ABSTRACT
SARS-CoV-2 is a novel positive-sense single-stranded RNA virus that has caused a recent pandemic. Most patients have a mild disease course, while approximately 20% have moderate to severe disease, often requiring hospitalization and, in some cases, care in the intensive care unit. By investigating a perceived increased rate of indeterminate QuantiFERON-TB Gold Plus results in hospitalized COVID patients, we demonstrate that severely ill COVID-19 patients have at least a 6-fold reduction of interferon gamma (IFN-γ) levels compared to control patients. What is more, over 60% of these severely ill COVID-19 patients’ peripheral T cells were found to be unable to produce measurable IFN-γ when stimulated with phytohemagglutinin (PHA), a potent IFN-γ mitogen, reflected by an indeterminate QuantiFERON-TB Gold Plus result. This defect of IFN-γ production was independent of absolute lymphocyte counts and immunosuppressive therapy. It was associated with increased levels of interleukin-6 (IL-6), which was a predictor of patient outcomes for our cohort when measured early in the course of disease. Finally, in a subset of COVID-19 patients, we found elevated IL-10 levels in addition to IL-6 elevation. In addition to finding a significant limitation of interferon-gamma release assay (IGRA) testing in severely ill COVID-19 patients, these data provide evidence that many of these patients demonstrate a focused Th2 immune response with inhibition of IFN-γ signaling and, in many cases, significant elevations of IL-6.
KEYWORDS: COVID-19, IGRA, indeterminate, QuantiFERON, SARS-CoV-2, interferon gamma
INTRODUCTION
QuantiFERON-TB Gold Plus is an enzyme-linked immunosorbent assay (ELISA) that is utilized to evaluate patients for latent tuberculosis (TB) infection. To carry out this test, peripheral blood is collected into specialized Vacutainer tubes. Two of the tubes contain Mycobacterium tuberculosis peptides that stimulate interferon gamma (IFN-γ) production in CD4 and CD8 T cells in the blood sample. In addition to these two antigen tubes, a positive control tube uses phytohemagglutinin (PHA) mitogen to induce an IFN-γ response and demonstrate that the patient’s T cells are functional. A fourth tube, the negative control (no stimulant), is used to adjust for background and preexisting serum IFN-γ. Patients with latent tuberculosis will induce T cells in the antigen tubes to produce IFN-γ that is quantitated by ELISA. A low response to PHA mitogen (<0.5 IU/ml) is classified as an indeterminate result when a blood sample also has a negative response to the TB antigens.
This study was initially undertaken because we found that the frequency of indeterminate results for the QuantiFERON-TB Gold Plus assay was increased during the months of March and April 2020 from a baseline of 8.7% to 15.50%. After analysis of patients that had indeterminate QuantiFERON-TB Gold Plus assay results, we found that many of these patients were immunosuppressed. However, after closer inspection, we identified a subpopulation of indeterminate patients that were positive for SARS-CoV-2 and had not been immunosuppressed. We subsequently evaluated all SARS-CoV-2-positive patients that had also been tested with the QuantiFERON-TB Gold Plus assay to determine the frequency of deficient IFN-γ responses in the COVID-19 population and assess additional laboratory correlates and clinical outcomes in this group.
MATERIALS AND METHODS
Study design.
This was a retrospective observational cohort study of patients with confirmed COVID-19 who were also tested with the QuantiFERON-TB Gold Plus assay between March 2020 and July 2020 at the University of North Carolina and associated regional hospitals. The control group consisted of samples obtained from June 2020 to September 2020 from patients who were similar in age. Data were obtained from the electronic medical record in accordance with the University of North Carolina internal review board (IRB)-approved study parameters (IRB number 20-2269).
Inclusion and exclusion criteria.
Subjects included outpatients and inpatients, aged 18 years or older, treated in the UNC Health Care system from March 2020 to September 2020 with a positive COVID-19 nasopharyngeal or tracheal aspirate sample using FDA-approved (EUA) SARS-CoV-2 PCR assays. Patients also needed to have a positive SARS-CoV-2 PCR test within 10 days of their QuantiFERON-TB Gold Plus test. Patients that tested positive for Mycobacterium tuberculosis antigen response with the QuantiFERON-TB Gold Plus assay were excluded. Overall, 48 patients met the inclusion criteria (Table 1).
TABLE 1.
Sociodemographic characteristics of the patient cohort
| Characteristic | COVID-19 patients (N = 48) | Healthy subjects (N = 76) | P value | COVID-19 IFN-γ normal (N = 17) | COVID-19 IFN-γ low (N = 31) | P value |
|---|---|---|---|---|---|---|
| Gender, no. (%) | <0.0001 | 0.7622 | ||||
| Female | 19 (40.0) | 61 (80.3) | 6 (35.3) | 13 (41.9) | ||
| Male | 29 (60.0) | 15 (19.7) | 11 (64.7) | 18 (58.1) | ||
| Age (yr), mean ± SD | 55.73 ± 15.96 | 48.72 ± 6.55 | 0.0054 | 59 ± 18.31 | 53.94 ± 15.20 | 0.2979 |
| Race, no. (%) | <0.0001 | 0.9709 | ||||
| African-American | 8 (16.7) | 15 (19.7) | 3 (17.6) | 5 (16.1) | ||
| Asian | 1 (2.1) | 5 (6.6) | 0 (0.0) | 1 (3.2) | ||
| Caucasian | 9 (18.8) | 37 (48.7) | 3 (17.6) | 6 (19.4) | ||
| Hispanic or Latino | 31 (64.6) | 4 (5.3) | 11 (64.7) | 17 (54.8) | ||
| Pacific Islander | 0 (0.0) | 1 (1.3) | 0 (0.0) | 0 (0.0) | ||
| Other or unspecified | 2 (4.2) | 14 (18.4) | 0 (0.0) | 2 (6.5) | ||
| Mortality, no. (%) | 16 (33.3) | 0 (0.0) | <0.0001 | 5 (29.4) | 9 (29.0) | >0.999 |
| CCI, median (range) | 3 (0–12) | 0 (0) | <0.0001 | 3 (0–9) | 3 (0–12) | 0.1992 |
| WHO ordinal scale, median (range) | 8 (1–10) | 0 (0) | <0.0001 | 6 (1–10) | 9 (5–10) | 0.0518 |
IGRA for Mycobacterium tuberculosis testing.
Interferon-gamma release assay (IGRA) testing was performed using the QuantiFERON-TB Gold Plus (Qiagen) chemiluminescence assay on the Liaison XL automated platform (DiaSorin) based on the manufacturer’s protocol. The Liaison XL (DiaSorin) was used to quantify the concentration of IFN-γ in each sample tube. A result of positive, negative, or indeterminate was made using an algorithm per the manufacturer’s instructions.
Additional clinical laboratory assays.
Lymphocyte and neutrophil absolute counts were obtained from patient peripheral blood drawn via venipuncture and collected in EDTA collection tubes using the ADVIA (Siemens) clinical hematology analyzer. C-reactive protein (CRP), total bilirubin, and lactate dehydrogenase (LDH) concentrations were measured from patient serum by chemiluminescence utilizing the VITROS 5600 (Ortho Clinical Diagnostics) clinical chemistry analyzer. Serum samples for cytokine analysis were frozen within 12 h postcollection and then allowed to thaw immediately prior to analysis. Serum concentrations of interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α) were measured by ELISA sandwich immune assay V-Plex (Mesoscale Discovery), which was performed at the Mayo Clinic (Rochester, MN). IL-2R, IL-2, IL-4, IL-5, IL-8, IL-10, IL-12, and IL-13 were measured using multiplex bead assay and multiplex array chemiluminescence at the Mayo Clinic (Rochester, MN).
Disease severity scores.
To measure patient COVID-GRAM critical illness risk score at the time of admission, several parameters were retrieved from the electronic medical record and used to calculate the risk of critical illness using the formula previously described (1). The severity of COVID-19-related illness was scored using the recently developed WHO clinical progression scale (2). The Charlson comorbidity index (CCI) score was used to estimate the underlying level of preexisting disease severity for each patient within our cohort. The scores were calculated using an online calculator (MDCalc) as previously described (3, 4).
Statistical analysis.
All statistical analyses were performed using GraphPad Prism version 9.0. Comparison between cohort groups, subgroups, and control groups was performed using Mann-Whitney U test, independent unpaired t test, or Welch’s t test, as appropriate. Comparison of group proportions was performed using Fisher’s exact test. Survival curve comparison was performed using the Gehan-Breslow-Wilcoxon test. Nested t test analysis was utilized to compare absolute lymphocyte count and absolute neutrophil count levels. P values of <0.05 were considered significant.
RESULTS
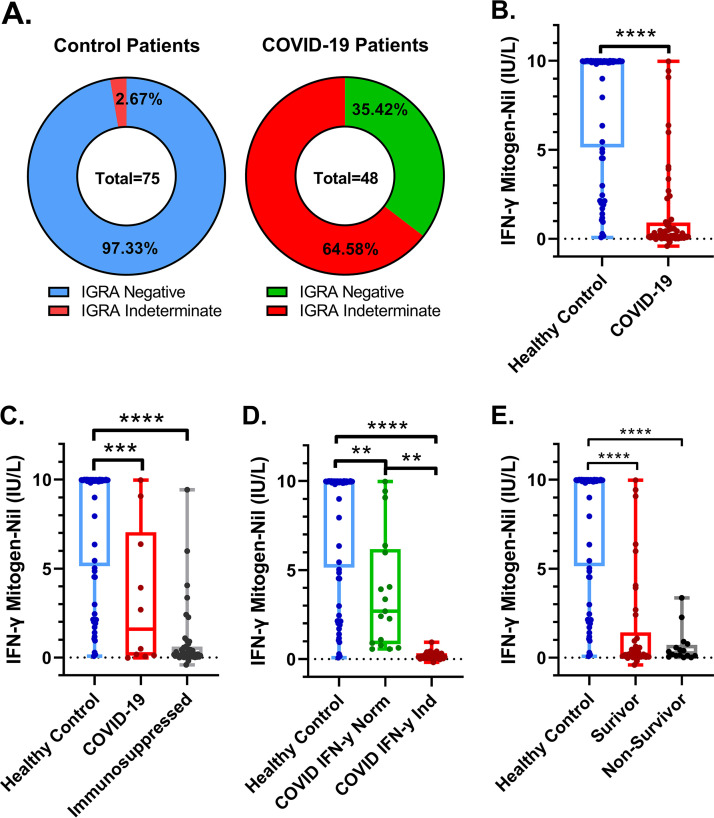
COVID-19 patients had more indeterminate QuantiFERON-TB Gold Plus interferon gamma release assay (IGRA) results than would be expected by chance (64.58% versus 2.97%) (Fig. 1A). Control patients had a 6-fold increase in mitogen-induced IFN-γ production compared to the COVID-19 patients in our cohort, with control patients measuring 7.87 (IU/liter) and COVID-19 patients measuring 1.29 (IU/liter) on average (Fig. 1B). The previous generation of the IGRA, the QuantiFERON-TB Gold assay, has been shown to be affected by immunosuppressive comorbidities and therapies (5, 6). Interestingly, when excluding all patients who had been treated with immunosuppressive medications or had preexisting immunosuppressive comorbidities, we found that the COVID-19 patients still had a significant decrease in IFN-γ produced by their peripheral blood T cells postmitogen stimulation (Fig. 1C). COVID-19 patients that had indeterminate test results had lower IFN-γ values from the mitogen minus the nil tube compared to COVID-19 patients that had negative results (acceptable IFN-γ control results) (Fig. 1D). It was interesting that in the COVID-19 patients that did have measurable IFN-γ, it was lower than what was typically observed from our control patients.
FIG 1.
Hospitalized patients with PCR-confirmed COVID-19 disease have peripheral blood T cells with a lower IFN-γ phenotype than closely aged healthy control patients. (A) COVID-19 patients have lower levels of IFN-γ produced in response to PHA stimulation than age-matched healthy patients. (B) COVID-19 patients have lower levels of IFN-γ produced by peripheral blood T cells. (C) COVID-19 patients that are not currently taking immunosuppressive medications have lower levels of IFN-γ produced by peripheral blood T cells, and those on immunosuppressive medications have extremely low levels of IFN-γ. (D) COVID-19 patients with indeterminate IGRA test results have extremely low IFN-γ production by peripheral blood T cells, while those with IGRA-negative test results have higher levels of IFN-γ produced but still lower than healthy controls. (E) COVID-19 patients with worse outcomes have similar IFN-γ produced by peripheral blood T cells. Comparisons of differences in continuous variables within these groups were performed using the independent unpaired t test or the Welch’s t test, as appropriate, where a difference in means was considered significant if the P value was less than 0.05. **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001.
We next sought to determine if there was a difference in survival outcomes in the COVID-19 patient cohort who had decreased ability to produce IFN-γ in response to mitogen stimulation compared to controls. We found that while there was a trend toward COVID-19 patients with increased IFN-γ response to mitogen stimulation and survival (Fig. 1E), it was not statistically significant (P = 0.067). However, as expected, there was a statistically significant difference in mitogen-induced IFN-γ in controls versus both the COVID-19 survivors and nonsurvivors.
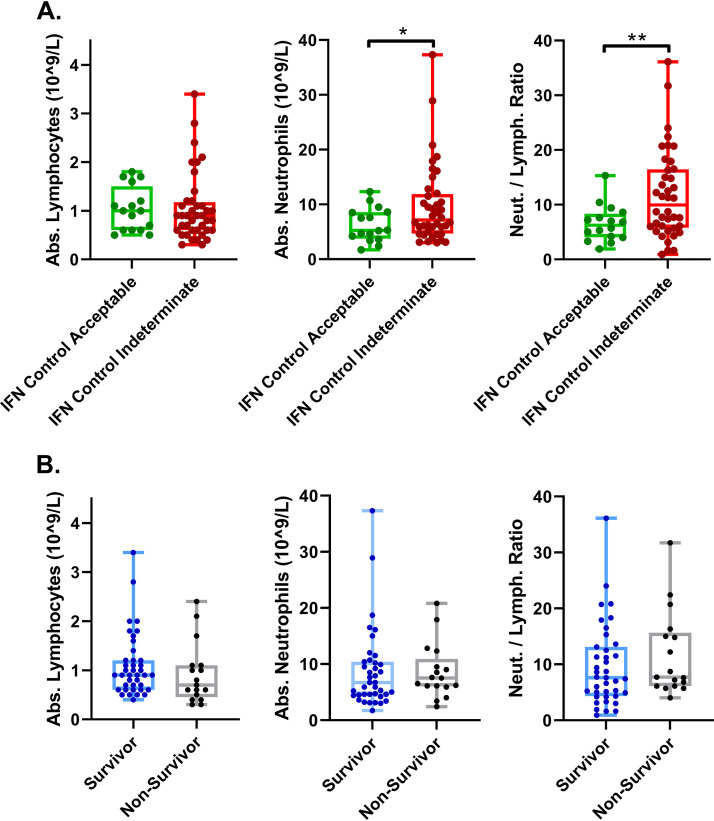
For further analysis, we divided our COVID-19 cohort into two subgroups, one containing the IGRA indeterminate patients (IFN-γ control indeterminate) and one with acceptable IFN-γ response. A previous study has shown that lack of ability to generate IFN-γ in response to mitogen in this assay can be secondary to lymphopenia and not due to impaired or altered peripheral T-cell status (7). We found that both the indeterminate and acceptable IFN-γ COVID-19 patients were lymphopenic, with no significant difference between their absolute lymphocyte counts (Fig. 2A), indicating that lymphopenia was not the cause of our COVID-19 patients’ IGRA indeterminate results. On the other hand, we found that there was a statistically significant difference between the absolute neutrophil count and the resulting neutrophil-to-lymphocyte ratio between the two groups (Fig. 2A). Absolute neutrophil-to-lymphocyte ratio has been correlated with severity of COVID-19 disease; however, we found no significant differences in absolute counts between survivors and nonsurvivors in our cohort of patients (Fig. 2B).
FIG 2.
COVID-19 patients with IFN-γ indeterminate controls have increased neutrophils but otherwise similar levels of lymphocytes and neutrophils during the course of hospitalization. (A) COVID-19 patients with indeterminate IFN-γ controls have slightly more neutrophils on average at the time IFN-γ testing was performed, leading to a higher neutrophil/lymphocyte ratio. Abs., absorbance. (B) COVID-19 patients with better outcomes had similar counts of lymphocytes and neutrophils at the time of IFN-γ testing. Comparisons of differences in continuous variables within these groups were performed using the independent unpaired t test or the Welch’s t test, as appropriate, where a difference in means was considered significant if the P value was less than 0.05. *, P < 0.05; **, P < 0.005.
Several recent studies proposed that an “immunologic collapse,” manifested by significant lymphopenia and reduced circulating T cells, characterizes severe COVID-19 disease (8, 9). Like these studies, we also observed a picture of leukopenia in all of the COVID-19 patients in our cohort (Fig. 2A). However, we did not observe a pattern of decreased absolute leukocytes between survivors and nonsurvivors in our cohort (see Fig. S1A in the supplemental material). Further, we investigated if there was a difference in absolute neutrophil count in survivors versus nonsurvivors over time, but there was no appreciable difference (Fig. S1B). Similarly, we compared the absolute lymphocyte and neutrophil counts (Fig. S1C and D) and found no significant differences.
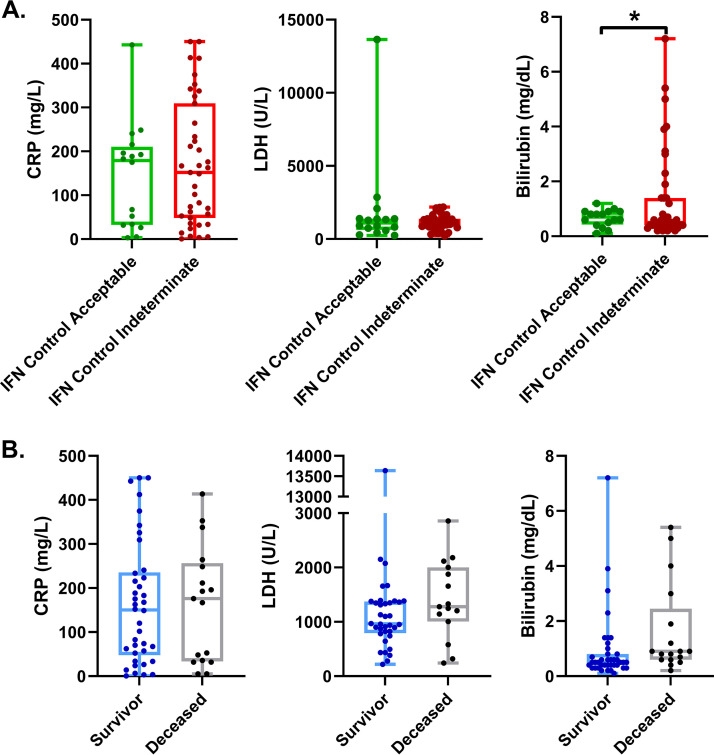
Other factors that have been associated with COVID-19 disease severity are serum CRP, LDH, and total bilirubin levels (10–12). We compared these serum levels between COVID-19 patients that had IFN-γ indeterminate and acceptable controls by IGRA (Fig. 3A). Interestingly, we found that a large portion of those with IFN-γ indeterminate controls had increased total bilirubin concentrations in peripheral serum. However, when we compared survivors to nonsurvivors (Fig. 3B) from our COVID-19 cohort, we did not see any differences in serum CRP, LDH, or bilirubin levels.
FIG 3.
Hospitalized COVID-19 patients with indeterminate IFN-γ controls and different clinical outcomes have similar markers of inflammation. (A) COVID-19 patients with decreased IFN-γ-producing T cells had similar levels of serum CRP and LDH; however, a portion did have significantly increased levels of total bilirubin. (B) COVID-19 patients with worse outcomes have similar serum concentrations of CRP, LDH, and total bilirubin. Comparisons of differences in continuous variables within these groups were performed using the independent unpaired t test or the Welch’s t test, as appropriate, where a difference in means was considered significant if the P value was less than 0.05. *, P < 0.05.
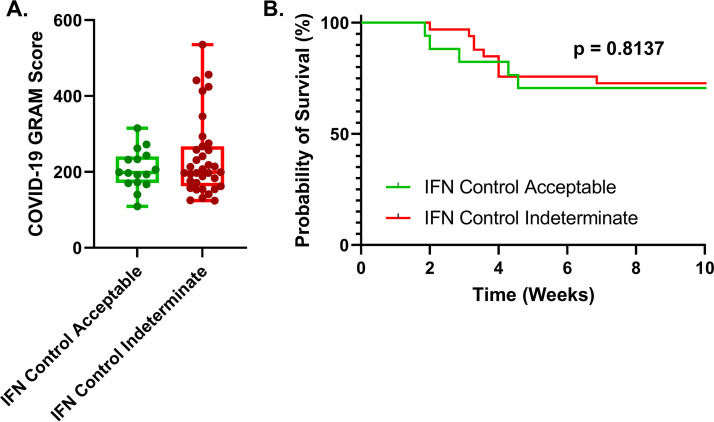
The COVID-GRAM critical illness risk score predicts the risk of critical illness in hospitalized COVID-19 patients (1). In our cohort, we found no appreciable difference between indeterminate or negative IGRA testing results for patients using this scoring system (Fig. 4A). The median COVID-GRAM critical illness risk score for both the indeterminate and acceptable IFN-γ control patients measured by IGRA was approximately 200, which is associated with a 92% chance of critical illness based on the risk scale, further indicating that our cohort was composed mainly of critically ill patients. To determine if an indeterminate IFN-γ control result was associated with overall mortality, we compared the overall probability of survival using a Kaplan-Meier estimator of survival and found there was no statistical difference between COVID-19 patient IFN-γ production measured by IGRA and survival outcome within our cohort (Fig. 4B). It is clear from the Kaplan-Meier estimate that most of these patients are critically ill, with a survival rate of 70.6% in the acceptable IFN-γ control patients versus 72.7% survival in the indeterminate IFN-γ control patient group over the course of 10 weeks, which is typically observed among critically ill COVID-19 patient cohorts (13, 14). Indeed, the median and average WHO clinical progression scale scores were six and seven, respectively, for the higher IFN-γ-producing patient and nine and eight, respectively, for the indeterminate IFN-γ control patients, indicating that nearly all of the patients in our cohort were in the intensive care unit (ICU) and received high-flow oxygen (Fig. S2A). When comorbidities and severity of illness were investigated in conjunction with WHO clinical progression scale scores, we found no significant differences in our COVID-19 patients (Fig. S2).
FIG 4.
COVID-19 patients with indeterminate IFN-γ controls have similar critical disease scores and survival outcomes. (A) Hospitalized COVID-19 patients with indeterminate IFN-γ controls have similar COVID-19 GRAM severity risk scores compared to patients with acceptable IFN-γ controls. (B) COVID-19 patients with indeterminate IFN-γ controls have similar survival outcomes during infection compared to patients with acceptable IFN-γ controls. Nonparametric scores between groups were performed using Mann-Whitney U test, and the survival curve comparison for COVID-19 patients between the IFN-γ groups was performed using the Gehan-Breslow-Wilcoxon test. The difference between the means was considered significant if the P value was less than 0.05.
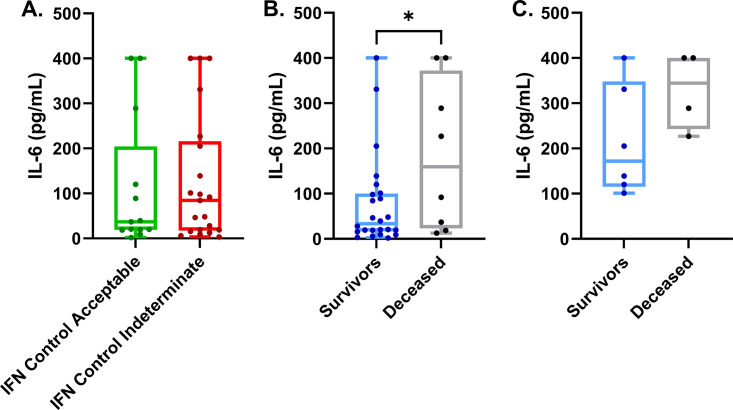
We wanted to determine if IL-6 and other cytokines were increased in our cohort of COVID-19 patients. Of the 48 patients in our cohort, 34 had at least one measurement of their serum IL-6 levels. As with previous reports, we found elevated concentrations of IL-6 in all the COVID-19 patients in our cohort. A significant minority (44%) had IL-6 concentrations greater than 100 pg/ml. These levels of IL-6 are often seen during overwhelming bacterial sepsis or in cytokine release syndrome (15, 16). Additionally, we found that nine of the 34 patients (27%) had cytokine levels greater than 400 pg/ml (the maximum measurable concentration for our assay). While not statically significant (P = 0.766), we found that COVID-19 patients who had indeterminate IFN-γ controls had a median concentration of serum IL-6 of 84.4 pg/ml and a mean of 128 pg/ml (standard deviation [SD], ±142; standard errors of the means [SEM], ±30.9) versus patients with acceptable IFN-γ. Controls had a median IL-6 level of 36.8 pg/ml and a mean of 113 pg/ml (SD, ±149; SEM, ±41.3) (Fig. 5A). While this difference may not be statistically significant, it may be physiologically significant, as those with higher IL-6 levels, above 100 pg/ml, may be far more likely to suffer ill effects.
FIG 5.
Serum concentrations of IL-6 are lower among COVID-19 survivors and similar between indeterminate and acceptable IFN-γ control COVID-19 patients. (A) Serum IL-6 concentrations measured for COVID-19 patients near collection time for IFN-γ production. (B) These same IL-6 levels were compared between COVID-19 patients who succumbed to disease versus patients who survived the course of disease. (C) Serum IL-6 concentrations measured near the time of collection for IFN-γ production that were >100 pg/ml for COVID-19 survivors versus nonsurvivors. Comparison of differences in continuous variables within these groups were performed using the independent unpaired t test or the Welch’s t test, as appropriate, where a difference in means was considered significant if the P value was less than 0.05. *, P < 0.05.
To investigate if concentration of serum IL-6 could be related to mortality in our cohort of patients, we compared the IL-6 concentrations measured near or at the collection time of our IFN-γ production assay in COVID-19 survivors versus nonsurvivors in our cohort (Fig. 5B). We found that our surviving patients had significantly (P = 0.039) lower levels of IL-6 at the time point near or at the time IGRA samples were drawn. This supports the hypothesis that IL-6 serum levels are good predictors of patient outcomes. Exploring further if high concentrations of IL-6 could be associated with mortality, we excluded patients with IL-6 levels below 100 pg/ml taken at the time of or near IGRA sample collection. As expected, there was no difference between these survival groups (Fig. 5C). Interestingly, there is a subset of survivors that also had IL-6 levels that were above 400 pg/ml at given time points during hospitalization. Many of these high IL-6 measurements above 400 pg/ml occurred later in the disease course, whereas the IL-6 levels measured in Fig. 5C in our deceased patients were typically measured early in the disease course. These analyses provide evidence that higher serum concentrations of IL-6 early in the course of disease, specifically those above 100 pg/ml, are associated with increased risk for COVID-19 mortality.
In order to evaluate the subtype of T-cell-mediated response and the potential presence of a cytokine storm, we identified patients within our cohort for whom the clinical team had ordered serum levels of multiple interleukins as part of a cytokine panel that also included direct IFN-γ levels and TNF-α. We identified seven patients in our study cohort that had this additional data (Table 2). Six of these seven patients demonstrated serum IL-6 levels above the normal reference range of 2.0 pg/ml (or 5 pg/ml, depending on the platform the assay was performed on). Similar to the previously discussed results, we found that patients with IL-6 levels higher than the reference range did worse, based on the WHO clinical progression scale for COVID-19, and that the severity of their disease was loosely associated with the degree of IL-6 elevation. Not surprisingly, all seven patients had IFN-γ levels below the threshold level of the test, including all those who tested negative with IGRA. Overall, the IFN-γ results further demonstrate that many patients with COVID-19 have diminished IFN-γ levels systemically. Of our patients for whom multiple cytokine measurements were performed, we found the only other cytokine that appeared to be elevated consistently was IL-10. Somewhat surprisingly, we found that none of the severely ill patients in our COVID-19 cohort had elevations in Th1 cytokines.
TABLE 2.
Cytokine panel results for cohort patients
| Cytokine | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Reference (pg/ml; patients 1–5) | Patient 6 | Patient 7 | Reference (pg/ml; patients 6–7) |
|---|---|---|---|---|---|---|---|---|---|
| Interleukin-2 | <2.1 | 2.2 | <2.1 | 2.4 | <2.1 | ≤2.1 | <5 | <5 | ≤12 |
| IL-2 Receptor | 673.4 | 4,328.2 | 1,804.4 | 3,720.5 | 774.8 | 175.3–858.2 | 98 | 959 | ≤1,033 |
| Interleukin-12 | <1.9 | <1.9 | <1.9 | <1.9 | <1.9 | ≤1.9 | <5 | <5 | ≤6 |
| Interferon Gamma | <4.2 | <4.2 | <4.2 | <4.2 | <4.2 | ≤4.2 | <5 | <5 | ≤5 |
| Interleukin-4 | <2.2 | <2.2 | <2.2 | <2.2 | <2.2 | ≤2.2 | <5 | <5 | ≤5 |
| Interleukin-10 | 3.1 | 17.9 | 45.2 | 38.9 | 3.5 | ≤2.8 | <5 | 29 | ≤18 |
| Interleukin-13 | <1.7 | <1.7 | <1.7 | 14 | <1.7 | ≤2.3 | <5 | <5 | ≤5 |
| IL-1 Beta | <6.5 | <6.5 | <6.5 | <6.5 | <6.5 | ≤6.7 | 6 | <5 | ≤36 |
| Interleukin-8 | <3.0 | <3.0 | <3.0 | 59.8 | <3.0 | ≤3.0 | <5 | <5 | ≤5 |
| TNF-alpha | <1.7 | <1.7 | <1.7 | 1.7 | 2.7 | ≤7.2 | <5 | <5 | ≤22 |
| Interleukin-5 | <2.1 | <2.1 | <2.1 | <2.1 | <2.1 | ≤2.1 | <5 | <5 | ≤5 |
| Interleukin-6 | 19.5 | 46.4 | 9.5 | 153.3 | <2.0 | ≤2.0 | 20.6 | >400 | ≤5 |
| WHO ordinal score | 6 | 6 | 8 | 9 | 4 | 6 | 9 | ||
| CCI score | 2 | 5 | 3 | 1 | 0 | 0 | 0 | ||
| IGRA result | Indeterminate | Indeterminate | Indeterminate | Negative | Negative | Negative | Indeterminate |
DISCUSSION
In this study, we demonstrate that our COVID-19 patient cohort had a 6-fold decrease, on average, in their ability to produce IFN-γ compared to healthy controls; the majority (62.5%) lacked the ability to generate a measurable IFN-γ level when stimulated with phytohemagglutinin. As a result, it is possible that SARS-CoV-2 infection, through some direct or indirect mechanism(s), subverts the ability of peripheral T cells to generate IFN-γ. Indeed, similar to previous reports (8), we found that both the indeterminate and acceptable IFN-γ COVID-19 patient groups in our study were lymphopenic; however, only the indeterminate group failed to generate adequate IFN-γ in response to phytohemagglutinin, indicating there is a qualitative difference in the population of the T cells between these two groups. As a cytokine, IFN-γ is important for humoral and innate immune responses and T-cell differentiation, and it has been hypothesized that regulation of the immune system is altered during COVID-19 infection (8, 9, 17). IGRAs have become one of the preferred methods for high-throughput screening of patients and staff for latent tuberculosis infection (LTBI) (18, 19). As such, it is a test of choice when patients exhibit respiratory disease symptomology, and tuberculosis is considered in the differential diagnosis. This study demonstrates that IGRA may be limited as an adequate tool for the detection of LTBI in patients with active COVID-19. Additionally, other IGRAs, such as T-spot.TB, might be similarly affected in COVID-19-infected patients.
We found that patients with IFN-γ indeterminate controls had elevated bilirubin on average. While there was not a significant difference in bilirubin levels between COVID-19 survivors and deceased patients, it is possible elevated bilirubin is an important marker that is negatively associated with survival in COVID-19 patients that our study did not have the power to identify. These data fit with several studies (12, 20, 21) that have shown a correlation with increased total bilirubin and severe COVID-19.
IFN-γ is well characterized to induce undifferentiated CD4-positive T helper cells (Th0 cells) into differentiated Th1 cells while inhibiting Th2 cell differentiation (18, 20, 22, 23). Decreased production or the inability to produce IFN-γ may have hindered these patients in our study from generating an effective Th1 immune response. This hypothesis is supported by our observations in the seven patients who had a cytokine panel performed that showed no measurable increase in Th1 cytokines, IFN-γ, IL-2, IL-2R, and IL-12. Thus, using a model of Th1 versus Th2 immune response, the COVID-19-positive patients in our cohort demonstrated a shift toward the Th2-based response, with most COVID-19 patients demonstrating an increase in the Th2 cytokine IL-6 and a subset of COVID-19 patients demonstrating increased IL-6 and IL-10 levels. However, it must be noted that this was not a robust Th2 response, since other Th2 cytokines, specifically IL-4 and IL-5, were not increased in any of the seven COVID-19 patients who had these cytokines measured. Nevertheless, it is interesting that a Th2-directed immune response is known to promote type 1 hypersensitivity (21, 24). Therefore, it is possible that promoting a Th2 immune phenotype leads to a hypersensitivity-like immune response that could promote lung and other organ injury, as observed in many severe COVID-19 cases (25). Future studies will need to be performed to better test this hypothesis.
Studies have reported (8, 16, 17, 23) that severely ill patients with COVID-19 have findings consistent with a cytokine storm. This has resulted in many COVID-19 patients being treated with pan-immunosuppression via steroids. Moreover, other studies have demonstrated that IL-6 is elevated in patients with COVID-19, and this has resulted in patients also being treated with IL-6 inhibitors such as tocilizumab (19, 26). Finally, IL-6 has been shown by several studies (27, 28) to be upregulated in COVID-19 patients, especially those who are critically ill. When comparing the levels of serum IL-6 between survivors and nonsurvivors (Fig. 5B), we found that patients who succumbed during the course of disease had significantly higher concentrations of IL-6. Based on data from our patients, it seems that serum IL-6 levels are most predictive of disease course outcome when taken early in the acute phase of infection.
Our study has several limitations. It is a retrospective study and limited by the tests ordered by the clinical care team. Our cohort is composed of COVID-19 patients with worse disease course, composed mainly of severely ill COVID-19 patients in the ICU. Our observations, especially those from the cytokine panel, are from a small sample size. While these findings provide additional data to the existing literature, future studies will need to be completed to confirm our findings. Finally, there is the confounding variable of immunosuppressive therapies or immunosuppressive conditions in the COVID-19 patients included in our study, as nearly all patients that had elevated IL-6 levels were treated with tocilizumab.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
We thank Andi Snyder, Christopher Parker, Bekah Lovern, George Nickolopoulos, Mary Le, Meghan Cleinmark, and Sally Speer from the UNC McLendon laboratory for assisting with questions and clinical laboratory testing. We appreciate the support received from the UNC School of Medicine and the Department of Pathology and Laboratory Medicine.
Footnotes
Supplemental material is available online only.
Contributor Information
John L. Schmitz, Email: John.Schmitz@unchealth.unc.edu.
Yi-Wei Tang, Cepheid.
REFERENCES
- 1.Liang W, Liang H, Ou L, Chen B, Chen A, Li C, Li Y, Guan W, Sang L, Lu J, Xu Y, Chen G, Guo H, Guo J, Chen Z, Zhao Y, Li S, Zhang N, Zhong N, He J, China Medical Treatment Expert Group for COVID-19. 2020. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med 180:1081–1089. 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall JC, Murthy S, Diaz J, Adhikari N, Angus DC, Arabi YM, Baillie K, Bauer M, Berry S, Blackwood B, Bonten M, Bozza F, Brunkhorst F, Cheng A, Clarke M, Dat VQ, de Jong M, Denholm J, Derde L, Dunning J, Feng X, Fletcher T, Foster N, Fowler R, Gobat N, Gomersall C, Gordon A, Glueck T, Harhay M, Hodgson C, Horby P, Kim YJ, Kojan R, Kumar B, Laffey J, Malvey D, Martin-Loeches I, McArthur C, McAuley D, McBride S, McGuinness S, Merson L, Morpeth S, Needham D, Netea M, Oh MD, Phyu S, Piva S, Qiu R, Salisu-Kabara H, et al. 2020. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis 20:e192–e197. 10.1016/S1473-3099(20)30483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlson ME, Pompei P, Ales KL, MacKenzie CR. 1987. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40:373–383. [3558716]. 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 4.Christensen DM, Strange JE, Gislason G, Torp-Pedersen C, Gerds T, Fosbøl E, Phelps M. 2020. Charlson comorbidity index score and risk of severe outcome and death in Danish COVID-19 patients. J Gen Intern Med 35:2801–2803. 10.1007/s11606-020-05991-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.La Jeon Y, Sun Nam Y, You E, Yang JJ, Kim MJ, Cho SY, Park TS, Lee HJ. 2013. Factors influencing discordant results of the QuantiFERON-TB Gold In-tube test in patients with active TB. J Infect 67:288–293. 10.1016/j.jinf.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Jeong SJ, Han SH, Kim CO, Hyeon Baek J, Jin SJ, Ku NS, Choi JY, Song YG, Kim HS, Kim JM. 2011. Predictive factors for indeterminate result on the QuantiFERON test in an intermediate tuberculosis-burden country. J Infect 62:347–354. 10.1016/j.jinf.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 7.González-Moreno J, García-Gasalla M, Losada-López I, Cifuentes Luna C, Mir Viladrich I, Fernández-Baca V, Serrano A, Juan Mas A, Riera-Oliver J, Payeras Cifre A. 2018. IGRA testing in patients with immune-mediated inflammatory diseases: which factors influence the results? Rheumatol Int 38:267–273. 10.1007/s00296-017-3852-9. [DOI] [PubMed] [Google Scholar]
- 8.Remy KE, Mazer M, Striker DA, Ellebedy AH, Walton AH, Unsinger J, Blood TM, Mudd PA, Yi DJ, Mannion DA, Osborne DF, Martin RS, Anand NJ, Bosanquet JP, Blood J, Drewry AM, Caldwell CC, Turnbull IR, Brakenridge SC, Moldwawer LL, Hotchkiss RS. 2020. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mudd PA, Crawford JC, Turner JS, Souquette A, Reynolds D, Bender D, Bosanquet JP, Anand NJ, Striker DA, Martin RS, Boon ACM, House SL, Remy KE, Hotchkiss RS, Presti RM, O’Halloran JA, Powderly WG, Thomas PG, Ellebedy AH. 2020. Distinct inflammatory profiles distinguish COVID-19 from influenza with limited contributions from cytokine storm. Sci Adv 6:eabe3024. 10.1126/sciadv.abe3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paliogiannis P, Zinellu A. 2020. Bilirubin levels in patients with mild and severe Covid‐19: a pooled analysis. Liver Int 40:1787–1788. 10.1111/liv.14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F, Li L, Xu MD, Wu J, Luo D, Zhu YS, Li BX, Song XY, Zhou X. 2020. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol 127:104370. 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu MY, Yao L, Wang Y, Zhu XY, Wang XF, Tang PJ, Chen C. 2020. Clinical evaluation of potential usefulness of serum lactate dehydrogenase (LDH) in 2019 novel coronavirus (COVID-19) pneumonia. Respir Res 21:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cummings MJ, Baldwin MR, Abrams D, Jacobson SD, Meyer BJ, Balough EM, Aaron JG, Claassen J, Rabbani LE, Hastie J, Hochman BR, Salazar-Schicchi J, Yip NH, Brodie D, O'Donnell MR. 2020. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet 395:1763–1770. 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wendel Garcia PD, Fumeaux T, Guerci P, Heuberger DM, Montomoli J, Roche-Campo F, Schuepbach RA, Hilty MP, Alfaro Farias M, Margarit A, Vizmanos-Lamotte G, Tschoellitsch T, Meier J, Cardona FS, Skola J, Horakova L, Aguirre-Bermeo H, Apolo J, Novy E, Losser M-R, Jurkolow G, Delahaye G, David S, Welte T, Wengenmayer T, Staudacher DL, Aslanidis T, Babik B, Korsos A, Gal J, Csaba H, Donati A, Carsetti A, Turrini F, Simonini MS, Ceriani R, Murrone M, Rezoagli E, Vitale G, Fogagnolo A, Spadaro S, Wu MA, Cogliati C, Colombo R, Catena E, Facondini F, Potalivo A, Gangitano G, Perin T, Bocci MG, et al. 2020. Prognostic factors associated with mortality risk and disease progression in 639 critically ill patients with COVID-19 in Europe: initial report of the international RISC-19-ICU prospective observational cohort. EClinicalMedicine 25:100449. 10.1016/j.eclinm.2020.100449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oberholzer A, Souza SM, Tschoeke SK, Oberholzer C, Abouhamze A, Pribble JP, Moldawer LL. 2005. Plasma cytokine measurements augment prognostic scores as indicators of outcome in patients with severe sepsis. Shock 23:488–493. [PubMed] [Google Scholar]
- 16.Sinha P, Matthay MA, Calfee CS. 2020. Is a “cytokine storm” relevant to COVID-19? JAMA Intern Med 180:1152. 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 17.Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. 2020. The COVID-19 cytokine storm; what we know so far. Front Immunol 11:1446. 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zwerling A, Van Den Hof S, Scholten J, Cobelens F, Menzies D, Pai M. 2012. Interferon-gamma release assays for tuberculosis screening of healthcare workers: a systematic review. Thorax 67:62–70. 10.1136/thx.2010.143180. [DOI] [PubMed] [Google Scholar]
- 19.Guaraldi G, Meschiari M, Cozzi-Lepri A, Milic J, Tonelli R, Menozzi M, Franceschini E, Cuomo G, Orlando G, Borghi V, Santoro A, Di Gaetano M, Puzzolante C, Carli F, Bedini A, Corradi L, Fantini R, Castaniere I, Tabbì L, Girardis M, Tedeschi S, Giannella M, Bartoletti M, Pascale R, Dolci G, Brugioni L, Pietrangelo A, Cossarizza A, Pea F, Clini E, Salvarani C, Massari M, Viale PL, Mussini C. 2020. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol 2:e474–e484. 10.1016/S2665-9913(20)30173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Li H, Guo X, Yoshida EM, Mendez-Sanchez N, Levi Sandri GB, Teschke R, Romeiro FG, Shukla A, Qi X. 2020. Incidence, risk factors, and prognosis of abnormal liver biochemical tests in COVID-19 patients: a systematic review and meta-analysis. Hepatol Int 14:621–637. 10.1007/s12072-020-10074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, Li R, Wang J, Jiang Q, Gao C, Yang J, Ge L, Hu Q. 2020. Correlation analysis between disease severity and clinical and biochemical characteristics of 143 cases of COVID-19 in Wuhan, China: a descriptive study. BMC Infect Dis 10.1186/s12879-020-05242-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosmann TR, Sad S. 1996. The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol Today 17:138–146. 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- 23.Mandalakas AM, Detjen AK, Hesseling AC, Benedetti A, Menzies D. 2011. Interferon-gamma release assays and childhood tuberculosis: systematic review and meta-analysis. Int J Tuberc Lung Dis 15:1018–1032. 10.5588/ijtld.10.0631. [DOI] [PubMed] [Google Scholar]
- 24.Abbas AK, Murphy KM, Sher A. 1996. Functional diversity of helper T lymphocytes. Nature 383:787–793.[8893001]. 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 25.Song YG, Shin HS. 2020. COVID-19, a clinical syndrome manifesting as hypersensitivity pneumonitis. Infection and chemotherapy. Korean Society of Infectious Diseases, Korean Society for Antimicrobial Therapy, Korean Society for AIDS, Seoul, South Korea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stone JH, Frigault MJ, Serling-Boyd NJ, Fernandes AD, Harvey L, Foulkes AS, Horick NK, Healy BC, Shah R, Bensaci AM, Woolley AE, Nikiforow S, Lin N, Sagar M, Schrager H, Huckins DS, Axelrod M, Pincus MD, Fleisher J, Sacks CA, Dougan M, North CM, Halvorsen Y-D, Thurber TK, Dagher Z, Scherer A, Wallwork RS, Kim AY, Schoenfeld S, Sen P, Neilan TG, Perugino CA, Unizony SH, Collier DS, Matza MA, Yinh JM, Bowman KA, Meyerowitz E, Zafar A, Drobni ZD, Bolster MB, Kohler M, D’Silva KM, Dau J, Lockwood MM, Cubbison C, Weber BN, Mansour MK. 2020. Efficacy of tocilizumab in patients hospitalized with Covid-19. N Engl J Med 383:2333–2344. 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mojtabavi H, Saghazadeh A, Rezaei N. 2020. Interleukin-6 and severe COVID-19: a systematic review and meta-analysis. Eur Cytokine Netw 31:44–49. 10.1684/ecn.2020.0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montalvo Villalba MC, Valdés Ramírez O, Muné Jiménez M, Arencibia Garcia A, Martinez Alfonso J, González Baéz G, Roque Arrieta R, Rosell Simón D, Alvárez Gainza D, Sierra Vázquez B, Resik Aguirre S, Guzmán Tirado MG. 2020. Interferon gamma, TGF-β1 and RANTES expression in upper airway samples from SARS-CoV-2 infected patients. Clin Immunol 220:108576. 10.1016/j.clim.2020.108576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2. Download JCM.00811-21-s0001.pdf, PDF file, 0.3 MB (326.9KB, pdf)