Abstract
The phylogeny of 46 geographically diverse Histoplasma capsulatum isolates representing the three varieties capsulatum, duboisii, and farciminosum was evaluated using partial DNA sequences of four protein coding genes. Parsimony and distance analysis of the separate genes were generally congruent and analysis of the combined data identified six clades: (i) class 1 North American H. capsulatum var. capsulatum, (ii) class 2 North American H. capsulatum var. capsulatum, (iii) Central American H. capsulatum var. capsulatum, (iv) South American H. capsulatum var. capsulatum group A, (v) South American H. capsulatum var. capsulatum group B, and (vi) H. capsulatum var. duboisii. Although the clades were generally well supported, the relationships among them were not resolved and the nearest outgroups (Blastomyces and Paracoccidioides) were too distant to unequivocally root the H. capsulatum tree. H. capsulatum var. farciminosum was found within the South American H. capsulatum var. capsulatum group A clade. With the exception of the South American H. capsulatum var. capsulatum group A clade, genetic distances within clades were an order of magnitude lower than those between clades, and each clade was supported by a number of shared derived nucleotide substitutions, leading to the conclusion that each clade was genetically isolated from the others. Under a phylogenetic species concept based on possession of multiple shared derived characters, as well as concordance of four gene genealogies, H. capsulatum could be considered to harbor six species instead of three varieties.
The pathogenic ascomycete species Histoplasma capsulatum Darling [teleomorph, Ajellomyces capsulatus (Kwon-Chung) McGinnis et Katz] occurs throughout the world and causes histoplasmosis in various mammalian species, including humans (46, 61). The fungus grows as a saprobe in nature and is acquired by the inhalation of airborne microconidia or hyphal fragments. Once inhaled, the fungus transforms from a mycelium to a pathogenic yeast form. Histoplasmosis primarily affects the host’s lungs, and its symptoms vary greatly. The vast majority of infected people are asymptomatic; however, the fungus can cause disseminated histoplasmosis in otherwise healthy people, and especially in immunocompromised individuals and AIDS patients (46).
Distinct genotypes and varieties of H. capsulatum which show different clinical manifestations and geographical distributions are known. Cases of histoplasmosis due to H. capsulatum var. capsulatum have been reported in at least 60 countries on all continents (1), but they are especially prevalent in the eastern half of the United States and most of Latin America (46). In North America, two prevalent groups (class 1 and class 2) of H. capsulatum isolates which showed differences in growth phenotype (53) and restriction fragment length polymorphisms in mitochondrial and genomic DNA have been identified (63, 68). The North American class 1 strains (NAm Hcc1; see Table 1 for this and other abbreviations) have been isolated mainly from patients with AIDS, whereas the North American class 2 strains (NAm Hcc2) have been isolated from patients both with and without AIDS (64). H. capsulatum var. duboisii Ciferri (1960) is the causal agent of African histoplasmosis and is endemic in the tropical areas of Africa (61). African histoplasmosis is characterized by the presence of lesions, primarily in cutaneous, subcutaneous, and osseous tissues, and by the larger size of the yeast cells. H. capsulatum var. farciminosum (Rivolta) Weeks et al. causes subcutaneous and ulcerated lesions of the skin in horses and mules (46). The disease is widespread throughout Europe, North Africa, India, and South Asia. The morphology of the yeast cell of H. capsulatum var. farciminosum resembles that of H. capsulatum var. capsulatum (59).
TABLE 1.
Abbreviations of varieties and geographical groups of H. capsulatum
| Abbreviation | Variety and population |
|---|---|
| NAm Hcc1 | Class 1 North American H. capsulatum var. capsulatum |
| NAm Hcc2 | Class 2 North American H. capsulatum var. capsulatum |
| Panama Hcc | Panamanian H. capsulatum var. capsulatum |
| SAm Hcc | South American H. capsulatum var. capsulatum |
Despite the clinical importance of the organism, the phylogenetic relationships among the varieties and geographical groups of H. capsulatum are presently unresolved. Leclerc et al. (49) and Guého et al. (37) have included representatives of the three H. capsulatum varieties in their phylogenetical studies of onygenalean fungi. However, the phylogeny of H. capsulatum varieties was not clearly resolved because there was not sufficient variation in the conserved rRNA gene sequences. In this research, we used 46 isolates comprising the three varieties and DNA sequences of four protein coding genes to analyze the evolutionary relationships of H. capsulatum varieties. In the process we also examined the mode of reproduction in isolates of one clade of H. capsulatum var. capsulatum.
MATERIALS AND METHODS
Fungal isolates, culture, and DNA isolation.
Table 2 lists the isolates used in the study. H82 and H83 were isolated from a single patient who lived in Panama (9). H87 and H91 were also isolated from a single patient, who was infected at the Guinea-Liberia border (23). Each of the remaining clinical and veterinary isolates was derived from different individuals. Living isolates were manipulated and cultivated under biohazzard level 3 containment. Mycelium was grown in liquid medium and heat killed for safety, and DNA was extracted as previously reported (14, 17).
TABLE 2.
List of fungal isolates used in this study
| Isolate | Variety/ population | Collection no.a
|
Location | Source | Year isolated | Provider of isolateb | |
|---|---|---|---|---|---|---|---|
| ATCC | Others | ||||||
| H2 | NAm Hcc2 | D14 | Indiana | Human with AIDS | 1990 | E. Keath from P. Connoly | |
| H5 | NAm Hcc2 | D20 | Indiana | Human with AIDS | 1989 | E. Keath from P. Connoly | |
| H6 | NAm Hcc2 | E14 | Indiana | Human | 1980 | E. Keath from P. Connoly | |
| H8 | NAm Hcc2 | 26032 | M. D. Berliner G217B | Louisiana | Human | 1973 or before | |
| H9 | NAm Hcc1 | 38904 | Downs | Missouri | Human | 1968 | E. Keath and G. Kobayashi |
| H10 | NAm Hcc1 | Missouri | Human with AIDS | 1987 | G. Kobayashi | ||
| H11 | NAm Hcc2 | 848 | Missouri | Human | 1993 or before | G. Kobayashi | |
| H18 | NAm Hcc2 | 5-1MD | Missouri | Human | 1993 or before | G. Kobayashi | |
| H59 | SAm Hcc B | H-0057-I-10 | Bogota, Colombia | Human | 1994 or before | A. Restrepo and J. McEwen | |
| H60 | SAm Hcc A | H-0057-I-11 | Bogota, Colombia | Human with AIDS | 1994 or before | A. Restrepo and J. McEwen | |
| H61 | SAm Hcc A | H-0057-I-14 | Bogota, Colombia | Human | 1994 or before | A. Restrepo and J. McEwen | |
| H62 | SAm Hcc A | H-0057-I-15 | Bogota, Colombia | Human | 1994 or before | A. Restrepo and J. McEwen | |
| H63 | SAm Hcc A | H-0057-I-18 | Bogota, Colombia | Human | 1994 or before | A. Restrepo and J. McEwen | |
| H64 | SAm Hcc A | H-0057-I-22 | Bogota, Colombia | Human | 1994 or before | A. Restrepo and J. McEwen | |
| H66 | SAm Hcc | 13594, GH | Medellin, Colombia | Human | 1986 | A. Restrepo and J. McEwen | |
| H67 | SAm Hcc A | 30177, JE | Medellin, Colombia | Human | 1993 | A. Restrepo and J. McEwen | |
| H68 | SAm Hcc B | 30318, CH | Medellin, Colombia | Human | 1993 | A. Restrepo and J. McEwen | |
| H69 | SAm Hcc | 21402, JVM | Medellin, Colombia | Human | 1991 | A. Restrepo and J. McEwen | |
| H70 | SAm Hcc B | 30956, WS | Medellin, Colombia | Human with AIDS | 1994 | A. Restrepo and J. McEwen | |
| H71 | SAm Hcc A | 21337, JJM | Medellin, Colombia | Human | 1989 | A. Restrepo and J. McEwen | |
| H73 | SAm Hcc A | H-0057-I-24 | Bogota, Colombia | Human | 1994 or before | A. Restrepo and J. McEwen | |
| H74 | SAm Hcc A | 26760, GM | Medellin, Colombia | Human | 1993 | A. Restrepo and J. McEwen | |
| H75 | SAm Hcc B | 14056, HC | Medellin, Colombia | Human | 1986 | A. Restrepo and J. McEwen | |
| H76 | SAm Hcc A | T29302, GC | Medellin, Colombia | Human | 1993 | A. Restrepo and J. McEwen | |
| H77 | NAm Hcc2 | 10886 | C. W. Emmons 6613 | Virginia | Brown rat | 1940 | |
| H79c | NAm Hcc1/2 | 11408 | C. W. Emmons 6617 | Georgia | Skunk | 1940 | |
| H81 | Panama Hcc | 26028 | M. D. Berliner G184B | Panama | Human | 1967 or before | |
| H82 | Panama Hcc | 26029 | M. D. Berliner G186A | Panama | Human | 1967 or before | |
| H83 | Panama Hcc | 26030 | M. D. Berliner G186B | Panama | Human | 1967 or before | |
| H84 | NAm Hcc2 | 26320 | C. W. Emmons 6623 | Georgia | Opossum | 1940 | |
| H85 | SAm Hcc B | 28308 | CDC B923 | Argentina | Soil | 1966 or before | |
| H86 | NAm Hcc2 | 32682 | A. F. DiSalvo SC74 | South Carolina | Soil | 1974? | |
| H87 | Hc var. duboisii | 24294 | D. Grigoriu 8107A | Guinea-Liberia border | Human | 1970 | |
| H88d | Hc var. duboisii | 32281 | RV26821 | Belgium | Human | 1975 or before | |
| H90 | Hc var. farcimi-nosum | 58332 | CDC B-3786 | Egypt | Horse | 1983 or before | |
| H91 | Hc var. duboisii | 24295 | D. Grigoriu 8123 | Guinea-Liberia border | Human | 1970 | |
| H95 | Hc var. farcimi-nosum | 58333 | CDC B-3787 | Egypt | Horse | 1983 or before | |
| H96 | Hc var. farci-minosum | 60358 | A. F. DiSalvo 85-1610 | India | Horse | 1985? | |
| H97 | NAm Hcc2 | 0001 | Alabama | Human | 1995 or before | W. Dismukes, S. Moser, and B. Hines | |
| H126 | NAm Hcc1 | Missouri | Human with AIDS | 1987 | G. Kobayashi | ||
| H127 | NAm Hcc1 | Missouri | Human with AIDS | 1987 | G. Kobayashi | ||
| H130 | NAm Hcc2 | 15 | Alabama | Human | 1995 or before | W. Dismukes, S. Moser, and B. Hines | |
| H137 | Hc var. duboisii | 28536 | Kwon-Chung Hd27 | Zaire | Human | 1962 | |
| H138 | NAm Hcc2 | 22635 | Kwon-Chung T-3-1 | Arkansas | Soil | 1975 | |
| H139 | NAm Hcc2 | 22636 | Kwon-Chung T-4-2 | Arkansas | Soil | 1975 | |
| H140e | SAm Hcc | MK9500885 | Maryland; Peru | Owl monkey | 1997 | G. Miller | |
| Blastomyces dermatitidis | 60915 | D. Stevens A | |||||
| Paracoccidioides brasiliensis | Pb18 | Z. Pires de Camargo and E. Bagagli | |||||
ATCC, American Type Culture Collection, Manassas, Va.; CDC, Centers for Disease Control and Prevention, Atlanta, Ga.; CBS, Centraalbureau voor Schimmelcultures, Baarn, The Netherlands.
E. Keath, St. Louis University, St. Louis, Mo.; P. Connoly, Indiana University Medical Center, Indianapolis, Ind.; G. Kobayashi, Washington University School of Medicine, St. Louis, Mo.; A. Restrepo and J. McEwen, Corporacion para Investigaciones Biologicas, Medellin, Colombia; W. Dismukes, S. Moser, and B. Hines, University of Alabama, Birmingham; G. Miller, National Institutes of Health, Bethesda, Md.; Z. Pires de Camargo, Universidade Federal de Sao Paulo, Sao Paulo, Brazil; E. Bagagli, Universidade Estadual Paulista, Botucatu, Brazil.
In this research, H79 was transferred from NAm Hcc2 to NAm Hcc1.
Almost all cases of histoplasmosis duboisii in Europe are attributable to acquisition in areas in Africa in which this variety is endemic.
DNA was directly isolated from a yeast-infested monkey liver. The monkey was caught in the wild in Peru and then kept in Maryland.
Design of PCR primers.
The DNA sequences of four nuclear genes of H. capsulatum available from GenBank were used to design PCR primers (Table 3). Internal transcribed spacer (ITS) primers were derived from White et al. (70). The primers (sequences) were as follows (5′ to 3′): arf1 (agaatatggggcaaaaagga) and arf2 (cgcaattcatcttcgttgag) (ADP-ribosylation factors); H-anti3 (cgcagtcacctccatactatc) and H-anti4 (gcgccgacattaaccc) (H antigen precursors); ole3 (tttaaacgaagcccccacgg) and ole4 (caccacctccaacagcagca) (delta-9 fatty acid desaturases); tub1 (ggtggccaaatcgcaaactc) and tub2 (ggcagctttccgttcctcagt) (alpha-tubulins); ITS4 (tcctccgcttattgatatgc) and ITS5 (ggaagtaaaagtcgtaacaagg) (internal transcribed spacers plus rRNA genes). PCRs were performed with 2 μl of diluted genomic DNA template in 50-μl reactions. Reactions consisted of 0.45 μM of each primer, 1.0 U of AmpliTaq DNA polymerase (Perkin-Elmer), 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, and 0.2 mM deoxynucleotide triphosphates with the following temperature profile: a 15-s DNA denaturation step at 94°C, a 30-s annealing step (see below), and a 1-min extension step at 72°C for 32 cycles, followed by a 5-min final extension step at 72°C. The annealing temperature in the first cycle was 65°C. This annealing temperature was subsequently reduced by 0.7°C/cycle for the next 12 cycles, and thereafter, the PCR was continued at an annealing temperature of 56°C for the remaining 20 cycles (Touchdown PCR [24]).
TABLE 3.
Lengths and maximum sequence divergence of the analyzed loci
| Locus | Encoding | Length (bp)a
|
Maximum sequence divergenceb
|
GenBank accession no. | Reference | ||
|---|---|---|---|---|---|---|---|
| Exons | Introns | Exons | Introns | ||||
| arf | ADP-ribosylation factor | 280 | 179 | 0.0107 (0.0558) | 0.0608 (0.4280) | L25112 | 50 |
| H-anti | H antigen precursor | 237 | 171 | 0.0253 | 0.0760 | U20346 | 22 |
| ole | Delta-9 fatty acid desaturase | 331 | 93 | 0.0423 | 0.0538 | X85962 | 32 |
| tub1 | Alpha-tubulin | 52 | 226 | 0.0578 (0.1047) | 0.0734 (0.5349) | M28356 | 39 |
| ITS | Internal transcribed spacers + rRNA genes | 224 | 384 | 0.0045 (0.0134) | 0.0584 (0.1620) | U18363 | 8 |
Lengths vary depending on the number of insertions or deletions. Lengths listed here are based on the isolate H8 (G217B).
For arf, H-anti, ole, and tub1, the maximum values of nucleotide substitutions per nucleotide obtained from pairwise comparisons of all isolates in Table 2 were used, whereas those of ITS were from comparison of representative isolates of each group, i.e., H2, H9, H62, H68, H71, H81, H88, and H90. Numbers in parentheses are the maximum distances between H. capsulatum and Blastomyces dermatitidis.
Sequencing.
Automated sequencing was done with an ABI dye terminator cycle sequencing ready reaction kit and PCR primers in accordance with the recommendations of the manufacturer (Applied Biosystems Division, Perkin-Elmer, Foster City, Calif.). Sequences were generated from both strands and were edited and initially aligned with the SEQUENCE NAVIGATOR (v1.01; Applied Biosystems) software package, and the alignments were then optimized visually.
Data analysis.
Phylogenetic analyses (both parsimony and neighbor joining) were performed by using PAUP* 4.0.0d62, a prerelease version generously provided by D. Swofford, Smithsonian Institute of Natural History. Most-parsimonious (MP) trees were generated by the heuristic search procedure using 1,000 replications of the random addition sequence option. Nucleotide sites were equally weighted, with character state transformations treated as unordered and of equal cost. Insertions and deletions were excluded from the data set. Indices of support (bootstrap values) for internal branches were generated by 500 replications of the bootstrap procedure (29). Neighbor-joining (NJ) trees were generated by using a maximum-likelihood correction for multiple hits with a transition/transversion ratio of 2 (40). Base deletions were treated as missing data. The Kimura two-parameter distance option gave the same topology as the maximum-likelihood option. The maximum sequence divergence value of each locus (Table 3) and the average and maximum sequence divergence values of combined loci (Table 4) correspond to the maximum and average values in the matrices of pairwise mean distances (nucleotide substitutions per nucleotide) of all isolates in Table 2.
TABLE 4.
Within- and between-population sequence divergence values from combined data of arf, H-anti, ole, and tub1a
| Population | No. of genotypes/ isolatesb | Avg (maximum) distance between isolates
|
|||||
|---|---|---|---|---|---|---|---|
| NAm Hcc1 | NAm Hcc2 | Panama | SAm Hcc | Hc farciminosum | Hc duboisii | ||
| NAm Hcc1 | 2/5 | 0.0006 (0.0006) | |||||
| NAm Hcc2 | 10/13 | 0.0317 (0.0332) | 0.0016 (0.0038) | ||||
| Panama Hcc | 1/3 | 0.0218 (0.0230) | 0.0267 (0.0238) | 0.0000 (0.0000) | |||
| SAm Hcc | 16/18 | 0.0282 (0.0329) | 0.0286 (0.0345) | 0.0226 (0.0269) | 0.0162 (0.0281) | ||
| Hc farciminosum | 1/3 | 0.0289 (0.0301) | 0.0293 (0.0294) | 0.0230 (0.0230) | 0.0149 (0.0243) | 0.0000 (0.0000) | |
| Hc duboisii | 3/4 | 0.0288 (0.0313) | 0.0337 (0.0351) | 0.0201 (0.0211) | 0.0257 (0.0307) | 0.0256 (0.0268) | 0.0055 (0.0102) |
Average (maximum) distances (nucleotide substitutions per nucleotide) between isolates, obtained with PAUP* 4.0.0d62, are listed.
Number of unique genotypes and number of isolates in each population.
RESULTS
Amplification and DNA sequence analysis of nuclear gene loci.
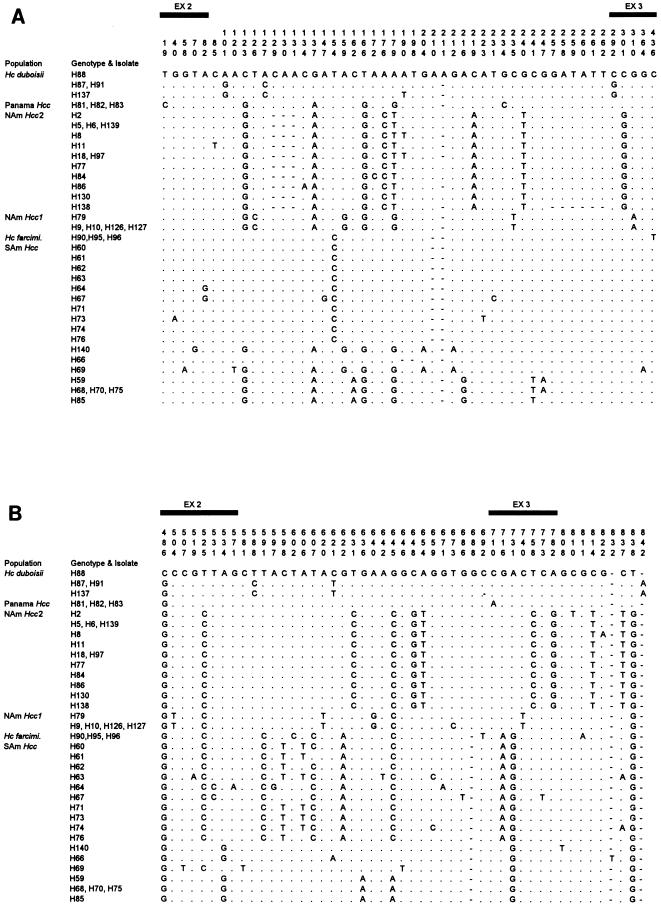
Forty-six isolates of H. capsulatum representing the three varieties taken from clinical and environmental sources of diverse geographical origin were used in this study (Table 2). DNA sequences of published protein coding genes of H. capsulatum were used to design PCR primers. Primers were located in exons to amplify exons as well as flanking introns. The ITS region in the rRNA array was also examined. To select genes with sufficient variation to address questions of Histoplasma evolution with statistical confidence, a representative isolate of each variety and geographical group was chosen for testing. Each candidate gene was PCR amplified from the tester isolates. DNA sequences of successfully amplified products were then determined. DNA sequence divergence values are shown in Table 3. Genes for proteins arf (50), H-anti (22), ole (32), and tub1 (39) contained moderate levels (3.0 to 7.0% substitution per nucleotide at maximum) of DNA polymorphisms, which were useful for phylogenetic analysis of H. capsulatum varieties. The ITS region in the rRNA gene array is widely used for taxonomic and phylogenetic studies of fungi at subgenus or subspecies level (8, 70). However, DNA polymorphism in the ITS region was not prevalent enough to resolve the varieties of H. capsulatum. The four protein genes were examined further for phylogenetic study by obtaining sequences for each gene from all 46 isolates (Table 2). Combining DNA sequence data for the four loci gave us 1,577 aligned sites, of which 208 were variable (Fig. 1). Introns were more variable than exons in all genes examined (Table 3). The 46 H. capsulatum isolates fell into 33 unique multilocus genotypes. Isolates with identical genotypes were found for NAm Hcc1 (four of five isolates), Panama Hcc (all three isolates), and H. capsulatum var. farciminosum (all three isolates). Among 13 isolates of NAm Hcc2, 10 genotypes were found, which were very homogeneous with a maximum diversity of 0.38% nucleotide substitutions per nucleotide (Table 4). Isolate H79, taken from a striped skunk in the 1940s (27), grouped with the four NAm Hcc1 isolates, from which it differed by a single T-C transition in 1,577 bp of sequence. H79 is therefore the first nonhuman NAm Hcc1 isolate and predates by 20 years what had been thought to be the initial class 1 isolate, the Downs strain (33). Among the four H. capsulatum var. duboisii isolates, three genotypes with a maximum divergence of 1.02% were found. The SAm Hcc isolates were the most variable. Of 18 isolates examined, 16 unique genotypes were identified, and the maximum sequence divergence among the group is 2.81%.
FIG. 1.
Polymorphic sites in combined data of arf (A), H-anti (B), ole (C), and tub1 (D) loci of H. capsulatum. The second column of each panel shows the name of the multilocus genotype, which was named after the isolate. When more than one isolate had same genotype, the isolate name with the smallest number was used for the genotype. The sequence of H88 is used as the master sequence and only nucleotides that differ from H88 sequence are shown; otherwise, nucleotides are shown as dots. A hyphen (-) denotes a gap. The shaded bars represent the locations of exons.
Phylogenetic analyses of protein coding genes.
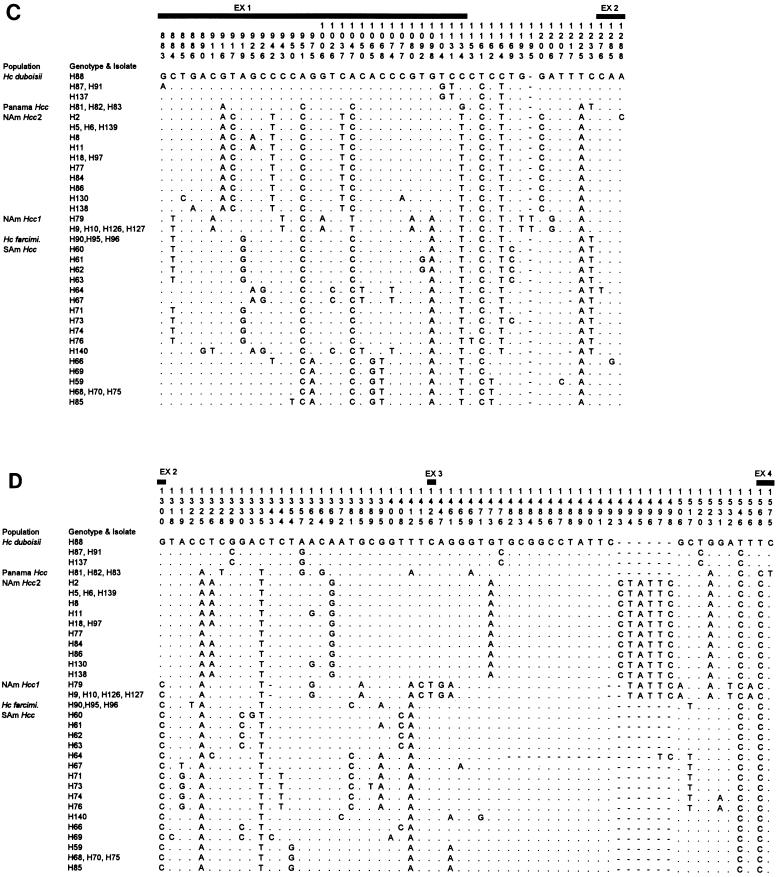
From 208 variable sites in the aligned 1,577 bp DNA, all gaps, of which there were 36, and all uninformative sites, of which there were 58, were excluded. The total of 114 informative sites were distributed among the protein coding genes as follows: 23 positions in arf, 27 in H-anti, 33 in ole, and 31 in tub1. The data for each gene were used with a parsimony analysis to analyze the phylogenetic relationships of the 33 genotypes (Fig. 2a through d).
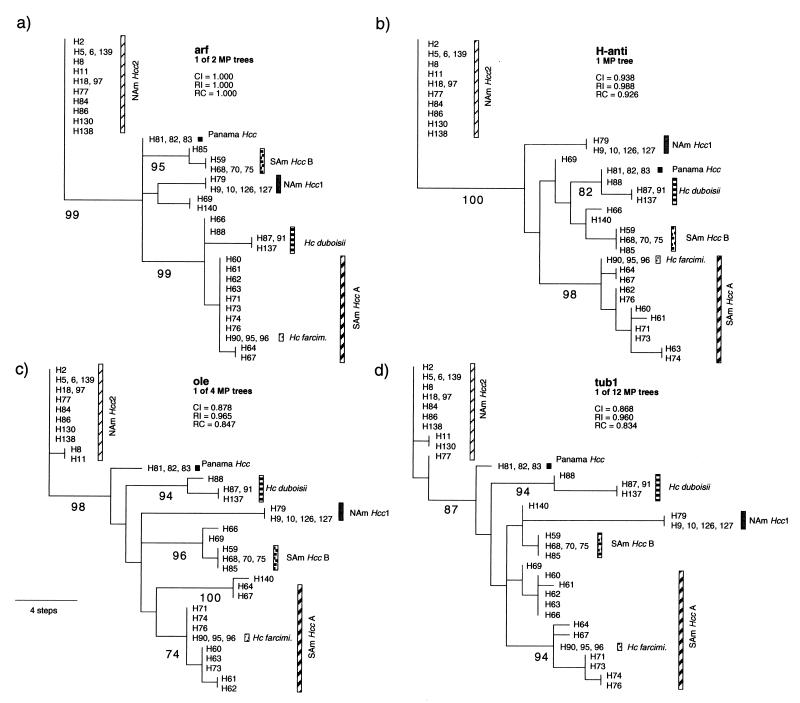
FIG. 2.
A single MP tree resulting from analysis of DNA sequences of the 33 multilocus genotypes from each of the four gene regions sequenced. Isolates with an identical genotype are shown together with the representative isolate for each of the 33 genotypes. CI, consistency index; RI, retention index; RC, rescaled consistency index. Numbers below branches represent indices of support based on 500 bootstrap replications of the parsimony procedure. Only values ≥70% are shown. Branch lengths are proportional to the numbers of changes in informative characters between nodes (scale at the lower left). SAm Hcc A, South American H. capsulatum group A; SAm Hcc B, South American H. capsulatum group B; Hc farcimi., H. capsulatum farciminosum. Abbreviations of other groups are listed in Table 1.
Parsimony analysis showed that isolates in the groups NAm Hcc1, NAm Hcc2, Panama Hcc, and H. capsulatum var. duboisii formed clades with all loci, with the exception of the location of genotypes H81 and H88 (Fig. 2a to d). On the other hand, SAm Hcc genotypes did not form such distinct clades. The majority of the Colombian H. capsulatum var. capsulatum genotypes (H60 to -64, -67, -71, -73, -74, and -76) formed a cluster (SAm Hcc group A), and Colombian H. capsulatum var. capsulatum genotypes H59, H68, and the Argentine genotype H85 formed the other cluster (SAm Hcc group B). Surprisingly, the horse pathogen H. capsulatum var. farciminosum was a member of SAm Hcc group A. South American H. capsulatum var. capsulatum isolates H66, H69, and H140 were distant from the two major SAm Hcc groups.
We attempted to root these parsimony trees with the fungal species thought to be the closest relatives of H. capsulatum, Blastomyces dermatitidis and Paracoccidioides brasiliensis (11, 37). Of the four protein genes, arf and tub1 genes from B. dermatitidis and P. brasiliensis were successfully amplified with primers designed based on H. capsulatum sequences. However, DNA sequences of both B. dermatitidis and P. brasiliensis appeared to be very distant from H. capsulatum (Table 3). We were unable to align the DNA sequences of B. dermatitidis and P. brasiliensis with those of H. capsulatum without significant ambiguity. Therefore, we could not use these sequences to locate the root on the tree of H. capsulatum varieties and populations. Nonetheless, it is noteworthy that no matter where the tree is rooted, H. capsulatum var. capsulatum cannot form a monophyletic group; that is, no clade can be found that contains all H. capsulatum var. capsulatum isolates, and only H. capsulatum var. capsulatum isolates.
We then examined the possibility of combining the data for the four protein coding genes into one phylogenetic analysis to improve the resolution. It is useful in phylogenetic analysis to test the congruence of separate data sets before combining them because incongruence of gene phylogenies may indicate problematic data sets (41, 58). However, when multiple individuals of a species are included in a phylogenetic analysis, incongruence among gene genealogies may be expected due to recombination (3). With this caveat in mind, we investigated congruence by two related methods, incongruence length difference (28, 54) and the partition homogeneity test (34, 41).
Incongruence length difference (I) was calculated as
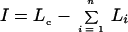 |
where Lc = the tree length of the summed data and Li = the tree length for the ith gene data set. I varies from 0 when all gene phylogenies are congruent to large values when they are incongruent. For the 33 genotypes given in Table 5, I = 35, a much larger value than the sum of homoplasy for the four gene phylogenies (12) and one that is indicative of incongruence.
TABLE 5.
Observed and minimum MP tree lengths for each of four loci among all groups used in this study
| Locus | Observed MP tree length | Minimum possible MP tree length | Excess steps or homoplasy |
|---|---|---|---|
| arf | 24 | 24 | 0 |
| H-anti | 32 | 30 | 2 |
| ole | 41 | 36 | 5 |
| tub1 | 38 | 33 | 5 |
| All four genes | 170 (Lc) | 135 (sum of Li) | 35 (I) |
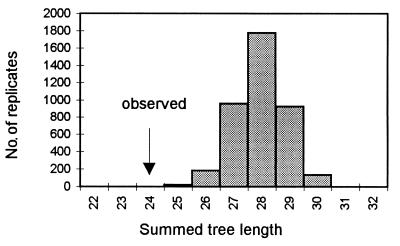
The partition homogeneity test compares the sum of lengths of the gene phylogenies to the distribution of such sums for 500 data sets where the variable positions for all the genes have been resampled without replacement to shuffle the sites among the genes while keeping the number of variable sites per gene constant. The null hypothesis is that all gene genealogies are congruent, and swapping sites among the gene data sets will not introduce homoplasy nor lead to longer trees. With Histoplasma, the sum of tree lengths for observed gene phylogenies (135) was significantly smaller than the sum for any of 500 shuffled data sets (range, 151 to 163; mode, 158; P < 0.002), demonstrating incongruence among the gene phylogenies (Fig. 3).
FIG. 3.
Partition homogeneity test in the total data set comparing the observed summed tree lengths with the distribution of summed tree lengths calculated for 500 randomized data sets.
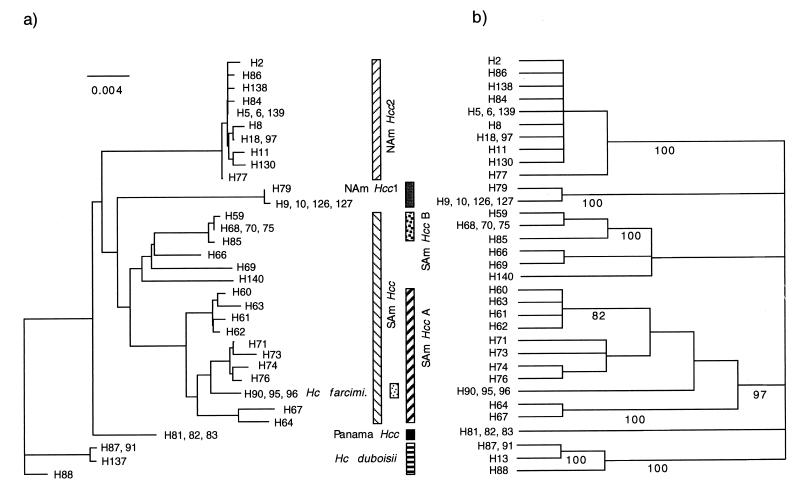
Given that H. capsulatum has been shown to recombine in nature (17), incongruence at the species level was to be expected. The next step was to determine if the incongruence lay above or at the species level. A neighbor-joining and a parsimony tree were constructed using the combined data set (Fig. 4a and b). As has been seen for the individual gene trees, the six clades, NAm Hcc 1, NAm Hcc 2, H. capsulatum var. duboisii, Panama Hcc, SAm Hcc group A, and SAm Hcc group B are distinct and are supported by high bootstrap values ranging from 97 to 100%. Because the locations of H66, H69, and H140 were inconsistent in individual protein gene trees, they were unresolved in the strict consensus tree of the combined data set. In the SAm Hcc group A clade, some of the internal branches were collapsed, and only two of the branches are supported by bootstrap values above 70%. Therefore, we conclude that the deep and well-supported branches are congruent among the gene trees and lead to the species, whereas the shallow and poorly supported branches are incongruent and lie within the species. Under this interpretation, all isolates are accommodated in the six clades, except H66, H69, and H140, which we provisionally have attached to the South American H. capsulatum var. capsulatum group B (Fig. 4).
FIG. 4.
(a) NJ tree from analysis of combined data of the four loci. Branch lengths are proportional to distance. (b) Strict consensus of 160 MP trees derived from analysis the same data of the four loci. Numbers below branches represent indices of support based on 500 bootstrap replications of the parsimony procedure; only values ≥70% are shown. Trees were rooted using H. capsulatum var. duboisii as the outgroup taxon based on the morphology of pathogenic yeasts.
SAm Hcc group A has a recombining population structure.
The observed poor resolution in the combined gene tree indicates genetic recombination in the SAm Hcc group A clade. Let us examine, for example, isolates H60, H71, and H73. In the arf and H-anti trees, these three isolates share an identical set of parsimony informative sites (Fig. 2). In the ole tree, H60 and H73 share identical sites and are one step distant from H71. In contrast, in the tub1 tree, H71 and H73 are identical and seven steps away from H60. All genotypes in the SAm Hcc group A were subjected to parsimony analysis, and the values obtained for Lc, Li, and I are listed in Table 6, which gives an I value of 6. The large I value is an indication of sexual recombination in the SAm Hcc group A population. The partition homogeneity test was also carried out by reconstructing 4,000 randomized data sets (Fig. 5). The observed sum of gene genealogy (tree) lengths of 24 was significantly smaller than those calculated from randomized data (range, 24 to 30; mean and mode, 28; P < 0.0005). This feature further corroborates the occurrence of genetic recombination in SAm Hcc group A. Note that this result does not address recombination in any of the other clades, where there are too few data (due to too few isolates or too little variation) to obtain a significant result. Of course, previous research on NAm Hcc2 using arbitrary loci has supported recombination in this group (17).
TABLE 6.
Observed and minimum MP tree lengths for each of four loci among South American H. capsulatum var. capsulatum group A
| Locus | Observed MP tree length | Minimum possible MP tree length | Excess steps or homoplasy |
|---|---|---|---|
| arf | 1 | 1 | 0 |
| H-anti | 5 | 5 | 0 |
| ole | 9 | 9 | 0 |
| tub1 | 9 | 9 | 0 |
| All four genes | 30 (Lc) | 24 (sum of Li) | 6 (I) |
FIG. 5.
Partition homogeneity test in the South American H. capsulatum var. capsulatum group A comparing the observed summed tree lengths with the distribution of summed tree lengths calculated for 4,000 randomized data sets.
DISCUSSION
Speciation in H. capsulatum.
Phylogenetic analysis revealed that H. capsulatum var. capsulatum was not monophyletic. In each of the four protein gene trees, there were at least six distinct clades in the H. capsulatum isolates we examined: NAm Hcc1, NAm Hcc2, H. capsulatum var. duboisii, Panama Hcc, SAm Hcc group A, and SAm Hcc group B. No clear association between geographical groups of H. capsulatum var. capsulatum was observed. In the combined gene tree, each clade had a deep branch that was supported by a high bootstrap value (97 to 100%). The concordance between the four gene genealogies at the deep branches and their lengths suggests that these six groups have been isolated reproductively from each other for a long time. Hence, each of the six groups corresponds to a phylogenetic species. Phylogenetic species have been defined as the smallest clade grouping populations or lineages diagnosable by a unique combination of character states, in this case, shared, derived nucleotide substitutions (57). However, this definition could be too narrow if the gene upon which the species concept was based proved to be polymorphic in a randomly mating population, because individuals would be grouped by alleles. Our use of four different gene fragments guards against this possibility by using branches that are congruent in the four gene trees to group individuals into species and by recognizing that conflict among gene genealogies would be expected to occur within recombining species (4, 6).
The North American H. capsulatum populations.
It was found that the genetic distance between the NAm Hcc1 and NAm Hcc2 is comparable to or larger than that of any other pair of groups, e.g., NAm Hcc2 versus H. capsulatum var. duboisii or NAm Hcc2 versus H. capsulatum var. farciminosum (Table 4). The two groups showed no close relationship in any of the four loci examined. This implies that the NAm Hcc1 and NAm Hcc2 are not historically closely related and that there is no genetic exchange between the groups. By comparison with Coccidioides immitis (43, 56), it seems certain that NAm Hcc1 and NAm Hcc2 have been genetically isolated for at least several million years (20 million years, assuming a substitution rate of third-base positions of μ = 10−9 bp/yr), far older than the date when humans first encountered H. capsulatum in the New World (no earlier than 65,000 years ago, and usually estimated to be about 12,000 years before the present) (16, 35, 36). In the laboratory however, H9 (NAm Hcc1) and H139 (NAm Hcc2) have been mated, and gymnothecia were observed (45, 48). The gymnothecia contained ascospores, but the ascospores did not look healthy and their viability was not determined (45). Carter et al. (17) showed that clinical isolates of NAm Hcc2 from Indianapolis formed a recombining population structure. Since NAm Hcc1 and Hcc2 have overlapping distributions and are sexually compatible, observation of hybrid genotypes in nature could be expected. However, there is no sign of hybridization in four isolates of NAm Hcc1 and two isolates of NAm Hcc2, all from St. Louis, Mo. It may be that interspecific mating does not occur in nature, or it may be that hybrid progeny are less fit than those from either species alone (12). Examining many more isolates may shed light on this problem.
Genetic diversities within both populations of NAm Hcc1 and Hcc2 were much smaller than that of SAm Hcc or H. capsulatum var. duboisii. This implies that each North American group has recently emerged from a small evolutionary bottleneck, in which alleles became fixed or nearly fixed in all loci. Rippon (61) suggested an association between starlings and the spread of H. capsulatum in North America. The saprobic phase of H. capsulatum lives on guano of bat and bird species. However, not all guano serves equally well as a substrate, and the guano of starlings (Sturnus vulgaris) is one of the infested sources of H. capsulatum. In North America, the areas in which H. capsulatum is most highly endemic are also the areas with the greatest concentrations of starlings (61). Starlings were introduced into New York from Europe in 1890 and spread westward (7, 20). The low genetic diversity of H. capsulatum in North America might be explained by the recent spread via starlings of H. capsulatum already in North America. Of course, the long branches separating North American and South American H. capsulatum var. capsulatum make it certain that these two groups were genetically isolated long before the introduction of starlings into North America.
The low-virulence race, NAm Hcc1, became conspicuous after the AIDS pandemic. So far, NAm Hcc1 has been isolated only from AIDS patients (64) and an 86-year-old woman (Downs isolate) (33). We have identified one additional strain, obtained from a striped skunk in the 1940s in Georgia, belonging to the NAm Hcc1, which suggests NAm Hcc1 existed in nature long before the AIDS pandemic, as had been suggested by Spitzer et al. (64). The absence of NAm Hcc1 isolates in collections of clinical isolates taken from patients with disseminated disease prior to the AIDS pandemic (68) suggests that these isolates seldom cause disease in otherwise healthy humans. As with NAm Hcc2, the genetic distance from NAm Hcc1 to isolates in any other clade is large.
H. capsulatum var. duboisii.
H. capsulatum var. duboisii is distinct from H. capsulatum var. capsulatum not only in epidemiology and medical manifestations but also in the morphology of the yeast phase (1). H. capsulatum var. duboisii was described originally as the species H. duboisii (26) but was redefined as a variety because the morphological and immunological differences between the two fungi were insufficient (18, 25). As with H. capsulatum var. capsulatum, H. capsulatum var. duboisii is also associated with bat guano (38). Kwon-Chung (44) demonstrated that the sexual state of H. capsulatum var. duboisii was identical to that of H. capsulatum var. capsulatum. Furthermore, she successfully obtained sexual crosses between isolates of H. capsulatum var. duboisii and the reciprocal mating types of NAm Hcc2 (H138 and H139). On agar media, the ascospores produced by the intervariety cross failed to germinate, but when the ascospore suspension was injected into mice, Histoplasma colonies were subsequently recovered from livers and spleens (47). Thus, H. capsulatum var. capsulatum and H. capsulatum var. duboisii were claimed to belong to a single biological species, in which the two varieties share a specific mate recognition system. However, Kwon-Chung (47) discussed the possibility that the progeny of the cross was an interspecies hybrid. In fact, the progeny isolates were sterile when backcrossed with the parents, suggesting that the hybrids had lost their sexual ability. As in other cases, the biological species concept is difficult to apply to allopatric populations (71), which would be the case when crossing NAm Hcc2 and H. capsulatum var. duboisii. Unlike sympatric species, allopatric species tend to lack mating suppression mechanisms and therefore are sexually compatible. Uniting taxa because they hybridize can lead to nonhistorical groups, which are problematic in speciation analysis (13, 71). Our data support Vanbreuseghem’s view (see reference 47) that H. capsulatum var. duboisii and H. capsulatum should be recognized as two independent species.
The South American H. capsulatum populations.
Isolates of SAm Hcc were extremely rich in polymorphisms, and most isolates fell into two well-supported clades. No significant differences in clinical manifestations of members of either group have been noted (52). The population genetic analysis revealed recombination in the SAm Hcc group A population. However, no recombinant between the SAm Hcc group A and B has yet been identified. As the Panama Hcc was very distant from the SAm Hcc population, there may be more clades or phylogenetic species existing in Central and South America.
DNA of H140 was isolated directly from the liver of an owl monkey that lived in a research facility in Maryland. The monkey was caught in the wild and was of Peruvian origin. An autopsy revealed massive numbers of budding yeast in the liver, but the yeast was unculturable. Several efforts to identify the yeast have been made by others (55). The morphology of the yeast resembled H. capsulatum var. duboisii and Loboa loboi. An immunohistochemistry assay was conducted, but the results were inconclusive. Our phylogenetic analysis, however, conclusively identified the yeast as H. capsulatum and furthermore demonstrated a close relationship of H140 to Colombian H. capsulatum. This result was consistent with the Peruvian origin of the monkey and provided evidence that the infection was not acquired in Maryland. Our PCR-based approach does not require a pure culture of the pathogen, but only a small amount of infected tissue, and it demonstrates how the phylogenetic approach can help us to understand epidemiology.
H. capsulatum var. farciminosum.
The most striking finding of this study is the phylogenetic position of H. capsulatum var. farciminosum. H. capsulatum var. farciminosum is deeply buried in the branch of SAm Hcc group A, both in the parsimony and NJ trees, looking as if it were an isolate of South American H. capsulatum var. capsulatum. H. capsulatum var. farciminosum is pathologically distinct; it infects horses and other Equidae and generally causes subcutaneous and ulcerated lesions of the skin. Primary infectious sites are thought to be skin injuries from harnesses, and infection through the pulmonary system is rare (46). H. capsulatum var. farciminosum was formerly described as an independent species but this assessment was changed to a variety of H. capsulatum due to the close morphological similarities of the mycelial and yeast forms (69). Antigenically, H. capsulatum var. farciminosum and H. capsulatum var. capsulatum are indistinguishable (65). We suggest that H. capsulatum var. farciminosum is not a separate taxon long adapted to Equidae but represents a recent infection of these animals caused by H. capsulatum var. capsulatum, presumably acquired in South America, transported to the Old World on an infected animal, and spread in the Old World from animal to animal via skin contact. Several observations pertain to this hypothesis. H. capsulatum var. farciminosum is prevalent in northern Egypt, where cases of histoplasmosis in humans are rare but do exist. Histoplasmin surveys showed that in Egypt and Israel, positive reaction of the skin test was infrequent or absent (2). However, Ajello et al. (2) successfully isolated H. capsulatum from soil samples from a bat-infested cave in Israel. These authors suggested the possibility that in these areas climatic conditions were not suitable for the proliferation and survival of H. capsulatum in the soil. The proposed scenario would explain why H. capsulatum var. farciminosum isolates from Egypt and India are genetically identical at the four gene sequences and more closely related to some South American isolates than to others. It would also explain why animals kept in crowded conditions, such as those at racehorse maintenance facilities, are likely to become infected (31). It would be valuable to challenge this scenario by including clinical and soil isolates from Egypt or Israel in future investigations.
Phylogeny for epidemiological studies.
The data presented here document the genetic differentiation of the different geographical and clinical groups of H. capsulatum. Each clade has many nucleotide positions that are unique shared characters in the language of phylogenetics, or fixed alleles in that of population genetics. This genetic differentiation indicates that the clades are phylogenetic species (5) and have the genetic differentiation expected of biological species (4). With the exception of the clade for H. capsulatum var. duboisii, members of the different clades do not show morphological differences. However, differences in pathogenicity in the case of members of the NAm Hcc1 clade indicate that phenotypic differentiation is following the genetic differentiation and we might expect that further differences will be found. The emergence of hosts with different susceptibilities due to the human immunodeficiency virus pandemic and voluntary immunosuppression and to the progressive intensification of international travel and immigration has increased the number of cases of histoplasmosis acquired in areas in which H. capsulatum is endemic by hosts not expected to contract this disease (51). From the clinical point of view, it may become increasingly important to identify pathogens to their genetical background and geographic origin. PCR-based phylogenetic analyses based on the genetic variation demonstrated in studies like ours can provide this identification. However, such identification and typing methods are only as good as the sampling of individual fungi and genes used to discover the genetically isolated groups or species (66). Acquisition of additional isolates in poorly sampled geographic locations will improve our ability to identify and type, as well as our knowledge of the world distribution of H. capsulatum.
In addition to H. capsulatum, several other fungal pathogens, such as Candida albicans, Cryptococcus neoformans, Aspergillus fumigatus, and Coccidioides immitis have become expanding threats due to immunosuppressive therapy and the AIDS pandemic. Genetic differentiation that correlates with geographic origin as well as allopatric species have been detected among populations of Coccidioides immitis (15, 43), H. capsulatum (42), and Cryptococcus neoformans var. gattii (10, 62). Conversely, genetic differentiation without geographic correlation has been seen in A. fumigatus (21, 60), Candida albicans (19, 67), and Cryptococcus neoformans var. neoformans (10, 30).
ACKNOWLEDGMENTS
We thank E. Keath, G. Kobayashi, J. McEwen, A. Restrepo, L. Wheat, P. Connolly, S. Moser, B. Hines, W. Dismukes, G. Miller, E. Bagagli, Z. Pires de Camargo, and K. Orle for supplying the isolates, DNA and associated clinical information; G. Koenig for technical assistance with fungal culture and DNA isolation; D. Geiser, S. Mack, and F. Harbinski for phylogenetic analysis; and J. Kwon-Chung for providing unpublished mating results.
Financial support for this work was provided by the National Institutes of Health (grant HL55953 to J.W.T.).
REFERENCES
- 1.Ajello L. Histoplasmosis—a dual entity: histoplasmosis capsulati and histoplasmosis duboisii. Ig Mod. 1983;79:3–30. [Google Scholar]
- 2.Ajello L, Kuttin E S, Beemer A M, Kaplan W, Padhye A. Occurrence of Histoplasma capsulatum Darling, 1906 in Israel, with a review of the current status of histoplasmosis in the Middle East. Am J Trop Med Hyg. 1977;26:140–147. doi: 10.4269/ajtmh.1977.26.140. [DOI] [PubMed] [Google Scholar]
- 3.Avise J C, Ball R M., Jr Principles of genealogical concordance in species concepts and biological taxonomy. Oxf Surv Evol Biol. 1990;7:45–67. [Google Scholar]
- 4.Avise J C, Wollenberg K. Phylogenetics and the origin of species. Proc Natl Acad Sci USA. 1997;94:7748–7755. doi: 10.1073/pnas.94.15.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum D A, Donoghue M J. Choosing among alternative “phylogenetic” species concepts. Syst Bot. 1995;20:560–573. [Google Scholar]
- 6.Baum D A, Shaw K L. Genealogical perspectives on the species problem. In: Hoch P C, Stephenson A G, editors. Experimental and molecular approaches to plant biosystematics. St. Louis, Mo: Missouri Botanical Garden; 1995. pp. 289–303. [Google Scholar]
- 7.Bent A C. Life histories of North American wagtails, shrikes, vireos and their allies, order Passeriformes. U.S. National Museum bulletin 197. U.S. Washington, D.C: Government Printing Office; 1950. [Google Scholar]
- 8.Berbee M L, Yoshimura A, Sugiyama J, Taylor J W. Is Penicillium monophyletic? An evaluation of phylogeny in the family Trichocomaceae from 18S, 5.8S and ITS ribosomal DNA sequence data. Mycologia. 1995;87:210–222. [Google Scholar]
- 9.Berliner M D. Primary subcultures of Histoplasma capsulatum. I. Macro and micro-morphology of the mycelial phase. Sabouraudia. 1968;6:111–118. [PubMed] [Google Scholar]
- 10.Boekhout T, van Belkum A, Leenders A C A P, Verbrugh H A, Mukamurangwa P, Swinne D, Scheffers W A. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int J Syst Bacteriol. 1997;47:432–442. doi: 10.1099/00207713-47-2-432. [DOI] [PubMed] [Google Scholar]
- 11.Bowman B H, White T J, Taylor J W. Human pathogenic fungi and their close nonpathogenic relatives. Mol Phylogenet Evol. 1996;6:89–96. doi: 10.1006/mpev.1996.0061. [DOI] [PubMed] [Google Scholar]
- 12.Brasier C M. The dynamics of fungal speciation. In: Rayner A D M, Brasier C M, Moore D, editors. Evolutionary biology of the fungi. Cambridge, United Kingdom: Cambridge University Press; 1987. pp. 83–95. [Google Scholar]
- 13.Brooks D R, McLennan D A. Phylogeny, ecology and behavior: a research program in comparative biology. Chicago, Ill: University of Chicago Press; 1991. [Google Scholar]
- 14.Burt A, Carter D A, Carter G L, White T J, Taylor J W. A safe method of extracting DNA from Coccidioides immitis. Fungal Genet Newsl. 1995;42:23. [Google Scholar]
- 15.Burt A, Dechairo B M, Koenig G L, Carter D A, White T J, Taylor J W. Molecular markers reveal differentiation among isolates of Coccidioides immitis from California, Arizona and Texas. Mol Ecol. 1997;6:781–786. doi: 10.1046/j.1365-294x.1997.00245.x. [DOI] [PubMed] [Google Scholar]
- 16.Cann R L. mtDNA and Native Americans: a southern perspective. Am J Hum Genet. 1994;55:7–11. [PMC free article] [PubMed] [Google Scholar]
- 17.Carter D A, Burt A, Taylor J W, Koenig G L, White T J. Clinical isolates of Histoplasma capsulatum from Indianapolis, Indiana, have a recombining population structure. J Clin Microbiol. 1996;34:2577–2584. doi: 10.1128/jcm.34.10.2577-2584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciferri R. Manuale di micologia media, Parte speciale. Vol. 2. Pavia, Italy: Casa Editrice Renzo Cortina; 1960. [Google Scholar]
- 19.Clemons K V, Feroze F, Holmberg K, Stevens D A. Comparative analysis of genetic variability among Candida albicans isolates from different geographic locales by three genotypic methods. J Clin Microbiol. 1997;35:1332–1336. doi: 10.1128/jcm.35.6.1332-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooke N T. The spread of the European starling in North America. U.S. Washington, D.C: Department of Agriculture; 1928. [Google Scholar]
- 21.Debeaupuis J P, Sarfati J, Chazalet V, Latge J P. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect Immun. 1997;65:3080–3085. doi: 10.1128/iai.65.8.3080-3085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deepe G S, Jr, Durose G G. Immunobiological activity of recombinant H antigen from Histoplasma capsulatum. Infect Immun. 1995;63:3151–3157. doi: 10.1128/iai.63.8.3151-3157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delacretaz J, Grigoriu D. Histoplasmose Africaine developpee sur une piqure d’insecte. Sabouraudia. 1972;10:24–25. [PubMed] [Google Scholar]
- 24.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drouhet E. Quelques aspect biologiques et mycologiques de l’histoplasmose. Pathol Biol (Paris) 1957;33:439–461. [Google Scholar]
- 26.Dubois A, Janssens P G, Brutsaert P. Un cas d’histoplasmose africaine avec une note mycologique sur Histoplasma duboisii n. sp., par R. Vanbreuseghem Ann Soc Belge Med Trop. 1952;32:569–584. [PubMed] [Google Scholar]
- 27.Emmons C W, Morlan H B, Hill E L. Histoplasmosis in rats and skunks in Georgia. Public Health Rep. 1949;64:1423–1430. [PubMed] [Google Scholar]
- 28.Farris J S, Kallersjo M, Kluge A G, Bult C. Testing significance of incongruence. Cladistics. 1995;10:315–319. [Google Scholar]
- 29.Felsenstein J. Confidence limits on phylogenies: an approach to using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 30.Franzot S P, Hamdan J S, Currie B P, Casadevall A. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J Clin Microbiol. 1997;35:2243–2251. doi: 10.1128/jcm.35.9.2243-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabal M A, Hassan F K, Siad A A, Karim K A. Study of equine histoplasmosis farciminosi and characterization of Histoplasma farciminosum. Sabouraudia. 1983;21:121–127. doi: 10.1080/00362178385380191. [DOI] [PubMed] [Google Scholar]
- 32.Gargano S, Di Lallo G, Kobayashi G S, Maresca B. A temperature-sensitive strain of Histoplasma capsulatum has an altered delta 9-fatty acid desaturase gene. Lipids. 1995;30:899–906. doi: 10.1007/BF02537480. [DOI] [PubMed] [Google Scholar]
- 33.Gass M, Kobayashi G S. Histoplasmosis: an illustrative case with unusual vaginal and joint involvement. Arch Dermatol. 1969;100:724–727. doi: 10.1001/archderm.100.6.724. [DOI] [PubMed] [Google Scholar]
- 34.Geiser D M, Pitt J I, Taylor J W. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc Natl Acad Sci USA. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbons A. The peopling of the Americas. Science. 1996;274:31–33. doi: 10.1126/science.274.5284.31. [DOI] [PubMed] [Google Scholar]
- 36.Greenberg J H, Turner C G, Zegura S L. The settlement of the Americas: a comparison of the linguistic, dental and genetic evidence. Curr Anthropol. 1986;27:483–496. [Google Scholar]
- 37.Guého E, Leclerc M C, de Hoog G S, Dupont B. Molecular taxonomy and epidemiology of Blastomyces and Histoplasma species. Mycoses. 1997;40:69–81. doi: 10.1111/j.1439-0507.1997.tb00191.x. [DOI] [PubMed] [Google Scholar]
- 38.Gugnani H C, Muotoe-Okafor F A, Kaufman L, Dupont B. A natural focus of Histoplasma capsulatum var. duboisii is a bat cave. Mycopathologia. 1994;127:151–157. doi: 10.1007/BF01102915. [DOI] [PubMed] [Google Scholar]
- 39.Harris G S, Keath E J, Medoff J. Characterization of alpha and beta tubulin genes in the dimorphic fungus Histoplasma capsulatum. J Gen Microbiol. 1989;135:1817–1832. doi: 10.1099/00221287-135-7-1817. [DOI] [PubMed] [Google Scholar]
- 40.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 41.Huelsenbeck J P, Bull J J, Cunningham C W. Combining data in phylogenetic analysis. Trends Ecol Evol. 1996;11:152–158. doi: 10.1016/0169-5347(96)10006-9. [DOI] [PubMed] [Google Scholar]
- 42.Keath E J, Kobayashi G S, Medoff G. Typing of Histoplasma capsulatum by restriction fragment length polymorphisms in a nuclear gene. J Clin Microbiol. 1992;30:2104–2107. doi: 10.1128/jcm.30.8.2104-2107.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koufopanou V, Burt A, Taylor J W. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc Natl Acad Sci USA. 1997;94:5478–5482. doi: 10.1073/pnas.94.10.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon-Chung K J. Perfect state (Emmonsiella capsulata) of the fungus causing large-form African histoplasmosis. Mycologia. 1975;67:980–990. [PubMed] [Google Scholar]
- 45.Kwon-Chung, K. J. 1998. Personal communication.
- 46.Kwon-Chung K J, Bennett J E. Medical mycology. Malvern, Pa: Lea & Febiger; 1992. [Google Scholar]
- 47.Kwon-Chung K J, Hill W B. Virulence of the two mating types of Emmonsiella capsulata and the mating experiments with Emmonsiella capsulata and Emmonsiella capsulata var. duboisii. In: Vanbreuseghem R, Vroey C D, editors. Sexuality and pathogenicity of fungi. New York, N.Y: Masson; 1981. pp. 48–56. [Google Scholar]
- 48.Kwon-Chung K J, Weeks R J, Larsh H W. Studies on Emmonsiella capsulata (Histoplasma capsulatum). II. Distribution of the two mating types in 13 endemic states of the United States. Am J Epidemiol. 1974;99:44–49. doi: 10.1093/oxfordjournals.aje.a121583. [DOI] [PubMed] [Google Scholar]
- 49.Leclerc M C, Philippe H, Guého E. Phylogeny of dermatophytes and dimorphic fungi based on large subunit ribosomal RNA sequence comparisons. J Med Vet Mycol. 1994;32:331–341. doi: 10.1080/02681219480000451. [DOI] [PubMed] [Google Scholar]
- 50.Lodge J K, Johnson R L, Weinberg R A, Gordon J I. Comparison of myristoyl-CoA:protein N-myristoyltransferases from three pathogenic fungi: Cryptococcus neoformans, Histoplasma capsulatum, and Candida albicans. J Biol Chem. 1994;269:2996–3009. [PubMed] [Google Scholar]
- 51.Manfredi R, Mazzoni A, Nanetti A, Chiodo F. Histoplasmosis capsulati and duboisii in Europe: the impact of the HIV pandemic, travel and immigration. Eur J Epidemiol. 1994;10:675–681. doi: 10.1007/BF01719280. [DOI] [PubMed] [Google Scholar]
- 52.McEwen, J., and A. Restrepo. 1998. Personal communication.
- 53.Medoff G, Maresca B, Lambowitz A M, Kobayashi G, Painter A, Sacco M, Carratu L. Correlation between pathogenicity and temperature sensitivity in different strains of Histoplasma capsulatum. J Clin Investig. 1986;78:1638–1647. doi: 10.1172/JCI112757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mickevich M F, Farris J S. The implications of congruence in Menidia. Syst Zool. 1981;30:351–370. [Google Scholar]
- 55.Miller, G. 1998. Personal communication.
- 56.Nei M. Molecular evolutionary genetics. New York, N.Y: Columbia University Press; 1987. [Google Scholar]
- 57.Nixon K C, Wheeler Q D. An amplification of the phylogenetic species concept. Cladistics. 1990;6:211–223. [Google Scholar]
- 58.O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 59.Panciera R J. Histoplasmic (Histoplasma capsulatum) infection in a horse. Cornell Vet. 1969;59:306–312. [PubMed] [Google Scholar]
- 60.Rinyu E, Varga J, Ferenczy L. Phenotypic and genotypic analysis of variability in Aspergillus fumigatus. J Clin Microbiol. 1995;33:2567–2575. doi: 10.1128/jcm.33.10.2567-2575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rippon J W. Medical mycology. W. B. Philadelphia, Pa: Saunders Company; 1988. [Google Scholar]
- 62.Sorrell T C, Chen S C A, Ruma P, Meyer W, Pfeiffer T J, Ellis D H, Brownlee A G. Concordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting. J Clin Microbiol. 1996;34:1253–1260. doi: 10.1128/jcm.34.5.1253-1260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spitzer E D, Lasker B A, Travis S J, Kobayashi G S, Medoff G. Use of mitochondrial and ribosomal DNA polymorphisms to classify clinical and soil isolates of Histoplasma capsulatum. Infect Immun. 1989;57:1409–1412. doi: 10.1128/iai.57.5.1409-1412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spitzer E D, Keath E J, Travis S J, Painter A A, Kobayashi G S, Medoff G. Temperature-sensitive variants of Histoplasma capsulatum isolated from patients with acquired immunodeficiency syndrome. J Infect Dis. 1990;162:258–261. doi: 10.1093/infdis/162.1.258. [DOI] [PubMed] [Google Scholar]
- 65.Standard P G, Kaufman L. Specific immunological test for the rapid identification of members of the genus Histoplasma. J Clin Microbiol. 1976;3:191–199. doi: 10.1128/jcm.3.2.191-199.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor J W, Geiser D M, Burt A, Koufopanou V. The evolutionary biology and population genetics underlying fungal strain typing. Clin Microbiol Rev. 1999;12:126–146. doi: 10.1128/cmr.12.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsang P C, Samaranayake L P, Philipsen H P, McCulloug M, Reichart P A, Schmidt-Westhausen A, Scully C, Porter S R. Biotypes of oral Candida albicans isolates in human immunodeficiency virus-infected patients from diverse geographic locations. J Oral Pathol Med. 1995;24:32–36. doi: 10.1111/j.1600-0714.1995.tb01126.x. [DOI] [PubMed] [Google Scholar]
- 68.Vincent R D, Goewert R, Goldman W E, Kobayashi G S, Lambowitz A M, Medoff G. Classification of Histoplasma capsulatum isolates by restriction fragment polymorphisms. J Bacteriol. 1986;165:813–818. doi: 10.1128/jb.165.3.813-818.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weeks R J, Padhye A A, Ajello L. Histoplasma capsulatum variety farciminosum: a new combination for Histoplasma farciminosum. Mycologia. 1985;77:964–970. [Google Scholar]
- 70.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M H, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 71.Zink R M, McKitrick M C. The debate over species concepts and its implications for ornithology. Auk. 1995;112:701–719. [Google Scholar]