Abstract
Cerebrospinal fluid (CSF) represents a promising source of cell-free DNA (cfDNA) for tumors of the central nervous system. A CSF-based liquid biopsy may obviate the need for riskier tissue biopsies and serve as a means for monitoring tumor recurrence or response to therapy. Spinal ependymomas most commonly occur in adults, and aggressive resection must be delicately balanced with the risk of injury to adjacent normal tissue. In patients with subtotal resection, recurrence commonly occurs. A CSF-based liquid biopsy matched to the patient’s spinal ependymoma mutation profile has potential to be more sensitive then surveillance MRI, but the utility has not been well characterized for tumors of the spinal cord. In this study, we collected matched blood, tumor, and CSF samples from three adult patients with WHO grade II intramedullary spinal ependymoma. We performed whole exome sequencing on matched tumor and normal DNA to design Droplet Digital™ PCR (ddPCR) probes for tumor and wild-type mutations. We then interrogated CSF samples for tumor-derived cfDNA by performing ddPCR on extracted cfDNA. Tumor cfDNA was not reliably detected in the CSF of our cohort. Anatomic sequestration and low grade of intramedullary spinal cord tumors likely limits the role of CSF liquid biopsy.
Keywords: Spinal ependymoma, Spinal cord tumor, Neurofibramatosis type 2 (NF2), Cell-free DNA (cfDNA), Cerebrospinal fluid (CSF), Circulating tumor DNA (ctDNA), Nucleic acid, Liquid biopsy, Droplet digital PCR
Introduction
Ependymomas are primary central nervous system (CNS) tumors, likely arising from radial glia cells [1]. They constitute 3–6% of all CNS tumors and 15% of all spinal cord tumors [2–4]. Spinal cord tumors are either intramedullary with normal tissue between the tumor and the CSF or intradural extramedullary with CSF in direct contact with the tumor. Intramedullary spinal cord tumors account for 60% of spinal ependymomas [5]. The presentation of spinal ependymoma may include back pain, gait ataxia, and sensory loss. Treatment for spinal ependymoma involves surgical gross total resection (GTR) with or without radiation; chemotherapy has not proven efficacious for ependymomas [6]. Achieving GTR without postoperative deficit requires clear planes of dissection between the tumor and normal spinal cord. Higher grade tumors frequently have unclear tumor margins, increasing the risk of spinal cord injury [7]. When GTR is achieved, recurrence is prevented in 90–100% of cases [8]. In patients with partial resection or radiation alone, recurrence occurs in 50–70% of cases [8]. The median progression-free survival and overall survival of spinal ependymomas are 82 months and 180 months, respectively [9]. Patients are followed for recurrence with MRI scans, however, the sensitivity of these scans are limited and do not provide biologic information about how residual tumor may escape standard therapy.
A more complete genetic and molecular understanding of spinal ependymomas is critical to improve treatment and outcomes for the patients with residual disease, yet spinal cord biopsies carry significant potential morbidity. An alternative would be to sample the CSF, which we and others have previously found to contain brain tumor associated cell-free DNA (cfDNA) [10, 11]. Circulating cfDNA is composed of short (70–200 base pairs) fragments of DNA thought to be shed by necrotic or apoptotic cells into a bodily fluid [12]. A blood-based liquid biopsy for tumors suffers from low sensitivity since blood contains many cell types making it difficult to detect tumor cfDNA from normal background cfDNA. By contrast, a CSF-based liquid biopsy may serve as a more useful approach for tumors of the CNS since there are fewer native cell populations compared to blood [13]. A lumbar puncture to access CSF has less risk then a CNS tissue biopsy and may aid in the characterization and monitoring of spinal cord tumors. Although a CSF-based liquid biopsy has been recently studied for brain tumors, there is a paucity of work for spinal cord tumors [10, 11, 14].
We sought to describe the genetic landscape of mutations found in three cases of intramedullary spinal ependymoma, and evaluate the utility of a liquid biopsy by searching for tumor-derived cfDNA in matched CSF.
Materials and methods
Study design
Three patients with spinal ependymoma underwent tumor resection at Stanford University. Matched blood, CSF, and tumor samples were obtained intraoperatively from each patient after an Institutional Review Board-approved informed consent process. Only tumor tissue not required for pathological diagnosis was utilized in this study. No procedures were performed for the exclusive purpose of research. Blood samples were obtained at the time of anesthetic induction, and CSF was obtained upon opening of the dura and prior to tumor dissection. The blood and CSF samples were centrifuged to separate cell-free and cellular DNA. Matched tumor and normal whole-exome sequencing was performed on DNA extracted from tumor tissue and the cellular component of blood. Fluorescent digital droplet PCR (ddPCR) probes were created for point mutations found in each patient’s tumor sample and used to interrogate the CSF for tumor-associated cfDNA.
Sample processing
Blood and CSF were centrifuged (1000×g, 10 min, 4 °C) within 2 h of collection to separate cell-free and cellular DNA. The initial supernatant from spun down blood samples underwent an additional centrifugation step (10,000×g, 10 min, 4 °C) to further separate cell-free plasma and blood cells. The supernatant and pellet components were transferred to separate tubes and stored at −80 °C until ready for DNA extraction. DNA from CSF was extracted with a QIAamp Circulating Nucleic Acid Kit (Qiagen), while DNA from blood cells was extracted with a QIAamp DNA Mini Kit (Qiagen). Cellular DNA extracted from the pellet component of blood samples was fragmented to approximately 300 base pairs by sonication (S220 focused ultrasonicator, Covaris). The size distribution of DNA was measured by a fragment analyzer (Advanced Analytical).
Whole exome sequencing
Indexed Illumina libraries were created (KAPA Hyper Prep Kit, Kapa Biosystems) from 200 nanograms of matched tumor (over 90% tumor content) and blood cellular DNA. Exonic DNA was captured using NimbleGen SeqCap EZ Human Exome Library (Roche). Approximately 100 million reads per library were sequenced with 150-bp paired end runs on a NextSeq sequencing system (Illumina).
Sequencing analysis was performed using RAVE v 2.7.9 (Bina Technologies, Inc.). In brief, alignment was performed on RAVE using BWA-MEM 0.7 to a human reference genome (hg19 assembly). The aligned reads were then realigned and recalibrated using GATK (version 3.3), following the best practices for secondary sequence analysis recommended by the Broad Institute. Somatic calling was performed on RAVE using JointSNVMix 0.7.5, MuTect 2014.3–24, SomaticSniper 1.0.4, and VarScan 2.3.7. Annotation was performed using wANNOVAR [15].
Confident mutations were first selected by filtering for mutations that were called by more than three of the four total variant calling tools used. We then further selected for highly confident mutations by filtering for mutant allele fractions greater than 20% in tumor and less than 5% in normal. Mutations were checked for listing in the Catalogue of Somatic Mutations in Cancer (COSMIC) database. The COSMIC database is an online database of somatic mutations that have been implicated in cancer [16]. All mutations that were selected for probe design were confirmed in Integrative Genomics Viewer.
Droplet digital™ PCR
Fluorophore labeled probes were made for mutant (FAM) and wild-type (HEX) alleles for each patient based on tumor-normal whole exome sequencing somatic calling. The probes, primers and extracted DNA were mixed together and ddPCR was performed using a QX100™ ddPCR System (Biorad). Extracted tumor and blood DNA was first interrogated to validate the mutant and wild type probes. This was followed by interrogation of cfDNA extracted from CSF. All ddPCR experiments were performed in duplicates with water serving as the appropriate negative control sample. The concentration of mutant and wild-type DNA was calculated automatically by QuantaSoft software (Biorad). Calculations of mutant and wild-type allele concentrations in CSF in copies per mL and nanograms per mL were back-calculated as described in Pan et al. [10].
Results
Case 1
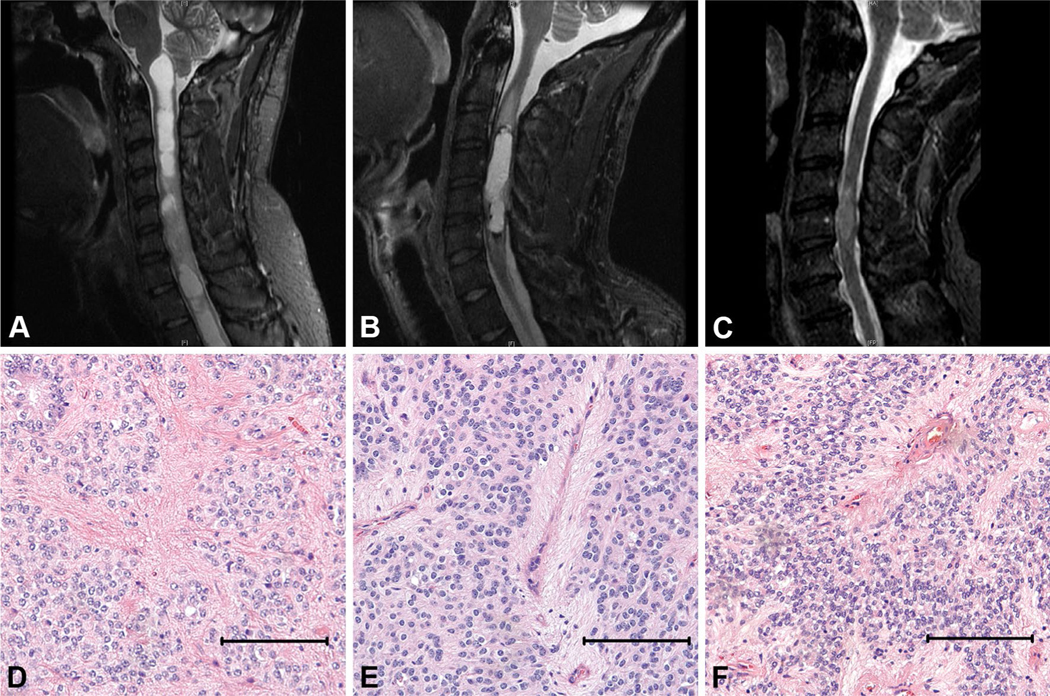
A 33 year-old male presented with sensory abnormalities and hyperreflexia of both lower extremities, and intermittent pain in the left arm and hand. MRI spine revealed an intramedullary spinal cord mass extending from C4 to C7, and associated hydromyelia (Fig. 1a). A C4–C7 laminectomy and gross total resection of the WHO Grade II ependymoma (Fig. 1d) was performed. The patient tolerated the procedure well. At 2 years of follow-up, the patient had some improvement in his symptoms with partial residual extremity numbness and no evidence of recurrence.
Fig. 1.
Cervical spine MRI (STIR sagittal sequence) showing intramedullary cervical spinal ependymoma in case 1 (a), 2 (b), and 3 (c). Associated hydromyelia is evident case 1 (a). Histology showing pseudovascular rosettes for patient 1, 2, and 3 (d–f). Scale bar 100 microns
Case 2
A 48 year-old male presented with intermittent neck pain, paresthesias of his upper extremities, and hyperreflexia. MRI spine revealed an intramedullary spinal cord mass extending from C3 to C6 (Fig. 1b). A C3–C7 laminectomy and gross total resection of the WHO Grade II ependymoma (Fig. 1e) was performed. The patient tolerated the procedure well. At 18 months of follow-up, the patient had some residual numbness in his extremities and no evidence of tumor recurrence.
Case 3
A 59 year-old male presented with gait difficulty, hand weakness and extremity paresthesias. MRI spine revealed an intramedullary spinal cord mass centered at C5 (Fig. 1c). A C4-C7 laminectomy and gross total resection of the WHO Grade II ependymoma (Fig. 1f) was performed. The patient tolerated the procedure well. At 18 months follow-up, the patient had some residual numbness in his hands and feet, and no evidence of tumor recurrence.
Exome sequencing
There were approximately 45,600,000 bases covered with a targeted coverage mean ranging from 72.8 to 88.2. Ensemble mutation calling, using the four tools mentioned previously, resulted in an average total of 2307 SNVs with an average total of 337 non-synonymous mutations. Confident mutations were filtered as described above. Of the nonsynonymous mutations, only GPR112, was listed in the COSMIC database (COSM363149). There were no confident ependymoma driver mutations found and no SNVs were shared between samples. Complete annotated mutation files for each patient are available in the supplementary file. A summary of the mutations selected for ddPCR fluorescent probes is summarized in Table 1 below. Because we collected approximately 5 mLs of CSF from case 1, we used a total of five probes. We were limited by sample volume (1–2 mLs) for case 2 and 3. Thus, we only used one and two probes respectively.
Table 1.
Summary of the highly confident mutations used for ddPCR fluorescent probes
| Probe | Sample | Gene | Chromosome | Position | Mutation type | Number of tools that called the mutation | Tumor |
Normal |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Reference allele reads (%) | Mutant allelle reads (%) | Reference allele reads (%) | Mutant allelle reads (%) | |||||||
| 1 | Case 1 | XKR7 | chr20 | 30585194 | Nonsynonymous | 4/4 | 63 (62%) | 39 (38%) | 59 (97%) | 2 (3%) |
| 2 | Case 1 | GPR112 | chrX | 135443724 | Nonsynonymous | 4/4 | 65 (76%) | 20 (24%) | 67 (100%) | 0 (0%) |
| 3 | Case 1 | OR5AC2 | chr3 | 97806176 | Nonsynonymous | 4/4 | 142 (75%) | 48 (25%) | 128 (100%) | 0 (0%) |
| 4 | Case 1 | GPR112 | chrX | 135429739 | Nonsynonymous | 4/4 | 60 (67%) | 29 (33%) | 59 (100%) | 0 (0%) |
| 5 | Case 1 | DDX41 | chr5 | 176943308 | Synonymous | 4/4 | 61 (59%) | 43 (41%) | 65 (100%) | 0 (0%) |
| 6 | Case 2 | FSCB | chrl4 | 44974189 | Nonsynonymous | 4/4 | 53 (56%) | 42 (44%) | 96 (96%) | 4 (4%) |
| 7 | Case 3 | CDH5 | chrl6 | 66426221 | Nonsynonymous | 4/4 | 24 (41%) | 34 (59%) | 46 (100%) | 0 (0%) |
| 8 | Case 3 | PICALM | chrll | 85692828 | Not exonic | 4/4 | 21 (57%) | 16 (43%) | 44 (100%) | 0 (0%) |
The gene for probe four (bolded), GPR112, was listed in the COSMIC database (COSM363149)
Validation of ddPCR probes with tumor and normal DNA
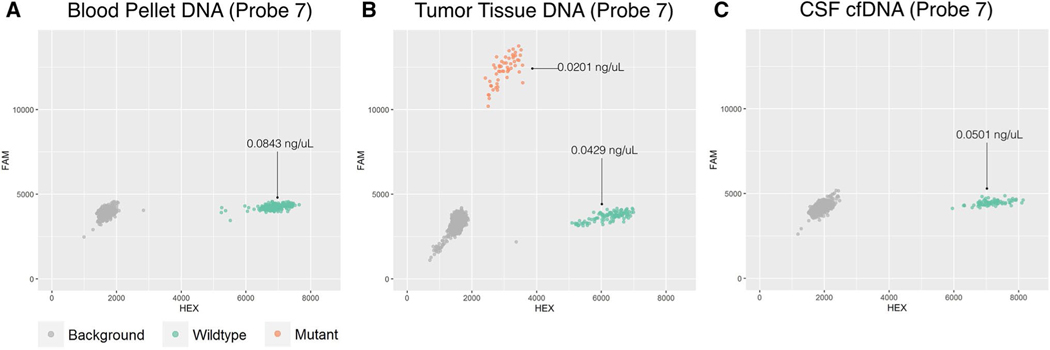
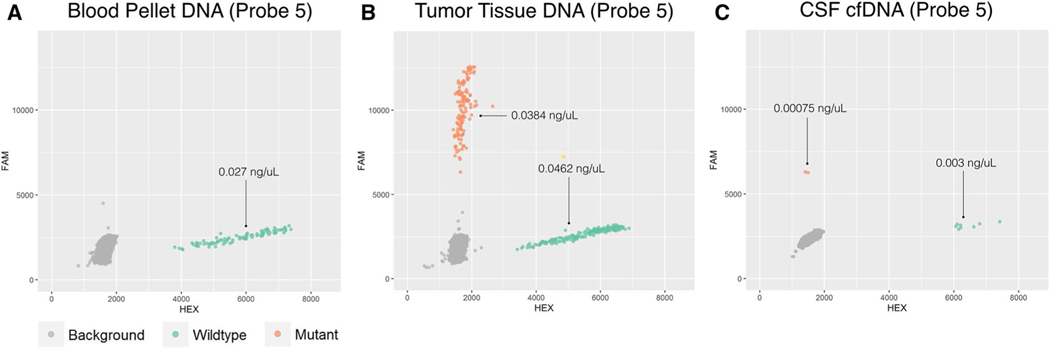
As a negative and positive control, we first tested the ddPCR probes on fragmented DNA extracted from blood pellet (negative control) and tumor tissue (positive control) samples (Figs. 2, 3a, b). All probes showed signal for both mutant and wild-type fluorophores when tested with DNA extracted from tumor tissue (Figs. 2b, 3b). When tested with DNA extracted from the blood pellet probes only showed a wild-type signal (Figs. 2a, 3a). We also performed a serial dilution experiment using fragmented tumor DNA from patient 1 to evaluate the detection limits of our probes (Supplementary Figure S1). The prepared dilution shown in panel C (reaction concentration of 0.0005 ng/uL) was unable to reliably detect DNA as it was only able to detect wild-type signal in two of four reaction replicates. Wild-type and mutant DNA from the prepared dilutions in panel A and B were detected in all four reaction replicates.
Fig. 2.
Representative ddPCR results for probe 7 (CDH5) are shown for DNA extracted from the blood pellet (a), tumor tissue (b), and CSF supernatant (c). Concentrations of the ddPCR reaction are given in ng/uL for mutant and wild-type. Only wild-type (green, HEX) signal is detected in DNA extracted from blood (a). Both wild-type (green, HEX) and mutant (orange, FAM) signals were detected in DNA extracted from tumor tissue (b). cfDNA extracted from CSF only contained wild-type signal (c)
Fig. 3.
Representative ddPCR results for probe 5 (DDX41) are shown for DNA extracted from the blood pellet (a), tumor tissue (b), and CSF supernatant (c). Concentrations of the ddPCR reaction are given in ng/uL for mutant and wild-type. Only wild-type (green, HEX) signal is detected in DNA extracted from blood (a). Both wild-type (green, HEX) and mutant (orange, FAM) signals were detected in DNA extracted from tumor tissue (b). cfDNA extracted from CSF contained wild-type signal and a small amount of mutant signal which was unreliably detected across experimental duplicates (c)
Interrogation of cfDNA in CSF
We then interrogated the CSF for tumor-derived cfDNA using the working probes. Altogether, we tested 5 probes for case 1, one probe for case 2, and two probes for case 3. For each set of probes, only wild-type DNA was found in CSF (Fig. 2c) with the exception of probe five from patient 1 (Fig. 3c). Probe 5 was able to detect a small amount of mutant signal in both reaction duplicates for patient 1. The calculated concentrations of the wild-type DNA were 2604 copies per mL (7.81 ng/mL) of CSF, 201,100 copies per mL (603.3 ng/mL) of CSF, and 33,493 copies per mL (100.48 ng/mL) of CSF for cases 1, 2, and 3 respectively (Table 2). This small amount of mutant cfDNA detected using probe 5 corresponded to approximately 380 copies per mL (1.14 ng/mL) of CSF or an approximate reaction well concentration of 0.0008 ng/uL. (Fig. 3c; Table 2). Additional controls from our group included in Supplementary Figure S2 confirm the ability of ddPCR to detect mutant cfDNA when the tumor is exposed to the CSF space.
Table 2.
Copies of wild-type cfDNA per mL of CSF were calculated
| Sample | Wild-type |
Mutant |
Number of probes tested | ||||
|---|---|---|---|---|---|---|---|
| Copies/mL of CSF (mean) | Estimated ng/ mL of CSF | Standard error | Copies/mL of CSF (mean) | Estimated ng/ mL of CSF | Standard error | ||
| Case 1 | 2304 | 7.81 | 317 | 76 | 1.14 | 76 | 5 |
| Case 2 | 201,100 | 603.3 | - | 0 | 0 | - | 1 |
| Case 3 | 33,493 | 100.48 | 1258 | 0 | - | - | 3 |
Patient 1, 2, and 3 had 2604, 201,100, and 33,493 wild-type copies per mL of CSF respectively. A small amount of mutant DNA was detected in case one in one of five probes
Discussion
The ability to monitor and target spinal ependymoma progression would be of great benefit in patients with subtotal resection due to the increased risks for recurrence [8]. These patients commonly undergo adjuvant radiotherapy in isolation, as chemotherapy has not shown benefit in the treatment of spinal ependymoma [6]. A better understanding of the molecular and genetic mechanisms of spinal ependymoma recurrence may help in the development of targeted therapeutics. A less invasive liquid biopsy would allow both scientific advancement of our understanding of resistance to therapy and act as a tumor marker for patients undergoing surveillance after subtotal resection of these tumors.
Although studies have shown genetic differences in supratentorial and infratentorial cranial ependymomas, fewer genetic studies have been performed on spinal ependymomas [17]. To date, research has revealed a relatively stable genome for spinal ependymoma. The only putative recurrent mutations have been in the NF2 gene which are thought to disrupt contact mediated growth inhibition mediated by the protein, merlin [18, 19]. Even so, this gene appears to be variably mutated in studies. Bettegowda et al., reported the presence of NF2 mutations in 9 of 19 spinal ependymoma samples while Wang and colleagues reported none in their cohort of five samples [11, 18]. Similarly, we did not detect any NF2 mutations, and patients with neurofibromatosis were not part of our cohort. After searching for mutated genes shared between our samples and those of Bettegowda et al. (three samples with supplemental data) and Wang et al. we only found one shared gene, CDH5. This gene mutation was only present in 1 patient in our study and in 1 patient in Wang et al. with a myxopapillary ependymoma. Mutations in HOX genes, which encode for multiple transcription factors playing a role in body segmentation, have also been described in spinal ependymomas [20]. No HOX genes were mutated in our cases nor in those of Bettegowda and Wang et al. In Patient 1 of our cohort, a nonsynonymous mutation in GPR112, which codes for a G protein coupled receptor, was detected and was listed in the COSMIC database (COSM363149). Although mutation of this gene and its relation to cancer progression has not been heavily researched, two studies have postulated that it may be a potential therapeutic target for carcinomas of the gastrointestinal tract [21, 22]. Losses of chromosome 22q and gain of chromosome 7 have also been previously reported [20]. Given the lack of overlap between whole exome sequencing results for spinal ependymoma in our cohort and others, it appears that these tumors are both genetically stable and mutationally diverse. This is supported by our results which revealed no known recurrent mutations and no shared SNVs between samples. This means that any attempt at a liquid biopsy for these patients would have to be personalized to known mutations in the patient’s primary tumor.
The ability of cfDNA to cross the CNS parenchyma and membranes remains largely uncharacterized. Jimenez et al. describes three barriers of the CNS—the blood–brain barrier, the blood–CSF barrier of the choroid plexus and arachnoid membrane, and the CSF-parenchyma barrier of the ependymal [23]. For intramedullary spinal ependymoma, the diffusive capacity through the CSF-parenchyma barrier (tortuosity of the extracellular space within the parenchyma, bulk flow of interstitial fluid, and degradation) represents the limiting factor for the outward shedding of tumor-derived cfDNA into CSF [24]. Although there is a paucity of research investigating outward diffusion of tumor-associated DNA from within the CNS parenchyma, inward diffusion of various substances from the CSF space has been heavily studied. Previous work has shown that various compounds (e.g. Vitamin C, sucrose, mannitol, and inulin) are able to distribute throughout the brain parenchyma and spinal cord from the CSF space [25–29].
Spinal ependymoma cfDNA could also access the CSF through the central canal of the spinal cord, although this structure may be obstructed in spinal cord tumors. Both this surface and the ventricular surface of the brain are lined by ependymal cells. Whish et al. concluded that the ability of various sized probes to diffuse through the ependyma increases with age and correlates with the progressive loss of ependymal strap junctions, adhesion molecules that are present in the ependyma of the developing brain [24].
Despite the potential diffusive capacity of intramedullary spinal cord tumor-associated cfDNA, we were unable to detect tumor-derived cfDNA reliably in our patients. In previous work by our group, we have detected low concentrations of brain tumor mutations in plasma which has much more genomic background noise compared to CSF using these techniques [10]. Because of this background noise, the use of ddPCR in plasma to detect tumor mutations is usually limited to cases with disseminated metastatic disease. CSF appears to a more reliable source for detecting low concentrations of tumor mutations due to reduced background noise. The methods employed in this study have been previously used by our group to identify mutated DNA in CSF samples of patients with known leptomeningeal disease [10, 30]. As comparison and as negative and positive controls, we have included ddPCR plots from a patient with a solid brain metastasis not exposed to CSF space (Supplementary Figure S2 A), a patient with a solid brain metastasis with exposure to the CSF space via the lateral ventricle (Supplementary Figure S2 B), and a patient with cytology positive leptomeningeal disease (Supplementary Figure S2 C). The plots show that both mutant cfDNA and wild-type cfDNA are detected when the tumor is in contact with the CSF space. Given the highly sensitive nature of ddPCR and the ability of the probes used in this study to detect mutant and wildtype DNA in tumor tissue DNA, a false negative result is unlikely. In serial dilutions (Supplementary Figure S1), ddPCR was able to detect DNA in the 0.0005 ng/uL prepared reaction in only two of the four replicates performed at this concentration. The representative plot shown in Supplementary Figure S1 C shows one wild-type DNA droplet corresponding to 0.00024 ng/uL suggesting that this concentration approaches the detection limit of ddPCR.
In our experiments, wild-type cfDNA was detected in CSF which is confirmation that our DNA extraction was performed successfully. We were able to detect a small amount of tumor-derived cfDNA using one of the 5 probes for patient 1 which corresponded to a reaction concentration of 0.00075 ng/uL. Interestingly, this concentration is similar to the detection limit seen in serial dilution ddPCR (0.00024 ng/uL) (Supplementary Figure S1 C). It is unclear why this point mutation was detected while others were not. Although the corresponding point mutation for this probe had the highest mutant allele fraction (41%) of all of the mutations for patient 1, it was not the highest overall. The CDH5 mutation in patient three had the highest mutant allele fraction (59%) overall. Surprisingly, this mutation was not detected in CSF. It is likely that for intramedullary spinal tumors that are anatomically sequestered and low grade with relatively little tissue turn over, meaning only a small fraction of tumor-derived cfDNA is able to escape into the CSF. Of this fraction, only a subset of mutations are present since the genome is not fully represented due to the fragmented nature of cfDNA. An additional reason for why we were not able to reliably detect tumor-derived cfDNA in CSF may be due to the relatively small size of these tumors, or due to the presence of CSF DNAse. The dynamic relationship of intramedullary tumor cfDNA efflux and their stability and concentration in CSF is a topic of ongoing study in our laboratory.
We were also able to derive the concentration of wild-type DNA in the CSF cell-free component. Patient 2 and 3 had much higher concentration of wild-type DNA. This is likely due to blood contamination during CSF collection as these CSF samples appeared slightly pink. The CSF specimen from patient 1 appeared clear and is more likely a truer reflection of native wild-type cfDNA concentrations in CSF. Thus, the wild-type DNA detected in CSF through ddPCR most likely represents a mixture of genomic DNA from blood and wild-type cfDNA. Since genomic DNA from blood contamination is normal, the mutant DNA detected using probe 5 (patient 1) is likely from tumor cfDNA.
Our current study, as well as prior studies by our group and others, support the hypothesis that anatomically sequestered tumors such as intramedullary spinal cord ependymomas may not be amenable to CSF liquid biopsy. In cases where the tumor is not surrounded by parenchyma or isolated by arachnoid membrannes, tumor-derived cfDNA can be detected. In our previous work, we found tumor-derived cfDNA in the CSF of six of seven brain tumor patients [10]. The single case where tumor cfDNA was not found was a patient with vestibular schwannoma, likely due to the tumor being encased in isolating arachnoid layers. A subsequent study by Wang and colleagues found tumor cfDNA in CSF in 21 of 24 cases where the tumor was in contact with a CSF space [11]. Of their cohort of 35 patients, there were 3 cases of WHO grade II spinal ependymoma. Tumor cfDNA was found in two of these cases, one of which was in definitive contact with a CSF space. In the third case, the tumor was not abutting a CSF space and no tumor cfDNA was found. In total, the study included 11 spinal tumor samples where the location of the tumor relative to a CSF space was available (spinal ependymomas, myxopapillary ependymomas, low-grade gliomas, and glioblastoma). Of the nine tumors that were abutting a CSF space, six had detectable levels of tumor cfDNA in CSF. In the 2 cases where the tumors did not abut a CSF space, no tumor cfDNA was detected in CSF. Our data are consistent with these results, suggesting CNS tumors should be in close contact with the CSF space in order for a liquid biopsy to sample tumor-associated cfDNA. Subsequent studies should also include recurrent spinal ependymoma where the tumor bed may be in continuity with the CSF given the prior surgical resection, and genetic changes after radiation may show mutation overlap between samples.
Limitations
The main limitation of our pilot study its small sample size. However, our results appear consistent with those of Wang and colleagues who also argued that anatomic sequestration may limit the use of a CSF based liquid biopsy for tumors that do not interface with the CSF [11]. Despite the high sensitivity of ddPCR, biological levels below the detection limit of this technique may be difficult to exclude entirely as well.
Conclusion
The dynamics and mechanisms of cfDNA excretion from tumors, the diffusive capacity of cfDNA, and the relative stability of cfDNA in CSF remain unclear. Whole exome sequencing performed on tumor-normal samples revealed no recurrent or driver mutations, as is typical for these tumors. Patient-specific cfDNA probes did not reliably find tumor-derived cfDNA in CSF, likely due to the barrier effect of the surrounding spinal cord parenchyma and pia mater. In genetically diverse and anatomically sequestered CNS tumors such as intramedullary spinal ependymoma, a CSF-based liquid biopsy may not be feasible. Continued work to determine the appropriate use a CSF liquid biopsy in CNS tumors will further enable the selected transition of this research tool into the clinic and scientific workflow.
Supplementary Material
Acknowledgements
This work was supported in part by The Hearst Foundation Grant, K08-NS901527, The Curci Foundation Grant, R21-CA 193046-01 (MHG), Stanford MedScholars funding (IDC), and a Stanford CHRI fellowship (YL).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11060-017-2557-y) contains supplementary material, which is available to authorized users.
References
- 1.Taylor MD, Poppleton H, Fuller C et al. (2005) Radial glia cells are candidate stem cells of ependymoma. Cancer Cell 8:323–335. doi: 10.1016/j.ccr.2005.09.001 [DOI] [PubMed] [Google Scholar]
- 2.Gilbert MR, Ruda R, Soffietti R (2010) Ependymomas in adults. Curr Neurol Neurosci Rep 10:240–247. doi: 10.1007/s11910-010-0109-3 [DOI] [PubMed] [Google Scholar]
- 3.Nagasawa DT, Smith ZA, Cremer N et al. (2011) Complications associated with the treatment for spinal ependymomas. Neurosurg Focus 31:E13. doi: 10.3171/2011.7.FOCUS11158 [DOI] [PubMed] [Google Scholar]
- 4.Tseng JH, Tseng MY (2007) Survival analysis of 459 adult patients with primary spinal cancer in England and Wales: a population-based study. Surg Neurol 67:53–58. doi: 10.1016/j.surneu.2006.04.011 [DOI] [PubMed] [Google Scholar]
- 5.Chamberlain MC (2003) Ependymomas. Curr Neurol Neurosci Rep 3:193–199. doi: 10.1007/s11910-003-0078-x [DOI] [PubMed] [Google Scholar]
- 6.Wu J, Armstrong TS, Gilbert MR (2016) Biology and management of ependymomas. Neuro Oncol 18:902–913. doi: 10.1093/neuonc/now016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tobin MK, Geraghty JR, Engelhard HH et al. (2015) Intramedullary spinal cord tumors: a review of current and future treatment strategies. Neurosurg Focus 39:E14. doi: 10.3171/2015.5.FOCUS15158 [DOI] [PubMed] [Google Scholar]
- 8.Rudà R, Gilbert M, Soffietti R (2008) Ependymomas of the adult: molecular biology and treatment. Curr Opin Neurol 21:754–761. doi: 10.1097/WCO.0b013e328317efe8 [DOI] [PubMed] [Google Scholar]
- 9.Gomez DR, Missett BT, Wara WM, et al. (2005) High failure rate in spinal ependymomas with long-term follow-up. Neuro Oncol 7:254–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan W, Gu W, Nagpal S et al. (2015) Brain tumor mutations detected in cerebral spinal fluid. Clin Chem 61:514–522. doi: 10.1373/clinchem.2014.235457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Springer S, Zhang M et al. (2015) Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci USA 112:9704–9709. doi: 10.1073/pnas.1511694112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarzenbach H, Hoon DSB, Pantel K (2011) Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11:426–437. doi: 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 13.Connolly ID, Li Y, Gephart MH, Nagpal S (2016) The “Liquid Biopsy”: the role of circulating DNA and RNA in central nervous system tumors. Curr Neurol Neurosci Rep 16:25. doi: 10.1007/s11910-016-0629-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Pan W, Connolly ID et al. (2016) Tumor DNA in cerebral spinal fluid reflects clinical course in a patient with melanoma leptomeningeal brain metastases. J Neurooncol. doi: 10.1007/s11060-016-2081-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H, Wang K (2015) Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 10:1556–1566. doi: 10.1038/nprot.2015.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forbes SA, Beare D, Gunasekaran P et al. (2015) COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43:D805–D811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreiuolo F, Puget S, Peyre M et al. (2010) Neuronal differentiation distinguishes supratentorial and infratentorial childhood ependymomas. Neuro Oncol 12:1126–1134. doi: 10.1093/neuonc/noq074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bettegowda C, Agrawal N, Jiao Y et al. (2013) Exomic sequencing of four rare central nervous system tumor types. Oncotarget 4:572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada T, Lopez-Lago M, Giancotti FG (2005) Merlin/NF-2 mediates contact inhibition of growth by suppressing recruitment of Rac to the plasma membrane. J Cell Biol 171:361–371. doi: 10.1083/jcb.200503165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connolly ID, Ali R, Li Y, Gephart MH (2015) Genetic and molecular distinctions in spinal ependymomas: a review. Clin Neurol Neurosurg 139:210–215. doi: 10.1016/j.clineuro.2015.10.011 [DOI] [PubMed] [Google Scholar]
- 21.Leja J, Essaghir A, Essand M et al. (2009) Novel markers for enterochromaffin cells and gastrointestinal neuroendocrine carcinomas. Mod Pathol 22:261–272. doi: 10.1038/modpathol.2008.174 [DOI] [PubMed] [Google Scholar]
- 22.Nilsson O (2013) Profiling of ileal carcinoids. Neuroendocrinology 97:7–18. doi: 10.1159/000343232 [DOI] [PubMed] [Google Scholar]
- 23.Jiménez AJ, Domínguez-Pinos M-D, Guerra MM et al. (2014) Structure and function of the ependymal barrier and diseases associated with ependyma disruption. Tissue barriers 2:e28426. doi: 10.4161/tisb.28426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whish S, Dziegielewska KM, Møllgård K et al. (2015) The inner csf-brain barrier: developmentally controlled access to the brain via intercellular junctions. Front Neurosci 9:1–15. doi: 10.3389/fnins.2015.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammarström L (1966) Autoradiographic studies on the distribution of c14-labelled ascorbic acid and dehydroascorbic acid. Acta Physiol Scand 70:1–83 [Google Scholar]
- 26.Lorenzo AV, Snodgrass SR (1972) Leucine transport from the ventricles and the cranial subarachnoid space in the cat1. J Neurochem 19:1287–1298 [DOI] [PubMed] [Google Scholar]
- 27.Spector R, Keep RF, Robert Snodgrass S et al. (2015) A balanced view of choroid plexus structure and function: Focus on adult humans. Exp Neurol 267:78–86. doi: 10.1016/j.expneurol.2015.02.032 [DOI] [PubMed] [Google Scholar]
- 28.Spector R (1981) Penetration of ascorbic acid from cerebrospinal fluid into brain. Exp Neurol 72:645–653 [DOI] [PubMed] [Google Scholar]
- 29.Cifuentes M, Fernández-LLebrez P, Perez J et al. (1992) Distribution of intraventricularly injected horseradish peroxidase in cerebrospinal fluid compartments of the rat spinal cord. Cell Tissue Res 270:485–494 [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Pan W, Connolly ID et al. (2016) Tumor DNA in cerebral spinal fluid reflects clinical course in a patient with melanoma leptomeningeal brain metastases. J Neurooncol 128:93–100. doi: 10.1007/s11060-016-2081-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.