Abstract
Background
mRNA vaccines hold great potential as therapeutic techniques against viral infections due to their efficacy, safety, and large-scale production. mRNA vaccines offer flexibility in development as any protein can be produced from mRNA without altering the production or application process.
Objective
This review highlights the iterative optimization of mRNA vaccine structural elements that impact the type, specificity, and intensity of immune responses leading to higher translational potency and intracellular stability.
Results
Modifying the mRNA structural elements particularly the 5′ cap, 5′-and 3′-untranslated regions (UTRs), the coding region, and polyadenylation tail help reduce the excessive mRNA immunogenicity and consistently improve its intracellular stability and translational efficiency.
Conclusion
Further studies regarding mRNA-structural elements and their optimization are needed to create new opportunities for engineering mRNA vaccines.
Keywords: mRNA vaccine; 5′ Cap; 5′-and 3′-UTRs; Poly(A) tail, structural modification
Introduction
A vaccine triggers the body’s immune system response prompting it to create antibodies. Most vaccines are derived from antiviral immunity (Rabinovich et al. 1994). At present, various virus-specific vaccines are administered for routine vaccination, resulting in substantial advancements in the prevention and treatment of viral illnesses (Wang et al. 2021). The first report of the successful use of in vitro transcribed (IVT) mRNA in animals was reported in 1990 (Pardi et al. 2018). However, coupled with significant technological advancements and research investment in the last 10 years, mRNA has emerged as a promising therapeutic option in the fields of vaccine development. The mRNA vaccine is a new advanced technique that integrates molecular biology and immunology. The basic concept underlying mRNA vaccines is to encode the antigen in mRNA and transfer the transcript to the host cell’s cytoplasm via a non-viral delivery route, permitting antigen expression and stimulation of an antigen-specific immune response (Jackson et al. 2020). The vaccine platform is significant as mRNA vaccines can be made for any pathogen with a known protein target. The mRNA also does not interact with the genome and only the parts directly required for the encoded protein expression are present in this basic genetic construct (Pardi et al. 2018). mRNA is created via a cell-free enzymatic transcription reaction that enables rapid and scalable production, as illustrated by the current pandemic's rapid pursuit of RNA vaccines (Verbeke et al. 2021).
The mRNA is susceptible to degradation and stabilizing it may lead to high expression. Several factors affect the expression and stability of mRNA vaccines. The structural characteristics of mRNA such as the CAP, poly(A) tail, and untranslated regions (UTRs) directly impact mRNA stability and translation efficiency (Orlandini von Niessen et al. 2019; Holtkamp et al. 2006). The Cap and poly(A) tail located at the 5′- and 3′-ends of mRNA are critical for stability of mRNA in the cytosol. mRNA also need 5′ and 3′ UTRs around the open reading frame (ORF) to increase half-life, vaccine mRNA expression levels, and translation (Conry et al. 1995). Using enzymes to cap the 5′ terminus of mRNA is more efficient than using cap analogs such as 7-methylguanosine (Jani and Fuchs 2012; Urbina and Morales-Pison 1633). Several studies have linked the length of the poly(A) tail to translation efficiency (Conry et al. 1995). Deleting the poly(A) region from mRNA results in mRNA being unstable compared to the intact gene (Whitelaw et al. 1986). The poly(A) tail is important for mRNA stability and translation success (Kudla et al. 2006). Furthermore, increasing the G-C fraction of mRNA improves mRNA stability (Homma et al. 2016). The 5′-UTR/3′-UTR, 5′ cap, the poly(A) tail, a rare codon and G:C content can be optimized to improve mRNA stability. The mRNA structure, translational efficiency, and protein-folding machinery are most likely to be altered by substituting the mRNA sequence with rare codons and inserting modified nucleotides (Zhong et al. 2017). The intensity and specificity of the immune responses are expected to be influenced by these changes. It is widely expected that the efficiency of an mRNA vaccination would increase as protein expression is extended; therefore, improved mRNA vaccines will emerge from improved mRNA formats. mRNA vaccines are a promising vaccine candidates against the pandemic due to their short production time, economical, variety in vaccine construct, and clinically validated ability to stimulate cellular and humoral immune responses (Wang et al. 2021; Verbeke et al. 2021). This review discussed the iterative optimization and significant modifications of the mRNA structure particularly the 5′ cap, 5′ and 3′UTRs, the coding region, and the poly(A) tail which improve the stability, translation capacity and increase the efficacy of mRNA vaccines to trigger the immune response.
mRNA structural elements design and modifications
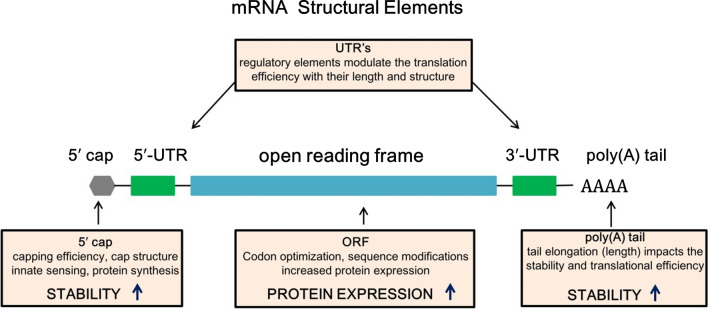
The basic structure of mRNA, same as eukaryotic mRNA, is made up of (i) protein encoding ORF, surrounded by (ii) 5′ and 3′ UTRs, and (iii) a 7-methyl guanosine 5′ cap structure and (iv) a 3′poly(A) tail at the end sides (Verbeke et al. 2019; Schlake et al. 2012; Sahin et al. 2014) (Fig. 1). These components can be modified or altered to increase the stability, translation efficiency, and immune-stimulatory feature of mRNA (Table 1). The translation and stability of mRNA can be improved and critical tasks have been undertaken to identify desirable mRNA elements.
Fig. 1.
Structural elements of mRNA vaccine include open reading frame (ORF) encoding protein, surrounded by 5′and 3′ untranslated regions (UTRs), 5′ cap structure and 3′poly(A) tail
Table 1.
mRNA structural elements and their effect of modifications
| Structural Element | Modification | Effect |
|---|---|---|
| Untranslated regions (UTR’s) | Length and structure | Modulate translation efficiency |
| 5′ Capping | Cap structure | Increase protein synthesis, stability |
| Open reading frame (ORF) | Codon optimization, sequence modification | Enhance protein expression |
| Poly(A) tail | Tail elongation | Increase Stability, translational efficiency |
Open reading frame (ORF)
Codon composition is known to influence translation efficiency. Translation efficiency, mRNA abundance, and protein folding may all be affected by codon optimization. Although high GC-content may cause problems for mRNA secondary structure, the higher GC sequence translates 100-fold higher than a low GC sequence (Kudla et al. 2006). The translation elongation rate is controlled by the accessibility of cognate tRNA species and the codon usage modification to prevent the match rare tRNA sequence of species and integrate sequences that match more common tRNA species (Hanson and Coller 2018). Furthermore, the rate of codon-dependent translation elongation has been a primary factor of mRNA stability, codon modification is critical for mRNA stability (Presnyak et al. 2015). Substituting rare codons with identical frequent codons boosts yield as reusing same tRNA speeds up translation due to amino-acylation of tRNAs near ribosomes (Cannarozzi et al. 2010). The translational elongation rate and efficiency are also affected by the neighboring nucleotides and codons (Bossi and Ruth 1980). The start codon should include Kozak sequence (Kozak 1986), and the stop codon's sequence can be modified (Liu 2005). In addition, there should be no upstream start codons in the mRNA preceding the correct start codon. Codon-optimized mRNAs have been effectively exploited in vaccine research against viral infections for expressing non-viral proteins, alike recombinant DNA-based methods (Kariko et al. 2012).
For appropriate folding, some proteins require delayed translation, which is guaranteed by uncommon codons (Kimchi-Sarfaty et al. 2007). mRNA sequence codon optimization can significantly impact the protein production rate and ribosome bide time. N1-methyl-pseudouridine (1mΨ) nucleotide substitution has been found to increase base pair stability, resulting in complex secondary structure and improved mRNA translation (Mauger et al. 2019). Furthermore, mRNA secondary structure design can be modified to improve mRNA stability against endonuclease cleavage and chemical degradation (Wayment-Steele et al. 2021).
mRNA transcripts containing modified nucleosides, also known as nucleoside-modified mRNAs, are created with lower immunostimulatory activity and a better safety profile. The modified nucleosides escape the type interferons (IFNs) induced antiviral pathways and are targeted to break and block invading mRNA; therefore they can be used to create mRNA vaccines with significantly improved translation efficiency (Verbeke et al. 2019). Kormann et al., demonstrated that 2 weeks following injection of nucleoside-modified mRNA containing 25% thio-uridine and 25% 5-methylcytidine resulted in Epo levels five times greater than that in untreated animals. Unmodified mRNA, on the other hand, showed no significant alterations and merely elicited a considerable immunological response (Kormann et al. 2011).
5-methylcytidine (m5C) and pseudouridine (ψ) nucleotide modifications are commonly preferred in base-pair alterations as they lower immunogenicity while increasing the efficiency of translation (Weng 2020). Pardi et al. constructed HPLC-purified and m1ψ modified mRNAs and delivered the mRNA with lipid nanoparticle (LNP). The findings revealed that mice administered mRNA-LNP produced protective antibodies, assisting them in overcoming HIV-1 infection (Pardi et al. 2017).
5ʹ- and 3ʹ-UTRs
The 5′- and 3′- UTRs comprising of regulatory sequence elements are known to regulate the mRNA stability. 5′-UTR regulatory elements and the length of the 3′-UTR aid translation of protein. They comprise various regulatory regions related to mRNA stability, ribosome recognition, interaction with translational machinery components, and mRNA secondary structures (Tanguay and Gallie 1996). By interacting with RNA-binding proteins, UTRs can also influence the rate of mRNA degradation and translation efficiency. The 5′ UTR sequence can be adjusted to improve mRNA stability and translation accuracy by (i) avoiding the presence of start codons and non-canonical start codons in the 5′ UTR which disrupts ORF translation, (ii) Ribosome recruitment and codon recognition can be prevented by avoiding the existence of highly stable secondary structures, (iii) Shorter 5′UTRs may be used, as past research has indicated that this type of 5′UTR is better for mRNA translation (Leppek et al. 2018).
The 3ʹ-UTRs of α- and β-globin mRNAs are found in many in vitro-transcribed (IVT) mRNA, and they contain sequence features that enhance the translation and stability of mRNA (Sahin et al. 2014; Karikó et al. 1999). The steady effect of human β-globin 3ʹ-UTR sequences is enhanced when two human β-globin 3ʹ-UTRs are placed in a head-to-tail orientation (Holtkamp et al. 2006). Furthermore, distinct areas of cellular and viral 5ʹ- and 3ʹ-UTRs improve mRNA stability and efficiency of translation. Destabilizing mRNA may be useful in some applications to reduce the time required for protein synthesis. This effect can be achieved by inserting AU-rich regions into 3′-UTRs ensuring fast mRNA breakdown and a short duration of protein expression (Chen and Shyu 1995).
5ʹ-capping of mRNA vaccines
For mRNA stability and efficient translation, mRNA undergo post-transcriptional changes such as 5′-capping and poly(A). A functional 5′ cap structure is required for robust mRNA translation. The 7-methylguanosine (m7G) moiety is found at the 5′ end of natural mRNA in the eukaryotic cytoplasm, followed by a triphosphate connection to the first nucleotide (m7GpppN) (Sahin et al. 2014). The 5′ cap regulates pre-mRNA splicing and nuclear export, works as a protective structure that shields RNA from exonuclease cleavage, and initiates mRNA translation. The cap 0 structure of m7GpppN is necessary for effective mRNA translation. The 5′-cap structure is a key factor in the host's ability to differentiate between self and non-self mRNA molecules (Devarkar et al. 2016). The bulk of the mRNA vaccines reported till date have an m7GpppNm cap inserted at the 5′-end during IVT (Corbett et al. 2020).
Screening mRNA against exonuclease activity, promoting pre-mRNA splicing, and functioning as binding site of eIF4F heterodimeric translation initiation complex are all important functions of 5′-capping (Izaurralde et al. 1994; Gallie 1991). The cap structure is similar to the most common eukaryotic cap form found in nature. The employment of “anti-reverse” cap analogs results in a cap that is in the correct orientation Anti-Reverse Cap Analog (ARCAs). The most prevalent ARCA enables only the insertion of a nucleotide at the non-methylated guanosine after 3′-O-methylation of base-methylated guanosine. When ARCA-capped mRNA was compared to mRNA capped by a conventional cap analog, translation efficiency was twice more than in rabbit reticulocyte lysate (Stepinski et al. 2001). Furthermore, mRNA transcribed in vitro using ARCA has been found to have a longer half-life in cultivated cells (Grudzien et al. 2006). ARCA-capped mRNA has been shown to boost and extend protein expression in cultured cells (Zohra et al. 2007).
The poly(A) tail
The poly(A) tail steadies mRNA and boosts protein translation, and the length of the poly(A) tail is proportional to translation efficiency. It is a critical factor in determining the longevity of mRNA molecules (Park et al. 2016; Jalkanen et al. 2014). The poly(A) tail is encoded in the template vector from which the mRNA is transcribed, or the IVT mRNA is extended enzymatically using recombinant poly(A)polymerase. The insertion of modified nucleotides into the poly(A) tail by recombinant poly(A) polymerase inhibits deadenylation by poly(A)-specific nucleases (Korner and Wahle 1997). The mammalian cells mRNA molecules contain poly(A) tails that are approximately 250 nucleotides (nt) long and gradually decreases from 3′ to 5′ during their lifetime in the cytoplasm. Poly(A) tails of approximately 100 nt are ideal for synthesizing mRNA therapies because tail size affects mRNA decay by modulating 3′ exonucleolytic degradation (Schlake et al. 2012). Arbuthnot et al. suggested recurrent restriction digestion with type IIS enzymes, ligation, and propagation to lengthen the homopolymeric sequence up to 100 bp in circular plasmids to overcome the difficulties in cloning (Arbuthnot et al. 2019). To overcome these constraints, Grier et al. advocated using linear plasmids as a template for mRNA synthesis (Grier 2016).
Enzymatic polyadenylation has the disadvantage of having a variety of RNA species with variable poly(A) tail lengths in each RNA preparation. Contrarily, in vitro transcription (IVT) of RNA from a DNA template produces RNA with a predetermined poly(A) tail length, which is preferable, particularly for clinical purposes.
Eliminating mRNA immunogenicity
IVT is a low-cost method of mRNA synthesis; however the immunogenicity of mRNA is the most challenging obstacle it faces (Sahin et al. 2014). Chemically altering nucleotides, adding poly(A) tails, and optimizing mRNA with a GC-rich sequence are efficient ways to minimize mRNA immunogenicity. 5-methylcytidine (m5C), and pseudouridine (ψ) are the most preferred base-pair alterations because they lower immunogenicity while increasing translation efficiency (Weng 2020). Adding poly(A) tails lowers mRNA immunogenicity by lowering U content and shielding mRNA in the sequence. The reduced efficacy of protein expression produced by high GC concentration needs to be addressed (Wang et al. 2021). mRNA purification is also required after IVT to eliminate immunogenicity. HPLC, anion exchange and affinity chromatography, are used in the purification process to remove truncated transcripts (Weissman et al. 2013).
Advantages of mRNA vaccines
Gene-based and protein-based vaccines are the two types of vaccinations available. To trigger adaptive and humoral immune responses, traditional protein-based vaccinations use attenuated or recombinant proteins administered directly as immunogens. Gene-based vaccines are supplied to host cells via a (DNA or RNA) vector, where they are expressed to produce antigens that elicit an immunological response in the host. Because of the ease and speed with which mRNA vaccines may be produced, they are often favored over alternative platforms. mRNA vaccines work on the principle of delivering a transcript that encodes a target antigen or immunogen. As soon as the sequence encoding the immunogen is available, RNA synthesis may begin on the same platform. The technique can be easily scaled requiring minimum platform changes during mRNA formulation and production (Pardi et al. 2018). After transfection, mRNA vaccine expresses the protein target (antigen) via translation from mRNA. Because antigen translation occurs in the cytoplasm rather than the nucleus; therefore mRNA vaccines have a greater biosafety than DNA vaccines as their antigens are translated into the cytoplasm rather than the nucleus, making it more difficult for the mRNA to integrate into the genome. Furthermore, mRNA is a transient molecule carries a short translation sequence it has no interaction with the host genome. Protein-based vaccinations are usually made from bacteria, however mRNA vaccines can be produced by the host translation machinery, resulting in an antigen that closely resembles the structure of the protein released from the viral genome (Park et al. 2021).
mRNA vaccines must be stored and carried at extremely low temperatures as compared to protein-based vaccinations that can be stored and transported at lower temperatures (Crommelin et al. 2020). Storing and transferring mRNA vaccines to and from warm countries is thus a major technical and financial challenge. However, with the advancement of lipid nanoparticle techniques, mRNA vaccine stability can be maintained under less rigorous settings (Reichmuth et al. 2016). Despite the advantages of mRNA vaccines there are still many risks involved such as long term effects of the mRNA vaccine are unknown. mRNA is also unstable and must be stored at high temperature making packaging and distribution of vaccine difficult.
COVID-19 mRNA vaccines
Vaccine development often requires years of research and testing to ensure efficacy and safety. In contrast, the two mRNA vaccines took less than a year to develop and deploy from start to finish. The extraction of neutralizing antibodies from COVID-19 patient’s serum verified the S protein’s high immunogenicity, whereas the receptor binding domain (RBD) and N-terminal domains were also equally immunogenic (NTD). Therefore, immunizing against the full S protein rather than just one of its structural units may lead to a better response that is not impacted by genetic drift (Liu et al. 2020).
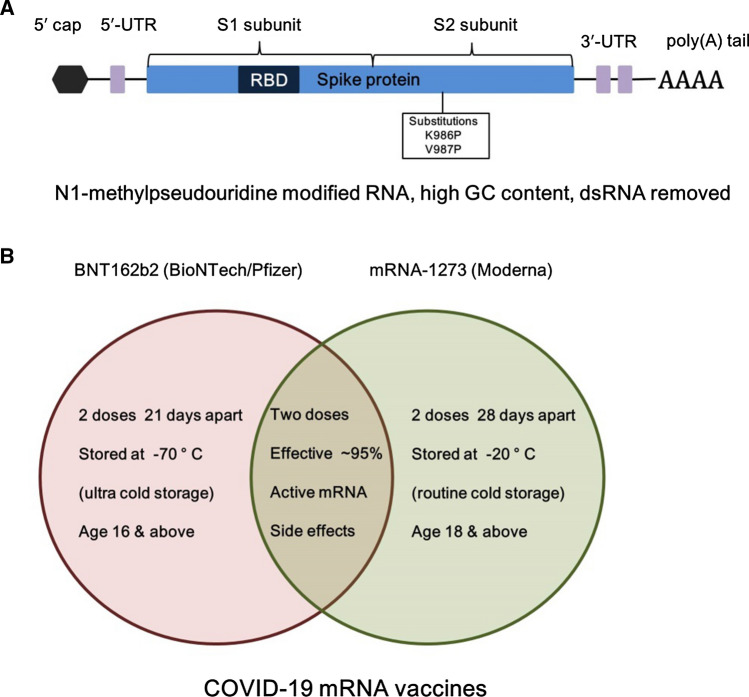
The COVID-19 vaccine candidates demonstrate the structurally modified mRNA approach allows for higher tolerated doses and may be more suitable for producing a fast antibody response (Verbeke et al. 2021). BNT162b2 (BioNTech/Pfizer) and mRNA-1273 (Moderna) used a mixture of altered nucleotide 1mΨ modifications and dsRNA fragment eviction in the mRNA manufacturing procedure to significantly lower innate immune signaling in response to mRNA (Walsh et al. 2020; Jackson et al. 2020b). Spike proteins with K986P, V987P substitutions are common in both vaccines. The BNT162b2 includes N1-methylpseudouridine, optimized codon; high GC sequence, removed dsRNA, and N1-methylpseudouridine modifications (Polack et al. 2020). In addition, 5′ UTR: human -globin RNA with optimized Kozak sequence, 3′ UTR: AES and mtRNR1 3′ UTR Motives, and 110 Poly(A) tail with nucleotide-linker (GCAUAUGACU) are also used (Orlandini von Niessen et al. 2019). Moderna on the other hand used N1-methylpseudouridine, removed dsRNA, and undisclosed structural elements (Baden et al. 2021) (Fig. 2A). The efficacy of both the mRNA vaccines is more than 90% (~ 95% to be precise). The storage and the dose information of these two mRNA vaccines are given in Fig. 2B. The COVID-19 pandemic has increased the interest in mRNA vaccinations. Their rapid creation, manufacture, and excellent effectiveness in preventing SARS-CoV-2 infection, show that mRNA-based therapies are ready for application in the future.
Fig. 2.
A Structural elements of the BNT162b2 (BioNTech/Pfizer) mRNA vaccine is shown. The figure is adapted from Vogel et al. (2021). B Comparison of the storage and the dose information of two currently used BNT162b2 and mRNA-1273 COVID-19 mRNA vaccines
Future perspective
mRNA is a promising although the challenging class of therapeutic molecules with the potential to become the foundation of a modern vaccination against infectious diseases. Modifying or improving the structural features can boost the mRNA’s potential to control innate immune responses, resulting in increased translation capacity and bioavailability. The final result can be dramatically improved by optimizing the structure of mRNA. The most difficult task that currently needs utmost attention is to strike a balance between translation efficiency and immune response to provide appropriate and safe immunogenicity.
Conclusion
The COVID-19 mRNA vaccine highlight that using structurally modified mRNA allows for increased dose tolerance and may be better for eliciting a rapid antibody response. mRNA vaccines provide flexibility in development as any protein can be made from mRNA without changing the production or application procedure. Optimizing the mRNA structural features particularly the 5′ cap, 5′ and 3′UTRs, the coding region, and the poly(A) tail increases the mRNA’s control of immune responses resulting in higher translational efficiency.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A1A06046235). This work was also supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (No. NRF-2019R1A2C1010860).
Author contributions
SCK, SSS, and W-RS wrote the review. All authors have read and approved the final manuscript.
Declarations
Conflict of interest
Author S.C.K. declares that he has no conflict of interest. Author S.S.S. declares that he has no conflict of interest. Author W.-R.S. declares that she has no conflict of interest. Author G.A. declares that she has no conflict of interest. Author B.-K.C. declares that she has no conflict of interest. Author J.-Y.A. declares that she has no conflict of interest. Author Y.-H.K. declares that she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Sun Chang Kim, Simranjeet Singh Sekhon and Woo-Ri Shin contributed equally to this work.
Contributor Information
Ji-Young Ahn, Email: jyahn@chungbuk.ac.kr.
Yang-Hoon Kim, Email: kyh@chungbuk.ac.kr.
References
- Arbuthnot P, Ely A, Bloom K. A convenient method to generate and maintain poly(A)-encoding DNA sequences required for in vitro transcription of mRNA. Biotechniques. 2019;66:37–39. doi: 10.2144/btn-2018-0120. [DOI] [PubMed] [Google Scholar]
- Baden LR, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L, Ruth JR. The influence of codon context on genetic code translation. Nature. 1980;286:123–127. doi: 10.1038/286123a0. [DOI] [PubMed] [Google Scholar]
- Cannarozzi G, et al. A role for codon order in translation dynamics. Cell. 2010;141:355–367. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Conry RM, et al. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995;55:1397–1400. [PubMed] [Google Scholar]
- Corbett KS, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crommelin DJA, Anchordoquy TJ, Volkin DB, Jiskoot W, Mastrobattista E. Addressing the cold reality of mRNA vaccine stability. J Pharm Sci. 2020;110:997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarkar SC, et al. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci U S A. 2016;113:596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Grier AE, et al. pEVL: a linear plasmid for generating mRNA IVT templates with extended encoded poly(A) sequences. Mol Ther Nucleic Acids. 2016;19:e306. doi: 10.1038/mtna.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudzien E, Kalek M, Jemielity J, Darzynkiewicz E, Rhoads RE. Differential inhibition of mRNA degradation pathways by novel cap analogs. J Biol Chem. 2006;281:1857–1867. doi: 10.1074/jbc.M509121200. [DOI] [PubMed] [Google Scholar]
- Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol. 2018;19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp S, et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- Homma K, Noguchi T, Fukuchi S. Codon usage is less optimized in eukaryotic gene segments encoding intrinsically disordered regions than in those encoding structural domains. Nucleic Acids Res. 2016;44:10051–10061. doi: 10.1093/nar/gkw899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, et al. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Jackson NAC, Kester KE, Casimiro D, Gurunathan S, DeRosa F. The promise of mRNA vaccine: a biotech and industrial perspective. Npj Vaccines. 2020;5:11. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LA, et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalkanen AL, Coleman SJ, Wilusz J. Determinants and implications of mRNA poly (A) tail size–does this protein make my tail look big? Semin Cell Dev Biol. 2014;34:24–32. doi: 10.1016/j.semcdb.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani B, Fuchs R. In vitro transcription and capping of Gaussia luciferase mRNA followed by HeLa cell transfection. J vis Exp. 2012;61:3702. doi: 10.3791/3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K, Kuo A, Barnathan E. Overexpression of urokinase receptor in mammalian cells following administration of the in vitro transcribed encoding mRNA. Gene Ther. 1999;6:1092–1100. doi: 10.1038/sj.gt.3300930. [DOI] [PubMed] [Google Scholar]
- Kariko K, et al. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol Ther. 2012;20:948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kormann MS, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- Korner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3ʹ-exoribonuclease. J Biol Chem. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4:e180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Das R, Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol. 2018;19:158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. Comparative analysis of base biases around the stop codons in six eukaryotes. Biosystems. 2005;81:281–289. doi: 10.1016/j.biosystems.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Liu L, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Mauger DM, et al. mRNA structure regulates protein expression through changes in functional half-life. Proc Natl Acad Sci USA. 2019;116:24075. doi: 10.1073/pnas.1908052116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandini von Niessen AG, et al. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3′ UTRs identified by cellular library screening. Mol Ther. 2019;27:824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, et al. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV- 1 challenge. Nat Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug Dis. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Yi H, Kim Y, Chang H, Kim VN. Regulation of Poly(A) tail and translation during the somatic cell cycle. Mol Cell. 2016;62:462–471. doi: 10.1016/j.molcel.2016.04.007. [DOI] [PubMed] [Google Scholar]
- Park JW, Lagniton PNP, Liu Y, Xu RH. mRNA vaccines for COVID-19: what, why and how. Int J Biol Sci. 2021;17:1446–1460. doi: 10.7150/ijbs.59233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack FP, et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, et al. codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich NR, McInnes P, Klein DL, Hall BF. Vaccine technologies: view to the future. Science. 1994;265:1401–1404. doi: 10.1126/science.7521064. [DOI] [PubMed] [Google Scholar]
- Reichmuth AM, Oberli MA, Jaklenec A, Langer R, Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics–developing a new class of drugs. Nat Rev Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- Schlake T, Thess A, Fotin-Mleczek M, Kallen K-J. Developing mRNA-vaccine technologies. RNA Biol. 2012;9:1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepinski J, Waddell C, Stolarski R, Darzynkiewicz E, Rhoads RE. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA. 2001;7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- Tanguay RL, Gallie DR. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol Cell Biol. 1996;16:146–156. doi: 10.1128/mcb.16.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina F, Morales-Pison S. Enzymatic protein biopolymers as a tool to synthetize eukaryotic messenger ribonucleic acid (mRNA) with uses in vaccination, immunotherapy and nanotechnology. Polymers (basel) 2020;12:1633. doi: 10.3390/polym12081633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeke R, Lentacker I, Smedt SC, Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28:100766. [Google Scholar]
- Verbeke R, Lentacker I, Smedt AC, Dewitte H. The dawn of mRNA vaccines: the COVID-19 case. J Control Release. 2021;333:511–520. doi: 10.1016/j.jconrel.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel AB, et al. A prefusion SARS-CoV-2 spike RNA vaccine is highly immunogenic and prevents lung infection in non-human primates. Biorxiv Preprint Server. 2021 doi: 10.1101/2020.09.08.280818v1. [DOI] [Google Scholar]
- Walsh EE, et al. Safety and immunogenicity of two RNA-based COVID-19 vaccine candidates. N Engl J Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. mRNA vaccine: a potential therapeutic strategy. Mol Cancer. 2021 doi: 10.1186/s12943-021-01311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayment-Steele HK, et al. Theoretical basis for stabilizing messenger RNA through secondary structure design. bioRxiv. 2021 doi: 10.1101/2020.08.22.262931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D, Pardi N, Muramatsu H, Karikó K. HPLC purification of in vitro transcribed long RNA. Methods Mol Biol. 2013;969:43–54. doi: 10.1007/978-1-62703-260-5_3. [DOI] [PubMed] [Google Scholar]
- Weng Y, et al. The challenge and prospect of mRNA therapeutics landscape. Biotechnol Adv. 2020;40:107534. doi: 10.1016/j.biotechadv.2020.107534. [DOI] [PubMed] [Google Scholar]
- Whitelaw E, Coates A, Proudfoot NJ. Globin gene transcripts can utilize histone gene 3′ end processing signals. Nucleic Acids Res. 1986;14:7059–7070. doi: 10.1093/nar/14.17.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C, Wei P, Zhang YP. Enhancing functional expression of codon optimized heterologous enzymes in Escherichia coli BL21 (DE3) by selective introduction of synonymous rare codons. Biotechnol Bioeng. 2017;114:1054–1064. doi: 10.1002/bit.26238. [DOI] [PubMed] [Google Scholar]
- Zohra FT, Chowdhury EH, Tada S, Hoshiba T, Akaike T. Effective delivery with enhanced translational activity synergistically accelerates mRNA-based transfection. Biochem Biophys Res Commun. 2007;358:373–378. doi: 10.1016/j.bbrc.2007.04.059. [DOI] [PubMed] [Google Scholar]