control group inoculated with PBS was included in the analysis. Under violet light illumination, porphyrin‐producing bacteria such as S. aureus produced visible red fluorescence as soon as 48 hours after inoculation; red fluorescence peaked at Day 4 or 5 and dissipated as the experiment progressed (Figure 1A). In contrast, P. aeruginosa produces pyoverdines that emit a cyan fluorescence,26, 45 and as such, a strong cyan fluorescent signal was detected in wounds inoculated with P. aeruginosa as early as Day 1 and up to the endpoint on Day 8 (Figure 1B). Two to three‐days after inoculation, red fluorescence was detectable from mouse wounds inoculated with either S. aureus or E. coli when illuminated by violet light, indicating that these porphyrin‐producing bacterial species produce red bacterial fluorescence in vivo (Figure 1C). In all mice inoculated with either S. aureus or E. coli, red fluorescence signals peaked 4 to 5 days post‐wound inoculation. As expected, mice inoculated with S. agalactiae and E. faecalis, which do not produce porphyrins,23 did not emit any red fluorescence throughout the time course of the experiment. These wounds effectively healed in the absence of any detectable red fluorescence over the course of 11 days, despite the inoculation and some obvious visual signs of bacterial presence (crust and exudate) in the wounds (Figure 1C). However, it is important to note that several mice inoculated with S. agalactiae developed systemic infection and were thus euthanised before the end of the experiment (Day 11), between Days 2 and 9, with unhealed, non‐fluorescing wounds. Systemic infection was diagnosed by a veterinarian and evidenced, in part, by severe weight loss and splenomegaly. Given the distinct differences in bacterial fluorescence observed between wounds inoculated with porphyrin‐producing bacterial species and wounds inoculated with non‐porphyrin producing species S. agalactiae and E. faecalis, these findings suggest that the host immune cell response does not appreciably contribute to the red fluorescence detected. The negative control wounds inoculated with PBS did not display any detectable red fluorescence throughout the course of the experiment.
FIGURE 1.
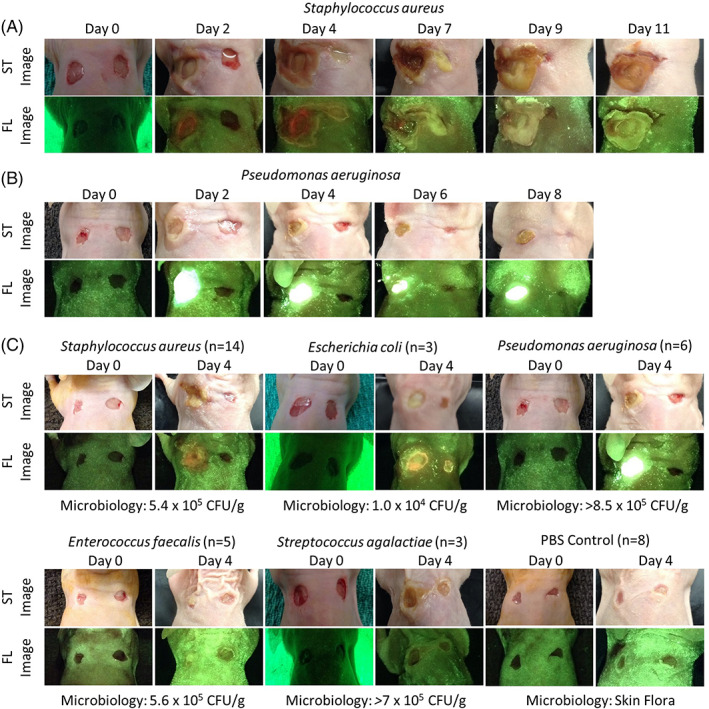
Full‐thickness skin wounds on NCr mice were inoculated with one of five bacterial species (Staphylococcus aureus, Enterococcus faecalis, Pseudomonas aeruginosa, Escherichia coli, Streptococcus agalactiae) or phosphate‐buffered saline as a control. These wounds were imaged with the fluorescence imaging device every 2 to 3 days under standard (ST) and fluorescent (FL) light for up 11 days. In each mouse, the wound on the right was intended to serve as a control, but contamination was observed in many mice. A, Representative images of a mouse inoculated with Staphylococcus aureus. Red fluorescence was observed by Day 2 and peaked at Day 4. B, Representative images of a mouse inoculated with Pseudomonas aeruginosa. Bright cyan fluorescence was observed by Day 1 and remained until the endpoint of the experiment on Day 8. C, Representative standard and fluorescence images of mice inoculated with each bacterial species or PBS negative control at Days 0 and 4 (peak fluorescence). The microbiology analysis shown for each bacterial species was completed at endpoint
Next, we sought to observe if there were gross differences in the immune response between the cohorts. A subset of animals inoculated with either S. aureus, E. faecalis, or PBS were euthanised on Day 5, at the peak of bacterial fluorescence intensity in the bacteria‐inoculated groups, to investigate bacterial load and immune infiltration. Quantitative microbiology culture data and Gram staining of these wound bed samples confirmed the presence of S. aureus and E. faecalis in their respective wounds. As a general indicator of immune response, we chose to examine macrophage recruitment via immunohistochemical staining. While macrophages are not the only immune cell population, they are heavily recruited to the wound site, and NCr mice lack T cells, another major immune cell population. Within the first 1.5 mm of tissue from the surface, there was no significant difference in macrophage recruitment to the wound in either the PBS negative control group or the wounds inoculated with E. faecalis compared with the red fluorescing wound inoculated with S. aureus.
3.2. In vitro detection of fluorescence from bacteria within biofilms
For bacteria within a biofilm to fluoresce red, the bacteria need to be able to take up ALA from the host through the biofilm matrix. To our knowledge, this capability had not been previously evaluated. In vitro experiments were performed to determine the behaviour and timeline of exogenous ALA uptake and biofilm‐encased bacterial fluorescence. Polymicrobial biofilms containing porphyrin‐producing species were grown in ALA (“ALA endogenous”) and transferred to chocolate agar (does not contain ALA) and then imaged every 24 hours (Figure 2A). Porphyrin red fluorescence was detected from these biofilms, but a decline in red signal was observed at 72 and 96 hours, most notably around the edges. In contrast, polymicrobial biofilms grown without ALA supplementation (“ALA exogenous”) produced no red fluorescence signature when they were initially transferred to Porphyrin Test Agar, (chocolate agar supplemented with ALA, Figure 2B). However, within 24 hours of ALA exposure, detectable red fluorescence was observed, and these biofilms continued to fluoresce bright red through the 96‐hour timepoint. Liquid from the biofilm spread into the surrounding media, resulting in the red fluorescing “halo” observed around the biofilm. Biofilms grown without ALA and transferred to normal chocolate agar without supplemented ALA served as negative controls (Figure 2C), and no red fluorescence was produced, supporting the notion that ALA and subsequent porphyrin production are required for the emission of detectable red fluorescence in vitro. The polymicrobial nature of the biofilms was confirmed through selective and differential plating of the homogenised biofilms (n = 4). The mean bacterial concentrations per biofilm were S. aureus (2.6 × 108 CFU), E. coli (4.0 × 104 CFU), and E. cloacae (7 × 105 CFU).
FIGURE 2.
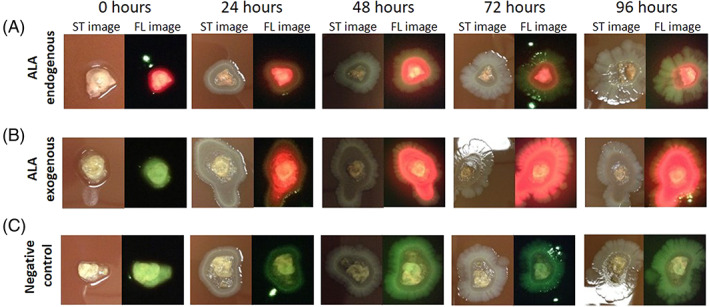
In vitro assessment of ALA uptake capability. A, Polymicrobial biofilms exposed to ALA for the last 24 hours of incubation (ALA endogenous) were transferred to chocolate agar and underwent standard (ST) and fluorescence (FL) imaging (n = 3). B, Polymicrobial biofilms lacking δ‐aminolaevulinic acid (ALA) (ALA exogenous) were transferred to Porphyrin Test Agar, which is supplemented with ALA, to observe uptake of ALA and production of porphyrin over time (n = 3). C, ALA‐negative biofilms were transferred onto chocolate agar as a negative control (n = 3). All biofilm plates were imaged every 24 hours up to 96 hours
3.3. In vivo detection of fluorescence from bacteria within biofilm
To assess whether polymicrobial bacteria encased in biofilms produce detectable red fluorescence in vivo, biofilms (1:1:1 ratio of S. aureus, E. coli and E. cloacae; 107 CFU/biofilm) were grown in vitro in the absence of ALA (“ALA negative”) and transplanted on to chronic mouse wounds using an established protocol.33, 34, 35, 38, 39, 40, 41, 42, 43, 44 These wounds then underwent daily fluorescence imaging up to 4 days. Under violet light illumination, red fluorescence was detected in all biofilm‐inoculated mouse wounds within 24 hours (Figure 3). Over the same time course, bright red fluorescence was observed in mice in the “ALA positive” control group containing polymicrobial biofilms that were grown in the presence of supraphysiological levels of ALA prior to transplantation. After euthanising the mice, a full‐thickness excision of wound bed biofilm material (resembling slough or a scab, see in vivo images) was conducted using scissors. This biofilm material from the wound beds was washed with 1X PBS to remove any planktonic bacterial cells. Under violet light, red fluorescence persisted from excised and washed wound bed material, indicating detectable fluorescence from bacteria encased within the biofilm matrix. These in vivo data confirm previous in vitro work28 indicating that the violet wavelength (405 nm) excitation light and emitted porphyrin red fluorescence can penetrate the bacteria‐derived biofilm EPS and/or host‐derived matrix surrounding these bacteria, and the porphyrin‐producing bacteria found within the EPS matrix can be detected by fluorescence imaging in vivo.
SEM imaging (n = 4) and histopathology (n = 4) were used to confirm the presence of biofilm (Figures 4 and 5). Both methods demonstrated close morphological co‐localisation of biofilm‐associated matrix with bacteria; association with both rod and cocci bacteria was evident on SEM images (Figure 4). The presence of a matrix surrounding these bacteria supports the presence of biofilm, consistent with other researchers who use these methods to determine the presence of biofilm in wounds.37, 46, 47 The polymicrobial nature of the biofilms was confirmed through selective and differential plating of the homogenised ex vivo wound bed (n = 3) and confirmed through culture‐based diagnostics at a CAP‐certified clinical laboratory. The final mean bacterial concentrations were S. aureus (4.62 x 107 CFU/g), E. coli (7.69 x 108 CFU/g), and E. cloacae (1.81 x 108 CFU/g); these species correspond with the rods and cocci observed in the SEM images. Our interpretation of bacterial cells or aggregates within the extracellular matrix as indicative of biofilm in clinical samples has previously been demonstrated by other groups using SEM47, 48, 49, 50, 51 and Transmission Electron Microscopy (TEM).52
FIGURE 4.
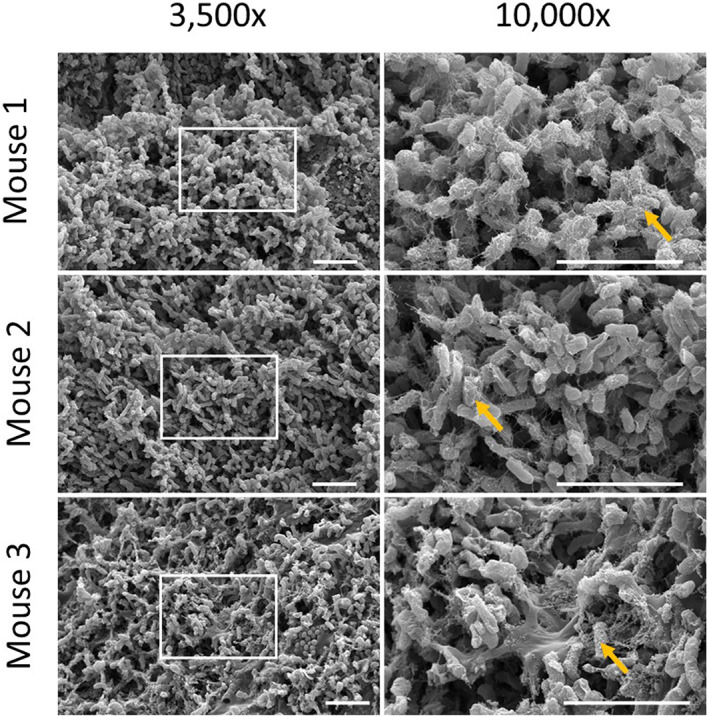
Scanning electron microscopy images of in vitro polymicrobial biofilms transplanted into in vivo murine chronic wound model. Left panel is at 3,500x magnification; right panel is at 10,000x magnification (n = 4). Images from three different mouse wounds are shown. Boxes indicate regions of magnification in right panel. Arrows denote regions of bacterial‐matrix interaction. Scale bars represent 50 μM
FIGURE 5.
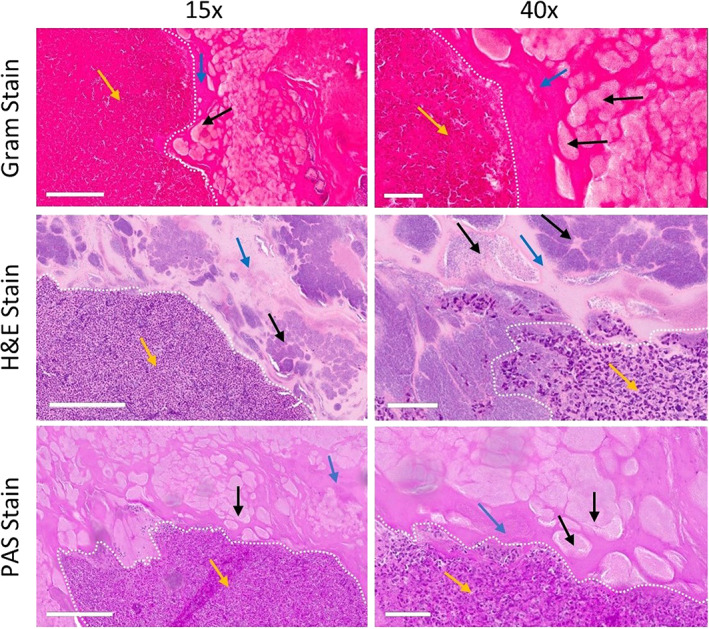
Histopathology confirms bacteria is encompassed in matrix of excised red fluorescing wound bed material. Four days after inoculation, wound bed material was excised from the wound bed and embedded in paraffin, and slides were prepared (n = 6). Slides were stained for Gram, haematoxylin and eosin (H&E), and Periodic acid‐Schiff (PAS). Gram staining appears dark pink because of a high prevalence of matrix. Yellow arrows indicate immune cells in regions of host‐derived matrix; black arrows indicate bacterial aggregates; blue arrows indicate bacterial‐derived matrix; white dashed line indicates delineation between host‐ and bacterial‐derived matrix. Both 15X (left panel) and 40X (right panel) images of the same region are shown. White scale bars represent 200 μm (left panel) or 50 μm (right panel). Representative images of each stain
Next, we aimed to see, through the histopathological analysis of biofilm‐inoculated mice, whether we could distinguish between a host and bacteria‐derived matrix. Histopathology staining showed regions of bacterial aggregates embedded in the matrix (both bacterial‐derived and host‐derived, see discussion; Figure 5). The histopathology images demonstrated a clear delineation between bacterial cells or aggregates embedded in the bacterial‐derived matrix versus the host‐derived matrix. The bacteria‐derived matrix was assumed to have formed in vitro as it was devoid of immune host cells; the host‐derived matrix surrounding this region was indicated by the infiltration of immune cells (assumed to be primarily neutrophils and macrophages).53, 54 Clinically, both bacteria‐ and host‐derived matrices contribute to the pathogenesis of biofilm‐associated infections.55, 56 Our results are in line with prior studies which suggest that the host immune cells (primarily neutrophils and/or macrophages) have difficulty penetrating the mechanical barrier of the bacteria‐derived biofilm EPS.57, 58, 59 Thus, the regions denoted in Figure 5 containing a high density of immune cells are indicative of host‐derived matrix compared with the bacteria‐derived matrix, which are devoid of immune cells and contain aggregates of bacteria. The interpretation of histopathology results reported here is consistent with previous in vivo and clinical studies that similarly used Gram stain,47, 58, 59, 60, 61 H&E,46, 61, 62, 63, 64 and PAS65, 66 to evaluate the presence of biofilm in clinical samples.
4. DISCUSSION
It is well established that biofilms contribute to the chronicity of wounds,67 yet detection of bacterial burden and biofilm at the point of care remains a challenge. This study is the first to demonstrate that red fluorescence is readily detected from biofilm‐associated bacteria in vivo using a point‐of‐care fluorescence imaging device. The in vivo detection of red fluorescence from bacteria embedded in biofilm reported here builds on previous in vitro work28 and confirms that fluorescence imaging can detect (but not distinguish between) both planktonic and biofilm‐encased bacteria.
One aim of this work was to establish that red or cyan fluorescence detected under violet light illumination is emitted from bacteria and not from an immune response or other host factors. It is well established that many bacteria fluoresce red when excited by violet light because of intrinsic porphyrin production20, 21, 28 and that Pseudomonas spp. uniquely produces a virulence factor, pyoverdine,26, 45, 68, 69 which is likely the compound responsible for the cyan fluorescence observed in this study. In vitro studies to assess red fluorescent porphyrin production require supplementing of media with an essential precursor, ALA.28 Based on clinical studies, ALA is readily available within tissues to facilitate endogenous porphyrin production and fluorescence detection.15, 27, 70 Our findings build on prior in vitro work28 by demonstrating that red and cyan fluorescent signals are readily detectable from bacteria‐inoculated mouse wounds illuminated with violet light. Consistent with these previous in vitro findings, mouse wounds inoculated with porphyrin‐producing bacteria produced a detectable red fluorescent signal that increased in intensity over time, up to 5 days, after which wound healing and clearance of the bacteria may have been responsible for reducing the signal. This increase in bacterial red fluorescence intensity observed over time may be caused by the uptake of ALA from host tissues, as well as increasing bacterial load as the infection progresses over time. As expected, cyan fluorescence was only observed when wounds were inoculated with P. aeruginosa. Non‐porphyrin‐producing bacterial species have been shown to not produce a red or cyan fluorescent signal in vitro.28 In line with this, we did not detect red or cyan fluorescent signals from wounds inoculated with known non‐porphyrin‐producing bacterial species (specifically S. agalactiae and E. faecalis). These findings confirm that the observed red fluorescence is emanating specifically from bacteria and not bacteria‐associated inflammation or other co‐localised host responses. These host responses would be similar between the cohorts of infected mice, independent of the bacteria's ability to produce porphyrins.
There is clinical consensus that biofilm matrix must be disrupted to enable bacterial removal71; however, there remains no consensus on how to readily and consistently identify a biofilm in a wound and monitor anti‐biofilm treatment efficacy. Bacterial aggregation with EPS matrix in close association is a hallmark of biofilm that is identified at high (~1,000X) magnifications with SEM.72 However, SEM imaging is a time‐ and labour‐intensive process that is not suitable for routine clinical diagnosis. Histopathology and fluorescence microscopy73 may be used to detect co‐location of bacterial and matrix stains, but similar to SEM, these methods are time intensive. Furthermore, these microscopic methods cannot distinguish between a bacterial and host‐derived matrix, although the importance of the EPS matrix being of bacterial origin has been questioned in recent years.74, 75 Next‐generation sequencing to confirm the presence of EPS matrix and wound blotting to identify a matrix74 have recently emerged as new technologies that may detect biofilm. However, none of these methods provide immediate identification of biofilm at the point of care, as well as distribution of the bacteria across the wound. The results of our study demonstrate point‐of‐care detection of bacterial fluorescence within a biofilm in a well‐established pre‐clinical model. Although the red fluorescence from porphyrins could not distinguish between planktonic and biofilm‐associated bacteria, it could provide immediate clinical information to guide treatments targeting bacterial eradication, such as mechanical disruption (eg, debridement), as well as providing immediate feedback on the effectiveness of those treatments.18, 27, 31, 32 The importance of identifying potential biofilm in wounds is driven by several factors, including: (1) the high prevalence of biofilm in chronic wounds (60%‐80%47, 76, 77) and its tremendous impact on wound healing,78, 79 (2) the need for clinicians to target regions of biofilm and bacterial bioburden to mechanically disrupt the bacterial and host‐derived matrices to facilitate access of antimicrobial agents for bacterial removal, and (3) the ability of biofilm to reform within 24 hours of aggressive debridement if the bacteria is not thoroughly removed.44 This biofilm reformation is likely enhanced because of regions of biofilm going undetected and is therefore not addressed during mechanical disruption efforts.71
Data from established biofilm models suggest that bacterial metabolism can be slowed when those bacteria are encased in a biofilm.80, 81 This has relevance to the current study as the detected fluorescent bacterial porphyrins are a by‐product of bacterial metabolism (the heme biosynthesis pathway); therefore, slowing of bacterial metabolism, when in a biofilm, could blunt porphyrin production relative to bacteria in a planktonic state. In this study, while both the polymicrobial biofilm and monomicrobial planktonic bacteria‐infected wounds produced red fluorescence when porphyrin‐producing bacteria were present, the relative intensity of the red fluorescence was not quantified. Uncontrolled variables such as final bacterial loads and clear differences in wound healing between the two models (bacteria in biofilm vs planktonic) along with the difference in immune status of the strains used for each model prevented a direct comparison and clear conclusions on this matter.
5. LIMITATIONS
In this study, the presence of biofilm was confirmed through both SEM and histopathology, which demonstrates close bacterial‐matrix interaction in the skin wounds. SEM is the gold standard for biofilm confirmation47, 48, 49, 50, 51; however, it should be acknowledged that it is not possible with any method to definitively distinguish between bacterial and host‐derived matrix, pre‐clinically or clinically, and even when attempted, this confirmation is typically used for research purposes72 and is not feasible for routine patient care and treatment planning.
As it is challenging to clinically establish what is and is not a biofilm, efforts to test fluorescence from definitive biofilm in this study needed to be performed pre‐clinically in a mouse model. There are inherent limitations to pre‐clinical studies that cannot completely recapitulate all factors, which may play a role in clinical biofilm fluorescence detection. These factors include many other potential bacterial species in the biofilm environment82; their various interactions and virulence factors; and the presence of host tissue, which emits its own fluorescent signals, for example, blood on the wound or in granulation tissue, which absorbs some of the violet excitation light.19 Therefore, studies to confirm fluorescence detection of bacterial biofilm in clinical samples are warranted. Despite the limitations of pre‐clinical work, in vitro and in vivo biofilm studies have been instrumental in our current knowledge of biofilm in wounds,35 including the foundational understanding of the tolerance of biofilm bacteria to antibiotics and antimicrobials,83 synergism of bacterial species within a biofilm enhancing virulence and antimicrobial tolerance,34, 76, 83, 84 biofilm's ability to impair wound healing,78 and methods of biofilm eradication.44, 85, 86 The current work represents further advances in developing methods of detecting bacteria in biofilm.
6. CONCLUSIONS AND CLINICAL IMPLICATIONS
The results of the current study show that bacteria within biofilm emit detectable fluorescence in a well‐established polymicrobial biofilm model, although this fluorescent signal does not distinguish between planktonic and biofilm‐associated bacteria. This has important clinical implications as identifying regions of potential biofilm may aid in more efficient detection and disruption via mechanical debridement and other methods. Wounds heal faster and patients have better outcomes with fewer complications when biofilm and bioburden are efficiently managed.71, 85, 87 This emphasises the clinical need to detect biofilm during wound management. Earlier detection of bacterial burden with fluorescence imaging—potentially within biofilm and certainly in regions of high bacterial load—has been shown to reduce reliance on antimicrobials and antibiotic prescribing27, 31, 88 and improve 12‐week healing rates in chronic wounds.88, 89 In these clinical studies, detection of bacterial burden at the point of care facilitated enhanced wound hygiene, primarily through aggressive, targeted cleansing and mechanical disruption (debridement).31, 70, 88 These findings, together with the high prevalence of biofilm in chronic wounds and therapeutic potential of mechanical disruption,71, 85, 86 suggest the possibility that targeting of bacteria through debridement may prevent biofilm from becoming well established in the wound or may facilitate biofilm disruption at an earlier stage when the bacteria can more easily be eradicated. The ability to detect potential biofilm and associated bacterial burden at the point of care would be a key component of that clinical strategy.
CONFLICT OF INTEREST
Laura M. Jones, Anna D'souza, and Monique Y. Rennie are salaried employees of MolecuLight Inc. Allie Clinton Smith has a sponsored research agreement between MolecuLight Inc. and Texas Tech University. Other authors have no conflicts to disclose.
ACKNOWLEDGEMENTS
The authors acknowledge technical support from Jean Kontogiannis and Rainerio De Guzman (University of Toronto, Division of Comparative Medicine, Donnelly Center for Cellular and Biomolecular Research), Jessica Bourke (Mount Sinai Hospital, Department of Microbiology), Jelisa Hernandez and Amanda Martinez (Clinical Laboratory Science Department, Covenant Medical Center), Warren Hatley (Department of Pathology, Texas Tech University Health Sciences Center), Mary C. Hastert and Bo Zhao (College of Arts and Sciences Microscopy, Texas Tech University), and Garrett S. Welch and Hui Hua (Department of Surgery, Texas Tech University Health Sciences Center).
Lopez AJ, Jones LM, Reynolds L, et al. Detection of bacterial fluorescence from in vivo wound biofilms using a point‐of‐care fluorescence imaging device. Int Wound J. 2021;18:626–638. 10.1111/iwj.13564
Funding information MolecuLight Inc.
Contributor Information
Monique Y. Rennie, Email: mrennie@moleculight.com
Allie Clinton Smith, Email: allie.c.smith@ttu.edu.
DATA AVAILABILITY STATEMENT
Data available on request from the authors.
REFERENCES
- 1.Bowler PG. Wound pathophysiology infection and therapeutic options. Ann Med. 2002;34(6):419‐427. [DOI] [PubMed] [Google Scholar]
- 2.Cutting KF, White RJ. Criteria for identifying wound infection‐revisited. Ostomy Wound Manage. 2005;51(1):28‐34. [PubMed] [Google Scholar]
- 3.Bianchi T, Wolcott RD, Peghetti A, et al. Recommendations for the management of biofilm: a consensus document. J Wound Care. 2016;25(6):305‐317. [DOI] [PubMed] [Google Scholar]
- 4.Xu L, McLennan SV, Lo L, et al. Bacterial load predicts healing rate in neuropathic diabetic foot ulcers. Diabetes Care. 2007;30(2):378‐380. [DOI] [PubMed] [Google Scholar]
- 5.Gardner SE, Frantz RA. Wound bioburden and infection‐related complications in diabetic foot ulcers. Biol Res Nurs. 2008;10(1):44‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turtiainen J, Hakala T, Hakkarainen T, Karhukorpi J. The impact of surgical wound bacterial colonization on the incidence of surgical site infection after lower limb vascular surgery: a prospective observational study. Eur J Vasc Endovasc Surg. 2014;47(4):411‐417. [DOI] [PubMed] [Google Scholar]
- 7.Copeland‐Halperin LR, Kaminsky AJ, Bluefeld N, Miraliakbari R. Sample procurement for cultures of infected wounds: a systematic review. J Wound Care. 2016;25(4):S4‐S6.S8‐10. [DOI] [PubMed] [Google Scholar]
- 8.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318‐1322. [DOI] [PubMed] [Google Scholar]
- 9.Clinton A, Carter T. Chronic wound biofilms: pathogenesis and potential therapies. Lab Med. 2015;46(4):277‐284. [DOI] [PubMed] [Google Scholar]
- 10.Flemming HC, Neu TR, Wozniak DJ. The EPS matrix: the "house of biofilm cells". J Bacteriol. 2007;189(22):7945‐7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balcazar JL, Subirats J, Borrego CM. The role of biofilms as environmental reservoirs of antibiotic resistance. Front Microbiol. 2015;6:1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anwar H, Dasgupta MK, Costerton JW. Testing the susceptibility of bacteria in biofilms to antibacterial agents. Antimicrob Agents Chemother. 1990;34(11):2043‐2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultz G, Bjarnsholt T, James GA, et al. Consensus guidelines for the identification and treatment of biofilms in chronic nonhealing wounds. Wound Repair Regen. 2017;25(5):744‐757. [DOI] [PubMed] [Google Scholar]
- 14.Hill R, Rennie MY, Douglas J. Using bacterial fluorescence imaging and antimicrobial stewardship to guide wound management practices: a case series. Ostomy Wound Manage. 2018;64(8):18‐28. [PubMed] [Google Scholar]
- 15.Hurley CM, McClusky P, Sugrue RM, Clover JA, Kelly JE. Efficacy of a bacterial fluorescence imaging device in an outpatient wound care clinic: a pilot study. J Wound Care. 2019;28(7):438‐443. [DOI] [PubMed] [Google Scholar]
- 16.Ottolino‐Perry K, Chamma E, Blackmore KM, et al. Improved detection of clinically relevant wound bacteria using autofluorescence image‐guided sampling in diabetic foot ulcers. Int Wound J. 2017;14(5):833‐841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rennie MY, Lindvere‐Teene L, Tapang K, Linden R. Point‐of‐care fluorescence imaging predicts the presence of pathogenic bacteria in wounds: a clinical study. J Wound Care. 2017;26(8):452‐460. [DOI] [PubMed] [Google Scholar]
- 18.Serena TE, Harrell K, Serena L, Yaakov RA. Real‐time bacterial fluorescence imaging accurately identifies wounds with moderate‐to‐heavy bacterial burden. J Wound Care. 2019;28(6):346‐357. [DOI] [PubMed] [Google Scholar]
- 19.Rennie MY, Dunham D, Lindvere‐Teene L, Raizman R, Hill R, Linden R. Understanding real‐time fluorescence signals from bacteria and wound tissues observed with the MolecuLight i:X(TM). Diagnostics (Basel). 2019;9(1):22. 10.3390/diagnostics9010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nitzan Y, Salmon‐Divon M, Shporen E, Malik Z. ALA induced photodynamic effects on gram positive and negative bacteria. Photochem Photobiol Sci. 2004;3(5):430‐435. [DOI] [PubMed] [Google Scholar]
- 21.Philipp‐Dormston WK, Doss M. Comparison of porphyrin and heme biosynthesis in various heterotrophic bacteria. Enzyme. 1973;16(1):57‐64. [DOI] [PubMed] [Google Scholar]
- 22.Anzaldi LL, Skaar EP. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect Immun. 2010;78(12):4977‐4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choby JE, Skaar EP. Heme synthesis and acquisition in bacterial pathogens. J Mol Biol. 2016;428(17):3408‐3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorchein A, Webber R. Delta‐aminolaevulinic acid in plasma, cerebrospinal fluid, saliva and erythrocytes: studies in normal, uraemic and porphyric subjects. Clin Sci (Lond). 1987;72(1):103‐112. [DOI] [PubMed] [Google Scholar]
- 25.Perez M, BR TS, Watanabe K, Miyanari S, Harrigan R. 5‐Aminolevulinic acid (5‐ALA): analysis of preclinical and safety literature. Food Nutr Sci. 2013;4(10):1009‐1013. [Google Scholar]
- 26.Schalk IJ, Guillon L. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: implications for metal homeostasis. Environ Microbiol. 2013;15(6):1661‐1673. [DOI] [PubMed] [Google Scholar]
- 27.Le LBM, Briggs P, Bullock N, et al. Diagnostic accuracy of point‐of‐care fluorescence imaging for the detection of bacterial burden in wounds: results from the 350‐patient fluorescence imaging assessment and guidance trial. Adv Wound Care. 2020. 10.1089/wound.2020.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones LM, Dunham D, Rennie MY, et al. In vitro detection of porphyrin‐producing wound bacteria with real‐time fluorescence imaging. Future Microbiol. 2020;15(5):319‐332. [DOI] [PubMed] [Google Scholar]
- 29.Warncke P, Fink S, Wiegand C, Hipler UC, Fischer D. A shell‐less hen's egg test as infection model to determine the biocompatibility and antimicrobial efficacy of drugs and drug formulations against Pseudomonas aeruginosa . Int J Pharm. 2020;585:119557. [DOI] [PubMed] [Google Scholar]
- 30.Wu YC, Kulbatski I, Medeiros PJ, et al. Autofluorescence imaging device for real‐time detection and tracking of pathogenic bacteria in a mouse skin wound model: preclinical feasibility studies. J Biomed Opt. 2014;19(8):085002. [DOI] [PubMed] [Google Scholar]
- 31.Cole W, Coe S. Use of a bacterial fluorescence imaging system to target wound debridement and accelerate healing: a pilot study. J Wound Care. 2020;29(suppl 7):S44‐s52. [DOI] [PubMed] [Google Scholar]
- 32.Moelleken M, Jockenhöfer F, Benson S, Dissemond J. Prospective clinical study on the efficacy of bacterial removal with mechanical debridement in and around chronic leg ulcers assessed with fluorescence imaging. Int Wound J. 2020;17:1011‐1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalton T, Dowd SE, Wolcott RD, et al. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One. 2011;6(11):e27317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, Rumbaugh KP. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun. 2014;82(11):4718‐4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabrilska RA, Rumbaugh KP. Biofilm models of polymicrobial infection. Future Microbiol. 2015;10(12):1997‐2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith AC, Rice A, Sutton B, et al. Albumin inhibits Pseudomonas aeruginosa quorum sensing and alters polymicrobial interactions. Infect Immun. 2017;85(9):e00116‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Y, Dowd SE, Smith E, Rhoads DD, Wolcott RD. In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 2008;16(6):805‐813. [DOI] [PubMed] [Google Scholar]
- 38.Brown RL, Greenhalgh DG. Mouse models to study wound closure and topical treatment of infected wounds in healing‐impaired and normal healing hosts. Wound Repair Regen. 1997;5(2):198‐204. [DOI] [PubMed] [Google Scholar]
- 39.Gawande PV, Clinton AP, LoVetri K, Yakandawala N, Rumbaugh KP, Madhyastha S. Antibiofilm efficacy of DispersinB(®) wound spray used in combination with a silver wound dressing. Microbiol Insights. 2014;7:9‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison F, Roberts AE, Gabrilska R, Rumbaugh KP, Lee C, Diggle SP. A 1,000‐year‐old antimicrobial remedy with antistaphylococcal activity. MBio. 2015;6(4):e01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rumbaugh KP, Diggle SP, Watters CM, Ross‐Gillespie A, Griffin AS, West SA. Quorum sensing and the social evolution of bacterial virulence. Curr Biol. 2009;19(4):341‐345. [DOI] [PubMed] [Google Scholar]
- 42.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 2014;10(7):e1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watters C, Everett JA, Haley C, Clinton A, Rumbaugh KP. Insulin treatment modulates the host immune system to enhance Pseudomonas aeruginosa wound biofilms. Infect Immun. 2014;82(1):92‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolcott RD, Rumbaugh KP, James G, et al. Biofilm maturity studies indicate sharp debridement opens a time‐dependent therapeutic window. J Wound Care. 2010;19(8):320‐328. [DOI] [PubMed] [Google Scholar]
- 45.Cueva AR, Pham O, Diaby A, Fleming D, Rumbaugh KP, Fernandes GE. Pyoverdine assay for rapid and early detection of Pseudomonas aeruginosa in burn wounds. ACS Appl Bio Mater. 2020;3(8):5350‐5356. [DOI] [PubMed] [Google Scholar]
- 46.Davis SC, Ricotti C, Cazzaniga A, Welsh E, Eaglstein WH, Mertz PM. Microscopic and physiologic evidence for biofilm‐associated wound colonization in vivo. Wound Repair Regen. 2008;16(1):23‐29. [DOI] [PubMed] [Google Scholar]
- 47.James GA, Swogger E, Wolcott R, et al. Biofilms in chronic wounds. Wound Repair Regen. 2008;16(1):37‐44. [DOI] [PubMed] [Google Scholar]
- 48.Cryer J, Schipor I, Perloff JR, Palmer JN. Evidence of bacterial biofilms in human chronic sinusitis. ORL J Otorhinolaryngol Relat Spec. 2004;66(3):155‐158. [DOI] [PubMed] [Google Scholar]
- 49.Hoa M, Tomovic S, Nistico L, et al. Identification of adenoid biofilms with middle ear pathogens in otitis‐prone children utilizing SEM and FISH. Int J Pediatr Otorhinolaryngol. 2009;73(9):1242‐1248. [DOI] [PubMed] [Google Scholar]
- 50.Nickel JC, Costerton JW. Bacterial localization in antibiotic‐refractory chronic bacterial prostatitis. Prostate. 1993;23(2):107‐114. [DOI] [PubMed] [Google Scholar]
- 51.Nickel JC, Reid G, Bruce AW, Costerton JW. Ultrastructural microbiology of infected urinary stone. Urology. 1986;28(6):512‐515. [DOI] [PubMed] [Google Scholar]
- 52.Chole RA, Faddis BT. Anatomical evidence of microbial biofilms in tonsillar tissues: a possible mechanism to explain chronicity. Arch Otolaryngol Head Neck Surg. 2003;129(6):634‐636. [DOI] [PubMed] [Google Scholar]
- 53.Hanke ML, Heim CE, Angle A, Sanderson SD, Kielian T. Targeting macrophage activation for the prevention and treatment of Staphylococcus aureus biofilm infections. J Immunol. 2013;190(5):2159‐2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamada KJ, Kielian T. Biofilm‐leukocyte cross‐talk: impact on immune polarization and Immunometabolism. J Innate Immun. 2019;11(3):280‐288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bowen WH, Burne RA, Wu H, Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018;26(3):229‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karygianni L, Ren Z, Koo H, Thurnheer T. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 2020;28(8):668‐681. [DOI] [PubMed] [Google Scholar]
- 57.Bylund J, Burgess LA, Cescutti P, Ernst RK, Speert DP. Exopolysaccharides from burkholderia cenocepacia inhibit neutrophil chemotaxis and scavenge reactive oxygen species. J Biol Chem. 2006;281(5):2526‐2532. [DOI] [PubMed] [Google Scholar]
- 58.Thurlow LR, Hanke ML, Fritz T, et al. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 2011;186(11):6585‐6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hansen CR, Pressler T, Nielsen KG, Jensen PO, Bjarnsholt T, Hoiby N. Inflammation in a xylosoxidans infected cystic fibrosis patients. J Cyst Fibros. 2010;9(1):51‐58. [DOI] [PubMed] [Google Scholar]
- 60.Oates A, Bowling FL, Boulton AJ, Bowler PG, Metcalf DG, McBain AJ. The visualization of biofilms in chronic diabetic foot wounds using routine diagnostic microscopy methods. J Diabetes Res. 2014;2014:153586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toth L, Csomor P, Sziklai I, Karosi T. Biofilm detection in chronic rhinosinusitis by combined application of hematoxylin‐eosin and gram staining. Eur Arch Otorhinolaryngol. 2011;268(10):1455‐1462. [DOI] [PubMed] [Google Scholar]
- 62.Hochstim CJ, Choi JY, Lowe D, Masood R, Rice DH. Biofilm detection with hematoxylin‐eosin staining. Arch Otolaryngol Head Neck Surg. 2010;136(5):453‐456. [DOI] [PubMed] [Google Scholar]
- 63.Sedghizadeh PP, Kumar SK, Gorur A, Schaudinn C, Shuler CF, Costerton JW. Microbial biofilms in osteomyelitis of the jaw and osteonecrosis of the jaw secondary to bisphosphonate therapy. J Am Dent Assoc. 2009;140(10):1259‐1265. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Z, Han D, Zhang S, et al. Biofilms and mucosal healing in postsurgical patients with chronic rhinosinusitis. Am J Rhinol Allergy. 2009;23(5):506‐511. [DOI] [PubMed] [Google Scholar]
- 65.Bulut F, Meric F, Yorgancilar E, et al. Effects of N‐acetyl‐cysteine and acetylsalicylic acid on the tonsil bacterial biofilm tissues by light and electron microscopy. Eur Rev Med Pharmacol Sci. 2014;18(23):3720‐3725. [PubMed] [Google Scholar]
- 66.Winther B, Gross BC, Hendley JO. Early SV. Location of bacterial biofilm in the mucus overlying the adenoid by light microscopy. Arch Otolaryngol Head Neck Surg. 2009;135(12):1239‐1245. [DOI] [PubMed] [Google Scholar]
- 67.Snyder RJ, Bohn G, Hanft J, et al. Wound biofilm: current perspectives and strategies on biofilm disruption and treatments. Wounds. 2017;29(6):S1‐S17. [PubMed] [Google Scholar]
- 68.Stintzi A, Cornelis P, Hohnadel D, et al. Novel pyoverdine biosynthesis gene(s) of Pseudomonas aeruginosa PAO. Microbiology. 1996;142(Pt 5):1181‐1190. [DOI] [PubMed] [Google Scholar]
- 69.Meyer JM, Stintzi A, De Vos D, et al. Use of siderophores to type pseudomonads: the three Pseudomonas aeruginosa pyoverdine systems. Microbiology. 1997;143(Pt 1):35‐43. [DOI] [PubMed] [Google Scholar]
- 70.Hill RWKA. Prospective multisite observational study incorporating bacterial fluorescence information into the upper/lower wound infection checklists. Wounds. 2020;32(11):299‐308. [PubMed] [Google Scholar]
- 71.Wolcott R. Disrupting the biofilm matrix improves wound healing outcomes. J Wound Care. 2015;24(8):366‐371. [DOI] [PubMed] [Google Scholar]
- 72.Hurlow J, Blanz E, Gaddy JA. Clinical investigation of biofilm in non‐healing wounds by high resolution microscopy techniques. J Wound Care. 2016;25(suppl 9):S11‐S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fromantin I, Seyer D, Watson S, et al. Bacterial floras and biofilms of malignant wounds associated with breast cancers. J Clin Microbiol. 2013;51(10):3368‐3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakagami G, Schultz G, Kitamura A, et al. Rapid detection of biofilm by wound blotting following sharp debridement of chronic pressure ulcers predicts wound healing: a preliminary study. Int Wound J. 2020;17(1):191‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cooper RA, Bjarnsholt T, Alhede M. Biofilms in wounds: a review of present knowledge. J Wound Care. 2014;23(11):570 2–4, 6–80 passim. [DOI] [PubMed] [Google Scholar]
- 76.Römling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med. 2012;272(6):541‐561. [DOI] [PubMed] [Google Scholar]
- 77.Malone M, Bjarnsholt T, McBain AJ, et al. The prevalence of biofilms in chronic wounds: a systematic review and meta‐analysis of published data. J Wound Care. 2017;26(1):20‐25. [DOI] [PubMed] [Google Scholar]
- 78.Metcalf DG, Bowler PG. Biofilm delays wound healing: a review of the evidence. Burns Trauma. 2013;1(1):5‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Omar A, Wright JB, Schultz G, Burrell R, Nadworny P. Microbial biofilms and chronic wounds. Microorganisms. 2017;5(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Booth SC, Workentine ML, Wen J, et al. Differences in metabolism between the biofilm and planktonic response to metal stress. J Proteome Res. 2011;10(7):3190‐3199. [DOI] [PubMed] [Google Scholar]
- 81.Wong EHJ, Ng CG, Goh KL, Vadivelu J, Ho B, Loke MF. Metabolomic analysis of low and high biofilm‐forming helicobacter pylori strains. Sci Rep. 2018;8(1):1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolcott RD, Hanson JD, Rees EJ, et al. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2016;24(1):163‐174. [DOI] [PubMed] [Google Scholar]
- 83.Burmolle M, Webb JS, Rao D, Hansen LH, Sorensen SJ, Kjelleberg S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol. 2006;72(6):3916‐3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. Mechanisms of synergy in polymicrobial infections. J Microbiol. 2014;52(3):188‐199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Attinger C, Wolcott R. Clinically addressing biofilm in chronic Wounds. Adv Wound Care. 2012;1(3):127‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolcott RD, Dowd SE. A rapid molecular method for characterising bacterial bioburden in chronic wounds. J Wound Care. 2008;17(12):513‐516. [DOI] [PubMed] [Google Scholar]
- 87.Wolcott RD, Kennedy JP, Dowd SE. Regular debridement is the main tool for maintaining a healthy wound bed in most chronic wounds. J Wound Care. 2009;18(2):54‐56. [DOI] [PubMed] [Google Scholar]
- 88.Price N. Routine fluorescence imaging to detect wound bacteria reduces antibiotic use and antimicrobial dressing expenditure while improving healing rates: retrospective analysis of 229 foot ulcers. Diagnostics. 2020;10:927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rahma S WJ, Nixon JE, Brown S, Russel DA.The use of Point‐of‐Care Bacterial Autofluorescence Imaging in the Management of Diabetic Foot Ulcers; A Pilot Randomised Controlled Trial. Poster presented at: Symposium on Advanced Wound Care 2020 Nov June 4, 2020; Virtual. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data available on request from the authors.


