Abstract
Background:
Increasing evidence has shown that gut microbiota may play a role in colorectal cancer. Diet, particularly fiber intake, may modify gut microbiota composition, which may consequently impact cancer risk.
Objective:
We investigated the relationship between total dietary fiber intake and gut microbiota in healthy humans.
Design:
Using 16S rRNA gene sequencing, we assessed gut microbiota in fecal samples from 151 healthy adults in two independent study populations: NCI, n= 75 (healthy controls from a colorectal cancer case-control study), and NYU, n=76 (polyp-free subjects from a cross-sectional colonoscopy study). We calculated energy-adjusted total dietary fiber intake of participants based on food frequency questionnaires. For each study population, we evaluated the relationship between quartiles of higher fiber intake as a continuous ordinal variable, and global gut microbiota community composition (via PERMANOVA of weighted UniFrac distance) and specific taxon abundance (via DESeq2).
Results:
Total fiber intake was significantly associated with overall microbial community composition in NYU (p=0.008) but not NCI (p=0.81), after adjustment for age, sex, race, body mass index, and cigarette smoking. In a taxonomy-based meta-analysis of these two study populations, higher fiber intake was associated with higher abundance of select genera from class Clostridia: SMB53 (fold change [FC]=1.04, p=0.04), Lachnospira (FC=1.03, p=0.05), and Faecalibacterium (FC=1.03, p=0.06), and lower abundance of genera Actinomyces (FC=0.95, p=0.002), Odoribacter (FC=0.95, p=0.03), and Oscillospira (FC=0.96, p=0.06). A species-level meta-analysis showed a marginal association between higher fiber intake and higher abundance of Faecalibacterium prausnitzii (FC=1.03, p=0.07) and lower abundance of Eubacterium dolichum (FC=0.96, p=0.04) and Bacteroides uniformis (FC=0.97, p=0.05).
Conclusions:
Our results suggest that increased dietary fiber intake may impact gut microbiota composition in healthy adults, particularly in favor of putatively beneficial bacteria such as Faecalibacterium prausnitzii. Given the potentially modifiable nature of gut microbiota through diet, these findings warrant further study of diet-microbiota based colorectal cancer prevention strategies.
Keywords: Gut Microbiome, diet fiber intake, cross-sectional study, epidemiology
Introduction
The human gastrointestinal tract hosts an estimated 100 trillion bacteria, which play a role in key physiologic activities, including gastrointestinal immune stimulation and fermentation of nutrients into beneficial metabolites (1). Disruption of this symbiotic relationship between human host and gut microbiota has been implicated in the development of intestinal pathology, including inflammatory bowel disease and colorectal cancer (2, 3). A growing number of epidemiologic studies have provided increasing evidence that perturbation of microbial community composition in the gut exists in colorectal cancer, with alterations of microbial taxa abundance in cancer cases compared to healthy controls (4, 5).
Dietary habits have been attributed to colorectal cancer risk development, with Western-style diets—low in fiber and high in red meat and fat—associated with higher risk for colorectal cancer (6, 7). Fiber intake, in particular, has remained an appealing modifiable dietary factor, given its protective biologic effects. Fiber undergoes fermentation by microbiota to yield short-chain fatty acid end-products, such as butyrate, which is not only essential for colon energy metabolism and epithelial proliferation, but in mouse models also exhibits tumor suppressive activity through histone deacetylase inhibition (8).
Consequently, there has been growing interest to understand the impact of dietary fiber on gut microbiota composition, which may ultimately affect cancer risk. While short-term dietary intervention trials have demonstrated that different amounts of fiber intake can significantly alter microbiota composition in a span of a few weeks (9, 10), there remain fewer studies evaluating the effect of long-term dietary habits of fiber intake on gut microbiota in humans (11).
We investigated the association between long-term dietary fiber habits and gut microbiota composition in fecal samples of healthy adults from two independent study populations: healthy controls from a case-control study of colorectal cancer and gut microbiome (5), and polyp-free adults from a cross-sectional colonoscopy study (12). We sought to examine the relationship between higher dietary fiber intake and overall gut microbiota composition, as well as specific taxa abundance.
Methods
Study Population
We assessed fecal samples of healthy adults from two independent study populations: control subjects from a National Cancer Institute (NCI) case-control study, hereafter referred to as NCI (5), and polyp-free adults from a cross-sectional colonoscopy study at New York University (NYU), called the NYU Human Microbiome and Colorectal Tumor study, hereafter referred to as NYU (12).
NCI enrolled participants from three Washington, DC area hospitals from 1985 to 1989 (13, 14). We included 75 control subjects who were awaiting elective surgery for non-oncologic, non-gastrointestinal conditions, and reported no antibiotic intake during the year prior to recruitment. Participants provided 2-day fecal samples that were freeze-dried, and samples with at least 100 mg of lyophilized fecal material available were included for analyses.
NYU enrolled participants from Kips Bay Endoscopy Center in New York City from 2012 to 2014. We included 76 polyp-free participants from this study. We excluded subjects with missing colonoscopy reports, history of inflammatory bowel disease, prior surgical anastomosis, prior history of colorectal cancer, history of familial adenomatous polyposis, and with most recent colonoscopy report >3 years prior to stool sample collection.
In both studies, participants provided written informed consent, reported no long-term antibiotic treatment, and completed diet and demographic questionnaires. We excluded subjects with less than 1000 microbial sequence reads, missing or extreme caloric intake (≤ 500 or >4000 kcal/day), and with a history of other cancers, for a final sample size of 151 (n=75 in NCI, n=76 in NYU). The NYU study was approved by the institutional review board (IRB) of NYU School of Medicine, and the NCI study was approved by the IRB of NYU School of Medicine and the NCI.
Demographic information and dietary fiber assessment
Information on age, sex, height, weight, race, and cigarette smoking status was collected by questionnaire at stool collection. BMI was calculated by dividing weight in kilograms by squared height in meters, and was then categorized as underweight or normal weight (<25kg/m2), overweight (25 ≤ BMI < 30 kg/m2), or obese (BMI ≥ 30 kg/m2), based on WHO definition (15). Cigarette smoking status was defined as never, current, or former smoker.
Usual dietary intake was calculated from self-administered food frequency questionnaires, which queried intake frequency and portion size of food types. Nutrient values per portion were multiplied by daily frequency of intake and summed across all relevant food items, using the US Department of Agriculture pyramid food group serving database (16). Nutrient data were standardized by total calorie intake (17). Study-specific quartiles of fiber intake were used (NCI: <11.21, 11.21-13.90, 13.91-16.50, ≥16.51 g/day; NYU: <20.07, 20.07-24.92, 24.93-30.79, ≥30.8 g/day).
Fecal samples
In NCI, fecal samples were collected by participants at home over a two day period, prior to hospitalization and treatment, and stored in a plastic container in a Styrofoam chest containing dry ice. Fecal samples were shipped to a USDA laboratory, lyophilized, and stored at a minimum of −40°C in sealed, air-tight containers. Sample aliquots were shipped to NYU for microbiome assay. In NYU, fecal samples were collected by participants onto two sections of Beckman Coulter Hemoccult II SENSA® cards (Beckman Coulter, CA) at home. Samples were shipped to NYU and stored immediately at −80°C.
Microbiota assay
In both NCI and NYU, DNA was extracted from fecal samples using the Mo Bio PowerSoil DNA Isolation Kit (Carlsbad, CA) with bead-beating, as previously reported (12). In NCI, 16S rRNA gene amplicons covering variable regions V3 to V4 were generated using the 347F-5′GG AGGCAGCAGTRRGGAAT′-3′ and 803R 5′-CTACCRGGGTATCTAATCC-3′ primer pair (5, 18). Amplicons were sequenced with the 454 Roche FLX Titanium pyrosequencing system, following the manufacturer’s protocol. In NYU, 16S rRNA gene amplicons covering the V4 region were generated using the F515-5’GTGCCAGCMGCCGCGGTAA’-3’ and R806- 5’GGACTACHVGGGTWTCTAAT-3’ primer pair (19). Amplicons were sequenced with the Illumina MiSeq platform.
Sequence data processing
Because two different sequencing platforms were used, we processed the sequence data separately. Sequences were demultiplexed, and poor-quality sequences excluded, using the default parameters of QIIME script split_libraries.py (for NCI) or split_libraries_fastq.py (for NYU) (20). Filtered sequence reads were clustered into de novo operational taxonomic units (OTUs) at 97% identity, and representative sequence reads for each OTU were assigned taxonomy based on fully sequenced microbial genomes (IMG/GG Greengenes) (20). Chimeric sequences were removed with ChimeraSlayer (21). Blinded quality control specimens in all sequencing batches showed good reproducibility: high intraclass correlation coefficients (ICCs) for the Shannon diversity index and abundances of bacterial phyla and genera have been previously reported (5, 12, 22).
Statistical analysis
Alpha-Diversity
We evaluated the association between quartiles of fiber intake and within-subject microbial diversity (α-diversity) indices of Shannon diversity and evenness (23). In both studies, these indices were calculated in 100 iterations of rarefied OTU tables of 1000 sequence reads per sample. We modeled the Shannon index and evenness as outcomes in linear regression, adjusting for age, sex, race, categorical BMI, and cigarette smoking status.
Beta-Diversity
We assessed the relationship of overall gut microbiota composition and quartiles of dietary fiber intake using weighted (quantitative) and unweighted (qualitative) phylogenetic UniFrac distance matrices (24). Permutational multivariate analysis of variance (PERMANOVA) of both weighted and unweighted UniFrac distances was used to evaluate whether fiber intake is associated with overall microbial community composition, after adjusting first for age, sex, race, categorical BMI, and cigarette smoking status (adonis function, ‘vegan’ package in R) (25). Principal coordinate analysis (PCoA) plots were generated using the first two principal coordinates (PCs), and labeled according to quartile of fiber intake.
Differential abundance testing
We assessed the relationship between higher quartiles of total fiber intake and specific taxa abundance using negative binomial generalized linear models, in the “DESeq2” package in R (22). Models were adjusted for age, sex, race, categorical BMI and cigarette smoking status. Nominal p-values and false discovery rate (FDR) adjusted q-values were calculated (26). DESeq2 default outlier replacement, independent filtering of low-count taxa, and filtering of count outliers were turned off. Taxa models with maximum Cook’s distance > 10 were removed prior to p-value adjustment for the FDR (27).
To identify similar taxa associations in both NCI and NYU, we then performed a taxonomy-based meta-analysis to evaluate for genera and species with concomitantly higher or lower abundance by fiber intake in the two study populations. In addition, we performed sub-analyses to examine associations between taxon abundance and higher intake of fiber from specific sources, such as fruits and vegetables, grains, and beans. We calculated nominal meta-analysis p-values based on Z-score methods (28).
All trends were tested using the median values of each quartile of fiber intake. All analyses were performed using R, version 3.3.2.
Results
Subject characteristics.
A total of 151 healthy adult subjects were included for analysis: n = 75 in NCI, and n = 76 in NYU. Subject characteristics are reported in Table 1. Among these participants, 73.3% in NCI and 51.3% in NYU were male, and 82.7% in NCI and 85.5% in NYU were white. Median age in both study groups generally increased with higher fiber intake, though the trends were not statistically significant.
Table 1.
Participant demographic characteristics
| NCI (n=75) | NYU (n=76) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Q1a | Q2a | Q3a | Q4a | p-value | Q1b | Q2b | Q3b | Q4b | p-value | |
| N | 19 | 19 | 18 | 19 | 19 | 19 | 19 | 19 | ||
| Age (median) | 50 | 55 | 61.5 | 62 | 0.11 | 56 | 57 | 62 | 61 | 0.34 |
| Sex, % | 0.08 | 0.16 | ||||||||
| Female | 31.6 | 21.1 | 50.0 | 57.9 | 36.8 | 36.8 | 52.6 | 68.4 | ||
| Male | 68.4 | 78.9 | 50.0 | 42.1 | 63.2 | 63.2 | 47.4 | 31.6 | ||
| Race, % | 0.46 | 0.10 | ||||||||
| White | 89.5 | 89.5 | 77.8 | 73.7 | 94.7 | 89.5 | 89.5 | 68.4 | ||
| Non-White | 10.5 | 10.5 | 22.2 | 26.3 | 5.3 | 10.5 | 10.5 | 31.6 | ||
| BMI (kg/m2), % | ||||||||||
| <25 | 57.9 | 68.4 | 33.3 | 68.4 | 0.19 | 36.8 | 63.2 | 57.9 | 63.2 | 0.29 |
| 25-30 | 36.8 | 15.8 | 38.9 | 21.1 | 42.1 | 31.6 | 26.3 | 36.8 | ||
| ≥30 | 5.3 | 15.8 | 27.8 | 10.5 | 21.1 | 5.3 | 15.8 | - | ||
| Smoking History, % | 0.10 | 0.13 | ||||||||
| Never | 26.3 | 36.8 | 61.1 | 57.9 | 63.2 | 84.2 | 47.4 | 63.2 | ||
| Former/Current | 73.7 | 63.2 | 38.9 | 42.1 | 36.8 | 15.8 | 52.6 | 36.8 | ||
NCI quartiles of fiber intake: Q1: < 11.25 g; Q2: 11.25-14.34 g; Q3: 14.35-16.68 g; Q4: ≥ 16.69 g
NYU quartiles of fiber intake: Q1: < 19.82 g; Q2: 19.82-23.82 g; Q3: 23.83-31.63 g; Q4: ≥ 31.64 g
Global diversity.
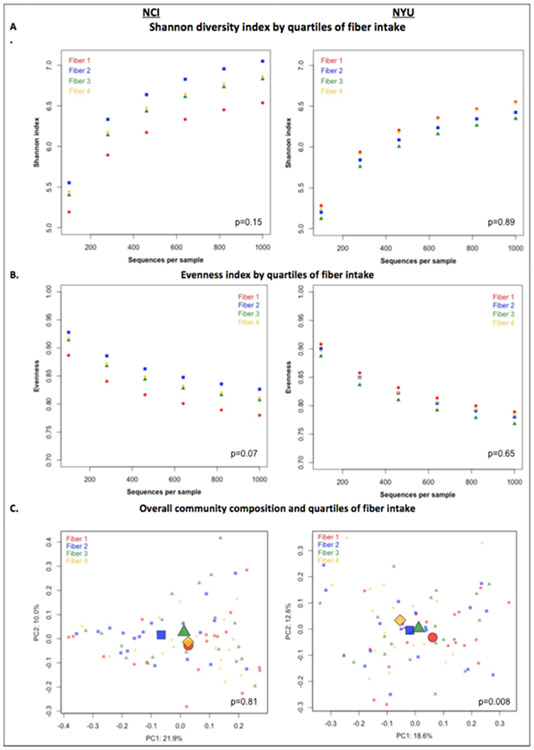
PERMANOVA analyses of between-sample UniFrac distances demonstrated that fiber intake was significantly associated with overall microbial community composition in NYU (weighted UniFrac p=0.008; unweighted UniFrac p=0.01) but not NCI (weighted UniFrac p=0.81; unweighted UniFrac p=0.75) after adjusting for covariates of age, sex, race, BMI, and cigarette smoking [Figure 1C]. However, in both NCI and NYU, total fiber intake was not significantly associated with microbial community diversity as measured by the Shannon diversity index or evenness (Figure 1A).
Figure 1. Alpha and beta diversity in relation to quartiles of fiber intake.
A) Shannon diversity index and B) Evenness index by quartiles of fiber intake are shown in N=151 healthy adult subjects from two independent study populations (NCI=75, NYU=76). These indices were calculated in 100 iterations of rarefied OTU tables of 1000 sequence reads per sample. Fiber 1, Fiber 2, Fiber 3, and Fiber 4 represent increasing quartiles of fiber intake. Shannon index and evenness were modeled as outcomes in linear regression, adjusting for age, sex, race, categorical BMI, and cigarette smoking status. P-values of fiber variable in regression analysis are reported in the figure. C) Principal Coordinate Analysis (PCoA) plots, based on weighted Unifrac phylogenetic distances, showed a difference between lowest and highest fiber intake in NYU. This relationship was not observed in NCI. PCoA plots were generated using the first two principal coordinates. P-values reported in the figure are based on PERMANOVA of weighted UniFrac distances evaluating the association between fiber intake and overall microbial community composition, after adjusting for age, sex, race, categorical BMI, and cigarette smoking status.
Taxon abundance.
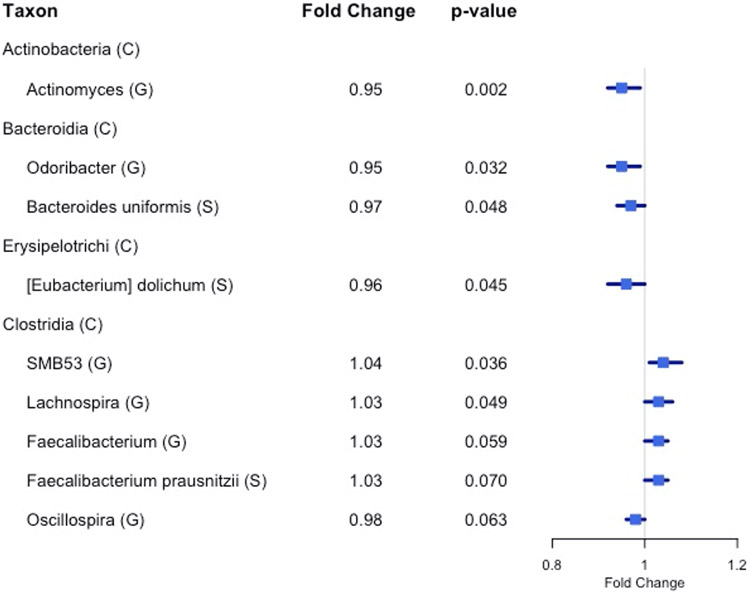
We found 14 genera with the same direction of association with fiber intake in NCI and NYU, out of 29 total genera observed overlapping in both studies. Higher total fiber intake was associated with lower abundance of genera Actinomyces (fold change [FC]=0.95, p=0.002) of class Actinobacteria, Odoribacter (FC=0.95, p=0.03) of class Bacteroidia, and Oscillospira (FC=0.96, p=0.06) of class Clostridia. Higher total fiber intake was associated with higher abundance of selected genera of class Clostridia: SMB53 (FC=1.04, p=0.04), Lachnospira (FC=1.03, p=0.05), and Faecalibacterium (FC=1.03, p=0.06) [Figure 2; Table 2].
Figure 2. Forest plot of fold change of select genera (G) and species (S) in both NYU and NCI with significant or marginally significant association with higher fiber intake, based on meta-anlysis of the two study populations.
Nominal meta-analysis p-values were calculated based on Z-score methods. (C) denotes class.
Table 2.
Meta-analysis of the association between total fiber intake and genera and species with concomitantly increasing or decreasing abundance in NCI and NYU study populations
| Phylum; Class; Order; Family; Genus; Species | NCI | NYU | Meta FC |
Meta 95% CI |
Meta p |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Base Mean |
FC | p- value |
p-adj | Base Mean |
FC | p- value |
p-adj | ||||
| Genus Level | |||||||||||
| Actinobacteria; Actinobacteria; Actinomycetales; Actinomycetaceae; Actinomyces | 2.16 | 0.884 | 0.003 | 0.110 | 0.75 | 0.969 | 0.130 | 0.454 | 0.951 | (0.917-0.986) | 0.0015 |
| Bacteroidetes; Bacteroidia; Bacteroidales; [Odoribacteraceae]; Odoribacter | 0.68 | 0.995 | 0.916 | 0.967 | 10.88 | 0.946 | 0.004 | 0.176 | 0.953 | (0.921-0.987) | 0.0318 |
| Firmicutes; Clostridia; Clostridiales; Clostridiaceae; SMB53 | 0.62 | 1.035 | 0.443 | 0.913 | 0.88 | 1.045 | 0.029 | 0.391 | 1.044 | (1.007-1.082) | 0.0360 |
| Firmicutes; Clostridia; Clostridiales; Lachnospiraceae; Lachnospira | 31.63 | 1.042 | 0.279 | 0.875 | 163.91 | 1.026 | 0.091 | 0.406 | 1.028 | (1.000-1.057) | 0.0497 |
| Firmicutes; Clostridia; Clostridiales; Ruminococcaceae; Faecalibacterium | 230.42 | 1.030 | 0.367 | 0.913 | 549.73 | 1.025 | 0.078 | 0.391 | 1.025 | (1.000-1.051) | 0.0589 |
| Firmicutes; Clostridia; Clostridiales; Ruminococcaceae; Oscillospira | 22.58 | 0.973 | 0.386 | 0.913 | 85.60 | 0.983 | 0.079 | 0.391 | 0.982 | (0.965-1.000) | 0.0630 |
| Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus | 55.61 | 0.957 | 0.299 | 0.875 | 57.84 | 0.985 | 0.443 | 0.797 | 0.980 | (0.947-1.015) | 0.2023 |
| Firmicutes; Clostridia; Clostridiales; Lachnospiraceae; Anaerostipes | 5.92 | 1.007 | 0.859 | 0.967 | 10.66 | 1.018 | 0.287 | 0.641 | 1.016 | (0.986-1.048) | 0.3772 |
| Firmicutes; Clostridia; Clostridiales; Lachnospiraceae; Dorea | 104.47 | 1.022 | 0.418 | 0.913 | 65.24 | 1.004 | 0.666 | 0.797 | 1.006 | (0.988-1.025) | 0.3815 |
| Firmicutes; Clostridia; Clostridiales; Veillonellaceae; Phascolarctobacterium | 18.58 | 1.009 | 0.832 | 0.967 | 34.43 | 1.012 | 0.558 | 0.797 | 1.012 | (0.975-1.049) | 0.5709 |
| Firmicutes; Clostridia; Clostridiales; Veillonellaceae; Dialister | 16.99 | 0.984 | 0.713 | 0.967 | 25.15 | 0.994 | 0.780 | 0.869 | 0.992 | (0.955-1.031) | 0.6476 |
| Proteobacteria; Betaproteobacteria; Burkholderiales; Alcaligenaceae; Sutterella | 4.32 | 1.009 | 0.849 | 0.967 | 85.39 | 1.008 | 0.653 | 0.797 | 1.008 | (0.976-1.040) | 0.6502 |
| Actinobacteria; Actinobacteria; Bifidobacteriales; Bifidobacteriaceae; Bifidobacterium | 84.01 | 1.001 | 0.973 | 0.973 | 90.68 | 1.004 | 0.850 | 0.925 | 1.003 | (0.967-1.041) | 0.8745 |
| Actinobacteria; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Eggerthella | 6.18 | 0.985 | NA | NA | 2.09 | 0.990 | 0.631 | 0.797 | 0.989 | (0.951-1.028) | NA |
| Species Level | |||||||||||
| Firmicutes; Erysipelotrichi; Erysipelotrichales; Erysipelotrichaceae; [Eubacterium]; dolichum | 7.70 | 0.933 | 0.198 | 0.770 | 3.34 | 0.963 | 0.122 | 0.540 | 0.958 | (0.917-1.000) | 0.045 |
| Bacteroidetes; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; uniformis | 32.05 | 0.948 | 0.202 | 0.770 | 305.56 | 0.972 | 0.128 | 0.540 | 0.968 | (0.936-1.001) | 0.048 |
| Firmicutes; Clostridia; Clostridiales; Ruminococcaceae; Faecalibacterium; prausnitzii | 230.05 | 1.032 | 0.354 | 0.884 | 540.38 | 1.024 | 0.103 | 0.540 | 1.026 | (0.999-1.053) | 0.070 |
| Firmicutes; Clostridia; Clostridiales; Ruminococcaceae; Ruminococcus; bromii | 24.12 | 0.966 | 0.489 | 0.951 | 1.65 | 0.969 | 0.161 | 0.578 | 0.968 | (0.930-1.008) | 0.138 |
| Firmicutes; Clostridia; Clostridiales; Lachnospiraceae; Dorea; formicigenerans | 1.92 | 1.033 | 0.447 | 0.915 | 20.11 | 1.018 | 0.323 | 0.730 | 1.020 | (0.988-1.054) | 0.216 |
| Bacteroidetes; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; ovatus | 5.59 | 0.987 | 0.790 | 0.951 | 12.37 | 0.982 | 0.309 | 0.730 | 0.982 | (0.950-1.016) | 0.363 |
| Actinobacteria; Actinobacteria; Bifidobacteriales; Bifidobacteriaceae; Bifidobacterium; adolescentis | 38.42 | 1.007 | 0.898 | 0.951 | 75.56 | 1.002 | 0.932 | 0.964 | 1.003 | (0.959-1.049) | 0.880 |
| Actinobacteria; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Eggerthella; lenta | 6.14 | 0.984 | NA | NA | 1.99 | 0.986 | 0.613 | 0.839 | 0.986 | (0.939-1.035) | NA |
Relationship between higher quartiles of fiber intake and differential taxon abundance was evaluated using negative binomial generalized linear models in the DESeq2 package in R. Models were adjusted for age, sex, race, categorical BMI and cigarette smoking status. This table reports results from a taxonomy-based meta-analysis to evaluate for genera and species with concomitantly higher or lower abundance by fiber intake (as determined by fold change [FC]) in both study populations.
At the species level, we found 8 species with the same direction of association with fiber intake in the two study populations, out of 17 total species observed overlapping in both studies. A meta-analysis at this taxonomic level showed a marginal association between higher total fiber intake and higher abundance of Faecalibacterium prausnitzii (FC=1.03, p=0.07), and lower abundance of Eubacterium dolichum (FC=0.96, p=0.04) and Bacteroides uniformis (FC=0.97, p=0.05) [Figure 2; Table 2].
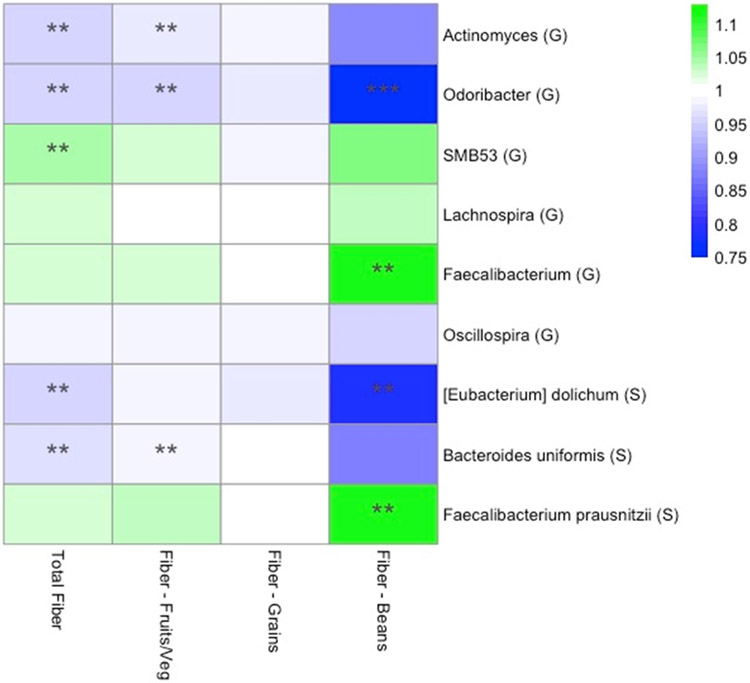
In a sub-analysis to identify associations between taxon abundance and fiber intake from specific dietary sources, we found that higher fiber intake from fruits and vegetables was associated with lower abundance of genera Actinomyces (FC=0.97, p=0.007), Odoribacter (FC= 0.96, p=0.04), and Oscillospira (FC=0.99, p=0.06). Higher abundance of genus Faecalibacterium was most significant specifically with higher fiber intake from beans (FC=1.11, p=0.01). At the species level, higher fiber intake from beans was associated with higher abundance of Faecalibacterium prausnitzii (FC=1.11, p=0.01), and lower abundance of Bacteroides uniformis (FC=0.87, p=0.08) [Figure 3].
Figure 3.
Heat map representing color-coded fold changes of select genera (G) and species (S), by total fiber and fiber from specific sources. ** denotes p<0.05
Although we observed similar associations with taxon abundance and fiber intake in both the NCI and NYU studies, we also noted some inconsistent associations [Supplementary Table 1]. For example, at the phylum level, higher total fiber intake was associated with higher abundance of Proteobacteria in NCI (p=0.03, q=0.10) but not in NYU (p=0.53, q=0.62). Higher total fiber intake was also marginally associated with higher abundance of phylum Bacteroidetes in NYU (p=0.07, q=0.26), but not in NCI (p=0.47, q=0.89). At the order level, higher total fiber intake was associated with lower abundance of Coriobacteriales (of phylum Actinobacteria) in NYU (p=0.008, q=0.10), but not in NCI (p=0.82, q=0.99).
Discussion
In this study, we examined the association of usual dietary fiber intakes with gut microbiota composition in healthy adults from two independent study populations. In a taxonomy-based meta-analysis, we found that higher total fiber intake is associated with specific taxon abundances, including higher abundance of select genera of Clostridia class. Some of these fiber and taxon abundance associations were consistent with specific fiber food sources, such as fruits and vegetables, grains, and beans.
The usual dietary intake of participants measured in our study provides additional insight into the potential effect of longer-term dietary patterns on gut microbiota composition, compared with controlled dietary intervention studies. Although Wu et al demonstrated that certain dietary modifications altered microbiome composition, they did not affect overall enterotype, possibly because this may be better correlated with long-term diet (11). Moreover, our study associations are more likely to represent microbial composition in an uncontrolled, real-world setting.
We found that higher fiber intake was associated with higher abundance of select genera of Clostridia class. This finding is notable given the particular role of Clostridium spp in colonocyte metabolism through production of short chain fatty acids (SCFAs) via fermentation. Butyrate, one of these SCFAs, serves as the preferred energy source for colonocytes (29, 30). Mouse models have shown that butyrate inhibits histone deacetylases and consequently affects gene expression and causes tumor suppression (8, 31), carrying implications for colorectal cancer treatment.
Within the Clostridia class, we noted a marginal association between higher fiber intake and higher abundance of species Faecalibacterium prausnitzii. The fiber and F. prausnitzii relationship has also been observed in a cross-sectional cohort of middle-age and older adults in Spain, in which greater adherence to a Mediterranean diet, rich in higher fiber content foods such as fruits, vegetables, and whole grains, correlated with higher levels of F. prausnitzii as well as Clostridium cluster XVIa (32). F. prausnitzii, one of the most abundant species found in the gut and a key producer of butyrate, has been associated with anti-inflammatory activity (33-35). Sokol and colleagues suggested that it exerts its anti-inflammatory effects on cellular and colitis mouse models, in part due to associated metabolites blocking NF-kB and IL-8 production (35). Moreover, a reduction in F. prausnitzii was associated with higher risk of recurrence of Crohn's disease in patients post-resection. In addition to inflammatory bowel disease, Lopez-Siles et al. reported that lower levels of F. prausnitzii were also found in patients with colorectal cancer compared to healthy controls (36). Thus, these results suggest a potential therapeutic and preventive role of F. prausnitzii in countering microbial dysbiosis in human disease. Our finding of higher abundance of F. prausnitzii particularly with higher fiber intake from beans lends support to future investigation of specific diet modifications that could impact disease states.
We also observed that higher fiber intake was associated with lower abundance of genera Odoribacter, Actinomyces, and Oscillospira, and higher abundance of genus Lachnospira. In mouse models, Zackular et al. reported significant microbial shifts found within stool samples of mice with colon tumors, specifically with enrichment of OTUs affiliated with members of Odoribacter (37). Along similar lines, Thomas and colleagues examined human tissue samples collected during colonoscopy from rectal cancer cases and non-cancer controls, and noted higher abundance of Odoribacter in the case tissues (38). Kasai et al noted higher proportions of several genera, including Actinomyces, in fecal samples from human subjects with colorectal carcinoma (39). Furthermore, higher abundance of Actinomyces has been reported in both colorectal adenoma and carcinoma cases (12, 39). Whether or not enrichment or depletion of these specific microbiota reflects cause or consequence of colon tumor development remains uncertain. Nonetheless, their opposite relationship with higher fiber intake in our healthy adult population is notable, and merits further examination of diet, microbiota, and disease associations.
Similarly, we previously reported an association between lower abundance of Clostridia and both colorectal carcinoma and adenoma (5, 12). In a case-control study of colorectal cancer cases and controls, stool samples from colorectal cancer cases were characterized by depletion of phylum Firmicutes, predominantly of class Clostridia, relative to controls (5). In addition, we noted a depleted abundance of members of Clostridia class in stool samples of colorectal adenoma cases, compared with controls (12). These findings suggest a potential relationship between this shift in Clostridia composition and both pre-cancerous and cancerous events. Thus, our finding that higher dietary fiber intake is associated with a higher abundance of select genera of Clostridia class carries implications for protective strategies against colorectal cancer risk.
Several potential limitations need to be considered. Measurement error is an inherent limitation in self-reported dietary assessment. Subjects in both study cohorts were mostly white, and thus our findings may not be generalizable to more racially diverse populations. The cross-sectional design of both studies limits assessment of the temporality of the diet-microbiota relationship. Nonetheless, strengths of this study include the relatively large sample size, excellent quality control of microbiome assays in both studies, and adjustment for potential confounders. Furthermore, our study hypothesis was tested in two independent study populations, and our meta-analysis findings confirmed similar taxon abundance associations across both populations.
In summary, we demonstrate that fiber intake may impact gut microbiota composition in healthy humans. Given the mounting evidence that microbial dysbiosis may impact human health and contribute to development of colorectal cancer, it is imperative to better elucidate the association of diet-microbiota relationships and their potential impact on colorectal cancer risk. This understanding may lay the groundwork needed for diet-microbiota based colorectal cancer prevention strategies.
Supplementary Material
Acknowledgments:
Research reported in this publication was supported in part by the US National Cancer Institute under award numbers U01CA182370, R01CA164964, R03 159414 and P30CA016087. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs or the United States Government.
Abbreviations List:
- NCI
National Cancer Institute
- NYU
New York Unviversity
- FC
fold change
- BMI
body mass index
Footnotes
Disclaimers: None
Conflict of Interest: The authors declare that there are no conflicts of interest.
Contributor Information
Daniel Lin, New York University Langone Medical Center, Perlmutter Cancer Center, New York, NY.
Brandilyn A. Peters, Department of Population Health, New York University School of Medicine, New York, NY.
Charles Friedlander, Concorde Medical Group, New York, NY; Department of Medicine, New York University School of Medicine, New York, NY.
Hal Frieman, Department of Medicine, New York University School of Medicine, New York, NY.
James J. Goedert, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD.
Rashmi Sinha, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD.
George Miller, New York University Langone Medical Center, Perlmutter Cancer Center, New York, NY; Department of Surgery, New York University School of Medicine, New York, NY.
Mitchell A. Bernstein, Department of Surgery, New York University School of Medicine, New York, NY.
Richard B. Hayes, New York University Langone Medical Center, Perlmutter Cancer Center, New York, NY; Department of Population Health, New York University School of Medicine, New York, NY.
Jiyoung Ahn, New York University Langone Medical Center, Perlmutter Cancer Center, New York, NY; Department of Population Health, New York University School of Medicine, New York, NY.
References
- 1.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hold GL, Smith M, Grange C, Watt ER, El-Omar EM, Mukhopadhya I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years? World J Gastroenterol. 2014;20(5):1192–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, Tjalsma H. Towards the human colorectal cancer microbiome. PLoS One. 2011;6(5):e20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6(1):e16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. Human gut microbiome and risk for colorectal cancer. Journal of the National Cancer Institute. 2013;105(24):1907–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouvard V, Loomis D, Guyton KZ, Grosse Y, Ghissassi FE, Benbrahim-Tallaa L, Guha N, Mattock H, Straif K, International Agency for Research on Cancer Monograph Working G. Carcinogenicity of consumption of red and processed meat. Lancet Oncol. 2015;16(16):1599–600. [DOI] [PubMed] [Google Scholar]
- 7.Aune D, Chan DS, Lau R, Vieira R, Greenwood DC, Kampman E, Norat T. Dietary fibre, whole grains, and risk of colorectal cancer: systematic review and dose-response meta-analysis of prospective studies. BMJ. 2011;343:d6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, Hillhouse A, Curry KP, Renner SW, Greenwalt A, Ryan EP, et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4(12):1387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tap J, Furet JP, Bensaada M, Philippe C, Roth H, Rabot S, Lakhdari O, Lombard V, Henrissat B, Corthier G, et al. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol. 2015;17(12):4954–64. [DOI] [PubMed] [Google Scholar]
- 11.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, Yuen E, Freiman H, Lustbader I, Salik J, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiffman MH, Van Tassell RL, Robinson A, Smith L, Daniel J, Hoover RN, Weil R, Rosenthal J, Nair PP, Schwartz S, et al. Case-control study of colorectal cancer and fecapentaene excretion. Cancer Res. 1989;49(5):1322–6. [PubMed] [Google Scholar]
- 14.Schiffman MH, Andrews AW, Van Tassell RL, Smith L, Daniel J, Robinson A, Hoover RN, Rosenthal J, Weil R, Nair PP, et al. Case-control study of colorectal cancer and fecal mutagenicity. Cancer Res. 1989;49(12):3420–4. [PubMed] [Google Scholar]
- 15.Organization WH. Classification BMI. Global Database on Body Mass Index. 2006. [Google Scholar]
- 16.Subar AF, Midthune D, Kulldorff M, Brown CC, Thompson FE, Kipnis V, Schatzkin A. Evaluation of alternative approaches to assign nutrient values to food groups in food frequency questionnaires. Am J Epidemiol. 2000;152(3):279–86. [DOI] [PubMed] [Google Scholar]
- 17.Willett WC, Howe GR, Kushi LH. Adjustment for total energy intake in epidemiologic studies. Am J Clin Nutr. 1997;65(4 Suppl):1220S–8S; discussion 9S-31S. [DOI] [PubMed] [Google Scholar]
- 18.Nossa CW, Oberdorf WE, Yang L, Aas JA, Paster BJ, Desantis TZ, Brodie EL, Malamud D, Poles MA, Pei Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J Gastroenterol. 2010;16(33):4135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108Suppl 1:4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon CE. The mathematical theory of communication. 1963. MD Comput. 1997;14(4):306–17. [PubMed] [Google Scholar]
- 24.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73(5):1576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MBHA MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82(1):290–7. [Google Scholar]
- 26.BYH Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. 1995;Series B (Methodological).(1):289–300. [Google Scholar]
- 27.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1-2):279–84. [DOI] [PubMed] [Google Scholar]
- 28.Evangelou E, Ioannidis JP. Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet. 2013;14(6):379–89. [DOI] [PubMed] [Google Scholar]
- 29.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217(2):133–9. [DOI] [PubMed] [Google Scholar]
- 30.Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, Bultman SJ. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13(5):517–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48(4):612–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez-Diaz I, Fernandez-Navarro T, Salazar N, Bartolome B, Moreno-Arribas MV, de Andres-Galiana EJ, Fernandez-Martinez JL, de Los Reyes-Gavilan CG, Gueimonde M, Gonzalez S. Adherence to a Mediterranean Diet Influences the Fecal Metabolic Profile of Microbial-Derived Phenolics in a Spanish Cohort of Middle-Age and Older People. J Agric Food Chem. 2017;65(3):586–95. [DOI] [PubMed] [Google Scholar]
- 33.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294(1):1–8. [DOI] [PubMed] [Google Scholar]
- 35.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Siles M, Martinez-Medina M, Suris-Valls R, Aldeguer X, Sabat-Mir M, Duncan SH, Flint HJ, Garcia-Gil LJ. Changes in the Abundance of Faecalibacterium prausnitzii Phylogroups I and II in the Intestinal Mucosa of Inflammatory Bowel Disease and Patients with Colorectal Cancer. Inflamm Bowel Dis. 2016;22(1):28–41. [DOI] [PubMed] [Google Scholar]
- 37.Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD. The gut microbiome modulates colon tumorigenesis. mBio. 2013;4(6):e00692–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas AM, Jesus EC, Lopes A, Aguiar S Jr., Begnami MD, Rocha RM, Carpinetti PA, Camargo AA, Hoffmann C, Freitas HC, et al. Tissue-Associated Bacterial Alterations in Rectal Carcinoma Patients Revealed by 16S rRNA Community Profiling. Frontiers in cellular and infection microbiology. 2016;6:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H, Tameda M, Shiraki K, Ito M, Takei Y, et al. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: Terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol Rep. 2016;35(1):325–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.