Abstract
Engineered cell-derived extracellular vesicles (EVs) such as exosomes and microvesicles hold immense potential as safe and efficient drug carriers due to their lower immunogenicity and inherent homing capabilities to target cells. In addition to innate vesicular cargo such as lipids, proteins, and nucleic acids, EVs are also known to contain functional mitochondria/mitochondrial DNA that can be transferred to recipient cells to increase cellular bioenergetics. In this proof-of-concept study, we isolated naïve EVs and engineered EVs loaded with an exogenous plasmid DNA encoding for brain-derived neurotrophic factor (BDNF-EVs) from hCMEC/D3, a human brain endothelial cell line, and RAW 264.7 macrophages. We tested whether mitochondrial components in naïve or engineered EVs can increase ATP levels in the recipient brain endothelial cells. EVs (e.g., exosomes and microvesicles; EXOs and MVs) were isolated from the conditioned medium of either untreated (naïve) or pDNA-transfected (Luc-DNA or BDNF-DNA) cells using a differential centrifugation method. RAW 264.7 cell line–derived EVs showed a significantly higher DNA loading and increased luciferase expression in the recipient hCMEC/D3 cells at 72 h compared with hCMEC/D3 cell line–derived EVs. Naïve EVs from hCMEC/D3 cells and BDNF-EVs from RAW 264.7 cells showed a small, but a significantly greater increase in the ATP levels of recipient hCMEC/D3 cells at 24 and 48 h post-exposure. In summary, we have demonstrated (1) differences in exogenous pDNA loading into EVs as a function of cell type using brain endothelial and macrophage cell lines and (2) EV-mediated increases in the intracellular ATP levels in the recipient hCMEC/D3 monolayers.
Keywords: exosomes and microvesicles, mitochondria and mtDNA, ATP increase, brain endothelial cells, ischemic stroke
INTRODUCTION
Extracellular vesicles (EVs) are a heterogeneous group of membrane-derived particles that are released from prokaryotic and eukaryotic cells under both physiological and pathophysiological conditions (1,2). EV membranes contain lipid bilayers with integrated surface receptors and membrane-bound proteins that allow binding to target cells and also protect the internal cargo from physiological degradation. The core of EVs is enriched with innate cargo that consists of soluble proteins, a variety of nucleic acids including ssDNA, dsDNA, siRNA, miRNA, mRNA, tRNA, rRNA, lipids, mitochondrial DNA, and/or organelles such as mitochondria. One of the key roles of EVs is to mediate intercellular communication between the donor and recipient cells via horizontal transfer of proteins, lipids, and nucleic acids (2-5). EVs have demonstrated a critical role in the normal physiological processes such as immune modulation and surveillance, cell signaling, cell maintenance, and tissue repair as well as pathophysiological processes during tumorigenesis, angiogenesis, neurodegenerative disorders, and inflammatory conditions (1). The inherent characteristics of EVs including their capabilities to contain nucleic acids and transfer them to the recipient cells make them an attractive choice as carriers for safe and efficient delivery of nucleic acids. Besides, EVs contain a lower abundance of MHC complexes compared with the parent cells (6) and have also shown inhibitory effects on immune responses (7). Thus, EVs may be less immunogenic than engineered cell-based therapies and synthetic drug carriers due to their innate origin. Furthermore, EVs have demonstrated a lower risk of tumorigenicity and latent viral pathogen transfer, a longer shelf life, and longer storage stability compared with live cell–based therapies (8,9). In contrast, commercial transfection platforms including but not limited to Lipofectamine 3000, Lipofectamine 2000, FuGENE, RNAiMAX, and Lipofectin complexed with oligonucleotides show higher toxicity that is correlated with their greater transfection efficiency (10). Moreover, the natural characteristics of EVs including their capability to home to specific cellular targets, avoidance of off-target effects, intrinsic ability to cross biological membranes, and overcome intracellular barriers to deliver cargo may allow them to outperform synthetic drug carriers (8). In contrast, the most extensively studied and FDA-approved liposomal formulation, Doxil for instance, is known to be rapidly cleared from the systemic circulation via reticuloendothelial system (RES) uptake and accumulates mainly in highly perfused organs such as the liver and spleen (11). Overall, the broad functionalities of cell-derived EVs and their numerous advantages compared with synthetic drug delivery vehicles hold immense promise for the delivery of biotherapeutics such as nucleic acids and proteins to treat human disorders.
Based on their biogenesis, EVs can be classified into three main groups i.e. exosomes, microvesicles, and apoptotic bodies. Exosomes (EXOs) are a homogenous class of endosome-derived, membrane-bound particles with diameters ranging from 30 to 150 nm (12). EXOs are generated from the inward budding of endosomal membranes and released to the extracellular space upon the fusion with the plasma membrane (5,13). Based on their biogenesis, exosomal membranes contain phosphatidylserine, phosphatidylethanolamine, phosphatidylinositol, sphingomyelin, ceramide, cholesterol, and lipid rafts bound to several proteins. The specific proteins expressed on exosomes include proteins from the tetraspanin family (CD9, CD63, CD81, and CD82), ESCRT family proteins (tumor susceptibility gene 101, TSG101, and Alix), and heat shock proteins (HSP60, HSP70, HSPA5, and HSP90) (2,14,15). Members of the heat shock protein family, in particular, are uniquely expressed in exosomes and may play an important role in modulating the cellular energetics and mitochondrial functions in the recipient cells. The mitochondrial HSP proteins maintain the mitochondrial membrane potential, preserve mitochondrial function, and protein import (16). Voloboueva et al. demonstrated that the overexpression of mitochondrial HSP70/HSP75 decreased reactive oxygen species, upregulated ATP levels, and the cell viability of astrocytes under oxygen-glucose deprivation (OGD) conditions (17). During OGD conditions, the inner mitochondrial membranes are damaged due to the ROS-mediated damage to lipids and proteins. HSPs can contribute to the preservation of ATP levels during stress conditions by interrupting cytochrome c translocation from mitochondria to the nucleus (16,17).
The discovery of mRNA and microRNA in exosomes and their ability to express new mouse proteins in the recipient human cells upon the transfer of mouse exosomal RNA catalyzed many studies to test their capability to serve as carriers for nucleic acids (18). Alvarez-Erviti et al. demonstrated that the systemic administration of EXOs modified with a neuron-specific RVG-peptide delivered exogenously loaded therapeutic BACE1 siRNA that resulted in ca. 60% knockdown of targeted mRNA and protein in a mouse model of Alzheimer’s disease (19). Ohn et al. reported that the intravenous administration of EXOs fused with a GE-11 peptide delivered let-7a miRNA targeted to the epidermal growth factor receptor and resulted in significant suppression of tumor growth in a mouse xenograft breast cancer model (20). Recently, Mendt et al. performed multiple in vitro and in vivo studies to determine the chemotherapeutic effect of clinical-grade EXOs loaded with exogenous oncogenic Kras siRNA and demonstrated increased survival in several mouse models of pancreatic cancer (21). Thakur et al. provided the evidence for the presence of double-stranded DNA in tumor-derived EXOs (22); however, there are only limited studies reporting the exosomal delivery of exogenous-loaded therapeutic DNA. Kanada et al. demonstrated limited loading of plasmid DNA in EXOs (23). Lamichhane et al. studied the effect of size of exogenous linear and circular; i.e. plasmid DNA on their loading into EXOs demonstrating that the pDNA loading efficiency in EXOs was less than 0.2% and a noted an upper size limit of about 750–1000 bp for efficient loading (13). The above results suggested that large-sized plasmid DNA constructs have limited loading into exosomes.
Microvesicles (MVs) are a heterogeneous population of membrane vesicles with particle diameters ranging from 100 to 1000 nm (24,25). The biogenesis of MVs involves sequential processes including sorting of cargo proteins, bulging out of a part of the membrane followed by the abscission of the section of the plasma membrane resulting in the budding of MVs followed by their release into the extracellular space (26). MV membranes contain a larger amount of phosphatidylserine, cholesterol, and diacylglycerol as compared with exosomes. MVs express characteristic surface marker proteins such as phosphatidylserine (23), integrin, selectins (P-selectin glycoprotein ligand-1 and E-selectin), and CD40 (2,26). A couple of studies have reported greater DNA loading into MVs and as a consequence, greater DNA transport into recipient cells compared with exosomes (13,23). Cha et al. reported that MVs derived from ischemic brain extract-pretreated human mesenchymal stem cells (hMSC) contained an abundance of cytokines and nucleic acids that led to the observed inflammatory, angiogenic, and strong neurogenic effects in hMSC culture (9). Importantly, MVs possess a unique feature related to the presence of mitochondria or mitochondrial DNA (mtDNA), which can be transferred between cells to facilitate cellular communication and/or increase cellular bioenergetics. In fact, mitochondrial transfer resulted in increased cellular bioenergetics leading to greater cell survival in alveolar macrophages under oxidative stress (27). Dozio et al. reported about 89 types of mitochondrial proteins in hCMEC/D3-derived MVs whereas EXOs contained a single type of mitochondrial protein, suggesting that mitochondria were selectively packaged in MVs when compared with EXOs (28). Under aerobic conditions, mitochondria provide the chemical energy in the form of ATP for the major intracellular metabolic pathways, and hence, critically affect cellular viability by regulating ATP-mediated cellular bioenergetics, calcium signaling, and apoptosis processes (29,30). Mutations in mtDNA or mitochondrial dysfunctions are often associated with neurodegenerative diseases, metabolic and age-related disorders, and ischemic injury in the heart and brain (29). Notably, it has been reported that the transfer of mitochondria from as few as 100 hMSC donor cells was sufficient to rescue cells depleted of mitochondrial DNA (31-34). Therefore, mitochondria-containing EVs (MVs and EXOs) can improve the cellular bioenergetics since mitochondria are known to have significant implications in the broad scope of ischemic repair (35).
Ischemic stroke, the world’s second leading cause of mortality, is caused by a reduction in blood flow to the affected brain tissues that leads to a series of interrelated and coordinated cellular processes. Such processes include cellular energy failure, neuron depolarization, increased intracellular calcium ions, and membrane, mitochondrial, and DNA damage caused by the generation of free radicals that eventually culminate in cell death (36). Post-ischemic injury, the delivery of oxygen and essential nutrients are greatly impaired which leads to a reduction in ATP levels in the endothelial cells lining the blood-brain barrier (BBB) leading to the generation of reactive oxygen species and subsequent mitochondrial dysfunction (37). As a result, intracellular cations accumulate in endothelial cells that can lead to endothelial cell swelling and disruption of the BBB (37). Protection of the brain endothelium without penetrating the BBB by increasing endothelial ATP levels and delivering neuroprotective agents such as neurotrophins is a viable strategy to decrease acute endothelial cell death and to restore the endothelial-neuronal trophic signaling pathways in ischemic stroke. While therapeutic studies have historically focused on delivering neuroprotectant drugs, the rising trend in studies delineating endothelial, and microglial cell death mechanisms in stroke (38) point to the importance of delivering therapeutics to decrease endothelial damage. We believe that protection of the brain endothelium is a critical aspect in decreasing ischemic damage because the endothelial cells are fundamentally neuroprotective (39) and decreasing acute cell death will preserve/rescue its protective functions.
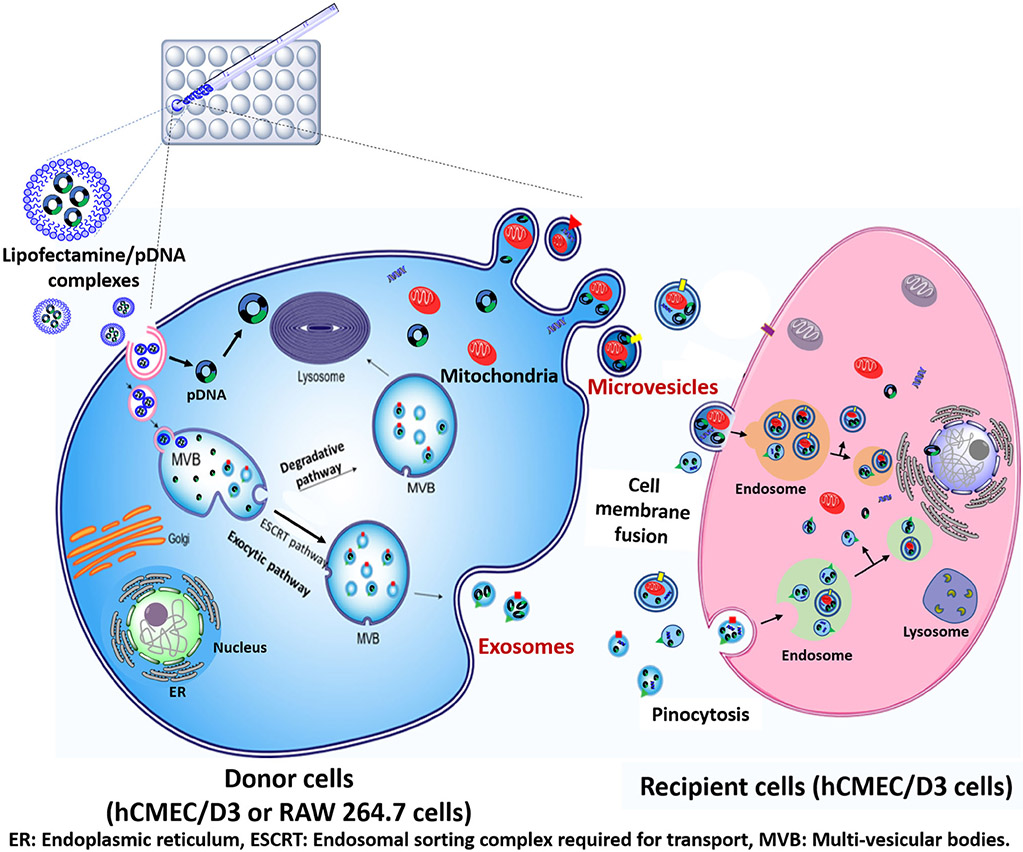
We wanted to study if EVs derived from the human brain endothelial cell line, hCMEC/D3, can be engineered as carriers for the delivery of a therapeutic plasmid DNA, for example, brain-derived neurotrophic factor (BDNF). Guo et al. demonstrated that the BDNF protein secreted from cerebral endothelial cells protected neurons against oxidative damage, hypoxia, oxygen-glucose deprivation, and decreased overall neuronal death (39). The lack of effective carriers limits the safe and efficient delivery of BDNF to the BBB. Our goal is to engineer EVs for BDNF-DNA delivery—coupled with its internal components, specifically, mitochondria, to decrease acute ischemic cell death. In this study, we hypothesized that the delivery of EVs derived from a brain endothelial cell line, hCMEC/D3, can increase the cellular ATP levels in the recipient endothelial cells and our proposed approach is depicted in Fig. 1. It should be pointed out that we collectively refer to EXOs and MVs as EVs, wherever applicable.
Fig. 1.
Schematic representation of lipofectamine/pDNA transfection in hCMEC/D3 (or) RAW264.7 cells and the subsequent internalization of DNA-loaded EVs into the recipient cells. hCMEC/D3 or RAW264.7 cells seeded in a 24-well plate were transfected with lipofectamine/pDNA complexes for 12 h. Once they are endocytosed, the DNA complexes can either be localized either in multivesicular bodies (MVB) to allow subsequent biogenesis into exosomes (exocytic pathway). It is also likely that the complexes in MVBs may progress along a degradative pathway for subsequent degradation in the lysosomes. DNA complexes that escape the endolysosomal degradation followed by release into the cytoplasm may be incorporated into microvesicles along with mitochondria that tend to migrate to the cell periphery. Upon addition to the recipient cells, DNA-EVs may internalize via pinocytosis or fuse with the cell membrane for subsequent intracellular processing
In this proof-of-concept study, we (1) tested if it is possible to load a therapeutic plasmid, BDNF pDNA for the subsequent development of engineered EVs as a carrier to deliver BDNF pDNA to ischemic brain endothelial cells and (2) tested if naïve/engineered EVs can increase cellular ATP levels in the recipient endothelial cells. To our knowledge, this is the first study utilizing EXOs and MVs to deliver an exogenous therapeutic pDNA (here, pBDNF) to human brain endothelial cells. In the context of developing novel stroke therapeutics, our strategy of delivering naïve- or BDNF-EVs to the brain endothelial cells is particularly significant because delivering mitochondrial components and stimulating BDNF production/secretion from the endothelial cells to promote cell viability without crossing the BBB explores a new paradigm (40) in treating cerebrovascular disorders. In this study, we isolated MVs and EXOs from the human brain endothelial; hCMEC/D3 and RAW 264.7 macrophage cell lines using a differential centrifugation method and characterized their physicochemical characteristics and marker proteins. Exogenous pDNA-loaded EVs were isolated from cells pre-transfected with lipofectamine/pDNA complexes. We utilized a Design of Experiments (DOE) approach to investigate the effect of experimental parameters (factors) on the transfection efficiency and resulting ATP levels in studies using Luc pDNA and BDNF pDNA-loaded EVs (Luc-EVs and BDNF-EVs), respectively. First, we performed a full factorial DOE to study the effects of the amount of Luc plasmid DNA transfected into the parent cells (either hCMEC/D3 endothelial or RAW 264.7 macrophage cells), type of EVs (EXOs or MVs), amount of total EV protein, and EV incubation time on the luciferase transgene expression mediated by Luc-EVs. In the second full factorial DOE, we evaluated the effects of the amount of BDNF pDNA transfected into the parent cells (either hCMEC/D3 endothelial or RAW 264.7 macrophage cells), type of EVs (EXOs or MVs), amount of total EV protein on the resulting intracellular ATP levels of hCMEC/D3 cells treated with BDNF-EVs. We demonstrated the capability of EVs to load an exogenous therapeutic pDNA; pBDNF and EV-mediated increases in intracellular ATP levels in the recipient hCMEC/D3 monolayers.
MATERIAL AND METHODS
Chemicals and Reagents
gWiz plasmid containing luciferase (gWiz-Luc) and plasmid DNA encoding BDNF (gWiz-BDNF) were purchased from Aldevron (Fargo, ND). Collagen type I was purchased from Corning (Discovery Labware Inc., Bedford, MA) and endothelial cell basal medium-2 (EBM-2) was procured from Lonza (Walkersville, MD). Hydrocortisone, human basic fibroblast growth factor, and ascorbic acid were purchased from Sigma-Aldrich (Saint Louis, MO). Dithiothreitol (DTT), glycylglycine, ethylenediaminetetraacetic acid (EDTA), HEPES, and magnesium chloride hexahydrate (MgCl2.6H2O) were received from Fisher Scientific (Pittsburgh, PA). Aprotinin solution was purchased from Fisher Bioreagents, New Zealand. The penicillin-streptomycin solution, Lipofectamine 3000 transfection reagent, and chemically defined lipid concentrate were procured from Invitrogen (Carlsbad, CA). Heat inactivated fetal bovine serum was bought from Hyclone Laboratories (Logan, UT). Pierce MicroBCA and BCA protein assay kits were purchased from Thermo Scientific (Rockford, IL). Quant-iT PicoGreen dsDNA Assay kit was obtained from Molecular Probes, Inc. (Eugene, OR). CellTiter-Glo 2.0 reagent (ATP assay), Beetle Luciferin, potassium salt, and cell culture lysis 5× Reagent were purchased from Promega (Madison, WI). Laemmli sodium dodecyl sulfate (SDS) sample buffer and RIPA buffer, 5×, were procured from Alfa Aesar (Ward Hill, MA). Adenosine-5-triphosphate (ATP) was obtained from MP Biomedicals (Illkirch, France). Odyssey blocking buffer was purchased from Li-COR (Lincoln, NE). Mouse monoclonal antibody to GRP94 and Alix were purchased from Santa Cruz Biotechnology, Inc. (Heidelberg, Germany). Mouse monoclonal antibodies against ATP5A and GAPDH were procured from Abcam. Mouse monoclonal antibody against CD9 was obtained from Life Technologies Corporation, whereas Alexa Fluor 790-conjugated donkey anti-mouse IgG was received from Jackson ImmunoResearch Lab Inc. (West Grove, PA). Calcein AM, flow cytometry sub-micron particle size reference kit, and an Attune NxT small particle side scatter filter were purchased from Invitrogen (Carlsbad, CA).
Cell Line and Cell Culture
Human cerebral microvascular endothelial cell line (hCMEC/D3, catalog number. 102114.3C) at passage number (P) 25 was purchased from Cedarlane Laboratories (Burlington, Ontario), and cells between P25 and P35 were used for experiments. The hCMEC/D3 cells were grown in tissue culture flasks or multiwell plates pre-coated using 0.15 mg/mL rat collagen I. The cells were cultured in a complete growth medium in a humidified 5% CO2 incubator at 37 ± 0.5 °C (Isotemp, Thermo Fisher Scientific). The complete growth medium contained endothelial cell basal medium (EBM-2) supplemented with 5% fetal bovine serum, penicillin (100 units/mL)-streptomycin (100 μg/mL) mixture, hydrocortisone (1.4 μM), ascorbic acid (5 μg/mL), chemically defined lipid concentrate (0.01%), 10 mM HEPES (pH 7.4), and bFGF (1 ng/mL). The complete growth medium was replenished every other day until the cells formed confluent monolayers. Prior to passage, the cells were washed using 1× phosphate buffer saline (PBS) and detached from the flask using 1× TrypLE Express Enzyme (Gibco, Denmark). Abelson murine leukemia virus-transformed macrophages, RAW 264.7 cell line (catalog number. ATCC TIB-71) was procured from ATCC (Manassas, VA). Cells were maintained in the complete growth medium containing glutamine-supplemented Dulbecco’s Modified Eagle’s Medium (DMEM (1×) + Glutamax, Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum.
Isolation of EVs from the hCMEC/D3 and RAW 264.7 Cells
The hCMEC/D3 and RAW 264.7 cells were cultured in complete growth medium in collagen-coated (for facilitating the adherence of hCMEC/D3 cells) or non-coated (RAW 264.7 cell line) 175 cm2 flasks (T175) for a minimum of 48 h until the cells formed a confluent monolayer (41). The complete growth medium was removed, the cells were washed with 25 mL of pre-warmed 1× PBS, and cultured in 25 mL of serum-free medium (conditioned medium) for 2 days in a humidified 5% CO2 incubator at 37 ± 0.5 °C. The EXOs and MVs present in the conditioned medium were isolated using a differential centrifugation method with slight modifications (23,28,41). Briefly, the conditioned medium from T175 flasks was transferred into polypropylene centrifuge tubes and centrifuged at 300×g for 10 min at 4 °C to remove dead cells and debris using a Sorvall RC-5C centrifuge (Kendro Laboratory Products, Newton, CT). The supernatant was carefully transferred into a new set of tubes and centrifuged at 2000×g for 20 min at 4 °C to remove large vesicles such as apoptotic bodies. Next, the supernatant was transferred into polycarbonate flanged tubes (Thermo Fisher Scientific) and centrifuged at 20,000×g for 45 min at 4 °C to pellet MVs using an XE-90 ultracentrifuge equipped with a 50.2 Ti rotor (Beckman Coulter, Indianapolis, IN). The resulting supernatant was filtered through 0.22-μm syringe filters to remove vesicles greater than 220 nm in diameter. Following that, EXOs were collected by centrifuging the supernatant at 120,000×g for 70 min at 4 °C. Lastly, EVs were suspended either in 1× RIPA buffer containing 3 μg/mL aprotinin for western blotting, in 10 mM HEPES buffer (pH 7.4) for zeta potential measurements, or 1× PBS for particle size measurements and transfection experiments. EV suspensions were stored at −80 °C until further use.
Total Protein Measurements of EVs Using BCA and MicroBCA Assays
Isolated EVs were quantified by measuring their total protein content using Pierce BCA protein assay for western blotting and MicroBCA protein assay for all other studies described herein. For BCA protein assay, a 25 μL volume of EV lysates, whole-cell lysates, and bovine serum albumin (BSA) standards at concentrations ranging from 20 to 2000 μg/mL were transferred into a 96-well plate and 200 μL BCA working reagent (WR, reagent A:reagent B = 50:1) was added to each well. The plate was incubated at 37 °C for 30 min in the dark, and the absorbance was measured at 562 nm using a SYNERGY HTX multi-mode reader (BioTek Instruments, Inc., Winooski, VT). For the MicroBCA protein assay, 10 μL of EVs suspended in PBS was lysed using 150 μL of 1× RIPA buffer containing 3 μg/mL aprotinin. A 150-μL volume of the EV lysates or BSA standards (2–40 μg/mL) was pipetted into a 96-well plate and an equal quantity of the MicroBCA WR (reagent A:reagent B:reagent C = 25:24:1) was added to each well. After 2 h of incubation at 37 °C, the absorbance was measured at 562 nm using a SYNERGY HTX multi-mode reader.
Particle Size Distribution and Zeta Potential of EVs
The average particle diameter and zeta potential of EVs were measured using Dynamic Light Scattering (DLS). After measuring the total protein concentration using MicroBCA assay, the EXOs and MVs were diluted to the final concentration of 0.7 mg/mL, using either 1× PBS at pH 7.4 for particle size distribution measurements or 10 mM HEPES buffer at pH 7.4 for zeta potential analysis. DLS was performed using a Zetasizer Nano (Malvern Panalytical Inc., Westborough, PA). Each sample was measured thrice, and three independent samples were analyzed. The data are represented as mean ± standard deviation (SD) of triplicate measurements.
Detection of EV Marker Proteins Using Western Blotting
The characteristic marker proteins in extracellular vesicles (MVs and EXOs) derived from the hCMEC/D3 cells were detected using the western blotting technique (42). Briefly, cellular and EV lysates containing 25 μg total protein were mixed with 4× Laemmli buffer and distilled water. The mixture was heated at 95 °C for 5 min using a heating block (Thermo Scientific). The samples and the premixed molecular weight markers (ladder, 250 kD-10 kD) were separated on a 4–10% gradient sodium dodecyl sulfate-polyacrylamide gel at 120 V for 90 min using a PowerPac Basic setup (Bio-Rad Laboratories, Inc.). The proteins were transferred onto a 0.45-μm nitrocellulose membrane using a transfer assembly (Fisher Scientific) at 75 V and 300 mA for 90 min. The membrane was washed using 0.1%-Tween 20 Tris Buffered- saline (T-TBS) and blocked with Odyssey blocking solution (Odyssey blocking buffer: T-TBS, 1:1) solution for an hour. The membrane was incubated with mouse anti-GRP94 (0.2 μg/mL), anti-ATP5A (1 μg/mL), anti-GAPDH (1 μg/mL), anti-Alix (1 μg/mL) and anti-CD-9 (1 μg/mL) primary antibodies prepared in Odyssey blocking solution at 4 °C overnight. The membrane was washed with T-TBS and incubated with anti-mouse AF790 (0.05 μg/mL) in Odyssey blocking solution for 1 h at room temperature. The membrane was washed and scanned under an 800-nm near-infrared channel using an Odyssey imager (LI-COR1Inc., Lincoln, NE) at intensity setting 5.
Confirmation of the Integrity of EVs After Ultracentrifugation and Resuspension Processes
To determine the integrity of EXOs and MVs after ultracentrifugation and resuspension, EVs were isolated from hCMEC/D3 and RAW 264.7 cell lines using a differential ultracentrifugation method described earlier. In order to confirm the instrument suitability for analyzing EVs, we first ran 0.1-, 0.2-, 0.5-, 1-, and 2-μm calibration beads (Molecular Probes, Life technologies) in the side scatter (SSC), forward scatter (FSC), and a small particle side scatter 488/10-nm filter (BL1) channels in an Attune flow cytometer. Next, the isolated EXOs and MVs were resuspended in PBS at 20 μg/mL and were labeled with 10 μM calcein AM dissolved in DMSO for 20 min at room temperature in dark. To demonstrate that the calcein AM signal intensity is specific to intact EVs, EXOs and MVs were lysed using 2% v/v Triton-X 100 solution in PBS for 20 min at room temperature. Lysed EVs were also incubated with 10 μM calcein AM as described earlier. A 100-μL aliquot of the sample was analyzed using Attune NxT Acoustic Focusing Cytometer (Invitrogen, Singapore) equipped with Attune NxT software. In each experiment, about 50,000 events from a 100-μL sample volume were recorded, and the fluorescence intensity of calcein-labeled EVs was detected at 488/10 nm. The percentage of calcein-labeled EVs were analyzed and are presented as density plots and histograms obtained from the Attune NxT software. PBS containing 10 μM calcein AM and PBS/2% Triton-X 100/calcein AM mixture were used as controls for gating the signal intensity associated with background noise.
Plasmid DNA Loading in EVs
Prior to isolating DNA-loaded EVs, we pre-transfected cells using lipofectamine/DNA complexes. We used gWiz-Luc pDNA to incorporate luciferase DNA into EVs (Luc-EVs) for luciferase transfection assay whereas gWiz-BDNF pDNA was used for generating BDNF-EVs. The hCMEC/D3 or RAW 264.7 cells were transfected with pDNA using Lipofectamine 3000 transfection reagent as described in the supplier’s protocol with slight modifications. Briefly, the hCMEC/D3 or RAW 264.7 cells were seeded in complete growth medium at a density of 75,000 cells/cm2 in 24-well plates and were cultured for 48 h in a humidified incubator (37 °C + 5% CO2). On the day of transfection, Lipofectamine 3000 diluted in the culture medium was mixed with 1 or 2 μL of P3000 reagent and 0, 0.5, or 1 μL of pDNA (1 μg/μL stock solution). The complexes were incubated for 15 min at room temperature. In each 24-well plate, the hCMEC/D3 or RAW 264.7 cells were transfected with Lipofectamine 3000/pDNA complexes containing either 0, 0.5, or 1 μg/well pDNA for 12 h corresponding to total amounts of 0, 12, or 24 μg of pDNA. The plate that received 0 μg/well indicates that the transfection mixture contained Lipofectamine only, no pDNA. The transfection medium was then removed, the cells were washed with 1× PBS, and were cultured with serum-free medium for 48 h. The collected supernatant from each 24-well plate was pooled into pre-labeled tubes marked with the respective pDNA dose for subsequent EV isolation. Plasmid DNA-loaded EVs (DNA-MVs and DNA-EXOs) were isolated using the differential centrifugation method as described above.
Quantification of DNA Loading in EVs Using PicoGreen Assay
The amount of double-stranded DNA (dsDNA) in EXOs and MVs was quantified using the Quant-iT Picogreen assay kit. Picogreen is an ultra-sensitive benzothiazole fluorochrome that selectively binds to dsDNA and can be excited at a wavelength of 480 nm and emits a maximum fluorescence intensity at 520 nm proportional to dsDNA concentration. The Luc pDNA-loaded EVs were lysed by adding 1× RIPA buffer containing 3 μg/mL aprotinin to a final volume of 100–300 μL. A Picogreen working reagent was freshly prepared by diluting Quant-iT Picogreen dsDNA reagent 200-fold with 1× Tris-EDTA (TE) buffer in the dark. For the DNA standard curve, gWiz-Luc pDNA (1 μg/μL) was diluted using 1× TE buffer to prepare the standards ranging from 1 to 150 ng/mL concentration. To each well in a 96-well plate, DNA standards or lysates and Picogreen working reagents were added at a 1:1 volume ratio (100 μL sample + 100 μL Picogreen reagent). The mixture was incubated for 2–5 min at room temperature in the dark. The relative fluorescence intensity of the mixture was measured at 485-nm excitation and 528-nm emission wavelength settings using a SYNERGY HTX multi-mode reader. It should be noted that the fluorescence values obtained from the 0 μg/well DNA-transfected cells (naïve EVs) were subtracted from the pDNA-loaded EVs (pDNA-EVs). The DNA loading (%) was calculated using the following equation (Eq. 1).
| (1) |
Cytocompatibility of hCMEC/D3 Cells with Luc-EVs Derived from hCMEC/D3 and RAW 264.7 Cell Lines
We performed cell viability studies to determine the cytocompatibility of engineered EVs and lipofectamine/pDNA complexes with hCMEC/D3 cells. Luc-EVs were isolated from hCMEC/D3 or RAW 264.7 cells pre-transfected with 0, 0.5, or 1 μg/well of pDNA (amounting to a total of 0, 12, or 24 μg of DNA per 24-well plate), and the total EV protein amount was quantified using a MicroBCA assay as described above. hCMEC/D3 cells were seeded at a density of 16,500 cells/well in collagen-coated 96-well plates and cultured in complete growth medium for 48 h in a humidified incubator (37 °C, 5% CO2). The culture medium was replaced with 60 μL of complete growth medium containing 6 μg or 12 μg/well total EXOs or MVs protein isolated from 0, 0.5, or 1 μg/well pDNA-transfected plates (Table I). Untreated cells were used as a control, and cells treated with branched polyethyleneimine (PEI, molecular weight 25 kDa) at 50 μg/mL were used as a positive control. Cells were also treated with lipofectamine/Luc-DNA complexes at a dose of 10 ng pDNA/well. It should be noted that the 10-ng dose is equivalent to the amount of DNA loaded in the EVs. Cells were incubated with the treatment mixture for 24, 48, or 72 h in a humidified incubator. Post-exposure, hCMEC/D3 cell viability was measured using a CellTiter-Glo luminescence assay (referred henceforth as an ATP assay). Briefly, 60 μL of fresh pre-warmed complete growth medium and an equal quantity of CellTiter-Glo 2.0 reagent was added to each well. The plate was incubated at room temperature for 15 min on an orbital shaker in dark. A 60-μL volume of the mixture was transferred into a white opaque-walled 96-well plate. Immediately, the relative luminescence units were measured at 1-s integration time using a SYNERGY HTX multi-mode reader. The relative cell viability (%) was calculated by normalizing the relative luminescence units (RLU) of treated cells to those treated with untreated cells as shown in the following equation (Eq. 2):
| (2) |
Table I.
Experimental Design to Determine the Effect of Factors on the Transfection Efficiency of Luc-MVs and Luc-EXOs
| Factors | Level 1 | Level 2 | Level 3 |
|---|---|---|---|
| Amount of Luc pDNA (μg/well)a | 0 | 0.5 | 1 |
| Total EV protein (μg)b | 6 | N/A | 12 |
| Type of EVs | EXOs | N/A | MVs |
| Incubation time (h)c | 24 | 48 | 72 |
Amount of Luc pDNA represents the amount of gWiz-Luc pDNA transfected into the donor cells using lipofectamine3000 in each well in a 24-well plate and the EVs were isolated from a single 24-well plate that was transfected with a total of either 0, 12, or 24μg pDNA.
Total EV protein represents the protein content in EXOs or MVs quantified by MicroBCA protein assay
Incubation time represents the time that recipient hCMEC/D3 cells were treated using Luc-EVs. N/A: not applicable
Transfection of Luc-EVs into the Recipient Cells and Luciferase Gene Expression Assay
Luc-EVs were isolated from hCMEC/D3 or RAW 264.7 cells pre-transfected with 0, 0.5, or 1 μg/well of pDNA (amounting to a total of 0, 12, or 24 μg DNA per 24-well plate), and their total protein amount was quantified using MicroBCA assay as described above. The hCMEC/D3 cells were seeded at a density of 50,000 cell/cm2 in a collagen-coated 48-well plate and cultured in a complete growth medium for 48 h in a humidified incubator (37 °C, 5% CO2). The culture medium was replaced with 250 μL of complete growth medium containing 6 μg or 12 μg/well total EV protein (Table I). Untreated cells were used as a control, and cells treated with lipofectamine/DNA complexes were used as a positive control. Cells were incubated with the indicated samples for either 24, 48, or 72 h (incubation time). Post-incubation, the culture medium was removed from wells, and the cells were washed with ice-cold 1× PBS, and the cells were lysed by adding 100 μL/well ice-cold 1× luciferase cell culture lysis reagent and was placed for 20 min on an orbital shaker. The plate was subjected to two freeze-thaw cycles (−20 °C for 30 min and 4 °C for 30 min) prior to the luciferase assay. For measurement of the luciferase protein content, 20 μL of cell lysates were mixed with 100 μL of luciferase assay buffer (20 mM glycylglycine (pH 8), 1 mM MgCl2, 0.1 mM EDTA, 3.5 mM DTT, 0.5 mM ATP, 0.27 mM coenzyme A) into a white opaque-walled 96-well plate. The luminescence was measured at 1 s integration time using a SYNERGY HTX multi-mode reader. The total protein in the cell lysates was measured using the BCA assay. The data is represented in mean ± SD luminescence/mg total protein of 9 replicates.
The effect of in vitro transfection parameters (factors) on hCMEC/D3 luciferase expression (response) was evaluated using a full factorial design. As shown in Table I, the four factors were the amount of Luc-pDNA used for the formation of lipofectamine/DNA complexes, type of EVs, amount of EVs (total protein content), and incubation time. The factors were studied at either three (1–3, or) or two levels (1 or 3). The permutation and combination of each factor at the levels listed in Table I led to 36 experimental conditions. We performed three independent experiments studying each experimental condition at n = 3. The effect of these experimental conditions on the luciferase expression was statistically evaluated using JMP Pro 14.
Effect of BDNF-EVs Derived from hCMEC/D3 and RAW 264.7 Cells on the Cell Viability of hCMEC/D3 Cells
EXOs and MVs from 0, 0.5, or 1 μg/well plasmid gWiz-BDNF-DNA-transfected hCMEC/D3 or RAW 264.7 cells (BDNF-EVs) were isolated, and their total protein amount was quantified using MicroBCA assay as described above. As noted earlier, EVs were isolated from a single 24-well plate that was transfected with a total of either 0, 12, or 24 μg BDNF pDNA. hCMEC/D3 cells were seeded at a density of 5000 cells/cm2 in a collagen-coated 96-well plate in complete growth medium and cultured for 72 h in the incubator (37 °C, 5% CO2). The old medium was replaced with 100 μL of complete growth medium containing either 6 μg or 12 μg/well BDNF-EVs. Cells treated with a mixture of complete growth medium and PBS were used as a control. The hCMEC/D3 cells were subsequently incubated with the indicated treatments for either 24, 48, or 72 h. The treatment mixture was then removed, and the cells were gently washed with pre-warmed 1× PBS buffer. The intracellular ATP levels in each well were measured using a CellTiter-Glo luminescence assay (ATP assay). Briefly, 75 μL of fresh pre-warmed complete growth medium and an equal quantity of CellTiter-Glo 2.0 reagent was added to each well. The plate was incubated at room temperature for 15 min on an orbital shaker in the dark. A 60-μL volume of the mixture was transferred into a white opaque-walled 96-well plate. Immediately, the relative luminescence units were measured at 1-s integration time using a SYNERGY HTX multi-mode reader. The relative ATP levels (%) were calculated by normalizing the luminescence of treated cells to those treated with 1× PBS buffer as shown in the following equation (Eq. 3):
| (3) |
The effect of treatment parameters (factors) on intracellular relative ATP levels (response) was evaluated using a full factorial design. We studied the effect of three treatment factors: the amount of BDNF pDNA used for transfecting the donor hCMEC/D3 cells, type of EVs, and the total protein content in EVs (Table II). The factors were studied at either three (1–3, or) or two levels (1 or 3). For the 24-h incubation time, we evaluated the effect of a total of 18 experimental conditions (3×2×3 factorial, Table II), whereas for the 48- and 72-h incubation times, we studied the effect of 12 experimental conditions (3×2×2* factorial, Table II) on the relative ATP levels (%) in the recipient hCMEC/D3 cells. The effect of the combination of two factors, the amount of EVs, and the type of carriers, was studied as follows: when the cells were incubated with EXOs or MVs alone, level 1 in Table II represents 6 μg protein and level 3 represents 12 μg protein, whereas when cells were incubated with a mixture of EXOs + MVs, level 1 represents 3 μg EXOs and 3 μg MVs (total protein 6 μg/well) and level 3 represent 6 μg EXOs and 6 μg MVs (total protein 12 μg/well). The relative ATP data is represented as mean ± SD levels of 6 replicates. The results were analyzed using JMP Pro 14.
Table II.
Experimental Design to Study the Effect of Factors on the ATP Levels of BDNF pDNA-Loaded MVs and EXOs
| Factors | Level 1 | Level 2 | Level 3 |
|---|---|---|---|
| Amount of BDNF pDNA (μg)a | 0 | 0.5 | 1 |
| Total EV protein (μg)b | 6 μg EXOs/MVs or 3 μg EXOs + 3 μg MVs | N/A | 12 μg EXOs/MVs or 6 μg EXOs + 6 μg MVs |
| Type of EVs | EXOs* | EXOs + MVs | MVs* |
The amount of DNA transfected into cells represents the total amount of BDNF pDNA that was transfected in each well of a 24-well plate using Lipofectamine 3000; EVs were isolated from a single 24-well plate that was transfected with a total of either 0, 12, or 24 μg pDNA.
The total EV protein represents the amount of EXOs or MVs that the cells were incubated with; N/A: not applicable. All EV types were tested at the 24-h incubation time and
indicates the EV types that were tested at the 48- and 72-h incubation times
Statistical Analysis
The matrix of experimental designs and their statistical analysis for luciferase and BDNF-DNA transfection studies were performed using the JMP Pro 14 (SAS Institute Inc., NC). The statistical differences between control and experimental conditions or within the different experimental conditions were compared using multiple regression analysis methods in JMP Pro 14 and GraphPad Prism 8.4.2. The levels of statistical difference are indicated using the following notations: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
RESULTS
Isolation and Characterization of EVs
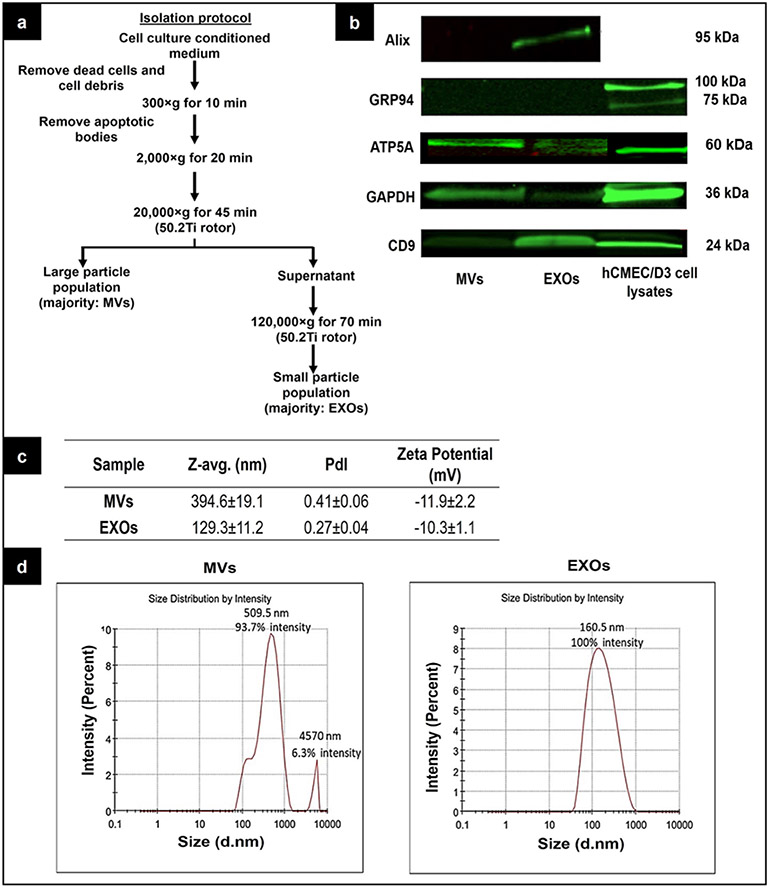
We utilized the widely used human cerebral microvascular endothelial cell line (hCMEC/D3) as a surrogate in vitro model of the blood-brain barrier (BBB) because it recapitulates many functions of the human BBB (43-47). EXOs and MVs were isolated from hCMEC/D3 conditioned medium using the differential ultracentrifugation method depicted in Fig. 2a. EXOs and MVs can be identified by their characteristic particle size distribution, surface charge, and marker proteins (1,2). We measured the particle sizes and zeta potentials using dynamic light scattering whereas the marker proteins were identified using western blotting. The Z-average particle diameter of EXOs was 129.3 ± 11.2 nm with a polydispersity index (PdI) of 0.27 ± 0.04, whereas MVs showed an average diameter of about 394.6 ± 19.1 nm and a broader PdI of 0.41 ± 0.06 (Fig. 2c). EXOs and MVs showed a negative zeta potential of ca. −10 to −12 mV (Fig. 2c). The particle size and zeta potential (Fig. 2c) values were consistent with published results (28,41,48). It is noteworthy that the particle size distribution of EXOs and MVs partially overlapped (Fig. 2d), indicating that particle size should not be used as the sole characteristic to distinguish EXOs and MVs. Western blotting was performed to further characterize these two EV populations (Fig. 2b). Alix (95 kDa) and tetraspanins CD9 (24 kDa) are commonly used as exosomal markers (49) because they are relatively enriched in EXOs but are inadequately expressed in MVs. The blot showed that the EXOs expressed Alix and CD9 but those proteins were hardly seen in the MV sample (Fig. 2b). ATP5A (53 kDa) is a subunit of mitochondrial ATP synthase (50) that produces ATP from ADP in the presence of a proton gradient across the membrane and was utilized as a mitochondrial marker for MVs (27,28). The blot confirmed the expression of ATP5A was present in MVs and EXOs (Fig. 2b). The enrichment of ATP5A in MVs and EXOs suggested that EVs may have the potential to increase cellular ATP levels in recipient cells. Dozio et al. (28) reported GRP94 as a MV marker at a molecular mass of 92 kDa. However, we did not observe this protein expressed in our MVs. A possible reason for this discrepancy could be due to the differences in the detection sensitivity of the used antibodies. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) protein is typically used as a loading control to verify equal protein loading among the different samples, but it should be pointed out here that we have used GAPDH as an additional protein in this study. The expression of GAPDH was lower in the EXOs than the MVs, which contradicted an earlier report that showed higher GAPDH expression in the EXOs than MVs (23,41). It should be pointed out that our EVs are derived from the hCMEC/D3 cell line whereas EVs were isolated from RAW 264.7 and HEK293FT cell lines in the reported studies (23,41). The differences in the biogenesis pathways of EXOs and MVs likely explains the differences in GAPDH expression among these two samples.
Fig. 2.
Characterization of EXO- and MV-derived from hCMEC/D3 cells. a A flowchart of the differential ultracentrifugation method that was used to isolate EXOs and MVs from the conditioned medium. b Detection of marker proteins in EXOs and MVs using western blotting. Each lane was loaded with 25 μg total protein (for ATP5A detection, the EV protein loading amount was 50 μg/lane) of the indicated sample, electrophoresed, and transferred to a nitrocellulose membrane prior to staining with antibodies to detect Alix, GRP94, ATP5A, CD9, and GAPDH proteins. The bands were imaged on the 800-nm channel using an Odyssey imager at intensity setting 5 and processed using ImageStudio 5.2 software. c Z-average particle diameter, polydispersity indices, the zeta potential of MVs and EXOs measured using dynamic light scattering (DLS). The samples at 0.7 mg/mL protein concentration were suspended in 10 mM HEPES buffer, pH 7.4 prior to DLS analysis. d The scattered light intensity-based particle size distribution of EXOs and MVs was measured by DLS on a Malvern Nano Zetasizer. Data are presented as mean ± SE of two independent experiments. The raw blot for ATP5A detection in hCMEC/D3 cell line–derived EVs was shown in SL. Fig. 5
The Integrity of EVs After Ultracentrifugation and Resuspension Processes
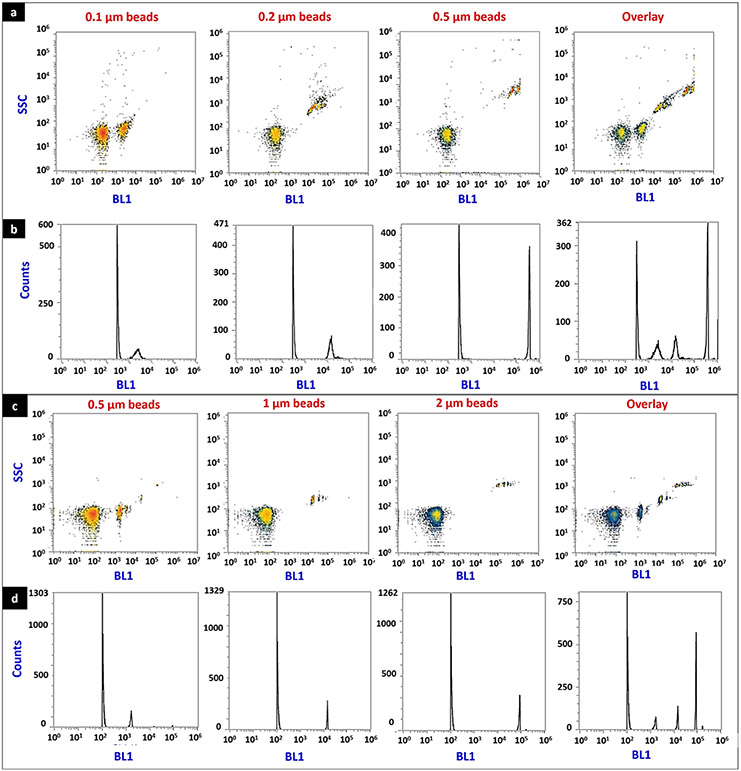
The integrity of the isolated EVs labeled using calcein AM was determined using a flow cytometry assay since this method allows quantifying the percentage (%) of intact vs. disrupted vesicles. Sub-micron sized populations were captured in SSC/BL1 dot plots using calibration beads of particle diameters ranging from 0.1 to 2 μm (Fig. 3). The relative position of the clusters on SSC/BLA plots (Fig. 3a and c) was directly proportional to the diameter of the bead. As the bead diameter increased, the clusters moved towards the right side of the plot. The overlay plots demonstrated the linearity between bead size and signal intensity for both calibration ranges i.e. (1) 0.1 to 0.5 μm (Fig. 3a and b), and (2) 0.5 to 2 μm (Fig. 3c and d). It should be noted that the average particle diameter of hCMEC/D3 cell line–derived EXOs and MVs were about 100–400 nm measured using dynamic light scattering (Fig. 2c), and hence, this flow protocol allowed us to analyze EXOs and MVs of our interest.
Fig. 3.
Flow cytometry analysis of calibration beads. Calibration beads with particle diameters ranging from 0.1 to 2 μm were diluted in PBS, and events were acquired in the side scatter (SSC), forward scatter (FSC), and a small particle side scatters 488/10-nm filter (BL1) channels in an Attune flow cytometer. Representative density plots of beads ranging from 0.1 to 0.5 μm (a), and 0.5–2 μm (c), histograms of beads ranging from 0.1 to 0.5 μm (b), and 0.5–2 μm (d). In density plots, X-axis represents the number of events captured in the side scatter 488/10-nm filter (BL1), whereas the Y-axis represents the number of events acquired in SSC. In histograms, X-axis represents the events captured by BL1, whereas the Y-axis represents the total count of the events. The axes scale in these plots were automatically adjusted by the Attune NxT software
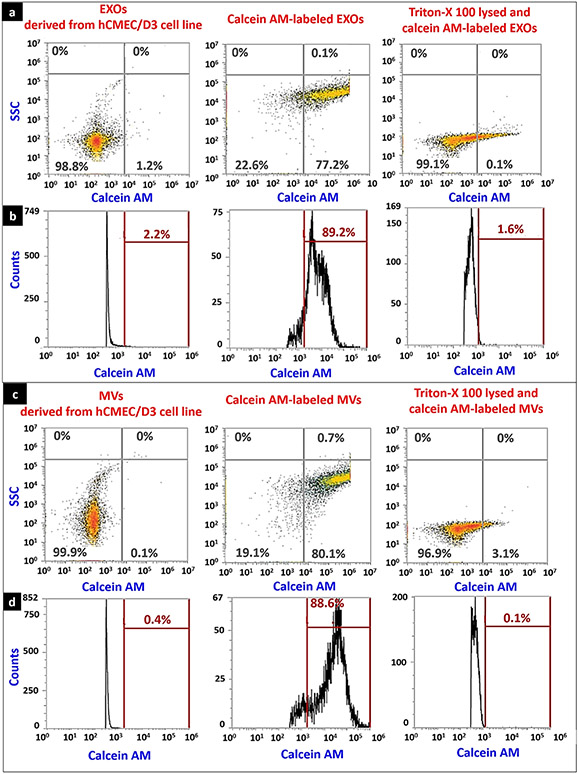
Prior to analyzing the calcein AM–labeled EVs using flow cytometry, the events associated with PBS/calcein AM, and PBS/Triton-X 100/calcein AM mixture (sample processing controls) was acquired on density plots (SL. Fig. 2a) and histograms (SL. Fig. 2b). The gates were created on density plots and histograms allowed us to eliminate the non-specific/background signals from PBS, Triton-X 100, and calcein AM and measure the signal intensities of calcein-labeled EVs. As can be seen from Fig. 4a-d, >85% of hCMEC/D3 cell line–derived EXOs and MVs were calcein-positive suggesting that at least 85% of hCMEC/D3 cell line–derived EXOs and MVs remained intact after the isolation and resuspension processes. EVs lysed with 2% Triton-X 100 showed <2% of calcein-positive EVs indicating that calcein signals are specific to intact EVs, and lysed EVs or cell debris did not contribute to calcein signal intensities. Furthermore, in a separate experiment, we confirmed the integrity of EXOs and MVs derived from RAW 264.7 cell line using the same method. The histograms showed that >90% of EXOs (SL. Fig. 3b) and MVs (SL. Fig. 3d) were calcein-positive suggesting that at least 90% of RAW 264.7 cell line–derived EVs remained intact after ultracentrifugation and resuspension processes. As earlier, the lysed EVs showed < 1% calcein signals confirming the specificity of calcein signal intensity to intact EVs.
Fig. 4.
Integrity of hCMEC/D3 cell line–derived EVs after ultracentrifugation and resuspension determined using flow cytometry. EVs were isolated from hCMEC/D3 cell line using sequential ultracentrifugation and the pellets were resuspended in PBS. EXOs and MVs at 20 μg/mL were labeled with 10 μM calcein AM. In a separate experiment, EVs were lysed with Triton-X 100 (2%) solution and were labeled with calcein AM. An aliquot of each sample was analyzed for calcein-positive EVs using an Attune NxT flow cytometer. The representative density plots (a), and histograms (b) of EXOs, calcein AM–labeled EXOs, and Triton-X 100-lysed calcein-labeled EXOs. The representative density plots (c), and histograms (d) of MVs, calcein AM–labeled MVs, and Triton-X 100-lysed calcein-labeled MVs. In density plots, X-axis represents the number of calcein-positive events captured in the side scatter 488/10-nm filter (BL1), whereas the Y-axis represents the number of events acquired in the side scatter (SSC). In histograms, X-axis represents the number of calcein-positive events captured in the side scatter 488/10-nm filter (BL1), whereas the Y-axis represents the total count of the events. The axes scale in these plots were automatically adjusted by the Attune NxT software. The data shown are representative plots of n = 3 samples
Luc pDNA Loading into EVs
We transfected hCMEC/D3 or RAW 264.7 cells using lipofectamine/Luc-pDNA complexes at 0, 0.5, and 1.0 μg pDNA/well into three 24-well plates (resulting in total DNA amounts of 0, 12, and 24 μg per plate) to evaluate the effect of the amount of Luc-pDNA transfected into hCMEC/D3 or RAW 264.7 cells on the extent of DNA loading in EVs. DNA-EVs were isolated using the differential centrifugation method and the extent of DNA loading in EXOs and MVs were quantified using the Quant-iT PicoGreen assay. The results in Table III showed that EXOs contained an average of 381.6, 480.8, and 390.8 ng pDNA when isolated from cells transfected with 0, 0.5, and 1 μg/well DNA dose, whereas the average DNA amounts in MVs isolated from the same groups were 33.6, 60.0, and 81.5 ng of pDNA, respectively. Interestingly, we observed that the amount of DNA in EXOs were significantly (p < 0.0001) higher compared with MVs for each pDNA dose group (Table III). The amount of DNA present in naïve EVs (pDNA dose 0 μg/well) was subtracted from other groups and normalized with the total amount of transfected pDNA to calculate the %DNA loading in EVs (Eq. 1). We observed that EXOs isolated from the 12 μg pDNA plate showed a 20-fold increase in %DNA loading compared with EXOs isolated from the 24 μg plate whereas there were no considerable differences in the MVs %DNA loading (Table III). We calculated the yield of DNA-EVs by measuring their total protein content using the MicroBCA assay. Total EV protein content ranged from ca. 130 to 220 μg, and there were no significant differences (p > 0.05) in the protein yield between naïve EXOs and MVs, or among the EVs isolated from cells transfected with different amounts of pDNA (Table III).
Table III.
Reporter Luciferase (Luc) DNA Loading Efficiency and Total Protein Content in Luc-EXOs and Luc-MVs Derived from hCMEC/D3 Cells
| Type of carrier | Amount of DNA transfected into cells (μg)a |
Amount of DNA in isolated EVs (ng) |
DNA loading (%)b | Total EV protein content (μg)c |
|---|---|---|---|---|
| EXOs | 0 | 381.6 ± 42.0 | N/A | 181.7 ± 70.5 |
| 12 | 480.8 ± 69.9 | 0.83 ± 0.23 | 172.0 ± 62.4 | |
| 24 | 390.8 ± 46.9 | 0.04 ± 0.02 | 187.0 ± 61.8 | |
| MVs | 0 | 33.6 ± 1.8 | N/A | 155.7 ± 63.0 |
| 12 | 60.0 ± 7.4 | 0.22 ± 0.08 | 152.3 ± 48.0 | |
| 24 | 81.5 ± 5.5 | 0.20 ± 0.02 | 192.4 ± 69.1 |
The amount of DNA transfected into cells indicates the total amount of Luc pDNA that was transfected into the donor cells using Lipofectamine 3000; EVs were isolated from a single 24-well plate that was transfected with a total of either 0, 12, or 24 μg pDNA.
The %DNA loading values were calculated after subtracting the fluorescence values obtained from the 0 μg/well DNA-transfected cells (naïve EVs) from the pDNA-loaded EVs (pDNA-EVs). The obtained values were divided by the amount of DNA transfected into cells and multiplied by 100 (Eq. 1).
The total protein amount in pDNA-EXOs or MVs was measured using a MicroBCA assay. Data are presented as mean ± SE of four independent experiments (n = 3/experiment)
To evaluate whether the source of EVs can alter the DNA loading in EXOs and MVs, we tested the DNA loading in Luc-EVs derived from RAW 264.7 cells (Table IV and Fig. 5). The results in Table IV demonstrated that RAW 264.7-derived EXOs showed averages of 5.9% and 1.9% pDNA loading when isolated from cells pre-transfected with 0.5 and 1 μg/well DNA dose, respectively, whereas the average DNA loading in MVs were 0.6 and 0.3% from equivalent treatments. Interestingly, the higher DNA loading in RAW 264.7 cell line–derived EXOs compared with MVs (Table IV) was consistent with the higher DNA loading observed in EXOs versus MVs derived from hCMEC/D3 cell line (Table III). Moreover, RAW 264.7-derived EXOs isolated from cells pre-transfected with 0.5 μg/well DNA showed significantly (p < 0.0001) greater DNA loading compared with hCMEC/D3- and RAW 264.7-derived MVs as well as hCMEC/D3-derived EXOs (Fig. 5).
Table IV.
Reporter Luciferase (Luc) DNA Loading Efficiency and Total Protein Content in Luc-EXOs and Luc-MVs derived from RAW 264.7 cells
| Type of carrier | Amount of DNA transfected into cells (μg)a |
Amount of DNA in isolated EVs (ng) |
DNA loading (%)b | Total EV protein content (μg)c |
|---|---|---|---|---|
| EXOs | 0 | 516.3 ± 5.2 | N/A | 2361.2 ± 139.7 |
| 12 | 586.8 ± 5.7 | 5.88 ± 0.48 | 2735.4 ± 71.3 | |
| 24 | 538.4 ± 4.1 | 1.85 ± 0.34 | 2090.7 ± 26.4 | |
| MVs | 0 | 22.2 ± 0.2 | N/A | 441.6 ± 56.4 |
| 12 | 29.6 ± 0.2 | 0.62 ± 0.01 | 460.0 ± 43.4 | |
| 24 | 28.3 ± 0.3 | 0.26 ± 0.01 | 375.2 ± 47.5 |
Fig. 5.
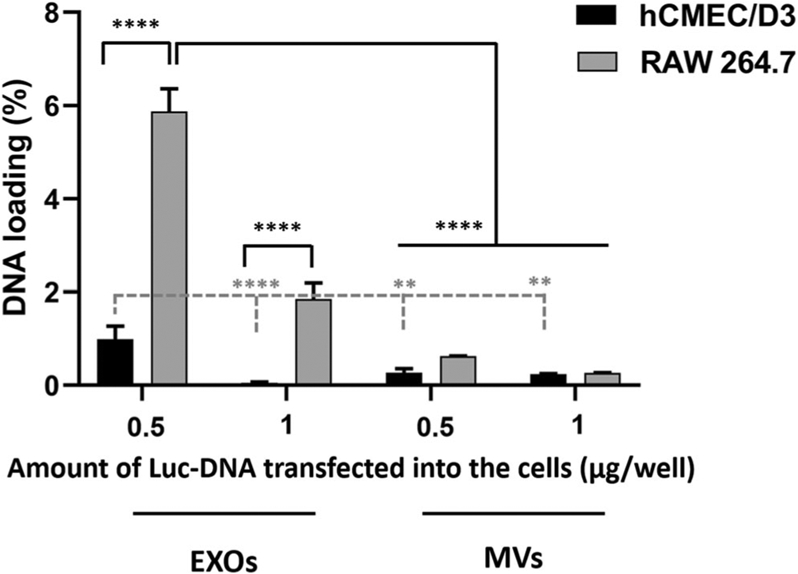
Luc-DNA loading in hCMEC/D3 and RAW 264.7 cell line–derived EXOs and MVs. Luc-DNA at 0, 0.5, and 1 μg/well were transfected into hCMEC/D3 and RAW 264.7 cell lines using Lipofectamine in 24-well plates. EVs from corresponding plates were isolated using sequential ultracentrifugation and resuspended in 1x PBS in volumes ranging from 300 to 1000 μL. Isolated EVs were lysed using 1X RIPA buffer and the total DNA content was measured using Quant-iT PicoGreen assay. DNA loading was calculated using Eq. 1 described in the method section. The differences in DNA loading between the groups were compared using one-way ANOVA and the significance levels are indicated as **p < 0.01, and ****p < 0.0001
Cytocompatibility of hCMEC/D3 Endothelial Cells with Luc-EVs Derived from hCMEC/D3 and RAW 264.7 Cell Lines
Prior to evaluating EV-mediated luciferase expression in the recipient cells, we performed cell viability studies to determine the cytocompatibility of engineered EVs and lipofectamine/pDNA complexes with hCMEC/D3 cells. The cell viabilities of hCMEC/D3-derived Luc-EVs was demonstrated in Fig. 6. The cell viability of hCMEC/D3 cells treated with 0, 0.5, and 1 μg/well Luc-DNA-loaded EXOs at 6 and 12 μg total EV protein per well was ≥ 100% at 24, 48, and 72 h (Fig. 6a). Similarly, hCMEC/D3 cells treated with Luc-MVs at 6 and 12 μg total EV protein/well showed cell viabilities ≥ 90% compared with untreated cells at 24, 48, and 72 h (Fig. 6b). Positive control, PEI at 50 μg/mL, showed a significant toxicity (p < 0.0001) confirming the sensitivity of the ATP assay (Fig. 6a and b). Lipofectamine/pDNA complexes at 10 ng pDNA/well were well tolerated (cell viability ≥ 100%) by hCMEC/D3 cells for 48 h (Fig. 6c). To summarize, Luc-EVs and lipofectamine/pDNA complexes were cytocompatible with hCMEC/D3 cells for 72 h and did not show any significant toxicity during the exposure time.
Fig. 6.
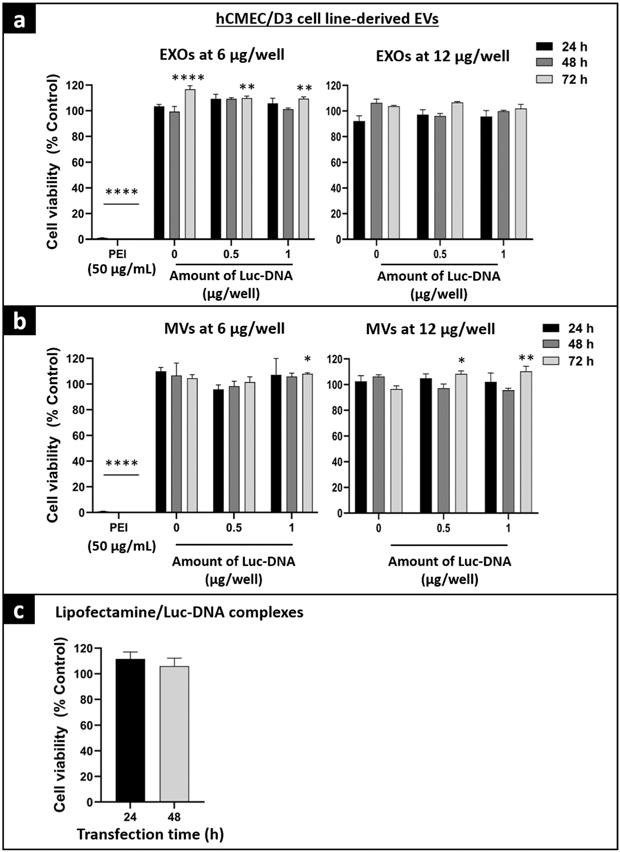
Cytocompatibility of hCMEC/D3 cell line–derived Luc-EVs with hCMEC/D3 cells using ATP assay. hCMEC/D3 cells were seeded in collagen-coated 96-well plates at 16,500 cells/well. At about 80% confluency, hCMEC/D3 cells were incubated with 6 or 12 μg of total EXO protein per well isolated from cells pre-transfected with 0, 0.5, and 1 μg Luc-DNA/well (a), 6 or 12 μg of total MV protein per well isolated from 0, 0.5, and 1 μg Luc-DNA/well plates (b), and lipofectamine/Luc-DNA complexes at 10 ng Luc-DNA per well (c). Cells were incubated with EVs for 24, 48, or 72 h, and with Lipofectamine/DNA complexes for the 24- or 48-h exposure time. PEI at 50 μg/mL was used as a positive control and untreated cells were also used as a control. Post-incubation, the CellTiter-Glo 2.0 reagent was added to an equal volume of cell culture media. The plate was measured for luminescence at 1 s integration time using a GloMAX luminometer. The % cell viability of the treated cells was calculated by the (relative luminescence unit (RLU) of treated cells/RLU of untreated cells) × 100. The significance of treated groups was compared against control using Student’s unpaired t test compared with control or one-way ANOVA and the significance levels are indicated as *p < 0.05, **p < 0.01 and ****p < 0.0001
We also performed cell viability studies to determine the cytocompatibility of hCMEC/D3 cells with RAW 264.7 cell line–derived Luc-EXOs. RAW 264.7-derived Luc-EVs were well tolerated by hCMEC/D3 cells for 72 h when incubated with 6 and 12 μg total EV protein per well (SL. Fig. 4). Luc-EXOs isolated from cells pre-transfected with 1 μg Luc-DNA/well showed significant toxicity (p < 0.05, SL. Fig. 4a) at 48 h, however, were well tolerated at 72 h. Similarly, RAW 264.7-derived Luc-MVs showed cell viabilities ≥ 90% during all transfection time points excluding the ca. <20% toxicity (p < 0.001, SL. Fig. 4b) observed at 48 h in the case of Luc-MVs isolated from cells pre-transfected with 0.5 and 1 μg Luc-DNA/well.
Transfection Activity of Luc pDNA-Loaded MVs and EXOs
The capability of EVs to deliver pDNA into the recipient hCMEC/D3 cells was evaluated by transfecting Luc-DNA-loaded EVs (Luc-EVs) and measuring the resulting luciferase expression using a standard luciferase assay. The emitted luminescence was measured as relative light units (RLU) and normalized to the total cellular protein (RLU/mg protein). The RLU/mg protein values obtained from the cells treated with either Luc-EVs or lipofectamine/pDNA complexes were normalized to the cells treated with PBS buffer (Fig. 7a-c).
Fig. 7.
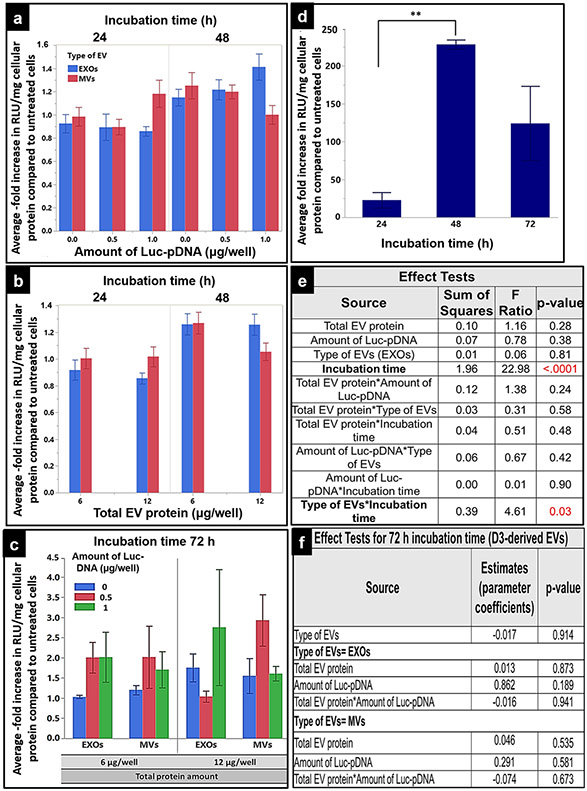
Effect of the transfection parameters of hCMEC/D3 cell line–derived Luc pDNA-EVs on luciferase gene expression. hCMEC/D3 cells were seeded at a density of 50,000 cell/cm2 in 24-well plates, cultured for 48 h, and incubated with Luc-EXOs or MVs derived from hCMEC/D3 cells pre-transfected with 0, 0.5, or 1 μg/well Luc-DNA. Cells were incubated with Luc-EXOs or Luc-MVs containing either 6 μg or 12 μg EV protein/well for 24, 48, or 72 h. Luciferase gene expression (Relative light units (RLU)) in the cell lysates were measured at 1 s integration time using a Synergy HTX multi-mode reader. RLU of the luciferase reagent (blank) was subtracted from the sample values and normalized to the total cellular protein and the data is presented as normalized RLU/mg protein. RLU/mg cellular protein of treated groups was further normalized to that of the control, untreated cells and is represented as an average -fold increase in the Y-axis. a-c The average -fold increase values in groups treated with DNA-EVs isolated from donor cells transfected with different amounts of Luc-pDNA (0, 0.5, or 1 μg/well) at different doses (6 or 12 μg EV protein/well) when incubated for 24, 48, 72 h. d The average-fold increase in cells treated with the positive control (lipofectamine/Luc-pDNA complexes) incubated for 24, 48, and 72 h. e, f Analysis of variance table from the effect test using multiple regression analysis depicting the sum of squares, F ratios, and p values of each transfection factor and their two-way interactions. Data are presented as mean ± SE of three independent experiments (n = 3/experiment). Statistical comparisons were made using multiple regression analyses, where p < 0.05, **p < 0.01, ****p < 0.0001
The effect of the following four transfection factors: the amount of Luc-pDNA, type of EVs, total EV protein content, and incubation time (Table I) on the luciferase gene expression was evaluated using multiple regression analysis in JMP Pro 14. The significance of each factor and their two-way interactions were determined using F-test in the regression analysis method at an alpha level of 0.05 (Fig. 7e). The ANOVA table (Fig. 7e) shows that the incubation time of EVs is statistically significant (p < 0.0001) compared with the other transfection factors. The data in Fig. 7a-d is presented as an average-fold increase in transfection (RLU/mg total protein) of lipofectamine-treated samples normalized to untreated cells. As shown in Fig. 7a, b, the average-fold increase in transfection at 48 h is ca. 40% higher compared with the values at 24 h. However, there were no significant differences observed among DNA-EVs isolated from different amounts of Luc-pDNA that was used to transfect the donor cells (Fig. 7a). Also, changing the amount of either EXOs or MVs from 6 to 12 μg EV protein/well did not affect the levels of luciferase gene expression (Fig. 7b). However, the interaction between the type of EVs and incubation time is statistically significant (p < 0.05, Fig. 7e). A significant interaction between the type of EVs and incubation time indicates that the effect of the incubation time on the observed luciferase expression is different for EXOs and MVs. As seen in Fig. 7a, MVs isolated from the 24 μg DNA plate showed the maximum luciferase expression at 24 h that then decreased at 48 h, in contrast, EXOs showed maximum transfection efficiency at 48 h compared with the 24-h incubation time. Overall, EXOs isolated from the 24 μg DNA plate incubated for 48 h resulted in the maximum luciferase expression irrespective of the treated EV dose, whereas, cells transfected with naïve MVs (0 μg/well Luc-pDNA amount) at 6 μg/well total EV amount for 48 h exhibited the highest transfection efficiency.
Cells transfected with lipofectamine/Luc-pDNA complexes (positive control) at 10 ng/well DNA dose for 24 h showed a 20-fold increase in transfection compared with untreated cells (Fig. 7d). It is important to note that the 10-ng dose in Lipofectamine complexes is equivalent to the amount of DNA in Luc-EVs. At 48 h, we noted about a 220-fold increase in luciferase expression compared with control, and a statistically significant increase (p < 0.01) in luciferase expression compared with 24 h (Fig. 7d). At 72 h, there was about a 125-fold increase in luciferase expression compared with the control, while the overall luciferase expression levels were lower compared with the 48 h time point (Fig. 7d). However, the decrease in the luciferase expression was statistically nonsignificant and was still higher compared with the 24 h time point. Importantly, no visual toxicities were noted under microscopic evaluation at 72 h post-incubation. Therefore, the observed changes in transfection are a direct result of luciferase transgene expression and are not caused by transfection-related toxicity.
As shown in Fig. 7e, there was a statistically significant effect of incubation time on the average fold increase in luciferase expression, therefore, we added the 72-h exposure time as an additional transfection time point. EXOs and MVs isolated from cells pre-transfected with 0.5 and 1 μg Luc-DNA/well showed ca. a two-fold increase in average luciferase expression compared with their naïve counterparts at 72 h (Fig. 7c and f). The increase in luciferase expression at 72 h was not statistically significant, however, was considerably higher compared with the 24-h and 48-h incubation times (Fig. 7a-c).
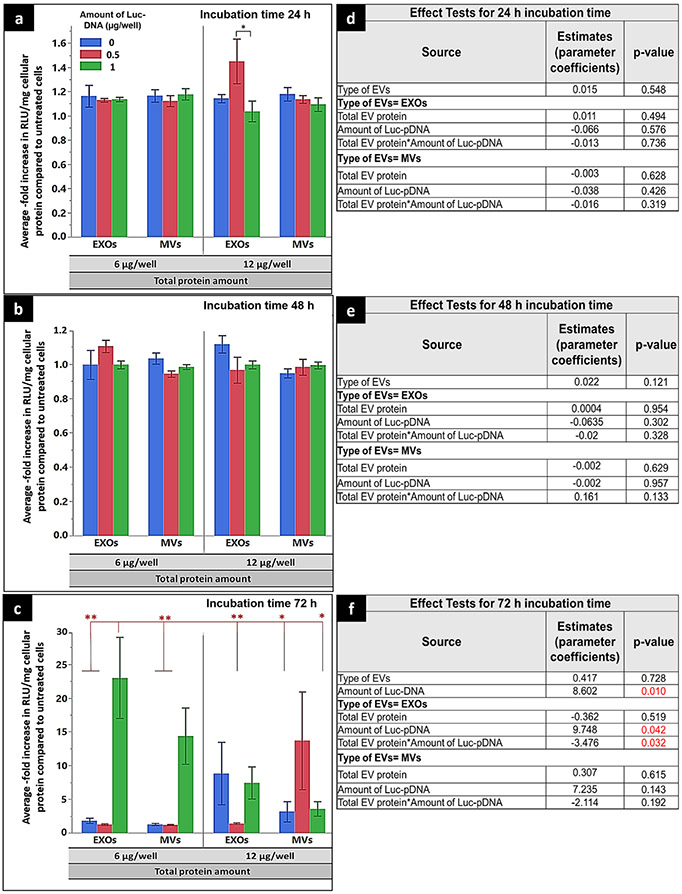
We further evaluated luciferase expression at 24, 48, and 72 h in hCMEC/D3 cells transfected with RAW 264.7-derived Luc-EVs at 6 and 12 μg total EV protein/well (Fig. 8). Post-transfection, there were no statistically significant (p > 0.05) increases in hCMEC/D3 cell luciferase expression at the 24- and 48-h incubation times (Fig. 8a and b). This lack of luciferase expression by RAW 264.7-derived Luc-EVs at 24 and 48 h was consistent with our observations from the experiments using hCMEC/D3-derived Luc-EVs (Fig. 7a and b). However, interestingly, Luc-EXOs isolated from 1 μg Luc-DNA group showed ca. 23-fold increase in luciferase expression compared with control, untreated cells, and the increased expression levels were statistically significant (p < 0.01) compared with naïve EXOs and MVs, as well as the EXOs and MVs isolated from cells pre-transfected with 0.5 μg Luc-DNA/well (Fig. 8c). Notably, Luc-MVs isolated from RAW 264.7 cells pre-transfected with 1 μg Luc-DNA/well showed a ca. 15-fold increase in luciferase expression compared with control, untreated cells. The luciferase expression in 12 μg total EV protein-treated hCMEC/D3 cells was lower than that of the 6 μg/well protein amount. The effect test table showed that the amount of Luc-DNA has a significant impact on luciferase expression at the 72-h incubation time (Fig. 8f). The higher luciferase transgene expression mediated by RAW 264.7 cell line–derived EVs can be correlated with higher DNA loading in those EVs compared with hCMEC/D3 cell-derived EVs (Table IV and Fig. 5).
Fig. 8.
Effect of the transfection parameters of RAW 264.7 cell line–derived Luc pDNA-EVs on luciferase gene expression. hCMEC/D3 cells were seeded at a density of 50,000 cell/cm2 in 24 well plates, cultured for 48 h, and incubated with Luc-EXOs or MVs derived from RAW 264.7 cells pre-transfected with 0, 0.5, or 1 μg/well Luc-DNA. Cells were incubated with Luc-EXOs or Luc-MVs containing either 6 μg or 12 μg EV protein/well for 24, 48, or 72 h. Luciferase gene expression (Relative light units (RLU)) in the cell lysates were measured at 1 s integration time using a Synergy HTX multi-mode reader. RLU of the luciferase reagent (blank) was subtracted from the sample values and normalized to the total cellular protein and the data is presented as normalized RLU/mg protein. RLU/mg cellular protein of treated groups was further normalized to that of the control, untreated cells. The average -fold increase in RLU/mg of cellular protein normalized to untreated cells values in groups treated with DNA-EVs isolated from donor cells transfected with different amounts of Luc-pDNA (0, 0.5, or 1 μg/well) at different doses (6 or 12 μg EV protein/well) when incubated for 24 (a), 48 (b), and 72 h (c). Analysis of variance table from the effect test using multiple regression analysis depicting the sum of squares, F ratios, and p values of each transfection factor and their two-way interactions for 24 (d), 48 (e), and 72 h (f). Data are presented as mean ± SE of three independent experiments (n = 3/experiment). Statistical comparisons were made using multiple regression analyses, where *p < 0.05, **p < 0.01, ****p < 0.0001
The Effect of EVs on Cellular ATP Levels in hCMEC/D3 Cells
It has been reported that extracellular vesicles containing mitochondria or mitochondrial DNA can participate in cell-signaling processes in the recipient cells (3). Mitochondria is an essential organelle that provides energy to the cells by synthesizing ATP through oxidative phosphorylation and electron transport chain pathways (51). Therefore, we hypothesized that treating hCMEC/D3 cells with EVs can increase their intracellular ATP levels. We measured the resulting intracellular ATP levels after treating the hCMEC/D3 cells with naïve EVs or BDNF pDNA (pBDNF)-loaded EVs using the luminescence-based CellTiter-Glo assay (ATP assay). The stoichiometric reaction between intracellular ATP and CellTiter-Glo reagent emits luminescence that is directly proportional to the cellular ATP levels and hence is proportional to the number of metabolically viable cells. The relative ATP levels in EV-treated cells was normalized to the ATP levels of 1× PBS-treated cells (Eq. 3).
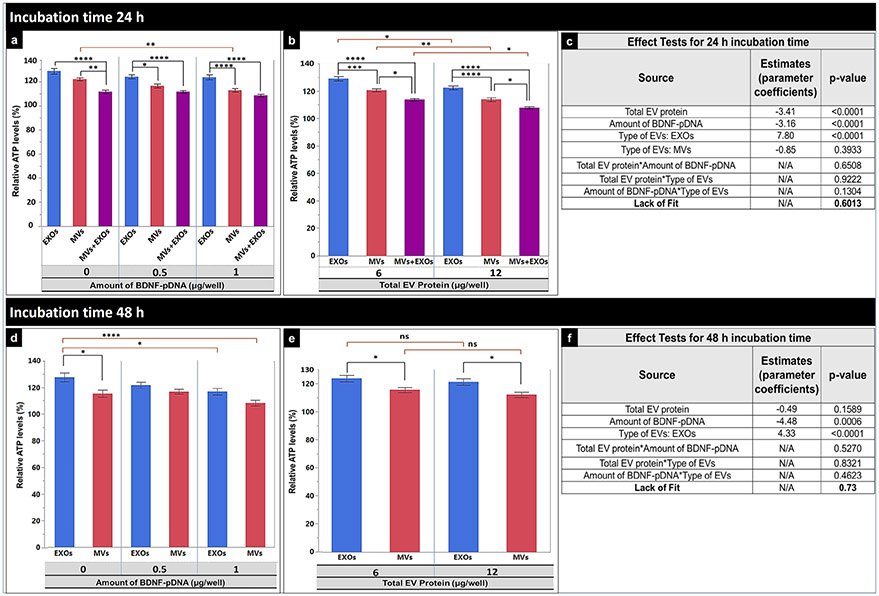
We evaluated the effect of the following three transfection factors: the amount of pBDNF transfected into the cells using Lipofectamine (0, 0.5, or 1 μg/well pBDNF corresponding to total amounts of 0, 12, or 24 μg per plate), type of EVs (EXOs, MVs, or EXOs + MVs), and the amount of EVs (6 or 12 μg total protein) on the resulting ATP levels using a full factorial design on JMP Pro 14 (Fig. 9). We evaluated the effect of these experimental conditions on the resulting ATP levels at either 24-h or 48-h incubation time. The significance of each factor and their two-way interaction at either 24 h or 48 h were determined by performing multiple regression analysis and F-test (ANOVA table) at α = 0.05. For the 24-h incubation time, it can be seen from Fig. 9c that the pDNA amount, type of EVs, and total EV protein showed a statistically significant (p < 0.0001) increase in intracellular ATP levels, whereas their two-way interaction was statistically insignificant (p > 0.05). EXOs showed significantly higher ATP levels compared with MVs and EXOs + MVs at all pBDNF amounts (Fig. 9a) and total EV protein amounts (Fig. 9b). Naïve EXOs or MVs (0 μg/well pBDNF) showed significantly higher ATP levels compared with EXOs isolated from the 12 or 24 μg pBDNF plates (Fig. 9a). The lower amount of total EV protein (6 μg/well) showed significantly (p < 0.01) higher ATP levels compared with 12 μg/well upon 24 h of incubation. The estimates (parameter coefficients) in Fig. 9c list the magnitude of the factor influencing the observed percent ATP levels wherein the positive coefficient values indicate the percent increase in the ATP levels when changing any factor level from 1 to 2 and vice versa for the negative coefficients. Notably, the negative estimates of total EV protein (− 3.41) and amount of pBDNF (− 3.16) suggested that increasing the amount of either EV protein or pBDNF would decrease the overall ATP levels. In contrast, the estimates for the type of EVs indicated that treatment with EXOs would increase the ATP levels by ca. 8% when compared with treating the cells using EXOs + MVs (Fig. 9c). The MV-mediated increase in ATP levels was insignificant (p > 0.05, Fig. 9c) compared with EXOs + MVs.
Fig. 9.
Effect of hCMEC/D3 cell line–derived naïve EVs or BDNF-EVs on cellular ATP levels in the recipient hCMEC/D3 monolayers. hCMEC/D3 cells seeded at a density of 5000 cell/cm2 in 96 -well plates were cultured for 48 h and incubated with BDNF-EXOs, MVs, or EXOs + MVs derived from hCMEC/D3 cells pre-transfected with 0 (naïve), 0.5, or 1 μg BDNF-DNA/well. EXOs, MVs, or EXOs + MVs were incubated for 24 h (a-c), or 48 h (d-f) at either 6 μg or 12 μg EV protein/well. Post-incubation, the relative ATP levels were measured using a CellTiter-Glo 2.0-based ATP assay. Relative light units (RLU) of the cell lysates were measured at 1 s integration time using a Synergy HTX multi-mode reader. The average RLU values from the recipient cells treated with DNA-EVs isolated from donor cells transfected with different amounts of BDNF pDNA (0, 0.5, or 1 μg/well) at different doses (6 or 12 μg EV protein/well) when incubated for 24 (a, b) or 48 h (d, e). The effect test and lack of fit test using multiple regression analysis presenting parameter estimates (coefficient), and p values of each transfection factor and their two-way interactions for 24-h (c) or 48-h incubation time (d). Data are presented as mean ± SE of three independent experiments (n = 3/experiment). Statistical comparisons were made using multiple regression analyses, where *p < 0.05, **p < 0.01, ***p <0.001, and ****p < 0.0001. ns: nonsignificant. N/A: not applicable
When incubated for 48 h (Fig. 9d-f), the type of EVs and amount of pBDNF showed a statistically significant (p < 0.05) effect on increasing ATP levels whereas the effect of total EV protein and two-way interactions between three factors were statistically insignificant (p > 0.05, Fig. 9f). Consistent with data from the 24-h incubation time, cells treated with EXOs for 48 h demonstrated significantly higher ATP levels compared with MVs at all pBDNF amounts (Fig. 9d) and total EV protein amounts (Fig. 9e). EXOs + MVs showed significantly lower ATP levels at the 24-h incubation time, and therefore, we did not evaluate the effect of EXOs + MVs on the ATP levels at the 48-h incubation time. Treatment with naïve EXOs showed significantly (p < 0.05) higher ATP levels compared with naïve MVs as well as EXOs or MVs treated isolated from cells transfected with 24 μg pBDNF (Fig. 9d). However, there was no effect of the amount of total EV protein (6 vs. 12 μg/well) on the ATP levels at the 48-h incubation time (Fig. 9e). The negative estimates of the amount of BDNF pDNA (− 4.48) suggested that increasing the amount of pBDNF from none to 1.0 μg/well pBDNF would decrease the ATP levels by ca. 4.5%, whereas treatment with EXOs would increase by ca. 4.3% of the ATP levels compared with MVs (Fig. 9f) at the 48-h incubation time. Lack of Fit test was insignificant (p < 0.05, Fig. 9c and f) for both the 24-h and 48-h incubation periods suggesting that the data fits the used regression model.
Overall, treatment with EXOs showed higher ATP levels at the 24- and 48-h incubation times suggesting that EXOs increased cell metabolic activity to a greater extent than MVs and MVs + EXOs. Naïve EXOs increased ATP levels to a higher extent when compared with EVs isolated from cells transfected with 12 or 24 μg pBDNF.
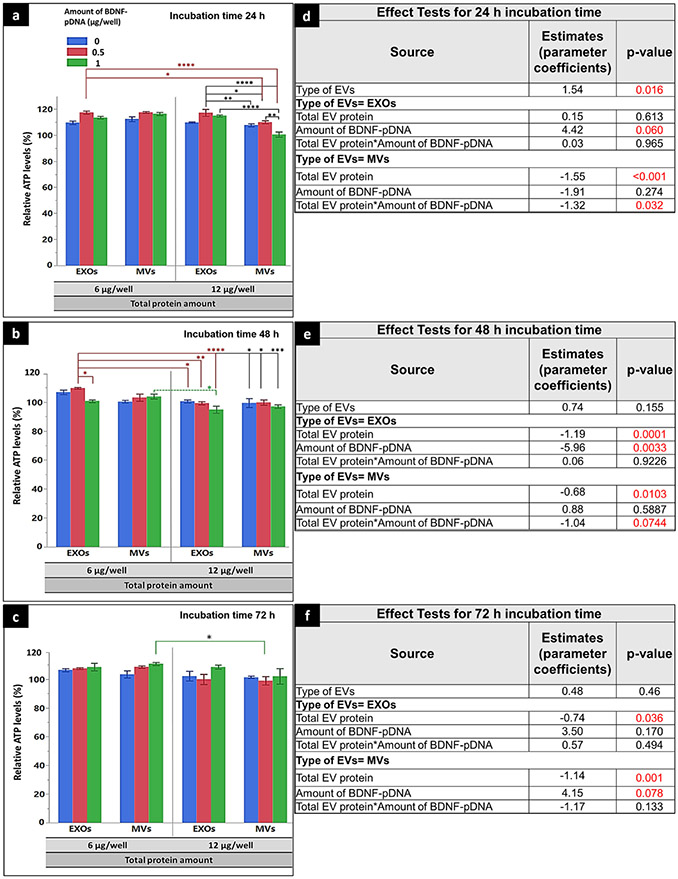
We additionally studied the effect of parent cell line source (endothelial vs. macrophage origin) of the isolated EVs on the resulting ATP levels in the recipient hCMEC/D3 cells using the same experimental setup. We tested if BDNF-EVs derived from RAW 264.7 cell line can increase relative ATP levels in the recipient hCMEC/D3 cells. BDNF-EVs were isolated from RAW 264.7 cells pre-transfected with 0, 0.5, or 1 μg/well pDNA (amounting to a total of 0, 12, or 24 μg DNA per 24-well plate), and their total protein amount was quantified using a MicroBCA assay as described above. RAW 264.7-derived BDNF-EVs were transfected into hCMEC/D3 cell line for 24, 48, or 72 h. At the 24-h exposure time, RAW 264.7-derived BDNF-EXOs isolated from cells pre-transfected using 0.5 μg pBDNF/well showed a statistically significant (p < 0.0001) increase in relative cellular ATP levels compared with naïve MVs and BDNF-MVs when treated at 12 μg total EV protein per well (Fig. 10a and d). At the 48-h exposure time, BDNF-EXOs with 0.5 μg/well pBDNF showed a statistically significant increase in ATP levels compared with 1 μg pDNA/well (p < 0.05), naïve and BDNF-EVs at 12 μg total EV protein/well (at least p < 0.05, Fig. 10b and e). There were no EV-mediated increases in ATP levels at 72 h excluding the BDNF-MV treatment group isolated from cells pre-transfected with 1 μg/well pBDNF (Fig. 10c and f). Interestingly, the effect of the amount of pBDNF loading is statistically significant for EXOs only at the 24- and 48-h exposure time, but was nonsignificant for MVs. In conclusion, RAW 264.7 cell line–derived EXOs isolated from cells pre-transfected with 0.5 μg pBDNF/well showed a potential to increase hCMEC/D3 cellular ATP levels compared with other treatment groups, however, the increase in ATP levels were < 20% compared with control, untreated cells under normoxic conditions.
Fig. 10.
Effect of RAW 264.7 cell line–derived naïve EVs or BDNF-EVs on cellular ATP levels in the recipient hCMEC/D3 monolayers. hCMEC/D3 cells seeded at a density of 5000 cell/cm2 in 96 well plates were cultured for 48 h and incubated with BDNF-EXOs, or MVs derived from RAW 264.7 cells pre-transfected with 0 (naïve), 0.5, or 1 μg BDNF-DNA/well. EXOs or MVs were incubated for 24 h (a and d), 48 h (b and e), or 72 h (c and f) at either 6 μg or 12 μg EV protein/well. Post-incubation, the relative ATP levels were measured using a CellTiter-Glo 2.0-based ATP assay. Relative light units (RLU) of the cell lysates were measured at 1 s integration time using a Synergy HTX multi-mode reader. The average relative ATP levels compared with control, untreated cells from the recipient cells treated with DNA-EVs isolated from donor cells transfected with different amounts of BDNF pDNA (0, 0.5, or 1 μg/well) at different doses (6 or 12 μg EV protein/well) when incubated for 24 (a, d), 48 h (b, e), and 72 h (c, f). The effect test using multiple regression analysis presenting parameter estimates (coefficient), and p values of each transfection factor and their two-way interactions for 24-h (d), 48-h (e), and 72-h incubation time (f). Data are presented as mean ± SE of three independent experiments (n = 3/experiment). Statistical comparisons were made using multiple regression analyses, where *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001
DISCUSSION
Extracellular vesicles (EVs) exert a critical role in the intercellular communication between parent and recipient cells via transferring their nucleic acids, proteins, organelles, and/or lipid components to the recipient cells (1,5,52-54). EVs have demonstrated the potential to carry and deliver exogenous nucleic acids as well as transfer their innate contents such as mitochondria to the target cells resulting in cell-signaling processes (3,13). The lack of ATP synthesis following oxygen and glucose deprivation in ischemic stroke is the first step that triggers the energy failure and loss of ionic gradients (38) resulting in the activation of multiple processes leading to cell death. Therefore, in the present study, we hypothesized that EXOs and MVs derived from a brain endothelial cell line, hCMEC/D3, can increase the cellular energetics via increasing ATP levels in the recipient hCMEC/D3 cells (Fig. 1). In this proof-of-concept study, we conducted these experiments in healthy endothelial cells to determine the initial feasibility of our approach. Our current ongoing studies in an in vitro oxygen-glucose deprivation model of stroke is determining the effects of EV delivery in injured endothelial cells whose results will be the subject of a forthcoming manuscript. We also sought to determine if we could load a therapeutic plasmid DNA; pBDNF, for future exploration of its delivery to ischemic endothelial cells. In this study, our objectives were (1) to isolate and characterize naïve EVs from hCMEC/D3 cells, (2) isolate luciferase pDNA-loaded EVs and evaluate the luciferase gene expression in the recipient cells to determine if EVs can transfer functional pDNA, and (3) to evaluate the resulting intracellular ATP levels in naïve or pBDNF-EV-treated hCMEC/D3 cells.
We used a differential ultracentrifugation method to isolate EVs from the conditioned culture medium of hCMEC/D3 monolayers, a human brain endothelial cell line (Fig. 2a). Differential centrifugation is the most commonly reported, economic isolation method for the separation of exosomes with a low risk of isolation reagents-mediated contamination and is also appropriate for the large volume preparations (55). Extracellular components in the conditioned medium can be sequentially separated based on their physical properties such as size, shape, and density under specific centrifugal forces. Exosomes are generally uniform, spherical membrane-covered structures with a sucrose density of 1.13–1.19 g/mL and particle diameters ranging from 30 to 150 nm (2). On the other hand, microvesicles are of various shapes, with broader particle diameters of about 100–1000 nm, and are relatively denser than exosomes (2). We isolated the MVs and EXOs by eliminating the larger bio-particles such as cell debris, protein aggregates, and apoptotic bodies from the conditioned medium using a couple of sequential low-speed centrifugation cycles (300–2000×g) and subsequent filtration through 0.22-μm filters. EXOs and MVs were pelleted in the ultracentrifugation tubes at 20,000 and 120,000×g centrifugal forces and resuspended in PBS. The average particle diameters of the isolated EVs aligned well with the published reports (28,41,48). A possible reason for the observed broad polydispersity index and bimodal distribution of MVs may be due to its aggregation whereas EXOs may not aggregate due to their relatively smaller particle size. The membranes of EXOs and MVs contain anionic lipids such as phosphatidylserine, phosphatidylinositol, and glycosylated lipid derivatives (1,2,23) that led to their negative zeta potential of about ca. − 11.2 mV (Fig. 2c).
EXOs and MVs represent a heterogeneous population of vesicles, and therefore, particle size data alone cannot be used to distinguish those two EV populations even when derived from the same cells. EXOs and MVs have defined protein compositions, and we determined their characteristic protein markers using western blotting (Fig. 2b). Tetraspanin, a superfamily of four transmembrane proteins, plays a critical role in EV biogenesis, selection of exosome cargos, binding, and uptake of exosomes by target cells, and mediate immune responses via their presentation on exosomal surfaces (14). Alix, a member of the endosomal sorting complex required for transport machinery family of proteins, is involved in the inward budding of endosomal membrane during the exosome biogenesis (15). Thus, Tetraspannin CD-9 and Alix are enriched in the membrane of exosomes. Our western blotting data indeed showed the presence of Tetraspannin CD-9 and Alix proteins in EXOs whereas these proteins were absent in the MVs suggesting the purity of the isolated EVs (Fig. 2b). Numerous studies have reported the entrapment of single, double-stranded mitochondrial DNA, or mitochondrial proteins in the MVs (3,5,27). Phinney et al. showed that the MVs derived from mesenchymal stem cells contained depolarized mitochondria (27). Mitochondria is a key organelle responsible for the ATP synthesis during the oxidative phosphorylation process using ATP5 synthase. MVs isolated from hCMEC/D3 cells demonstrated the presence of ATP5A protein (Fig. 2b) suggesting the presence of mitochondrial components in MVs. In contrast, we observed the presence of ATP5A in the EXOs sample derived from RAW 264.7 macrophage cells (SL. Fig. 1). Hence, EXOs or MVs enriched with mitochondrial components may have the potential to increase cellular ATP levels in the recipient cells.
The integrity of the isolated EVs labeled using calcein AM was determined using a flow cytometry assay since this method allows quantifying the percentage (%) of intact vs. disrupted vesicles. Calcein AM, an acetoxymethyl derivative of the fluorescent molecule calcein, is membrane-permeant and non-fluorescent until activated by intravesicular esterases (56). This strategy can be applied for live cell detection, cell tracking, and to determine vesicle integrity (i.e., differentiate intact vs. disrupted vesicles) that is likely to be affected due to factors such as mechanical forces, enzymatic degradation, freeze/thaw cycles, osmolarity, pH, and time (56). The acetoxymethyl ester moiety of calcein AM is hydrolyzed by intracellular esterases and converted into a membrane-impermeant green fluorescent calcein. A few studies have reported calcein-based assessments of exosome and microvesicle leakage as well as cell tracking (57-59). Mitchell et al. investigated the effect of urine osmosis on the integrity of B cell line–derived exosomes using flow cytometry analysis of calcein-loaded exosome bead complexes (57). Gray et al. developed a sensitive and precise flow cytometry method for determining the integrity of EVs using calcein AM (56). The results of the integrity of EVs derived from two different origins (human brain endothelial cells and mouse macrophages) demonstrated that our isolation process does not result in significant damage to EV membrane integrity. Our analysis indicated that at least 90% of the vesicles are calcein-positive (Fig. 4 and SL. Fig. 3) and thus gave us the confidence that the observed effects of EVs arise from intact vesicles.
In order to determine the capability of EVs as vectors to deliver exogenous DNA into recipient cells, we first pre-transfected hCMEC/D3 or RAW264.7 cells using lipofectamine/DNA complexes and quantified the amount of Luc pDNA in the isolated EVs. We then evaluated the transfection efficiency of Luc pDNA-loaded EVs (Luc-EXOs/MVs) in the recipient hCMEC/D3 cells. We used the Picogreen assay to quantify the amount of dsDNA entrapped in the isolated EVs. While this assay is not specific to the exogenously loaded pDNA construct, it quantifies the amount of total double-stranded nucleic acids entrapped into the EVs. Therefore, as expected, we observed a considerable amount of DNA loading in the naïve EVs (0 μg/well pDNA, Table III) as they are enriched with a variety of nucleic acids (1,2,5). We subtracted the naïve EVs DNA content and the resulting DNA loading data showed that EXOs contained a significantly (p < 0.05) higher amount of Luc pDNA compared with MVs. However, the DNA loading efficiency of both EXOs and MVs were less than ca. 1%, indicating that a limited amount of the exogenous Luc-DNA was packaged into EXOs and MVs and was subsequently secreted to the extracellular environment. We speculate that the following are likely reasons for the observed low DNA loading: First, despite using the cationic Lipofectamine reagent for transfection of the parent hCMEC/D3 cells, only a fraction of exogenous DNA from the lipofectamine/pDNA complexes may have entered the cells as brain endothelial cells inherently show low rates of pinocytic uptake (60,61). Second, post-entry into the cells, a portion of the endogenous DNA may have degraded in the acidic lysosomes, and/or a part of DNA may have delivered into the nuclei. It is also important to note that the exosome biogenesis pathway is endosomal in origin and thus, a proportion of the packaged DNA may have been routed to lysosomal degradation. As a combined effect, only small amounts of the delivered DNA may have been packaged in EXOs and MVs and secreted into the extracellular environment. It is also important to account for the cell line-specific effects of EV biogenesis and release. We would also like to emphasize that the transfection levels of non-viral gene carriers such as Lipofectamine used in the current study are largely cell line-dependent (62). Lamichhane et al. (13) used an electroporation method to entrap DNA and reported less than 2% of exogenous DNA loading into extracellular vesicles, in line with our observations. The extent of DNA loading is dependent on the molecular size of dsDNA, particle diameters of EXOs and MVs, the specific transfection methods, and the experimental parameters used. Lamichhane et al. reported that the DNA loading decreased from about 35 ng/3 × 108 EVs for a 750 bp DNA size to nearly 3 ng/3 × 108 EVs for a 1000 bp DNA size. It should be pointed out that the size of gWiz-Luc-DNA we used is 6732 bp. Therefore, there is a need for further optimization of the transfection protocol to increase the DNA loading efficiency. Importantly, RAW 264.7 cell line–derived EXOs isolated from cells pre-transfected with 0.5 μg/well DNA showed significantly (p < 0.0001) higher DNA loading compared with hCMEC/D3 and RAW 264.7 cell line–derived MVs as well as hCMEC/D3 cell-derived EXOs (Fig. 5).
Noteworthy, although an earlier work (23) demonstrated functional delivery of pDNA via MVs but not EXOs, the extent of DNA loading into the different EVs were not discussed and thus does not allow a comparison with our DNA loading values. We compared the protein yields of DNA-loaded EVs and EVs isolated from cells treated with Lipofectamine alone (no DNA) with that of naïve EVs. There was no significant difference in the total EV protein content or EV yield among the three groups transfected with different DNA amounts indicating that the biogenesis and release of EVs were not affected by the transfection process. We have demonstrated the partial specificity of BCA or MicroBCA assay for determining the total EV protein content in the discussion section in supplemental 1 (SL. Fig. 6 and SL. Fig. 7).
We evaluated the effect of the amount of Luc pDNA, total EV protein, type of EVs, and incubation time on the transfection activity of Luc-EVs in hCMEC/D3 cells (Fig. 7). Incubation time and the two-way interaction between type of EVs and incubation time showed a significant (p < 0.0001) effect on the luciferase gene expression in hCMEC/D3 cells. A significant interaction between the type of EVs and incubation time indicates that the extent of change in luciferase expression at 24 h or 48 by Luc-EVs was different for EXOs or MVs (Fig. 7d). Kanada et al. measured luciferase expression in units of photon flux (p/s) and reported peak luciferase expression levels when Lipofectamine 2000/Luc-pDNA complexes were transfected for 48 h, whereas the Luc-MVs derived from HEK293FT cells showed the maximum expression at 72 h (23). This suggests that EVs took longer to express the delivered gene compared with cationic lipids. The difference in the observed gene expression kinetics could be due to the differences in subcellular trafficking and release of pDNA from the lipofectamine/DNA complexes vs. DNA-EVs. Lipofectamine/DNA complexes are taken up by cellular endocytosis and the cationic lipid subsequently fuses with the endosomal membrane allowing the escape of DNA into the cytosol (63). On the other hand, confocal laser microscopy and flow cytometry studies revealed that EVs also undergo energy-dependent endocytic or macropinocytic uptake in hCMEC/D3 cells (41,64) and colocalize with early endosomes within 2 h of incubation. Using confocal fluorescence microscopy, Kanada et al. demonstrated that fluorescently labeled EVs internalized by cells were still localized within the endocytic compartments even at 48 h post-incubation suggesting that the EV-mediated delivery is a slower process compared with cationic lipid transfections (23). Importantly, lipofectamine/DNA complexes showed significantly (p < 0.0001) higher gene expression compared with Luc-EVs at 48 h of incubation (Fig. 7d). If cellular uptake occurs via endocytosis, it is also likely that the uptake of negatively charged pDNA-EVs could be hindered by the anionic cell membranes, whereas the cationic lipofectamine/DNA complexes generally show greater levels of cellular uptake and hence, a higher transfection efficiency. We would like to point out that the Luc-EVs did not considerably increase luciferase gene expression compared with untreated cells at the 24- or 48-h incubation time (Fig. 7b). Besides, the increase in luciferase expression at 72 h was not statistically significant, however, considerably higher compared with 24-h and 48-h incubation time points (Fig. 7a-c). It is important to note that the transfection activity can be cell line- and/or EV source-dependent and hence, we were not surprised to notice low transfection levels in the hCMEC/D3 cell line which inherently possess low pinocytic capabilities (60).
Luc-EVs derived from RAW 264.7 cell line showed a statistically significant (p < 0.01) increase in luciferase expression in hCMEC/D3 cells at the 72-h incubation time. EV-mediated transfection of pDNA took at least 72 h to express luciferase proteins in the recipient cells. The luciferase expression was significantly higher in hCMEC/D3 cells transfected with RAW 264.7 cell-derived Luc-EVs compared with hCMEC/D3 cell line–derived Luc-EVs suggesting that the cell source of EV play a critical role in EV-mediated gene expression in recipient cells. We speculate that the resulting membrane composition of EVs derived from a low pinocytic cell line like hCMEC/D3 contributed to the low transfection activity of those EVs. However, EVs derived from a RAW264.7, a macrophage cell line, showed greater transfection levels in the recipient cells, likely due to their improved migratory/cell entry capabilities. It should also be noted that under conditions like stroke, macrophages and other immune cells breach the brain endothelial cells in a process known as diapedesis (65). Thus, engineered EVs derived from macrophage cells can be used for therapeutic drug delivery in diseases like stroke and in other diseases where BBB permeability is implicated in the pathogenesis. Yuan et al. (41) have demonstrated that naïve EVs derived from RAW 264.7 macrophages utilize the integrin lymphocyte function-associated antigen 1 (LFA-1), intercellular adhesion molecule 1 (ICAM-1), and, the carbohydrate-binding C-type lectin receptors to interact with hCMEC/D3 monolayers. It is also known that the upregulation of ICAM-1, a common process in inflammation, promotes the uptake of macrophage-derived exosomes in the hCMEC/D3 monolayers. The authors also demonstrated that intravenously administered RAW-derived exosomes crossed the BBB and delivered a cargo protein to the brain.
Despite their moderate transfection efficiency, EV-mediated gene transfer is still intriguing due to their rich vesicular cargo that may enhance the biologic effects of the delivery and result in functional changes such as increased cellular energetics. Thus, we evaluated if EVs can be loaded with a therapeutic payload, BDNF pDNA, and studied their effects on the resulting intracellular ATP levels in recipient hCMEC/D3 cells using the ATP assay (Fig. 9). It has been reported that MVs package mitochondria and mitochondrial DNA (mtDNA) from the parent cells (27,28). We confirmed the presence of ATP5A, the enzyme responsible for ATP synthesis from ADP in the mitochondria, in MVs and EXOs derived from hCMEC/D3 cells (Fig. 2b) and in the EXOs derived from the RAW 264.7 macrophage cells (SL. Fig. 1). ATP produced by mitochondria provides the chemical energy required for overall cellular activity and metabolism, calcium homeostasis, induction of cell apoptosis, regulation of innate immunity, and maintenance of cell differentiation and reprogramming (29). Therefore, EVs containing mitochondria may have a strong potential to mediate cell survival by increasing the ATP levels during cell duress (27,32). MVs and EXOs differ in their biogenesis and as a result, contain different protein and gene materials (5,28). We speculated that a combination of EXOs and MVs (EXOs + MVs) may include a menagerie of proteins and nucleic acids that could likely result in higher cellular ATP levels compared with treatment with MVs or EXOs alone. Therefore, we evaluated the effect of type of EVs (EXOs, MVs, or EXOs + MVs) and naïve vs. BDNF-EVs isolated from cells transfected with different amounts of pBDNF (12 vs. 24 μg) on the relative ATP levels in hCMEC/D3 cells incubated for 24 or 48 h (Fig. 9).
EXOs showed the maximum and significant (p < 0.05) increase in the relative ATP levels compared with MVs or EXOs + MVs at the 24-h incubation time. The relatively reduced efficiency of EXOs + MVs may be due to the reduced amount of EXOs (by 50%) in the combination compared with EXOs or MVs alone. Thus, we did not use the EXOs + MVs combination treatment group for studying the effect of the factors at the 48-h incubation time. Despite the presence of a mitochondrial marker in the MVs (Fig. 2b), our data showed that treatment with EXOs resulted in higher cellular ATP levels compared with treatments with MVs and MVs + EXOs. Hough et al. demonstrated that packaged mitochondria in exosomes isolated from the myeloid-derived regulatory donor cells that were transferred to the recipient T cells facilitated oxidative phosphorylation and pro-inflammatory signaling (66).
Apart from the type of EVs and amount of transfected pBDNF, the effect of the amount of total EV protein on cellular ATP levels was statistically (p < 0.0001) significant at 24 h, however insignificant (p > 0.05) at 48 h. A possible reason could be the incubation time-dependent aggregation of EVs, denaturation or degradation of vesicular proteins as can be deduced by the reduction in parameter estimates from 7.8 at 24 h to 4.33 at 48 h (Fig. 9c and f). The highest cellular ATP levels were noted in cells treated with naïve-EXOs at a dose of 6 μg protein/well instead of 12 μg protein/well suggesting that an optimal dose of EVs is required to increase the cellular metabolic activity. Besides, EXOs have a wide range of proteins such as heat shock proteins and pyruvate kinase that regulate functions such as cellular growth and migration in recipient cells (28,67,68). Naïve EVs (0 μg pDNA) showed significantly (p > 0.05) higher ATP levels compared with pBDNF-EVs isolated from transfected cells. A possible reason for the relatively lower enhancement in ATP levels in the BDNF-EV-treated cells may be related to the qualitative changes in vesicular content (but not the overall quantitative total protein content) as a result of DNA transfection. Our current studies are comparing vesicular content in EVs isolated from naïve vs. transfected cells using proteomic analyses and will be presented in a forthcoming manuscript. Notably, the interactions between any two factors (amount of BDNF pDNA, type of EV protein, or type of EVs) at either incubation times were statistically insignificant suggesting that the effect of any one factor on the ATP levels was independent and not influenced by the presence of the other two factors. RAW 264.7 cell line–derived EXOs isolated from cells pre-transfected with 0.5 μg pBDNF/well showed the potential to increase intracellular ATP levels in hCMEC/D3 monolayers compared with other treatment groups, however, the increase in ATP levels were < 20% compared with control, untreated cells under normoxic conditions. We have demonstrated (D’Souza....Manickam; manuscript under preparation) that the exposure of EVs to brain endothelial cells under hypoxic conditions (using cells subjected to oxygen-glucose deprivation; OGD) rescues the loss in endothelial cell viability, compared with untreated cells. Thus, our future works will explore the therapeutic utility of these engineered EVs under hypoxic conditions.
Collectively, hCMEC/D3 cell line–derived naïve EVs but not BDNF-EVs mediated a significant increase in intracellular ATP levels, and the presence of ATP5A in naïve EVs suggested that cell-derived naïve EXOs and MVs contain ATP5A and/or mitochondria/mtDNA that can also be transferred to the endothelial cells during ischemic conditions to increase their bioenergetics and survival. In our studies, we did not directly establish the relationship between the transfected amount of DNA and the resulting ATP levels with ATP5A expression. However, we would like to point out here that the BDNF-DNA in hCMEC/D3 cell line–derived EVs is per se not expected to contribute to increased cellular ATP levels. We wish to emphasize that we utilized plasmid BDNF-DNA as a model therapeutic gene to evaluate the efficiency of EVs to carry a therapeutic pDNA and used the ATP assay as a readout to determine if these BDNF-EVs can also result in increased cellular energetics via the transfer of their innate functional components such as mitochondria/mtDNA. We wish to additionally emphasize that the observed changes in ATP levels were not used here as a readout for the direct therapeutic activity of BDNF-loaded EVs.
The effects between DNA-MVs and naïve MVs are subtle and are likely due to the limited amount of DNA loading observed under the current experimental conditions. Notwithstanding these observations, we believe that the innate, mitochondrion-related cargo in EVs can be beneficially exploited to increase cellular energetics in injured/ischemic endothelial cells. To this effect, we have engineered EVs for the delivery of a cationic protein, ATP5A, and have demonstrated that the exposure of EVs or ATP5A-EVs to OGD-subjected endothelial cells can increase their ATP levels and consequently, their overall cell viability (D’Souza....Manickam; manuscript under preparation).
The observed increase in ATP levels is highly significant because during post-ischemic injury, the lack of oxygen supply to the affected brain tissues leads to the mitochondrial dysfunction-mediated reduction in ATP levels in the endothelial cells lining the BBB. Thus, restoring the cellular ATP levels in injured ischemic endothelial cells has a strong potential to decrease acute cell death and consequently preserve the protective functions of the BBB. Cell-derived naïve EXOs and MVs contain mitochondria and/or mtDNA that can also be transferred to the damaged endothelial cells during oxidative stress to increase their bioenergetics and survival. Further optimization of the engineering of BDNF-EVs will allow its delivery to facilitate the synthesis/release of BDNF from endothelial cells thus enabling the protective effects of the delivered BDNF without crossing the BBB.
Additional discussion in Supplemental 1: For any interested readers, we have discussed and presented data on modifying EVs using synthetic cationic polymers that can render an overall positive charge to the formed EV/polymer complexes. We have discussed how EVs can be modified for purposing their delivery to specific diseases such as ischemic stroke. We have also discussed routes of EV administration that have been used in pre-clinical and clinical experiments.
CONCLUSION
In the present study, we explored the capability of EVs derived from a brain endothelial and a mouse macrophage cell line to load exogenous plasmid DNAs and deliver the external payload along with their innate vesicular content to the recipient human brain endothelial cells. Our results showed that EXOs and MVs can load exogenous luciferase or BDNF plasmid DNA. Increased ATP levels in naïve- and BDNF-EV-treated hCMEC/D3 cells supported our hypothesis that innate EV components such as mitochondria/mtDNA can increase cellular energetics in the recipient cells even under normoxic conditions. Design of experiment analyses revealed that the type of EVs, amount of DNA in isolated EVs, and total EV protein are statistically significant factors that affected the observed changes in endothelial cell ATP levels. Naïve EXOs increased ATP levels to a greater extent than naïve MVs and EVs isolated from cells transfected with pBDNF. An optimum amount of the total EV protein, here 6 μg/well, was required to increase the cellular ATP levels at the 24-h incubation time; however, at 48 h, varying the total EV protein did not change the cellular ATP levels. Further optimization of the EV engineering process is expected to lead to the development of this exciting class of new therapies for decreasing acute cell death in ischemic stroke.
Supplementary Material
ACKNOWLEDGMENTS
The authors express their deep appreciation to Dr. Lauren O’Donnell (DU), Mss. Manisha Chandwani, and Yashika Kamte for flow cytometry support. We thank Dr. Jelena Janjic for allowing use of the Malvern Zetasizer Nano.
FUNDING
The study was supported using start-up funds for the Manickam laboratory from the School of Pharmacy at Duquesne University (DU). We acknowledge funding for CH through the Neurodegeneration Undergraduate Research Program NIH R25NS100118 (Drs. Benedict Kolber, Kevin Tidgewell, Michael Cascio, and Rita Mihailescu, DU).
Footnotes
Conflict of Interest The authors declare that they have no conflicts of interest.
SUPPLEMENTARY INFORMATION
The online version contains supplementary material available at https://doi.org/10.1208/s12249-020-01892-w.
REFERENCES
- 1.El Andaloussi S, Mäger I, Breakefield XO, Wood MJA. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–57. [DOI] [PubMed] [Google Scholar]
- 2.Konoshenko MY, Lekchnov EA, Vlassov AV, Laktionov PP. Isolation of extracellular vesicles: general methodologies and latest trends. Biomed Res Int. 2018;2018:8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puhm F, Afonyushkin T, Resch U, Obermayer G, Rohde M, Penz T, et al. Mitochondria are a subset of extracellular vesicles released by activated monocytes and induce type I IFN and TNF responses in endothelial cells. Circ Res. 2019;125(1):43–52. [DOI] [PubMed] [Google Scholar]
- 4.Nawaz M, Fatima F, Vallabhaneni KC, Penfornis P, Valadi H, Ekström K, et al. Extracellular vesicles: evolving factors in stem cell biology. Stem Cells Int. 2016;2016:1073140–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalra H, Drummen GP, Mathivanan S. Focus on extracellular vesicles: introducing the next small big thing. Int J Mol Sci. 2016;17(2):170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong S-G, Wu JC. Exosomes as potential alternatives to stem cell therapy in mediating cardiac regeneration. Circ Res. 2015;117(1):7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shigemoto-Kuroda T, Oh JY, Kim D-K, Jeong HJ, Park SY, Lee HJ, et al. MSC-derived extracellular vesicles attenuate immune responses in two autoimmune murine models: type 1 diabetes and uveoretinitis. Stem Cell Rep. 2017;8(5):1214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy DE, de Jong OG, Brouwer M, Wood MJ, Lavieu G, Schiffelers RM, et al. Extracellular vesicle-based therapeutics: natural versus engineered targeting and trafficking. Exp Mol Med. 2019;51(3):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha JM, Shin EK, Sung JH, Moon GJ, Kim EH, Cho YH, et al. Efficient scalable production of therapeutic microvesicles derived from human mesenchymal stem cells. Scientific reports. 2018;8(1):1171–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T, Larcher LM, Ma L, Veedu RN. Systematic screening of commonly used commercial transfection reagents towards efficient transfection of single-stranded oligonucleotides. Molecules (Basel, Switzerland). 2018;23(10):2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barenholz Y Doxil® — the first FDA-approved nano-drug: lessons learned. J Control Release. 2012;160(2):117–34. [DOI] [PubMed] [Google Scholar]
- 12.Hood JL, Wickline SA. A systematic approach to exosome-based translational nanomedicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012;4(4):458–67. [DOI] [PubMed] [Google Scholar]
- 13.Lamichhane TN, Raiker RS, Jay SM. Exogenous DNA loading into extracellular vesicles via electroporation is size-dependent and enables limited gene delivery. Mol Pharm. 2015;12(10):3650–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang T, Deng C-X. Current progresses of exosomes as cancer diagnostic and prognostic biomarkers. Int J Biol Sci. 2019;15(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JY, Kim JW, Yenari MA. Heat shock protein signaling in brain ischemia and injury. Neurosci Lett. 2020;715:134642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voloboueva LA, Duan M, Ouyang Y, Emery JF, Stoy C, Giffard RG. Overexpression of mitochondrial Hsp70/Hsp75 protects astrocytes against ischemic injury in vitro. J Cereb Blood Flow Metab. 2008;28(5):1009–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. [DOI] [PubMed] [Google Scholar]
- 19.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJA. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–5. [DOI] [PubMed] [Google Scholar]
- 20.S-i O, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendt M, Kamerkar S, Sugimoto H, McAndrews KM, Wu C-C, Gagea M, et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018;3(8):e99263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanada M, Bachmann MH, Hardy JW, Frimannson DO, Bronsart L, Wang A, et al. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc Natl Acad Sci U S A. 2015;112(12):E1433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otero-Ortega L, Laso-Garcia F, Gomez-de Frutos M, Fuentes B, Diekhorst L, Diez-Tejedor E, et al. Role of exosomes as a treatment and potential biomarker for stroke. Transl Stroke Res. 2018. [DOI] [PubMed] [Google Scholar]
- 25.van Dommelen SM, Vader P, Lakhal S, Kooijmans SAA, van Solinge WW, Wood MJA, et al. Microvesicles and exosomes: opportunities for cell-derived membrane vesicles in drug delivery. J Control Release. 2012;161(2):635–44. [DOI] [PubMed] [Google Scholar]
- 26.Liu M-L, Williams KJ. Microvesicles: potential markers and mediators of endothelial dysfunction. Curr Opin Endocrinol Diabetes Obes. 2012;19(2):121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phinney DG, Di Giuseppe M, Njah J, Sala E, Shiva S, St Croix CM, et al. Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle microRNAs. Nat Commun. 2015;6:8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dozio V, Sanchez JC. Characterisation of extracellular vesicle-subsets derived from brain endothelial cells and analysis of their protein cargo modulation after TNF exposure. J Extracell Vesicles. 2017;6(1):1302705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osellame LD, Blacker TS, Duchen MR. Cellular and molecular mechanisms of mitochondrial function. Best Pract Res Clin Endocrinol Metab. 2012;26(6):711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonckheere AI, Smeitink JAM, Rodenburg RJT. Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis. 2012;35(2):211–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayakawa K, Esposito E, Wang X, Terasaki Y, Liu Y, Xing C, et al. Transfer of mitochondria from astrocytes to neurons after stroke. Nature. 2016;535(7613):551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spees JL, Olson SD, Whitney MJ, Prockop DJ. Mitochondrial transfer between cells can rescue aerobic respiration. Proc Natl Acad Sci U S A. 2006;103(5):1283–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King MP, Attardi G. Injection of mitochondria into human cells leads to a rapid replacement of the endogenous mitochondrial DNA. Cell. 1988;52(6):811–9. [DOI] [PubMed] [Google Scholar]
- 34.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246(4929):500–3. [DOI] [PubMed] [Google Scholar]
- 35.Anne Stetler R, Leak RK, Gao Y, Chen J. The dynamics of the mitochondrial organelle as a potential therapeutic target. J Cerebr Blood Flow Metab Off J Int Soc Cerebr Blood Flow Metab. 2013;33(1):22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodruff TM, Thundyil J, Tang S-C, Sobey CG, Taylor SM, Arumugam TV. Pathophysiology, treatment, and animal and cellular models of human ischemic stroke. Mol Neurodegener. 2011;6(1):11–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdullahi W, Tripathi D, Ronaldson PT. Blood-brain barrier dysfunction in ischemic stroke: targeting tight junctions and transporters for vascular protection. Am J Phys Cell Phys. 2018;315(3):C343–C56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lo EH, Moskowitz MA, Jacobs TP. Exciting, radical. Suicidal Stroke. 2005;36(2):189–92. [DOI] [PubMed] [Google Scholar]
- 39.Guo S, Kim WJ, Lok J, Lee S-R, Besancon E, Luo B-H, et al. Neuroprotection via matrix-trophic coupling between cerebral endothelial cells and neurons. Proc Natl Acad Sci. 2008;105(21):7582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banks WA. From blood–brain barrier to blood–brain interface: new opportunities for CNS drug delivery. Nat Rev Drug Discov. 2016;15:275–92. [DOI] [PubMed] [Google Scholar]
- 41.Yuan D, Zhao Y, Banks WA, Bullock KM, Haney M, Batrakova E, et al. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials. 2017;142:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dave KMAL, Manickam DS. Characterization of the SIM-A9 cell line as a model of activated microglia in the context of neuropathic pain. PLoS ONE. 2020;15(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cucullo L, Couraud PO, Weksler B, Romero IA, Hossain M, Rapp E, et al. Immortalized human brain endothelial cells and flow-based vascular modeling: a marriage of convenience for rational neurovascular studies. J Cereb Blood Flow Metab. 2008;28(2):312–28. [DOI] [PubMed] [Google Scholar]
- 44.Wolff A, Antfolk M, Brodin B, Tenje M. In vitro blood-brain barrier models-an overview of established models and new microfluidic approaches. J Pharm Sci. 2015;104(9):2727–46. [DOI] [PubMed] [Google Scholar]
- 45.Weksler B, Romero IA, Couraud PO. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids Barriers CNS. 2013;10(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohtsuki S, Ikeda C, Uchida Y, Sakamoto Y, Miller F, Glacial F, et al. Quantitative targeted absolute proteomic analysis of transporters, receptors and junction proteins for validation of human cerebral microvascular endothelial cell line hCMEC/D3 as a human blood-brain barrier model. Mol Pharm. 2013;10(1):289–96. [DOI] [PubMed] [Google Scholar]
- 47.Tornabene E, Brodin B. Stroke and drug delivery–in vitro models of the ischemic blood-brain barrier. J Pharm Sci. 2016;105(2):398–405. [DOI] [PubMed] [Google Scholar]
- 48.Agrahari V, Agrahari V, Burnouf PA, Chew CH, Burnouf T. Extracellular microvesicles as new industrial therapeutic Frontiers. Trends Biotechnol. 2019;37(7):707–29. [DOI] [PubMed] [Google Scholar]
- 49.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteome. 2010;73(10):1907–20. [DOI] [PubMed] [Google Scholar]
- 50.ATP5F1A ATP synthase F1 subunit alpha: NCBI; 2019. [Available from: https://www.ncbi.nlm.nih.gov/gene/498.
- 51.Midzak AS, Chen H, Aon MA, Papadopoulos V, Zirkin BR. ATP synthesis, mitochondrial function, and steroid biosynthesis in rodent primary and tumor Leydig cells. Biol Reprod. 2011;84(5):976–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Javeed N, Mukhopadhyay D. Exosomes and their role in the micro-/macro-environment: a comprehensive review. J Biomed Res. 2017;31(5):386–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makridakis M, Roubelakis MG, Vlahou A. Stem cells: insights into the secretome. Biochim Biophys Acta (BBA) - Proteins Proteomics. 2013;1834(11):2380–4. [DOI] [PubMed] [Google Scholar]
- 54.Nawaz M, Fatima F, Vallabhaneni KC, Penfornis P, Valadi H, Ekstrom K, et al. Extracellular vesicles: evolving factors in stem cell biology. Stem Cells Int. 2016;2016:1073140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang D, Zhang W, Zhang H, Zhang F, Chen L, Ma L, et al. Progress, opportunity, and perspective on exosome isolation - efforts for efficient exosome-based theranostics. Theranostics. 2020;10(8):3684–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gray WD, Mitchell AJ, Searles CD. An accurate, precise method for general labeling of extracellular vesicles. MethodsX. 2015;2:360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell PJ, Welton J, Staffurth J, Court J, Mason MD, Tabi Z, et al. Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med. 2009;7(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warren D, Gray KMF, Ghosh-Choudhary S, Maxwell JT, Brown ME, Platt MO, et al. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ Res. 2015;116(2):255–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexy T, Rooney K, Weber M, Gray WD, Searles CD. TNF-α alters the release and transfer of microparticle-encapsulated miRNAs from endothelial cells. Physiol Genomics. 2014;46(22):833–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cipolla MJ, Crete R, Vitullo L, Rix RD. Transcellular transport as a mechanism of blood-brain barrier disruption during stroke. Front Biosci (Schol Ed). 2004;9:777–85. [DOI] [PubMed] [Google Scholar]
- 61.Deeken JF, Löscher W. The blood-brain barrier and cancer: transporters, treatment, and Trojan horses. Clin Cancer Res. 2007;13(6):1663–74. [DOI] [PubMed] [Google Scholar]
- 62.von Gersdorff K, Sanders NN, Vandenbroucke R, De Smedt SC, Wagner E, Ogris M. The internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex type. Mol Ther. 2006;14(5):745–53. [DOI] [PubMed] [Google Scholar]
- 63.Stegmann T, Legendre J-Y. Gene transfer mediated by cationic lipids: lack of a correlation between lipid mixing and transfection. Biochim Biophys Acta Biomembr. 1997;1325(1):71–9. [DOI] [PubMed] [Google Scholar]
- 64.Ajikumar A, Long MB, Heath PR, Wharton SB, Ince PG, Ridger VC, et al. Neutrophil-derived microvesicle induced dysfunction of brain microvascular endothelial cells in vitro. Int J Mol Sci. 2019;20(20):5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopes Pinheiro MA, Kooij G, Mizee MR, Kamermans A, Enzmann G, Lyck R, et al. Immune cell trafficking across the barriers of the central nervous system in multiple sclerosis and stroke. Biochim Biophys Acta (BBA) - Mol Basis Dis. 2016;1862(3):461–71. [DOI] [PubMed] [Google Scholar]
- 66.Hough KP, Trevor JL, Strenkowski JG, Wang Y, Chacko BK, Tousif S, et al. Exosomal transfer of mitochondria from airway myeloid-derived regulatory cells to T cells. Redox Biol. 2018;18:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev. 2015;34(4):474–90. [DOI] [PubMed] [Google Scholar]
- 68.Ferguson SW, Nguyen J. Exosomes as therapeutics: the implications of molecular composition and exosomal heterogeneity. J Control Release. 2016;228:179–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.