SUMMARY
Although the roles of initiation factors, RNA binding proteins, and RNA elements in regulating translation are well defined, how the ribosome functionally diversifies remains poorly understood. In their human hosts, poxviruses phosphorylate serine 278 (S278) at the tip of a loop domain in the small subunit ribosomal protein RACK1, thereby mimicking negatively charged residues in the RACK1 loops of dicot plants and protists to stimulate translation of transcripts with 5′ poly(A) leaders. However, how a negatively charged RACK1 loop affects ribosome structure and its broader translational output is not known. Here, we show that although ribotoxin-induced stress signaling and stalling on poly(A) sequences are unaffected, negative charge in the RACK1 loop alters the swivel motion of the 40S head domain in a manner similar to several internal ribosome entry sites (IRESs), confers resistance to various protein synthesis inhibitors, and broadly supports noncanonical modes of translation.
Graphical Abstract
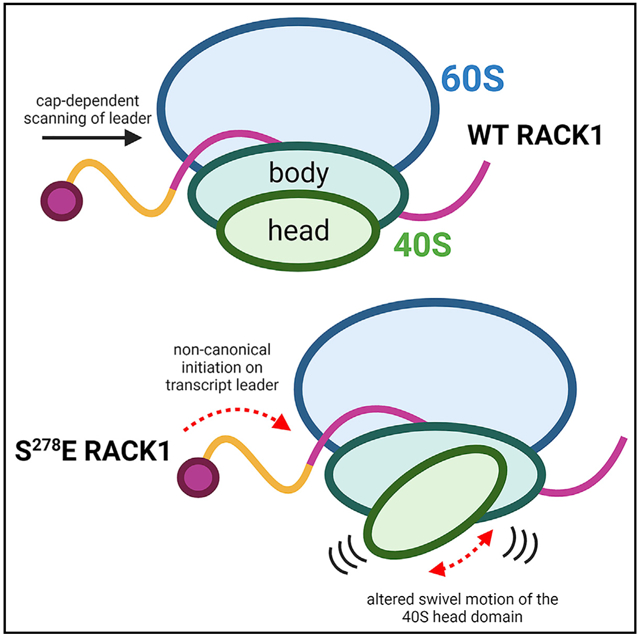
In brief
How ribosomes functionally diversify to selectively control translation is only beginning to be understood. Rollins et al. show that negative charge in a loop domain of the small subunit ribosomal protein RACK1 increases the swiveling motion of the 40S head and broadens the translational capacity of the human ribosome.
INTRODUCTION
There is growing evidence that the ribosome can structurally and functionally diversify to regulate translation (Gilbert, 2011; Sulima and Dinman, 2019; Xue and Barna, 2012). For example, cell-type-specific expression of the large ribosomal subunit protein L38 (RPL38) (Kondrashov et al., 2011; Xue et al., 2015) and ribosomal expansion segments (Leppek et al., 2020) regulate homeobox (Hox) mRNA translation during cytoskeletal patterning. Ribosomal protein (RP) paralogs diversify ribosome activity during gonad development (Hopes et al., 2021), and intracellular heterogeneity in ribosomes regulates translation (Shi et al., 2017). However, the structural basis by which these subunit differences alter ribosome specificity remains unclear.
Beyond subunit differences, post-translational modifications (PTMs) to RPs control ribosome activity. Several RPs are mono- or polyubiquitinated during cell stress and ribosome quality control (RQC) (Higgins et al., 2015; Matsuo et al., 2017; Saito et al., 2015; Silva et al., 2015). RQC senses aberrant translation events or mis-processed transcripts (Sitron and Brandman, 2020). For example, ribosomes are inherently designed to stall on poly(A) stretches to detect mRNAs that are erroneously internally polyadenylated; therefore, poly(A) tracts are heavily selected against outside of the 3′ untranslated region (UTR) (Arthur et al., 2015; Koutmou et al., 2015). Upon encountering poly(A) stretches, ribosomes stall, collide, and activate stress signals, along with destruction of the mRNA and nascent peptide (Brandman and Hegde, 2016; Brandman et al., 2012; Komander and Rape, 2012; Wu et al., 2020). In the earliest stages of stalling, the ubiquitin E3 ligase zinc finger protein 598 (ZNF598), with the aid of receptor for activated C kinase 1 (RACK1), monoubiquitinates several small RP subunits (RPSs) (Garzia et al., 2017; Juszkiewicz et al., 2018; Juszkiewicz and Hegde, 2017; Matsuo et al., 2017; Simms et al., 2017; Sundaramoorthy et al., 2017). RACK1 also prevents stalled ribosomes from frameshifting and enables endonucleolytic cleavage on mRNA lacking stop codons (Ikeuchi and Inada, 2016; Wang et al., 2018; Wolf and Grayhack, 2015). Structures of stall-inducing sequences (Chandrasekaran et al., 2019; Tesina et al., 2020) and ribosomes in various RQC stages have been solved (Sitron and Brandman, 2020), and extensive polyubiquitination traps the ribosome in a rotated and inactive state (Zhou et al., 2020). However, beyond the broad inactivation effects of ubiquitination during RQC, how other PTMs to RPs affect ribosome structure and customize its output remains unknown.
Beyond its role in RQC, RACK1 regulates several other aspects of translation. RACK1 is a conserved Trp-Asp (WD) repeat protein that largely consists of seven β-propeller blades that mediate protein binding (Murzin, 1992; Xu and Min, 2011). RACK1 is a core RP that is located on the head domain of the 40S subunit near the mRNA exit channel, with much of its surface solvent exposed (Coyle et al., 2009; Sengupta et al., 2004). This enables RACK1 to act as a docking site for eukaryotic translation initiation factors (eIFs) and kinases, thereby integrating signaling with translational output (Gandin et al., 2013; Nielsen et al., 2017). There is strong evidence that in many cell types and like other RPs, extra-ribosomal RACK1 is degraded to restrict its signaling and other activities to the ribosome (DiGiuseppe et al., 2020; Dobrikov et al., 2018a, 2018b; Gallo et al., 2019; Jha et al., 2017; Nielsen et al., 2017; Rollins et al., 2019; Sung et al., 2016). In terms of effects on translation, RACK1 can stimulate overall rates of protein synthesis (Nielsen et al., 2017), as well as control translation of specific mRNA subsets (Kim et al., 2017; Thompson et al., 2016). RACK1 also contributes to noncanonical cap-independent initiation by viral internal ribosome entry sites (IRESs) (Jackson, 2013; Johnson et al., 2019; LaFontaine et al., 2020; Majzoub et al., 2014; Qin and Sarnow, 2004).
RACK1 also contains a short interconnecting loop between blades six and seven that is not required for ribosome binding and whose amino acid sequence varies across species (Coyle et al., 2009; Jha et al., 2017; Rollins et al., 2019; Sengupta et al., 2004). The human loop sequence consists of uncharged amino acids, but during poxvirus infection, a viral kinase introduces negative charge into the loop through single-site phosphorylation at serine 278 (S278) to enhance translation of viral mRNAs that harbor unusual 5′ poly(A) leaders (Jha et al., 2017; Rollins et al., 2019). This phosphorylation of human RACK1 mimics negatively charged amino acids that are present in the RACK1 loops of dicot plants and protists (Jha et al., 2017; Rollins et al., 2019), which unlike mammals also encode adenosine-rich 5′ UTRs (Guo et al., 2016; Steel and Jacobson, 1991). Expression of RACK1 in which S278 is replaced with a glutamic acid (S278E), which mimics poxvirus phosphorylation of human RACK1, as well as the negatively charged loops of many other species, is sufficient to enhance translation of mRNAs with adenosine-rich 5′ UTRs (Jha et al., 2017; Rollins et al., 2019). But beyond this, how a charged RACK1 loop affects ribosome structure and translational output remains unknown. Here, we show that negative charge in the RACK1 loop does not affect its ability to transmit ribotoxin signals but alters the swivel motion of the 40S head domain and enables the human ribosome to broadly support noncanonical modes of translation.
RESULTS
WT and S278E RACK1 ribosomes form stable interactions with eEF2, SERBP1, and Ebp1
Understanding how S278E RACK1 influences ribosome structure and function necessitated the development of a new cell system. Our prior approaches involved expression of exogenous forms of RACK1 against a background of competition with endogenous RACK1 for ribosome binding and protein stabilization, which results in a 50:50 expression ratio in primary normal human fibroblasts (Jha et al., 2017; Rollins et al., 2019). Although this was sufficient to study enhancer effects of S278E RACK1 on the specific 5′ poly(A) transcripts of interest, the continued presence of endogenous RACK1 confounded attempts to understand its broader impact on translation. Indeed, in this system, negatively charged RACK1 does not impair overall translation as measured by 35S-methionine/cysteine pulse labeling or luciferase expression from a β-actin reporter; yet RiboTag as-says, specifically isolating green fluorescent protein (GFP)-tagged wild-type (WT) or S278E RACK1 forms away from endogenous RACK1, suggested that S278E RACK1 had reduced affinity for β-actin mRNA compared with WT RACK1. However, commonly used ribosome profiling and RiboTag RNA affinity assays do not discern transcripts associated with active versus inactive ribosomes. Moreover, our subsequent studies revealed that negative charge in the loop weakens RACK1’s association with the ribosome in a buffer-dependent manner, which further confounds the interpretation of such in vitro RiboTag assays while hinting at the potential structural impact of a charged RACK1 loop (Jha et al., 2017; Rollins et al., 2019). Stemming from these biochemical observations, clash modeling suggested that negatively charged RACK1 loops create electrostatic repulsive interactions with the negatively charged phosphate backbone of the 18S rRNA (Rollins et al., 2019). From this, we hypothesized that these electrostatics may alter local contacts that RACK1 makes on the 40S. As such, key questions as to whether S278E RACK1 truly affects ribosome structure and regarding its broader effects on translation remain unanswered.
To address this, we developed a HAP1 cell-based knockout and rescue system to enable both global analysis of effects on translation in the absence of endogenous RACK1 and large-scale ribosome isolation for cryoelectron microscopy (cryo-EM). We chose HAP1 cells because they are fibroblast-like, are not as translationally hyper-activated as many commonly used cell lines, and recapitulate the strict ribosome association and homeostatic control of RACK1 expression that we observe in primary fibroblasts (DiGiuseppe et al., 2020; Jha et al., 2017; Rollins et al., 2019) (Figure S1A). Moreover, we previously generated RACK1 knockout HAP1 cells (Jha et al., 2017) that have been shown by others to be phenotypically rescued using FLAG-tagged RACK1 (Johnson et al., 2019, 2020; LaFontaine et al., 2020). We therefore generated RACK1 knockout rescue pools expressing FLAG-tagged WT or S278E forms of RACK1 (Figure S1B). The minimal impact of RACK1 knockout, here called no-rescue cells, on polysome profiles (Figure S1C) validates the nonessentiality of RACK1 to global translation and mirrors phenotypes reported by others (Johnson et al., 2019; LaFontaine et al., 2020). Furthermore, the rescue lines reproduce known phenotypes of primary fibroblasts expressing GFP-tagged forms of RACK1 (Rollins et al., 2019), such as monosome and disome accumulation induced by S278E RACK1, which we observe in both polysome profiles (Figure S1C) and cryo-EM micrographs (Figures S1D and S1E). Finally, both WT and S278E RACK1 forms are restricted to the ribosome, being detectable, together with other RPs on the 40S subunit, monosomes, and disomes, but not in free fractions (Figure 1A). As expected, translation factors like eIF4G and eukaryotic elongation factor 2 (eEF2), as well as RNA binding proteins like Serpine mRNA binding protein 1 (SERBP1) and ErbB3-binding protein 1 (Ebp1), are present in both ribosomal and free fractions (Figure 1A).
Figure 1. Effects of S278E RACK1 on ribosome rotation and translational output.
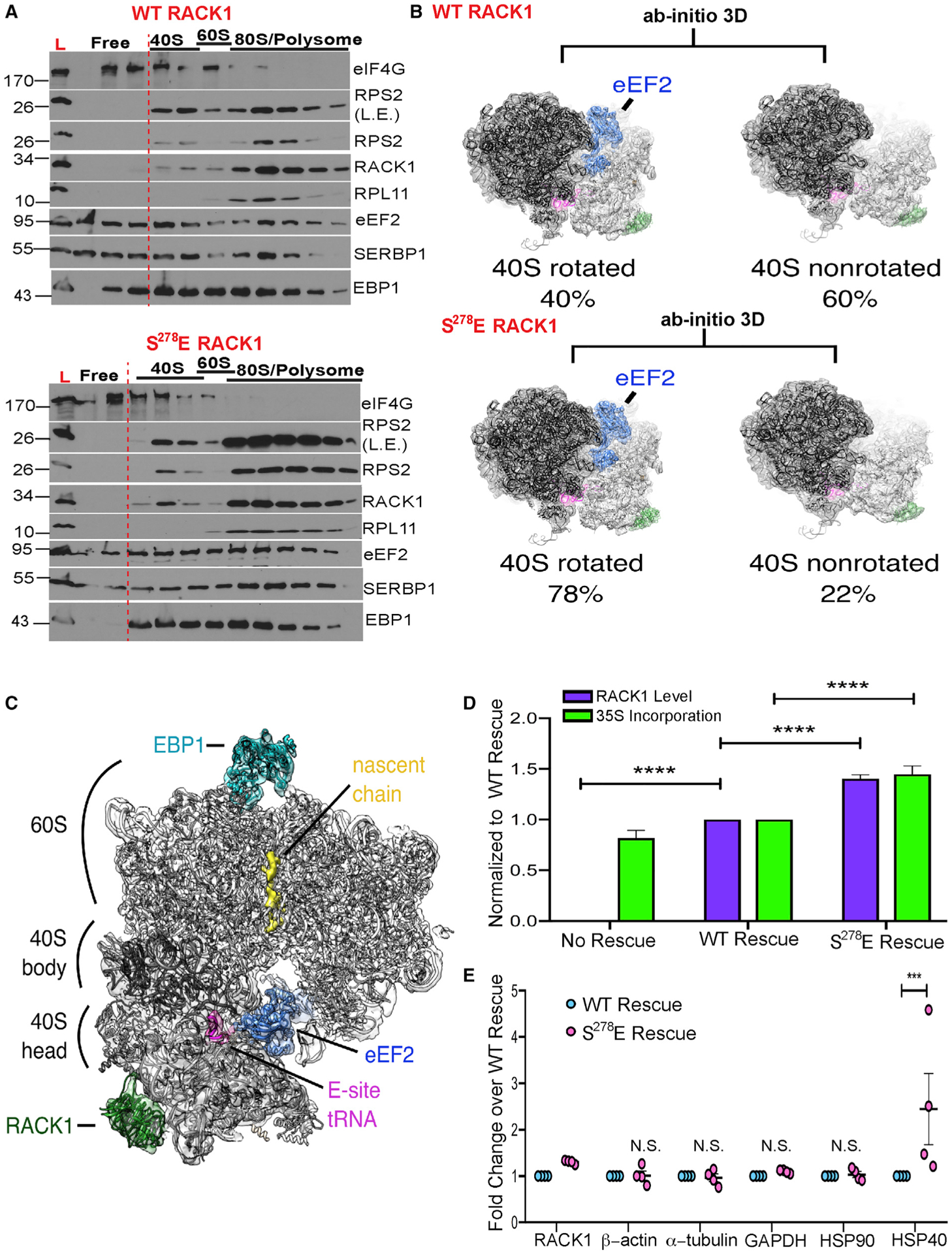
(A) Western blot analysis of free and ribosomal fractions. L, lysate; L.E., long exposure. Representative of 3 independent biological replicates.
(B) Ab initio 3D classification of 80S ribosomes from WT RACK1 and S278E RACK1 purifications reveals a shift toward 40S-rotated, eEF2-bound particles in the presence of S278E RACK1.
(C) Reconstruction of rotated ribosomes from S278E RACK1 purifications reveal densities ascribed to eEF2, E-site tRNA, EBP1, and a nascent chain.
(D) Quantification of RACK1 protein levels (n = 22) and 35S-methionine/cysteine (35S-Met/Cys) incorporation (n ≥ 4). Bars represent SEM; ****p ≤ 0.0001; two-way ANOVA with Sidak’s multiple comparison test.
(E) Densitometry-based quantification of the indicated protein levels (n = 4). Bars represent ± SEM; ***p = 0.0004; N.S., not significant; two-way ANOVA with Sidak’s multiple comparison test.
See also Figures S1–S5.
We first determined effects on the large-scale ratcheting rotation of the 40S relative to the 60S that occurs during elongation (Cornish et al., 2008; Frank and Agrawal, 2000; Zhang et al., 2009). Cryo-EM structure analysis revealed that of the FLAG-RACK1-bound ribosome particles recovered, an expected balance of 40S rotated (40%) and 40S nonrotated (60%) ribosomes was observed in WT RACK1 cells (Figures 1B and S2). In contrast, 78% of S278E RACK1-bound ribosomes were found to be in a 40S rotated state (Figures 1B and S3). Additional focused subclassification of particles did not reveal heterogeneous mixtures of particles with other translation factors (Figure S4). 40S rotated ribosomes were associated with eEF2, E-site tRNA, and Ebp1 (Figure 1C), with well-defined SERBP1 density also observed on both WT and S278E RACK1 ribosomes (Figure S5A). In line with this, polysome analyses suggested that SERBP1, Ebp1, and eEF2 shifted distribution from free fractions to ribosome fractions (Figure 1A). Previous reports classify SERBP1-eEF2-Ebp1-80S complexes as inactive because SERBP1 obstructs the 40S aminoacyl (A) and peptidyl (P) tRNA binding sites, as well as the mRNA entry channel, making them incompatible with elongation (Anger et al., 2013; Ben-Shem et al., 2011; Brown et al., 2018; Wells et al., 2020). Although loss of tRNA and mRNA in our rapid purification approach prevents us from distinguishing translationally active versus inactive ribosomes, or further subclassifying ribosome states, we do observe nascent chain density in our ribosome reconstructions (Figure 1C). This suggests that these ribosomes were translationally active at or near the time of isolation. Furthermore, increased levels of these ribosome populations did not correlate with translational suppression, because 35S-methionine/cysteine labeling demonstrated that rescue of RACK1 knockout cells with either WT or S278E RACK1 stimulates overall translation and that each form does so in proportion to the level of RACK1 expression (Figure 1D). In addition, despite modest differences in overall translation rates, there were no significant differences in the steady-state levels of housekeeping proteins tested, although elevated heat shock protein 40 (HSP40) levels in the S278E RACK1-expressing cells hinted that negative charge in the loop may selectively regulate certain transcripts (Figure 1E). Moreover, SERBP1 depletion did not stimulate translation in either cell line, suggesting that it does not have a substantial repressive effect (Figure S5B). Although SERBP1 can repress translation in vitro in some contexts, it does not do so in others (Abaeva et al., 2020; Balagopal and Parker, 2011; Hayashi et al., 2018; Zinoviev et al., 2015). By contrast, SERBP1 is required for translation in living yeast and can help create ribosome pools that are rapidly reactivated during stress recovery (Balagopal and Parker, 2011; Coppolecchia et al., 1993; Van Dyke et al., 2006, 2009, 2013). SERBP1 is also abundant on mammalian ribosomes and acts as an oncogene (Muto et al., 2018), suggesting that its function in vivo is likely to modulate ribosome availability rather than purely inactivate translation. Although our structures align with previously described structures of inactive ribosomes, our functional data suggest that in at least some contexts, these inactive states may be dynamic intermediates in resolving, for example, termination events but have little impact on overall translational output. As such, the increased abundance of this ribosome pool is likely an indirect consequence of broader changes in ribosome dynamics, also evident in changes in polysome versus disome abundance (Figure S1C), that are caused by RACK1 loop charge as part of how it modulates translation.
Negative charge in the RACK1 loop remodels the ribosome A and E sites
To explore the extent to which RACK1 loop charge alters ribosome activity, we determined the sensitivity of these cells to ribosome-targeting drugs (Figure 2A). We first treated cells with anisomycin, an elongation inhibitor that binds to the 60S A site. 35S-methionine/cysteine labeling showed that anisomycin effectively repressed translation in our no-rescue and WT RACK1 rescue cells (Figure 2B). In the S278E RACK1 rescue cells, anisomycin treatment similarly impaired the synthesis of most proteins (Figure 2B). However, the translation of a subset of cellular transcripts was sustained even with a ten-fold increase in drug concentration (Figure 2B). Similar results were obtained using cycloheximide, an elongation inhibitor that binds to the 60S E site (Figure 2C), and emetine, another E-site-targeting elongation inhibitor that binds to the 40S subunit (Figure 2D). The continued sensitivity of many proteins to all three inhibitors, along with the insensitivity of specific proteins to a ten-fold increase in drug concentration, demonstrates that this phenomenon is not simply an inhibitor-dosing effect or a reflection of modest differences in RACK1 expression but rather is a specific and selective effect of S278E RACK1 on ribosome activity. In line with earlier data suggesting that increases in SERBP1-associated ribosome pools are reflective rather than causal of effects of S278E RACK1, depletion of SERBP1 did not affect the synthesis of cycloheximide-resistant proteins (Figure S5C). Furthermore, cells expressing either WT or S278E RACK1 exhibited a similar ribotoxic stress response (RSR) that is activated to varying extents by 60S- and 40S-targeting drugs, resulting in phosphorylation of stress kinases p38 and JNK (Iordanov et al., 1997; Laskin et al., 2002; Vind et al., 2020; Wu et al., 2020). We find that the RSR is not activated in RACK1 knockout cells (Figures 2B–2D), which is in line with studies in other cell types (Kim et al., 2019) and demonstrates that RACK1 mediates these ribosome-centric stress signals. The RSR was restored in WT or S278E RACK1 rescue lines, and in line with prior studies, the most potent response was elicited by anisomycin and to a lesser extent by cycloheximide (Figures 2B and 2C). Further in line with other systems (Wu et al., 2020), only lower concentrations of emetine induced modest activation of p38 above the effects of solvent controls (Figure 2D). In the absence of differential effects on RSR signaling, these results suggested that inhibitor resistance likely resulted from effects of RACK1 loop charge on the ribosome.
Figure 2. Negative charge in the RACK1 loop confers resistance to ribosome-targeting drugs.
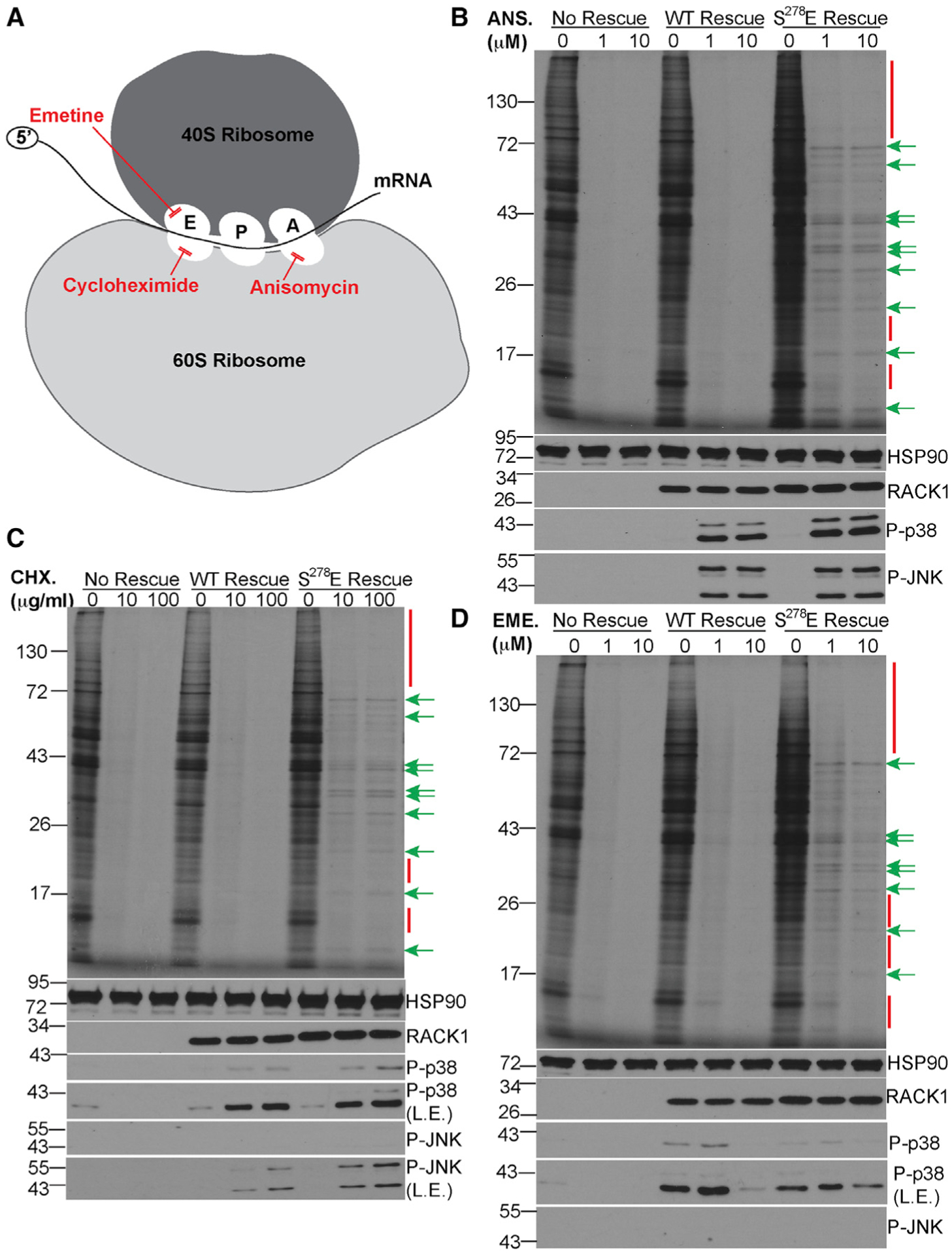
(A) Schematic of the ribosome and target sites of inhibitors used in (B)–(D).
(B–D) 35S-Met/Cys-labeling gels (top panel) and western blot analysis (bottom panels) of cells treated with the indicated concentrations of anisomycin (ANS; B), cycloheximide (CHX; C), or emetine (EME; D). Red bars/arrows highlight examples of proteins whose synthesis is repressed by inhibitors. Green arrows highlight examples of proteins whose synthesis is sustained. P-p38, phosphorylated p38; P-JNK, phosphorylated JNK; L.E., long exposure. Representative of 3 independent biological replicates.
See also Figure S5.
To test this, we performed rigid-body fitting of emetine- and anisomycin-bound ribosome structures into our reconstructions. For emetine fitting, densities of unknown origin at guanine 961 (G961) of 18S rRNA, a key residue of the 40S E-site binding pocket (Meng et al., 2010), partially occlude the emetine binding pocket through a pronounced interaction with the E-site tRNA compared with the reconstruction of WT RACK1-bound ribosomes (Figure 3A). Anisomycin fitting also reveals unidentified densities at uridine 4452 (U4452) and pseudouridine 4531 (ψ4531) of the 60S subunit 28S rRNA that overlaps with anisomycin in the A-site binding pocket (Figure S5D). U4452 and ψ4531 are also key functional residues of the 60S peptidyl transferase center (PTC) (Dao Duc et al., 2019; Shanmuganathan et al., 2019; Yanshina et al., 2015). To examine PTC activity, we treated cells with puromycin, which is incorporated into nascent chains in the PTC. Puromycin treatment effectively halted translation in all lines tested, as represented by the smear of puromycin-terminated peptides in samples treated with lower concentrations (Figure 3C). However, a persistent protein roughly 72 kDa in size continued to be synthesized in the puromycin-treated S278E RACK1 samples. Densities, including that of the nascent chain, may also affect puromycin binding to some extent (Figure S5E), which may explain the continued synthesis of this specific protein. However, S278E RACK1-bound ribosomes are mostly puromycin sensitive and PTC activity is not grossly altered. Given the competitive nature by which these elongation inhibitors operate, certain mRNAs and nascent peptides likely escape their effects because of the altered densities we observe in the A and E sites of S278E RACK1-containing ribosomes that likely reduce inhibitor efficacy.
Figure 3. A negatively charged RACK1 loop affects the ribosomal E-site and RQC reporter activity.
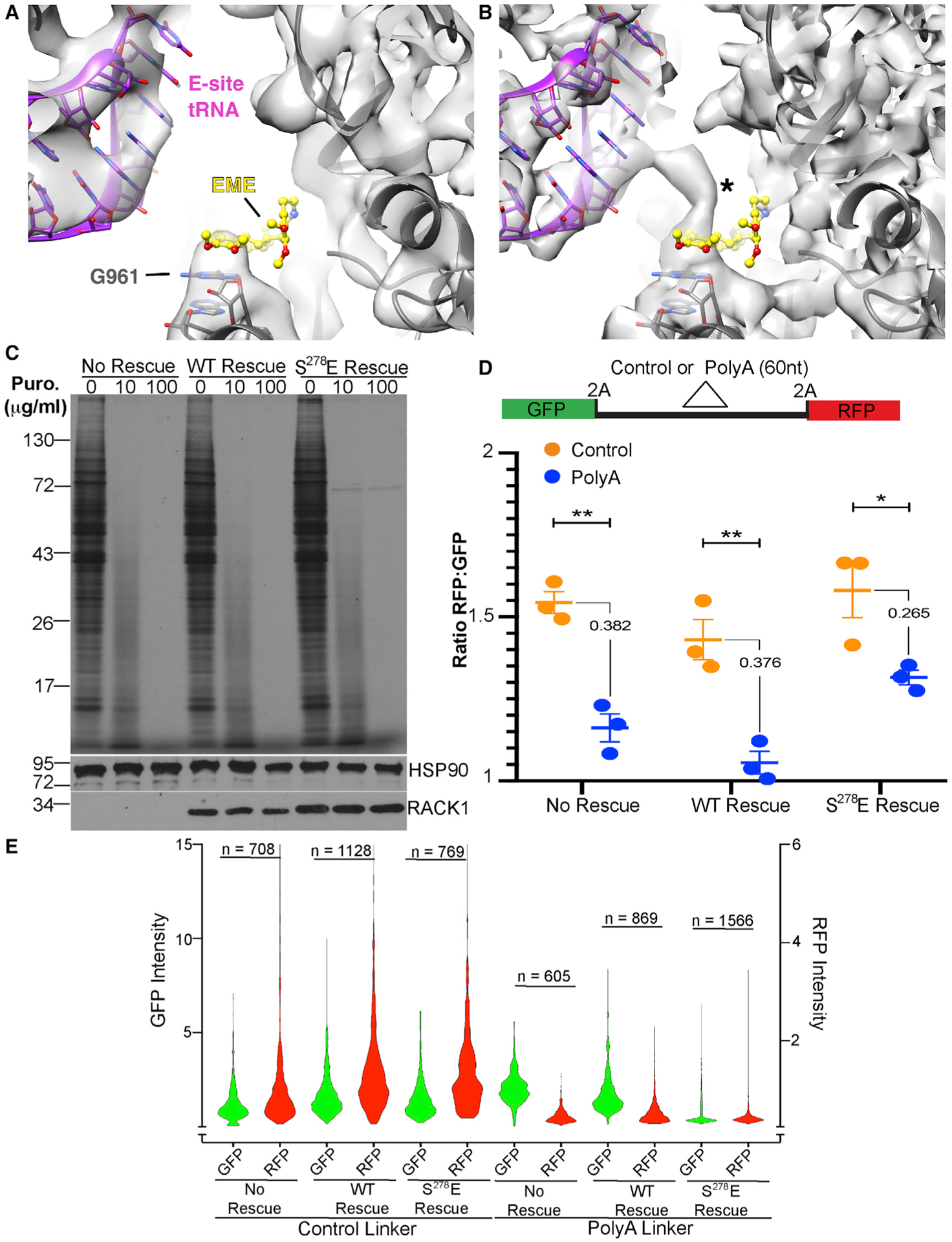
(A and B) Views of the EME binding site WT RACK1 (A) and S278E RACK1 (B) 80S reconstructions (rotated state shown). In S278E RACK1, an unidentified density connects G961 of the 18S rRNA with the E-site tRNA (asterisk). EME modeling based on PDB: 3J7A (Wong et al., 2014).
(C) 35S-Met/Cys-labeling gels (top panel) and western blot analysis (bottom panels) of cells treated with the indicated concentrations puromycin (Puro). Representative of 3 independent biological replicates.
(D) Top: schematic of control or poly(A) RQC reporters, with 2A protease and linker sites indicated. Bottom: densitometry-based quantification of GFP and RFP from western blot analysis of cells transfected with RQC reporters, presented as the RFP:GFP ratio. n = 3; no rescue **p = 0.002, WT rescue **p = 0.006, S278E rescue *p = 0.037; unpaired t test between control and poly(A) reporter. The numeric difference in ratio between each reporter is also shown.
(E) Fluorescence intensity measurements of GFP or RFP (reported as arbitrary units) in cells transfected with RQC reporters, presented as violin plots. n = number of fluorescent cells analyzed over 3 independent biological replicates.
See also Figures S5 and S6.
Negative charge in the RACK1 loop alters ribosome behavior toward poly(A) sequences and enables eIF4A-independent translation
Given its effects on A-site inhibitors, to determine whether S278E RACK1 also influences stall resolution on poly(A) tracts that interact with the A site (Chandrasekaran et al., 2019; Tesina et al., 2020), we transfected our no-rescue and rescue cells with dual fluorescence translational stall reporters (Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017). The reporter contains an N-terminal GFP and C-terminal red fluorescent protein (RFP) flanked by 2A protease sites to generate individual as opposed to fusion proteins, which are separated by either a control linker or a 60 adenosine stall sequence (Figure 3D). Normally, both GFP and RFP are made from the control reporter, but due to ribosome stalling, more GFP than RFP is made from the poly(A) construct. Densitometry of GFP and RFP detected by western blotting revealed that as expected, the poly(A) stall reporter produced less RFP relative to GFP than the control linker reporter in both no-rescue and WT RACK1 rescue lines (Figure 3D). Although we do not explore the potential for differences in frameshifting on these poly(A) constructs, prior studies using the same reporter and readouts found a requirement for RACK1 in regulating RFP levels from this stall reporter in HEK293T cells. However, these studies also showed that stalling depended on the levels of poly(A) reporter expression and is more reliant on ZNF598 than RACK1 (Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017). As such, our failure to observe a significant requirement for RACK1 in HAP1 cells likely reflects differences in translation rates or cell-type-specific differences in requirements for RACK1 for robust stalling on poly(A) sequences.
By contrast, the difference in GFP to RFP expression with the poly(A) reporter and between control linker and poly(A) linker transfections was notably smaller in S278E RACK1 rescue cells (Figure 3D). To explore this in more detail, we performed single-cell fluorescence analysis of GFP and RFP intensity. Presented as violin plots, we observe that WT and S278E RACK1 increase GFP and RFP expression from the control reporter plasmid compared with no-rescue cells (Figure 3E), which aligns with the increased translation detected using 35S-methionine/cysteine labeling earlier. Results also confirm that the relative expression of RFP to GFP is reduced in no-rescue and WT RACK1 rescue cells with the poly(A) reporter. However, although GFP and RFP levels were more equivalent in S278E RACK1 cells transfected with the poly(A) reporter, in line with densitometry-based assessment of overall GFP:RFP ratios, this effect appeared to at least partly result from reduced GFP expression (Figure 3E). Presenting each cell as a single data point based on its GFP and RFP fluorescence intensity would suggest that the S278E RACK1 cells behave normally in stalling, because we observe a reduced slope in plots in all three lines transfected with the poly(A) reporter (Figure 4A). This is similar to results observed in other cell types using fluorescence-activated cell sorting (FACS) analysis and is indicative of stalling (Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017). However, more detailed analysis of this data showed that although some cells expressed high levels of GFP and low levels of RFP in line with conventional stalling, a larger fraction of S278E RACK1-expressing cells produced RFP with relatively little or no GFP (Figures 4B, S6A, and S6B). Altogether, these data suggest that although S278E RACK1 does not affect ribotoxin signaling or poly(A)-based stalling, it may favor internal initiation on the poly(A) sequence. Several viruses that infect dicot plants encode long poly(A) sequences that support internal initiation (Dorokhov et al., 2006; May et al., 2017; Várallyay et al., 2010). However, unlike mammalian IRESs, these poly(A) elements are unstructured, and their ability to act in an IRES-like manner may relate to dicot plant RACK1 loops being negatively charged.
Figure 4. A negatively charged RACK1 loop broadly enables eIF4A-independent translation.
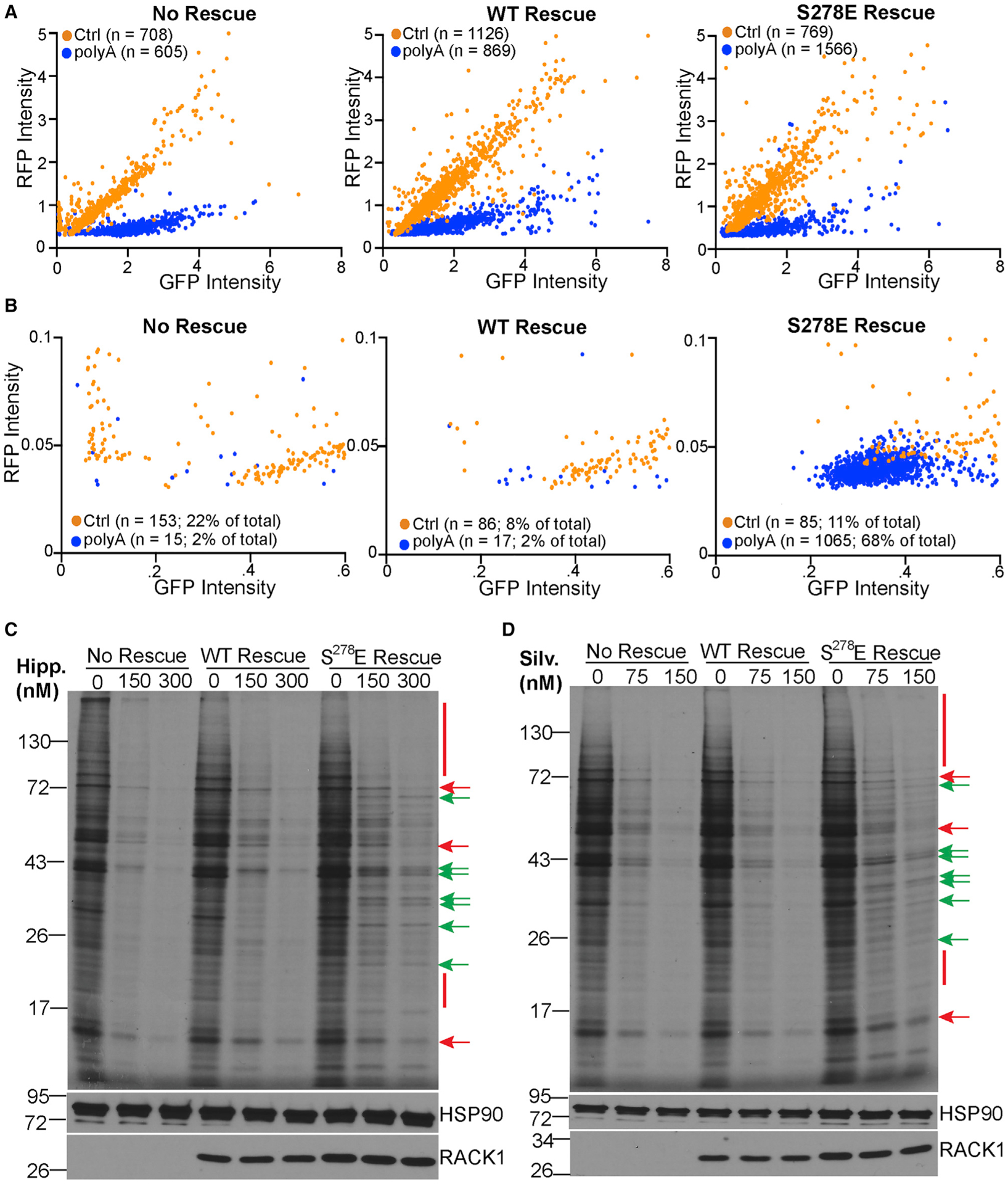
(A and B) Fluorescence intensity measurements of GFP and RFP (reported as arbitrary units) in cells transfected with control (Ctrl) or poly(A) RQC reporters as in Figure 3E. Each cell is presented as an individual data point. n = number of fluorescent cells analyzed over 3 independent biological replicates. The whole dataset is shown in (A). The zoomed dataset in (B) highlights the large population of cells in S278E RACK1 rescue lines that express RFP but little GFP.
(C and D) 35S-Met/Cys-labeling gels (top panel) and western blot analysis (bottom panels) of cells treated with the indicated concentrations of hippuristanol (Hipp; C) or silvestrol (Silv; D). Red bars/arrows highlight examples of proteins whose synthesis is repressed by inhibitors. Green arrows highlight examples of proteins whose synthesis is sustained. Representative of 3 independent biological replicates.
See also Figure S6.
Because these GFP:RFP assays are indirect measures of translation and are limited in scope, to determine whether negative charge in the loop more broadly facilitates alternative initiation, we treated cells with either hippuristanol or silvestrol, two inhibitors that block eIF4A activity in distinct manners (Cencic and Pelletier, 2016; Liu et al., 2012). eIF4A is an RNA helicase essential for cap-dependent scanning, the primary mode of initiation in eukaryotes (Hinnebusch, 2011, 2014; Pestova and Kolupaeva, 2002). Both inhibitors repressed translation in the no-rescue and WT RACK1 rescue cells (Figures 4C and 4D), exhibiting a dose-dependent profile that would be expected for increasingly impaired eIF4A activity. Synthesis of many proteins was equivalently suppressed in a dose-dependent manner in S278E RACK1-expressing cells. However, S278E RACK1 sustained the synthesis of several proteins even at higher inhibitor concentrations (Figures 4C, 4D, and S6C). These results indicate that negative charge in the RACK1 loop fundamentally alters the ribosome to enable eIF4A-independent translation of subsets of cellular mRNAs.
Negative charge in the RACK1 loop affects 40S head rotation
We analyzed our cryo-EM datasets to determine whether other structural changes to S278E RACK1-bound ribosomes occur and potentially explain the ability to support eIF4A-independent initiation. The structures of our WT RACK1-bound 80S particles are consistent with published structures of the human 80S ribosome in rotated (PDB: 6Z6M) and nonrotated states (PDB: 4UG0) (Khatter et al., 2015; Wells et al., 2020). In addition, ribosomes in the rotated state are structurally superimposed between WT and S278E RACK1 datasets (Figure 5A). As such, the addition of negative charge to the RACK1 loop does not alter the normal trajectory of ribosome ratcheting, because the rotated states of both WT and S278E RACK1 80S are superimposable and consistent with published structures of the 80S ribosome (Brown et al., 2018). However, within nonrotated datasets, an overlay of the WT and S278E RACK1-bound ribosomes revealed striking differences. The structures overlaid well at the 60S subunit and 40S body, but not at the 40S head region (Figure 5B). The nonrotated S278E RACK1 reconstruction exhibits an unusually greater degree of swiveling, in which the 40S head shifts toward the 60S subunit and altered contacts with other RPs could be detected (Figures 5B–5D).
Figure 5. The 40S head is displaced in S278E RACK1-containing ribosomes.
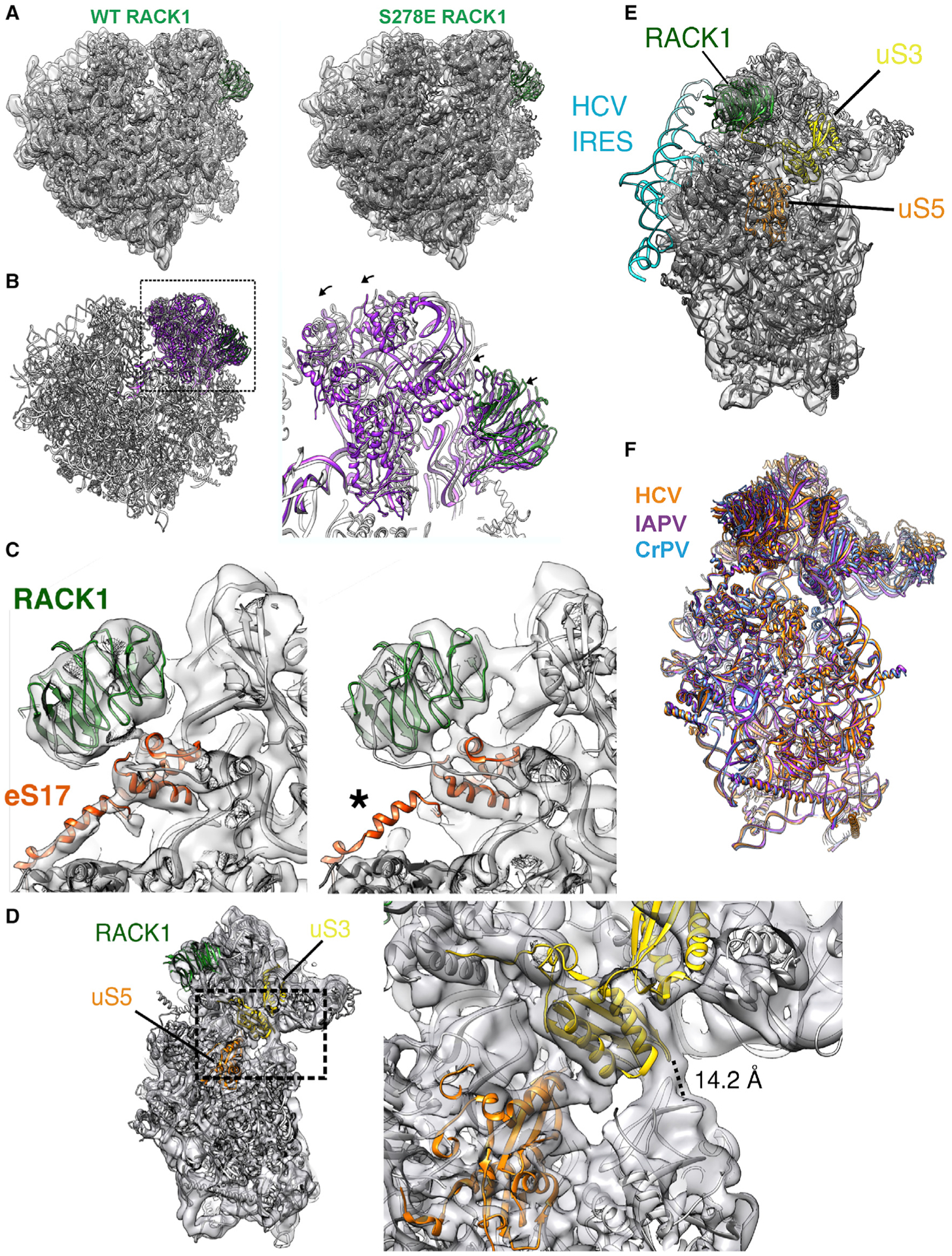
(A) Rigid-body fits of the human 80S ribosome in the nonrotated state (PDB: 4UG0) show agreement in the 60S and 40S body for both WT and S278E RACK1 reconstructions.
(B) In contrast, the fitting is inconsistent at the 40S head between the two reconstructions (WT, gray; S278E, purple). Arrows indicate the direction of S278E 40S head displacement toward the 60S.
(C) Closeup views of the RACK1-eS17 interface in WT (left) and S278E (right) reconstructions of nonrotated 80S particles. eS17 contains a connecting helix between the 40S body and the 40S head, which is less pronounced in the S278E reconstruction (asterisk).
(D) Reconstruction of 40S particles isolated from S278E RACK1 purifications (left). Zoomed-in view of the latch separating the 40S head and body. The distance between Q179 of uS3 and G610 of the 18S rRNA is indicated and consistent with the 40S latch in the closed conformation.
(E) Reconstruction of S278E 40S particles shows agreement with the rigid-body fit of the 40S ribosome bound to HCV IRES (PDB: 5A2Q).
(F) Overlaid models of IRES-bound 40S subunits are generally superimposable (HCV, orange, PDB: 5A2Q; IAPV, purple, PDB: 6P4G; CrPV, blue, PDB: 7JQC; IRES models removed for clarity).
See also Figures S2 and S3.
Intriguingly, the type III or IV IRES elements of RNA viruses such as hepatitis C virus (HCV), Israeli acute paralysis virus (IAPV), or cricket paralysis virus (CrPV) manipulate 40S head rotation to enable cap- and scanning-independent initiation (Acosta-Reyes et al., 2019; Murray et al., 2016; Quade et al., 2015; Spahn et al., 2001; Yamamoto et al., 2015). These IRESs also interact with RACK1 and require RACK1 for their translation (Jackson, 2013; Johnson et al., 2019; LaFontaine et al., 2020; Majzoub et al., 2014; Qin and Sarnow, 2004). Given that IRESs are not widely prevalent in cellular mRNAs and eIF4A-independent translation was specific to S278E RACK1-expressing cells, we examined whether the 40S head rotation induced by S278E RACK1 mimics that induced by IRES elements. Our superimpositions revealed that the S278E RACK1 40S-only reconstructions are consistent with the structures of 40S subunits bound to HCV, CrPV, and IAPV IRES (Figures 5D–5F). Altogether, these data suggest that similar to certain IRESs, negative charge in the RACK1 loop alters 40S head swivel, which may unlock atypical modes of initiation.
DISCUSSION
Although the finer structural and mechanistic details remain to be determined, our findings reveal the broad extent to which a single phosphomimetic or charged residue in the RACK1 loop can alter ribosome structure, dynamics, and translational capacity.
The breadth of effects of S278E RACK1 on ribosome structure and function was somewhat unexpected but likely originates from effects on the behavior of the 40S head, where RACK1 is positioned. Charge in the RACK1 loop altered local contacts in the latch and increased 40S head swivel. This may destabilize the nonrotated state and drive the rate of formation of SERBP1-eEF2-EBP1-associated rotated 80S ribosomes. Alternatively, broader effects of RACK1 loop charge on ribosome dynamics may influence binding of eEF2 and indirectly drive 40S rotation, which is difficult to test experimentally given eEF2’s essentiality. Beyond these direct or indirect effects on the levels of rotated 80S ribosomes, RACK1 loop charge specifically altered 40S head swivel on nonrotated 80S and free 40S subunits. This increase in 40S head swivel motion may alter 40S and 60S contacts that affect the organization and function of A and E sites, leading to the inhibitor resistance that we observe.
Viruses often evolve strategies to dysregulate tightly controlled processes, and in this case, negative charge in the human RACK1 loop appears to dysregulate and broaden the functionality of the human ribosome to support the noncanonical modes of translation that many RNA viruses use. More directly related to effects on 40S head swivel, S278E RACK1 mimics the 40S remodeling induced by structurally complex HCV, CrPV, and IAPV IRES elements that drive 80S assembly with minimal dependence on eIFs (Acosta-Reyes et al., 2019; Neupane et al., 2020; Quade et al., 2015; Spahn et al., 2001; Yamamoto et al., 2015). By contrast, poxviruses are DNA viruses that generate mRNAs with very short 5′ poly(A) leaders (Meade et al., 2019a). Early studies reported that such leaders have reduced dependence on eIFs or scanning but lack the structural complexity of true IRES elements (Chandrasekaran et al., 2019; Dhungel et al., 2017; Dorokhov et al., 2002; Mulder et al., 1998; Shirokikh and Spirin, 2008; Tang and Passmore, 2019). Poly(A) leaders are foreign to their mammalian hosts, and their maximal activity requires either poxvirus infection or expression of phosphomimetic S278E RACK1 (Dhungel et al., 2017; Dorokhov et al., 2002; Jha et al., 2017; Mulder et al., 1998; Rollins et al., 2019; Shirokikh and Spirin, 2008; Sundaramoorthy et al., 2021; Tang and Passmore, 2019). Our findings suggest that S278E RACK1 likely primes 40S subunits in a similar way to IRESs to initiate on mRNAs with little to no scanning. Beyond our earlier focus on 5′ poly(A) leaders, data here show that a negatively charged RACK1 loop more broadly enables eIF4A-independent translation of many cellular mRNAs. Given that human mRNAs do not contain 5′ poly(A) tracts and bona-fide IRESs are rare, this suggests that poxviruses introduce negative charge to the RACK1 loop not to control a process unique to their poly(A) leaders but to maximize the capacity of ribosomes to accommodate alternative modes of translation that benefit different leader types. Overall, our findings suggest that in lieu of more complex IRES structures, modifications to RPs such as RACK1 can achieve similar effects to unlock noncanonical modes of translation by the human ribosome.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Please direct any requests for further information or reagents to the lead contact, Derek Walsh (derek.walsh@northwestern.edu)
Materials availability
Plasmids and cell lines generated in this study are available upon request from the lead contact.
Data and code availability
Electron microscopy maps and locally refined maps generated in this manuscript have been deposited in Electron Microscopy Data Bank (EMDB) and are publicly available. Accession numbers are listed in the Key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-HSP90 (C45G5) | Cell Signaling Technology | Cat# 4877; RRID: AB_2233307 |
| Rabbit polyclonal anti-HSP40 | Cell Signaling Technology | Cat# 4868, RRID: AB_2094249 |
| Rabbit monoclonal anti-phospho-SAPK/JNK (Thr183/Tyr185) (81E11) | Cell Signaling Technology | Cat# 4668; RRID: AB_823588 |
| Rabbit monoclonal anti-RACK1 (D59D5) | Cell Signaling Technology | Cat# 5432; RRID: AB_10705522 |
| Rabbit anti-phospho-p38 MAPK (Thr180/Tyr182) | Cell Signaling Technology | Cat# 9211; RRID: AB_331641 |
| Rabbit anti-eIF4G | (Walsh and Mohr, 2006) | N/A |
| Rabbit anti-PABP | (Walsh and Mohr, 2006) | N/A |
| Rabbit polyclonal anti-eEF2 | Bethyl Laboratories | Cat# A301-688A; RRID: AB_1210953 |
| Rabbit polyclonal anti-SERBP1 | Bethyl Laboratories | Cat# A303-938A; RRID: AB_2620287 |
| Rabbit polyclonal anti-Ebp1 | Bethyl Laboratories | Cat# A303-084A; RRID: AB_10923145 |
| Rabbit monoclonal anti-RPL11 (D1P5N) | Cell Signaling Technology | Cat# 18163; RRID: AB_2798794 |
| Rabbit polyclonal anti-RPS2 (N2C3) | GeneTex | Cat# GTX114734; RRID: AB_10620358 |
| Rabbit monoclonal anti-GFP (D5.1) XP | Cell Signaling Technology | Cat# 2956; RRID: AB_2798794 |
| Mouse monoclonal anti-RFP [6G6] | ChromoTek | Cat# 6g6-100; RRID: AB_2631395 |
| Mouse monoclonal anti-β-actin (8H10D10) | Cell Signaling Technology | Cat# 3700; RRID: AB_2242334 |
| Mouse monoclonal anti-α-tubulin (DM1A) | Sigma-Aldrich | Cat# T6199; RRID: AB_477583 |
| Mouse monoclonal anti-GAPDH (0411) | Santa Cruz Biotechnology | Cat# sc-47724; RRID: AB_627678 |
| Rabbit monoclonal anti-Raptor (24C12) | Cell Signaling Technology | Cat# 2280; RRID: AB_561245 |
| Mouse monoclonal anti-FLAG M2 | Sigma Aldrich | Cat# A2220; RRID: AB_10063035 |
| Bacterial and virus strains | ||
| Subcloning Efficiency DH5α Competent Cells | Thermo Fisher Scientific | Cat# 18265017 |
| Chemicals, peptides, and recombinant proteins | ||
| Hygromycin B | Thermo Fisher Scientific | Cat# 10687010; CAS: 31282-04-9 |
| Hippuristanol | Laboratory of Jerry Pelletier | N/A |
| Silvestrol | MedChemExpress | Cat# HY-13251; CAS: 697235-38-4 |
| Anisomycin from Streptomyces griseolus | Millipore Sigma | Cat# 9789; CAS: 22862-76-6 |
| Cycloheximide | Research Products International | Cat# 81040; CAS: 66-81-9 |
| Emetine hydrochloride hydrate | Cayman Chemical | Cat# 21048; CAS: 7083-71-8 |
| Puromycin, dihydrochloride | Millipore Sigma | Cat# 540222; CAS: 58-58-2 |
| Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | Cat# L3000-008 |
| Lipofectamine RNAiMAX Transfection Reagent | Thermo Fisher Scientific | Cat# 13778-075 |
| Pierce ECL Western Blotting Reagent | Thermo Fisher Scientific | Cat# 32106 |
| SuperSignal West Femto Maximum Sensitivity Substrate | Thermo Fisher Scientific | Cat# 34096 |
| Diethyl pyrocarbonate | MP Biomedicals | Cat# 02150902-CF; CAS: 1609-47-8 |
| Dithiothreitol (DTT) | Acros Organics | Cat# 32719-0100; CAS: 3483-12-3 |
| Triton X-100 | Fisher Scientific | Cat# BP151-500; CAS: 9002-93-1 |
| RiboLock RNase Inhibitor | Thermo Fisher Scientific | Cat# EO0382 |
| Pierce Protease Inhibitor Mini Tablets, EDTA-free | Thermo Fisher Scientific | Cat# A32955 |
| Acetone | Fisher Scientific | Cat# A949-1; CAS: 67-64-1 |
| Trichloroacetic acid | Fisher Scientific | Cat# A322-500; CAS: 76-03-9 |
| FluorSave Reagent | Millipore Sigma | Cat# 345789 |
| Hoescht 33342 | Thermo Fisher Scientific | Cat# 62249 |
| SpeI | New England BioLabs | Cat# R0133S |
| NotI-HF | New England BioLabs | Cat# R3189S |
| Cryo-EM data | ||
| WT 80S rotated state | This paper | EMD-24545 |
| WT 80S nonrotated state | This paper | EMD-24548 |
| S278E 80S rotated state | This paper | EMD-24541 |
| S278E 80S nonrotated state | This paper | EMD-24542 |
| S278E 40S | This paper | EMD-24544 |
| Locally refined maps | ||
| WT 80S rotated state (60S) | This paper | EMD-24554 |
| WT 80S rotated state (40S body) | This paper | EMD-24556 |
| WT 80S rotated state (40S head) | This paper | EMD-24557 |
| S278E 80S rotated state (60S) | This paper | EMD-24549 |
| S278E 80S rotated state (40S body) | This paper | EMD-24552 |
| S278E 80S rotated state (40S head) | This paper | EMD-24553 |
| Experimental models: Cell lines | ||
| Human: HAP1 RACK1 knockout (no rescue) cells | (Jha et al., 2017) | N/A |
| Human: HAP1 RACK1-WT-Flag rescue cells | This paper | N/A |
| Human: HAP1 RACK1-S278E- Flag rescue cells | This paper | N/A |
| Human: HEK293-T | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| Oligonucleotides | ||
| Primer: RACK1 SpeI Forward AAAAAACTAGTCTCAAGCTTATGACTGAGCAGATG | (Jha et al., 2017) | N/A |
| RACK1-Flag NotI Reverse ACCGAGCGGCCGCC TACTTGTCGTCATCGTCTTTGTAGTCGCCGCTG CCGCGTGTGCCAATGGT | This paper | N/A |
| Silencer Negative Control No. 1 siRNA | Thermo Fisher Scientific | Cat# AM4635 |
| Silencer Select SERBP1 siRNA - #1 | Thermo Fisher Scientific | Cat# 4392420, ID: s25142 |
| Silencer Select SERBP1 siRNA - #2 | Thermo Fisher Scientific | Cat# 4392429, ID: s25143 |
| Recombinant DNA | ||
| pLVX-IRES-Hygromycin plasmid | Takara Bio USA, Inc. | Cat# 632185 |
| WT-RACK1 FLAG | This paper | N/A |
| S278E-RACK1-FLAG | This paper | N/A |
| pmGFP-P2A-K0-P2A-RFP | (Juszkiewicz and Hegde, 2017) | RRID: Addgene_105686 |
| pmGFP-P2A-K(AAA)20-P2A-RFP | (Juszkiewicz and Hegde, 2017) | RRID: Addgene_105688 |
| Software and algorithms | ||
| Chimera or Chimera X | (Pettersen et al., 2004) | RRID: SCR_004097 https://www.cgl.ucsf.edu/chimera/ |
| FIJI | (Schindelin et al., 2012) | RRID: SCR_002285; https://fiji.sc/ |
| GraphPad Prism (version 8.0) | GraphPad Software Inc. | RRID:SCR_002798; https://www.graphpad.com/scientific-software/prism/ |
| Metamorph Microscopy Automation and Image Analysis Software | Molecular Devices | RRID:SCR_002368; https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy |
| CellProfiler Image Analysis Software | (McQuin et al., 2018) | RRID:SCR_007358; https://cellprofiler.org/releases |
| cryoSPARC | (Punjani et al., 2017) | RRID:SCR_016501; https://cryosparc.com |
| RELION | (Scheres, 2012) | RRID:SCR_016274; https://cam.ac.uk/relion |
| pyem | Daniel Asarnow, Eugene Palovcak, Yifan Cheng | https://zenodo.org/record/3576630 |
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and culture conditions
Parental HAP1 cells (Horizon, C859), HAP1 RACK1 knockout cells (Jha et al., 2017), HAP1 RACK1-WT-Flag rescue cells and HAP1 RACK1-S278E-Flag rescue cells were grown in Iscove’s Modified Dulbecco’s medium with 4mM L-glutamine and HEPES (IMDM; Fisher Scientific, SH3022801) that was supplemented with 5% fetal bovine serum (FBS). 293T cells used to propagate lentivirus were obtained from the ATCC. 293T cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM; Fisher Scientific, MT15017CV) supplemented with 2 mM L-glutamine and 5% FBS. For maintenance of RACK1-WT-Flag and RACK1-S278E-Flag rescue HAP1 cells, IMDM was supplemented with 200 μg/ml Hygromycin B; however, Hygromycin B was removed from the media for all experiments performed. All cell cultures were maintained at 37°C with 5% CO2 and confirmed negative for mycoplasma using Hoechst staining.
Generation of cell lines
Generation of HAP1 RACK1 knockout cells was previously described (Jha et al., 2017). Plasmids for rescue with either WT or S278E phosphomimetic RACK1 with a C-terminal Flag were generated by first PCR amplifying RACK1 forms from existing pLVX-IRES hygro plasmids expressing wild-type RACK1 or S278E RACK1 (Jha et al., 2017). Primers incorporated the appropriate restriction sites and the C-terminal Flag tag (RACK1 SpeI Forward: AAAAAACTAGTCTCAAGCTTATGACTGAGCAGATG; RACK1-Flag NotI Reverse: ACCGAGCGGCCGCCTACTTGTCGTCATCGTCTTTGTAGTCGCCGCTGCCGCGTGTGCCAATGGT). Using standard cloning procedures, amplicons were purified (QIAGEN PCR purification, 28104), digested with SpeI and NotI (New England Biolabs), and ligated into empty pLVX-IRES-Hygromycin vector (Takara Bio USA, Inc.) to generate the Flag-tagged constructs. All constructs were verified by sequencing at ATGC, Inc.
Lentivirus vectors were produced by co-transfection of 293T cells with the Flag-tagged constructs, together with p8.91 (gag-pol) and p-VSVG (envelope). Supernatants containing lentivirus were then filtered and used to transduce the HAP1 RACK1 knockout cells. The transduced cultures were then selected with 1mg/ml Hygromycin B to generate pools of HAP1 RACK1 knockout cells rescued with C-terminally Flag-tagged WT RACK1 or S278E RACK1.
METHOD DETAILS
Sucrose gradient centrifugation and TCA precipitation
Sucrose gradient centrifugation and polysome analysis was performed as previously described (Jha et al., 2017). Briefly, HAP1 RACK1 knockout and rescue cells were grown to confluency, pre-treated with cycloheximide for 10 min and washed twice in ice-cold PBS containing 100 μg/ml cycloheximide prior to collection. Cells were harvested in ice-cold 1X polysome lysis buffer (1% Triton X-100, 200 U/ml RiboLock RNase Inhibitor, complete EDTA-free protease inhibitor tablet, 20 mM Tris-Cl pH 7.5, 10 mM MgCl2, 100 mM potassium acetate, 1 mM DTT, 100 μg/ml cycloheximide) and lysed for 20 min at 4°C. Lysates were clarified by centrifugation (15,000×g for 5 min at 4°C) and 500 μg total RNA were run through 10 mL of 5%–50% sucrose gradient prepared in 1X polysome buffer (20 mM Tris-Cl pH 7.5, 10 mM MgCl2, 100 mM potassium acetate, 1 mM DTT, 100 μg/ml cycloheximide) using a Beckmann Coulter SW 41 Ti rotor at 36,000 rpm at 4°C for 2 h. Following centrifugation, sucrose gradients were fractionated using an automated Density Gradient Fractionation System (Brandel Biomedical Research & Development Laboratories, Inc.) with continuous monitoring at 254nm using an UA-6 absorbance detector and recorded using PeakChart software.
For western blot analysis, the 500 μl fractions were precipitated in TCA at a final concentration of 10% at 4°C overnight. The precipitate was spun down at 10,000×g for 15 min. The supernatant was removed and the pellet was washed twice in a 1:4 solution of 1X polysome buffer:acetone followed by centrifugation at 10,000×g for 15 min. Following removal of the supernatant, protein pellets were air-dried for 10 min, re-suspended in 1X Laemmli buffer and boiled for 3 min. Total lysate samples were not TCA precipitated.
RNAi
Pre-designed siRNAs were acquired from Thermo Fisher Scientific: control non-targeting siRNA (Cat No. AM4635), SERBP1 siRNA #1 (Cat No. 4392420, ID: s25142), SERBP1 siRNA #2 (Cat No. 4392420, ID: s25143). When at approximately 60% confluency, cells were transfected with siRNA (100 pmol/ml) using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific). Complete IMDM was added to the 500 μl OptiMEM used for RNAi approximately 4 h post-transfection. At 48 h post-transfection, cells were trypsinized and re-suspended in IMDM to minimize clumping. At 72 h post-transfection, cells were metabolically labeled and harvested.
Inhibitor treatment
Inhibitor stocks were prepared as follows with the appropriate vehicle noted in parentheses: 10 mM anisomycin (DMSO), 100 mg/ml cycloheximide (70% ethanol), 10 mM emetine (dIH2O), 100 μM hippuristanol (DMSO), 100 μM silvestrol (DMSO), and 100 mg/ml puromycin (PBS). Prior to metabolic labeling, cells were pre-treated with the inhibitors for either 30 min (anisomycin, cycloheximide, emetine and puromycin) or 2h (hippuristanol and silvestrol) and in all cases, inhibitors remained present during labeling. The final concentrations of the inhibitors used for treatment are indicated in the figures.
35S-Methionine/Cysteine labeling and liquid scintillation counting
For metabolic labeling, cells were incubated in methionine/cysteine (Met/Cys)-free DMEM (Corning, 17-204-CL) supplemented with 40 mM HEPES, 2 mM L-glutamine and a 35S-L-methionine and 35S-L-cysteine mix (PerkinElmer, NEG072007MC) for 30 min prior to cell lysis. For each ml of labeling media prepared, 0.035 mCi of the 35S Met/Cys mix was added. Where inhibitors were used, inhibitors were present during labeling. After in-well lysis in Laemmli buffer, samples were resolved by SDS-PAGE and gels were then fixed in 10% acetic acid/25% methanol solution for 30 min. The fixed gels were then dried at 80°C for 2 h using a Model 583 Gel Dryer (Biorad) and exposed to audioradiography film.
To quantify the radioactivity of 35S present in the label samples, 20 μl of radiolabeled sample was incubated with 10 μl of 10 mg/ml BSA and 1 mL of ice-cold 10% TCA solution for 30 min on ice. Precipitated proteins were vacuum filtered using a 1225 Sampling Manifold (Millipore Sigma) onto glass microfiber filters (GE Life Sciences, 1822-025), and washed twice each with ice-cold 10% TCA solution and 95% ethanol. Filter counting was performed by immersing the filters into 3 mL of Complete Counting Cocktail 3a70B (Research Products International Corp., 111154). The number of counts registered per minute (CPM) was measured using a Beckman LS 6500 liquid scintillation counter with a counting time of 5 min.
Western blotting
Western blotting was performed as previously described (Jha et al., 2017). Briefly, whole cell lysates were resolved using 10% Trisglycine SDS-PAGE and transferred to a 0.2 μm pore-size nitrocellulose membrane using a wet electroblotting system. Membranes were blocked in 5% non-fat milk dissolved in 1X Tris-buffered saline and 0.1% Tween (TBS-T) for 1 h at room temperature and then washed in 1X TBS-T before incubation overnight at 4°C with primary antibody diluted in 3% BSA dissolved in 1X TBS-T. For primary antibodies used, please see the Key resources table; all antibodies were used at a 1:1000 dilution except for α-tubulin (1:4000), β-actin (1:10,000), eIF4G (1:5000) and PABP (1:5000). Following primary antibody incubation, membranes were washed and incubated with HRP-conjugated secondary antibody diluted 1:3000 in 5% non-fat dry milk in 1X TBS-T for 1 h at room temperature followed by another washing. For detection, membranes were incubated with Pierce ECL Western Blotting Substrate for 2 min before exposure to audioradiography film. If standard ECL produced low level protein detection, membranes were incubated with Pierce SuperSignal West Femto Maximum Sensitivity Substrate for 2 min. Western blot were quantified using densitometry as described previously (Procter et al., 2018).
Dual fluorescence translation stall assay
HAP1 cells were seeded onto 12-well plates with or without coverslips, depending on downstream analysis. Dual fluorescence reporter plasmids (1 μg) were transfected into cells using Lipofectamine 3000 (Thermo Fisher) according to the manufacturer’s protocol. For transfection experiments where cells were treated with either 300 nM hippuristanol or DMSO solvent control, the vehicle or inhibitor were added 12 h post-transfection. 48 h post-transfection, cells seeded onto plates without coverslips were harvested for western blotting and densitometry was used to measure GFP and RFP levels, as previously described (Meade et al., 2019b). Cells seeded onto plates with coverslips were used to measure cellular GFP and RFP fluorescence and perform single-cell fluorescence analysis.
Fluorescence microscopy and single-cell fluorescence analysis
Immunofluorescence microscopy was performed as previously described (Meade et al., 2018). Briefly, glass coverslips were used to seed cells in a 12-well plate and transfection was carried out as described above. Cells were rinsed in PBS and fixed in 4% paraformaldehyde in PBS at room temperature for direct imaging of GFP and RFP signals. Samples were rinsed in PBS and blocked (10% FBS and 0.25% saponin in PBS) for 1h at 37°C. Samples were then washed (0.025% saponin in PBS) before and after staining with Hoescht 3342 (Thermo Fisher Scientific, 62249). Coverslips were mounted on glass slides using FluorSave (Millipore Sigma, 345780). A Leica DMI6000B-AFC microscope with 100X objective (HC PL APO 100x/1.44NA OIL), X-Cite XLED1 illumination, ORCA FLAH 4.0 cMOS camera was used to acquire wide-field images. Metamorph software using the multi-dimensional acquisition function ensured that acquisition settings remained consistent for all acquired images. Image analysis was completed using Metamorph and compiled using the Fiji distribution of ImageJ (Schindelin et al., 2012). All images within a given dataset were processed equivalently.
For single cell RFP and GFP intensity measurements, randomized images were entered into the CellProfiler pipeline (McQuin et al., 2018; Procter et al., 2020) and resized to 256×256 pixels. The pipeline was used to identify target objects using the images in the RFP channel. Locations were marked based on RFP intensity relative to the background. Once the target object locations were identified, CellProfiler measured the RFP and GFP frequency at the location of each object. The mean intensity measurements of the object locations were used to generate violin plots and single-cell scatterplots.
Ribosome purification for Cryo-EM
In brief, WT RACK1 or S278E RACK1 rescue cells were cultured in IMDM supplemented with 10% (v/v) FBS and 1X penicillin-streptomycin. Cells were grown to 80% confluency in 6 × 150mm plates before discarding media by aspiration and washed with ice cold PBS thereafter. After aspiration, the residual PBS was used to scrape the cells from the dishes and the collected cells were pelleted by centrifugation at 2500 × g for 5 min. Freshly harvest cells were resuspended in IP buffer (100 mM KOAc, 10 mM MgCl2, 25 mM HEPES-KOH pH 7.4, 5% glycerol, 0.2% Igepal CA-630, 1 mM DTT, and a protease inhibitor cocktail comprising 0.5 μg/mL leupeptin, 0.5 μg/mL aprotinin, 0.7 μg/mL pepstatin, and 16.67 μg/mL PMSF) in a 4:1 (w/v) ratio and supplemented with benzonase (2.5 U/mL). Cells were lysed using a Dounce homogenizer submerged in ice with ~60 continuous strokes. Lysates were clarified by centrifugation, and the supernatant was incubated with anti-FLAG M2 affinity resin (Sigma) for 1 hour at 4°C. Resin was washed thoroughly with IP buffer followed by several washes using the same buffer without detergent and glycerol. Ribosome complexes were recovered from the resin by competitive elution with synthetic 3×FLAG peptide (APExBIO) for 1 hour at 4°C with mild agitation. Eluted samples were immediately used for cryo-EM grid preparation.
Electron cryo-microscopy
UltrAuFoil R1.2/1.3 Au300 mesh grids (Quantifoil) were glow discharged using a Pelco easiGlow (Ted Pella, Inc.) for 25 s at 25 mA. 3.5 μL of sample were applied to the glow discharged grid, and grids were vitrified using a Mk. II Vitrobot (Thermo Fisher Scientific) operating at ~85% relative humidity, 4°C, and 2.5 s blot time, and then plunge frozen into liquid ethane.
A total of 3,842 cryo-EM movies of S278E RACK1 samples were recorded using a 300kV Titan Krios G3 (Thermo Fisher Scientific) equipped with a K2 Summit direct detector (Gatan, Inc.) at a nominal magnification of 105K x, corresponding to 1.348 Å pixel size. Movies were recorded using SerialEM with a defocus range of −1.0 to −3.0 μm and at a dose rate of 1.0 e−/Å2/frame with a total exposure of 40 frames and each movie recording time was 8 s (Mastronarde, 2005).
For WT RACK1 samples, a total of 2,141 movies were recorded on a 300kV Titan Krios G3 equipped with a K3 direct detector (Gatan) at a nominal magnification of 81,000× corresponding to 1.058 Å pixel size. Data were collected in super-resolution mode using SerialEM with a defocus range of from −0.8 to −1.8 μm and at a dose rate of 1.1 e−/Å2/frame with a total exposure of 40 frames and each movie recorded for 2.53 s.
Image Processing
Cryo-EM movie frames were dose weighted, motion corrected and summed using MotionCor2 (Zheng et al., 2017). All downstream steps were performed in cryoSPARC, including CTF estimation, particle selection, 2D class averaging, 3D classification, and 3D refinement (Punjani et al., 2017). Micrographs with poor CTF estimates or crystalline ice were discarded. For RACK1S278E images, a total of 191,242 particles were selected from 3,522 micrographs. A total of 103,133 particles were sorted into well-defined classes after two rounds of 2D classification (K = 50) and used for ab-initio 3D reconstruction (K = 5). Resulting classes revealed the separation of 80S particles in 40S rotated and nonrotated states (52,603 and 14,878 particles, respectively), a 40S class (16,758 particles), and two junk classes. The 80S and 40S classes were processed separately using non-uniform refinement followed by local resolution estimation and local filtering. Final resolution estimates were 3.1 Å for the 80S rotated state, 4.0 Å for the 80S nonrotated state, and 5.2 Å for the 40S particle.
For processing of WT RACK1 dataset, a total of 1,985 dose weighted, motion corrected, and summed movie frames (generated using MotionCor2) were used after discarding poor micrographs on the basis of CTF estimation and ice quality. A total of 30,650 particles were resolved into clear 2D classes after two rounds of 2D classification from an initial number of 73,284 boxed particles. Particles were subject to ab-initio 3D reconstruction (K = 3). Resulting classes revealed the separation of 80S particles in 40S head rotated and nonrotated states (10,413 and 15,813 particles, respectively) and one junk class. No 40S classes were recovered. The 80S classes were refined separately using non-uniform refinement, as described above. Final resolution estimates were 4.2 Å for the 80S rotated state and 5.0 Å for the nonrotated state.
In order to address variable local resolutions, local 3D refinement was performed on RACK1S278E and RACKWT 80S rotated particles as implemented in cryoSPARC (Punjani et al., 2017). Separate soft-edged masks were generated for 60S, 40S body, and 40S head components and signal subtraction was carried out to generate three new image stacks that contain only signal from each component (Figures S2 and S3). The signal-subtracted image stacks were then used as the basis for local 3D refinement, leading to maps with more consistent local resolutions across the entire ribosome.
In order to probe for compositional heterogeneity within our dataset, we performed focused 3D classification as implemented in RELION (Scheres, 2012). The 52,603 images of S278E RACK1 rotated 80S particles and 14,878 images of S278E RACK1 non-rotated 80S particles were exported from cryoSPARC to RELION format using pyem (csparc2star.py) (Asarnow et al., 2019). Separate soft-edged masks were generated over the P-site tRNA binding site and the 40S head (Figure S4). Focused classification was performed by applying the masks and disabling particle re-alignment (K = 3–5, tau factor = 6). In each classification run, most particles classified into a single, well-resolved class and remaining particles were classified into low-resolution or junk classes.
Visualization and segmentation of density maps for both datasets were carried out in UCSF Chimera (Pettersen et al., 2004). All models were fitted with published ribosome structures using rigid-body fitting in UCSF Chimera.
QUANTIFICATION AND STATISTICAL ANALYSIS
No statistical methods were used to pre-determine sample size. Investigators were neither blinded to sample treatment allocation during experiments nor outcome assessment. GraphPad Prism 8 software was used for all statistical analyses. All scatter and bar plots are displayed as the mean ± standard error of the mean (SEM) unless otherwise noted. Unpaired t tests or analysis of variance (ANOVA) were performed to determine statistical significance (ns, p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001 and ****p ≤ 0.0001). ANOVA was followed by Sidak’s multiple comparison post hoc test. Additional statistical details can be found in the figure legends.
Supplementary Material
Highlights.
Negative charge in the RACK1 loop skews the rotation state of 80S ribosomes
RACK1 loop charge does not affect ribotoxic stress response signaling or poly(A) stalling
RACK1 loop charge increases swiveling of the 40S head domain similar to several IRESs
RACK1 loop charge enables human ribosomes to support eIF4A-independent translation
ACKNOWLEDGMENTS
We thank the Arnold and Mabel Beckman Center for Cryo-EM and Center for High Performance Computing at the University of Utah for support in data collection and data processing, respectively. This work was supported by grants from the NIH to M.G.R. (F31 AI152548), P.S.S. (R35 GM133772), and D.W. (R01 AI127456 and Diversity Supplement R01 AI127456-S).
INCLUSION AND DIVERSITY STATEMENT
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109663.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abaeva IS, Vicens Q, Bochler A, Soufari H, Simonetti A, Pestova TV, Hashem Y, and Hellen CUT (2020). The Halastavi árva Virus Intergenic Region IRES Promotes Translation by the Simplest Possible Initiation Mechanism. Cell Rep. 33, 108476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta-Reyes F, Neupane R, Frank J, and Fernández IS (2019). The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs. EMBO J. 38, e102226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, and Beckmann R (2013). Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85. [DOI] [PubMed] [Google Scholar]
- Arthur L, Pavlovic-Djuranovic S, Smith-Koutmou K, Green R, Szczesny P, and Djuranovic S (2015). Translational control by lysine-encoding A-rich sequences. Sci. Adv 1, e1500154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asarnow D, Palovcak E, and Cheng Y (2019). UCSF pyem v0.5. Zenodo. [Google Scholar]
- Balagopal V, and Parker R (2011). Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. RNA 17, 835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, and Yusupov M (2011). The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529. [DOI] [PubMed] [Google Scholar]
- Brandman O, and Hegde RS (2016). Ribosome-associated protein quality control. Nat. Struct. Mol. Biol 23, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. (2012). A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Baird MR, Yip MC, Murray J, and Shao S (2018). Structures of translationally inactive mammalian ribosomes. eLife 7, e40486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, and Pelletier J (2016). Hippuristanol—A potent steroid inhibitor of eukaryotic initiation factor 4A. Translation (Austin) 4, e1137381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran V, Juszkiewicz S, Choi J, Puglisi JD, Brown A, Shao S, Ramakrishnan V, and Hegde RS (2019). Mechanism of ribosome stalling during translation of a poly(A) tail. Nat. Struct. Mol. Biol 26, 1132–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppolecchia R, Buser P, Stotz A, and Linder P (1993). A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J. 12, 4005–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, and Ha T (2008). Spontaneous intersubunit rotation in single ribosomes. Mol. Cell 30, 578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle SM, Gilbert WV, and Doudna JA (2009). Direct link between RACK1 function and localization at the ribosome in vivo. Mol. Cell. Biol 29, 1626–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Duc K, Batra SS, Bhattacharya N, Cate JHD, and Song YS (2019). Differences in the path to exit the ribosome across the three domains of life. Nucleic Acids Res. 47, 4198–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhungel P, Cao S, and Yang Z (2017). The 5’-poly(A) leader of poxvirus mRNA confers a translational advantage that can be achieved in cells with impaired cap-dependent translation. PLoS Pathog. 13, e1006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiuseppe S, Rollins MG, Astar H, Khalatyan N, Savas JN, and Walsh D (2020). Proteomic and mechanistic dissection of the poxvirus-customized ribosome. J. Cell Sci 134, jcs246603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikov MI, Dobrikova EY, and Gromeier M (2018a). Ribosomal RACK1:Protein Kinase C bII Modulates Intramolecular Interactions between Unstructured Regions of Eukaryotic Initiation Factor 4G (eIF4G) That Control eIF4E and eIF3 Binding. Mol. Cell. Biol 38, e00306–e00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikov MI, Dobrikova EY, and Gromeier M (2018b). Ribosomal RACK1:Protein Kinase C bII Phosphorylates Eukaryotic Initiation Factor 4G1 at S1093 To Modulate Cap-Dependent and -Independent Translation Initiation. Mol. Cell. Biol 38, e00304–e00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov YL, Skulachev MV, Ivanov PA, Zvereva SD, Tjulkina LG, Merits A, Gleba YY, Hohn T, and Atabekov JG (2002). Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc. Natl. Acad. Sci. USA 99, 5301–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov YL, Ivanov PA, Komarova TV, Skulachev MV, and Atabekov JG (2006). An internal ribosome entry site located upstream of the crucifer-infecting tobamovirus coat protein (CP) gene can be used for CP synthesis in vivo. J. Gen. Virol 87, 2693–2697. [DOI] [PubMed] [Google Scholar]
- Frank J, and Agrawal RK (2000). A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–322. [DOI] [PubMed] [Google Scholar]
- Gallo S, Ricciardi S, Manfrini N, Pesce E, Oliveto S, Calamita P, Mancino M, Maffioli E, Moro M, Crosti M, et al. (2019). Correction for Gallo et al., “RACK1 Specifically Regulates Translation through Its Binding to Ribosomes”. Mol. Cell. Biol 39, e00544–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Senft D, Topisirovic I, and Ronai ZA (2013). RACK1 Function in Cell Motility and Protein Synthesis. Genes Cancer 4, 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T, and Sonenberg N (2017). The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat. Commun 8, 16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WV (2011). Functional specialization of ribosomes? Trends Biochem. Sci 36, 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Spinelli M, Liu M, Li QQ, and Liang C (2016). A Genome-wide Study of “Non-3UTR” Polyadenylation Sites in Arabidopsis thaliana. Sci. Rep 6, 28060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Nagai R, Abe T, Wada M, Ito K, and Takeuchi-Tomita N (2018). Tight interaction of eEF2 in the presence of Stm1 on ribosome. J. Biochem 163, 177–185. [DOI] [PubMed] [Google Scholar]
- Higgins R, Gendron JM, Rising L, Mak R, Webb K, Kaiser SE, Zuzow N, Riviere P, Yang B, Fenech E, et al. (2015). The Unfolded Protein Response Triggers Site-Specific Regulatory Ubiquitylation of 40S Ribosomal Proteins. Mol. Cell 59, 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2011). Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev 75, 434–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2014). The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem 83, 779–812. [DOI] [PubMed] [Google Scholar]
- Hopes T, Norris K, Agapiou M, McCarthy CGP, Lewis PA, O’Connell MJ, Fontana J, and Aspden JL (2021). Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching. Nucleic Acids Res. Published online July 20, 2021. 10.1093/nar/gkab606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K, and Inada T (2016). Ribosome-associated Asc1/RACK1 is required for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA. Sci. Rep 6, 28234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanov MS, Pribnow D, Magun JL, Dinh TH, Pearson JA, Chen SL, and Magun BE (1997). Ribotoxic stress response: activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol. Cell. Biol 17, 3373–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ (2013). The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb. Perspect. Biol 5, a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Rollins MG, Fuchs G, Procter DJ, Hall EA, Cozzolino K, Sarnow P, Savas JN, and Walsh D (2017). Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature 546, 651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AG, Lapointe CP, Wang J, Corsepius NC, Choi J, Fuchs G, and Puglisi JD (2019). RACK1 on and off the ribosome. RNA 25, 881–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AG, Flynn RA, Lapointe CP, Ooi YS, Zhao ML, Richards CM, Qiao W, Yamada SB, Couthouis J, Gitler AD, et al. (2020). A memory of eS25 loss drives resistance phenotypes. Nucleic Acids Res. 48, 7279–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, and Hegde RS (2017). Initiation of Quality Control during Poly(A) Translation Requires Site-Specific Ribosome Ubiquitination. Mol. Cell 65, 743–750.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, and Hegde RS (2018). ZNF598 Is a Quality Control Sensor of Collided Ribosomes. Mol. Cell 72, 469–481.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK, and Klaholz BP (2015). Structure of the human 80S ribosome. Nature 520, 640–645. [DOI] [PubMed] [Google Scholar]
- Kim HD, Kong E, Kim Y, Chang JS, and Kim J (2017). RACK1 depletion in the ribosome induces selective translation for non-canonical autophagy. Cell Death Dis. 8, e2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Kim HD, Park YJ, Kong E, Yang HW, Jung Y, Kim Y, and Kim J (2019). JNK activation induced by ribotoxic stress is initiated from 80S monosomes but not polysomes. BMB Rep. 52, 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, and Rape M (2012). The ubiquitin code. Annu. Rev. Biochem 81, 203–229. [DOI] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Ishijima J, Shiroishi T, and Barna M (2011). Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, and Green R (2015). Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife 4, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFontaine E, Miller CM, Permaul N, Martin ET, and Fuchs G (2020). Ribosomal protein RACK1 enhances translation of poliovirus and other viral IRESs. Virology 545, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskin JD, Heck DE, and Laskin DL (2002). The ribotoxic stress response as a potential mechanism for MAP kinase activation in xenobiotic toxicity. Toxicol. Sci 69, 289–291. [DOI] [PubMed] [Google Scholar]
- Leppek K, Fujii K, Quade N, Susanto TT, Boehringer D, Lenarčič T, Xue S, Genuth NR, Ban N, and Barna M (2020). Gene- and Species-Specific Hox mRNA Translation by Ribosome Expansion Segments. Mol. Cell 80, 980–995.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Nair SJ, Lescarbeau A, Belani J, Peluso S, Conley J, Tillotson B, O’Hearn P, Smith S, Slocum K, et al. (2012). Synthetic silvestrol analogues as potent and selective protein synthesis inhibitors. J. Med. Chem 55, 8859–8878. [DOI] [PubMed] [Google Scholar]
- Majzoub K, Hafirassou ML, Meignin C, Goto A, Marzi S, Fedorova A, Verdier Y, Vinh J, Hoffmann JA, Martin F, et al. (2014). RACK1 controls IRES-mediated translation of viruses. Cell 159, 1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, et al. (2017). Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun 8, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May J, Johnson P, Saleem H, and Simon AE (2017). A Sequence-Independent, Unstructured Internal Ribosome Entry Site Is Responsible for Internal Expression of the Coat Protein of Turnip Crinkle Virus. J. Virol 91, e02421–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, et al. (2018). CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 16, e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade N, Furey C, Li H, Verma R, Chai Q, Rollins MG, DiGiuseppe S, Naghavi MH, and Walsh D (2018). Poxviruses Evade Cytosolic Sensing through Disruption of an mTORC1-mTORC2 Regulatory Circuit. Cell 174, 1143–1157.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade N, DiGiuseppe S, and Walsh D (2019a). Translational control during poxvirus infection. Wiley Interdiscip. Rev. RNA 10, e1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade N, King M, Munger J, and Walsh D (2019b). mTOR Dysregulation by Vaccinia Virus F17 Controls Multiple Processes with Varying Roles in Infection. J. Virol 93, e00784–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Jackson NL, Shcherbakov OD, Choi H, and Blume SW (2010). The human IGF1R IRES likely operates through a Shine-Dalgarno-like interaction with the G961 loop (E-site) of the 18S rRNA and is kinetically modulated by a naturally polymorphic polyU loop. J. Cell. Biochem 110, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder J, Robertson ME, Seamons RA, and Belsham GJ (1998). Vaccinia virus protein synthesis has a low requirement for the intact translation initiation factor eIF4F, the cap-binding complex, within infected cells. J. Virol 72, 8813–8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J, Savva CG, Shin BS, Dever TE, Ramakrishnan V, and Fernández IS (2016). Structural characterization of ribosome recruitment and translocation by type IV IRES. eLife 5, e13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG (1992). Structural principles for the propeller assembly of beta-sheets: the preference for seven-fold symmetry. Proteins 14, 191–201. [DOI] [PubMed] [Google Scholar]
- Muto A, Sugihara Y, Shibakawa M, Oshima K, Matsuda T, and Nadano D (2018). The mRNA-binding protein Serbp1 as an auxiliary protein associated with mammalian cytoplasmic ribosomes. Cell Biochem. Funct 36, 312–322. [DOI] [PubMed] [Google Scholar]
- Neupane R, Pisareva VP, Rodriguez CF, Pisarev AV, and Fernández IS (2020). A complex IRES at the 5′-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate. eLife 9, e54575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MH, Flygaard RK, and Jenner LB (2017). Structural analysis of ribosomal RACK1 and its role in translational control. Cell. Signal 35, 272–281. [DOI] [PubMed] [Google Scholar]
- Pestova TV, and Kolupaeva VG (2002). The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16, 2906–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Procter DJ, Banerjee A, Nukui M, Kruse K, Gaponenko V, Murphy EA, Komarova Y, and Walsh D (2018). The HCMV Assembly Compartment Is a Dynamic Golgi-Derived MTOC that Controls Nuclear Rotation and Virus Spread. Dev. Cell 45, 83–100.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procter DJ, Furey C, Garza-Gongora AG, Kosak ST, and Walsh D (2020). Cytoplasmic control of intranuclear polarity by human cytomegalo-virus. Nature 587, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryo-SPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Qin X, and Sarnow P (2004). Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem 279, 13721–13728. [DOI] [PubMed] [Google Scholar]
- Quade N, Boehringer D, Leibundgut M, van den Heuvel J, and Ban N (2015). Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nat. Commun 6, 7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins MG, Jha S, Bartom ET, and Walsh D (2019). RACK1 evolved species-specific multifunctionality in translational control through sequence plasticity within a loop domain. J. Cell Sci 132, jcs228908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Horikawa W, and Ito K (2015). Inhibiting K63 polyubiquitination abolishes no-go type stalled translation surveillance in Saccharomyces cerevisiae. PLoS Genet. 11, e1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, and Frank J (2004). Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat. Struct. Mol. Biol 11, 957–962. [DOI] [PubMed] [Google Scholar]
- Shanmuganathan V, Schiller N, Magoulopoulou A, Cheng J, Braunger K, Cymer F, Berninghausen O, Beatrix B, Kohno K, von Heijne G, and Beckmann R (2019). Structural and mutational analysis of the ribosome-arresting human XBP1u. eLife 8, e46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Fujii K, Kovary KM, Genuth NR, Röst HL, Teruel MN, and Barna M (2017). Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol. Cell 67, 71–83.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokikh NE, and Spirin AS (2008). Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proc. Natl. Acad. Sci. USA 105, 10738–10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GM, Finley D, and Vogel C (2015). K63 polyubiquitination is a new modulator of the oxidative stress response. Nat. Struct. Mol. Biol 22, 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Yan LL, and Zaher HS (2017). Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol. Cell 68, 361–373.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitron CS, and Brandman O (2020). Detection and Degradation of Stalled Nascent Chains via Ribosome-Associated Quality Control. Annu. Rev. Biochem 89, 417–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, and Frank J (2001). Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291, 1959–1962. [DOI] [PubMed] [Google Scholar]
- Steel LF, and Jacobson A (1991). Sequence elements that affect mRNA translational activity in developing Dictyostelium cells. Dev. Genet 12, 98–103. [DOI] [PubMed] [Google Scholar]
- Sulima SO, and Dinman JD (2019). The Expanding Riboverse. Cells 8, 1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, and Bennett EJ (2017). ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol. Cell 65, 751–760.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy E, Ryan AP, Fulzele A, Leonard M, Daugherty MD, and Bennett EJ (2021). Ribosome quality control activity potentiates vaccinia virus protein synthesis during infection. J. Cell Sci 134, jcs257188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MK, Reitsma JM, Sweredoski MJ, Hess S, and Deshaies RJ (2016). Ribosomal proteins produced in excess are degraded by the ubiquitin-proteasome system. Mol. Biol. Cell 27, 2642–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TTL, and Passmore LA (2019). Recognition of Poly(A) RNA through Its Intrinsic Helical Structure. Cold Spring Harb. Symp. Quant. Biol 84, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesina P, Lessen LN, Buschauer R, Cheng J, Wu CC, Berninghausen O, Buskirk AR, Becker T, Beckmann R, and Green R (2020). Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts. EMBO J. 39, e103365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MK, Rojas-Duran MF, Gangaramani P, and Gilbert WV (2016). The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. eLife 5, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke N, Baby J, and Van Dyke MW (2006). Stm1p, a ribosome-associated protein, is important for protein synthesis in Saccharomyces cerevisiae under nutritional stress conditions. J. Mol. Biol 358, 1023–1031. [DOI] [PubMed] [Google Scholar]
- Van Dyke N, Pickering BF, and Van Dyke MW (2009). Stm1p alters the ribosome association of eukaryotic elongation factor 3 and affects translation elongation. Nucleic Acids Res. 37, 6116–6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke N, Chanchorn E, and Van Dyke MW (2013). The Saccharomyces cerevisiae protein Stm1p facilitates ribosome preservation during quiescence. Biochem. Biophys. Res. Commun 430, 745–750. [DOI] [PubMed] [Google Scholar]
- Várallyay E, Válóczi A, Agyi A, Burgyán J, and Havelda Z (2010). Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 29, 3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vind AC, Genzor AV, and Bekker-Jensen S (2020). Ribosomal stress-surveillance: three pathways is a magic number. Nucleic Acids Res. 48, 10648–10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, and Mohr I (2006). Assembly of an active translation initiation factor complex by a viral protein. Genes Dev. 20, 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhou J, Yang Q, and Grayhack EJ (2018). Multi-protein bridging factor 1(Mbf1), Rps3 and Asc1 prevent stalled ribosomes from frameshifting. eLife 7, e39637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JN, Buschauer R, Mackens-Kiani T, Best K, Kratzat H, Berninghausen O, Becker T, Gilbert W, Cheng J, and Beckmann R (2020). Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes. PLoS Biol. 18, e3000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AS, and Grayhack EJ (2015). Asc1, homolog of human RACK1, prevents frameshifting in yeast by ribosomes stalled at CGA codon repeats. RNA 21, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Bai XC, Brown A, Fernandez IS, Hanssen E, Condron M, Tan YH, Baum J, and Scheres SH (2014). Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. eLife 3, e03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Peterson A, Zinshteyn B, Regot S, and Green R (2020). Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 182, 404–416.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, and Min J (2011). Structure and function of WD40 domain proteins. Protein Cell 2, 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, and Barna M (2012). Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol 13, 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Tian S, Fujii K, Kladwang W, Das R, and Barna M (2015). RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517, 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Collier M, Loerke J, Ismer J, Schmidt A, Hilal T, Sprink T, Yamamoto K, Mielke T, Bürger J, et al. (2015). Molecular architecture of the ribosome-bound Hepatitis C Virus internal ribosomal entry site RNA. EMBO J. 34, 3042–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanshina DD, Bulygin KN, Malygin AA, and Karpova GG (2015). Hydroxylated histidine of human ribosomal protein uL2 is involved in maintaining the local structure of 28S rRNA in the ribosomal peptidyl transferase center. FEBS J. 282, 1554–1566. [DOI] [PubMed] [Google Scholar]
- Zhang W, Dunkle JA, and Cate JH (2009). Structures of the ribosome in intermediate states of ratcheting. Science 325, 1014–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Kastritis PL, Dougherty SE, Bouvette J, Hsu AL, Burbaum L, Mosalaganti S, Pfeffer S, Hagen WJH, Förster F, et al. (2020). Structural impact of K63 ubiquitin on yeast translocating ribosomes under oxidative stress. Proc. Natl. Acad. Sci. USA 117, 22157–22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Hellen CUT, and Pestova TV (2015). Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol. Cell 57, 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Electron microscopy maps and locally refined maps generated in this manuscript have been deposited in Electron Microscopy Data Bank (EMDB) and are publicly available. Accession numbers are listed in the Key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-HSP90 (C45G5) | Cell Signaling Technology | Cat# 4877; RRID: AB_2233307 |
| Rabbit polyclonal anti-HSP40 | Cell Signaling Technology | Cat# 4868, RRID: AB_2094249 |
| Rabbit monoclonal anti-phospho-SAPK/JNK (Thr183/Tyr185) (81E11) | Cell Signaling Technology | Cat# 4668; RRID: AB_823588 |
| Rabbit monoclonal anti-RACK1 (D59D5) | Cell Signaling Technology | Cat# 5432; RRID: AB_10705522 |
| Rabbit anti-phospho-p38 MAPK (Thr180/Tyr182) | Cell Signaling Technology | Cat# 9211; RRID: AB_331641 |
| Rabbit anti-eIF4G | (Walsh and Mohr, 2006) | N/A |
| Rabbit anti-PABP | (Walsh and Mohr, 2006) | N/A |
| Rabbit polyclonal anti-eEF2 | Bethyl Laboratories | Cat# A301-688A; RRID: AB_1210953 |
| Rabbit polyclonal anti-SERBP1 | Bethyl Laboratories | Cat# A303-938A; RRID: AB_2620287 |
| Rabbit polyclonal anti-Ebp1 | Bethyl Laboratories | Cat# A303-084A; RRID: AB_10923145 |
| Rabbit monoclonal anti-RPL11 (D1P5N) | Cell Signaling Technology | Cat# 18163; RRID: AB_2798794 |
| Rabbit polyclonal anti-RPS2 (N2C3) | GeneTex | Cat# GTX114734; RRID: AB_10620358 |
| Rabbit monoclonal anti-GFP (D5.1) XP | Cell Signaling Technology | Cat# 2956; RRID: AB_2798794 |
| Mouse monoclonal anti-RFP [6G6] | ChromoTek | Cat# 6g6-100; RRID: AB_2631395 |
| Mouse monoclonal anti-β-actin (8H10D10) | Cell Signaling Technology | Cat# 3700; RRID: AB_2242334 |
| Mouse monoclonal anti-α-tubulin (DM1A) | Sigma-Aldrich | Cat# T6199; RRID: AB_477583 |
| Mouse monoclonal anti-GAPDH (0411) | Santa Cruz Biotechnology | Cat# sc-47724; RRID: AB_627678 |
| Rabbit monoclonal anti-Raptor (24C12) | Cell Signaling Technology | Cat# 2280; RRID: AB_561245 |
| Mouse monoclonal anti-FLAG M2 | Sigma Aldrich | Cat# A2220; RRID: AB_10063035 |
| Bacterial and virus strains | ||
| Subcloning Efficiency DH5α Competent Cells | Thermo Fisher Scientific | Cat# 18265017 |
| Chemicals, peptides, and recombinant proteins | ||
| Hygromycin B | Thermo Fisher Scientific | Cat# 10687010; CAS: 31282-04-9 |
| Hippuristanol | Laboratory of Jerry Pelletier | N/A |
| Silvestrol | MedChemExpress | Cat# HY-13251; CAS: 697235-38-4 |
| Anisomycin from Streptomyces griseolus | Millipore Sigma | Cat# 9789; CAS: 22862-76-6 |
| Cycloheximide | Research Products International | Cat# 81040; CAS: 66-81-9 |
| Emetine hydrochloride hydrate | Cayman Chemical | Cat# 21048; CAS: 7083-71-8 |
| Puromycin, dihydrochloride | Millipore Sigma | Cat# 540222; CAS: 58-58-2 |
| Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | Cat# L3000-008 |
| Lipofectamine RNAiMAX Transfection Reagent | Thermo Fisher Scientific | Cat# 13778-075 |
| Pierce ECL Western Blotting Reagent | Thermo Fisher Scientific | Cat# 32106 |
| SuperSignal West Femto Maximum Sensitivity Substrate | Thermo Fisher Scientific | Cat# 34096 |
| Diethyl pyrocarbonate | MP Biomedicals | Cat# 02150902-CF; CAS: 1609-47-8 |
| Dithiothreitol (DTT) | Acros Organics | Cat# 32719-0100; CAS: 3483-12-3 |
| Triton X-100 | Fisher Scientific | Cat# BP151-500; CAS: 9002-93-1 |
| RiboLock RNase Inhibitor | Thermo Fisher Scientific | Cat# EO0382 |
| Pierce Protease Inhibitor Mini Tablets, EDTA-free | Thermo Fisher Scientific | Cat# A32955 |
| Acetone | Fisher Scientific | Cat# A949-1; CAS: 67-64-1 |
| Trichloroacetic acid | Fisher Scientific | Cat# A322-500; CAS: 76-03-9 |
| FluorSave Reagent | Millipore Sigma | Cat# 345789 |
| Hoescht 33342 | Thermo Fisher Scientific | Cat# 62249 |
| SpeI | New England BioLabs | Cat# R0133S |
| NotI-HF | New England BioLabs | Cat# R3189S |
| Cryo-EM data | ||
| WT 80S rotated state | This paper | EMD-24545 |
| WT 80S nonrotated state | This paper | EMD-24548 |
| S278E 80S rotated state | This paper | EMD-24541 |
| S278E 80S nonrotated state | This paper | EMD-24542 |
| S278E 40S | This paper | EMD-24544 |
| Locally refined maps | ||
| WT 80S rotated state (60S) | This paper | EMD-24554 |
| WT 80S rotated state (40S body) | This paper | EMD-24556 |
| WT 80S rotated state (40S head) | This paper | EMD-24557 |
| S278E 80S rotated state (60S) | This paper | EMD-24549 |
| S278E 80S rotated state (40S body) | This paper | EMD-24552 |
| S278E 80S rotated state (40S head) | This paper | EMD-24553 |
| Experimental models: Cell lines | ||
| Human: HAP1 RACK1 knockout (no rescue) cells | (Jha et al., 2017) | N/A |
| Human: HAP1 RACK1-WT-Flag rescue cells | This paper | N/A |
| Human: HAP1 RACK1-S278E- Flag rescue cells | This paper | N/A |
| Human: HEK293-T | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| Oligonucleotides | ||
| Primer: RACK1 SpeI Forward AAAAAACTAGTCTCAAGCTTATGACTGAGCAGATG | (Jha et al., 2017) | N/A |
| RACK1-Flag NotI Reverse ACCGAGCGGCCGCC TACTTGTCGTCATCGTCTTTGTAGTCGCCGCTG CCGCGTGTGCCAATGGT | This paper | N/A |
| Silencer Negative Control No. 1 siRNA | Thermo Fisher Scientific | Cat# AM4635 |
| Silencer Select SERBP1 siRNA - #1 | Thermo Fisher Scientific | Cat# 4392420, ID: s25142 |
| Silencer Select SERBP1 siRNA - #2 | Thermo Fisher Scientific | Cat# 4392429, ID: s25143 |
| Recombinant DNA | ||
| pLVX-IRES-Hygromycin plasmid | Takara Bio USA, Inc. | Cat# 632185 |
| WT-RACK1 FLAG | This paper | N/A |
| S278E-RACK1-FLAG | This paper | N/A |
| pmGFP-P2A-K0-P2A-RFP | (Juszkiewicz and Hegde, 2017) | RRID: Addgene_105686 |
| pmGFP-P2A-K(AAA)20-P2A-RFP | (Juszkiewicz and Hegde, 2017) | RRID: Addgene_105688 |
| Software and algorithms | ||
| Chimera or Chimera X | (Pettersen et al., 2004) | RRID: SCR_004097 https://www.cgl.ucsf.edu/chimera/ |
| FIJI | (Schindelin et al., 2012) | RRID: SCR_002285; https://fiji.sc/ |
| GraphPad Prism (version 8.0) | GraphPad Software Inc. | RRID:SCR_002798; https://www.graphpad.com/scientific-software/prism/ |
| Metamorph Microscopy Automation and Image Analysis Software | Molecular Devices | RRID:SCR_002368; https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metamorph-microscopy |
| CellProfiler Image Analysis Software | (McQuin et al., 2018) | RRID:SCR_007358; https://cellprofiler.org/releases |
| cryoSPARC | (Punjani et al., 2017) | RRID:SCR_016501; https://cryosparc.com |
| RELION | (Scheres, 2012) | RRID:SCR_016274; https://cam.ac.uk/relion |
| pyem | Daniel Asarnow, Eugene Palovcak, Yifan Cheng | https://zenodo.org/record/3576630 |
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


