ABSTRACT
Plant pathogen effector proteins are key to pathogen virulence. In susceptible host Brassicas, the clubroot pathogen, Plasmodiophora brassicae, induces the production of nutrient-sink root galls, at the site of infection. Among a list of 32 P. brassiae effector candidates previously reported by our group, we identified SSPbP53 as a putative apoplastic cystatin-like protein highly expressed during the secondary infection. Here we found that SSPbP53 encoding gene is conserved among several P. brassicae pathotypes and that SSPbP53 is an apoplastic protein able to directly interact with and inhibit cruciferous papain-like cysteine proteases (PLCPs), specifically Arabidopsis XYLEM CYSTEINE PEPTIDASE 1 (AtXCP1). The severity of clubroot disease is greatly reduced in the Arabidopsis xcp1 null mutant (AtΔxcp1) after infection with P. brassicae resting spores, indicating that the interaction of P. brassicae SSPbP53 with XCP1 is important to clubroot susceptibility. SSPbP53 is the first cystatin-like effector identified and characterized for a plant pathogenic protist.
KEYWORDS: Cysteine protease inhibitor, papain-like cysteine proteases (plcps), apoplast, clubroot, plant defense, plasmodiophora brassicae
Introduction
Clubroot is caused by the obligate biotrophic, soil-borne, parasitic protist Plasmodiophora brassicae member of the order Rhizaria [1] that can infect most plants in the Brasicaceae family, including Arabidopsis. A P. brassicae primary infection of a susceptible host plant, through primary zoospores intrusion of root hairs, leads to the production of secondary zoospores that, upon secondary infection of cortical cells, results in the reprogramming of phloem and xylem development and the establishment of nutrient-sink galls to support the production of new resting spores [2,3]. To complete the life cycle P. brassicae needs to escape or down-regulate the host plant microbe-associated molecular patterns (MAMPs)-triggered immunity (MTI) [4]. Many biotrophic and necrotrophic plant pathogens regulate MTI by manipulating plant salicylic acid (SA) metabolism [5–7]. Similarly, P. brassicae produces a methyltransferase, PbBSMT, that methylates SA, benzoic and anthranilic acids, thereby reducing SA content in the host cell and attenuating plant defense responses [8,9]. A key component of the sophisticated plant immune system are papain-like cysteine proteases (PLCPs). Aimed at the detection of MAMPS and the induction of the plant defense response, PLCPs bind to and degrade pathogen effector proteins [10,11]. Recent reports suggest that PLCPs are also involved in the induction of systemic immunity and the promotion of cell death during an infection [12,13].
To counter PLCP activity, many evolutionary unrelated biotrophic pathogens produce PLCP inhibitors during colonization of the plant host [14–21]. For example, Avr2 from the extracellular pathogen C. fulvum, is able to inhibit the Arabidopsis PLCPs CPR1, XCP1, and XCP2 as well as tomato Rcr3 and Pip1 [22]; EPI1 and EPIC2B from Phytophthora infestans are inhibitors of the tomato PLCP PIP1 [23]; SDE1 from the Huanglongbing (HLB) pathogen Candidatus Liberibacter asiaticus inhibits citrus RD19a, RD21a, and SAG12 PLCPs [21]; and Pit2 from Ustilago maydis inhibits CP2, a maize PLCP member of the aleurain like cysteine protease family [19], suggesting a key role for PLCPs in the plant immune response and a key target by pathogens.
Using genome-wide transcriptomic analysis we have previously identified, small secreted P. brassicae protein 53 (SSPbP53: PBRA_008207), to be a cystatin-like PLCP inhibitor, with a functional signal peptide, that is over-expressed during secondary infection and subsequent gall formation in P. brassicae infected Arabidopsis roots [24]. In this study, further analyses of SSPbP53 during clubroot disease development show that it inhibits PLCP activity from six susceptible cruciferous plant hosts, with a clear affinity for the xylogenesis-related XCP1 in Arabidopsis. This characterization of the P. brassicae effector protein and its corresponding plant PLCP provides a possible target to be exploited for clubroot management and the generation of increased resistant Brassicaceae varieties. This is the first description of such a mechanism for a plant pathogenic member of the order Rhizaria.
Materials and methods
Plant material and growth conditions
Arabidopsis, rapeseed (B. napus), arugula (Eruca sativa), broccoli (B. oleracea var. italica), cabbage (B. oleracea var. oleracea), wild mustard (B. kaber), and Nicotiana benthamiana seeds, disinfected with 70% ethanol, 95% ethanol, 10% bleach and sterile water, were sown on agar (1%, w/v) plates containing 1/2 Murashige and Skoog (MS) (Sigma-Aldrich, CAD) salts with 1% sucrose. Plates were sealed with parafilm, placed at 4°C for 4 days and then transferred to a growth chamber (Conviron E8, CMP6050 control system; 100 μmol photons m2 s−1; 16 h/8 h light/dark cycle; 22°C) for germination and growth. The Arabidopsis mutants AtΔrd19 (SALK_031088) and AtΔxcp1 (SALK_084789) were obtained from the Arabidopsis Biological Resource Center (TAIR, Ohio State University), while the AtΔrd21 mutant was kindly provided by Prof. Robert Fluhr [25]. All homozygous mutants were confirmed by RT-PCR to ensure the lack of expression of their respective gene.
Infection and disease indexing
Ten-day old cruciferous seedlings were transferred to Sunshine Mix #4 soil (Sun Gro Horticulture Inc., BC) with four plants in each “square pot” (3.5 x 3.5 inch) of an 18-pot sheet. Seedlings were allowed to acclimatize in these pots for four additional days before 400 uL of 5 × 107 resting spores mL−1 of a Saskatchewan field isolate of P. brassicae pathotype 3 H, was applied to each plant where the stem entered the soil. Control plants were inoculated with 400 μL of distilled water and grown in separate trays in the same growth chamber.
Disease index (DI) was determined for AtCol-0, AtΔxcp1, AtΔrd19 and AtΔrd21 infected plants at 21 dpi as previously described [26]. Root infection was assessed on a 0–4 scale where 0, no symptoms; 1, small galls mainly on lateral roots; 2, small galls on the main and lateral roots; 3, medium-sized galls with possible negative effect on plant growth and 4, severe galls on both main and lateral roots, deformed roots and impaired growth [26]. The experiment, repeated three times, was arranged in a completely randomized design with the DI of infected plants determined from 68 plants per experimental unit.
Spore quantification
Spore production was determined for clubroot infected AtCol-0 and AtΔxcp1 plants at 21 dpi. The soil in each of the 51 square pots for each plant line was air-dried at room temperature (21 ± 2°C), thoroughly mixed, weighed and 0.5 g of soil was collected for resting spore extraction [27]. Soil was mixed, using a blender, with 20 mL of sterile distilled water for 1 min and filtered through eight layers of cheesecloth. The pass-through was centrifuged at 3900 g (AllegraTM 25 R Centrifuge) for 15 min and the supernatant was discarded. The pellet was suspended in 6 mL of 50% (w/v) sucrose by vortex for 2 min and then centrifuged for 5 min at 1700 g. The supernatant was transferred to a 50 mL tube, brought to 50 mL with sterile distilled water, mixed with a vortex and centrifuged at 3900 g for 15 min. Pellets were resuspended in 5 mL of sterile distilled water, vortexed and centrifuged and the supernatant was again discarded and the pellet suspended in 2 mL of sterile distilled water. Spores were counted using a hemocytometer and expressed as the number of spores per plant.
Structural analysis of SSPbP53
Model structures for SSPbP53 and CHIKCYS were created using SWISS-MODEL [28,29], based on crystal structures in the PDB database with the highest structural similarity to each query protein. The models of SSPbP53 and CHIKCYS were modeled using PyMOL [30].
Pathotype SSPbP53 analysis
To amplify SSPbP53, genomic DNAs (gDNA) of Plasmodiophora brassicae pathotype 3 H (Pb3H) obtained from Dr. Gary Peng (AAFC-Saskatoon Research Center) and single spore isolates SACAN-SS3 (Pb2), ORCA-SS3 (Pb5), AbotJE-SS3 (Pb6), and CDCN-SS1 (Pb8) [31], were used. PCR amplification used Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, CAD) in a 50 μL final volume containing 300 nM of each primer (gSSPbP53F/gSSPbP53R, Supplementary Table S1) and 2 to 4 ng of gDNA. The 530 bp amplicon generated for each pathotype was purified using GeneJET PCR Purification Kit (Fermentas Life Science, CAD) and directly sequenced with the amplification primers (Eurofins Scientific, CAD).
Plasmid construction
For plasmid construction, the Gateway cloning system (Thermo Fisher Scientific, CAD) was used following the manufacturer´s recommendations. All plasmids used and generated in this study are listed in Supplementary Table S2.
To amplify all of the coding sequences cloned in this study, total RNA was extracted from Pb3 infected (when pathogen cDNA needed), or uninfected Arabidopsis Col-0 roots using a Trizol based extraction method [32]. cDNA was synthesized from 2 ug of total RNA using the QuantiTect® Reverse Transcription Kit (Qiagen, Canada) following the manufacturer’s recommendations. Each cDNA was used as a template to amplify SSPbP53 (SSPbP53 is minus the signal peptide sequence and with the stop codon at the C-terminus, PBRA_008207), AtSAG12-cys (AT5G45890), AtRD19-cys (AT4G39090), AtRD21-cys (AT1G47128), AtAALP-cys (AT5G60360), AtCATHB3-cys (AT4G01610), AtXBCP3-cys (AT1G09850), and AtXCP1-cys (AT4G35350) coding sequences, adding the attB1 and attB2 recombination sites at the 5ʹ and 3ʹ ends, respectively, with the designated primer pairs (Supplementary Table S1). The attB-flanked DNA fragments were cloned into pDON R™/Zeo (Thermo Fisher Scientific, CAD) using BP Clonase™ (Thermo Fisher Scientific, CAD) following the manufacturer´s recommendations, generating entry clones.
To study the interaction between SSPbP53 and AtPLCPs through yeast two-hybrid (Y2H) assay, the coding sequences previously cloned into pDON R™/Zeo were cloned into pDEST32 (Thermo Fisher Scientific, CAD) and pDEST22 (Thermo Fisher Scientific, CAD), respectively, using LR Clonase™ (Thermo Fisher Scientific, CAD) following the manufacturer´s recommendations. To express SSPbP53 in planta, the coding sequence was cloned under the control of the CaMV 35S promoter into pEarlyGate100 (without fluorescent tag) and pEarlyGate103 (with N-terminus GFP) (Thermo Fisher Scientific, CAD), using LR Clonase™ (Thermo Fisher Scientific, CAD) and the pDONZeo_SSPbP53 entry vector following the manufacturer´s recommendations. Constructs generated are listed in Supplementary Table S2. To use as an empty vector (EV) control, SSPbP53 signal peptide sequence followed by a C-terminal stop codon was cloned in frame into pEarlyGate103 to remove the toxic ccdB selection marker.
To obtain His-SSPbP53, the coding sequence was cloned into pDEST17 using LR Clonase™ (Thermo Fisher Scientific, CAD) and the SSPbP53_pDONR/Zeo entry vector following the manufacturer´s recommendations. The same procedure was followed to obtain GST-tagged AtAALP-cys, AtCATHB3-cys, AtXBCP3-cys, and AtXCP1-cys, from coding sequences that were previously cloned into their corresponding entry vector and transferred into pDEST15 using LR Clonase™ (Thermo Fisher Scientific, CAD) following the manufacturer´s recommendations. To use as an empty vector (EV) control, SSPbP53 signal peptide sequence followed by a C-terminal stop codon was cloned in frame into pDEST15 to remove the toxic ccdB selection marker.
To obtain the His-SSPbP53ΔL1 mutant (the deletion of loop 1), the coding sequence for SSPbP53ΔL1 was synthesized (GenScript, USA) and cloned into pET-14b (GenScript, USA). The same procedure was followed to obtain His-AtXCP1-cys.
Yeast two-hybrid (Y2H) assay
The recombinant plasmid pDEST32_SSPbP53 was transformed into the Saccharomyces cerevisiae strain AH109 to generate the bait. The pDEST22_ AtSAG12-cys, _AtRD19-cys, _AtRD21-cys, AtAALP-cys, _AtCATHB3-cys, _AtXBCP3-cys, and AtXCP1-cys plasmids were transformed into the bait and growth into on SD-2 media to confirm yeast transformation. The interaction was assessed through selection on SD-3 media. Yeast transformed with the empty vectors served as negative controls. The experiments were repeated three times with similar results.
Production of recombinant proteins
For protein expression, recombined pDEST15 and pDEST17 vectors were used to transform E. coli BL2E cells (Thermo Fisher Scientific, CAD). E. coli transformants carrying the respective expression plasmids (Supplementary Table S2) were grown in 50 mL LB medium supplemented with ampicillin (100 μg mL−1) at 37°C, 200 rpm for 3 h. Protein expression was induced by the addition of 20% arabinose to a final concentration of 0.02% and growth was continued at 37°C, 200 rpm for 2 h. After centrifugation at 3000 g for 15 min, cell pellets were lysed using a sonicator VirTis equipped with a microtip (VirSonic, USA). Pellets expressing GST tagged AtAALP-cys, AtCATHB3-cys, AtXBCP3-cys, and AtXCP1-cys were resuspended in 8 mL B-PERTM bacterial cell lysis reagent (Thermo Fisher Scientific, CAD), sonicated on ice using six 10-second bursts at high intensity with a 10-second cooling period between each burst and the lysate was centrifuged at 3000 g for 15 min. The supernatant was used in in vitro pull-down assays (see below). Pellets containing His-SSPbP53, His-SSPbP53ΔL1, and His-AtXCP1-cys were resuspended in 8 mL of guanidinium lysis buffer, sonicated as described above, and purified with Ni-NTA agarose (Thermo Fisher Scientific, CAD) through hybrid conditions following the manufacturer´s recommendations. Protein concentration was measured using a Qubit protein assay kit (Thermo Fisher Scientific, CAD).
In vitro inhibition assay
The papain inhibition assay was performed as previously described [21]. Briefly, fluorescein-labeled casein (FITC-casein, Pierce™ fluorescent protease assay kit) (Thermo Fisher Scientific, CAD) was used as substrate. Recombinant His-SSPbP53, protease inhibitor E-64 (Thermo Fisher Scientific, CAD), or BSA (Sigma-Aldrich, CAD) (100 and/or 500 nM), were mixed with 100 μg mL−1 papain (Sigma-Aldrich, CAD) in 96-well plates (Thermo Fisher Scientific, CAD) in the dark. Papain was also mixed with MES buffer as the negative control. After 1 h at room temperature in the dark, fluorescence was measured using a microplate fluorometer Fluoroskan Ascent® (Thermo Fisher Scientific, CAD) at 485/538 nm excitation/emission, with a gain value of 40, following the manufacturer´s recommendations. Each experiment was repeated three times with similar results.
For SSPbP53 and SSPbP53ΔL1 inhibition of AtXCP1, a similar experiment using fluorescein-labeled casein as substrate and the recombinant proteins at 500 nM was performed. Fluorescence was measured using a microplate fluorometer EPOCH 2 (BioTek, USA) as described above. This experiment was repeated three times.
Western blot analysis
Proteins were separated by 15% SDS-PAGE and transferred to 0.2 μm PVDF membranes (Bio-Rad, CAD) for 1 h at 0.4 V cm−1 in a Mini-PROTEAN® Tetra Cell (Bio-Rad, CAD). His-tagged proteins were detected using 1:1000 diluted monoclonal anti-polyHis-HRP conjugate (Sigma-Aldrich, CAD). GFP and GST tagged proteins were visualized with 1:1500 diluted monoclonal anti-GFP and anti-GST-HRP conjugates, respectively (Santa Cruz Biotechnology, USA). Biotinylated proteins were detected with 1:2000 diluted HRP-conjugated Streptavidin (Thermo Fisher Scientific, CAD). Membranes were blocked with 5% skim-milk (BD Difco, CAD) and HPR activity was detected using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, CAD) with a ChemiDoc™ Imaging System (Bio-Rad, CAD) able to detect the chemiluminiscent signal.
Activity-based protein profiling
Apoplastic fluid was obtained following the previously described protocol of de Wit and Spikman [33]. Papain at 100 μg mL−1 (Sigma-Aldrich, CAD), Arabidopsis, rapeseed, arugula, broccoli, cabbage, wild mustard, and N. benthamiana apoplastic fluids were pretreated with either MES buffer (negative control), 100 nM E-64 (positive control), or 500 nM SSPbP53. For rapeseed cotyledons expressing SSPbP53, apoplastic fluids were pretreated only with MES buffer or E-64 (100 nM). Following 1 h pretreatment at room temperature, the samples were incubated with a final concentration of 2.5 μM DCG-04 (Syntides, CHN) for 3 h at room temperature, followed by acetone precipitation of the proteins. Precipitated proteins were re-suspended in 50 μL of 50 mM Tris buffer (pH 6.4) and enriched using streptavidin magnetic beads (Thermo Fisher Scientific, CAD). Resuspended proteins were incubated with 25 μL streptavidin magnetic beads at room temperature for 1 h, washed twice with 1% SDS, and eluted by heating for 5 min at 95°C in Laemmli sample buffer with 13% β-mercaptoethanol as previously described [21]. Biotinylated proteins were detected by western blot as described above. Each experiment was repeated three times with similar results. Two-week old rapeseed cotyledons were infiltrated with A. tumefaciens containing untagged SSPbP53 to assess SSPbP53 inhibitory ability.
Microscopy
Two-week-old rapeseed cotyledons and 4-week-old N. benthamiana leaves were infiltrated with 0.3 OD600 A. tumefaciens GV3101 (pMP90) containing pEG103_SSPbP53-GFP. Fluorescence was observed using a Zeiss LSM-510 confocal microscope with a PL APO 40×/1.3 water-dipping objective and 488/500-530 nm excitation/emission after no more than 2 days post infiltration. To assess SSPbP22 localization, 4-week-old N. benthamiana leaves were infiltrated with 0.3 OD600 A. tumefaciens containing pEG103_SSPbP22-GFP [24] and fluorescence was observed using a Zeiss Axioskop 2 plus fluorescence microscope and 488/500-530 nm excitation/emission after 3 days post infiltration.
Roots of A. thaliana Col-0 and xcp1 Pb+ at 20 dpi and xcp1 Pb- were fixed using FAA solution (formaldehyde 37%: ethanol 99%: acetic acid, 10%:50%:5% + 35% water) and immobilized in parafilm wax for 5 μm sectioning using a Microm microtome.
Gene expression analysis by qRT-PCR
cDNAs for P. brassicae infected and non-infected root tissue at 14, 21 and 28 dpi were diluted 1:80 (v:v) in RNase-free water and used to detect the expression of Arabidopsis PLCP genes. Real-time qPCR was performed using SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad, Canada) in a 20 μL final volume containing 300 nM of each primer, and 2 μL of cDNA diluted 1:5 (v:v) in RNase-free water. Amplification was carried out using a C1000 thermocycler base with a CFX96 real-time system (Bio-Rad) and reactions were quantified using BioRad CFX manager software (v.3.1). Each amplification used three technical replicates, the results of which were averaged to give the value for a single biological replicate. Three biological replicates were prepared for each time point using material harvested from 12 plants in each replicate, grown on three separate occasions. Results are expressed as LOG2 expression relative to P. brassicae ELONGATION FACTOR-LIKE (PbEFL, PBRA_001540) expression using the comparative quantification method as previously described [34]. Primers used are presented in Table S2.
In vitro pull-down assays
Three hundred microliters of E. coli cell lysate supernatant of GST tagged AtAALP-cys, AtCATHB3-cys, AtXBCP3-cys, and AtXCP1-cys were incubated with 25 μL glutathione resin (Thermo Fisher Scientific, CAD) for 1 h at 4°C and washed with TKET buffer (20 mM Tris-HCl, 200 mM KCl, 0.1 mM EDTA, 0.05% Triton X-100, pH 6.0) as previously described [19]. His-SSPbP53-expressing cell lysate was added to the AtPLCP-bound resins and incubated for 3 h at 4°C, followed by washing with TKET buffer. Resins were then boiled in 25 μL Laemmli sample buffer and the supernatants were analyzed through western blot using anti-His-HRP and anti-GST-HRP as described above.
Phylogenetic analysis of host PLCPs
The 32 PLCPs from Arabidopsis (TAIR; https://www.arabidopsis.org/) were used to identify orthologs in the B. napus genome (Genbank accession number PRJNA237736) using BLAST (NCBI) and phylogenetic analysis was carried out using MUSCLE v3 [35] and MEGA v6 [36]. The phylogenetic tree was constructed using maximum-likelihood, the James–Taylor–Thorthon model and a bootstrap value of 1000. Sequences used in the analysis are provided in supplementary Table S3.
Statistical analysis
The data were analyzed using the R Statistical Package [37]. Test and control groups, were compared using a two-sided Student’s t-test, while a one-way ANOVA followed by Tukey’s HSD post hoc test was used to compare the mean of multiple groups.
Results
SSPbP53 is conserved among P. brassicae pathotypes
Among a number of SSPbPs, SSPbP53 was previously identified by our group as a cystatin-like protein [24]. SSPbP53 shares a common tertiary structure, an alpha helix lying on top of an anti-parallel beta sheet, with that of the extracellular protease inhibitor with cystatin-like domain (EPIC) proteins (Figure 1a), containing many of the signature sequences of cystatin-like protease inhibitors, including the conserved Gly residue in the N-terminal region and the highly conserved Gln-Xaa-Val-Xaa-Gly motif in the first binding loop (with Xaa represented by Val and Ser, respectively, in both SSPbP53 and EPIC1). However, a conserved Trp in the second binding loop [38] is not present in SSPbP53 (Figure 1b).
Figure 1.
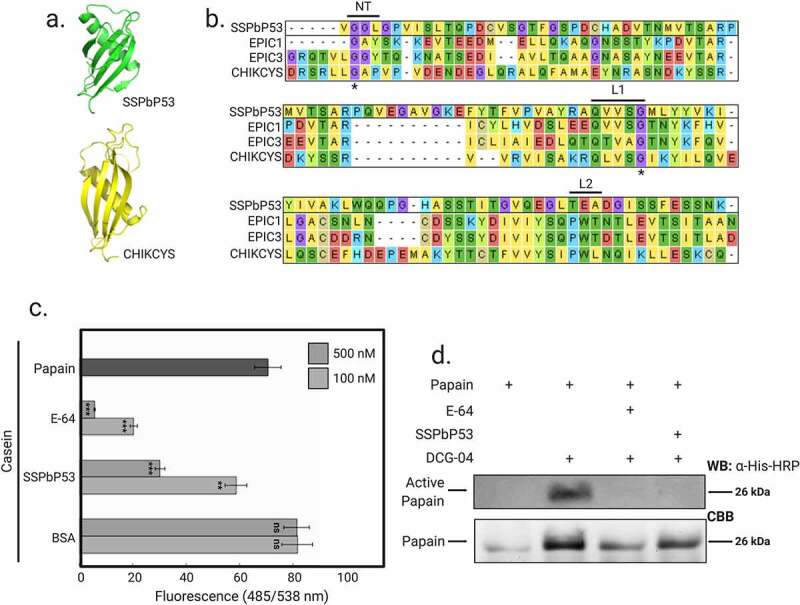
SSPbP53 is a putative cysteine protease inhibitor. A. Comparison of the 3D structure of SSPbP53 with the extracellular protease inhibitor with cystatin-like domain CHIKCYS. B. Sequence alignment of SSPbP53 and four other cysteine protease inhibitors showing the putative active site of the cystatins, including the N-terminal trunk (NT) and binding loops 1 and 2 (L1 and L2). Asterisks (*) indicate conserved amino acids in NT and L1. C. Proteolytic activity of papain, measured by digestion of a fluorescent casein substrate, is inhibited by E-64 (positive control) and purified His-SSPbP53 protein, but not BSA (negative control). The proteolytic activity of papain without inhibitors was also measured and included in the analysis. Fluorescence was measured at 485/538 nm excitation/emission. Mean ± standard deviation (n = 3). Statistically significant differences based on the two-tailed Student’s t-test are indicated by asterisks (*) where p < 0.01 is represented by (**), p < 0.001 by (***). ns – no significant difference. D. Activity-based protein profiling (ABPP) showing inhibition of papain by SSPbP53, preventing the subsequent binding of DCG-04 to papain
SSPbP53 (PBRA_008207) was amplified from the Canadian P. brassicae pathotypes Pbc2, Pbc3, Pbc5, Pbc6, and Pbc8 and the nucleotide sequences of the resulting 500 bp amplicons were identical among all five pathotypes. The corresponding sequences, retrieved from the P. brassicae European pathotype e3 (GCA_001049375) and eH (GCA_003833335) genomes and the Chinese pathotype ZJ1 (GCA_002093825), also showed 100% nucleotide identity, resulting in a 100% shared amino acid sequence for SSPbP53 in all tested P. brassicae pathotypes.
SSPbP53 inhibits plant papain-like cysteine proteases
To study SSPbP53 interactions with plant papain-like cysteine proteases (PLCPs), SSPbP53, minus the signal peptide and with an N-terminal His tag, was expressed in E. coli (Supplementary Fig. S1). SSPbP53 activity was first assessed using fluorescein-labeled casein as substrate, papain as the model PLCP and the chemical cysteine protease inhibitor E-64 as previously described [18,19,33]. SSPbP53 inhibited casein degradation by papain (Figure 1c). Inhibition of papain activity was dose-dependent with a > 50% reduction of activity achieved with 500 nM purified SSPbP53, comparable to that of 100 nM E-64 (Figure 1c, Supplementary Table S4). These concentrations for SSPbP53 and E-64 were used for all further analyses.
Using an activity-based protein profiling (ABPP) assay, together with E-64 and DCG-04, a biotinylated derivative of E-64, we further analyzed SSPbP53 activity [39]. The assay is based on the ability of E-64 or SSPbP53 to bind to papain or PCLPs, thereby blocking further binding of DCG-04 to the papain/PLCPs. Streptavidin beads were used to bind the biotin tag on DCG-04 and thereby measure the binding ability of SSPbP53 to papain/PCLPs, as indicated by a reduced streptavidin-horseradish peroxidase (HRP) signal on subsequent western blots. The pre-incubation of papain with either SSPbP53 or E-64 completely blocked any subsequent binding of DCG-04 to the papain (Figure 1d). These results show SSPbP53 to be a functional inhibitor of the model cysteine protease, papain.
SSPbP53 localizes to the apoplast and inhibits cruciferous PLCP activity
ApoplastP [40] analysis predicted (P = 0.82) that SSPbP53 is an apoplastic protein. To confirm this prediction, SSPbP53:GFP was expressed in B. napus cotyledons (Supplementary Fig S2a). SSPbP53-GFP transcript was confirmed through RT-PCR and SSPbP53-GFP expression was confirmed through western blot using anti GFP-HRP (Supplementary Fig S2c-d). SSPbP53-GFP localized in the apoplastic space, with clear visualization of the cell wall dividing neighboring plant cells. Plasmolization of B. napus cotyledons further confirmed this apoplastic localization for SSPbP53-GFP (Figure 2e), when compared to SSPbP22-GFP, a previously characterized nuclear and cytoplasmic kinase [24], that remains in the cytoplasm and nucleus after plasmolysis (Supplementary Fig. S3). Similar apoplastic localization was observed for SSPbP53-GFP in cotyledons of N. benthamiana (Supplementary Fig S2b-e).
Figure 2.
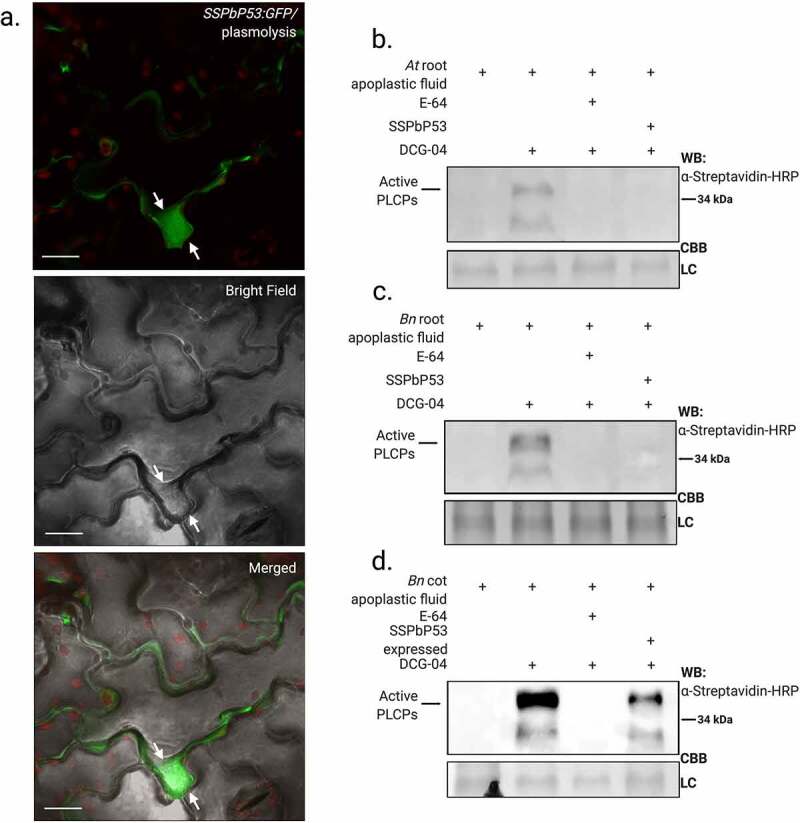
SSPbP53 is an apoplastic protein and inhibits apoplastic cruciferous PLCPs. A. Confocal images showing SSPbP53-GFP localized to the apoplast of rapeseed cotyledons, after salt-induced plasmolysis (Scale bars, 20 μM). Arrows indicate the retraction of the membrane from the cell wall. B. His-SSPbP53 inhibits Arabidopsis apoplastic PLCPs. Apoplastic proteins from Arabidopsis Col-0 roots were labeled via ABPP in the presence of purified His-SSPbP53. WB, western blot; CBB, coomassie brilliant blue; LC, Loading control. C. His-SSPbP53 inhibits apoplastic PLCPs in rapeseed roots. D. His-SSPbP53 transiently expressed in rapeseed cotyledons inhibits apoplastic PLCPs, A. tumefaciens containing empty vector (EV) was used as a negative control, together with non-treated cotyledons (Full figure in Supplementary Fig. S5)
ABPP assays were carried out on root apoplastic fluid from Arabidopsis, B. napus, wild mustard (Brassica kaber), broccoli (Brassica oleracea var. italica), cabbage (Brassica oleracea var. capitate), and arugula (Eruca vesicaria). SSPbP53 almost completely inhibited biotinylation of PLCPs in all cruciferous root apoplastic fluids (Figure 3b-c, Supplementary Fig. S4). An ABPP assay of apoplastic fluid from B. napus cotyledons transiently expressing SSPbP53 without a fluorescence tag, also showed a reduced level of active apoplastic PLCPs (Figure 3d, Supplementary Fig. S5a). SSPbP53 transcript, in agroinfiltrated B. napus cotyledons and 21 dpi B. napus roots, was confirmed through RT-PCR (Supplementary Fig. S5b). SSPbP53 did not inhibit N. benthamiana PLCPs (Supplementary Fig. S6), suggesting crucifer specificity for this P. brassicae effector.
Figure 3.
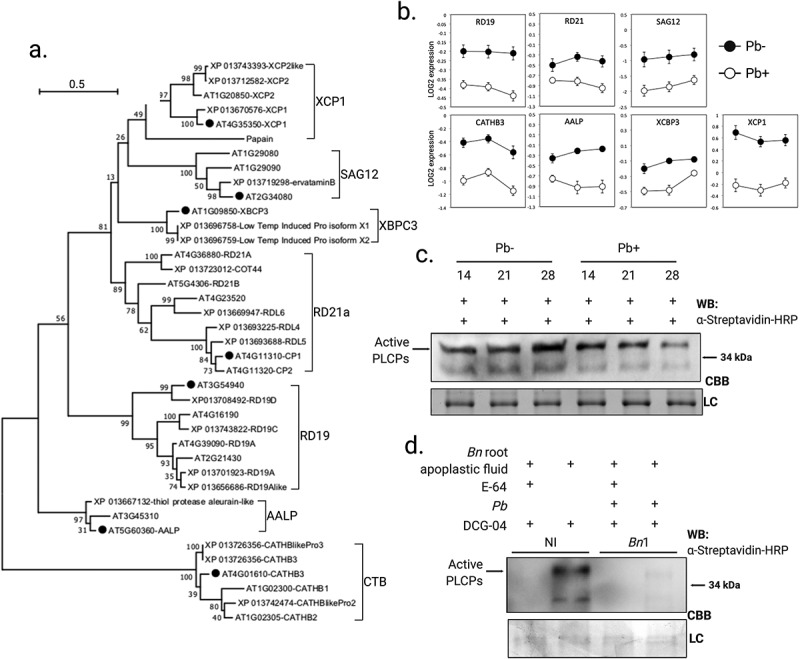
Levels of Arabidopsis and rapeseed PLCPs are reduced during P. brassicae infection. A. Phylogenetic tree of Arabidopsis and rapeseed orthologous PLCPs and subfamily classification. B. qRT-PCR amplification of Arabidopsis PLCPs, for which there are orthologs in the rapeseed genome, at 14, 21 and 24 dpi with P. brassicae (Pb+) or mock-inoculated (Pb-). Results are LOG2 expression relative to P. brassicae ELONGATION FACTOR-LIKE gene (PbEF1), normalized to expression in uninfected plants, and are the average of three biological replicates ±SE. C. Reduction of active apoplastic PLCPs in Pb+ Arabidopsis roots at 14, 21 and 24 dpi relative to Pb-. D. P. brassicae infection resulted in decreased availability of active apoplastic PLCPs in rapeseed roots at 21 dpi. NI, non-infected negative control
Expression of PLCPs is down regulated in roots infected by P. brassicae
The Arabidopsis genome encodes 32 PLCPs classified into eight main groups [41], of which, only seven orthologous groups are found in the B. napus genome (Genbank accession number PRJNA237736) (Figure 3a). To examine the expression of Arabidopsis PLCPs in P. brassicae infected roots a member from each of the seven orthologous groups was selected and transcript levels determined at 14, 21, and 28 dpi. Transcript levels for all investigated PLCPs were decreased in P. brassicae infected plants in comparison with mock-inoculated plants (Figure 3b). The down regulation was significant for ALEURAIN-LIKE PROTEASE (AtAALP), CATHEPSIN B-LIKE PROTEASE 3 (AtCATHB3), XYLEM BARK CYSTEINE PEPTIDASE 3 (AtXBCP3), and XYLEM CYSTEINE PEPTIDASE 1 (AtXCP1) in P. brassicae infected plants (Figure 3b). Furthermore, active apoplastic PLCPs were also reduced in the roots of P. brassica infected (Pb+) Arabidopsis plants when compared to mock inoculated (Pb-) plants (Figure 3c). ABPPs were carried out on the apoplastic fluids from B. napus, arugula, broccoli, cabbage, and wild mustard roots at 21 dpi with P. brassicae, when SSPbP53 transcript was highly expressed. Active apoplastic PLCPs were lower in P. brassicae infected cruciferous plants, when compared to non-infected (NI) plants (Figure 3d, Supplementary Fig. S7), suggesting that P. brassicae infection decreased total apoplastic PLCPs.
SSPbP53 interacts with xylem-associated PLCPs and the interaction contributes to plant host susceptibility to P. brassicae
To determine if one or more of these Arabidopsis PLCPs can interact with SSPbP53, a pair-wise Y2H analysis was performed. The results showed that the bait yeast expressing SSPbP53 co transformed with AtAALP, AtXBCP3 and AtXCP1 cysteine protease domains (AtPLCP-cys) grew on SD-3 selective media (Figure 4a), while empty vector or the bait yeast only transformed with SSPbP53 was not able to grow. To confirm the results obtained through Y2H analysis, in vitro pull-down assays were carried out now for four of the initial PLCPs candidates evaluated. Each of AtAALP-cys, AtCATHB3-cys, AtXBCP3-cys and AtXCP1-cys cysteine protease domains with a GST tag at the N-terminus, was expressed in E. coli. The recombinant GST-PLCP-cys(s) were incubated separately with SSPbP53 and immunoprecipitated using glutathione agarose. SSPbP53 co-precipitated with AtXCP1-cys and to a lesser degree AtXBCP3-cys (Figure 4b). The interaction with AtAALP-cys was not confirmed through pull-down, a more restrictive method than Y2H. Pre-incubation of AtXCP1-cys with E-64 successfully prevented the AtXCP1-SSPbP53 interaction (Figure 4c). The ability of SSPbP53 to inhibit AtXCP1 activity and the importance of the loop 1 QVVAQ sequence (Figure 1b, Figure 4d), previously identified as essential for activity in cysteine proteases [42], to this interaction was assessed using fluorescein-labeled casein as substrate, SSPbP53ΔL1 (SSPbP53 mutant lacking the QVVAQ loop 1 sequence – Supplementary Fig. S8) and recombinant His-AtXCP1-cys. SSPbP53 substantially inhibited casein degradation by AtXCP1 whereas SSPbP53ΔL1 had no inhibitory effect on AtXCP1 activity (Figure 4e, Supplementary Table S5), confirming the role of loop 1 in the inhibitory activity of SSPbP53.
Figure 4.

SSPbP53 interacts with xylem PLCPs. A. Yeast-two-hybrid (Y2H) assays using the clubroot pathogen SSPbP53 as the bait and Arabidopsis PLCP-cys domains representing different subfamilies as the prey. Growth of yeast cells on SD-3 selective media represents protein–protein interaction, while growth of the same cells on SD-2 media confirms yeast transformation. Yeast transformed with the empty vectors served as negative controls. B. In vitro pull-down assay using GST-AtPLCP-cys to immunoprecipitate His-SSPbP53 protein. Input and output (immunoprecipitated proteins) were detected by western blotting (WB) using anti-GST and anti-His antibodies. E. coli transformed with empty GST-vector was used as negative control. Immunoprecipitated fraction was visualized through Ponceau stain (PS). C. Pre-incubation of GST-XCP1-cys with E-64 inhibits the interaction with His-SSPbP53. Input and output (immunoprecipitated proteins) were visualized by western blotting (WB) using anti-GST and anti-His antibodies. Immunoprecipitated fraction was visualized through Ponceau stain (PS). D. Schematic representation of SSPbP53 and SSPbP53ΔL1 interaction with XCP1. E. Proteolytic activity of His-XCP1-cys, measured by digestion of a fluorescent casein substrate, is inhibited by purified His-SSPbP53 protein, but not by SSPbP53ΔL1. Fluorescence was measured at 485/538 nm excitation/emission. Mean ± standard deviation (n = 3). Statistically significant differences based on the two-tailed Student’s t-test are indicated by asterisks (*) where p < 0.001 is represented by (***)
To explore the in planta role of the AtXCP1-SSPbP53 interaction, the clubroot disease index [24] was scored in the Arabidopsis xcp1 null mutant (AtΔxcp1) at 21 dpi when, in infected AtCol-0 plants, SSPbP53 is highly expressed and AtXCP1is down regulated. Although XCP1 has been identified as involved in xylem maturation, AtΔxcp1mutants do not show abnormal vessel anatomy when compared with AtCol-0 [43]. Transverse sections of the hypocotyl region of AtΔxcp1 mutant and AtCol-0 plants during secondary infection with P. brassicae did not show any major anatomical differences between the two sets of plant roots (Figure 5a). The only observed difference was that the beginning of the expansive phase [44] of the secondary infection was more evident in AtCol-0 than in AtΔxcp1 (Figure 5a). In AtΔxcp1, the cell wall destruction of neighbor cells that is characteristic of the expansion [24], was delayed compared to AtCol-0 (Figure 5a). This was not observed in AtΔxcp1 mutants Pb- (Supplementary Fig. S9a). A detailed response time course should be performed to confirm these observations.
Figure 5.

SSPbP53-AtXCP1 interaction is required for Arabidopsis immunity. A. Light microscopy images of Pb+ AtΔxcp1 and AtCol-0 hypocotyl transverse sections at 21 dpi. Right upper corner amplification of expanded vessels indicated by yellow stars for AtCol-0 and red arrowheads for AtΔxcp1 (Scale bar – 50 μm). The observations presented are representative of 10 independent plants. B. Representation of the above and below ground of Pb+ AtΔxcp1 mutant and AtCol-0 at 21 dpi. C. Disease index for each group and graphic representation of the percentage of plants in each symptomatology for both groups, where NT is the total number of plants analyzed
Infected AtΔxcp1 plants showed reduced susceptibility to P. brassicae infection and/or development of symptoms when compared to infected AtCol-0 plants (Figure 5b-c, Supplementary Table S6). Furthermore, a lower number of resting spores was produced in P. brassicae infected AtΔxcp1 plants compared to infected AtCol-0 plants (Supplementary Fig. S9b, Supplementary Table S7), suggesting that a successful secondary infection and subsequent production of resting spores requires the inhibition of AtXCP1 activity by SSPbP53. Null mutants of AtΔrd19 and AtΔrd21 did not show a decreased susceptibility to P. brassicae (Supplementary Fig. S9c-d, Supplementary Table S6), indicating that the AtXCP1-SSPbP53 interaction is a specific requirement for P. brassicae pathogenicity.
Discussion
Plant proteolytic enzymes are important players in the arms race between plants and their pathogens. Here, we report on the role of the P. brassicae effector SSPbP53 as a cysteine protease, specifically XCP1, inhibitor. We have shown that SSPbP53 is an apoplastic cystatin-like protein that inhibits the enzymatic activity of the Arabidopsis PLCP, AtXCP1 (Figures 2 – 4). The results obtained from pull down experiments demonstrate that SSPbP53 interacts with two closely related xylem PLCPs, AtXCPB3 and AtXCP1 (Figure 4), but the interaction is clearly stronger with AtXCP1. Xylem cysteine proteases like XCP1 and XCP2, members of the same PLCP group, play key roles in the pathogenicity of biotrophic pathogens [17,19]. In a maize-U. maydis compatible interaction (resulting in smut disease), the inhibition of maize XCP2 activity, that unhindered would result in plant cell death, by the endogenous cysteine protease inhibitor CC9, transcriptionally induced during epidermal penetration in the compatible interaction, is key to suppressing host immunity during infection [18]. U. maydis also secretes Pit2, a cystatin that mimics CC9 and targets XCP2 [19]. Pit2 is active in the apoplastic space where it inhibits host cysteine proteases and contributes to the suppression of host immunity [19]. In Arabidopsis, XCP2 activity is inhibited by the endogenous protein PRN2, that stabilizes XCP2 and prevents its autocatalytic degradation, even though PRN2 does not possess any known protease inhibitor domains such as those present in serpins and cystatins [45]. Arabidopsis prn2, xcp2, and prn2-xcp2 null mutants are less susceptible than wild type to the vascular pathogen Ralstonia solanacearum, suggesting an essential role for XCP2 in plant susceptibility to this pathogen [45]. Interestingly, increased susceptibility to pathogens has also been reported when other PLCPs such as AtRD19, a cysteine protease required for RRS1-R-mediated resistance [46], AtRD21, an ortholog of the tomato immune protease C14 [47] and Solanum lycopersicum PIP1, an apoplastic PLCP [48] were knocked out.
A P. brassicae infection results in a reduction in active salicylic acid (SA) and its precursors in part due to the methylation of SA by PbBSMT [8]. We suggest that this reduced SA activity induces a down regulation of plant PLCP-encoding genes through a mechanism possibly similar to that recently described in maize [49]. SSPbP53 inhibits XCP1, XBCP3, and probably other PLCPs in the XCP1 group, thereby could be further contributing to the suppression of plant immunity. A very recent study has identified XCP1 as a caspase that proteolyzes Pathogenesis related protein 1, leading to the activation of systemic immunity [13]. SSPbP53 activity would be of particular importance during secondary infection, with its associated cell wall and vascular disruption [44,50] that would leave extracellular P. brassicae effectors susceptible to the protease activity of plant apoplastic PLCPs and cysteine proteases normally found in tertiary elements including XCP1. AtΔxcp1 plants were less susceptible to P. brassicae than wild type plants or plants lacking other PLCPs suggesting that the XCP1-SSPbP53 interaction is important in cortical disruption and the development of the nutrient sink galls associated with clubroot disease. Under normal growth conditions, the xcp1 mutation is complemented by increased expression of other PLCPs in the XCP1 group [43]. In Pb+ AtΔxcp1 plants we observed incomplete breaking of the cell wall of neighbor cells, a normal process of the expansive phase of secondary infection (Figure 5). That SSPbP53 does not inhibit other PLCPs would allow a degree of normal root growth in infected xcp1 plants. Something also very interesting in this study was the activity of SSPbP53 late during the secondary infection. Secondary infection is a vital infection stage for P. brassicae because it is when the pathogen initiates the expansion of cortical cells and the formation of galls, the nutrient rich niche required for completion of the life cycle and the formation of resting spores [44]. Other cysteine inhibitors with a similar function may act to protect the pathogen during primary infection, however, our study focused on secondary infection.
While it would be interesting to assess clubroot progression in 35S:XCP1 Arabidopsis lines, it has been reported that the over expression of XCP1 is detrimental to plant growth [51]. It will also be of interest to investigate the response of Arabidopsis 35S:SSPbP53 lines to clubroot disease. However, we are confident that the results presented here are a good characterization of the effector and its role in clubroot progress.
This study is an important step toward understanding the mechanisms used by P. brassicae to induce galls during secondary infection and one of the very few studies characterizing an effector protein from P. brassicae, a plant pathogen that has proved to be a mystery for the plant pathology community. We hope this study will contribute to the generation of increased clubroot-resistant germplasm.
Supplementary Material
Acknowledges
Thanks to Dr. Dwayne D. Hegedus from AAFC-Saskatoon Research Centre for providing plasmids PEG100 and PEG103. Thanks to Professor Renier van der Hoorn from the University of Oxford and Prof. Robert Malinowski from the Institute of Plant Genetics of the Polish Academy of Sciences for the critical review and recommendations.
Funding Statement
This work was supported by Saskatchewan Agriculture Development Fund (SADF) and the Saskatchewan Canola Development Commission (SaskCanola) funding to PCBS, YW and CDT, and NSERC-Discovery Grant funding to EPL; Ministry of Agriculture - Saskatchewan [20160138]; Saskatchewan Canola Development Commission [20160138].
Author contribution
EPL, CDT, YW, and PCBS designed the research; EPL and MMH performed the research; EPL, CDT, YW, and PCBS analyzed the data; EPL, MMH, CDT, YW, and PCBS wrote and/or edited the manuscript.
Disclosure statement
There are no relevant financial or non-financial competing interests to report.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Burki F, Kudryavtsev A, Matz MV, et al. Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol Biol. 2010;10(1):377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Walerowski P, Gündel A, Yahaya N, et al. Clubroot disease stimulates early steps of phloem differentiation and recruits SWEET sucrose transporters within developing galls. Plant Cell. 2018;30(12):3058–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Botero A, García C, Gossen BD, et al. Clubroot disease in Latin America: distribution and management strategies. Plant Pathol. 2019;68(5):827–833. [Google Scholar]
- [4].Dodds PN, Rathjen JP.. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet. 2010;11(8):539–548. [DOI] [PubMed] [Google Scholar]
- [5].Rabe F, Ajami-Rashidi Z, Doehlemann G, et al. Degradation of the plant defence hormone salicylic acid by the biotrophic fungus Ustilago maydis. Mol Microbiol. 2013;89(1):179–188. [DOI] [PubMed] [Google Scholar]
- [6].Lowe-Power TM, Jacobs JM, Ailloud F, et al. Degradation of the plant defense signal salicylic acid protects Ralstonia solanacearum from toxicity and enhances virulence on tobacco. mBio. 2016;7(3):e00656–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Han X, Kahmann R. Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front Plant Sci. 2019;10:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ludwig-Müller J, Jülken S, Geib K, et al. A novel methyltransferase from the intracellular pathogen Plasmodiophora brassicae methylates salicylic acid. Mol Plant Pathol. 2015;16(4):349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bulman S, Richter F, Marschollek S, et al. Arabidopsis thaliana expressing Pb BSMT, a gene encoding a SABATH-type methyltransferase from the plant pathogenic protist Plasmodiophora brassicae, show leaf chlorosis and altered host susceptibility. Plant Biol. 2019;21(S1):120–130. [DOI] [PubMed] [Google Scholar]
- [10].Misas-Villamil JC, Hoorn RA, Doehlemann G. Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 2016;212(4):902–907. [DOI] [PubMed] [Google Scholar]
- [11].Balakireva AV, Zamyatnin AA. Indispensable role of proteases in plant innate immunity. Int. J. of Mol. 2018;19(2):E629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Paulus JK, Kourelis J, Ramasubramanian S, et al. Extracellular proteolytic cascade in tomato activates immune protease Rcr3. Proc Natl Acad Sci. 2020;117(29):17409–17417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen Y-L, Lin F-W, Cheng K-T, et al. XCP1 is a caspase that proteolyzes Pathogenesis related protein 1 to produce the cytokine CAPE9 for systemic immunity in Arabidopsis. Res Square. 2021. DOI: 10.21203/rs.3.rs-155784/v1 [DOI] [Google Scholar]
- [14].Rooney H, Van der Hoorn RAL, Joosten MHAJ, et al. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science. 2005;308(5729):1783–1789. [DOI] [PubMed] [Google Scholar]
- [15].Shabab M, Shindo T, Gu C, et al. Fungal effector protein AVR2 targets diversifying defense-related cys proteases of tomato. Plant Cell. 2008;20(4):1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Song J, Win J, Tian M, et al. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Nat. Acad. Sci. USA. 2009;106(5):1654–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lozano-Torres JL, Wilbers RHP, Gawronski P, et al. Dual disease resistance mediated by immune receptor Cf-2 in tomato require a common virulence target of a fungus and a nematode. Proc. Nat. Acad. Sci. USA. 2012;109(25):10119–10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Van der Linde K, Hemetsberger C, Kastner C, et al. A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell. 2012;24(3):1285–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Müller AN, Ziemann S, Treitschke S, et al. Compatibility in the Ustilago maydis – maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PloS Pathog. 2013;9(2):e1003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gumtow R, Wu D, Uchida J, et al. A Phytophthora palmivora extracellular cystatin-like protease inhibitor targets papain to contribute to virulence on papaya. Molecular Plant-Microbe Interactions®. 2018;31(3):363–373. [DOI] [PubMed] [Google Scholar]
- [21].Clark K, Franco JY, Schwizer S, et al. An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat Commun. 2018;9(1):1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Krüger J, Thomas CM, Golstein C, et al. A tomato cysteine protease required for Cf-2 -dependent disease resistance and suppression of auto necrosis. Science. 2002;296(5568):744–747. [DOI] [PubMed] [Google Scholar]
- [23].Kaschani F, Shabab M, Bozkurt T, et al. An effector-targeted protease contributes to defense against phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 2010;154(4):1794–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pérez-López E, Hossain MM, Tu J, et al. Transcriptome analysis identifies Plasmodiophora brassicae secondary infection effector candidates. J. Euk. Microbiol. 2020;67(3):337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lampl N, Alkan N, Davydov O, et al. Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant J. 2013;74(3):498–510. [DOI] [PubMed] [Google Scholar]
- [26].Siemens J, Nagel M, Ludwig-Müller J, et al. The interaction of Plasmodiophora brassicae and Arabidopsis thaliana: parameters for disease quantification and screening of mutant lines. J Phytopathol. 2002;150(11–12):592–605. [Google Scholar]
- [27].Hwang SF, Ahmed HU, Strelkov SE, et al. Seedling age and inoculum density affect clubroot severity and seed yield in canola.Can. J. Plant Sci. 2011;91:183–190. [Google Scholar]
- [28].Arnold K, Bordoli L, Kopp J, et al. The SWISSMODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. [DOI] [PubMed] [Google Scholar]
- [29].Kiefer F, Arnold K, Kunzli M, et al. The SWISS-MODEL repository and associated resources. Nuc. Acid Res. 2009;37(Database):387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].De Lano WL. Pymol: an open-source molecular graphics tool. CCP4 newsletter on protein crystallography. 2002;40:82–92. [Google Scholar]
- [31].Xue S, Cao T, Howard RJ, et al. Isolation and variation in virulence of single-spore isolates of Plasmodiophora brassicae from Canada. Plant Dis. 2008;92(3):456–462. [DOI] [PubMed] [Google Scholar]
- [32].Chomczynski P, Mackey K. Modification of the TRIZOL reagent procedure for isolation of RNA from polysaccharide-and proteoglycan- rich sources. Biotechniques. 1995;19(6):942–945. [PubMed] [Google Scholar]
- [33].De Wit PJGM, Spikman G. Evidence for the occurrence of race and cultivar-specific elicitors of necrosis in intercellular fluids of compatible interactions of Cladosporium fulvum and tomato. Physiological Plant Pathology. 1982;21(1):1–11. [Google Scholar]
- [34].Warton K, Foster NC, Gold WA, et al. A novel gene family induced by acute inflammation in endothelial cells. Gene. 2004;342(1):85–95. [DOI] [PubMed] [Google Scholar]
- [35].Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].R Core Team . R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013: http://www.R-project.org/ [Google Scholar]
- [38].Nagata K, Kudo N, Abe K, et al. Three-dimensional solution of oryzacystatin-I, a cysteine proteinase inhibitor of rice, Oryza sativa L. japonica. Biochemistry. 2000;39:14753–14760. [DOI] [PubMed] [Google Scholar]
- [39].Powers JC, Asgian JL, Ekici ÖD, et al. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem Rev. 2002;102(12):4639–4750. [DOI] [PubMed] [Google Scholar]
- [40].Sperschneider J, Dodds PN, Singh KB, et al. ApoplastP: prediction of effectors and plant proteins in the apoplast using machine learning. New Phytol. 2017;217(4):1764–1778. [DOI] [PubMed] [Google Scholar]
- [41].Grudkowska M, Zagdanska B. Multifunctional role of plant cysteine proteinases. Acta Biochim Pol. 2004;51(3):609–624. [PubMed] [Google Scholar]
- [42].Björk I, Brieditis I, Raub-Segall E, et al. The importance of the second hairpin loop of cystatin C for proteinase binding. characterization of the interaction of trp-106 variants of the inhibitor with cysteine proteinases†. Biochem. 1996;35(33):10720–10726. [DOI] [PubMed] [Google Scholar]
- [43].Avci U, Petzold HE, Ismail IO, et al. Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J. 2008;56(2):303–315. [DOI] [PubMed] [Google Scholar]
- [44].Malinowski R, Truman W, Blicharz S. Genius Architect or Clever Thief—How Plasmodiophora brassicae reprograms host development to establish a pathogen-oriented physiological sink. Mol. Plant-Microb. Interact. 2019;32(10):1259–1266. [DOI] [PubMed] [Google Scholar]
- [45].Zhang B, Tremousaygue D, Denance N, et al. PIRIN 2 stabilizes cysteine protease XCP2 and increases susceptibility to the vascular pathogen Ralstonia solanacearum in Arabidopsis. Plant J. 2014;79(6):1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bernoux M, Timmers T, Jauneau A, et al. RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell. 2008;20(8):2252–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shindo T, Misas-Villamil JC, Hörger A, et al. A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS One. 2012;7(1):e29317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ilyas M, Hörger AC, Bozkurt TO, et al. Functional divergence of two secreted immune proteases of tomato. Curr Biol. 2015;25(17):2300–2306. [DOI] [PubMed] [Google Scholar]
- [49].Ziemann S, Van Der Linde K, Lahrmann U, et al. An apoplastic peptide activates salicylic acid signalling in maize. Nat Plants. 2018;4(3):172–180. [DOI] [PubMed] [Google Scholar]
- [50].Jubault M, Lariagon C, Taconnat L, et al. Partial resistance to clubroot in Arabidopsis is based on changes in the host primary metabolism and targeted cell division and expansion capacity. Funct Integr Genomics. 2013;13(2):191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Funk V, Kositsup B, Zhao C, et al. The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 2002;128(1):84–94. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


