Abstract
Eukaryotic cells express a myriad of circular RNAs (circRNAs), many of them displaying tissue-specific expression patterns. They arise from linear precursor RNAs in which 5′ and 3′ ends become covalently ligated. Given these features, biochemical and computational approaches traditionally used to study linear RNA must be adapted for analysis of circular RNAs. Such circRNA-specific methodologies are allowing the systematic identification of circRNAs and the analysis of their biological functions. Here, we review the resources and molecular methods currently utilized to quantify circRNAs, visualize their distribution, identify interacting partners, and elucidate their function. We discuss the challenges of analyzing circRNAs and propose alternative approaches for studying this unique class of transcripts.
This article is characterized under:
RNA Structure and Dynamics > RNA Structure, Dynamics, and Chemistry
RNA Methods > RNA Analyses in vitro and In Silico
RNA Methods > RNA Analyses in Cells
Keywords: backsplice junction, circRNAs, divergent primers, ribonucleoprotein complex
1 |. INTRODUCTION
The ENCODE Project Consortium has discovered that while >80% of the human genome is transcribed, only ~2% of the transcribed RNAs are translated into proteins, indicating that vast portions of RNA are noncoding and may play regulatory roles (Atianand & Fitzgerald, 2014). Much of this noncoding (nc)RNA is linear, including ribosomal (r)RNAs, long noncoding (lnc)RNAs, transfer (t)RNAs, and micro (mi)RNAs. However, a vast class of ncRNAs with a circular structure (circRNAs) has emerged over the past few years through high-throughput RNA sequencing (RNA-seq) coupled with novel bioinformatic analyses. Circular RNAs are ubiquitous and highly abundant (tens of thousands in a given cell type), display tissue specificity, and are more evolutionarily conserved than lncRNAs (Memczak et al., 2013; Rybak-Wolf et al., 2015; Salzman, Chen, Olsen, Wang, & Brown, 2013).
Most circRNAs are believed to be generated from precursor (pre-)RNAs through backsplicing of exonic sequences involving the canonical splicing machinery or from intronic lariat precursors (Cocquerelle, Mascrez, Hetuin, & Bailleul, 1993; Jeck et al., 2013; Kristensen et al., 2019; Liang & Wilusz, 2014; T. Li et al., 2018; Nigro et al., 1991). Due to the lack of free ends, circRNAs are substantially more stable than their linear counterparts (Enuka et al., 2016). Although the molecular functions of only a small fraction of circRNAs have been elucidated to-date (discussed in Section 1.2), they generally influence gene expression patterns by binding proteins and nucleic acids. By affecting gene expression programs, circRNAs modulate key cellular processes, including proliferation, differentiation, senescence, innate immune response, and the maintenance of stem cell pluripotency (Bachmayr-Heyda et al., 2015; Gu, Li, Jin, Liu, & Wei, 2017; C. X. Liu et al., 2019; Panda et al., 2017; Yu et al., 2017). Accordingly, circRNAs have been implicated in several pathologies, including cancer, Alzheimer’s disease, Parkinson’s disease, cardiovascular disease, and atherosclerosis (Floris, Zhang, Follesa, & Sun, 2017; Holdt et al., 2016; Lukiw, 2013; D. Yang, Yang, & Yang, 2018; Zhao & Shen, 2017). In addition, as efforts intensify to find new biomarkers for a range of conditions, circRNAs are emerging as promising candidates (Y. Li et al., 2015). Here, we review the current analytical tools and molecular biology approaches available to investigate circRNA levels, distribution, sequence, and interaction partners.
1.1 |. Biogenesis
There has been substantial progress in elucidating the mechanisms of circRNA biogenesis (Jeck et al., 2013; Liang & Wilusz, 2014; T. Li et al., 2018; Kristensen et al., 2019). CircRNAs can be generated from exons (ecircRNAs), introns (ciRNAs) or a combination of both (eiciRNAs) (Zhao & Shen, 2017). Spliceosome-dependent backsplicing uses the canonical spliceosome machinery to join the downstream 5′ donor site of an exon and the upstream 3′ acceptor site (L. L. Chen & Yang, 2015; Vicens & Westhof, 2014). Intron-pairing and lariat-driven circularization rely on reverse complementary motifs of introns to promote the circularization through alternative 5′ to 3′ splicing (Ivanov et al., 2015; X. O. Zhang et al., 2014). In addition, circularization can also be mediated by RNA-binding proteins (RBPs) forming circRNAs under particular conditions or stimuli. These RBPs bind to specific RNA sequences and bring donor—acceptor ends in close proximity, enabling circularization (Ashwal-Fluss et al., 2014; Errichelli et al., 2017; X. Li et al., 2017).
1.2 |. Functions
1.2.1 |. microRNA sponge
circRNAs that possess binding sites for microRNAs can function as competing endogenous RNAs (ceRNAs), as they can act as microRNA sponges and thereby prevent microRNAs from binding to target mRNAs. One of the earliest circRNAs identified, ciRS-7, possesses over 50 binding sites for miR-7, while the mouse circRNA sex-determining region Y, Sry, has 16 binding sites for miR-138; by binding to these microRNAs, ciRS-7 and Sry reduced miRNA interactions with their mRNA targets (Hansen et al., 2013). Other circRNAs with similar functions have been described, including circHIPK3, which can potentially sponge miR-124, miR-30a-3p, and miR-558 (Hansen et al., 2013; Y. Li et al., 2017; Shan et al., 2017; Zheng et al., 2016). By sponging microRNAs, several circRNAs have been found to modulate cell growth, proliferation, and differentiation (Hansen et al., 2013).
1.2.2 |. Protein sponge
CircRNAs can also bind proteins and these interactions can modulate RBP activity. CircMbl acts as a sponge for muscle blind (MBL) protein, an essential factor for the alternative splicing of Mbl pre-mRNA, reducing Mbl pre-mRNA alternative splicing. When MBL is abundant, a feedback loop mechanism is triggered whereby Mbl pre-mRNA preferentially yields circMbl, which in turn binds to MBL and decreases its bioavailability (Ashwal-Fluss et al., 2014). In another example, HuR binding to circPABPN1 prevents HuR from binding PABPN1 mRNA, in turn leading to a reduction in PABPN1 production and diminished proliferation (Abdelmohsen et al., 2017). Recently, endogenous circRNAs were found to form 16–26 bp imperfect RNA duplexes that bind the innate immune dsRNA receptor (PKR) and repress its activity (C. X. Liu et al., 2019). These findings further support the notion that circRNAs can regulate protein homeostasis.
1.2.3 |. Transcriptional regulation
Some nuclear circRNAs, particularly eiciRNAs, can interact with RNA Pol II at the promoter region of select host genes. Illustrating this function, circEIF3J and circPAIP2 bind the U1 snRNP in the nucleus (Z. Li et al., 2015), and the resulting complex then interacts with RNA Pol II in the promoter region of the host genes (EIF3J and PAIP2), promoting transcription.
1.2.4 |. Translation potential
There is increasing evidence that some circRNAs can be translated partially. For example, circZNF609 in mammalian myoblasts and circMbl in fruit fly were recently shown to have coding potential (Legnini et al., 2017; M. Zhang et al., 2018). Each circRNA has an IRES (internal ribosome entry site) through which ribosomes translate segments of the circRNA independently of the 5′-cap required for linear RNA translation. In addition, Y. Yang et al. (2017) reported that N6-methyladenosine (m6A) was capable of mimicking an IRES to trigger the initiation of translation. In a more high-throughput manner, a translational landscape analysis of the human heart has identified 40 circRNAs encoding novel microproteins from 39 genes (van Heesch et al., 2019).
2 |. APPROACHES TO STUDY circRNAs
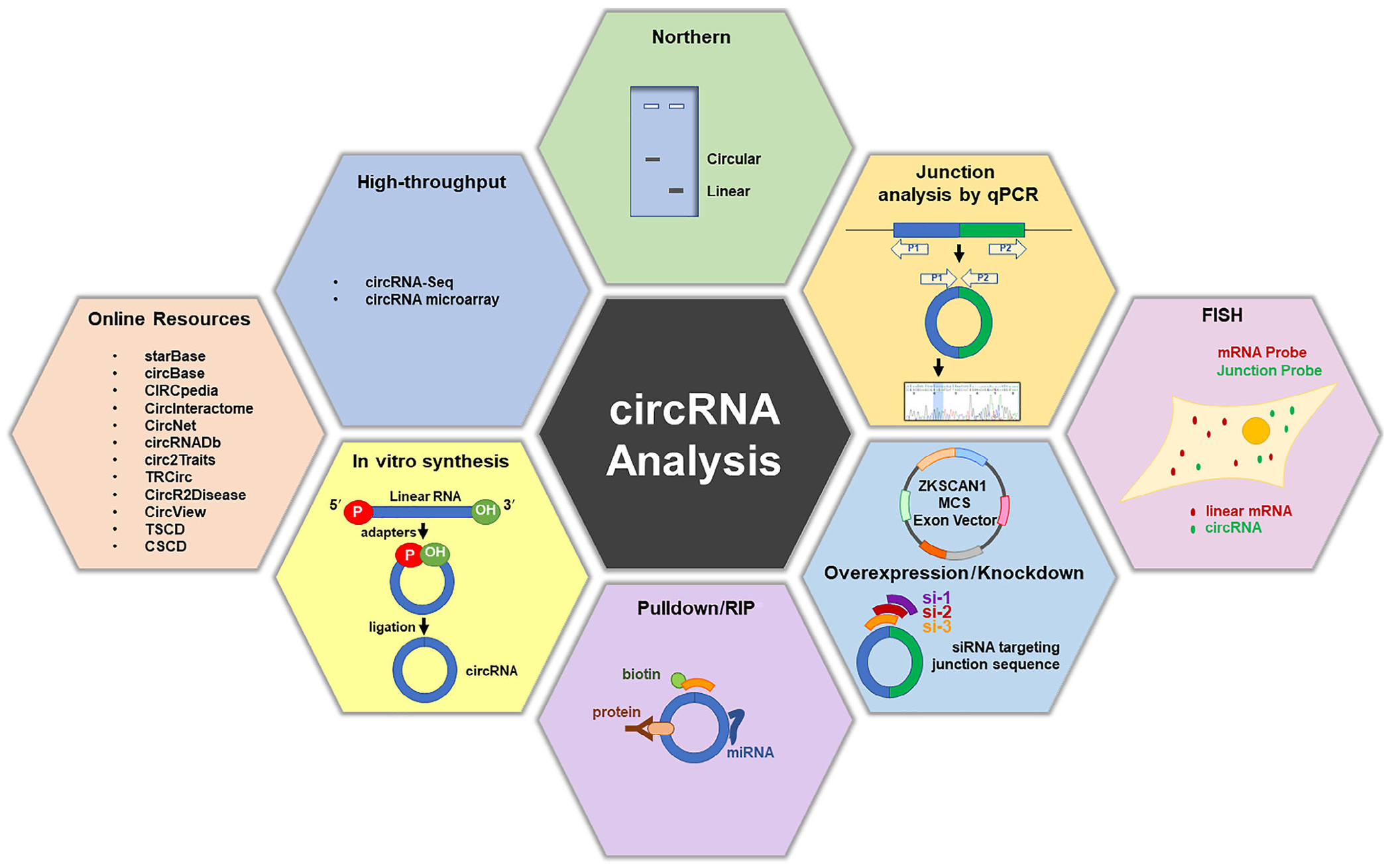
With increasing need to investigate circRNAs with depth and precision, it has become essential to utilize tools and methodologies that investigate accurately circRNA sequence, length, subcellular localization, biological function, disease implication, interacting molecules, and therapeutic potential. In this section we summarize the resources and techniques available at present to study circRNAs, and we discuss the strengths and limitations of each method.
2.1 |. Online circRNA resources
Several databases and tools have been established to provide key bioinformatic knowledge about circRNAs. For instance, circBase informs about circRNA identity and tissue specificity based on RNA-seq data (Memczak et al., 2013) and CircInteractome can be used to design divergent primers and siRNAs (Dudekula et al., 2016). CircInteractome, starBase, CircView, and TRcirc can identify potential interacting factors, including microRNAs, RBPs, and transcription factors (TFs) (Dudekula et al., 2016; Feng et al., 2018; J. H. Li, Liu, Zhou, Qu, & Yang, 2014; Tang et al., 2018). Similarly, circRNABase and CircNet can provide information on potential interactions with microRNAs based on global sequencing approaches like HITS-CLIP (crosslinking immunoprecipitation), PAR-CLIP, iCLIP, and CLASH (Y. C. Liu et al., 2016). Given that circRNAs are highly conserved, CIRCpedia can be exploited to find circRNA conservation in several species, cell types, tissues, and disease conditions (Dong, Ma, Li, & Yang, 2018). In addition, ecircRNAs potentially translated into proteins can be studied using CircRNADb. Given their potential link to diseases, several databases have been developed to explore circRNAs in diseases, including Circ2Traits, CSCD, TSCD, and CircR2Disease (Fan, Lei, Fang, Jiang, & Wu, 2018; Ghosal, Das, Sen, Basak, & Chakrabarti, 2013; Xia et al., 2018). These resources, compiled in Table 1, can be used to obtain valuable details for the experimental analysis of circRNAs, as indicated below.
TABLE 1.
Databases and online resources for circular RNA research
| Database | Website | References | Suitable for analysis of |
|---|---|---|---|
| circBase | http://www.circbase.org | Glazar, Papavasileiou, and Rajewsky, 2014 | circRNA expression levels and sequence |
| CircInteractome | http://circinteractome.nia.nih.gov | Dudekula et al., 2016 | Identification of interacting miRNAs and RBPs, design of divergent primers and siRNAs |
| CircNet | http://circnet.mbc.nctu.edu.tw/ | Y. C. Liu et al., 2016 | circRNA–miRNA interactions |
| CIRCpedia | http://www.picb.ac.cn/rnomics/circpedia | Dong et al., 2018 | circRNAs expressed in different species, cell types, and tissues |
| circRNADb | http://reprod.njmu.edu.cn/circrnadb | X. Chen et al., 2016 | Human exonic circRNAs |
| CircRNABase | http://www.hzrna.com/circrna-shujuku/circrnabase | Y. C. Liu et al., 2016 | circRNAs associated with other molecules |
| CircR2Disease | http://bioinfo.snnu.edu.cn/CircR2Disease/ | Fan et al., 2018 | circRNAs associated with disease conditions |
| CircView | http://gb.whu.edu.cn/CircView/ | Feng et al., 2018 | Identification of interacting miRNAs and RBPs |
| Circ2Traits | http://gyanxet-beta.com/circdb/ | Ghosal et al., 2013 | circRNAs associated with disease conditions |
| CSCD | http://gb.whu.edu.cn/CSCD/ | Xia et al., 2018 | circRNAs expressed, interacting miRNAs and RBPs, circRNAs with coding potential |
| starBase | http://starbase.sysu.edu.cn/ | J. H. Li et al., 2014 | circRNA–miRNA interactions |
| TSCD | http://gb.whu.edu.cn/TSCD | Xia et al., 2017 | Tissue-specific expression (human, mouse), circRNA markers of organogenesis |
| TRcirc | http://www.licpathway.net/TRCirc/ | Tang et al., 2018 | circRNA-transcription factor regulatory network |
2.2 |. circRNA sequencing
Advances in high-throughput circRNA sequencing (Circ-seq) analysis have enabled the identification of thousands of circRNAs. Starting from a pool of mixed RNAs, circRNAs can be highly enriched by digestion with RNase R to eliminate most linear transcripts while circRNAs are left intact (Jeck et al., 2013; Salzman et al., 2013). As many linear RNAs, particularly those with strong secondary structures, are refractory to RNase R digestion, Circ-seq can be improved by increasing the purity of the circRNA preparation through an additional polyadenylation and poly(A)+ RNA depletion (the RPAD technique; Panda, De, et al., 2017; Pandey, Rout, Das, Gorospe, & Panda, 2019) and by improving buffer conditions (Li+ instead of K+) to render RNAs containing G-quadruplexes and structured 3′ ends susceptible to degradation by RNase R (Xiao & Wilusz, 2019).
Pretreatments to enrich in circRNAs the RNA preparation are not necessary if sequencing is performed at high depth (i.e., 100–150 million reads), since the analysis mainly relies on junction reads that do not exist in canonically spliced linear transcripts. Although Circ-seq analysis detects circRNA junctions, it does not reveal the full sequence of a given circRNA nor the number of circRNA transcripts with the same junction within a given preparation (Szabo & Salzman, 2016). Other assays (below) have been employed to overcome these limitations.
It is important to note that circRNA annotations and analysis algorithms are still evolving. Current annotation strategies begin with the identification of the circularizing junction, and the body of the circRNA is then inferred from known sequences between the circularizing ends. Other annotation methods, based on de novo assembly of the circRNA, are less accurate, as the remnants of linear RNA molecules (possibly also circRNAs with shared junctions) can confound the assembly. In addition, the nomenclature of circRNAs is also not standardized. CircRNAs are computationally annotated and can be named by default after the circularizing junction; however, in some cases, they are named according to the parent gene annotation (e.g., circPVT1), regardless of the actual circularizing junction. CircBase and the cancer-specific circRNAs database (CSCD) have tried to give known circRNAs a unique circRNA ID, but this strategy is not widely used.
2.3 |. circRNA microarrays
Microarrays are useful tools for high-throughput analysis and multiple comparisons of the expression levels of specific circRNAs. Microarray probes specifically designed to recognize the junction sequences of circRNAs are immobilized on a solid support and incubated with mixed populations of circular and linear RNA. As with Circ-seq, microarray analysis typically includes prior treatment of the RNA sample with RNase R to reduce the presence of linear RNAs and enhance circRNA detection and quantification. This method was successfully employed for investigating several circRNAs, including circPABPN1, circEPSTI1, and circMTO1 (Abdelmohsen et al., 2017; B. Chen et al., 2018; Han et al., 2017). Microarray use for circRNA profiling has some drawbacks, including the facts that microarray platforms include only a limited number of known circRNAs, identify circRNAs based only on the junction sequences, do not inform on the internal sequence of a given circRNA, and cannot distinguish among circRNAs with shared junction sequences.
2.4 |. Northern blotting
To investigate circRNAs using Northern blot analysis, short probes can be employed that span the splice junction, although longer probes complementary to the entire circRNA can also be used, as shown for circHIPK3 (Zheng et al., 2016). In some cases, Northern blot analysis can be employed to distinguish between a circRNA and its linear counterpart, as electrophoresis through denaturing polyacrylamide gels mobilizes linear RNAs at the expected size, while circRNAs tend to run more slowly. Polyacrylamide pore spaces limit the sizes of circRNAs that can be analyzed on polyacrylamide gels to those in the range of 0.2–1 kb, although longer circRNAs can be studied using agarose gels (Tabak et al., 1988). With appropriate adaptations, Northern blot analysis can be used to assess circRNA size, isoforms, processing, sequence, and abundance (Pamudurti et al., 2017).
2.5 |. PCR-based analyses
2.5.1 |. RT-qPCR analysis
Circ-seq analysis provides essential information regarding circRNA junctions and annotation, but it is important to perform RT-qPCR (reverse transcription followed by quantitative polymerase chain reaction) analysis to validate high-throughput results. RT-qPCR analysis of circRNA is performed using divergent primers spanning the circRNA junction, which can be designed using online resources such as CircInteractome (Dudekula et al., 2016). In some cases, circRNAs extracted from cells must be enriched by RNase R treatment followed by RT-qPCR analysis, but often this step can be avoided if the circRNA is abundant, as divergent primers do not amplify linear RNA. Another advantage of RT-qPCR analysis is that the products can be sequenced to further verify the junction sequence and may reveal the full sequence of the circRNA of interest (Panda, Grammatikakis, Kim, et al., 2017). Furthermore, it offers a quantitative estimation of circRNA abundance under different conditions such as circRNA depletion, stress, and disease conditions (Huang et al., 2017; Y. Li et al., 2015; Panda, Abdelmohsen, & Gorospe, 2017) as well as circRNA abundance in different subcellular compartments. Among its limitations, RT-qPCR analysis is designed to be low-throughput.
2.5.2 |. Digital droplet PCR analysis
Digital droplet PCR (ddPCR) is a relatively new technology that can be used to quantify circRNA copy number. It partitions nucleic acids into nanoliter-sized droplets containing the target sequence for PCR amplification. The ratio of positive to negative droplets is analyzed to assess RNA concentration (Hindson et al., 2011; Quan, Sauzade, & Brouzes, 2018). X. Li, Yang, and Chen (2018) successfully used ddPCR to measure the absolute levels of circRNAs in the plasma of patients with gastric cancer relative to healthy controls, identifying two circRNAs, hsa_circ_0001017 and hsa_circ_0061276, as potential biomarkers of gastric cancer. Similarly, Panda, Grammatikakis, Kim, et al. (2017) calculated the number of circPVT1 molecules per cell by ddPCR analysis in senescent and proliferating fibroblasts. While ddPCR and conventional RT-qPCR analyses follow the same principles, ddPCR is more accurate, provides absolute numbers, and measures low-abundance RNAs, which may be particularly valuable for circRNA quantification. However, it requires special instrumentation, software, and proprietary reagents, and therefore its use is somewhat limited.
2.6 |. RNA fluorescence in situ hybridization
RNA fluorescence in situ hybridization (RNA-FISH) coupled with high-resolution microscopy is widely used to visualize the abundance and subcellular localization of RNA molecules in cells and tissues (Itzkovitz & van Oudenaarden, 2011). RNA-FISH probes targeting circRNA junctions have been used to detect and quantify several circRNAs, including cdr1AS and circHIPK3 (Jeck & Sharpless, 2014; Piwecka et al., 2017; Zheng et al., 2016). Combined with immunohistochemistry, FISH may also be used to assess the colocalization of circRNAs with proteins. A major limitation of this technique is that FISH probe design is restricted to the backspliced junction in order to avoid recognition of the linear RNA counterpart. Thus, unlike the strong FISH signals that often identify linear RNAs thanks to the use of multiple FISH probes recognizing long RNA segments, circRNA FISH signals tend to be weaker, as only one or a small number of FISH probes on the junction sequence can be used for visualization.
2.7 |. Depletion and overexpression of circRNAs
A major challenge to study circRNA function is reducing circRNA levels effectively, given that, in principle, only the circRNA junction can be targeted by the siRNA without affecting the linear mRNA counterpart. Accordingly, siRNAs/shRNAs are designed specifically to target the circRNA junction, evenly targeting each side of the junction or slightly on one side or the other (Figure 1). This strategy successfully knocked down circRNAs like circPVT1 in gastric cancer and circQKI in primary myoblasts (J. Chen et al., 2017; Legnini et al., 2017). While siRNAs can be manually designed around the junctions, the CircInteractome tool provides multiple options for circRNA siRNAs (Dudekula et al., 2016). In addition, the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology can be used to delete intronic complementary regions and potentially generate circRNA knockouts (Abudayyeh et al., 2017; Cox et al., 2017). Although most circRNA knockouts are not feasible because the linear counterparts would be disrupted, a Cdr1as knockout mouse was successfully created because the linear counterpart was not expressed (Piwecka et al., 2017).
FIGURE 1.
Schematic representation of the different approaches and tools to study circular RNA
Since circRNA-directed siRNAs may target the linear mRNA or other circRNAs with the same junction sequence, it is essential to perform gain-of-function approaches through overexpression. Backsplicing is triggered when intronic repeats, which usually complement each other, are base-paired to bring the intervening splice sites in close proximity (L. L. Chen & Yang, 2015; Ivanov et al., 2015). This strategy has been used to design expression vectors for the ectopic expression of circRNAs using the cellular splicing machinery (Kramer et al., 2015). For example, the vector pcDNA3.1(+) ZKSCAN1 has been successfully used to express specific circRNAs (Kramer et al., 2015) and was used to investigate the coding potential of circ-ZNF609 (Legnini et al., 2017). Additionally, circR (pcDNA3.1 His C vector backbone 6.3 kbp) contains an upstream intron and its reverse complementary sequence downstream (~800 bp) to promote circularization. The circRNA of interest and its endogenous flanking sequence, which contains the splicing sites (~200 bp), can be cloned between the two complementary introns. This strategy has been used to overexpress the brain-specific cdr1AS and the testis-specific circSry (Hansen et al., 2013). Collectively, these tools have been instrumental for studying circRNA functions.
2.8 |. circRNA affinity pulldown
Several studies have shown that circRNAs form ribonucleoprotein complexes with RBPs, TFs, and miRNAs (Panda, Grammatikakis, Munk, Gorospe, & Abdelmohsen, 2017). CircRNA pulldown can be performed using biotinylated antisense oligomers (ASOs) that specifically recognize the junction sequence of a target circRNA. Using streptavidin-coated beads, the biotinylated probes are pulled down together with interacting RNA molecules and associated proteins. The protein component of the pulldown can be analyzed by mass spectrometry and further assessed by Western blot analysis. The RNA component can be studied by sequencing and RT-qPCR analysis. When studying low-abundance circRNAs, overexpression followed by pulldown may increase the coverage of the interacting partners. Recently, this method was successfully used to investigate several circRNAs. For example, an ASO against circPVT1 (Panda, Grammatikakis, Kim, et al., 2017) revealed its interaction with let-7. Similarly, ASOs against circFoxo3 uncovered its association with ID1, E2F1, FAK, and HIF-1α (Du et al., 2016, 2017), circMbl ASOs revealed that the circRNA interacted with MBL1 (Ashwal-Fluss et al., 2014), and ASOs that pulled down circPABPN1 revealed its interaction with HuR (Abdelmohsen et al., 2017).
This technique can provide essential knowledge at the molecular level of the network of circRNA-interacting molecules. The limitations of this approach include background detection of other molecules associated with the ASOs, and inaccessible junction sequences due to circRNA folding.
2.9 |. Immunoprecipitation of circRNA-RBP complexes
Ribonucleoprotein (RNP) immunoprecipitation (RIP) analysis is another strategy for assessing circRNA interactions with particular proteins. In this assay, an antibody is used to immunoprecipitate an RBP of interest, and the RBP-associated RNAs, including circRNAs, are then detected by RNA-seq, Northern blot, or RT-qPCR analyses. The major advantage of this method is that it allows the study of endogenous complexes in cells or tissues. Using this approach, insulin-like growth factor 2 binding protein 3 (IMP3) was found to bind 34 circRNAs, and HuR was found to bind several circRNAs including circPABPN1 (Abdelmohsen et al., 2017; Schneider et al., 2016). While RIP followed by RT-qPCR analysis or sequencing can identify target circRNAs, the sites of interaction are not identified. To overcome this limitation, CLIP methods can be employed to determine the specific binding sequences.
2.10 |. In vitro circRNA synthesis
In vitro synthesized circRNAs have a wide range of applications in molecular biology and potentially in therapeutic applications. They can be synthesized chemically using cyanogen bromide or enzymatically using T4 RNA ligases as well as by using an RNA cyclase ribozyme (L. L. Chen & Yang, 2015; Petkovic & Muller, 2015). In vitro synthesized circRNAs have been introduced into cells to study the innate immune response, as well as to explore ways to synthesize protein in eukaryotic cells (Y. G. Chen et al., 2017; Wesselhoeft, Kowalski, & Anderson, 2018).
3 |. CONCLUDING REMARKS
Recent advances in RNA-seq and computational analyses have critically increased our knowledge of circRNA biology through the identification and annotation of a large number of circRNAs. However, virtually all circRNAs remain to be characterized in molecular and functional detail. In this review, we highlight key online resources and tools that facilitate the initial steps of circRNA analysis, including circRNA sequence, design of silencing molecules, and association with other molecules. We also summarize and discuss common techniques to assess circRNA identity, abundance, subcellular localization, biochemical interactions, and functions, as well as the challenges and limitations of each approach (Table 2).
TABLE 2.
Approaches to study circRNAs
| Method | Pros | Cons |
|---|---|---|
| Online resources | Allow the identification, annotation and potential associations with trans-acting factors and diseases | Experimental validation required |
| circRNA sequencing (Circ-seq) | Allows identification and annotation, high-throughput screening, and profiling under different conditions | Does not inform on variants and may include linear RNAs resistant to RNase R |
| Microarrays | Assess differential expression and circRNA profiling under different conditions | Analysis is limited to the probes on the array, relies on junction sequences, and does not inform on mature sequences |
| Northern blot analysis | Detects circular versus linear, informs roughly on size and abundance | Not suitable for high-throughput screening |
| RT-qPCR | Permits Circ-seq validation, may be used to elucidate full sequences | May recognize linear trans-spliced RNAs, not suitable for high-throughput screening |
| Digital droplet PCR (ddPCR) | Informs on absolute copy number and measures low-abundance circRNAs | Not cost-effective |
| RNA-FISH | Aids in circRNA visualization, subcellular localization, quantification | Possible detection of linear RNAs |
| Circ siRNA | Loss of function analysis | May silence linear RNAs |
| Overexpression of circRNAs | Gain of function analysis | May generate concatemer products |
| Pulldown with ASOs | Detects trans-acting factors | May recognize linear mRNA counterparts |
| Immunoprecipitation (RIP) | Detects interaction with RBPs | Does not identify binding site(s) |
| Synthetic circRNAs | Can be used to study circRNAs and develop therapeutic applications | Are expensive to produce, may contain linear byproducts |
Abbreviations: ASO, antisense oligomer; RBP, RNA-binding protein; RIP, ribonucleoprotein immunoprecipitation; RNA-FISH, RNA fluorescence in situ hybridization; RT-qPCR, reverse transcription followed by quantitative polymerase chain reaction.
Going forward, it will be important to understand in depth how circRNA biogenesis is regulated, including the pathways and factors governing the tissue-specific expression of circRNAs, the kinetics and developmental cues driving biogenesis, the specific RNA segments included, and other mechanistic details. With rising recognition that some circRNAs may have important diagnostic, prognostic and possibly therapeutic value, it will be critical to develop more specific and sensitive detection methods, advanced tools to produce them in tissues and in vitro, and animal models to investigate their full impact in physiology and disease.
ACKNOWLEDGMENTS
This work was supported entirely by the National Institute on Aging Intramural Research Program, National Institutes of Health.
Funding information
National Institutes of Health
Footnotes
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
REFERENCES
- Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, … Gorospe M (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biology, 14(3), 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, … Zhang F (2017). RNA targeting with CRISPR-Cas13. Nature, 550(7675), 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, … Kadener S (2014). circRNA biogenesis competes with pre-mRNA splicing. Molecular Cell, 56(1), 55–66. [DOI] [PubMed] [Google Scholar]
- Atianand MK, & Fitzgerald KA (2014). Long non-coding RNAs and control of gene expression in the immune system. Trends in Molecular Medicine, 20(11), 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, … Pils D (2015). Correlation of circular RNA abundance with proliferation—Exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Scientific Reports, 5, 8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, … Xie X (2018). circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics, 8(14), 4003–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, … Huang S (2017). Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Letters, 388, 208–219. [DOI] [PubMed] [Google Scholar]
- Chen LL, & Yang L (2015). Regulation of circRNA biogenesis. RNA Biology, 12(4), 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Han P, Zhou T, Guo X, Song X, & Li Y (2016). circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Scientific Reports, 6, 34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, … Chang HY (2017). Sensing self and foreign circular RNAs by intron identity. Molecular Cell, 67(2), 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C, Mascrez B, Hetuin D, & Bailleul B (1993). Mis-splicing yields circular RNA molecules. FASEB Journal, 7(1), 155–160. [DOI] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, & Zhang F (2017). RNA editing with CRISPR-Cas13. Science, 358(6366), 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Ma XK, Li GW, & Yang L (2018). CIRCpedia v2: An updated database for comprehensive circular RNA annotation and expression comparison. Genomics, Proteomics & Bioinformatics, 16(4), 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, … Yang BB (2017). Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. European Heart Journal, 38(18), 1402–1412. [DOI] [PubMed] [Google Scholar]
- Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, & Yang BB (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Research, 44(6), 2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, & Gorospe M (2016). CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biology, 13(1), 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, & Yarden Y (2016). Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Research, 44(3), 1370–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errichelli L, Dini MS, Laneve P, Colantoni A, Legnini I, Capauto D, … Bozzoni I (2017). FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nature Communications, 8, 14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Lei X, Fang Z, Jiang Q, & Wu FX (2018). CircR2Disease: A manually curated database for experimentally supported circular RNAs associated with various diseases. Database, 2018. 10.1093/database/bay044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Xiang Y, Xia S, Liu H, Wang J, Ozguc FM, … Han L (2018). CircView: A visualization and exploration tool for circular RNAs. Briefings in Bioinformatics, 19(6), 1310–1316. [DOI] [PubMed] [Google Scholar]
- Floris G, Zhang L, Follesa P, & Sun T (2017). Regulatory role of circular RNAs and neurological disorders. Molecular Neurobiology, 54(7), 5156–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal S, Das S, Sen R, Basak P, & Chakrabarti J (2013). Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Frontiers in Genetics, 4, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazar P, Papavasileiou P, & Rajewsky N (2014). circBase: A database for circular RNAs. RNA, 20(11), 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Li M, Jin Y, Liu D, & Wei F (2017). Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genetics, 18(1), 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Li J, Wang H, Su X, Hou J, Gu Y, … Cao X (2017). Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology, 66(4), 1151–1164. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, & Kjems J (2013). Natural RNA circles function as efficient microRNA sponges. Nature, 495(7441), 384–388. [DOI] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, … Colston BW (2011). High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical Chemistry, 83(22), 8604–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, … Teupser D (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nature Communications, 7, 12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Zhang Y, Han B, Bai Y, Zhou R, Gan G, … Yao H (2017). Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy, 13(10), 1722–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, & van Oudenaarden A (2011). Validating transcripts with probes and imaging technology. Nature Methods, 8(Suppl. 4), S12–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, … Rajewsky N (2015). Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Reports, 10(2), 170–177. [DOI] [PubMed] [Google Scholar]
- Jeck WR, & Sharpless NE (2014). Detecting and characterizing circular RNAs. Nature Biotechnology, 32(5), 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, … Sharpless NE (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 19(2), 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, & Wilusz JE (2015). Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes & Development, 29(20), 2168–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, & Kjems J (2019). The biogenesis, biology and characterization of circular RNAs. Nature Reviews Genetics. 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, … Bozzoni I (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Molecular Cell, 66(1), 22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JH, Liu S, Zhou H, Qu LH, & Yang JH (2014). starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Research, 42(D1), D92–D97. 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, … Guo J (2018). Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. Journal of Molecular Medicine (Berlin), 96(1), 85–96. [DOI] [PubMed] [Google Scholar]
- Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, … Chen LL (2017). Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Molecular Cell, 67(2), 214–227. [DOI] [PubMed] [Google Scholar]
- Li X, Yang L, & Chen LL (2018). The biogenesis, functions, and challenges of circular RNAs. Molecular Cell, 71(3), 428–442. [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, … Jiang G (2017). CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Reports, 18(9), 1646–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, … Huang S (2015). Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Research, 25(8), 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, … Shan G (2015). Exon–intron circular RNAs regulate transcription in the nucleus. Nature Structural & Molecular Biology, 22(3), 256–264. [DOI] [PubMed] [Google Scholar]
- Liang D, & Wilusz JE (2014). Short intronic repeat sequences facilitate circular RNA production. Genes & Development, 28(20), 2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, … Chen LL (2019). Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell, 177(4), 865–880. [DOI] [PubMed] [Google Scholar]
- Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, … Huang HD (2016). CircNet: A database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Research, 44(D1), D209–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ (2013). Circular RNA (circRNA) in Alzheimer’s disease (AD). Frontiers in Genetics, 4, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, … Rajewsky N (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature, 495(7441), 333–338. [DOI] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, … Vogelstein B (1991). Scrambled exons. Cell, 64(3), 607–613. [DOI] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, … Kadener S (2017). Translation of CircRNAs. Molecular Cell, 66(1), 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda AC, Abdelmohsen K, & Gorospe M (2017). RT-qPCR detection of senescence-associated circular RNAs. Methods in Molecular Biology, 1534, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda AC, De S, Grammatikakis I, Munk R, Yang X, Piao Y, … Gorospe M (2017). High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Research, 45(12), e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda AC, Grammatikakis I, Kim KM, De S, Martindale JL, Munk R, … Gorospe M (2017). Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Research, 45(7), 4021–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda AC, Grammatikakis I, Munk R, Gorospe M, & Abdelmohsen K (2017). Emerging roles and context of circular RNAs. WIREs RNA, 8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey PR, Rout PK, Das A, Gorospe M, & Panda AC (2019). RPAD (RNase R treatment, polyadenylation, and poly(A)+ RNA depletion) method to isolate highly pure circular RNA. Methods, 155, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovic S, & Muller S (2015). RNA circularization strategies in vivo and in vitro. Nucleic Acids Research, 43(4), 2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, … Rajewsky N (2017). Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science, 357(6357), eaam8526. [DOI] [PubMed] [Google Scholar]
- Quan PL, Sauzade M, & Brouzes E (2018). dPCR: A technology review. Sensors (Basel), 18(4), e1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, … Rajewsky N (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Molecular Cell, 58(5), 870–885. [DOI] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, & Brown PO (2013). Cell-type specific features of circular RNA expression. PLoS Genetics, 9(9), e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Hung LH, Schreiner S, Starke S, Eckhof H, Rossbach O, … Bindereif A (2016). CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Scientific Reports, 6, 31313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, … Yan B (2017). Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation, 136(17), 1629–1642. [DOI] [PubMed] [Google Scholar]
- Szabo L, & Salzman J (2016). Detecting circular RNAs: Bioinformatic and experimental challenges. Nature Reviews Genetics, 17(11), 679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak HF, Van der Horst G, Smit J, Winter AJ, Mul Y, & Groot Koerkamp MJ (1988). Discrimination between RNA circles, interlocked RNA circles and lariats using two-dimensional polyacrylamide gel electrophoresis. Nucleic Acids Research, 16(14A), 6597–6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Li X, Zhao J, Qian F, Feng C, Li Y, … Li C (2018). TRCirc: A resource for transcriptional regulation information of circRNAs. Briefings in Bioinformatics. 10.1093/bib/bby083. [DOI] [PubMed] [Google Scholar]
- van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, … Hubner N (2019). The translational landscape of the human heart. Cell, 178(1), 242–260. [DOI] [PubMed] [Google Scholar]
- Vicens Q, & Westhof E (2014). Biogenesis of circular RNAs. Cell, 159(1), 13–14. [DOI] [PubMed] [Google Scholar]
- Wesselhoeft RA, Kowalski PS, & Anderson DG (2018). Engineering circular RNA for potent and stable translation in eukaryotic cells. Nature Communications, 9(1), 2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, … He C (2018). CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Research, 46(D1), D925–D929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, … He C (2017). Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Briefings in Bioinformatics, 18(6), 984–992. [DOI] [PubMed] [Google Scholar]
- Xiao MS, & Wilusz JE (2019). An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3′ ends. Nucleic Acids Research. 10.1093/nar/gkz576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Yang K, & Yang M (2018). Circular RNA in aging and age-related diseases. Advances in Experimental Medicine and Biology, 1086, 17–35. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, … Wang Z (2017). Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Research, 27(5), 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Li TC, Wu YY, Yeh CH, Chiang W, Chuang CY, & Kuo HC (2017). The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nature Communications, 8(1), 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, … Zhang N (2018). A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nature Communications, 9(1), 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, & Yang L (2014). Complementary sequence-mediated exon circularization. Cell, 159(1), 134–147. [DOI] [PubMed] [Google Scholar]
- Zhao ZJ, & Shen J (2017). Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biology, 14(5), 514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, … Huang S (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature Communications, 7, 11215. [DOI] [PMC free article] [PubMed] [Google Scholar]


