Abstract
In cilia and flagella, dyneins form complexes which give rise to the inner and outer axonemal arms. Defects in the dynein arms are the leading cause of primary ciliary dyskinesia (PCD), which is characterized by chronic respiratory infections, situs inversus, and sterility. While the pathological features associated with PCD are increasingly well characterized, many of the causative genetic lesions remain elusive. Using Drosophila, here we analyze genetic requirements for wampa (wam), a previously uncharacterized component of the outer dynein arm. While homozygous mutant animals are viable and display no morphological defects, loss of wam results in complete male sterility. Ultrastructural analysis further reveals that wam mutant spermatids lack the axonemal outer dynein arms, which leads to a complete loss of flagellar motility. In addition to a role in outer dynein arm formation, we also uncover other novel microtubule-associated requirements for wam during spermatogenesis, including the regulation of mitochondrial localization and the shaping of the nuclear head. Due to the conserved nature of dyneins, this study advances our understanding of the pathology of PCD and the functional role of dyneins in axonemal formation and other aspects of spermatogenesis.
Keywords: Axoneme, Spermatogenesis, Sterile, Transport, Dynein, Cilia
Introduction
Cilia and flagella represent evolutionarily ancient organelles and their components are highly conserved across eukaryotes. These microtubule based structures are found on the surface of nearly every mammalian cell and are composed of hundreds of proteins that collectively form an interior structure known as the axoneme [1]. Due to the abundance of cilia within diverse cell types, disruption of axonemal assembly can lead to severe and pleiotropic defects. In humans, these defects are collectively referred to as ciliopathies or primary ciliary dyskinesia (PCD) for ciliopathies associated with motile cilia. Chronic respiratory issues, congenital heart defects, situs inversus, and sterility are some of most common phenotypes resulting from PCD and can be found singularly or in combination, depending on the genes affected [2, 3]. In PCD patients, up to 76% of affected males exhibit reduced sperm motility, resulting in decreased fertility [4]. Disruption of the axonemal outer dynein arms is the primary cause of PCD and is found in nearly 80% of all diagnosed patients [5, 6]. However, to date only about 30 different genetic lesions have been associated with PCD. For example, only 50% of the causative genes have been identified in Joubert’s syndrome, a PCD disease which affects the development of the brain [7, 8]. Due to the complexity of axonemal assembly and the pleiotropic nature of ciliary defects, determining additional genes essential for the proper formation of cilia is a daunting yet essential task [7, 9].
In Drosophila, motile cilia are assembled in a stepwise fashion that requires the coordinated pre-assembly and continual transport of components from the cytoplasm to the elongating axoneme. Disruption of any of these concurrent processes may disrupt the function of the cilia and potentially cause a cascade of other defects as development proceeds [10]. This is particularly true in the assembly of sperm flagella, one of three major microtubule-dependent remodeling events that occurs during the final stages of spermatogenesis. In addition to axonemal assembly, microtubules are also essential for the reshaping of the nucleus and for the elongation of the mitochondria that support the developing axoneme [10]. Indeed, several microtubule associated genes are reported to cause pleiotropic effects during spermatogenesis when perturbed. For example, male sterile lesions in centrosomin, haywire, and β-tubulin all affect axonemal formation and meiosis, as well as nuclear shaping and mitochondrial formation in the latter two [10–12]. Mutations in other microtubule-associated factors can result in sterility without disrupting the architecture of the axoneme. For instance, Lissenchepaly-1, asunder, and abnormal spindle display defects affecting various combinations of meiosis, centrosome positioning, mitochondrial remodeling, and individualization of sperm [13–15]. A better understanding of the precise role of genes required for these processes has the potential to contribute to our understanding of both spermatogenesis and ciliogenesis in diverse cell types.
Sensory neurons and sperm are the only ciliated cell types in Drosophila, which facilitates loss-of-function genetic analysis of axonemal assembly without the confounding effects of lethality or pleiotropic phenotypes arising from defective ciliogenesis in multiple tissues. Here, using Drosophila, we characterize a previously unidentified component of the axoneme, CG17083, which we named wampa (wam). Based on sequence similarities with Chlamydomonas, wam was predicted to be a component of the outer dynein arm docking complex, which anchors the outer dynein arm complexes to the microtubule doublets of the axoneme [16]. By generating mutant alleles of wam using CRISPR/CAS9 mutagenesis, we demonstrate that wam is indeed essential for the assembly of the outer dynein arms onto the axoneme in developing sperm flagella. Correlated with these defects, we also observed a complete loss of flagellar motility. In addition to the loss of the outer dynein arms, wamKO mutants were fully sterile and displayed additional unexpected defects during spermiogenesis, including mis-localization of mitochondria and malformation of the nucleus. These phenotypes bring to light the complexities of spermatogenesis and tie together the disparate phenotypes observed in wam’s mammalian homologs, CCDC114 and CCDC63 [17, 18]. Additionally, these phenotypes bear a striking similarity to defects observed in mammalian models that disrupt intra-manchette transportation during spermatogenesis, suggesting the possibility that a similar transportation mechanism may operate during Drosophila spermatogenesis.
Results
Wam is essential for male fertility
Spermatogenesis in Drosophila begins at the apical tip of the testes, where the germline stem cells reside. Approximately every 10 hours, these stem cells asymmetrically divide to produce two daughter cells, one of which will remain in contact with the hub cells in the niche, while the other will move away in order to differentiate into an immature spermatogonia [10, 19]. This spermatogonia will undergo four rounds of mitotic divisions and, after an extended growth phase, will undergo two rounds of meiotic divisions to produce 64 interconnected spermatids (Figure 1A). Upon the completion of meiosis, the spermatid will then enter a phase known as spermiogenesis, which consists of 3 main parts: The formation of the axoneme, drastic morphological re-modeling of the mitochondria, and re-shaping of the nucleus. These morphological steps are all dependent on microtubules and are necessary in order to finalize the production of the mature sperm [10, 20]. During this phase, hundreds of mitochondria aggregate around the basal body, which is embedded into the nuclear membrane. The mitochondria then fuse to form two separate masses that intertwine around one another forming one large sphere [21]. The resulting structure is referred to as a nebenkern. As the axoneme begins to elongate, the two mitochondrial derivatives will unfurl from one another and grow with the developing axoneme, supporting it during an extreme elongation phase [22]. During axonemal elongation, the nuclear head also undergoes a dramatic remodeling that results in a nearly 200-fold reduction in size [10, 23]. At the end of the elongation and nuclear compaction stages, an actin complex individualizes all 64 sperm and cytoplasmic debris is removed, resulting in fully mature, individualized, and highly coiled spermatozoa that can be transferred to the seminal vesicle (Figure 1B).
Figure 1. Wampa is essential for male fertility.
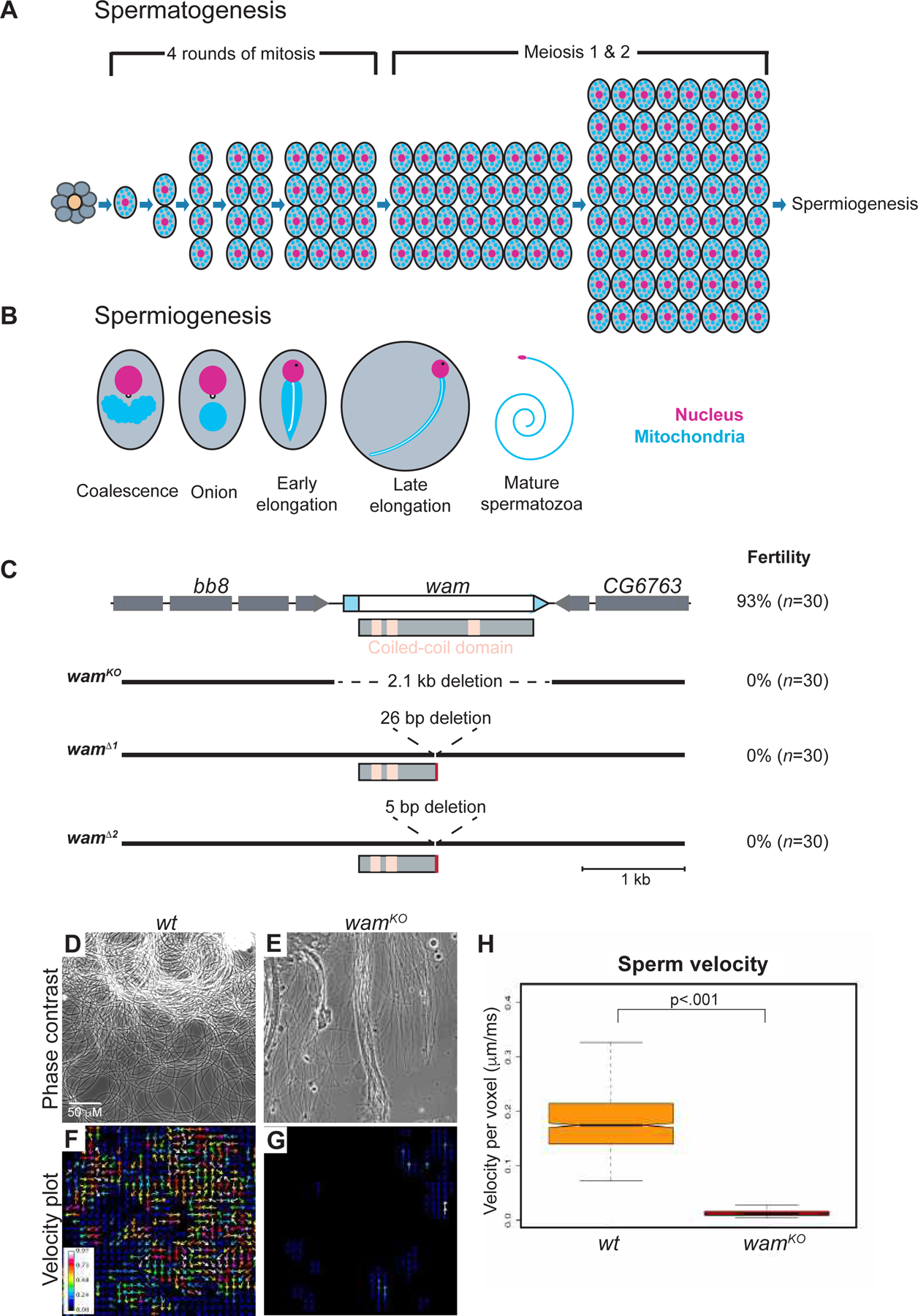
(A) Schematic of spermatogenesis through the development of spermatids. The germline stem cells surround the hub cells (orange) at the apical tip. The stem cells produce both a new stem cell as well as a gonialblast (gray). The gonialblast will undergo 4 rounds of mitotic divisions, followed by two rounds of meiosis, resulting in 64 spermatids.
(B) Spermiogenesis. Immediately following meiosis, mitochondria coalesce around the basal body, which is embedded in the nuclear membrane, and fuse to form a large sphere-like structure known as the nebenkern. By the onion stage, the basal body is visible on the nucleus and axonemal assembly has initiated. As elongation proceeds, a phase-dark protein spot becomes apparent on the nucleus, the mitochondrial derivatives elongate with and support the developing axoneme (white line on mitochondria), and nuclear remodeling begins to compact the nucleus. The final steps in spermiogenesis will individualize and coil the sperm, forming mature spermatozoa (adapted from Hales and Fuller, 1997) [47].
(C) Graphic of the wampa locus and CRISPR-generated alleles. CDR and UTRs are represented by white and blue boxes, respectively. The three coiled-coil domains are denoted by pink boxes on the protein schematic. wamKO contains a large 2.1kb deletion that removes the entire gene region, while wamΔ1 and wamΔ2 both generated small indels, which resulted in premature stop codons. Fertility quantification is labeled on the right of the schematic for each genotype. See also Figure S1.
(D–G) Representative frame from a movie of live wildtype (D) and wamKO/KO (E) sperm and a corresponding velocity heat map displaying actively moving sperm in wildtype denoted by warmer colors (F) and immotile sperm in wamKO/KO denoted by cooler or lack of color (G). See also movie S1. Scale is the same for wildtype and wamKO.
(H) Box plot of the average sperm velocity per pixel, for both wildtype (orange, n=12 animals) and wamKO/KO (red, n=10 animals) showing a lack of movement in sperm from wamKO/KO males compared to wildtype.
Based on sequence similarity with Chlamydomonas DC2, wam is a highly conserved gene that encodes a 550 amino acid (aa) coiled-coiled domain containing protein predicted to be involved in tethering outer dynein arms to the axoneme (Figure 1C) [16, 17]. In order to assess its role in Drosophila, we employed CRISPR/CAS9 mutagenesis to create three mutant alleles. We first generated a null allele, wamKO, by utilizing two gRNAs targeting either sides of the coding region, removing the entire gene locus. Additional alleles, wamΔ1 and wamΔ2, were generated using a single gRNA to target the middle of the coding region, which resulted in a truncated protein at 250aa that lacks the last coiled-coiled domain (Figure 1C and Figure S1). Phenotypically, homozygous animals from each allele appeared normal and displayed no obvious defects. However, when assayed for fertility, homozygous mutant males exhibited complete sterility while females produced similar amounts of progeny as wildtype (Figure 1C). All three wam alleles resulted in identical phenotypes in every assay and did not show any apparent defects in sensory behavior. For simplicity, only wamKO is utilized from here on. Using both complementation and rescue experiments, we confirmed that the sterility was due to the loss of wam and not bb8, a neighboring gene also required for male fertility (Figure S2) [24]. Taken together, these data indicate that wam is required for male fertility.
Wam is required for axonemal assembly
To investigate the cause of sterility in wamKO homozygous males, we first examined the testes and associated seminal vesicles using phase contrast microscopy. This revealed that while wam mutants generated mature sperm, they exhibited a complete loss of flagellar motility and failed to fully individualize (Figure 1D–1H and movie S1). We also observed that, while the overall morphology of the testes appeared indistinguishable from wildtype, the seminal vesicles of wamKO/KO animals were devoid of sperm and significantly reduced in size compared to wildtype (Figure 2A–G). To quantify this observation and examine the phenotypic progression, we held males as virgins for two weeks and examined the morphology and content of the testes. At day one post-eclosion, the gross morphology of testes and associated structures were indistinguishable between wamKO/KO and wildtype animals (Figure 2A and D). However, as the animals aged, the seminal vesicle of wamKO/KO animals failed to fill with sperm and increase in size as observed in wildtype (Figure 2, black arrows). Instead wam mutant animals showed an enlargement of the basal end of the testes (Figure 2C and E–F, white arrows). Opening the seminal vesicles confirmed that they were indeed devoid of sperm, consistent with a failure of sperm to enter the seminal vesicles. We next examined the individualization defect using Sir-actin to visualize individualization complexes. In wildtype, an average of 5.4 complexes can be observed per testis, however, only 1.9 were observed in wamKO/KO (Figure 2H–L). Additionally, 64% of the individualization complexes that were observed in wam mutants appeared abnormal, compared to only 8% in wildtype (Figure 2J–L). This data suggests that in the absence of wam sperm individualization stalls soon after complex formation at the basal end of the testes.
Figure 2. Wampa is required for the assembly of the outer dynein arms.
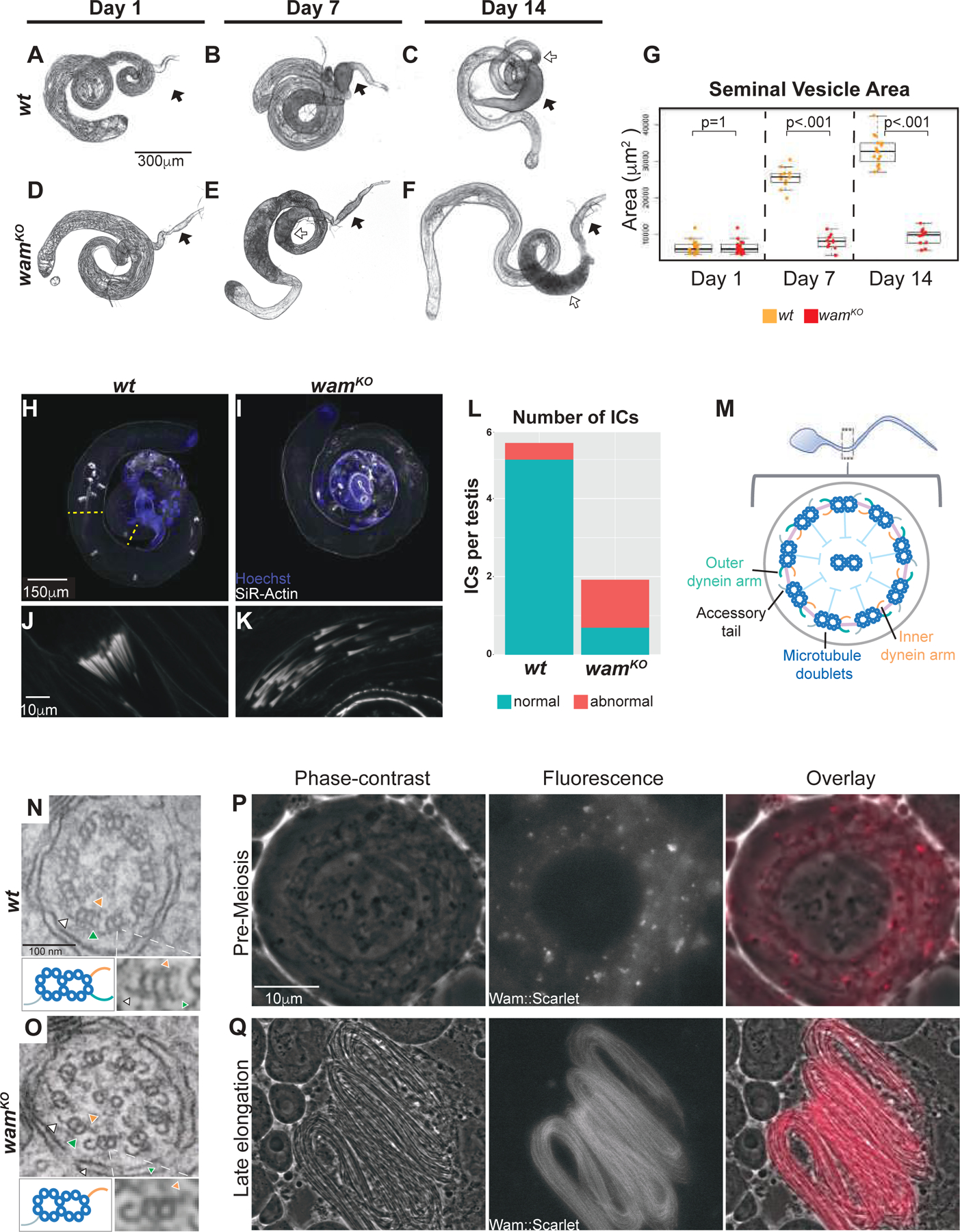
(A–F) Wildtype (A–C) and wamKO (D–F) homozygous males were held as virgins for two weeks and their testes were examined for morphological differences during this time. At day 1, testes from wildtype (A) and wamKO (D) are comparable. However, by day 7, the differences in the seminal vesicles can be detected as wildtype seminal vesicles begin to enlarge (B, black arrow), whereas wamKO remains approximately the same size (E, black arrow). At day 14, seminal vesicle size remains the only difference observed (C and F, black arrows), indicating a failure for sperm to be loaded and, as a result, the basal end of the testes becomes enlarged in wamKO (E and F, white arrow) compared to wildtype (C, white arrow). Scale is the same for wildtype and wamKO.
(G) Box plot of the average area of the seminal vesicles for both wildtype and wamKO homozygotes at day 1 (wt, n=18 and wamKO, n=18), day 7 (wt, n=11 and wamKO, n=10), and day 14 (wt, n=16 and wamKO, n=14). wildtype seminal vesicles gradually increase in size over time, while wamKO seminal vesicles stay approximately the same size.
(H–K) Whole testes labeled with Hoechst (blue) and SiR-Actin (white) showing the distribution of actin cones that form the individualization complexes throughout the testis of wildtype (H) and wamKO (I). Normal ICs from wildtype are clustered (J), however, wamKO ICs (K) are fragmented and typically found closer to the basal end. Yellow lines mark the region between the formation and termination of ICs. Scale is the same for wildtype and wamKO.
(L) Stacked box plot of the average number of individualization complexes (ICs) observed in wildtype and wamKO homozygotes. Wildtype contained an average of 5.4 ICs that were 92% normal (n=14), whereas wamKO contained an average of 1.9 ICs where only 36% appeared normal (n=13). ICs were quantified only between their formation at basal end and their termination at apical end (Figure H, yellow lines).
(M–O) Schematic of the flagellar axoneme (M). TEM images of a wildtype axoneme showing the inner (orange arrow) and outer (green arrow) dynein arms on the microtubule doublet, opposite from the position of the accessory tail (white arrow) (P). Outer dynein arms are notably absent in wamKO homozygotes (Q). A blowup of an individual doublet is displayed below the EM images on the right, with a corresponding cartoon on the left. Scale is the same for wildtype and wamKO.
(P–Q) Live phase contrast and fluorescent images from testes expressing the Wam::mScarlet-I tag, demonstrating endogenous Wam localization in spermatocytes is observed in the cytoplasm (P) and in elongating spermatids, Wam can be visualized along the entire length of the flagella of a developing spermatid bundle during mid-elongation stage of spermiogenesis (Q). Scale is the same for all panels.
Since Wam is predicted to be a component of the outer dynein arm docking complex and wamKO/KO mutants displayed complete immotility of the sperm flagellum, we next examined the ultrastructure of the axoneme. In normal wildtype sperm, outer dynein arms are an essential axonemal substructure known to be required for motility [9]. Strikingly, transmission electron microscopy (TEM) revealed that wamKO/KO sperm flagella lacked outer dynein arms on the microtubule doublets along the length of the axoneme (Figure 2M–O), indicating that Wam was essential for outer dynein arm complexes on the axoneme.
Dynein arms are large, multisubunit structures that are preassembled in the cytoplasm before being transported onto the growing axoneme [16, 25]. We next sought to determine whether wam is involved in the cytoplasmic pre-assembly of the outer dynein arms or whether it is a component of the flagellum that is incorporated onto the axoneme [26, 27]. To address this question, we generated a transgenic line expressing the fusion protein Wam::mScarlet-I under the control of the endogenous wam promoter. Analysis revealed that Wam does in fact accumulate in the cytoplasm in premeiotic cells, where the dynein arms and associated docking complexes are being assembled (Figure 2P) [16]. As elongation proceeded, we observed strong localization of Wam along the entire length of the axoneme, suggesting more copies of Wam were progressively incorporated (Figure 2Q and Figure S3A). Consistent with this, co-localization of the Wam::mScarlet-I construct and tubulin revealed a delay in the incorporation of Wam onto the newly polymerized tubulin (Figure S3A). To test the functionality of the Wam::mScarlet-I construct, we performed a rescue assay in wamKO homozygous males which revealed that the fusion protein was able to fully restore fertility to wildtype levels (Figure S2B). The localization of Wam and the dependency of the outer dynein arm complex on Wam is consistent with the interpretation that Wam is an essential component of the sperm flagellum that is required for attachment of the outer dynein arms onto the axoneme. While it remains unclear whether the individualization phenotype in the absence of Wam is a primary defect or perhaps secondary to the crowding and disorganization of sperm that are unable to exit the testes (Figure 2E–F), it is known that some axonemal components are required for proper individualization [28, 29]. This suggests the possibility that Wam may also be required for proper formation or function of the individualization complex.
Mitochondrial localization and elongation defects are observed in wamKO
We next investigated whether further defects in spermatogenesis might be associated with loss of wam function. In addition to the lack of flagellar motility, sperm failed to fully individualize and coil in wamKO/KO testes (Figure 1D–E and Figure 2H–L). Failure to complete these last steps of spermiogenesis could be an indicator of defects in earlier stages in spermatogenesis [29]. To examine whether wam functions at other stages, we generated a His2AV-mRFP1; wamKO, sqh>EYFP-mito stock to analyze nuclear morphology and mitochondrial localization throughout spermatogenesis. In homozygous mutants, pre-meiotic stages of spermatogenesis were indistinguishable from wildtype. During meiosis, however, we began to observe mitochondrial aggregation defects. Normally wildtype mitochondria align evenly along the microtubules that form the meiotic spindle in order to ensure equal segregation during cytokinesis (Figure 3A). In contrast, mitochondria failed to evenly align in approximately 44% of cells in wamKO/KO mutants (Figure 3A–B and Figure S4).
Figure 3. Wampa is required for proper mitochondrial remodeling.
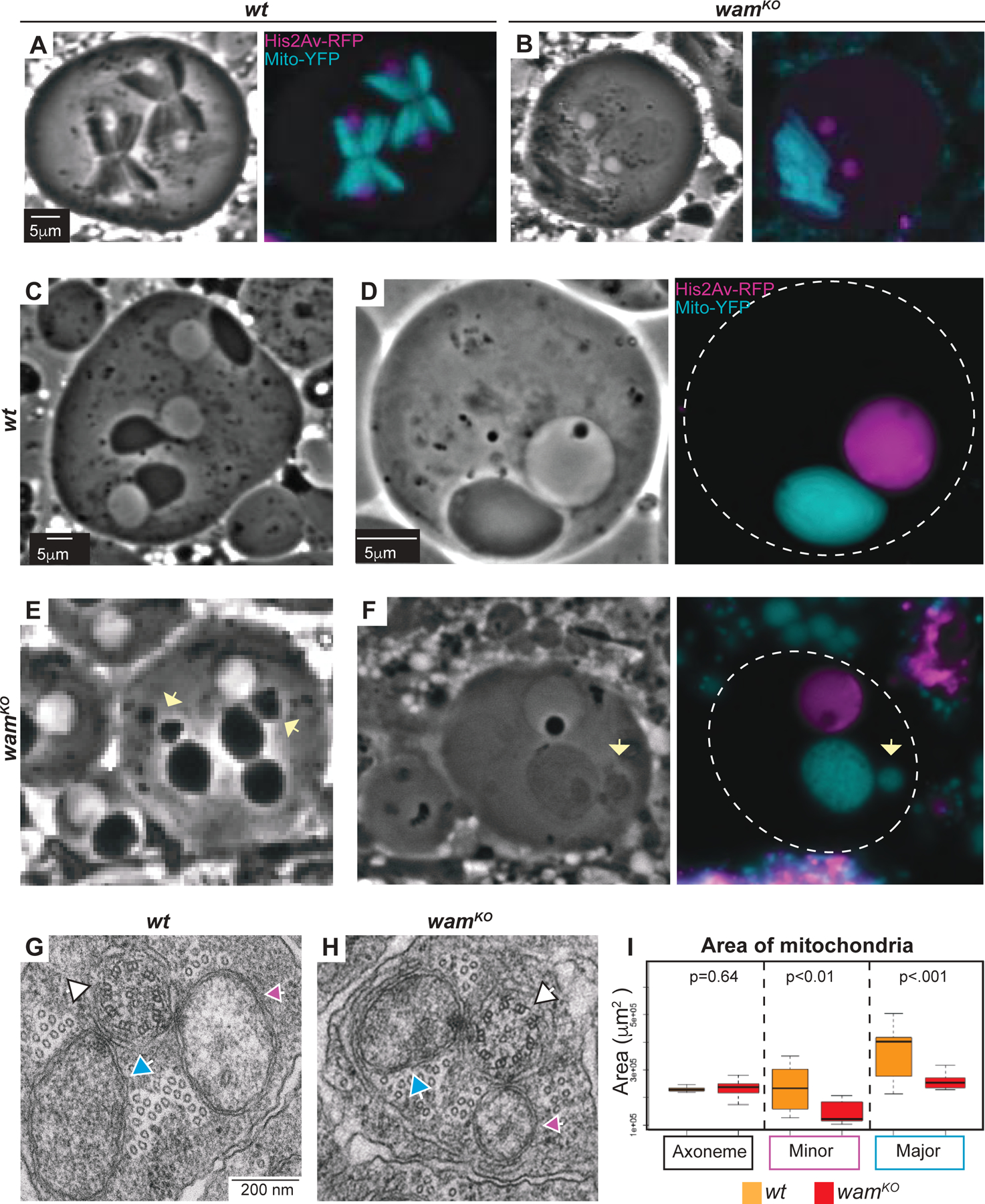
(A–B) Phase contrast and fluorescent images of meiotic cells from wildtype (A) and wamKO/KO (B) testes displaying the unequal alignment of mitochondria during meiosis. Scale is the same for all panels.
(C–F) Phase contrast and fluorescent images of onion stage spermatids from wildtype (C–D) and wamKO/KO (E–F) testes at 20x (C and E) and 100x (D and F). Fragmentation of the nebenkerne can be observed in wamKO spermatids (E and F, yellow arrow). Scale is the same for wildtype and wamKO.
(G–H) EM images of spermatids during elongation for both wildtype (G) and wamKO/KO (H). The axoneme (white arrow) is visible with major (blue arrow) and minor (pink arrow) mitochondrial derivatives on either side. Scale is the same for wildtype and wamKO.
(I) Quantification of the area of the major and minor mitochondrial derivatives and axonemes for wildtype (orange, n=17) and wamKO/KO spermatids (red, n=24). The axoneme is approximately the same size for both wildtype and wamKO/KO, but the major and the minor mitochondrial derivatives in wamKO/KO show a decrease in size compared to wildtype.
Following the meiotic divisions, mitochondria associated with each developing spermatid aggregate around the basal body and are then fused into the two separate derivatives that form the nebenkern, a process that is dependent on cytoplasmic microtubules and is speculated to be dynein dependent [10, 30]. While the majority of onion- and elongation-stage cells appeared normal in wamKO/KO mutants, a small subset displayed a fragmentation phenotype where small clusters of unincorporated mitochondria were observed adjacent to the nebenkern (Figure 3C–F, arrows and Figure S4). Additionally, TEM analysis revealed a surprising defect in the mitochondrial derivatives that was not obvious by brightfield microscopy. At the beginning of the elongation phase, the two mitochondrial derivatives that form the nebenkern unfurl from one another and extend along with the growing axoneme, providing both physical and metabolic support [11]. In wamKO/KO testes, the mitochondrial derivatives were consistently and significantly smaller than in wildtype (Figure 3G–I). As it is known that the mitochondria provide structural support for the developing axoneme and are essential for the formation of the flagellum, a disruption in their elongation could further destabilize the developing axoneme [10, 31].
wam is required for nuclear remodeling
During spermiogenesis, every cell within a 64-cell cyst should contain a nucleus and a nebenkern. This ratio is always 1:1 and deviations from the expected ratio are typically due to meiotic defects, most commonly during cytokinesis [11, 32]. In wamKO/KO testes examined by phase contrast microscopy, numerous onion-stage cells appeared to lack a nucleus, containing only a nebenkern. To examine these onion stage nuclei more precisely we again used the His2AV-mRFP1; wamKO, sqh>EYFP-mito stock. Unexpectedly, rather than a skewed ratio of nuclei to nebenkern, wamKO/KO cells presented with severe defects in nuclear morphology, suggesting errors during the reshaping of the nucleus that occurs after meiosis (Figure 4A–E). Indeed, analysis of over 100 onion staged nuclei from both wildtype and wamKO/KO testes revealed that even superficially normal nuclei were in fact smaller than their wildtype counterparts (Figure 4E). This phenotype is very similar to that reported following disruption of Hay, a microtubule associated protein [11]. Cytoplasmic microtubules are known to be required for proper shaping of the nucleus during spermiogenesis [11]. Taken together with the defects observed in the other microtubule dependent processes, we conclude that Wam plays important axonemal and cytoplasmic roles during spermiogenesis.
Figure 4. Wampa regulates shaping of the nuclear head.
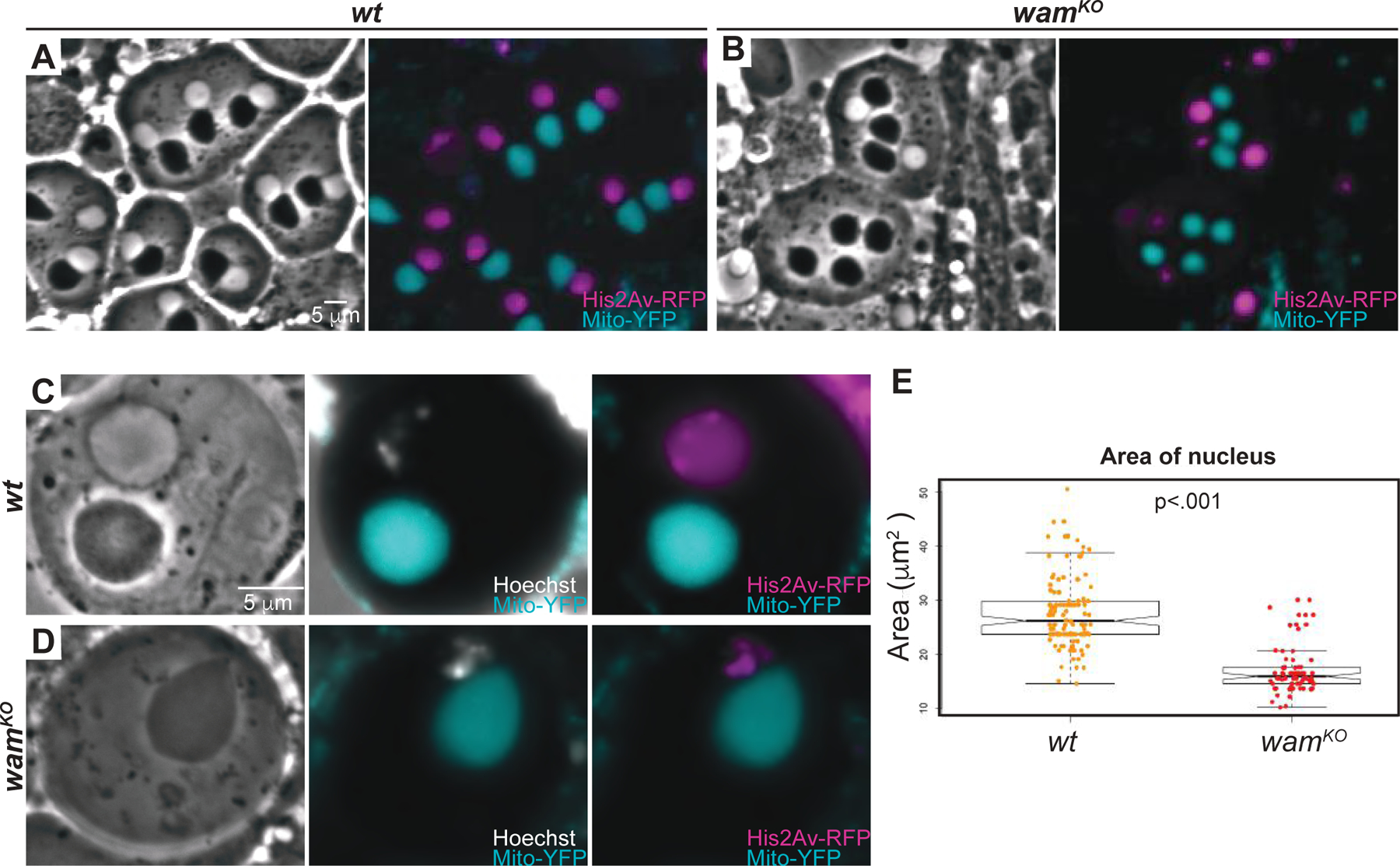
(A–D) Phase-contrast and florescent images of onion stage spermatids from wildtype (A and C) and wamKO/KO testes (B and D) shown at 20x (A–B) and 100x (C–D). Wildtype cells display the expected 1:1 ratio of nebenkern to nucleus (A and C), whereas a corresponding is not observed by phase-contrast in wamKO/KO for some cells. However, fluorescently labeled DNA reveals the presence of the DNA (B and D) which corresponds to the Hoechst signal (C–D). Scale is the same for wildtype and wamKO.
(E) Graph displaying quantification of the area of nucleuses of developing spermatids at the onion stage for wildtype (orange, n=144) and wamKO (red, n=116) revealing a reduction in size in nucleuses after meiosis for wamKO in the cells that contain a seemingly normal nucleus by phase-contrast.
Discussion
Ciliogenesis requires the coordinated activity of hundreds of different proteins to ensure proper form and function. Disruption of any one of these factors risks the potential failure of the tissues associated with the aberrant cilia. Here we show that the Drosophila dynein-associated protein wam is essential for proper axonemal assembly as well as successful completion of spermatogenesis. In the absence of wam function, outer dynein arms were lost along the entire length of the axoneme, which was in turn correlated with defects in individualization, mitochondrial remodeling, and nuclear shaping. Since microtubules are broadly required during spermatogenesis and dyneins are well known to play critical roles in cell cycle progression [13], it is not surprising that dynein-related genes might have a profound effect on multiple different stages of spermatogenesis. The defects observed in wam, while seemingly incongruent, all involve known microtubule-dependent processes. Indeed, lesions in the spermatogenesis-specific isoform of tubulin, B2t, and the microtubule associated protein, Haywire, both display similar defects as wam mutants in meiosis, flagellar formation, and nuclear shaping [11]. Mutations in Lis-1, a regulator of cytoplasmic dynein, also display similar mitochondrial phenotypes to wam, and mutations in the cytoplasmic dyneins dlc90F and ddlc1 disrupt formation of the individualization complex and shaping of the nucleus [13, 33–35]. Together, these observations highlight the essential axonemal and non-axonemal roles that microtubules and their associated proteins play during spermatogenesis.
Based on sequence similarity, wam is predicted to be a homolog of the Chlamydomonas outer dynein arm docking complex subunit DC2, which directly interacts with the outer dynein arm complex linking it to the microtubules of the axoneme (Figure S3A) [16, 26]. Consistent with this, we found that Wam localizes along the entire length of the axoneme and that outer dynein arms fail to assemble onto the microtubule doublets of the elongating axoneme in wam mutants (Figure 2). This indicates that wam is an essential component of the axoneme that is necessary for the incorporation of the outer dynein arms. Interestingly, wam has two mammalian orthologs, CCDC114 and CCDC63. Like wam, these genes are predicted to encode homologs of DC2 [16]. CCDC114 mutations have been identified in patients presenting with PCD related defects including chronic respiratory issues, heart defects, and situs inversus [2, 17, 36]. While fertility issues have not been reported in these patients, the authors show that CCDC114, like Wam, localizes along the entire length of the axoneme and is required for outer dynein arm assembly [17]. Intriguingly, mice mutant for the other homolog, CCDC63, exhibit male sterility but still retain integrity of the outer dynein arms [18]. The authors speculate that the lack of outer dynein arm phenotypes might be due to compensation from CCDC114, which is also expressed in the testes [18]. Nevertheless, while the outer dynein arms were intact in CCDC63 mutants, spermiogenesis was associated with abnormal morphology of the flagellum, a misshapen nuclear head, and probable mitochondrial defects, all consistent with the cytoplasmic defects we observed in wamKO/KO testes [18].
While wampa also has a paralog, CG14905, its expression is restricted to the sensory cilia, which uses different variants for the assembly of the outer dynein arms [26]. This demonstrates the conserved nature of the single copy of Drosophila wam and its ability to phenocopy defects observed in both mammalian orthologs and supports the theory that mammals, like Drosophila, might also exhibit cell type specificity with regards to components of the axoneme [26].
In addition to its role in the docking of the outer dynein arms to the axonemal microtubules, wam is also involved in cytoplasmic functions that are known to be microtubule-dependent, such as mitochondrial remodeling and nuclear shaping. During our analysis of wam mutants, we observed that the mitochondria were not properly aligned during meiosis resulting in abnormal segregation. This led to a small subset of nebenkerne that appeared to be fragmented during the onion stage. Interestingly, this phenotype bares a strong resemblance to phenotypes observed in lis-1 mutants, which is a consequence of the disruption of its dynein function [13]. Further TEM analysis also uncovered a defect in the mitochondrial derivatives associated with the elongating axoneme in wam mutants, which were consistently and significantly smaller than those observed in wildtype. This was an intriguing finding as it correlated with the observation of the fragmented nebenkerne, suggesting the possibility that not all of the mitochondria were properly incorporated into the mitochondrial derivatives, resulting in smaller derivatives at elongation. While the derivatives were consistently smaller than wildtype, it remains unclear why a larger percentage of nebenkerne did not appear fragmented. It is possible that the unfused fragments are in close enough proximity to the nebenkern so as to be indistinguishable or perhaps they are being degraded, resulting in less mitochondria forming the nebenkern and thus, smaller derivatives. It is also plausible that there is an unidentified defect which stunts the growth of the mitochondrial derivatives during elongation. While the precise cause of the elongation defect is not clear, the observation of fragmented nebenkerne and misaligned mitochondria during meiosis strongly suggests that wam is involved in mitochondrial localization during coalescence. It has previously been suggested that the movement of mitochondria during spermatogenesis is dependent on microtubules, and specifically on dynein, and disruption of dynein-associated genes have been shown to perturb the mitochondrial dynamics [10, 13, 20]. Indeed, our imaging reveals that the mitochondria and microtubules co-localize during spermatogenesis (Figure S3B). The disruption of the nebenkerne and of the elongation of the mitochondrial derivatives are strong indicators that wam is involved in the dynamic movement and localization of mitochondria during spermatogenesis.
Perhaps the most striking defect associated with wam alleles is the disruption of nuclear morphology. Our data shows that the morphogenesis of the nuclear head was perturbed in wam mutants. Similarly, hay, a microtubule associated gene, results in phenotypes that are very similar to those observed in wam, including a nucleus that is not visible by phase-contrast but is visible when stained with florescent DNA dye. [11]. In mammals, this nuclear remodeling process is known to be facilitated by the manchette, a microtubule-based structure which links the developing spermatid to the cell surface through microtubule interactions [37, 38]. The manchette is describe as “skirt like” structure that slides along the nucleus during remodeling, suggesting a zipper-like action during reshaping [37, 38]. Defects in either the formation of the nuclear head or the flagellum can result in reciprocal abnormalities in the other process. This has been shown to be a result of disruptions in intra-manchette transportation of proteins essential for spermatogenesis in mammals [39, 40]. In fact, over a dozen genes in mammals have recently been identified to be involved in intra-manchette transportation that perturb both the remodeling of the nuclear head as well as the formation of the flagellum [41]. While the transport function of the manchette remains to be fully elucidated, it is suggested to be very similar to intraflagellar transportation [40]. Interestingly, we observe a manchette-like microtubule based structure in Drosophila that transiently attaches to the nucleus, forming during early elongation of the axoneme and disassembles by late elongation, as is reported in mammals (Figure S3B, green arrows) [40]. While the manchette is not well established in Drosophila, it has been postulated to exist [20]. Indeed, a structure referred to as the dense complex has been reported as a candidate, which is shown with Anillin staining to localize to the ciliary cap [20]. However, more functional investigation needs to be performed to identify if these structures are related or whether either is analogous to a true manchette.
Given that intraflagellar transport has been shown to not be utilized during Drosophila spermatogenesis [42, 43], it is conceivable that Drosophila, like mammals, requires intra-manchette transportation in order to move essential proteins to the nucleus, mitochondria, and axoneme during these key remodeling events. It is well documented that the assembly of all three of these structures are dependent on microtubules and that disruption of intra-manchette transportation can result in defects in their assembly [10, 37, 41, 44]. Since dynein accessory proteins are not exclusively axonemal or cytoplasmic [33, 45], we hypothesize that wam may associate with a cytoplasmic dynein complex in a similar fashion as it is known to with the axonemal dynein complex, to aid in transporting remodeling proteins during spermiogenesis (Figure S3). This would explain the defects observed in wam mutants and suggests that insects and mammals have a more similar approach to completing spermatogenesis than previously recognized.
In summary, here we identified Wam as an essential component of axonemal assembly and uncovered roles throughout spermatogenesis, which are reminiscent of a role during intra-manchette transportation. Disruption of the mammalian orthologs of wam, CCDC63 and CCDC114, results in PCD associated with sterility and severe respiratory distress, heart malformations, and laterality defects, respectively. These syndromes are due to errors either in spermiogenesis that disrupt nuclear remodeling and the formation of the flagellum or in loss of the outer dynein arms during ciliogenesis. Since there are hundreds of genes affiliated with ciliogenesis, many PCD-related diseases are considered rare and often difficult to diagnose. Despite extensive effort, at least a third of genes that cause PCD remain elusive and those that are known do not always have a clear functional role [46]. Here we show that wam is able to recapitulate the phenotypes that result in PCD defects observed in both of its mammalian orthologs. As such, further analysis of ciliary proteins in Drosophila could be a productive avenue for investigating molecular mechanisms underlying the pathology of diseases associated with ciliogenesis.
Contact for reagent and resource sharing:
Further information and requests for resources and reagents should be directed to Matt Gibson (MG2@stowers.org).
Drosophila melanogaster stocks:
All stocks were maintained at 25°C on syrup food. The wildtype stock used was w; His2A-mRFP;EYFP-Mito for experiments with fluorescent imaging. w1118 was used for all other experiments.
Generation of wampa transgenic lines
gRNA 1 and 2, which were used in the construction of wamKO, were cloned into pCFD2-dU6.2gRNA and gRNA 3, which was used for creating wamΔ1 and wamΔ2 was cloned into pCFD3-dU6.3gRNA [48]. The following primers (5’−3’) were annealed and cloned into the BbsI site:
gRNA1F: CTT CGA TAT TGG GCT TTG ATA CAC T
gRNA1R: AAA CAG TGT ATC AAA GCC CAA TAT C
gRNA2F: CTT CGG ATT TAA ATA TTC ATC AGT
gRNA2R: AAA CAC TGA TGA ATA TTT AAA TCC
gRNA3F: GTC GAA GGC GGT GTT ATC ACC ATC
gRNA3R: AAA CAT GGT GAT AAC ACC GCC TTC
To generate a donor plasmid for creating wamKO, we used HIFI DNA assembly master mix to assemble 4 fragments that were generated from the following primers (5’−3’):
w1118 genomic DNA
(5’ homology arm F) CCT GCA GGT CGA CTC TAG AGA AGA CGG TCA TAG TGC AGG G
(5’ homology arm R) CCA AGC TTG GCG AAT AAC TTC GTA TAA TGT ATG CTA TAC GAA GTT ATA
GTG TAT CAA AGC CCA ATA TTT ATG CC
(3’ homology arm F) GTA TGT AAG TTA AAT AAC TTC GTA TAG CAT ACA TTA TAC GAA GTT ATA
CTG ATG AAT ATT TAA ATC CGT T
(3’ homology arm R) ATTCGAGCTCGGTACCCGGGTACCATCGTCGGACGTGCAT
Ubi-mCherry DNA
(mCherryF) ATA TTG GGC TTT GAT ACA CTA TAA CTT CGT ATA GCA TAC ATT ATA CGA AGT TAT
TCG CCA AGC TTG GGC TGC ATC ACG TAA TAA G
(mCherryR) GGA TTT AAA TAT TCA TCA GTA TAA CTT CGT ATA ATG TAT GCT ATA CGA AGT TAT
TTA ACT TAC ATA CAT ACT AGA ATT GAT CGG C
pHSG98 DNA
CCCGGGTACCGAGCTCGAATTCG
CTCTAGAGTCGACCTGCAGGCATGCAAG
The fragments were gel extracted using Zymoclean Gel DNA Recovery kit, assembled at 50°C for 3 hours, and then transformed into electrocompetent cells. The gRNA and donor templates were sequence verified, ethanol precipitated, and injected into , y2 cho2 v1; attP40(nos-Cas9)/+ embryos. For wamKO, the donor and guides 1 and 2 were mixed and injected at 250 ng/µl each (total concentration of 750 ng/µl). wamΔ1 and wamΔ2 were generated by injecting 250 ng/µl of gRNA 3. For all injections, y2 cho2 v1; attP40(nos-Cas9)/CyO males were crossed to w1118 virgins and kept at 25°C in egg laying cages on grape plates during injections and the embryos were injected at 18°C. Transformants were screened by visualization of mCherry for wamKO and by homozygous male sterility for wamΔ1 and wamΔ2.
The lox-P sites that flank the m-Cherry selection cassette were removed by Cre/loxP-mediated recombination, generating the wamKO line [49]. This line was recombined with sqh-EYFP-Mito and crossed to His2Av-mRFP1 to generate the w; His2Av-mRFP1 II.2/CyO; wamKO, sqh-EYFP-Mito/TM3,Sb line that was used during fluorescent imaging analysis.
The Wam::mScarlet-I fusion construct and the wamSR2 rescue construct were created by generating a 6 kb fragment encompassing the wampa locus plus 2 kb on either side. The tag was placed at the C-terminus with a flexible 10x GLY linker that was incorporated into the primers. The following primers were used (5’−3’):
CH322–169E18
5’ homology arm F for tag and rescue: GCC AAG GCA AGT ATT AAA ACG T
5’ homology arm R for tag: ACC GCC TCC TCC ACC TCC GCC ACC ACC ACC TCC GCC ACC ACC ACC
CAT GTT GCG CTT GGC GGC CAG GAG ACG
5’ homology arm R for rescue: TGA ATT CCG ACG GGT GCA C
3’ homology arm F for rescue: AGA TGC AGG GTA TTA TGC
3’ homology arm F for tag: TAG TTC TGG AAT ATA TGA TG
3’ homology arm R for tag and rescue: CAA CAG CAG GCG GTA TAA ATC CA
pmScarlet-i_C1
Scarlet-I F: GCG GAG GTG GAG GAG GCG GTA TGG TGA GCA AGG GCG AG
Scarlet-I R: CAT CAT ATA TTC CAG AAC TAC TTG TAC AGC TCG TCC ATG C
The fragments were gel extracted using Zymoclean Gel DNA Recovery kit, assembled using HIFI DNA assembly master mix for an hour at 50°C and transformed into electrocompetent cells. The insert was then cloned into the w+attB plasmid by digesting with BamHI and ApaI, for the tag construct and BamHI and NotI for the rescue construct. The ends were blunted using klenow fragment and gel purified. w+attB donor plasmid was digested with XhoI, blunted with klenow fragment, treated with shrimp alkaline phosphatase, and purified. The insert and vector were then ligated, transformed, screened, and sequence verified. Positive colonies were miniprepped and the DNA was injected into y[1] M(vas-int.Dm)ZH-2A w[*]; PBac(y[+]-attP-3B)VK00001 embryos at 1:200 ng/µl. These constructs lack the 5’UTR and the first 47bp of the neighboring gene, bb8.
To test the functionality of the tag and rescue constructs, they were double balanced on the second and third chromosomes and crossed wamKO, wamΔ1, and wamΔ2. Stocks were generated that contained the either the tag or the rescue with the wam mutations. Homozygous males from this stock were collected and assayed for fertility.
Fertility assays
For all fertility assays, 30+ single males were crossed to 3–5 wt (w1118) virgins for all indicated genotypes. Vials with progeny or pupae after 10 days were considered fertile.
Live imaging
Sperm motility was monitored by dissecting testes of 2–3 day old wt and wamKO males and transferring them to a clean slide in a drop of PBS. The testes and seminal vesicles were pierced to release the sperm and imaged every 2 ms for 100 frames on a Zeiss Axiovert 200M microscope using a 20× 0.8 Phase Plan-Apochromat objective.
Testis and seminal vesicle morphology were analyzed by collecting several newly eclosed males from both wt and wamKO and holding them as virgins for two weeks. Every few days males were analyzed. Their testes and associated seminal vesicles were dissected in PBS and transferred to a clean slide into a drop of PBS and imaged on a Leica CTR 5000 compound microscope.
Testes squashes were performed as previously described [32]. In brief, testes were dissected from 0–2 day-old wt (w;His2Av,RFP;Mito,YFP) and wamKO (w;His2Av-mRFP;wamKO,Mito-YFP) homozygous males and transferred to a clean poly-L-lysine coated slide into a drop of PBS. SiR-tubulin was added at 1:250 and Hoechst was used at 1:1000. The testes were pierced 2–3 times to release the cells. Occasionally cells merge as an artifact of this process. The squashes were imaged on a Zeiss Axiovert 200M microscope using either a 20× 0.8 Phase Plan-Apochromat or 100× 1.4 Phase Plan-Apochromat objective. Brightness was occasionally adjusted for clarity and to avoid saturation.
Fixed imaging
Whole testes from 0–2 day old wt and wamKO males were dissected in PBS and fixed in 4% paraformaldehyde for 25min, samples were then washed three times in 0.1% PBS+Triton x-100 for 15min each. Hoechst was added at 1:1000 and incubated overnight at 4°C. Samples were again washed three times in 0.1% PBS+Triton x-100 for 15min each and then incubated in SiR-actin for 3 hours at room temperature. Samples were then mounted in vectashield and imaged on a Leica TCS SP5 confocal microscope.
EM analysis
For TEM analysis, testes were dissected and fixed with a buffer containing 2.5% paraformaldehyde, 2% glutaraldehyde, 1% sucrose and 50 mM sodium cacodylate (PH7.4). After a brief rinse, the tissue was post-fixed with 1% OsO4 for 90 min and then stained with 1% uranium acetate en bloc overnight. Thereafter, the samples were dehydrated through an ethanol gradient to 100%, equilibrated with propylene oxide, and infiltrated with 50% propylene oxide/50% Epon, then 100% Epon resin (EMS, Fort Washington, PA) for 3 times over a day. After polymerizing at 60°C for 48hr, the sample block was sectioned with a Leica Ultra microtome (Leica UC-6) using diamond knives. The sections were post-stained with uranyl acetate and lead citrate, and then imaged with a FEI transmission electron microscope (Tecnai Bio-TWIN 12, FEI).
Image analysis and quantification
All analysis was performed using imageJ [50]. Brightness was adjusted as needed to avoid saturation. Area quantifications were performed on images acquired on the same scope, with the same volume of media, with the same settings using the ‘Measure’ function. The velocity analysis was performed using the ‘STICS’ (space-time image correlation spectroscopy) function as previously described [51].
Statistical analysis were obtained using two sample t-test, n values are listed in the figure legends.
Supplementary Material
(A) Schematic displaying general design and confirmation for the generation of wamKO allele. Two gRNAs were used to create double strand breaks on either side of the wampa locus (red arrows). The breaks were repaired via homologous recombination off a donor template that contained 1 kb homology upstream (green line) and 1 kb homology downstream of wampa (peach line), as well as a mCherry cassette that was flanked with loxP sites (purple arrows). This generated the wamKO-mc line. We then used CRE-LOX recombination to remove the mCherry cassette, generating the wamKO line. This line was validated with sanger sequencing using primers in the 5’ and 3’ homology arms that resided outside of the gene region and showed a full removal of entire coding region for wampa and for the mCherry cassette, leaving only the expected single loxP site.
(B) Schematic displaying the design and confirmation for the generation of wamΔ1 and wamΔ2 alleles. A single gRNA was used to create breaks in the middle of the coding region for wampa. The break was repaired via non-homologous end joining. Due to the inefficiency of this repair process, indels were created generating two separate lines. Sequencing confirmed that wamΔ1 had indels that resulted in 3 extra aa and an approximately 26bp deletion, while wamΔ2 resulted in a smaller deletion of only 5bp with indels that generated 10 extra aa. Both alleles had an early stop site that terminated at 250aa, just under halfway through the 550aa protein.
(A) Trans-heterozygotes were generated by crossing wamKO/TM3 allele to the bb8MB10362/TM3 allele. All Progeny were collected and screened for fertility, as well homozygotes from the parental stocks for controls. Males from both bb8MB10362 and wamKO are sterile as homozygotes, however, as trans-heterozygotes, they are fertile indicating that bb8MB10362 is not contributing to the sterility observed in wamKO.
(B) Genomic rescue constructs of Wam (wamSR2) and the mScarlet-I fusion protein (Wam::mScarlet-I) were tested for fertility in the background of wamKO allele. Both were able to fully restore fertility to wildtype levels.
(A) Schematic displaying how Wam is added to axonemal microtubules during elongation. Drosophila docking complex (red circles) is loaded on to the newly polymerized microtubules (gray) of the axoneme (blue). Once the docking complex is attached to the microtubules, the outer dynein arm complex (green) can then be loaded onto the elongating axoneme. Florescent images during late elongation of a developing spermatid showing tubulin compared to Wam. Wam signal appears shorter in length than tublin, indicating it has not yet been loaded on to the newly polymerized microtubules. Scale bar is 17µm. Green box depicts an enlargement of the outer dynein arm complex (adapted from Lage et al., 2019) [26]. Docking complex (red; Wam and Gudu), Light chains (brown; Dlc90F, Robl, Ctp, CG7276, CG8407, CG10839, CG8800), Intermediate chains (orange; CG9319, CG10859, CG1571), Heavy Chains (blue; Kl-3, Kl-5).
(B) Phase-contrast and florescent live images of spermatids during early elongation of wildtype and wamKO. The elongating axoneme extending from the basal body (white arrow) can be seen sandwiched in-between the mitochondrial derivates as they grow along with it. SiR-tubulin dye reveals the presence of microtubules that are co-localizing with the mitochondrial derivatives in the cell (blue arrows) and a manchette-like microtubule bundle (green arrow) can be observed sitting on top of the nucleus as it is undergoing remodeling. No observable differences are detected in these microtubule-based structures. Green box depicts a purposed model of a cytoplasmic dynein complex that Wampa might interact with during its cytoplasmic functions. Scale is the same for wildtype and wamKO.
(A) Pie graph displaying the percent of abnormal mitochondrial arrangements along the meiotic spindle. In wildtype cells, 82.1% appeared normal during meiosis (n=112), whereas only 55.8% appeared normal in wamKO (n=77).
(B) Pie graph displaying the percent of mitochondria that appear fragmented during the onion stage. In wildtype, fragmentation of the nebenkerne was only observed in 1.8% of cells at the onion stage (n=112), however fragmentation was observed in 9.8% of cells in wamKO (n=113).
(A–B) Movie displaying the motility of mature sperm from wildtype (A) males compared to the lack of motility, which is observed in wamKO/KO males (B).
Acknowledgements
We thank the Bloomington Stock Center for fly stocks, Eric Hill for a critical reading of the manuscript, Steve Hoffman for assisting with microscopy, Brian Slaughter for aiding with the STICS analysis, Cindi Staber for teaching testes dissection and a critical reading of the manuscript, and Takuya Akiyama for extensive discussion and suggestions. This work was supported by funding from the Stowers Institute for Medical Research and NIH Grant R01-GM111733 to M.G..
Footnotes
Declaration of Interests
No competing interests
References
- 1.Pazour GJ, et al. , Proteomic analysis of a eukaryotic cilium. J Cell Biol, 2005. 170(1): p. 103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li P, et al. , CCDC114 is mutated in patient with a complex phenotype combining primary ciliary dyskinesia, sensorineural deafness, and renal disease. J Hum Genet, 2019. 64(1): p. 39–48. [DOI] [PubMed] [Google Scholar]
- 3.Sanders CD, et al. , The prevalence of the defining features of primary ciliary dyskinesia within a cri du chat syndrome cohort. Pediatr Pulmonol, 2018. 53(11): p. 1565–1573. [DOI] [PubMed] [Google Scholar]
- 4.Auguste Y, et al. , Loss of Calmodulin- and Radial-Spoke-Associated Complex Protein CFAP251 Leads to Immotile Spermatozoa Lacking Mitochondria and Infertility in Men. Am J Hum Genet, 2018. 103(3): p. 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hjeij R, et al. , CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am J Hum Genet, 2014. 95(3): p. 257–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fassad MR, et al. , Mutations in Outer Dynein Arm Heavy Chain DNAH9 Cause Motile Cilia Defects and Situs Inversus. Am J Hum Genet, 2018. [DOI] [PMC free article] [PubMed]
- 7.Nevers Y, et al. , Insights into Ciliary Genes and Evolution from Multi-Level Phylogenetic Profiling. Mol Biol Evol, 2017. 34(8): p. 2016–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horani A, et al. , Genetics and biology of primary ciliary dyskinesia. Paediatr Respir Rev, 2016. 18: p. 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badano JL, et al. , The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet, 2006. 7: p. 125–48. [DOI] [PubMed] [Google Scholar]
- 10.Fuller MT, Spermatogenesis The Development of Drosophila melanogaster., ed. Bate M AA. Vol. 1. 1993, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 11.Regan CL and Fuller MT, Interacting genes that affect microtubule function in Drosophila melanogaster: two classes of mutation revert the failure to complement between haync2 and mutations in tubulin genes. Genetics, 1990. 125(1): p. 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li K, et al. , Drosophila centrosomin protein is required for male meiosis and assembly of the flagellar axoneme. J Cell Biol, 1998. 141(2): p. 455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sitaram P, et al. , Regulation of dynein localization and centrosome positioning by Lis-1 and asunder during Drosophila spermatogenesis. Development, 2012. 139(16): p. 294–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen LB, et al. , The lissencephaly protein Lis1 is present in motile mammalian cilia and requires outer arm dynein for targeting to Chlamydomonas flagella. J Cell Sci, 2007. 120(Pt 5): p. 858–67. [DOI] [PubMed] [Google Scholar]
- 15.Casal J, et al. , Abnormal meiotic spindles cause a cascade of defects during spermatogenesis in asp males of Drosophila. Development, 1990. 108(2): p. 251–60. [DOI] [PubMed] [Google Scholar]
- 16.Takada S, et al. , The outer dynein arm-docking complex: composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol Biol Cell, 2002. 13(3): p. 1015–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onoufriadis A, et al. , Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am J Hum Genet, 2013. 92(1): p. 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young SA, et al. , CRISPR/Cas9-Mediated Rapid Generation of Multiple Mouse Lines Identified Ccdc63 as Essential for Spermiogenesis. Int J Mol Sci, 2015. 16(10): p. 24732–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsley DT, Tokuyasu KT , Spermatogenesis, in The Genetics and Biology of Drosophila, Ashburner M.a.W., T.R.F., Eds., Editor. 1980, Academic Press: London. p. 225–294. [Google Scholar]
- 20.Fabian L and Brill JA, Drosophila spermiogenesis: Big things come from little packages. Spermatogenesis, 2012. 2(3): p. 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokuyasu KT, Dynamics of spermiogenesis in Drosophila melanogaster. VI. Significance of “onion” nebenkern formation. J Ultrastruct Res, 1975. 53(1): p. 93–112. [DOI] [PubMed] [Google Scholar]
- 22.Pitnick S, Markow TA, and Spicer GS, Delayed male maturity is a cost of producing large sperm in Drosophila. Proc Natl Acad Sci U S A, 1995. 92(23): p. 10614–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tates AD, Cytodifferentiation during spermatogenesis in Drosophila melanogaster: an electron microscope study. 1971, University of Leiden. [Google Scholar]
- 24.Vedelek V, et al. , Testis-Specific Bb8 Is Essential in the Development of Spermatid Mitochondria. PLoS One, 2016. 11(8): p. e0161289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King SM, Axonemal Dynein Arms. Cold Spring Harb Perspect Biol, 2016. 8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zur Lage P, et al. , Ciliary dynein motor preassembly is regulated by Wdr92 in association with HSP90 co-chaperone, R2TP. J Cell Biol, 2018. 217(7): p. 2583–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakabayashi K, et al. , Transport and arrangement of the outer-dynein-arm docking complex in the flagella of Chlamydomonas mutants that lack outer dynein arms. Cell Motil Cytoskeleton, 2001. 48(4): p. 277–86. [DOI] [PubMed] [Google Scholar]
- 28.Fatima R, Drosophila Dynein intermediate chain gene, Dic61B, is required for spermatogenesis. PLoS One, 2011. 6(12): p. e27822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinhauer J, Separating from the pack: Molecular mechanisms of Drosophila spermatid individualization. Spermatogenesis, 2015. 5(2): p. e1041345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson MA, et al. , Asunder is a critical regulator of dynein-dynactin localization during Drosophila spermatogenesis. Mol Biol Cell, 2009. 20(11): p. 2709–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoyle HD and Raff EC, Two Drosophila beta tubulin isoforms are not functionally equivalent. J Cell Biol, 1990. 111(3): p. 1009–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White-Cooper H, Spermatogenesis: analysis of meiosis and morphogenesis. Methods Mol Biol, 2004. 247: p. 45–75. [DOI] [PubMed] [Google Scholar]
- 33.Li MG, et al. , The Drosophila tctex-1 light chain is dispensable for essential cytoplasmic dynein functions but is required during spermatid differentiation. Mol Biol Cell, 2004. 15(7): p. 3005–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh-Roy A, et al. , Cytoplasmic dynein-dynactin complex is required for spermatid growth but not axoneme assembly in Drosophila. Mol Biol Cell, 2004. 15(5): p. 2470–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh-Roy A, Desai BS, and Ray K, Dynein light chain 1 regulates dynamin-mediated F-actin assembly during sperm individualization in Drosophila. Mol Biol Cell, 2005. 16(7): p. 3107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knowles MR, et al. , Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am J Hum Genet, 2013. 92(1): p. 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Donnell L and O’Bryan MK, Microtubules and spermatogenesis. Semin Cell Dev Biol, 2014. 30: p. 45–54. [DOI] [PubMed] [Google Scholar]
- 38.Russell LD, et al. , Linkage of manchette microtubules to the nuclear envelope and observations of the role of the manchette in nuclear shaping during spermiogenesis in rodents. Am J Anat, 1991. 192(2): p. 97–120. [DOI] [PubMed] [Google Scholar]
- 39.Kierszenbaum AL, Intramanchette transport (IMT): managing the making of the spermatid head, centrosome, and tail. Mol Reprod Dev, 2002. 63(1): p. 1–4. [DOI] [PubMed] [Google Scholar]
- 40.Gunes S, et al. , Microtubular Dysfunction and Male Infertility. World J Mens Health, 2018. [DOI] [PMC free article] [PubMed]
- 41.Lehti MS and Sironen A, Formation and function of the manchette and flagellum during spermatogenesis. Reproduction, 2016. 151(4): p. R43–54. [DOI] [PubMed] [Google Scholar]
- 42.Han YG, Kwok BH, and Kernan MJ, Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr Biol, 2003. 13(19): p. 1679–86. [DOI] [PubMed] [Google Scholar]
- 43.Sarpal R, et al. , Drosophila KAP interacts with the kinesin II motor subunit KLP64D to assemble chordotonal sensory cilia, but not sperm tails. Curr Biol, 2003. 13(19): p. 1687–96. [DOI] [PubMed] [Google Scholar]
- 44.Pleuger C, et al. , CBE1 Is a Manchette- and Mitochondria-Associated Protein With a Potential Role in Somatic Cell Proliferation. Endocrinology, 2019. 160(11): p. 2573–2586. [DOI] [PubMed] [Google Scholar]
- 45.Hook P and Vallee RB, The dynein family at a glance. J Cell Sci, 2006. 119(Pt 21): p. 4369–71. [DOI] [PubMed] [Google Scholar]
- 46.Zietkiewicz E, et al. , CFAP300: Mutations in Slavic Primary Ciliary Dyskinesia Patients and a Role in Ciliary Dynein Arms Trafficking. Am J Respir Cell Mol Biol, 2019. [DOI] [PubMed]
- 47.Hales KG and Fuller MT, Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell, 1997. 90(1): p. 121–9. [DOI] [PubMed] [Google Scholar]
- 48.Port F, et al. , Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A, 2014. 111(29): p. E2967–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akiyama T, User SD, and Gibson MC, Somatic clones heterozygous for recessive disease alleles of BMPR1A exhibit unexpected phenotypes in Drosophila. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider CA, Rasband WS, and Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nat Methods, 2012. 9(7): p. 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hebert B, Costantino S, and Wiseman PW, Spatiotemporal image correlation spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys J, 2005. 88(5): p. 3601–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic displaying general design and confirmation for the generation of wamKO allele. Two gRNAs were used to create double strand breaks on either side of the wampa locus (red arrows). The breaks were repaired via homologous recombination off a donor template that contained 1 kb homology upstream (green line) and 1 kb homology downstream of wampa (peach line), as well as a mCherry cassette that was flanked with loxP sites (purple arrows). This generated the wamKO-mc line. We then used CRE-LOX recombination to remove the mCherry cassette, generating the wamKO line. This line was validated with sanger sequencing using primers in the 5’ and 3’ homology arms that resided outside of the gene region and showed a full removal of entire coding region for wampa and for the mCherry cassette, leaving only the expected single loxP site.
(B) Schematic displaying the design and confirmation for the generation of wamΔ1 and wamΔ2 alleles. A single gRNA was used to create breaks in the middle of the coding region for wampa. The break was repaired via non-homologous end joining. Due to the inefficiency of this repair process, indels were created generating two separate lines. Sequencing confirmed that wamΔ1 had indels that resulted in 3 extra aa and an approximately 26bp deletion, while wamΔ2 resulted in a smaller deletion of only 5bp with indels that generated 10 extra aa. Both alleles had an early stop site that terminated at 250aa, just under halfway through the 550aa protein.
(A) Trans-heterozygotes were generated by crossing wamKO/TM3 allele to the bb8MB10362/TM3 allele. All Progeny were collected and screened for fertility, as well homozygotes from the parental stocks for controls. Males from both bb8MB10362 and wamKO are sterile as homozygotes, however, as trans-heterozygotes, they are fertile indicating that bb8MB10362 is not contributing to the sterility observed in wamKO.
(B) Genomic rescue constructs of Wam (wamSR2) and the mScarlet-I fusion protein (Wam::mScarlet-I) were tested for fertility in the background of wamKO allele. Both were able to fully restore fertility to wildtype levels.
(A) Schematic displaying how Wam is added to axonemal microtubules during elongation. Drosophila docking complex (red circles) is loaded on to the newly polymerized microtubules (gray) of the axoneme (blue). Once the docking complex is attached to the microtubules, the outer dynein arm complex (green) can then be loaded onto the elongating axoneme. Florescent images during late elongation of a developing spermatid showing tubulin compared to Wam. Wam signal appears shorter in length than tublin, indicating it has not yet been loaded on to the newly polymerized microtubules. Scale bar is 17µm. Green box depicts an enlargement of the outer dynein arm complex (adapted from Lage et al., 2019) [26]. Docking complex (red; Wam and Gudu), Light chains (brown; Dlc90F, Robl, Ctp, CG7276, CG8407, CG10839, CG8800), Intermediate chains (orange; CG9319, CG10859, CG1571), Heavy Chains (blue; Kl-3, Kl-5).
(B) Phase-contrast and florescent live images of spermatids during early elongation of wildtype and wamKO. The elongating axoneme extending from the basal body (white arrow) can be seen sandwiched in-between the mitochondrial derivates as they grow along with it. SiR-tubulin dye reveals the presence of microtubules that are co-localizing with the mitochondrial derivatives in the cell (blue arrows) and a manchette-like microtubule bundle (green arrow) can be observed sitting on top of the nucleus as it is undergoing remodeling. No observable differences are detected in these microtubule-based structures. Green box depicts a purposed model of a cytoplasmic dynein complex that Wampa might interact with during its cytoplasmic functions. Scale is the same for wildtype and wamKO.
(A) Pie graph displaying the percent of abnormal mitochondrial arrangements along the meiotic spindle. In wildtype cells, 82.1% appeared normal during meiosis (n=112), whereas only 55.8% appeared normal in wamKO (n=77).
(B) Pie graph displaying the percent of mitochondria that appear fragmented during the onion stage. In wildtype, fragmentation of the nebenkerne was only observed in 1.8% of cells at the onion stage (n=112), however fragmentation was observed in 9.8% of cells in wamKO (n=113).
(A–B) Movie displaying the motility of mature sperm from wildtype (A) males compared to the lack of motility, which is observed in wamKO/KO males (B).


