ABSTRACT
Mycobacterium leprae is the predominant cause of leprosy worldwide, and its genotypes can be classified into four single-nucleotide polymorphism (SNP) types and 16 subtypes. Determining M. leprae drug resistance and genotype is typically done by PCR and Sanger DNA sequencing, which require substantial effort. Here, we describe a rapid method involving multiplex PCR in combination with nested amplification and next-generation sequence analysis that allows simultaneous determination of M. leprae drug resistance and SNP genotype directly from clinical specimens. We used this method to analyze clinical samples from two paucibacillary, nine multibacillary, and six type-undetermined leprosy patients. Regions in folP1, rpoB, gyrA, and gyrB that determine drug resistance and those for 84 SNP-InDels in the M. leprae genome were amplified from clinical samples and their sequences determined. The results showed that seven samples were subtype 1A, three were 1D, and seven were 3K. Three samples of the subtype 3K had folp1 mutation. The method may allow more rapid genetic analyses of M. leprae in clinical samples.
KEYWORDS: Mycobacterium leprae, single-nucleotide polymorphism, nested multiplex PCR
INTRODUCTION
Leprosy is a chronic infectious disease caused by Mycobacterium leprae that mainly affects the skin and peripheral nerves (1). In 2019, 202,185 new cases of leprosy were reported worldwide (2). Leprosy is also caused by Mycobacterium lepromatosis closely related to M. leprae (3, 4). The first case of drug-resistant M. leprae was reported in 1964 for dapsone (DDS) and in 1976 for rifampin (RIF) (5, 6). In 1981, multidrug therapy (MDT) against leprosy comprising three drugs, DDS, RIF, and clofazimine, was recommended by the World Health Organization (WHO). The MDT strategy has significantly reduced the number of leprosy patients, but several multidrug-resistant M. leprae strains, such as 92008 and Zensho-4, have been reported (7, 8).
Since M. leprae still cannot be cultivated on artificial media, drug susceptibility testing of M. leprae is performed via a time-consuming mouse footpad assay (9). DNA diagnosis to detect missense mutations in the drug resistance-determining regions (DRDRs) of M. leprae folP1, rpoB, and gyrA is used to determine the potential for resistance to DDS, RIF, and fluoroquinolones, respectively (10–14). WHO Guidelines for Global Surveillance of Drug Resistance in Leprosy recommend that mutation detection of DRDRs in the three drug resistance genes be performed using PCR and DNA sequencing (15).
Several complete genome sequences of M. leprae, including strain TN isolated from a patient in Tamil Nadu, India, and strain Br4923 isolated from a patient in Brazil, have been reported (16, 17). Based on screening of >600 M. leprae isolates collected from different regions around the world, M. leprae can be classified into four single-nucleotide polymorphism (SNP) types (1 to 4) and 16 subtypes (A to P) (18, 19). The SNP genotyping was defined by surveying 78 informative SNPs and six single-base insertion/deletions (InDels) (18) and is useful for identifying sources of M. leprae and tracking clonal transmission patterns.
DNA diagnosis of M. leprae drug resistance and SNP genotyping are usually performed by PCR and Sanger sequencing or whole-genome sequence (WGS) analysis (10–14, 17–19). Several computer programs to predict drug resistance of Mycobacterium tuberculosis strains based on WGS data have been developed (20), but obtaining WGS data for M. leprae from clinical specimens is challenging due to the presence of contaminating human DNA that is often present at much greater abundance relative to the amount of bacterial DNA, and that can preclude amplification of target M. leprae DNA by standard PCR (21, 22). Nested PCR, a modification of PCR that can be used to reduce nonspecific binding in products, addresses this challenge and is useful for rare templates or PCR with high background, but this method requires larger numbers of total cycles (23).
To monitor drug resistance trends and understand transmission patterns and genetic diversity in leprosy, we developed a rapid method that uses multiplex PCR in combination with nested amplification (nested multiplex PCR, or nmPCR) and amplicon sequencing to facilitate rapid and straightforward prediction of M. leprae drug resistance and determination of SNP genotype in clinical specimens.
MATERIALS AND METHODS
Ethics statement.
This study was approved by the medical research ethics committee of the National Institute of Infectious Diseases for the use of human subjects (approval number 1172). All procedures in the study, including biological sample collection and testing involving human subjects, were performed in accordance with the Helsinki Declaration, ethical guidelines for medical research involving human subjects.
Clinical specimens.
Seventeen clinical samples were randomly selected from our collections of skin biopsy specimens and slit-skin smears obtained from multibacillary (MB), paucibacillary (PB), and type-undetermined leprosy patients between 2013 and 2019. Patient characteristics are listed in Table 1.
TABLE 1.
Clinical information for leprosy patients in this studya
| National origin | Age (yr) | Sex | WHO classification | Clinical specimen | Type of case | yr sample was obtained | Sample name |
|---|---|---|---|---|---|---|---|
| Brazil | 30–39 | F | Unknown | Skin biopsy | New case | 2014 | ML015 |
| Bangladesh | 50–59 | M | PB | Skin biopsy | New case | 2015 | ML007 |
| East Timor | 30–39 | M | Unknown | Skin biopsy | New case | 2014 | ML026 |
| Indonesia | 20–29 | M | MB | Skin biopsy | New case | 2014 | ML025 |
| Japan | 80–89 | F | MB | Skin biopsy | New case | 2016 | ML003 |
| 70–79 | F | MB | Skin-slit smear | Relapse | 2014 | ML009 | |
| 60–69 | M | Unknown | Skin-slit smear | Relapse | 2013 | ML010 | |
| 80–89 | F | Unknown | Skin biopsy | Relapse | 2014 | ML024 | |
| 70–79 | M | MB | Skin biopsy | New case | 2015 | ML028 | |
| Nepal | 20–29 | M | MB | Skin biopsy | New case | 2018 | ML001 |
| 20–29 | F | Unknown | Skin biopsy | New case | 2018 | ML002 | |
| 30–39 | M | MB | Skin biopsy | New case | 2016 | ML004 | |
| 30–39 | M | PB | Skin biopsy | New case | 2016 | ML012 | |
| Philippines | 50–59 | F | MB | Skin biopsy | New case | 2015 | ML005 |
| 40–49 | F | MB | Skin biopsy | New case | 2015 | ML008 | |
| 50–59 | M | Unknown | Skin biopsy | New case | 2019 | ML013 | |
| 50–59 | M | MB | Skin biopsy | New case | 2014 | ML023 |
MB, multibacillary leprosy; PB, paucibacillary leprosy; unknown, not classified. F, female; M, male.
DNA preparations from clinical specimens.
DNA was purified from clinical specimens using a QIAamp DNA micro kit (Qiagen, Valencia, CA). Approximately 10 mg skin tissue was used as starting material and the extraction was carried out according to the manufacturer’s protocol. The DNA was then eluted with 50 μl AE buffer.
Primer design.
The primers used in this study are shown in Tables S1 and S2 in the supplemental material. Primers for DRDRs of the drug resistance genes and SNP genotyping in nmPCR were designed based on the genome sequence of M. leprae strain TN (GenBank accession number AL450380.1). The length of the outer and inner primers ranged from 20 to 25 bases. The guanine-cytosine contents of the primers were between 50 and 60%, except for several primers targeting the GC-rich regions. Amplicons ranging from 230 bp to 260 bp were expected in the first round of PCR, whereas the second round of PCR was expected to generate amplicons ranging from 130 to 150 bp.
nmPCR.
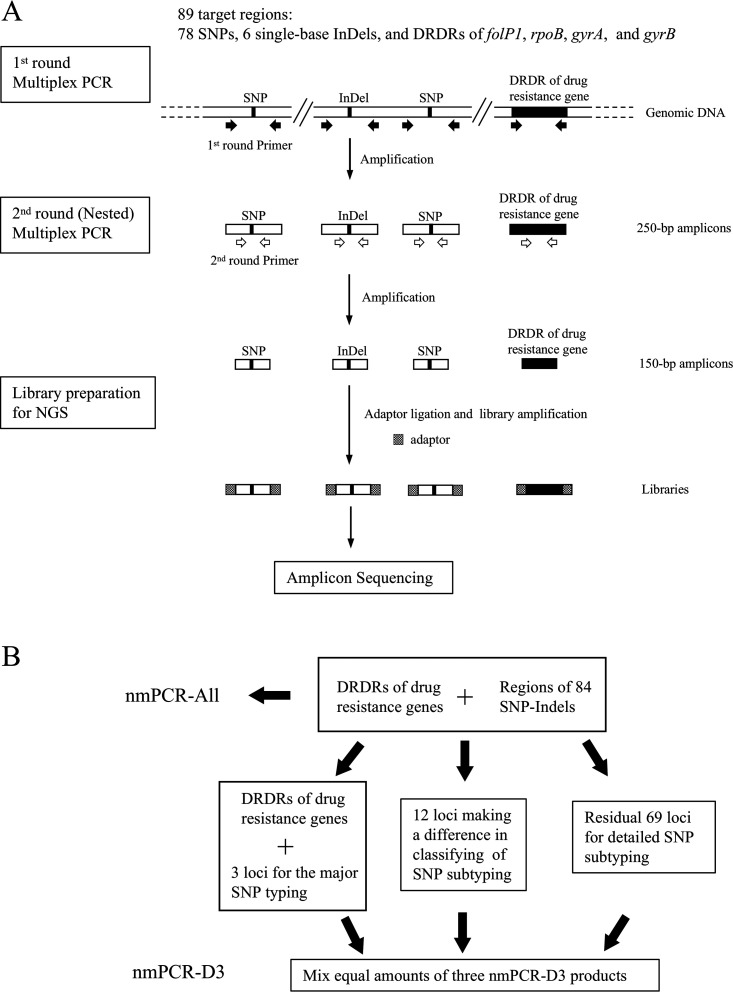
The scheme for nmPCR is shown in Fig. 1. Two nmPCR procedures to amplify target regions were designed. One procedure amplified all 89 target regions in one tube (nmPCR-All). The other divided the target regions into three groups that were amplified in three separate tubes (nmPCR-D3). The three groups included the following: group 1, three loci for the major SNP typing and five loci for drug resistance determination; group 2, 12 loci that allowed classification of SNP subtype; and group 3, 69 residual loci for detailed SNP subtyping.
FIG 1.
Scheme for nmPCR amplification of 89 target regions (A) and two nmPCR procedures (nmPCR-All and nmPCR-D3) to amplify target regions (B).
The first-round nmPCR-All was performed with PrimeSTAR HS DNA polymerase and GC buffer (TaKaRa Bio Inc., Shiga, Japan) in a 50-μl reaction volume, comprising 25 μl 2× GC buffer, 4 μl deoxynucleoside triphosphate (dNTP) mix (2.5 mM each dNTP), 0.25 μl PrimeSTAR HS DNA polymerase (2.5 U/μl), 8 μl of equimolar primer mixture containing 92 outer primer pairs (16 μM in total), and 100 ng DNA extracted from clinical specimens. For SNP14676, SNP1642875, and DRDR of folp1, two sets of primer pairs were used in the first-round PCR to overcome difficulties in amplification. The cycling conditions were 98°C for 5 min in the initial denaturing step, followed by 40 cycles of 98°C for 10 s, 55°C for 5 s, 72°C for 15 s, and finally one cycle of extension at 72°C for 5 min. The reactions were carried out in a TaKaRa PCR Thermal Cycler Dice Touch (TaKaRa Bio Inc., Shiga, Japan). The second-round multiplex PCR was conducted with the same polymerase as that used in the first-round mPCR in a 50-μl reaction volume, using the equimolar primer mixture containing 89 internal primer pairs (16 μM total) and 1 μl of the first-round PCR mixture as the template. The temperature and cycling conditions were the same as those for the first round of PCR.
For nmPCR-D3, the same conditions as those for nmPCR-All were used, except for the primer mixture. In multiplex PCR for major SNP typing and drug resistance determination, 11 primer pairs for the first-round PCR and 8 primer pairs for the second-round PCR were used (Tables S1 and S2). In the remaining nmPCRs for 12 loci used for subtyping and 69 loci used for detailed subtyping, 12-12 and 69-69 sets of outer-inner primer pairs were used, respectively.
The amplified products were electrophoresed on 2% agarose gels containing 0.5 μg/ml ethidium bromide and were visualized with a UV transilluminator.
Preparation of amplicon libraries.
After electrophoresis, DNA bands of around 150 bp were excised from the agarose gels. Purification of the amplified products was performed using Quantum Prep Freeze ‘N Squeeze DNA gel extraction spin columns (Bio-Rad Laboratories, Hercules, CA, USA) with the following modification: DNA was eluted in 200 μl Tris-EDTA buffer (pH 8.0). The extracted DNA was treated with phenol-chloroform (1:1) and precipitated with ethanol. The products of nmPCR-All and mixtures of the same amounts of the three nmPCR-D3 products were subjected to next-generation sequencing (NGS) analysis. For library preparation, an NEB Next Ultra II DNA library preparation kit for Illumina with NEBNext Singleplex Oligos for Illumina (New England Biolabs Inc., Ipswich, MA, USA) was used with the manufacturer’s protocol.
Sequencing and data analysis for drug resistance determination and SNP genotyping.
HiSeq2500 (Illumina, Inc., San Diego, CA, USA) was used for sequencing of PCR products from ML028 in nmPCR-All. NovaSeq 6000 (Illumina, Inc., San Diego, CA, USA) was used for sequencing of all other PCR products. The workflow of the NGS data analysis is summarized in Fig. S1. The data processing was performed on MacOS (version 10.4). The quality of each read was checked by using FastQC (version 0.11.9) (24). Low-quality reads in the raw reads obtained from amplicon sequencing were trimmed using Sickle (version 1.33) with basically the default setting and threshold for trimming based on average quality in a window modified to 30 (25). The trimmed reads were mapped on the reference genome (M. leprae strain TN) using the BWA-MEM algorithm with default parameters (26). The output SAM file was converted to a BAM file, and the file information was corrected using SAMtools with default settings (27). SAMtools mplieup and bcftools call with the default parameters were used to generate the VCF files containing SNP information (27, 28). Target region sequences of the BAM files and VCF files were confirmed with IGV software (29).
RESULTS
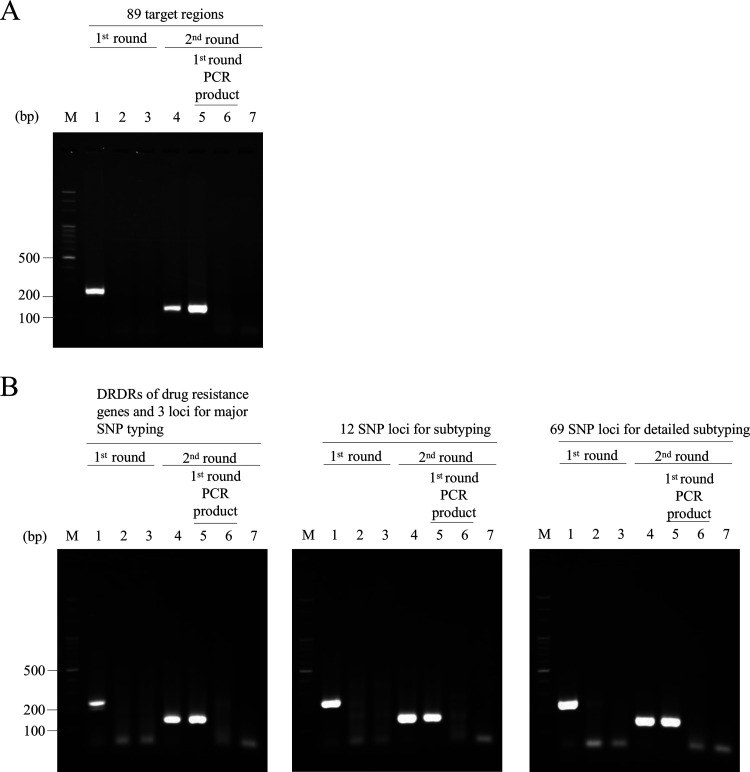
The nmPCR was optimized first. A total of 89 equimolar primer mixtures having various concentrations (4 μM, 8 μM, 16 μM, and 32 μM in total) were tested in the first- and second-round PCR using ML005 DNA as a template. The primer concentration that yielded the maximum amplification was 16 μM (data not shown). Thus, all nmPCR experiments were performed using 16 μM total primer mixtures. Representative images of results for nmPCR-All and nmPCR-D3 are shown in Fig. 2. In the first-round PCR, bands corresponding to an expected PCR product of approximately 250 bp were not seen. However, in the second-round PCR, products of around 150 bp were amplified in both nmPCRs.
FIG 2.
Agarose gel electrophoresis of nmPCR-All (A) and nmPCR-D3 (B). Lane 1, M. leprae DNA plus human DNA (100 ng + 100 ng, positive control); lane 2, ML002 clinical sample; lane 3, human DNA (100 ng, negative control); lane 4, M. leprae DNA plus human DNA (100 ng + 100 ng); lane 5, first PCR products for ML002; lane 6, first PCR products for human DNA; lane 7, water; M, 100-bp DNA ladder (Watson, Tokyo, Japan). In panel B, the gels show targeted DRDRs of drug resistance genes and 3 loci for major SNP typing (left), 12 SNP loci for subtyping (middle), and 69 SNP loci for detailed subtyping (right).
DNA from 10 clinical samples (ML001 to ML005, ML007 to ML010, and ML028) yielded amplification products having the expected size of approximately 150 bp by nmPCR-All and nmPCR-D3. These PCR products were subjected to amplicon sequencing by NGS, and results for nmPCR-All and nmPCR-D3 were compared (see Table S3 in the supplemental material). The results showed that nmPCR-D3 amplified the target regions more efficiently than nmPCR-All. In 10 clinical specimens, nmPCR-All and nmPCR-D3 yielded sufficient reads (≥26,989,826 raw reads and ≥25,916,222 trimmed and mapped reads). However, in seven samples, nmPCR-All obtained no reads for one or two loci in the major SNP typing (Table S3). In four samples (ML001, ML003, ML004, and ML028; Table S4), nmPCR-D3 yielded reads for all 89 loci. In addition, nmPCR-D3 yielded reads for the three loci for SNP typing and for five DRDRs in drug resistance genes in all samples. nmPCR-D3 also obtained reads for the 12 subtyping loci in all the samples, except for SNP313361 in ML015.
Based on these results, we used nmPCR-D3 to test the remaining 7 clinical samples (ML012, ML013, ML015, and ML023 to ML026). Overall, >97% of the reads from the amplicons were mapped to the reference genome, with the exception of ML012 and ML015, for which 64.11% and 65.77% of reads, respectively, could be mapped to the reference (Table S3). Positive rates for sequence determination and depths of the 84 SNP-InDels and DRDRs calculated using SAMtools are shown in Table S5. For determination of the major SNP type and drug resistance gene mutations in nmPCR-D3, the positive rates were >88.2% in all 17 samples. For the 12 subtyping loci, the positive rates for nmPCR-D3 were also >88%. However, some of the 69 loci (e.g., SNP1324009, SNP298572, and SNP379804) for detailed subtyping had low positive rates for sequence determination.
SNP type, subtype, and drug resistance gene mutations in 17 clinical samples were next investigated by nmPCR and amplicon sequencing (Table 2). Seven samples, including two from PB patients, were subtype 1A, three were subtype 1D, and seven were subtype 3K. Seven samples containing M. leprae subtype 1A were from patients from countries in Southeast Asia such as the Philippines and Bangladesh, while one sample, ML015, was from a patient from Brazil. Three samples with M. leprae subtype 1D were from patients from Nepal. Among the M. leprae subtype 3K samples, 5 of 7 were from patients from Japan, and there was one each from patients living in Indonesia and the Philippines. The results of this study are generally consistent with previous data for the geographical association of M. leprae genotypes (Table 3). Of the 5 type 3K samples from Japanese patients, 3 had a mutation in the folp1 gene. Of these, two (ML003 and ML010) had an A-to-G transition at nucleotide 157 (Thr53Ala) in folp1, and one (ML009) had a C-to-T transition at nucleotide 164 (Pro55Leu); these results were consistent with those obtained by standard PCR amplification and Sanger sequencing (data not shown). None of the samples had mutation in the DRDRs of rpoB, gyrA, and gyrB.
TABLE 2.
Genotype and DRDR mutation in 17 clinical samples
| SNP type | SNP subtypea | Origin | DRDR mutation | Sample name |
|---|---|---|---|---|
| 1 | A | Brazil | —b | ML015 |
| 1 | A | Bangladesh | —b | ML007 |
| 1 | A | East Timor | —b | ML026 |
| 1 | A | Nepal | —b | ML012 |
| 1 | A | Philippines | —b | ML008 |
| 1 | A | Philippines | —b | ML013 |
| 1 | A | Philippines | —b | ML023 |
| 1 | D | Nepal | —b | ML004 |
| 1 | D | Nepal | —b | ML001 |
| 1 | D | Nepal | —b | ML002 |
| 3 | K | Indonesia | —b | ML025 |
| 3 | K | Japan | folp1 (T53A)c | ML003 |
| 3 | K | Japan | folp1 (P55L)c | ML009 |
| 3 | K | Japan | folp1 (T53A)c | ML010 |
| 3 | K | Japan | —b | ML024 |
| 3 | K | Japan | —b | ML028 |
| 3 | K | Philippines | —b | ML005 |
TABLE 3.
Correlation between SNP genotype and geographical location
| SNP type | Countries where the M. leprae genotype has been reporteda | Reference(s) |
|---|---|---|
| 1A (branch 1) | Bangladesh (B-105), Guyana (I487), India (TN), Indonesia (Indonesia-03), Japan (Japon-35), South Korea (Coree-04), New Caledonia (N-Cal-94003), Nepal (Nepal-02), Philippines (Phi-01), Thailand (Thai53) | 18 |
| 1B (branch 1) | Antilles (S2_95034), French West Indies (89001), India (Inde-825) | 18, 38 |
| 1C | Bangladesh (B-107), New Caledonia (N-Cal-97021), Nepal (Nepal-B14230) | 18 |
| 1D (branch 1) | Bangladesh (B-112), Brazil (Br2016-18), French West Indies (88056), India (S11_Inde2), Japan (Japon-27), Madagascar (Mdg-B107), Malawi (Malawi-02), Nepal (Nepal-21A), Pakistan (Pak), Venezuela (V-17) | 18, 19, 38 |
| 2E (branch 2E) | Ethiopia (ARLP-08), Malawi (Malawi-01) | 18, 19 |
| 2F (branch 2F) | Iran (Iran-10), Denmark (Refshale_16),b Sweden (3077),b Turkey (Turc-1-6), UK (SK8)b | 18, 38 |
| 2G | New Caledonia (N-Cal-82061), Nepal (Nepal-B5008) | 18 |
| 2H (branch 2H) | Ethiopia (Afr-Eth) | 18 |
| 3I (branch 3) | Brazil (Brazil-01), Denmark (Jorgen_625),b French West Indies (85054), UK (SK2),b Morocco (Maroc-01), Mexico (NHDP98), USA (Armadillo-260,c NHDP10), Venezuela (V-01) | 18, 38 |
| 3J | New Caledonia (N-Cal-96008) | 18 |
| 3K-0 (branch 0) | China (S10_Ch-04), Indonesia (Indonesia-16), Hungary (KD271), b Japan (Zensho-4), South Korea (Korea-3-2), New Caledonia (S9_96008), Philippines (CM1)d | 19, 32, 38, 39 |
| 3K-1 (branch 5) | Japan (Zensho-9), South Korea (K02), Marshall Islands (US57), Philippines (PS04), Turkey (Turc2.3) | 19 |
| 3L (branch 4) | New Caledonia (N-Cal-92041) | 18 |
| 3M (branch 4) | France (Fr-2310), French West Indies (1261) | 18 |
| 4N (branch 4) | Benin (Bn7-39), Brazil (Brazil-05), Guinea (Gu4-17), Ivory Coast (C-I-07), French West Indies (81030), Mali (S13_Ml-3-28), Morocco (Maroc-2704), Senegal (Senegal-88063), Venezuela (V-06) | 18, 19 |
| 4O (branch 4) | Guinea (C30), Ivory Coast (C-I-02), Mali (S14_Ml-2-7), Senegal (Senegal-2662), Venezuela (V-07) | 18, 19 |
| 4P (branch 4) | Brazil (Br4923), French West Indies (98007), Mali (Mali-C13), Venezuela (V-10) | 18 |
Representative strain is in parentheses.
Ancient DNA extracted from medieval skeletons.
M. leprae strains found in wild armadillo.
M. leprae strains found in Cynomolgus macaque.
DISCUSSION
In this study, we developed a method using nmPCR followed by amplicon sequencing to simultaneously determine the presence of mutations in drug resistance genes and the SNP genotype of M. leprae in clinical specimens. Using this method, we demonstrated that the 84 informative SNP-InDels and mutations in DRDRs in folP1, rpoB, gyrA, and gyrB of M. leprae in 17 clinical specimens from MB, PB, and type-undetermined leprosy patients could be detected.
M. leprae SNP genotypes are typically determined by performing separate PCR amplifications for the SNPs, followed by Sanger sequencing or complete genome sequence analysis (18, 19, 30, 31). A previous report showed that successful coverage for complete genome analysis of M. leprae could be achieved even if the bacillary index (BI) was as low as 1+, but the sequencing success rates were 23% for BI 1+ and 33% for BI 2+ (19). Therefore, PCR amplification would be a good solution to maximize the amount of information that can be obtained for M. leprae genomes in clinical specimens.
The results of our study are generally consistent with previous data for the geographical association of M. leprae genotypes (Table 3). Recently, phylogenetic analysis of M. leprae has defined six branches (branches 0 to 5) (32–34). Branch 1, including SNP subtypes 1A, 1B, and 1D, is mainly distributed in Southeast Asia. Branch 2 has branches 2E, 2F, and 2H, including subtypes 2E, 2F, and 2H, respectively (33). Branches 2E and 2H are mainly distributed in East Africa, and branch 2F is mainly in Europe. Branch 3 (subtype 3I) is distributed mainly in North and South America, while branch 4 (subtypes 3J, 3L, 4M, 4O, and 4P) is distributed in West Africa and South America (33). Branch 0 (subtype 3K-0) is found in Far East Asia and branch 5 (subtype 3K-1) in the Pacific Islands (32–34). In Brazil, subtypes 3I and 4P are predominant, and subtype 1D has been reported (19). In the present study, subtype 1A was detected from a Brazilian patient for the first time.
In two samples (ML012 and ML015), read depths were low in multiple loci, probably because of preferential amplification to the specific loci. The primer sequences may need to be considered. Goulart et al. reported that, compared with larger amplicons, the 130-bp amplicon improved M. leprae DNA detection by PCR in samples from skin lesions of tuberculoid (TT) and borderline-tuberculoid (BT) leprosy patients (35). Thus, design of amplicons shorter than 150 bp could improve the positive rates for target amplification. In addition, if the sample preparation for the targeted NGS is made easier, the utility value of our method will increase.
Nested PCR is useful for studies on certain human tissue microbiota, as this approach allows amplification of target DNA, having concentrations that are severalfold lower than that needed for standard PCR (23, 36). Meanwhile, NGS is a powerful tool that enables collection of several gigabases of nucleotide sequence in total. Using the method described in the present study, we could amplify and sequence any M. leprae genome region of interest by using appropriate primer pairs. Although NGS analysis currently requires specialized skills, the nmPCR with the sequence determination approach will be valuable for determining the SNP genotype and drug resistance of M. leprae in clinical specimens.
ACKNOWLEDGMENTS
This research was supported by JSPS KAKENHI grant numbers JP19K08785 and JP20K08663.
We thank Satoko Machida for help with data aggregation.
Footnotes
Supplemental material is available online only.
Contributor Information
Noboru Nakata, Email: n-nakata@nih.go.jp.
Erin McElvania, NorthShore University HealthSystem.
REFERENCES
- 1.Britton WJ, Lockwood DNJ. 2004. Leprosy. Lancet 363:1209–1219. doi: 10.1016/S0140-6736(04)15952-7. [DOI] [PubMed] [Google Scholar]
- 2.WHO. 2020. Global leprosy (Hansen disease) update, 2019: time to step-up prevention initiatives. Wkly Epidemiol Rec 95:417–440. [Google Scholar]
- 3.Han XY, Seo YH, Sizer KC, Schoberle T, May GS, Spencer JS, Li W, Nair RG. 2008. A new Mycobacterium species causing diffuse lepromatous leprosy. Am J Clin Pathol 130:856–864. doi: 10.1309/AJCPP72FJZZRRVMM. [DOI] [PubMed] [Google Scholar]
- 4.Sharma R, Singh P, McCoy RC, Lenz SM, Donovan K, Ochoa MT, Estrada-Garcia I, Silva-Miranda M, Jurado-Santa Cruz F, Balagon MF, Stryjewska B, Scollard DM, Pena MT, Lahiri R, Williams DL, Truman RW, Adams LB. 2020. Isolation of Mycobacterium lepromatosis and development of molecular diagnostic assays to distinguish Mycobacterium leprae and M. lepromatosis. Clin Infect Dis 71:E262–E269. doi: 10.1093/cid/ciz1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pettit JH, Rees RJ. 1964. Sulphone resistance in leprosy. An experimental and clinical study. Lancet 2:673–674. doi: 10.1016/s0140-6736(64)92482-1. [DOI] [PubMed] [Google Scholar]
- 6.Jacobson RR, Hastings RC. 1976. Rifampin-resistant leprosy. Lancet 2:1304–1305. doi: 10.1016/s0140-6736(76)92071-7. [DOI] [PubMed] [Google Scholar]
- 7.Maeda S, Matsuoka M, Nakata N, Kai M, Maeda Y, Hashimoto K, Kimura H, Kobayashi K, Kashiwabara Y. 2001. Multidrug resistant Mycobacterium leprae from patients with leprosy. Antimicrob Agents Chemother 45:3635–3639. doi: 10.1128/AAC.45.12.3635-3639.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cambau E, Perani E, Guillemin I, Jamet P, Ji B. 1997. Multidrug-resistance to dapsone, rifampicin, and ofloxacin in Mycobacterium leprae. Lancet 349:103–104. doi: 10.1016/S0140-6736(05)60888-4. [DOI] [PubMed] [Google Scholar]
- 9.Shepard CC. 1967. A kinetic method for the study of activity of drugs against Mycobacterium leprae in mice. Int J Lepr Other Mycobact Dis 35:429–435. [PubMed] [Google Scholar]
- 10.Honore N, Cole ST. 1993. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrob Agents Chemother 37:414–418. doi: 10.1128/AAC.37.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cambau E, Jarlier V. 1996. Resistance to quinolones in mycobacteria. Res Microbiol 147:52–59. doi: 10.1016/0923-2508(96)80204-x. [DOI] [PubMed] [Google Scholar]
- 12.Kai M, Matsuoka M, Nakata N, Maeda S, Gidoh M, Maeda Y, Hashimoto K, Kobayashi K, Kashiwabara Y. 1999. Diaminodiphenylsulfone resistance of Mycobacterium leprae due to mutations in the dihydropteroate synthase gene. FEMS Microbiol Lett 177:231–235. doi: 10.1111/j.1574-6968.1999.tb13737.x. [DOI] [PubMed] [Google Scholar]
- 13.Williams DL, Gillis TP. 2004. Molecular detection of drug resistance in Mycobacterium leprae. Lepr Rev 75:118–130. doi: 10.47276/lr.75.2.118. [DOI] [PubMed] [Google Scholar]
- 14.Kim S-K, Lee S-B, Kang T-J, Chae G-T. 2003. Detection of gene mutations related with drug resistance in Mycobacterium leprae from leprosy patients using Touch-Down (TD) PCR. FEMS Immunol Med Microbiol 36:27–32. doi: 10.1016/S0928-8244(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 15.WHO. 2009. Guidelines for global surveillance of drug resistance in leprosy. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 16.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honoré N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 17.Monot M, Honoré N, Garnier T, Araoz R, Coppée J-Y, Lacroix C, Sow S, Spencer JS, Truman RW, Williams DL, Gelber R, Virmond M, Flageul B, Cho S-N, Ji B, Paniz-Mondolfi A, Convit J, Young S, Fine PE, Rasolofo V, Brennan PJ, Cole ST. 2005. On the origin of leprosy. Science 308:1040–1042. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- 18.Monot M, Honoré N, Garnier T, Zidane N, Sherafi D, Paniz-Mondolfi A, Matsuoka M, Taylor GM, Donoghue HD, Bouwman A, Mays S, Watson C, Lockwood D, Khamesipour A, Khamispour A, Dowlati Y, Jianping S, Rea TH, Vera-Cabrera L, Stefani MM, Banu S, Macdonald M, Sapkota BR, Spencer JS, Thomas J, Harshman K, Singh P, Busso P, Gattiker A, Rougemont J, Brennan PJ, Cole ST. 2009. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet 41:1282–1289. doi: 10.1038/ng.477. [DOI] [PubMed] [Google Scholar]
- 19.Benjak A, Avanzi C, Singh P, Loiseau C, Girma S, Busso P, Fontes ANB, Miyamoto Y, Namisato M, Bobosha K, Salgado CG, da Silva MB, Bouth RC, Frade MAC, Filho FB, Barreto JG, Nery JAC, Bührer-Sékula S, Lupien A, Al-Samie AR, Al-Qubati Y, Alkubati AS, Bretzel G, Vera-Cabrera L, Sakho F, Johnson CR, Kodio M, Fomba A, Sow SO, Gado M, Konaté O, Stefani MMA, Penna GO, Suffys PN, Sarno EN, Moraes MO, Rosa PS, Baptista IMFD, Spencer JS, Aseffa A, Matsuoka M, Kai M, Cole ST. 2018. Phylogenomics and antimicrobial resistance of the leprosy bacillus Mycobacterium leprae. Nat Commun 9:352. doi: 10.1038/s41467-017-02576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schleusener V, Köser CU, Beckert P, Niemann S, Feuerriegel S. 2017. Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: comparison of automated analysis tools. Sci Rep 7:46327–46329. doi: 10.1038/srep46327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stearns JC, Lynch MDJ, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. 2011. Bacterial biogeography of the human digestive tract. Sci Rep 1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coldham T, Rose K, O'Rourke J, Neilan BA, Dalton H, Lee A, Mitchell H. 2013. Detection of Helicobacter species in the gastrointestinal tract of ringtail possum and koala: possible influence of diet, on the gut microbiota. Vet Microbiol 166:429–437. doi: 10.1016/j.vetmic.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, He J, Liu J, You Y, Yuan L, Wen Y. 2019. Nested PCR and the TaqMan SNP genotyping assay enhanced the sensitivity of drug resistance testing of Mycobacterium leprae using clinical specimens of leprosy patients. PLoS Negl Trop Dis 13:e0007946. doi: 10.1371/journal.pntd.0007946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews S. 2018. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 25.Joshi NA, Fass JN. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.33).
- 26.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997v2 [q-bio.GN].
- 27.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma R, Singh P, Loughry WJ, Lockhart JM, Inman WB, Duthie MS, Pena MT, Marcos LA, Scollard DM, Cole ST, Truman RW. 2015. Zoonotic leprosy in the Southeastern United States. Emerg Infect Dis 21:2127–2134. doi: 10.3201/eid2112.150501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Truman RW, Singh P, Sharma R, Busso P, Rougemont J, Paniz-Mondolfi A, Kapopoulou A, Brisse S, Scollard DM, Gillis TP, Cole ST. 2011. Probable zoonotic leprosy in the southern United States. N Engl J Med 364:1626–1633. doi: 10.1056/NEJMoa1010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendum TA, Taylor GM, Donoghue HD, Wu H, Szalontai C, Marcsik A, Molnár E, Pálfi G, Stewart GR. 2018. The genome sequence of a SNP type 3K strain of Mycobacterium leprae isolated from a seventh-century Hungarian case of lepromatous leprosy. Int J Osteoarchaeol 28:439–447. doi: 10.1002/oa.2673. [DOI] [Google Scholar]
- 33.Schuenemann VJ, Avanzi C, Krause-Kyora B, Seitz A, Herbig A, Inskip S, Bonazzi M, Reiter E, Urban C, Dangvard Pedersen D, Taylor GM, Singh P, Stewart GR, Velemínský P, Likovsky J, Marcsik A, Molnár E, Pálfi G, Mariotti V, Riga A, Belcastro MG, Boldsen JL, Nebel A, Mays S, Donoghue HD, Zakrzewski S, Benjak A, Nieselt K, Cole ST, Krause J. 2018. Ancient genomes reveal a high diversity of Mycobacterium leprae in medieval Europe. PLoS Pathog 14:e1006997. doi: 10.1371/journal.ppat.1006997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blevins KE, Crane AE, Lum C, Furuta K, Fox K, Stone AC. 2020. Evolutionary history of Mycobacterium leprae in the Pacific Islands: M. leprae in the Pacific. Philos Trans R Soc Lond B Biol Sci 375:20190582. doi: 10.1098/rstb.2019.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goulart IMB, Cardoso AM, Santos MS, Gonçalves MA, Pereira JE, Goulart LR. 2007. Detection of Mycobacterium leprae DNA in skin lesions of leprosy patients by PCR may be affected by amplicon size. Arch Dermatol Res 299:267–271. doi: 10.1007/s00403-007-0758-5. [DOI] [PubMed] [Google Scholar]
- 36.Yu G, Fadrosh D, Goedert JJ, Ravel J, Goldstein AM. 2015. Nested PCR biases in interpreting microbial community structure in 16S rRNA gene sequence datasets. PLoS One 10:e0132253. doi: 10.1371/journal.pone.0132253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh P, Cole ST. 2011. Mycobacterium leprae: genes, pseudogenes and genetic diversity. Future Microbiol 6:57–71. doi: 10.2217/fmb.10.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuenemann VJ, Singh P, Mendum TA, Krause-Kyora B, Jäger G, Bos KI, Herbig A, Economou C, Benjak A, Busso P, Nebel A, Boldsen JL, Kjellström A, Wu H, Stewart GR, Taylor GM, Bauer P, Lee OY-C, Wu HHT, Minnikin DE, Besra GS, Tucker K, Roffey S, Sow SO, Cole ST, Nieselt K, Krause J. 2013. Genome-wide comparison of medieval and modern Mycobacterium leprae. Science 341:179–183. doi: 10.1126/science.1238286. [DOI] [PubMed] [Google Scholar]
- 39.Honap TP, Pfister L-A, Housman G, Mills S, Tarara RP, Suzuki K, Cuozzo FP, Sauther ML, Rosenberg MS, Stone AC. 2018. Mycobacterium leprae genomes from naturally infected nonhuman primates. PLoS Negl Trop Dis 12:e0006190. doi: 10.1371/journal.pntd.0006190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download JCM.00814-21-s0001.xlsx, XLSX file, 0.03 MB (26.2KB, xlsx)
Table S2. Download JCM.00814-21-s0002.xlsx, XLSX file, 0.02 MB (24KB, xlsx)
Table S3. Download JCM.00814-21-s0003.xlsx, XLSX file, 0.02 MB (18.4KB, xlsx)
Table S4. Download JCM.00814-21-s0004.xlsx, XLSX file, 0.05 MB (55.9KB, xlsx)
Table S5. Download JCM.00814-21-s0005.xlsx, XLSX file, 0.02 MB (21.7KB, xlsx)
Fig. S1. Download JCM.00814-21-s0006.pdf, PDF file, 0.2 MB (211.2KB, pdf)