ABSTRACT
The purpose of this study was to characterize the diagnostic performance of a newly developed enzyme-linked immunosorbent assay (ELISA) for detection of SARS-CoV-2 nucleocapsid protein (NP) in blood. Blood samples were collected during hospitalization of 165 inpatients with PCR-confirmed SARS-CoV-2 infection and from 505 outpatients predominantly with relevant symptoms of COVID-19 simultaneously with PCR testing. For the 143 inpatients who had their first blood sample collected within 2 weeks after PCR-confirmed infection, the diagnostic sensitivity of the ELISA was 91.6%. The mean NP concentration of the 131 ELISA-positive blood samples was 1,734 pg/ml (range, 10 to 3,840 pg/ml). An exponential decline in NP concentration was observed for 368 blood samples collected over the first 4 weeks after PCR-confirmed SARS-CoV-2 infection, and all blood samples taken later had an NP concentration below the 10-pg/ml diagnostic cutoff. The diagnostic sensitivity of the ELISA was 81.4% for the 43 blood samples collected from outpatients with a simultaneous positive PCR test, and the mean NP concentration of the 35 ELISA-positive samples was 157 pg/ml (range, 10 to 1,377 pg/ml). For the 462 outpatients with a simultaneous negative PCR test, the diagnostic specificity of the ELISA was 99.8%. In conclusion, the SARS-CoV-2 NP ELISA is a suitable laboratory diagnostic test for COVID-19, particularly for hospitals, where blood samples are readily available and screening of serum or plasma by ELISA can facilitate prevention of nosocomial infections and reduce the requirement for laborious swab sampling and subsequent PCR analysis to confirmatory tests only.
KEYWORDS: COVID-19, SARS-CoV-2, diagnostics, nucleocapsid protein
INTRODUCTION
The pandemic coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) virus has led to the rapid development and widespread application of many laboratory diagnostic tests (1). According to the World Health Organization (WHO), the standard confirmation of acute infections with SARS-CoV-2 is based on a nucleic acid amplification test, such as real-time reverse transcription-PCR, for the presence of unique sequences of SARS-CoV-2 RNA (2). Testing for genomic RNA by PCR is widely supplemented by two other major diagnostic test principles: testing for specific virus antigens and humoral immune response to the infection. Like PCR, analyses for SARS-CoV-2 antigens are typically employed before the onset of clinical symptoms of COVID-19 or during the anticipated acute phase of infection. In contrast, immunoassays for humoral antibodies directed against components of SARS-CoV-2 should not be applied until about 10 days after symptom onset, when the expected humoral immune response has matured sufficiently to reach a detectable level (1).
These three fundamental in vitro diagnostic test principles have their individual advantages and limitations, which partially are associated with their respective sampling techniques for appropriate test material. For almost all PCR analyses for SARS-CoV-2 RNA and immunoassays for SARS-CoV-2 antigen, the hitherto preferred test material is extracted from swabs collected from the upper respiratory tract (URT). In contrast to this heterogeneously composed, individually fluctuating, and somewhat ill-defined test material, immunoassays for antibodies to SARS-CoV-2 rely on a blood sample. In general, blood samples are by far the most used biological material in laboratory diagnostic procedures, and consistencies and variations of this sample material are very well characterized.
Shortly after the severe acute respiratory syndrome (SARS) epidemic in 2002 to 2004, it was reported that the nucleocapsid protein (NP) of the original SARS coronavirus (SARS-CoV) could be detected by enzyme-linked immunosorbent assay (ELISA) in serum samples collected from 95% of infected patients 3 days after symptom onset (3). The SARS-CoV-2 NP is highly conserved and 90.5% identical to the primary structure of SARS-CoV NP, whereas the full proteome identity of these two viruses is 77.1% (4). Inspired by these observations, a new ELISA has been developed and tested for detection of SARS-CoV-2 NP antigen in blood samples collected from COVID-19 patients during the early stages of SARS-CoV-2 infection (5–7). By using PCR analysis of URT swabs as a reference, the present clinical study reports the laboratory diagnostic characteristics and performance of this NP ELISA when used for SARS-CoV-2 antigen quantification in serum and plasma samples.
MATERIALS AND METHODS
Patients and blood samples.
Venous blood was collected from patients at two Danish university hospitals and prepared as either serum or EDTA plasma according to the standard operating procedures of Bio- and GenomeBank, Denmark (8). All blood samples were collected from patients who had not been COVID-19 vaccinated.
Serum samples were obtained from two different patient groups, inpatients with symptoms of COVID-19 and a confirmatory PCR-positive test result admitted to a COVID-19-specific department at University Hospital Rigshospitalet, Copenhagen, and outpatients referred for testing for SARS-CoV-2 infection at an outpatient testing facility at University Hospital Rigshospitalet. Only outpatients with symptoms of upper respiratory tract infection (e.g., fever, sore throat, and cough) were included in the study.
Plasma samples were obtained from two different patient groups, inpatients with symptoms of COVID-19 and a confirmatory PCR-positive test result admitted to COVID-19-specific departments at Aalborg University Hospital and outpatients referred for testing for SARS-CoV-2 infection at an outpatient testing facility at Aalborg University Hospital, including persons with and without symptoms of SARS-CoV-2 and persons who had been exposed.
For each inpatient, 1 to 10 blood samples collected within the interval from the day of their first PCR-positive URT swab (day 0) until day 201 were included in the study. For outpatients, only the blood sample collected simultaneously with their URT swab (day 0) was included in the study.
The serum and plasma samples were stored at −20°C or −80°C until testing by ELISA.
All patients provided written statements for being part of Bio- and Genome Bank, Denmark. For participants under the age of 18 years, a parent or legal guardian provided consent. The present methodology study was approved by the Central Denmark Region Committees on Biomedical Research Ethics (IORG-number 0005129).
PCR analysis.
For all in- and outpatients included in this study, the reference laboratory diagnosis of COVID-19 was performed at the involved hospitals by PCR analysis of URT swabs for the presence of unique sequences of SARS-CoV-2 RNA. All URT swabs were collected according to Danish national guidelines (9) by health professionals and taken as oropharyngeal samples. The swabs were then processed as routine samples and analyzed using a real-time reverse-transcription PCR assay. Two different PCR test kits were used, either the Cobas SARS-CoV-2 test on a Cobas 6800 system (Roche, Basel, Switzerland) or the RealStar SARS-CoV-2 RT-PCR kit (Altona, Hamburg, Germany). The result of PCR analysis was reported as positive or negative for SARS-CoV-2 genomic RNA.
Quantification of NP in blood samples.
The quantification of NP concentration [NP] in serum or plasma was accomplished in approximately 2 h by the Solsten SARS-CoV-2 antigen ELISA kit from Solsten Diagnostics International (Aarhus, Denmark) according to the manufacturer’s guidelines. Up to 12 strips of each 8 wells precoated with antibody to SARS-CoV-2 NP were mounted in each 96-well frame. First, 50 μl of biotin-conjugated antibody was added to each well and then directly supplemented with 50 μl/well either internal NP calibrator, serum, or plasma. The wells were incubated for 1 h at 37°C, washed, incubated with 100 μl/well peroxidase-conjugated streptavidin for 30 min at 37°C, washed, and then incubated with the provided substrate for 15 min at 37°C before stopping the chromogenic enzyme reaction and measuring the absorbance photometrically. Standard curves based on ELISA results of the 5 internal calibrators were used for quantification of [NP] between 0 and 160 pg/ml, as defined by the manufacturer.
All samples were analyzed twice by the ELISA on different days. The first NP quantification was done blinded for the characteristics of the individual sample, except for being serum or plasma. Similarly, the second ELISA analysis was done blinded for COVID-19 status but with insight into the [NP] determined by the first ELISA run. This allowed the appropriate dilution of samples with previously determined [NP] higher than 100 pg/ml. Serum and plasma samples that, after a 24-fold dilution, produced an ELISA absorbance value higher than the highest NP calibrator (160 pg/ml), were not further diluted for precise quantification but registered as having an [NP] of 3,840 pg/ml.
Statistical analysis.
The statistical uncertainty of the estimates of diagnostic accuracy for the SARS-CoV-2 NP ELISA, including sensitivity, specificity, and predictive values of positive and negative results, were reported as 95% confidence intervals (95% CI). The mean [NP] ± standard deviation (SD) from serum and plasma were compared by two-tailed t tests. A P value of less than 0.05 was considered statistically significant.
RESULTS
Patients and blood samples.
The 670 individuals included in this study comprised 414 females aged 14 to 102 years (mean ± SD, 52 ± 19 years) and 256 males aged 20 to 100 years (mean ± SD, 62 ± 20 years). According to PCR analysis of URT swabs, 208 of these individuals were infected with SARS-CoV-2. The COVID-19 patients comprised 97 females aged 22 to 96 years (mean ± SD, 62 ± 19 years) and 111 males aged 28 to 100 years (mean ± SD, 70 ± 16 years).
A total of 914 human blood samples were collected from the 670 individuals between 3 March 2020 and 2 February 2021 and prepared as either serum (n = 439) or plasma (n = 475). Of these, 447 (49%) blood samples were from 165 COVID-19 inpatients and 43 COVID-19 outpatients, including 173 serum samples and 231 plasma samples collected from 38 and 127 inpatients, respectively, between 0 and 201 days after their first confirmatory PCR-positive URT swab. Furthermore, 15 serum samples and 28 plasma samples were collected from outpatients simultaneously with their first confirmatory PCR-positive URT swab. Most of the blood samples from COVID-19 patients (n = 324, 72%) were collected within 14 days of the first PCR-positive URT swab, confirming the patient’s infection with SARS-CoV-2, and the remaining blood samples (n = 123, 28%) were collected from COVID-19 inpatients more than 2 weeks after their first PCR-positive test. All other blood samples (n = 467, 51%) were collected from 462 outpatients without COVID-19 according to a simultaneously collected PCR-negative URT swab and comprised 251 serum samples from 251 outpatients and 216 plasma samples from 211 outpatients.
A schematic overview of patients and samples of the study is provided in the supplemental material (Fig. S1).
Diagnostic performance of the SARS-CoV-2 NP ELISA.
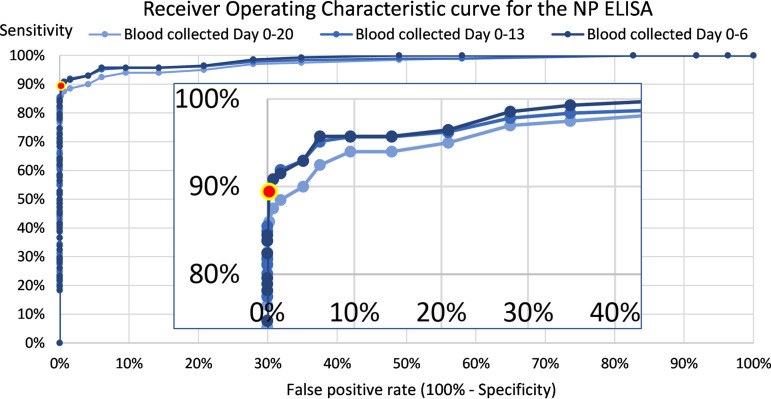
Based on the receiver operating characteristic (ROC) curve in Fig. 1 and prioritization of a low false-positive rate, the manufacturer’s recommended diagnostic cutoff value of 10 pg/ml NP was confirmed. When using this cutoff value, the specificity of the SARS-CoV-2 NP ELISA was 99.8% (95% CI, 99.4% to 100%), as 1 of 462 outpatients without COVID-19 had a false-positive blood sample with an [NP] of 12 pg/ml. All 462 blood samples were collected simultaneously with a PCR-negative URT swab.
FIG 1.
ROC curves for the SARS-CoV-2 NP ELISA according to the time gap from the first PCR-positive URT swab to first blood sampling. A zoom of the upper left corner of the curves was inserted. The area under the curve was 0.986 for the 604 blood samples collected within a time gap of 1 week (day 0 to 6), 0.982 within 2 weeks (648 blood samples collected day 0 to 13), and 0.975 within 3 weeks (662 blood samples collected day 0 to 20). In compliance with the recommendations by the manufacturer, a diagnostic cutoff at 10 pg/ml secured a combination of very low false-positive rate and high sensitivity (red point with yellow halo).
The diagnostic sensitivity of the SARS-CoV-2 NP ELISA was determined at the patient level by using only the [NP] measured for the first blood sample after collection of the confirmatory PCR-positive URT swab. According to results for 160 COVID-19 inpatients, the ELISA sensitivity varied with the time gap from confirmatory PCR-positive URT swab to blood sampling (Fig. 2).
FIG 2.
Variation in the diagnostic sensitivity of the SARS-CoV-2 NP ELISA according to the time gap from the first PCR-positive URT swab to first blood sampling. The blood sample was collected from each of 160 COVID-19 inpatients within 5 weeks after their confirmatory PCR-positive URT swab (red circle) and from each of 43 COVID-19 outpatients simultaneously with their PCR-positive URT swab (blue square). The data point area is proportional to the number of inpatients contributing to the data point.
When the first blood sample was collected from COVID-19 inpatients within 1 and 2 weeks from PCR-confirmed infection, the diagnostic sensitivity of the SARS CoV-2 NP ELISA was 92.9% (n = 99; 95% CI, 87.9% to 98.0%) and 91.6% (n = 143; 95% CI, 87.1% to 96.2%), respectively (Table 1). The average [NP] (±SD) of the true ELISA-positive blood samples collected within the first 2 weeks from 131 COVID-19 inpatients was 1,734 ± 1,560 pg/ml (range, 10 to 3,840 pg/ml; median, 1,184 pg/ml). The average [NP] (±SD) of the true ELISA-positive blood samples collected at day 0 from 35 COVID-19 outpatients was 157 ± 294 pg/ml (range, 10 to 1,377 pg/ml; median, 52 pg/ml).
TABLE 1.
Diagnostic performance and clinical relevance of the SARS-CoV-2 NP ELISA (n = 604 patients)
| No. of patients with SARS-CoV-2 RNA PCR test result |
||||||
|---|---|---|---|---|---|---|
| Time of blood collection | NP ELISA result | Positive, inpatients | Positive, outpatients | Negative, outpatients | Total | Relevance (prediction) |
| Days 0–6 | Positive | 92 | 35 | 1 | 128 | 99.2% |
| Negative | 7 | 8 | 461 | 476 | 96.8% | |
| Total | 99 | 43 | 462 | 604 | ||
| Sensitivity | 92.9% | 81.4% | ||||
| Specificity | 99.8% | |||||
| Days 7–13 | Positive | 39 | 39 | |||
| Negative | 5 | 5 | ||||
| Total | 44 | 44 | ||||
| Sensitivity | 88.6% | |||||
According to all patients (n = 520) with a blood sample collected simultaneously with the URT swab, i.e., at day 0, the PCR-defined point prevalence of COVID-19 was 11.2%: 15 PCR-positive inpatients and 505 outpatients, including 43 PCR positives. For the SARS-CoV-2 NP ELISA, at this time point and prevalence, the specificity and sensitivity were 99.8% (95% CI, 99.4 to 100%) and 82.8% (95% CI, 73.0 to 92.5%), respectively, whereas the positive predictive value (PPV) and negative predictive value (NPV) were 98.0% (95% CI, 94.0 to 100%) and 97.9% (95% CI, 96.6 to 99.2%), respectively.
For COVID-19 outpatients, who had their blood sample collected at day 0, i.e., simultaneously with the confirmatory PCR-positive URT swab, the diagnostic sensitivity of the SARS-CoV-2 NP ELISA was 81.4% (n = 43; 95% CI, 69.8% to 93.0%). For the present study, where the point prevalence of COVID-19 for the analyzed group of 505 outpatients according to PCR analysis was 8.5%, the probability of having COVID-19 was 97.2% for ELISA-positive outpatients (n = 36; 95% CI, 91.9% to 100.0%), and the probability of not being infected with SARS-CoV-2 was 98.3% for ELISA-negative outpatients (n = 469; 95% CI, 97.1% to 99.5%).
Serum and plasma analysis by the SARS-CoV-2 NP ELISA.
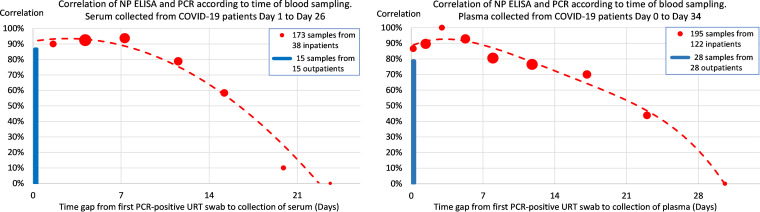
According to 368 blood samples from 160 COVID-19 inpatients, the correlation between SARS-CoV-2 ELISA and PCR confirmed SARS-CoV-2 infection varied with the time gap from confirmatory PCR-positive URT swab to blood sampling (Fig. 3).
FIG 3.
Correlation between the results of SARS-CoV-2 NP ELISA and PCR analysis according to the time gap from the confirmatory PCR-positive URT swab to collection of serum (A) or plasma (B). The 368 blood samples were collected from 160 COVID-19 inpatients within 5 weeks after their confirmatory PCR-positive URT swab (red circle), and 43 blood samples were collected from COVID-19 outpatients simultaneously with their PCR-positive URT swab (blue square). Each inpatient data point area is proportional to the number of blood samples contributing to the data point.
For 131 serum and 150 plasma samples collected from COVID-19 inpatients within 2 weeks after their confirmatory PCR-positive URT swab, the correlation was 89.3% (95% CI, 84.0% to 94.6%) and 86.7% (95% CI, 81.2% to 92.1%), respectively (Table 2). For collection within the first week only, the corresponding correlations were higher: 92.3% (n = 65; 95% CI, 85.8% to 98.8%) for serum and 91.7% (n = 84; 95% CI, 85.8% to 97.6%) for plasma.
TABLE 2.
Correlation between measurement of SARS-CoV-2 RNA by PCR in URT swabs collected at day 0 and measurement of NP by ELISA in serum (n = 397) or plasma (n = 394) collected 0 to 13 days thereafter
| Sample type and NP ELISA result | SARS-CoV-2 RNA PCR test result (no.) |
Relevance (prediction) | |||
|---|---|---|---|---|---|
| Positive, inpatients | Positive, outpatients | ||||
| Negative, outpatients | Total | ||||
| Serum | |||||
| Positive | 117 | 13 | 1 | 131 | 99.2% |
| Negative | 14 | 2 | 250 | 266 | 94.0% |
| Total | 131 | 15 | 251 | 397 | |
| Performance (correlation) | 89.3% | 86.7% | 99.6% | ||
| Plasma | |||||
| Positive | 130 | 22 | 0 | 152 | 100% |
| Negative | 20 | 6 | 216 | 242 | 89.3% |
| Total | 150 | 28 | 216 | 394 | |
| Performance (correlation) | 86.7% | 78.6% | 100% | ||
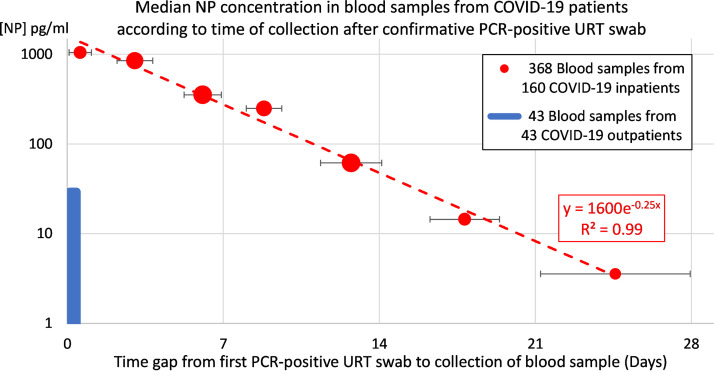
The average [NP] (±SD) of 65 serum samples and 84 plasma samples collected from COVID-19 inpatients within the first week from their confirmatory PCR-positive URT swab was 1,041 ± 1,332 pg/ml (range, 3 to 3,840 pg/ml; median, 337 pg/ml) and 1,631 ± 1,553 pg/ml (range, 3 to 3,840 pg/ml; median, 1,036 pg/ml), respectively. The median [NP] decreased exponentially with the time gap from collection of the PCR-positive URT swab to blood sampling (Fig. 4). No systematic difference was observed in the [NP] levels between serum and plasma samples collected within the first week from COVID-19 inpatients (P = 0.0577).
FIG 4.
Median [NP] declined exponentially over time for 368 blood samples collected from 160 COVID-19 inpatients within 5 weeks after their confirmatory PCR-positive URT swab (red circle). The median [NP] of blood samples collected at day 0 to 1 for 32 COVID-19 inpatients (1,045 pg/ml) was 36 times higher than the median [NP] (29 pg/ml) of blood samples collected from 43 COVID-19 outpatients simultaneously with their PCR-positive URT swab (blue square). For each inpatient data point, the time of blood sample collection is illustrated as mean ± SD number of days after the first PCR-positive URT swab. Each inpatient data point area is proportional to the number of blood samples contributing to the data point.
For outpatients with SARS-CoV-2 infection according to PCR, the average apparent [NP] (±SD) from 15 serum samples and 28 plasma samples were 54 ± 69 pg/ml (range, 2 to 274 pg/ml; median, 28 pg/ml) and 169 ± 328 pg/ml (range, 3 to 1,377 pg/ml; median, 31 pg/ml), respectively. No systematic difference was observed in the [NP] levels between serum and plasma samples of COVID-19 outpatients (P = 0.418).
For outpatients without SARS-CoV-2 infection according to PCR, the average apparent [NP] (±SD) from 251 serum samples and 216 plasma samples were 2.1 ± 1.3 pg/ml (range, 0 to 12 pg/ml; median, 2.4 pg/ml) and 2.5 ± 1.3 pg/ml (range, 0 to 7.4 pg/ml; median, 2.4 pg/ml), respectively.
Individual dynamics in [NP] levels of blood samples.
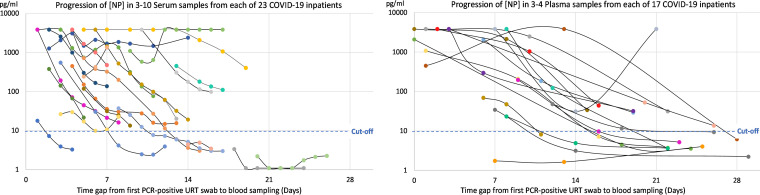
The individual progression in [NP] during the first month after PCR-based diagnosis was observed for 40 COVID-19 inpatients who had at least 3 blood samples collected within 30 days from their first PCR-positive URT swab (Fig. 5). For 4 of these inpatients (10%), none of their blood samples (total n = 20) reached an [NP] above the diagnostic cutoff value of 10 pg/ml. In two of these cases, the earliest blood samples were collected more than 2.5 weeks after their PCR-based diagnosis, when a substantial humoral immune response to infection was measured (see Table S1 and Fig. S2 in the supplemental material). For all the remaining 36 COVID-19 inpatients (90%), at least their first blood sample had an [NP] above the diagnostic cutoff value. Despite the clear individual tendency of decline in [NP] over time, all blood samples (total n = 122) collected from 23 of the 40 COVID-19 inpatients (58%) were positive according to the NP ELISA.
FIG 5.
Individual dynamics in [NP] of 40 COVID-19 inpatients within 1 month from first PCR-positive URT swab (total number of blood samples, n = 200). (A) Three to 10 serum samples collected from each of 23 inpatients. (B) Three to 4 plasma samples collected from each of 17 inpatients.
None of the 34 plasma samples collected from 10 COVID-19 inpatients 27 to 201 days after their first PCR-positive URT swab had an [NP] above the diagnostic cutoff value of 10 pg/ml (Fig. 6), even though 1 to 3 plasma samples collected earlier from each of these patients were clearly positive for NP (mean ± SD, 1,444 ± 1,448 pg/ml; range, 34 to 3,840 pg/ml).
FIG 6.
Plasma [NP] above the diagnostic cutoff value (10 pg/ml) was only observed for COVID-19 inpatients within the first 26 days of detection of SARS-CoV-2 infection by PCR.
DISCUSSION
The outbreak of COVID-19 has caused an unparalleled worldwide requirement for laboratory diagnostic tests for virus infection, and PCR analysis for genomic RNA of SARS-CoV-2 in extracts of swabs collected from the upper respiratory tract has proven very suitable for early detection of infection, even in patients with mild or no clinical symptoms. Still, the characterization of PCR as the gold standard laboratory diagnostic test for COVID-19 (1) and its wide application as a reference test in performance evaluation of other laboratory diagnostic methods is debated (10). This is particularly due to concerns of false‐negative PCR results caused by a low viral load at the chosen time and site of URT sample collection, inadequate URT swabbing technique of some operators, failing storage conditions during specimen transportation, laboratory error, and/or mutation of the viral target RNA (11). These concerns have intensified the search for improved and less resource-demanding laboratory test procedures for COVID-19 and have led to the development of complementing and supplementing screening methods, which will contribute to diagnostic triage procedures relying on a final confirmation of positive results by PCR analysis.
In the present study, we have characterized the first ELISA kit for quantification of the SARS-CoV-2 NP antigen in serum and plasma samples. When used for blood samples collected from COVID-19 inpatients within 2 weeks after PCR-confirmed SARS-CoV-2 infection, the diagnostic sensitivity of the ELISA was 91.6% (95% CI, 85.6% to 95.2%).
The group of 505 outpatients in this study had an 8.5% point prevalence of SARS-CoV-2 infection, and for outpatients with an ELISA-positive blood sample, the probability of having COVID-19 was 97.2% (n = 36; 95% CI, 91.9% to 100.0%), whereas those with an ELISA-negative blood sample had a 98.3% probability of not being infected with SARS-CoV-2 (n = 469; 95% CI, 97.1% to 99.5%). During this early stage of infection, the SARS-CoV-2 NP ELISA thereby proved to be a very reliable predictor of COVID-19.
The individual NP concentrations of COVID-19 patients varied considerably, even for blood samples collected within the first week of PCR-confirmed SARS-CoV-2 infection, and probably reflected the disease severity. The [NP] in blood samples collected at day 0 from 15 COVID-19 inpatients (median, 1,237 pg/ml; mean ± SD, 1,792 ± 1,687 pg/ml; range, 3 to 3,840 pg/ml) was at a substantially higher level than the [NP] in blood samples collected from 43 COVID-19 outpatients simultaneously with their PCR-positive URT swab (median, 29 pg/ml; mean ± SD, 129 ± 271 pg/ml; range, 2 to 1,377 pg/ml).
Despite the variability in [NP] between patients and over time, the individual progressions in [NP] were systematic and declining for almost all the 40 inpatients in the present study who had at least 3 blood samples collected within the first month. For the total of 368 blood samples collected from 160 COVID-19 inpatients during the first month, the median [NP] declined exponentially with time and then consistently remained below the diagnostic cutoff value of 10 pg/ml for all samples collected during the succeeding 6 months after infection.
When verifying the diagnostic performance of an antigen test by using PCR analysis of a URT swab as the reference, all misclassifications (false negatives and false positives) by definition will be ascribed to the antigen test, no matter whether the test material is matching or different. Almost all rapid antigen tests for COVID-19 are lateral-flow immunoassays for the qualitative detection of SARS-CoV-2 NP in extracts of URT swabs. Possibly more rightfully characterized as tests of individual infectiousness (12, 13), their diagnostic performance is typically evaluated by comparison to the outcome of PCR analysis of the same or a simultaneously collected URT swab and thereby is affected by the same risks of a sampling-associated false-negative result as PCR. The diagnostic performance of the quantitative ELISA for SARS-CoV-2 NP investigated in the present study also relied on using PCR analysis of a URT swab as the reference. Despite the distinct sampling techniques and test materials of these two laboratory diagnostic procedures, the analysis of blood samples by the NP ELISA highly confirmed the laboratory diagnosis of SARS-CoV-2 infection based on PCR.
According to the observed performance data, we conclude that the SARS-CoV-2 NP ELISA is suitable for laboratory diagnosis of COVID-19 when used for testing serum or plasma early after infection.
Towards the end of the outbreak of SARS in 2002 to 2004, an ELISA was developed with an analytical detection limit of approximately 50 pg/ml SARS-CoV NP (14). Using a diagnostic cutoff at 100 pg/ml, its diagnostic sensitivity increased from 65% 1 to 2 days after onset of SARS symptoms to over 95% at 3 to 5 days after first symptoms (3). In comparison, the present ELISA for SARS-CoV-2 NP (5) has a substantially improved analytical sensitivity, with a detection limit of around 2 pg/ml, which, in combination with the assay’s high resistance to irregular hemolytic reactions and potentially interfering blood substances, such as rheumatoid factors (5), allows the recommended low and robust diagnostic cutoff at 10 pg/ml SARS-CoV-2 NP. Although differences in shedding of NP into circulation and time of symptom onset after infection may vary between SARS and COVID-19, the 10-fold reduction in diagnostic cutoff contributes decisively to the very early detection of SARS-CoV-2 infection achieved by the novel NP ELISA investigated in the present study.
WHO has concluded that early laboratory diagnosis of SARS-CoV-2 infection can aid clinical management and outbreak control of COVID-19, and that the standard confirmation of acute infection should be based on a nucleic acid amplification test (2). However, URT swab collection followed by PCR analysis is a tedious and expensive method for COVID-19 screening. In contrast, in settings such as hospitals and blood banks, where blood samples are collected anyway, the SARS-CoV-2 NP ELISA provides a simple and economical screening tool for COVID-19. For example, serum and plasma samples prepared at hospitals for biochemical and other clinical laboratory analyses also may be systematically examined by ELISA for the presence of SARS-CoV-2 NP and thereby contribute importantly to reduce the risk of nosocomial COVID-19 infection (15).
Our study has strengths and limitations. First, only a subset of the included participants had blood samples collected within 0 to 1 days after their first PCR-positive URT swab, although this period is the most clinically relevant for early detection of SARS-CoV-2. However, the wide range of collection of blood samples after the first PCR-positive URT swab allowed us to investigate the individual progress of NP concentration in blood for participants with numerous samples available. Second, we did not have information on onset or duration of symptoms for in- and outpatients. Instead, the first PCR-positive URT swab was used as the confirmatory test for COVID-19, although the infection with SARS-CoV-2 may have started days before the PCR test was performed. Theoretically, in a setting where this information was available, the false-negative rate would be even lower, as participants with longer duration of symptoms before blood sampling could be excluded from the main analyses. Third, oropharyngeal swabs were utilized for URT sampling according to Danish national guidelines (9), and although appropriate for clinical testing, as stated by the FDA, they may be less sensitive than nasopharyngeal swabs (16). The strengths of the study include a large sample size and a standardized and highly reproducible method for quantification of SARS-CoV-2 NP.
Although being a recommended subject for further investigations, we propose that automated routine screening of blood samples by the NP ELISA will be a suitable procedure for early identification of inpatients who bring or acquire SARS-CoV-2 infection while hospitalized. As indicated by the projected confusion matrix (see Table S2 in the supplemental material), even for hospitals with a low prevalence of COVID-19 among inpatients treated for other diseases and a high number of routinely analyzed blood samples, the observed 99.8% specificity of the SARS-CoV-2 NP ELISA will ensure a low number of false positives and an acceptable positive predictive value and thereby lead to substantial reductions in the requirement for laborious swab sampling and subsequent confirmatory PCR analysis.
ACKNOWLEDGMENTS
We thank all patients who participated in the study.
The Danish COVID-19Biobank (D19B at the Bio- and Genome Bank, Denmark) is acknowledged for biological material and for data information. CGI Danmark is thanked for providing the IT solution (intellectual property) and competences at no cost for the biobank solution used in the Danish COVID-19Biobank. We thank BioHit Healthcare (Hefei), Anhui Province, People’s Republic of China, for cooperation on the two ELISAs for quantification of the SARS-CoV-2 NP antigen and for measurement of titers of neutralizing antibodies to the ACE-2 receptor binding domain of the S1 subunit of SARS-CoV-2 spike protein.
A.P. and N.T.F. are employees of Solsten Diagnostics International, Aarhus, Denmark, which is the company providing the Solsten SARS-CoV-2 Antigen ELISA kit.
R.F.T. reports a grant from Rigshospitalet Research Council. S.D.N. has unrestricted research grants from Novo Nordisk Foundation and Danish Council for Independent Research, Medical Sciences (FSS). The present methodology study was cofinanced by Innovation Fund Denmark and Eureka via grants 0173-00085 and 0221-00005, respectively.
Footnotes
Supplemental material is available online only.
Contributor Information
Niels T. Foged, Email: ntf@solsten-diag.com.
Angela M. Caliendo, Rhode Island Hospital
REFERENCES
- 1.Da Silva SJR, Da Silva CTA, Guarines KM, Mendes RPG, Pardee K, Kohl A, Pena L. 2020. Clinical and laboratory diagnosis of SARS-CoV-2, the virus causing COVID-19. ACS Infect Dis 6:2319–2336. 10.1021/acsinfecdis.0c00274. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2020Diagnostic testing for SARS-CoV-2: interim guidance, Sep 2020. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Di B, Hao W, Gao Y, Wang M, Wang Y, Qiu LW, Wen K, Zhou DH, Wu XW, Lu EJ, Liao ZY, Mei YB, Zheng BJ, Che XY. 2005. Monoclonal antibody-based antigen capture enzyme-linked immunosorbent assay reveals high sensitivity of the nucleocapsid protein in acute-phase sera of severe acute respiratory syndrome patients. Clin Diagn Lab Immunol 12:135–140. 10.1128/CDLI.12.1.135-140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ceraolo C, Giorgi FM. 2020. Genomic variance of the 2019-nCoV coronavirus. J Med Virol 92:522–528. 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li T, Wang L, Wang H, Li X, Zhang S, Xu Y, Wei W. 2020. Serum SARS-COV-2 nucleocapsid protein: a sensitivity and specificity early diagnostic marker for SARS-COV-2 infection. Front Cell Infect Microbiol 10:470. 10.3389/fcimb.2020.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Hingrat Q, Visseaux B, Laouenan C, Tubiana S, Bouadma L, Yazdanpanah Y, Duval X, Burdet C, Ichou H, Damond F, Bertine M, Benmalek N, Choquet C, Timsit J-F, Ghosn J, Charpentier C, Descamps D, Houhou-Fidouh N, Diallo A, Le Mestre S, Mercier N, Paul C, Petrov-Sanchez V, Malvy D, Chirouze C, Andrejak C, Dubos F, Rossignol P, Picone O, Bompart F, Gigante T, Gilg M, Rossignol B, Levy-Marchal C, Beluze M, Hulot JS, Bachelet D, Bhavsar K, Bouadma L, Chair A, Couffignal C, Da Silveira C, Debray MP, Descamps D, Duval X, Eloy P, Esposito-Farese M, Ettalhaoui N, Gault N, Ghosn J, Gorenne I, et al. 2020. Detection of SARS-CoV-2 N-antigen in blood during acute COVID-19 provides a sensitive new marker and new testing alternatives. Clin Microbiol Infect 27:789.e1–789.e5. 10.1016/j.cmi.2020.11.025.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahava M, Kurkela S, Kuivanen S, Lappalainen M, Jarva H, Jääskeläinen A. 2021. Detection of SARS-CoV-2 nucleocapsid antigen from serum can aid in timing of COVID-19 infection. medRxiv 10.1101/2021.01.08.20248771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bio- and GenomeBank Denmark. 2020. Instruks for håndtering af standardsæt blod Regionernes Bio- og GenomBank. Bio- and GenomeBank Denmark, Copenhagen, Denmark. [Google Scholar]
- 9.Danish Health Authorities. 2020. Retningslinjer for håndtering af COVID-19 i sundhedsvaesenet, 24th ed. Danish Health Authorities, Copenhagen, Denmark. [Google Scholar]
- 10.Dramé M, Tabue Teguo M, Proye E, Hequet F, Hentzien M, Kanagaratnam L, Godaert L. 2020. Should RT-PCR be considered a gold standard in the diagnosis of COVID-19? J Med Virol 92:2312–2313. 10.1002/jmv.25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Silva SJR, Pena LJ. 2021. A word of caution in interpreting COVID-19 diagnostics tests. J Med Virol 93:717–718. 10.1002/jmv.26531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guglielmi G. 2021. Rapid coronavirus tests: a guide for the perplexed. Nature 590:202–205. 10.1038/d41586-021-00332-4. [DOI] [PubMed] [Google Scholar]
- 13.Mina MJ, Peto TE, García-Fiñana M, Semple MG, Buchan IE. 2021. Clarifying the evidence on SARS-CoV-2 antigen rapid tests in public health responses to COVID-19. Lancet 397:1425–1427. 10.1016/S0140-6736(21)00425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Che XY, Qiu LW, Pan YX, Wen K, Hao W, Zhang LY, Wang Y, Liao ZY, Hua X, Cheng VCC, Yuen KY. 2004. Sensitive and specific monoclonal antibody-based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. J Clin Microbiol 42:2629–2635. 10.1128/JCM.42.6.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbas M, Robalo Nunes T, Martischang R, Zingg W, Iten A, Pittet D, Harbarth S. 2021. Nosocomial transmission and outbreaks of coronavirus disease 2019: the need to protect both patients and healthcare workers. Antimicrob Resist Infect Control 10:7–13. 10.1186/s13756-020-00875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee RA, Herigon JC, Benedetti A, Pollock NR, Denkinger CM. 2021. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: a systematic review and meta-analysis. J Clin Microbiol 59:e02881-20. 10.1128/JCM.02881-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2, Tables S1 and S2, and additional information. Download JCM.01001-21-s0001.pdf, PDF file, 0.2 MB (218.9KB, pdf)