ABSTRACT
Determinants of protective immunity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection require the development of well-standardized, reproducible antibody assays. This need has led to the emergence of a variety of neutralization assays. Head-to-head evaluation of different SARS-CoV-2 neutralization platforms could facilitate comparisons across studies and laboratories. Five neutralization assays were compared using 40 plasma samples from convalescent individuals with mild to moderate coronavirus disease 2019 (COVID-19): four cell-based systems using either live recombinant SARS-CoV-2 or pseudotyped viral particles created with lentivirus (LV) or vesicular stomatitis virus (VSV) packaging and one surrogate enzyme-linked immunosorbent assay (ELISA)-based test that measures inhibition of the spike protein receptor binding domain (RBD) binding its receptor human angiotensin converting enzyme 2 (hACE2). Vero cells, Vero E6 cells, HEK293T cells expressing hACE2, and TZM-bl cells expressing hACE2 and transmembrane serine protease 2 were tested. All cell-based assays showed 50% neutralizing dilution (ND50) geometric mean titers (GMTs) that were highly correlated (Pearson r = 0.81 to 0.89) and ranged within 3.4-fold. The live virus assay and LV pseudovirus assays with HEK293T/hACE2 cells showed very similar mean titers, 141 and 178, respectively. ND50 titers positively correlated with plasma IgG targeting SARS-CoV-2 spike protein and RBD (r = 0.63 to 0.89), but moderately correlated with nucleoprotein IgG (r = 0.46 to 0.73). ND80 GMTs mirrored ND50 data and showed similar correlation between assays and with IgG concentrations. The VSV pseudovirus assay and LV pseudovirus assay with HEK293T/hACE2 cells in low- and high-throughput versions were calibrated against the WHO SARS-CoV-2 IgG standard. High concordance between the outcomes of cell-based assays with live and pseudotyped virions enables valid cross-study comparison using these platforms.
KEYWORDS: antibody, COVID-19, neutralization assay, SARS-CoV-2
INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused more than 100 million confirmed infections and over 2.4 million deaths worldwide as of February 15, 2021 (https://www.worldometers.info/coronavirus). Despite governmental regulations designed to minimize virus transmission and reduce mortality, such as mask use and social distancing guidelines, vaccines are required to limit the spread of the virus and the burden of COVID-19.
Most efficacious licensed vaccines would elicit pathogen-neutralizing antibodies (nAb) (1). Humans can mount nAb responses against SARS-CoV-2 during natural infection (2–5). Epidemiologic data suggest that reinfection rates are low, albeit increasing numbers of sporadic reinfections are being reported (6, 7). A crucial unknown at this time is what immune responses are associated with protective immunity. While there is mixed evidence supporting the efficacy of convalescent-phase sera infusion for disease shortening, recent studies suggest that passive infusion of monoclonal antibodies can alter COVID-19 progression (8, 9). In order to determine what constitutes protective immunity, well-standardized, reproducible antibody assays are required to establish correlates of risk and protection. Efficacy data for several SARS-CoV-2 vaccines have been already published, but analyses of correlates of protection are yet to come (10–12). For that, massive serological measurements, including virus neutralization, are under way. In this regard, it is important to understand how results obtained with different virus neutralization platforms can be compared.
The plaque reduction neutralization test (PRNT) is considered a “gold standard” to assess virus neutralizing potency of a serum or antibody sample. However, a variety of live virus neutralization assays that use recombinant SARS-CoV-2 (rSARS-CoV-2) containing a reporter gene at the ORF7 locus of the viral genome have been suggested as alternatives (13, 14). These recombinant viruses replicate similar to SARS-CoV-2 clinical isolates in vitro and successfully infect primary airway epithelial cell cultures. A fluorescence-based rSARS-CoV-2 neutralization assay yielded comparable results to PRNT in nAb detection from convalescent patient plasma (13). With a shorter turnaround time (24 to 48 h for reporter virus versus 3 days for PRNT), rSARS-CoV-2 provides a useful high-throughput (HTS) platform to study nAb responses but unfortunately still requires biosafety level 3 (BSL-3) containment for assay set-up and readout.
Reporter assays with pseudotyped viruses restricted to a single round of replication allow nAb experiments to be performed in BSL-2 laboratories. Pseudotyped viral particles created with lentivirus (LV) and vesicular stomatitis virus (VSV) (15–18) packaging platforms have already been adapted for SARS-CoV-2 (19–21). Several cell lines endogenously or exogenously expressing angiotensin converting enzyme 2 (ACE2), the host receptor for the SARS-CoV-2 spike protein, have been tested, and Vero cells were among the most susceptible to VSV pseudovirus entry (22–24). HEK 293T cells transfected to express ACE2 have also been developed for use in pseudovirus neutralization assays (25). In addition to ACE2, transmembrane serine protease 2 (TMPRSS2) has been shown to prime the spike protein for viral cell entry (24).
Because the receptor binding domain (RBD) of the spike protein is the major target for nAbs (26–28), surrogate enzyme-linked immunosorbent assay (ELISA)-based assays were introduced to evaluate antibodies that compete with ACE2 for RBD binding (20, 29, 30). Major advantages of these assays include low cost, speed, and safety. As opposed to measuring actual virus neutralization, surrogate assays report percent binding inhibition between RBD and ACE2, which is then interpreted as percent neutralization. While they provide inexpensive and rapid detection of RBD-targeting nAbs, surrogate assays cannot measure neutralization via non-RBD spike protein epitopes. The importance of this issue has increased with the increasing prevalence of escape resistant variants of SARS CoV-2 (31–33) (https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html).
The global pandemic led to the unprecedented rapid development and implementation of many SARS-CoV-2 neutralization assays. However, interassay comparison and validation is needed to better understand antibody kinetics and longevity of humoral immune responses, correlates of immune protection, and vaccine efficacy (34). In the current study, we aimed to fill this gap by evaluating the same set of plasma samples from convalescent individuals with mild to moderate COVID-19 with five SARS-CoV-2 neutralization assays, including (i) a live rSARS-CoV-2 assay on Vero E6 cells, (ii) VSV pseudotyped with SARS-CoV-2 spike on Vero cells, (iii) LV pseudotyped with SARS-CoV-2 spike on HEK293T cells expressing hACE2 in a regular and HTS format, (iv) LV pseudovirus on TZM-bl cells expressing hACE2 and TMPRSS2, and (v) a surrogate, ELISA-based test that measures inhibition of binding between RBD and ACE2. We also examined the correlation between neutralization and the plasma concentration of SARS-CoV-2 nucleoprotein-, spike-, and RBD-specific IgG.
MATERIALS AND METHODS
Detailed descriptions of reagents and procedures are available in the supplemental material.
Study population and specimen collection.
Plasma samples used for this study were obtained from participants (≥18 years of age) of a seroepidemiology study following a county-wide outbreak of SARS-CoV-2 in Blaine County, ID, in March to April 2020. Study participants were randomly selected after stratification by ZIP code and within ZIP code, age, gender, and race/ethnicity. All volunteers signed electronic consent forms. Demographic information and symptom histories since January 15, 2020, were collected.
Blood was collected in 10-ml vials with acid citrate dextrose and shipped overnight to the laboratory (Fred Hutch, Seattle, WA) where plasma was separated by centrifugation. One aliquot was submitted for the Architect SARS-CoV-2 IgG assay (Abbott, Abbott Park, IL). Other aliquots were heat inactivated for 30 min at 56°C, frozen at −80°C, and distributed to testing laboratories for SARS-CoV-2 neutralization assays. Study participants were informed of the qualitative results of the IgG serology assay via email within 1 week of obtaining test results. This study was approved by the Fred Hutchinson Cancer Research Center Institutional Review Board, and all study materials were provided in both English and Spanish.
Cell lines.
Vero cells (CCL-81; ATCC, Manassas, VA) are kidney epithelial cells of Cercopithecus aethiops; Vero E6 (CRL-1586; ATCC) is a cloned variant of Vero cells. Human embryonic kidney cells (CRL-3216; ATCC), HEK293T, expressing hACE2 (293T/ACE2.MF) were kindly provided by Mike Farzan and Huihui Mu at Scripps (La Jolla, CA). TZM-bl cells (also called JC53BL-13; NIH AIDS Research and Reference Reagent Program, no. 8129) are a HeLa cell derivative engineered by amphotropic retroviral transduction to express CD4, CXCR4, and CCR5 (35) and to contain Tat-responsive reporter genes for firefly luciferase (Luc) and Escherichia coli β-galactosidase (36) and are additionally engineered to express both ACE2 and TMPRSS2 (TZM-bl/ACE2/TMPRSS2 cells); these were kindly provided by Mike Farzan and Huihui Mu at Scripps.
Viruses.
All assays were performed under BSL-2 conditions unless noted differently.
(i) Live SARS-CoV-2. Live recombinant SARS-CoV-2-nanoLuc virus (rSARS-CoV-2-nLuc) was prepared as described elsewhere (14).
(ii) VSV pseudovirus. VSV pseudovirus was prepared using a codon-optimized SARS-CoV-2 spike protein (YP_009724390.1) and a VSV(G*ΔG-luciferase) system purchased from Kerafast (Boston, MA) (18, 37). VSV(G*ΔG-luciferase) pseudotyped with SARS-CoV-2 spike (PsVSV-Luc-D19) was produced in 293T cells and stored at −80°C. Median tissue culture infectious dose (TCID50) was measured using Vero cells (CCL-81; ATCC) with serial 2-fold dilutions of the prepared pseudovirus.
(iii) LV pseudoviruses. An expression plasmid encoding codon-optimized full-length spike of the Wuhan-1 strain (VRC7480) was provided by Barney Graham and Kizzmekia Corbett at the Vaccine Research Center, National Institutes of Health (USA). The D614G mutation was introduced into VRC7480 by site-directed mutagenesis using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) (LV-pseudo). Pseudovirions were produced in HEK 293T/17 cells (CRL-11268; ATCC). Culture supernatants from transfections were clarified of cells by low-speed centrifugation and filtration (0.45-μm filter) and were stored in 1-ml aliquots at −80°C.
For the HTS format of the LV pseudovirus assay, the pseudovirus was prepared in 293T cells using a five-plasmid system as described in (38). Lentiviral backbone plasmids and SARS-CoV-2 spike (Wuhan-1, D614G) vector were provided by Jessy Bloom at Fred Hutch.
Detection of IgG antibodies to SARS-CoV-2 using a commercial serologic assay.
Plasma samples were tested at the Clinical Laboratory Improvement Amendments (CLIA)-certified University of Washington Virology lab using the Architect SARS-CoV-2 IgG assay (Abbott) under the FDA’s emergency use authorization. The assay is a chemiluminescent microparticle immunoassay that measures IgG antibodies to the SARS-CoV-2 nucleocapsid protein. Qualitative results and index values reported by the instrument were used in analyses. A recommended index value cutoff of 1.40 was used for determining positivity (39).
Luminex SARS-CoV-2 IgG binding antibody assay.
Detailed descriptions can be found in the supplemental material. Two replicate dilutions of plasma were incubated with MagPlex beads conjugated with SARS-CoV-2 spike, RBD, nucleoprotein, and tetanus toxoid followed by incubation with anti-human IgG Fc-phycoerythrin (Fc-PE) (Southern Biotech, Birmingham, AL). Background was established by measuring the mean fluorescence intensity (MFI) of beads conjugated to antigens incubated in assay buffer and was subtracted from all readings. Pooled sera collected in 2015 to 2016 from normal human donors was included as the negative control for SARS-CoV-2 antigens. Convalescent plasma from a subject with PCR-confirmed severe COVID-19 was used as a positive control.
Concentration of antigen-specific IgG was estimated using a standard curve based on the measurement of MFI for serial dilutions of standard IgG (Sigma, St. Louis, MO) captured by MagPlex beads conjugated with goat anti-human Ig Fab-specific antibody (Southern Biotech). MFI readings and associated IgG concentrations were fitted to a four-parameter logistic curve (4PL) using the R packages nCal and drc.
Live SARS-CoV-2 neutralization assay.
All the live virus experiments were performed under BSL-3 conditions at negative pressure by operators in Tyvek suits wearing personal powered air-purifying respirators. Vero E6 cells were seeded at 2 × 104 cells/well in a 96-well plate 24 h before the assay was performed. An 8-point, 3-fold dilution curve was generated for each sample with a starting concentration of 1:50. Seventy-five PFU of rSARS-CoV-2-nLuc (14) was mixed with individual patient plasma at a 1:1 ratio and incubated at 37°C for 1 h; after that, virus was added to cells and incubated at 37°C in 5% CO2 for 48 h. Luciferase was measured as relative luminescence units (RLU) by a Nano-Glo luciferase assay system (Promega, Madison, WI) following manufacturer protocols using a SpectraMax M3 luminometer (Molecular Devices, San Jose, CA). Percent neutralization was calculated by the following equation: (1 − [RLU with sample/RLU with mock treatment]) × 100. Mouse serum produced by BALB/c mice immunized with SARS-CoV-2 spike was used as a positive control (14).
VSV pseudovirus neutralization assay.
Vero cells were seeded at 2 × 104 cells/well in black-walled 96-well plates 24 h before the assay was performed. A 7-point, 3-fold dilution curve was generated with a starting sample dilution of 1:20. PsVSV-Luc-D19 (3.8 × 102 TCID50) was mixed with the plasma dilutions, incubated at 37°C in 5% CO2 for 30 min, and then transferred onto Vero cells. Cells were incubated for 18 to 20 h. Luciferase activity was measured by a Bio-Glo luciferase assay system (Promega) using a 2030 Victor X3 multilabel reader (PerkinElmer, Waltham, MA). Percent virus neutralization was calculated as in the live virus assay. Plasma collected from a subject with severe, PCR-confirmed SARS-CoV-2 infection collected after the person was released from the hospital was used as a positive control. Pooled human serum collected in 2015 to 2018 was used as a negative control.
LV pseudovirus neutralization assays.
(i) 293T/ACE2 cells pseudovirus assay. A pretitrated dose of LV-pseudo was incubated with serial 3-fold dilutions of plasma in duplicate for 1 h at 37°C in 96-well plates. Freshly trypsinized 104 293T/ACE2 cells were added to each well. One set of control wells received cells + virus (virus control) and another set received cells only (background control). After 68 to 72 h of incubation, 100 μl of cell lysate was transferred to a 96-well plate for measurements of luminescence using the Promega luciferase assay system (Promega).
(ii) ACE2/TMPRSS2 TZM-bl cells pseudovirus assay. The assay was carried out similar to the 293T/ACE2 cell pseudovirus assay with the exception that the growth medium used for infection of TZM-bl/ACE2/TMPRSS2 cells contained 75 μg/ml DEAE dextran. After 68 to 72 h of incubation, 100 μl of cell lysate was transferred to a 96-well plate (Costar) for luminescence measurement using the BriteLite luminescence reporter gene assay system (PerkinElmer). Percent virus neutralization was calculated as previously mentioned.
For both LV pseudovirus assays, SARS-CoV-2 neutralizing monoclonal antibody COVA1-18 (40) was used as a positive control, and normal human serum collected in 2016 was used as a negative control.
(iii) HTS version of 293T/ACE2 cells pseudovirus assay. An HTS SARS-CoV-2 neutralization assay was performed in the CLIA-certified University of Washington Virology lab using a Mantis liquid handler (Formulatrix, Bedford, MA) to dispense growth medium, virus, and luciferase substrate. The 293T/ACE2 cells were seeded in 96-well black-walled plates manually at 12,500 cells/well and were incubated for 16 to 18 h. Various amounts of growth medium were dispensed into 96-well plates using Mantis according to the plate map. In the plates with growth medium, patient sera were manually diluted 10-fold followed by six 3-fold serial dilutions for a total of seven dilution points at 60 μl of sample per well. Mantis was then used to dispense 60 μl of diluted pseudovirus at 4 × 105 RLU/well into the 96-well plates with serially diluted serum samples. After incubating at 37°C for 1 h, 100 μl of the pseudovirus and serum mixture was manually added to the 293T/ACE2 cells in 96-well plates. At 52 to 58 h postinfection, 100 μl of medium was manually removed from each well and 30 μl of Bright-Glo luciferase substrate was added by the Mantis. The plates were read with a Victor Nivo multimode microplate reader (PerkinElmer).
For all neutralization assays, neutralization titers are the reciprocal of the plasma dilution at which RLU were reduced by 50% (ND50) and 80% (ND80) compared to virus control wells after subtraction of background RLU.
SARS-CoV-2 surrogate virus neutralization test.
The SARS-CoV-2 surrogate virus neutralization test (sVNT) was performed in a BSL-1 laboratory and was performed according to manufacturer’s (GenScript, Piscataway, NJ) protocol recommendations. Briefly, capture plates were incubated with plasma samples diluted 1:10, washed, and probed with secondary antibody conjugated to horseradish peroxidase. Plates were developed with 3,3′,5,5′-tetramethylbenzidine (ThermoFisher, Waltham, MA), and optical density (OD) at 450 nm was measured using a SpectraMax M2 reader (Molecular Devices). Positive and negative controls were provided in the kit. Binding inhibition was determined using the following formula: inhibition = (1 – [OD of sample/OD of negative control]) × 100. Percent binding inhibition was interpreted as a percent neutralization. In order to determine ND50, plasma samples were serially diluted starting from 1:10, and the assay was performed as described above.
Assay calibration with the WHO anti-SARS-CoV-2 immunoglobulin standard.
The First WHO International Standard for anti-SARS-CoV-2 antibodies developed and distributed by the National Institute for Biological Standards and Control (NIBSC) of the United Kingdom (number 20/136) was used to establish calibrating factors for VSV-pseudo/Vero, LV-pseudo/293T, and HTS-LV-pseudo/293T assays as follows. The lyophilized standard was reconstituted in ultrapure water as per NIBSC instructions. The resulting serum was stored at 4°C for no longer than 1 week and was used in the assays similar to as described above for patient samples via serial dilutions starting at 1:20. The ratio between the assigned neutralization unitage (1,000 IU/ml) and measured ND50 and ND80 for the standard sample was used as a calibrating factor to convert assay-derived ND50 and ND80 readouts into IU/ml.
Statistical analysis and visualization.
Neutralization titers were defined as the plasma dilution that reduced RLU by 50% (ND50) or 80% (ND80) relative to virus control wells (cells + virus only) after subtraction of background RLU in cell-only control wells (see supplemental material for details). Correlations were estimated between pairs of neutralization or binding antibody readouts using Pearson’s correlation coefficient (r), and group means were compared using a paired two-sample t test; measures in units of neutralization and IgG concentration were logged before estimating correlation and comparing group means. Association of neutralization and IgG concentration with age and body mass index (BMI) were conducted using Spearman’s rank correlation. Statistical significance was based on a P value of <0.05.
RESULTS
Cohort characteristics, demographics, survey participation, and serological testing.
To characterize and compare different platforms of SARS-CoV-2 nAb assays, we used plasma samples obtained from a seroepidemiology study conducted May 4 to 19, 2020, following a county-wide outbreak of SARS-CoV-2 in Blaine County, ID. Of 967 participants, 222 (22.8%) had IgG antibodies to SARS-CoV-2 nucleoprotein as measured by the Abbott Architect test (index value of ≥1.40) indicative of prior infection with SARS-CoV-2. From these 222 samples, we randomly selected 40 plasma samples for use in evaluating SARS-CoV-2 neutralization assays. Selected participants had a median age of 51.5 years (range, 23 to 81), and 60% identified as female (Table 1). Only one participant reported being hospitalized, and four participants (10%) were self-described as asymptomatic. Among participants reporting different symptoms, 57.5% had fever, while fatigue (87.5%), cough (72.5%), headache (67.5%), and chills (65%) were more prevalent (Table S1 in the supplemental material). The majority of participants reported COVID-19 symptoms occurring in March of 2020. Based on this, our cohort can be categorized as representing mild to moderate symptomatic COVID-19 infections with samples collected at about 1.5 to 2 months after disease onset.
TABLE 1.
Demographic and exposure/symptom characteristics of study participants
| Characteristic | n | % |
|---|---|---|
| Age (yrs) | ||
| 23–40 | 8 | 20 |
| 41–50 | 11 | 27.5 |
| 51–60 | 11 | 27.5 |
| 61–70 | 6 | 15 |
| >70 | 4 | 10 |
| Median | 51.5 | |
| Range | 23–81 | |
| Gender | ||
| Female | 16 | 40 |
| Male | 24 | 60 |
| Exposures/symptoms | ||
| Tested positive | 8 | 20 |
| Symptomatic contact of known positive | 9 | 22.5 |
| Symptomatic without confirmation | 19 | 47.5 |
| Asymptomatic contact of someone symptomatic | 2 | 5 |
| Asymptomatic, no exposures | 2 | 5 |
| Travel outside United States since 1 December 2019 | 7 | 17.5 |
| Other | ||
| Essential worker | 6 | 15 |
| Lives with children | 14 | 35 |
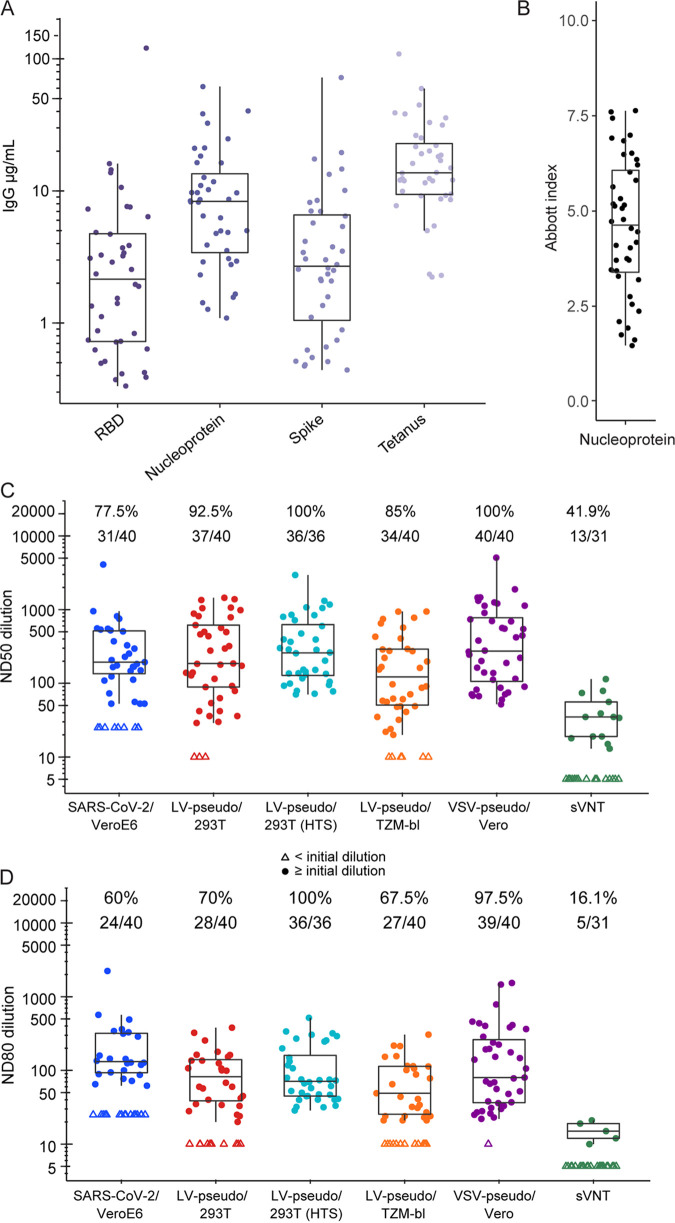
We measured the concentration of IgG in participant sera targeting SARS-CoV-2 spike, RBD, and nucleoprotein via a quantitative, Luminex-based immunoassay. IgG to tetanus toxoid was measured as a proxy for overall IgG level. IgG to spike and RBD were detected in all 40 plasma samples, indicating a seroconversion on all SARS-CoV-2 antigens. The mean plasma concentrations for spike- and RBD-specific IgG were 2.8 μg/ml (95% confidence interval [CI], 1.9 to 4.1) and 2.1 μg/ml (95% CI, 1.4 to 3.3), respectively, which were considerably lower than those to nucleoprotein (7.3 μg/ml [95% CI, 5.3 to 10]) (Fig. 1A). The concentration of tetanus-specific IgG was higher than IgG targeting SARS-CoV-2 antigens for all individuals (mean, 14.5; 95% CI, 11.1 to 18.9). Although the Abbott SARS-CoV-2 IgG CLIA test is designed and used for qualitative detection of IgG against the SARS-CoV-2 nucleoprotein, the instrument reports index values that can be used in quantitative analyses (Fig. 1B) (39).
FIG 1.
SARS-CoV-2 neutralization and binding antibody concentration from COVID-19 convalescent patients. (A) Concentration of IgG against SARS-CoV-2 spike, RBD, nucleoprotein, and tetanus toxoid measured in the Luminex binding antibody assay. (B) Indexes reported by the Abbott Architect nucleoprotein IgG test. (C) ND50 and (D) ND80 neutralization titer measured using five SARS-CoV-2 neutralization assays for 40 plasma samples from 40 participants. Each assay defined its own lower limit of detection (LOD) based on the initial dilution: 50-fold for SARS-CoV-2/VeroE6, 20-fold for the LV and VSV pseudovirus assays, and 10-fold for the sVNT. Data below the LOD (open triangles) are plotted at LOD/2. Number and percentage of samples above the LOD are indicated above each plot. For each assay, the box represents the extent of the interquartile range (IQR) with a line indicating the median; whiskers extend to 1.5 times the IQR.
Cell-based assays provided comparable estimates of neutralization activity.
Forty selected plasma samples were distributed across four laboratories conducting different SARS-CoV-2 neutralization assays (Table 2). Serial plasma dilutions were used in cell-based assays to generate titration curves (Fig. S1) and estimate the 50% and 80% neutralizing dilutions (ND50 and ND80, respectively). In the sVNT, only 22 of 40 samples showed neutralization above 50% when analyzed according to the manufacturer’s protocol in a single 1:10 dilution (Fig. S2A), and 13 samples representing different neutralization capacity were selected for ND50 measurement using serial dilutions (Fig. S2B).
TABLE 2.
SARS-CoV-2 neutralization assay platforms used in the study
| SARS-CoV-2/VeroE6 | VSV-pseudo/Vero | LV-pseudo/293T | LV-pseudo/TZM-bl | HTS-LV-pseudo/293T | Surrogate virus neutralization test (sVNT) | |
|---|---|---|---|---|---|---|
| Lab | Baric | Corey | Montefiori | Montefiori | Huang/Jerome | Corey |
| Cell line | Vero E6 | Vero | HEK293T | TZM-bl | HEK293T | None |
| ACE2 expression | Endogenous | Endogenous | Engineered | Engineered | Engineered | Recombinant |
| TMPRSS2 expression | No | No | No | Engineered | No | NAa |
| Virus shorthand | rSARS-CoV-2-nLuc | VSV-pseudo | LV-pseudo | LV-pseudo | HTS-LV-pseudo | NA |
| Virus type | Live recombinant | VSV(G*ΔG-luciferase) pseudotyped | pCMV-ΔR8.2 lentiviral packaging with pHR'-CMV-Luc | pSG3ΔEnv lentiviral packaging | pHDM lentiviral packaging | NA |
| SARS-CoV-2 strain/isolate | WA-CDC-WA1-A12/2020 | Wuhan-Hu-1 | Wuhan-Hu-1 (VRC7480) D614G | Wuhan-Hu-1 (VRC7480) D614G | Wuhan-Hu-1 D614G | Unknown |
| GenBank | MT020880.1 | MN908947.3 | MN908947.3 | MN908947.3 | MN908947.3 | NA |
| Amino acid 614 | D | D | G | G | G | Unknown |
| Biosafety level | 3 | 2 | 2 | 2 | 2 | 1 |
NA, not available.
Overall, the cell-based assays showed comparable estimated ND50 geometric mean titers (GMTs) with considerable overlap in the interquartile range and 95% CI among pairs of assays (Fig. 1C; see also Fig. S3A and Table S2). The SARS-CoV-2/VeroE6 and LV-pseudo/293T assays yielded very similar ND50 GMT values, 141 (95% CI, 93 to 214) versus 178 (95% CI, 112 to 283), respectively. The estimated mean ND50 values for other cell-based assays were also comparable; however, ND50 GMTs for VSV-pseudo/Vero test and HTS-LV-pseudo/293T were the highest among cell-based assays (310 [95% CI, 211 to 454] and 272 [95% CI, 267 to 643], respectively; Table S2). The live virus assay and all three LV pseudovirus assays yielded ND50 values within a 2-fold range, indicating high concordance. Notably, rSARS-CoV-2-nLuc and PsVSV-Luc-D19 contained the spike protein with an aspartate residue at position 614 (Wuhan-1 strain), while the LV pseudoviruses contained spike protein with the D614G mutation. Nevertheless, the difference between outcomes of LV-pseudo/293T assays in regular and HTS formats and the VSV-pseudo/Vero assay were within 2-fold. The lowest ND50 GMT (from the LV-pseudo/TZM-bl assay) was 3.4-fold lower than that of the highest-yielding assay (VSV-pseudo/Vero).
Despite overlapping distributions, it was possible to detect shifts in ND50 for each cell-based assay using a two-sample, paired t test (P < 0.05); the exceptions were SARS-CoV-2/VeroE6 versus LV-pseudo/293T and HTS-LV-pseudo/293T versus VSV-pseudo/Vero. The ND50 GMTs in these two assay pairs were not significantly different (P = 0.112 and 0.856, respectively). Taken together, these data demonstrate that the variability across different cell-based assays is low, and it suggests that results of cell-based assays could be adjusted for head-to-head comparability, as we show more in the next section. In contrast, the sVNT yielded significantly different ND50 values, up to 26-fold lower than cell-based assays.
Differences and similarities among the cell-based assay ND50 values were generally recapitulated using the ND80 values (Fig. 1D; see also Fig. S3B and Table S2). The sVNT and VSV-pseudo/Vero assays yielded the lowest and the highest ND80 GMTs, respectively. However, the overall difference between ND80 values was less dramatic than for ND50. For all cell-based assays, it was within a 3-fold range, and sVNT ND80 was only 6- to 17-fold lower than cell-based assays. As expected, the ND80 titers were consistently lower than the ND50 titers (Fig. S4, Table S3). For other pseudovirus assays, the difference between ND50 and ND80 was greater and ranged between 3-fold and 4.6-fold (Table S3). Interestingly, the live virus assay showed the smallest difference between ND50 and ND80 GMTs, 1.95-fold (Table S3), a direct consequence of the steeper titration curves observed for this assay (Fig. S1A). Indeed, the slope parameter from the live virus neutralization curves was higher than in other assays (slope B = 3.3 versus 0.6, 1.4, and 1.5 for LV-pseudo/293T, LV-pseudo/TZM-bl, and VSV-pseudo/Vero, respectively; all P < 0.001).
The neutralization assay response rate was in agreement with estimated ND50 and ND80 values. The highest response rates came from the VSV-pseudo/Vero assay (100% of ND50 and 97.5% of ND80 titers) and HTS-LV-pseudo/293 assays (100% of ND50 and ND80 titers) (Fig. 1C and D). The lowest response rate among the cell-based assays was measured via the SARS-CoV-2/VeroE6 assay, and the sVNT was the lowest overall. For the SARS-CoV-2/VeroE6 assay, the lower response rate than other cell-based assays could be due to the starting plasma dilution, which was 1:50 versus 1:20 used in the pseudovirus assays.
Strong correlation among neutralization assays.
We conducted a correlation analysis of the ND50 and ND80 values derived from each of the five neutralization assays (Fig. 2; Fig. S5). The live virus and all four pseudovirus neutralization assays generated ND50 values that were highly correlated across samples (Pearson r = 0.78 to 0.89), with the highest correlation observed between the three LV pseudovirus assays (r = 0.89; 95% CI, 0.81 to 0.94; P < 0.001). The readout with the lowest correlation with the cell-based assays was the sVNT ND50 (r = 0.32 to 0.6), although sVNT percent neutralization tended to be more highly correlated (r = 0.73 to 0.8). Similar correlations were observed for ND80 outcomes for cell-based assays (r = 0.69 to 0.88) (Fig. 2B).
FIG 2.
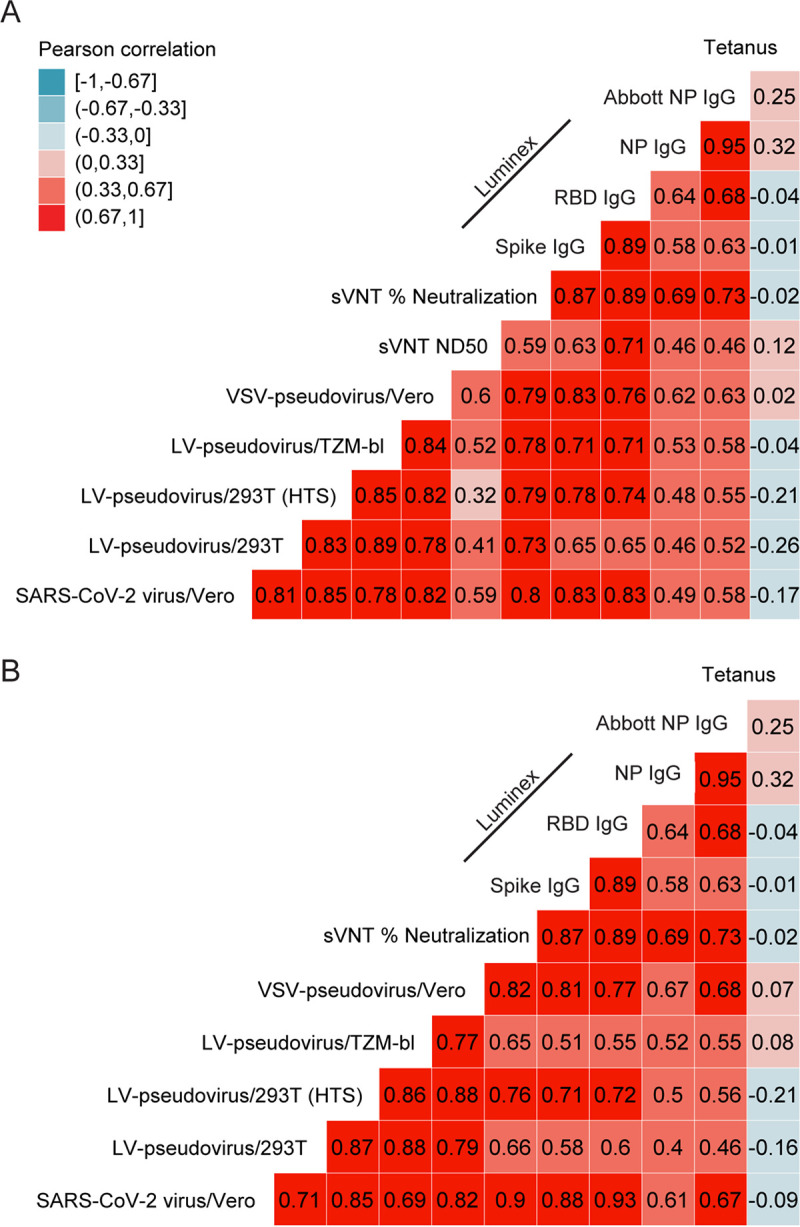
Correlation among assay readouts measuring neutralization or antigen-specific IgG concentration in plasma. Heat map color is determined by the Pearson’s correlation coefficient (r, annotations). Each panel includes either ND50 titers (A) or ND80 titers (B) and their correlation with sVNT percent neutralization, SARS-CoV-2-specific IgG concentration (Luminex bead-based assay), the quantitative index of the Abbott nucleoprotein assay, and tetanus toxoid-specific IgG concentration. ND50 and ND80 values below 50 were truncated at 25.
Plasma neutralization potency correlated with concentration of SARS-CoV-2 binding IgG.
Correlation analyses revealed a strong association between levels of IgG to spike and RBD (r = 0.89; 95% CI, 0.81 to 0.94) (Fig. 2). Luminex immunoassay-measured nucleoprotein-specific IgG highly correlated with the quantitative index of the Abbott SARS-CoV-2 IgG assay (r = 0.95; 95% CI, 0.91 to 0.97), which is based on detection of nucleoprotein-specific IgG, but both parameters only moderately correlated with IgG targeting the other viral antigens (r = 0.58 to 0.68). There was no significant correlation between tetanus-specific IgG and IgG to SARS-CoV-2 antigens (all P > 0.05).
Next, we examined the relationship between virus neutralization and IgG levels to spike, RBD, and nucleoprotein. IgG concentrations to each antigen positively correlated with the ND50 titer measured by each neutralization assay (r = 0.46 to 0.83) (Fig. 2A; Fig. S6). The strongest correlation was observed between sVNT percent neutralization and concentration of RBD IgG (r = 0.89). Among the cell-based assays, the live virus ND50 titer showed the strongest correlation with IgG against spike and RBD (r = 0.83 for both), followed by the VSV-pseudovirus/Vero assay (r = 0.83 and 0.76, respectively). Notably, nucleoprotein-specific IgG only moderately correlated with ND50 titers from the cell-based assays but showed a strong correlation with sVNT percent neutralization. Tetanus-specific IgG did not correlate with any of the SARS-CoV-2-associated IgG concentrations or neutralization titers.
With the caveat that our cohort is rather small for such analyses, we found a moderately positive correlation between age and concentration of spike-specific IgG (Spearman’s rho = 0.37, P = 0.02), RBD-specific IgG (rho = 0.39, P = 0.013), and nucleoprotein-specific IgG (rho = 0.45, P = 0.003) (Table S4). Similarly, there were positive correlations between age and neutralization titer (Table S4, Fig. S7), although the correlations tended to be higher with ND80 titer (rho = 0.51, P = 0.001) than with ND50 titer (rho = 0.28, P = 0.075).
Assay calibration with the WHO anti-SARS-CoV-2 immunoglobulin standard.
To evaluate readout conversion between assays, we calibrated VSV-pseudo/Vero, LV-pseudo/293T, and HTS-LV-pseudo/293T using the First WHO International Standard for anti-SARS-CoV-2 antibodies (Table 3). After conversion, the regular and HTS version of the LV-pseudo assay reported the same ND50 GMT of 58.4 IU/ml. Of note, raw ND50 titers for these assays also showed high concordance and had a less than 2-fold difference. GMT ND50 from the VSV-pseudo/Vero assay was found at 205 IU/ml after calibration. If before calibration the difference in ND50 titers was about 2-fold between VSV and LV pseudovirus assays, after calibration, it increased up to 3.5-fold. Conversion of ND80 GMTs into IU/ml format produced perplexing results. The ND80 value after calibration became greater than ND50 for both LV-pseudo assays and was almost equal to ND50 for the VSV-pseudo assay (Table 3).
TABLE 3.
Calibration of SARS-CoV-2 neutralization assays using first WHO standard for anti-SARS-CoV-2 immunoglobulin
| First WHO International Standard for anti-SARS-CoV-2 immunoglobulin | VSV-pseudo/Vero |
LV-pseudo/293T |
HTS-LV-pseudo/293T |
|||
|---|---|---|---|---|---|---|
| ND50 | ND80 | ND50 | ND80 | ND50 | ND80 | |
| WHO standard, GMT | 1,511 | 557 | 3,047 | 567 | 4,650 | 1,396 |
| Calibration factor (1,000 IU/ml ÷ standard GMT) | 0.662 | 1.795 | 0.328 | 1.764 | 0.215 | 0.716 |
| GMT neutralization titer among participants | 309.7 | 102.8 | 177.9 | 41.96 | 271.7 | 86.3 |
| Calibrated readout (IU/ml) | 205 | 184.5 | 58.4 | 74.0 | 58.4 | 61.8 |
To provide context for our data, we accessed the WHO report that established their reference standard (41) and analyzed the GMTs that were contributed by different research groups using a range of assays and reference samples. We pooled measurements from LV pseudovirus and VSV pseudovirus assays and calculated their respective ND50 GMT values. Of note, most of the LV and VSV pseudovirus assays used for establishing the WHO standard contained the Wuhan-1 D614 spike (41). The calculated GMT for the WHO standard was 1,347 for the VSV pseudovirus assay and 3,406 for the LV pseudovirus assay and were similar to the cognate values from our study (Table 3).
To test possible influence of D614G mutation on the assay readout, we tested VSV pseudovirus carrying D614 versus G614 using the WHO standard and found no difference in ND50 or ND80 titers between virus variants (Fig. S8). Therefore, the difference in readouts between VSV and LV platforms is either due to the target cells or the virus used for pseudotyping.
DISCUSSION
In this study, we conducted a detailed comparison of four cell-based and one ELISA-based SARS-CoV-2 neutralization assays using a set of 40 plasma samples collected from SARS-CoV-2 convalescent individuals with mild to moderate disease. Our data show a high level of congruency among cell-based assays, suggesting that the results obtained with any of the tested pseudovirus platforms accurately reflect the potency of the sample to neutralize the Wuhan-Hu-1 strain of SARS-CoV-2. The 50% and 80% neutralization titers strongly correlate between different assays as well as between the neutralization assays and plasma concentration of RBD and spike-specific IgG, which is consistent with other studies (19, 42–46). Although the correlation was modest in comparison, the ELISA-based sVNT results also positively correlated with the other neutralization assays. The demonstrated differences in ND50 and ND80 GMTs between assays should be considered when conducting SARS-CoV-2 natural history studies and vaccine trials.
Although levels of spike-specific IgG highly correlated with neutralization, our data do not confirm that all IgG targeting the spike protein have neutralization activity. Rather, the results imply that individuals who produce spike-specific binding antibodies are also likely to make neutralizing IgG. The correlation between nucleoprotein-specific IgG and neutralization was consistently lower than the correlations between spike- and RBD-specific IgG with neutralization. This is not surprising, as much of the immunodominant response associated with neutralization involves binding and/or blocking the spike RBD to inhibit viral entry to host cells (26, 28, 47).
The association of age with both spike-specific IgG and neutralization titer suggests that the previously reported association of high neutralization titer among older individuals may be mediated by higher concentrations of spike and RBD-specific IgG (48, 49). However, this is not a result of cross-reactive humoral responses to prior infections with seasonal coronaviruses (50, 51). Whether this is a direct effect of age on the developing immune response to SARS-CoV-2 or a result of cross-reacting T-cell immunity remains unclear (52, 53). Although our cohort was well balanced by sex and age and a positive correlation between age and neutralizing titers was detected, the influence of other demographic and environmental factors cannot be excluded due to a small sample size collected in a limited geographic origin (54, 55). As such, larger, geographically distinct cohorts are required for proper analyses as, for example, a recent publication showing good congruency across geographically distant laboratories with the VSV pseudovirus assay (56).
Our study shows that ELISA-based surrogate assays have two major limitations: (i) inability to account for synergistic action of antibodies targeting different epitopes, and (ii) limit detection to only antibodies that block the RBD/ACE2 interaction, thus missing other antibodies that neutralize via non-RBD sites on the virus glycoprotein (27, 57). In fact, synergistic action of antibodies targeting the RBD and S2 domain has been reported (58). Thus, surrogate assays have a lower sensitivity than cell-based assays and can lead to more false-negative results. These results contradict the use of sVNT as a rapid assay to select positive samples for further screening with cell-based assays, as was recently suggested (59).
TMPRSS2 was shown to be essential for SARS-CoV-2 infectivity of different cell types, although there was no significant difference observed in virus titer at 48 h postinfection between wild-type Vero cells and Vero cells expressing furin (14, 24). Our comparison revealed that the presence of TMPRSS2 is not critical for assay performance, as TZM-bl cells expressing both ACE2 and TMPRSS2 showed no significant difference compared to 293T cells expressing only ACE2.
In addition to lower safety requirements than assays using replication-competent SARS-CoV-2, pseudotyped virus assays are well positioned for HTS testing of antibody responses elicited by natural infection, vaccination, and now, critically, to new viral variants of concern (VOC), as reported for other viruses (60–62). One limitation of the current study was that only two strains, Wuhan and D614G, were tested. The D614G mutation has been shown to be moderately more susceptible to neutralization (63, 64), while in other reports no difference was observed (65, 66). Our results indicate no pattern between D614G and neutralization capability, and thus the marginal differences observed between assays are likely due to assay sensitivity rather than viral sequence. Neutralization of recently emerging VOCs (67) by serum and monoclonal antibodies has been measured using several assay platforms (68–70); however, assay standardization and validation will be required for proper comparisons.
Use of the First WHO International Standard for anti-SARS-CoV-2 immunoglobulin clearly demonstrated that ND50 GMTs measured with the same assay platform in different laboratories and at different throughputs are highly concordant despite pseudotyped virions using different spike proteins. However, comparisons across assay platforms are not straightforward, and the discordance of GMTs between LV pseudovirus and VSV pseudovirus neutralization even after calibration demonstrates that direct conversion from one to the other may require further calibration using multiple samples covering a broad range of neutralization potency.
Compliance with good clinical laboratory practices is required to ensure that assay results are as reliable as possible (71). Therefore, further assay optimization and subsequent validation addressing how a range of test conditions affect assay specificity, precision, linearity, accuracy, limit of detection, limit of quantitation, and robustness will be required before all or one of the methods evaluated in our study will be transferable between laboratories and suitable for even greater throughput in a 384-well format used for clinical trial testing (62, 72, 73).
SARS-CoV-2 is predicted to remain circulating in the global population for many years due to emerging new strains and incomplete vaccine delivery and uptake (74). Therefore, monitoring of acute and convalescent infection and the broad spectrum of immunity against SARS-CoV-2 both in natural infection and after vaccination will become a routine task for clinical microbiology/virology facilities. Selection of a SARS-CoV-2 neutralization assay and the ability to compare results obtained using different assays will remain a crucial issue.
ACKNOWLEDGMENTS
We thank Bill McLaughlin, Hollie Bearce, and Terry O’Connor for sample collection and Sara Thiebaud for assistance with data management and analysis.
This work was funded by NIAID Service Agreement 225472-99 to R.K.S., R01AI134878 and UM1AI068614 to L.C., and a Fred Hutch Evergreen grant to A.M.S.
H.K., A.P., and F.A.L. are employees of Vir Biotechnology and may hold shares in Vir Biotechnology. L.C. is a founder of Vir Biotechnologies. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Anton M. Sholukh, Email: asholukh@fredhutch.org.
Lawrence Corey, Email: lcorey@fredhutch.org.
Angela M. Caliendo, Rhode Island Hospital
REFERENCES
- 1.Plotkin SA. 2010. Correlates of protection induced by vaccination. Clin Vaccine Immunol 17:1055–1065. 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KH, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SP, Carnahan RH, CroweJE, Jr., Bloom JD. 2021. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 29:44–57. 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F, Wang A, Liu M, Wang Q, Chen J, Xia S, Ling Y, Zhang Y, Xun J, Lu L, Jiang S, Lu H, Wen Y, Huang J. 2020. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv 10.1101/2020.03.30.20047365. [DOI] [Google Scholar]
- 4.Huang AT, Garcia-Carreras B, Hitchings MDT, Yang B, Katzelnick LC, Rattigan SM, Borgert BA, Moreno CA, Solomon BD, Trimmer-Smith L, Etienne V, Rodriguez-Barraquer I, Lessler J, Salje H, Burke DS, Wesolowski A, Cummings DAT. 2020. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun 11:14704. 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, Liao P, Qiu JF, Lin Y, Cai XF, Wang DQ, Hu Y, Ren JH, Tang N, Xu YY, Yu LH, Mo Z, Gong F, Zhang XL, Tian WG, Hu L, Zhang XX, Xiang JL, Du HX, Liu HW, Lang CH, Luo XH, Wu SB, Cui XP, Zhou Z, Zhu MM, Wang J, Xue CJ, Li XF, Wang L, Li ZJ, Wang K, Niu CC, Yang QJ, Tang XJ, Zhang Y, Liu XM, Li JJ, Zhang DC, Zhang F, Liu P, Yuan J, Li Q, Hu JL, Chen J, Huang AL. 2020. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 26:845–848. 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 6.Larson D, Brodniak SL, Voegtly LJ, Cer RZ, Glang LA, Malagon FJ, Long KA, Potocki R, Smith DR, Lanteri C, Burgess T, Bishop-Lilly KA. 2020. A case of early re-infection with SARS-CoV-2. Clin Infect Dis 2020:ciaa1436. 10.1093/cid/ciaa1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.To KK-W, Hung IF-N, Ip JD, Chu AW-H, Chan W-M, Tam AR, Fong CH-Y, Yuan S, Tsoi H-W, Ng AC-K, Lee LL-Y, Wan P, Tso E, To W-K, Tsang D, Chan K-H, Huang J-D, Kok K-H, Cheng VC-C, Yuen K-Y. 2020. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis 2020:ciaa1275. 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, Perry C, Pan C, Hosain R, Mahmood A, Davis JD, Turner KC, Hooper AT, Hamilton JD, Baum A, Kyratsous CA, Kim Y, Cook A, Kampman W, Kohli A, Sachdeva Y, Graber X, Kowal B, DiCioccio T, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD, Trial Investigators. 2021. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med 384:238–251. 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levi-Schaffer F, de Marco A. 2021. COVID-19 and the revival of passive immunization: antibody therapy for inhibiting SARS-CoV-2 and preventing host cell infection: IUPHAR review: 31. Br J Pharmacol 10.1111/bph.15359. [DOI] [PubMed] [Google Scholar]
- 10.Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, Bibi S, Briner C, Cicconi P, Collins AM, Colin-Jones R, Cutland CL, Darton TC, Dheda K, Duncan CJA, Emary KRW, Ewer KJ, Fairlie L, Faust SN, Feng S, Ferreira DM, Finn A, Goodman AL, Green CM, Green CA, Heath PT, Hill C, Hill H, Hirsch I, Hodgson SHC, Izu A, Jackson S, Jenkin D, Joe CCD, Kerridge S, Koen A, Kwatra G, Lazarus R, Lawrie AM, Lelliott A, Libri V, Lillie PJ, Mallory R, Mendes AVA, Milan EP, Minassian AM, McGregor A, et al. 2021. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 397:99–111. 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T, COVE Study Group. 2021. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 384:403–416. 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, C4591001 Clinical Trial Group. 2020. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 383:2603–2615. 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muruato AE, Fontes-Garfias CR, Ren P, Garcia-Blanco MA, Menachery VD, Xie X, Shi P, Fontes-Gar CR, Ren P, Garcia-Blanco MA, Menachery VD, Xie X, Shi P. 2020. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat Commun 11:4059. 10.1038/s41467-020-17892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, Kato T, Lee RE, Yount BL, Mascenik TM, Chen G, Olivier KN, Ghio A, Tse LV, Leist SR, Gralinski LE, Schäfer A, Dang H, Gilmore R, Nakano S, Sun L, Fulcher ML, Livraghi-Butrico A, Nicely NI, Cameron M, Cameron C, Kelvin DJ, de Silva A, Margolis DM, Markmann A, Bartelt L, Zumwalt R, Martinez FJ, Salvatore SP, Borczuk A, Tata PR, Sontake V, Kimple A, Jaspers I, O’Neal WK, Randell SH, Boucher RC, Baric RS. 2020. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182:429–446. 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q, Liu Q, Huang W, Li X, Wang Y. 2018. Current status on the development of pseudoviruses for enveloped viruses. Rev Med Virol 28:e1963. 10.1002/rmv.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carnell GW, Ferrara F, Grehan K, Thompson CP, Temperton NJ. 2015. Pseudotype-based neutralization assays for influenza: a systematic analysis. Front Immunol 6:161. 10.3389/fimmu.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao G, Du L, Ma C, Li Y, Li L, Poon VKM, Wang L, Yu F, Zheng B-J, Jiang S, Zhou Y. 2013. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol J 10:266. 10.1186/1743-422X-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitt MA. 2010. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods 169:365–374. 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang R, Huang B, A R, Li W, Wang W, Deng Y, Tan W. 2020. Development and effectiveness of pseudotyped SARS-CoV-2 system as determined by neutralizing efficiency and entry inhibition test in vitro. Biosaf Health 2:226–231. 10.1016/j.bsheal.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker SN, Chokkalingam N, Reuschel EL, Purwar M, Xu Z, Gary EN, Kim KY, Helble M, Schultheis K, Walters J, Ramos S, Muthumani K, Smith TRF, Broderick KE, Tebas P, Patel A, Weiner DB, Kulp DW. 2020. SARS-CoV-2 assays to detect functional antibody responses that block ACE2 recognition in vaccinated animals and infected patients. J Clin Microbiol 58:e01533-20. 10.1128/JCM.01533-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann MAG, Bar-On Y, Yang Z, Gristick HB, Gnanapragasam PNP, Vielmetter J, Nussenzweig MC, Bjorkman PJ. 2020. Nanoparticles presenting clusters of CD4 expose a universal vulnerability of HIV-1 by mimicking target cells. Proc Natl Acad Sci U S A 117:18719–18728. 10.1073/pnas.2010320117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, Nagata N, Sekizuka T, Katoh H, Kato F, Sakata M, Tahara M, Kutsuna S, Ohmagari N, Kuroda M, Suzuki T, Kageyama T, Takeda M. 2020. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A 117:7001–7003. 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren X, Glende J, Al-Falah M, de Vries V, Schwegmann-Wessels C, Qu X, Tan L, Tschernig T, Deng H, Naim HY, Herrler G. 2006. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J Gen Virol 87:1691–1695. 10.1099/vir.0.81749-0. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J, Gao Q, He C, Huang A, Tang N, Wang K. 2020. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis 7:551–557. 10.1016/j.gendis.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seydoux E, Homad LJ, MacCamy AJ, Parks KR, Hurlburt NK, Jennewein MF, Akins NR, Stuart AB, Wan YH, Feng J, Whaley RE, Singh S, Boeckh M, Cohen KW, McElrath MJ, Englund JA, Chu HY, Pancera M, McGuire AT, Stamatatos L. 2020. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity 53:98–105. 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He WT, Limbo O, Smith C, Song G, Woehl J, Yang L, Abbott RK, Callaghan S, Garcia E, Hurtado J, Parren M, Peng L, Ramirez S, Ricketts J, Ricciardi MJ, Rawlings SA, Wu NC, Yuan M, Smith DM, Nemazee D, Teijaro JR, Voss JE, Wilson IA, Andrabi R, Briney B, Landais E, Sok D, Jardine JG, Burton DR. 2020. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369:956–963. 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J, Lan J, Yuan J, Wang H, Zhao J, Zhang S, Wang Y, Shi X, Liu L, Zhao J, Wang X, Zhang Z, Zhang L. 2020. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584:115–119. 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 29.Abe KT, Li Z, Samson R, Samavarchi-Tehrani P, Valcourt EJ, Wood H, Budylowski P, Dupuis A, Girardin RC, Rathod B, Colwill K, McGeer A, Mubareka S, Gommerman JL, Durocher Y, Ostrowski M, McDonough KA, Drebot MA, Drews SJ, Rini JM, Gingras A-C, Dupuis Ii AP, Girardin RC, Rathod B, Wang J, Barrios-Rodiles M, Colwill K, McGeer A, Mubareka S, Gommerman JL, Durocher Y, Ostrowski M, McDonough KA, Drebot MA, Drews SJ, Rini JM, Gingras A-C. 2020. A simple protein-based SARS-CoV-2 surrogate neutralization assay. JCI Insight 5:e142362. 10.1172/jci.insight.142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan CW, Chia WN, Qin X, Liu P, Chen MI-C, Tiu C, Hu Z, Chen VC-W, Young BE, Sia WR, Tan Y-J, Foo R, Yi Y, Lye DC, Anderson DE, Wang L-F. 2020. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol 38:1073–1078. 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 31.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Lambson BE, Vermeulen M, van den Berg K, Rossouw T, Boswell M, Ueckermann V, Meiring S, von Gottberg A, Cohen C, Morris L, Bhiman JN, Moore PL. 2021. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 27:622–625. 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 32.Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann HH, Michailidis E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Luchsinger L, Hillyer CD, Caskey M, Robbiani DF, Rice CM, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2020. Escape from neutralizing antibodies 1 by SARS-CoV-2 spike protein variants. eLife 9:e61312. 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resende PC, Bezerra JF, de Vasconcelos RHT, Arantes I, Appolinario L, Mendonça AC, Paixao AC, Rodrigues ACD, Silva T, Rocha AS, Pauvolid-Correa A, Motta FC, Florentino Teixeira DL, de Oliveira Carneiro TF, Freire Neto FP, Herbster ID, Leite AB, Riediger IN, do Carmo Debur M, Gomes Naveca F, Almeida W, Livorati M, Bello G, Siqueira MM. 2021. Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil. Virological https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584.
- 34.Gundlapalli AV, Salerno RM, Brooks JT, Averhoff F, Petersen LR, McDonald LC, Iademarco MF, CDC COVID-19 Response. 2021. SARS-CoV-2 serologic assay needs for the next phase of the US COVID-19 pandemic response. Open Forum Infect Dis 8:ofaa555. 10.1093/ofid/ofaa555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. 10.1128/JVI.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao X, Howell KA, He S, Brannan JM, Wec AZ, Davidson E, Turner HL, Chiang C-I, Lei L, Fels JM, Vu H, Shulenin S, Turonis AN, Kuehne AI, Liu G, Ta M, Wang Y, Sundling C, Xiao Y, Spence JS, Doranz BJ, Holtsberg FW, Ward AB, Chandran K, Dye JM, Qiu X, Li Y, Aman MJ. 2017. Immunization-elicited broadly protective antibody reveals ebolavirus fusion loop as a site of vulnerability. Cell 169:891–904. 10.1016/j.cell.2017.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, Pettie D, King NP, Balazs AB, Bloom JD. 2020. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12:513. 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryan A, Pepper G, Wener MH, Fink SL, Morishima C, Chaudhary A, Jerome KR, Mathias PC, Greninger AL. 2020. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J Clin Microbiol 58:e00941-20. 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, Bentlage AEH, van Haaren MM, Guerra D, Burger JA, Schermer EE, Verheul KD, van der Velde N, van der Kooi A, van Schooten J, van Breemen MJ, Bijl TPL, Sliepen K, Aartse A, Derking R, Bontjer I, Kootstra NA, Wiersinga WJ, Vidarsson G, Haagmans BL, Ward AB, de Bree GJ, Sanders RW, van Gils MJ. 2020. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369:643–650. 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattiuzzo G, Bentley EM, Hassall M, Routley S. 2020. Establishment of the WHO International Standard and reference panel for anti-SARS-CoV-2 antibody. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 42.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer DS, Lugo LA, Kojic EM, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, García-Sastre A, Caplivski D, Cheng AC, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, Krammer F. 2020. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 26:1033–1036. 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Padoan A, Zuin S, Cosma C, Basso D, Plebani M, Bonfante F. 2020. Clinical performances of an ELISA for SARS-CoV-2 antibody assay and correlation with neutralization activity. Clin Chim Acta 510:654–655. 10.1016/j.cca.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt F, Weisblum Y, Muecksch F, Hoffmann H-H, Michailidis E, Lorenzi JCC, Mendoza P, Rutkowska M, Bednarski E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Caskey M, Robbiani DF, Nussenzweig MC, Rice CM, Hatziioannou T, Bieniasz PD. 2020. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med 217:e20201181. 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Case JB, Rothlauf PW, Chen RE, Liu Z, Zhao H, Kim AS, Bloyet LM, Zeng Q, Tahan S, Droit L, Ilagan MXG, Tartell MA, Amarasinghe G, Henderson JP, Miersch S, Ustav M, Sidhu S, Virgin HW, Wang D, Ding S, Corti D, Theel ES, Fremont DH, Diamond MS, Whelan SPJ. 2020. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe 28:475–485. 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazzini L, Martinuzzi D, Hyseni I, Benincasa L, Molesti E, Casa E, Lapini G, Piu P, Trombetta CM, Marchi S, Razzano I, Manenti A, Montomoli E. 2021. Comparative analyses of SARS-CoV-2 binding (IgG, IgM, IgA) and neutralizing antibodies from human serum samples. J Immunol Methods 489:112937. 10.1016/j.jim.2020.112937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, Agudelo M, Barnes CO, Gazumyan A, Finkin S, Hägglöf T, Oliveira TY, Viant C, Hurley A, Hoffmann HH, Millard KG, Kost RG, Cipolla M, Gordon K, Bianchini F, Chen ST, Ramos V, Patel R, Dizon J, Shimeliovich I, Mendoza P, Hartweger H, Nogueira L, Pack M, Horowitz J, Schmidt F, Weisblum Y, Michailidis E, Ashbrook AW, Waltari E, Pak JE, Huey-Tubman KE, Koranda N, Hoffman PR, West AP, Rice CM, Hatziioannou T, Bjorkman PJ, Bieniasz PD, Caskey M, Nussenzweig MC. 2020. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584:437–442. 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlickeiser S, Schwarz T, Steiner S, Wittke K, Al Besher N, Meyer O, Kalus U, Pruß A, Kurth F, Zoller T, Witzenrath M, Sander LE, Müller MA, Scheibenbogen C, Volk H-D, Drosten C, Corman VM, Hanitsch LG. 2020. Disease severity, fever, age, and sex correlate with SARS-CoV-2 neutralizing antibody responses. Front Immunol 11:628971. 10.3389/fimmu.2020.628971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang HS, Costa V, Racine-Brzostek SE, Acker KP, Yee J, Chen Z, Karbaschi M, Zuk R, Rand S, Sukhu A, Klasse PJ, Cushing MM, Chadburn A, Zhao Z. 2021. Association of age with SARS-CoV-2 antibody response. JAMA Netw Open 4:e214302. 10.1001/jamanetworkopen.2021.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Legros V, Denolly S, Vogrig M, Boson B, Siret E, Rigaill J, Pillet S, Grattard F, Gonzalo S, Verhoeven P, Allatif O, Berthelot P, Pélissier C, Thiery G, Botelho-Nevers E, Millet G, Morel J, Paul S, Walzer T, Cosset FL, Bourlet T, Pozzetto B. 2021. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell Mol Immunol 18:318–327. 10.1038/s41423-020-00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson EM, Goodwin EC, Verma A, Arevalo CP, Bolton MJ, Weirick ME, Gouma S, McAllister CM, Christensen SR, Weaver J, Hicks P, Manzoni TB, Oniyide O, Ramage H, Mathew D, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, Alanio C, D’Andrea K, Kuthuru O, Dougherty J, Pattekar A, Kim J, Han N, Apostolidis SA, Huang AC, Vella LA, Kuri-Cervantes L, Pampena MB, Betts MR, Wherry EJ, Meyer NJ, Cherry S, Bates P, Rader DJ, Hensley SE. 2021. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell 184:1858–1864. 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, Burger ZC, Rawlings SA, Smith DM, Phillips E, Mallal S, Lammers M, Rubiro P, Quiambao L, Sutherland A, Yu ED, da Silva Antunes R, Greenbaum J, Frazier A, Markmann AJ, Premkumar L, de Silva A, Peters B, Crotty S, Sette A, Weiskopf D. 2020. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 370:89–94. 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lipsitch M, Grad YH, Sette A, Crotty S. 2020. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat Rev Immunol 20:709–713. 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altman DG. 1999. Practical statistics for medical research. Chapman & Hall/CRC, Boca Raton, FL. [Google Scholar]
- 55.Hajian-Tilaki K. 2014. Sample size estimation in diagnostic test studies of biomedical informatics. J Biomed Inform 48:193–204. 10.1016/j.jbi.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 56.Oguntuyo KY, Stevens CS, Hung C-T, Ikegame S, Acklin JA, Kowdle SS, Carmichael JC, Chiu H-P, Azarm KD, Haas GD, Amanat F, Klingler J, Baine I, Arinsburg S, Bandres JC, Siddiquey MN, Schilke RM, Woolard MD, Zhang H, Duty AJ, Kraus TA, Moran TM, Tortorella D, Lim JK, Gamarnik AV, Hioe CE, Zolla-Pazner S, Ivanov SS, Kamil JP, Krammer F, Lee B. 2020. Quantifying absolute neutralization titers against SARS-CoV-2 by a standardized virus neutralization assay allows for cross-cohort comparisons of COVID-19 sera. mBio 12:e02492-20. 10.1128/mBio.02492-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wec AZ, Wrapp D, Herbert AS, Maurer DP, Haslwanter D, Sakharkar M, Jangra RK, Dieterle ME, Lilov A, Huang D, Tse LV, Johnson NV, Hsieh C-L, Wang N, Nett JH, Champney E, Burnina I, Brown M, Lin S, Sinclair M, Johnson C, Pudi S, Bortz R, Wirchnianski AS, Laudermilch E, Florez C, Fels JM, O’Brien CM, Graham BS, Nemazee D, Burton DR, Baric RS, Voss JE, Chandran K, Dye JM, McLellan JS, Walker LM. 2020. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 369:731–736. 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schäfer A, Muecksch F, Lorenzi JCC, Leist SR, Cipolla M, Bournazos S, Schmidt F, Gazumyan A, Baric RS, Robbiani DF, Hatziioannou T, Ravetch JV, Bieniasz PD, Nussenzweig MC, Sheahan TP. 2021. Antibody potency, effector function and combinations in protection from SARS-CoV-2 infection in vivo. J Exp Med 218:e20201993. 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valcourt EJ, Manguiat K, Robinson A, Chen JCY, Dimitrova K, Philipson C, Lamoureux L, McLachlan E, Schiffman Z, Drebot MA, Wood H. 2021. Evaluation of a commercially-available surrogate virus neutralization test for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Diagn Microbiol Infect Dis 99:115294. 10.1016/j.diagmicrobio.2020.115294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tseng YT, Wang SM, Huang KJ, I-Ru Lee A, Chiang CC, Wang CT. 2010. Self-assembly of severe acute respiratory syndrome coronavirus membrane protein. J Biol Chem 285:12862–12872. 10.1074/jbc.M109.030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tasker S, Wight O’Rourke A, Suyundikov A, Jackson Booth P-G, Bart S, Krishnan V, Zhang J, Anderson KJ, Georges B, Roberts MS. 2021. Safety and immunogenicity of a novel intranasal influenza vaccine (NasoVAX): a phase 2 randomized, controlled trial. Vaccines 9:224. 10.3390/vaccines9030224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin C, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, Mascola JR, Montefiori DC. 2014. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods 409:131–146. 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen HT, Zhang S, Wang Q, Anang S, Wang J, Ding H, Kappes JC, Sodroski J. 2020. Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects. J Virol 95:e02304-20. 10.1128/JVI.02304-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weissman D, Alameh M-G, de Silva T, Collini P, Hornsby H, Brown R, LaBranche CC, Edwards RJ, Sutherland L, Santra S, Mansouri K, Gobeil S, McDanal C, Pardi N, Hengartner N, Lin PJC, Tam Y, Shaw PA, Lewis MG, Boesler C, Şahin U, Acharya P, Haynes BF, Korber B, Montefiori DC. 2020. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe 29:23–31. 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, Baum A, Diehl WE, Dauphin A, Carbone C, Veinotte K, Egri SB, Schaffner SF, Lemieux JE, Munro JB, Rafique A, Barve A, Sabeti PC, Kyratsous CA, Dudkina NV, Shen K, Luban J. 2020. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183:739–751. 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, on behalf of the Sheffield COVID-19 Genomics Group, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Centers for Disease Control and Prevention. 2021. SARS-CoV-2 variant classifications and definitions. CDC, Atlanta, GA. [Google Scholar]
- 68.Supasa P, Zhou D, Dejnirattisai W, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Nutalai R, Tuekprakhon A, Wang B, Paesen GC, Slon-Campos J, López-Camacho C, Hallis B, Coombes N, Bewley KR, Charlton S, Walter TS, Barnes E, Dunachie SJ, Skelly D, Lumley SF, Baker N, Shaik I, Humphries HE, Godwin K, Gent N, Sienkiewicz A, Dold C, Levin R, Dong T, Pollard AJ, Knight JC, Klenerman P, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert S, Hall DR, Williams MA, Paterson NG, James W, Carroll MW, Fry EE, Mongkolsapaya J, Ren J, et al. 2021. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell 184:2201–2211. 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, Porrot F, Planchais C, Buchrieser J, Rajah MM, Bishop E, Albert M, Donati F, Prot M, Behillil S, Enouf V, Maquart M, Smati-Lafarge M, Varon E, Schortgen F, Yahyaoui L, Gonzalez M, De Sèze J, Péré H, Veyer D, Sève A, Simon-Lorière E, Fafi-Kremer S, Stefic K, Mouquet H, Hocqueloux L, van der Werf S, Prazuck T, Schwartz O. 2021. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med 27:917–924. 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 70.Wang G-L, Wang Z-Y, Duan L-J, Meng Q-C, Jiang M-D, Cao J, Yao L, Zhu K-L, Cao W-C, Ma M-J. 2021. Susceptibility of circulating SARS-CoV-2 variants to neutralization. N Engl J Med 384:2354–2356. 10.1056/NEJMc2103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.International Conference on Harmonisation. 2005. Validation of analytical procedures: text and methodology Q2(R1). ICH, Geneva, Switzerland. [Google Scholar]
- 72.Stiles T, Grant V, Mawbey N. 2003. Good clinical laboratory practice (GCLP): a quality system for laboratories that undertake the analyses of samples from clinical trials. BARQA, Ipswich, UK. [Google Scholar]
- 73.Sarzotti-Kelsoe M, Cox J, Cleland N, Denny T, Hural J, Needham L, Ozaki D, Rodriguez-Chavez IR, Stevens G, Stiles T, Tarragona-Fiol T, Simkins A. 2009. Evaluation and recommendations on good clinical laboratory practice guidelines for phase I–III clinical trials. PLoS Med 6:e1000067. 10.1371/journal.pmed.1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phillips N. 2021. The coronavirus is here to stay—here’s what that means. Nature 590:382–384. 10.1038/d41586-021-00396-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods, Tables S1 to S4, and Fig. S1 to S8. Download JCM.00527-21-s0001.pdf, PDF file, 1.4 MB (1.4MB, pdf)