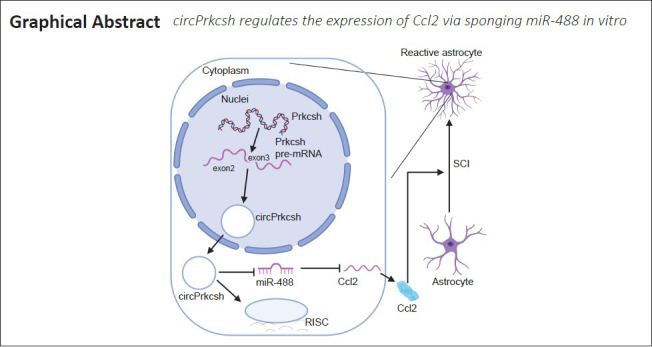
Key Words: astrocyte, Ccl2, ceRNA, circRNA, inflammation, miR-488, ncRNA, Prkcsh, RNA sequencing, spinal cord injury
Abstract
Circular RNAs (circRNAs) are a class of conserved, endogenous non-coding RNAs that are involved in transcriptional and post-transcriptional gene regulation and are highly enriched in the nervous system. They participate in the survival and differentiation of multiple nerve cells, and may even promote the recovery of neurological function after stroke. However, their role in the inflammatory response after spinal cord injury remains unclear. In the present study, we established a mouse model of T9 spinal cord injury using the modified Allen’s impact method, and identified 16,013 circRNAs and 960 miRNAs that were differentially expressed after spinal cord injury. Of these, the expression levels of circPrkcsh were significantly different between injured and sham-treated mice. We then treated astrocytes with tumor necrosis factor-α in vitro to simulate the inflammatory response after spinal cord injury. Our results revealed an elevated expression of circPrkcsh with a concurrent decrease in miR-488 expression in injured cells. We also found that circPrkcsh regulated the expression of the inflammation-related gene Ccl2. Furthermore, in tumor necrosis factor-α-treated astrocytes, circPrkcsh knockdown decreased the expression of Ccl2 by upregulating miR-488 expression, and reduced the secretion of inflammatory cytokines in vitro. These findings suggest that differentially expressed circRNAs participate in the inflammatory response after spinal cord injury and act as the regulators of certain microRNAs. Furthermore, circPrkcsh may be used as an miR-488 sponge to regulate Ccl2 expression, which might provide a new potential therapy for SCI. The study was approved by the Animal Ethics Committee of Shandong University of China (approval No. KYLL-20170303) on March 3, 2017.
Chinese Library Classification No. R446; R744; R364.5
Introduction
Spinal cord injury (SCI) is a severe physical injury that induces a loss of function. In accordance with its etiology, it can be further categorized into traumatic and non-traumatic SCI, the former of which accounts for the larger component (Fehlings, 2013). There are approximately 2700,000 cases of SCI worldwide, caused by car accidents, falling from buildings, violent injuries, and many other reasons. SCI causes much physical trauma to patients and places an economic burden on their families and society. Traumatic SCI can be divided into either the primary or secondary injury (Ahuja et al., 2017). A primary injury represents damage to the spinal cord caused by direct mechanical trauma, and involves the death of neurons, astrocytes, oligodendrocytes, and microglia, among other cell types (Ahuja et al., 2017). Ischemia, inflammation, and the destruction of the blood–spinal cord barrier caused by the primary injury are termed the secondary injury, and result in glial scars and cystic cavities (Silva et al., 2014). Inflammation, coupled with the weakening of the inherent regenerative ability of axons, results in the poor recovery of spinal cord nerve function; inflammation plays a key role in pathological changes after SCI (Alilain et al., 2011). Tumor necrosis factor-α (TNF-α), which is a highly expressed inflammatory cytokine, is one of the most clearly altered proteins at the injury site after SCI (Olmos and Lladó, 2014; Oh et al., 2020). TNF-α regulates the expression of various types of inflammatory cytokines via the nuclear factor kappa-B pathway and the mitogen-activated protein kinase (MAPK) pathway, and actively participates in the process of SCI (Paterniti et al., 2014; Zhang et al., 2015).
Non-coding RNA is the general term used for RNA that does not encode proteins (Mattick, 2001). There are many kinds of non-coding RNA, including circular RNA (circRNA) and microRNA (miRNA), which have been reported as important in many pathophysiological processes (Geng et al., 2018; Slack and Chinnaiyan, 2019; Ding et al., 2020). Of these, circRNA, which is a newly identified non-coding RNA that is generated by back-splicing and has no poly-A tail or 5′ cap, forms a more stable covalent closed-loop structure compared with linear RNA (Memczak et al., 2013). Moreover, circRNAs are conserved among species and are highly enriched in the nervous system (Memczak et al., 2013). Thus, it might be more meaningful to regulate circRNAs than to regulate miRNAs in neurological diseases. In the central nervous system (CNS), He et al. (2020) reported that circMAPK4 regulates glioma cell survival and apoptosis through miR-125a-3p via the p38/MAPK signaling pathway. In addition, Wang et al. (2020) demonstrated that small interfering (si)-circHIPK2 can regulate neural stem cell differentiation, and that neural stem cells with microinjection of si-circHIPK2 can induce functional recovery following stroke, which may serve as a prospective therapeutic strategy. However, the functional role of circRNAs in SCI remains unclear.
Astrocytes constitute approximately 30% of mammalian CNS cells, and are generally recognized as either housekeeping cells or inert scaffold cells. Nevertheless, astrocytes have also been suggested to be involved in the immune response of the CNS (Rothhammer and Quintana, 2015; Guo et al., 2021). Chemokine CC motif ligand 2 (CCL2, also known as monocyte chemoattractant protein 1) is a subfamily of CC chemokines. It has been identified as a key medium in both internal and external inflammatory responses of the CNS (Ambrosini and Aloisi, 2004; Yadav et al., 2010). It is generally believed that CCL2 is produced mainly by cultured astrocytes (Park et al., 2018). Furthermore, studies have shown that CCL2 produced by astrocytes in chronic experimental autoimmune encephalomyelitis can recruit and activate microglia (Kim et al., 2014; Mayo et al., 2014).
The aim of the current study was to identify any differentially expressed circRNAs after SCI. We then aimed to elucidate the role of circPrkcsh, which was a circRNA with significantly different levels after SCI, by treating astrocytes with TNF-α in vitro. The relationship between circPrkcsh, miR-488, and CCL2 was also investigated.
Materials and Methods
Animals and surgery
One hundred and thirty male C57BL/6 mice (8 weeks old, 18–22 g weight) were purchased from the Jinan Pengyue Laboratory Animal Breeding Company (license No. SCXK (Lu) 2019 0003). The animals were raised in a clean-grade animal room at Shandong University, with free access to food and water, under a 12-hour light/dark cycle at 23 ± 1°C. To eliminate the influence of estrogen and any other sex-related physiological differences, male mice only were used. Relevant procedures were approved by the Animal Ethics Committee of Shandong University (Jinan, China) (approval No. KYLL-20170303) on March 3, 2017.
Mice were anesthetized by intraperitoneal injection of 3% pentobarbital (30 mg/kg; Sigma-Aldrich, Merck KGaA, Darmstadt, Germany). A laminectomy was conducted at T8–10 to expose the thoracic spinal cord. For the circRNA sequencing experiments, 60 mice were randomly allocated to either the SCI or sham group (30 mice per group). In addition, for the miRNA sequencing experiments, 60 mice were randomly allocated to the SCI and sham groups (30 mice per group). Furthermore, 10 mice were used for hematoxylin and eosin (HE) staining. All mice in the SCI group then underwent SCI at T9 using the modified Allen’s weight-drop apparatus (8 g × 50 mm; Shinva, Zibo, China) (Wang et al., 2018). Mice in the sham group underwent the laminectomy procedure but did not undergo SCI. Manual bladder expression was conducted twice per day until the recovery of spontaneous voiding. Post-surgery, all mice were sacrificed in accordance with the different experiments.
Tissue preparation
Mice were anesthetized by intraperitoneal injection of 3% pentobarbital (30 mg/kg; Sigma-Aldrich) at 3 days post-surgery. After transcardial perfusion with 4% paraformaldehyde, spinal cord samples from 5 mm above and below the injured lesion were carefully removed. The samples for RNA sequencing and polymerase chain reaction (PCR) were rapidly transferred into cryotubes for storage in liquid nitrogen. Fresh spinal cord tissue was removed and placed directly in 4% paraformaldehyde overnight for fixation, dehydrated through graded ethanol solutions and xylene, and paraffin embedded. Serial 5-μm sections were then prepared from the paraffin blocks using a microtome (Leica (China), Shanghai, China).
HE staining
A series of spinal cord sections were immersed in xylene, rehydrated through decreasing concentrations of ethanol solutions, rinsed with distilled water, and then stained with hematoxylin for 6 minutes. After a 30-second wash with distilled water, the sections were stained with eosin for 1 minute, followed by dehydration through increasing concentrations of ethanol solutions and xylene. The images were then observed and captured using an Eclipse Ni-U Upright Microscope (Nikon, Shanghai, China).
circRNA library preparation and sequencing
The total RNA was extracted from spinal cord tissue using TRIzol (Invitrogen, Carlsbad, CA, USA). The RNA integrity was examined using a LabChip Kit (Agilent, Santa Clara, CA, USA). Ribosomal RNA was depleted with a Ribo-Zero™-rRNA Removal Kit (Illumina Inc., San Diego, CA, USA) in accordance with the manufacturer’s protocol. Linear RNA in the remaining RNA were removed by RNase R treatment (Epicentre, Madison, WI, USA). Finally, paired-end sequencing was conducted using an Illumina HiSeq 4000 system according to the vendor’s recommended instructions (GeneChem, Shanghai, China).
circRNA sequencing data analysis
First, any reads containing undetermined bases, low quality bases, or adaptor contamination were removed using Cutadapt (version 1.9, https://cutadapt.readthedocs.io/en/stable/) (Kechin et al., 2017). After processing the aligned reads using in-house scripts, the fragments per kilobase of exon per million mapped fragments model was used to assess relative abundance. CIRCexplorer (v2-2.2.6, http://yanglab.github.io/CIRCexplorer/) was then used to quantify circRNA expression.
Small RNA library preparation and sequencing
Total RNA from the spinal cord tissue was extracted using TRIzol. The RNA integrity was examined using the LabChip Kit. Next, a small RNA library was constructed with a TruSeq Small RNA Sample Prep Kit (Illumina Inc.) using 1 μg of total RNA from each individual sample, in accordance with the manufacturer’s instructions. Finally, single-end sequencing was conducted based on the Illumina HiSeq 2500 system according to the vendor’s recommended instructions (GeneChem).
miRNA sequencing data analysis
Raw sequence reads containing junk and low complexity sequences were removed using an in-house program. Deep-sequencing counts were then used to assess the differential expression of miRNAs using DESeq (version 1.12.0, Bioconductor, Boston, MA, USA). The computational target prediction software packages miRanda (http://www.microrna.org/microrna/getDownloads.do) and TargetScan (http://www.targetscan.org/mmu_72/) were used to identify miRNA binding sites and predict the gene targets of the miRNAs with the highest abundance. The data from both software were combined and the overlaps were calculated.
Gene Ontology analysis
Gene Ontology (GO) analysis was performed for the differentially expressed circRNA. GO analysis includes biological processes, cellular components, and molecular functions (http://www.geneontology.org) (Gene Ontology Consortium, 2015).
Sanger sequencing
The total RNA was treated with 3 U/μg RNase R for 20 minutes at 37°C and purified to obtain the circRNA. To validate the junction sequences of circRNAs, Sanger sequencing was then performed according to the manufacturer’s instructions, provided by Thermo Fisher Scientific (Shanghai, China).
Cell culture and transfection of miR-488 mimics and inhibitor
Primary mouse astrocytes (M1800-57) were purchased from ScienCell Research Laboratories (San Diego, CA, USA). Cells were maintained in Dulbecco’s Modified Eagle’s Medium (Gibco, Shanghai, China) with 100 IU/mL penicillin–streptomycin (Solarbio, Beijing, China) plus 10% fetal bovine serum (Gibco, Brisbane, Australia) supplementation, at 37°C under a humidified atmosphere with 5% CO2. Astrocytes were then seeded onto six-well plates. When the cells reached approximately 80% confluency, they received a 6-hour siRNA or plasmid transfection. To regulate the expression of miR-488, cells were transfected with mimics and an inhibitor (RiboBio Co., Guangzhou, China), as well as with a negative control. Cells were treated with Opti-MEM (Gibco, Shanghai, China) and Lipo2000 (Invitrogen, Shanghai, China) for 6 hours in accordance with the manufacturer’s instructions. The LV-mmu-miR-488-sponge (miR-488 sponge lentivirus; miR-488 KD) and its negative control (miR-488 sponge lentivirus negative control; KD NC), as well as the LV-mmu-miR-488 (miR-488 overexpression lentivirus; miR-488 OE) and its negative control (miR-488 overexpression lentivirus negative control; OE NC), were synthesized by Shanghai OBiO (Shanghai, China). After preliminary experiments, the transfection was conducted by adding lentivirus at a multiplicity of infection = 20 combined with polybrene (50 μg/mL, OBiO).
Fluorescence in situ hybridization assay
The fluorescence in situ hybridization (FISH) assay was conducted using primary mouse astrocytes. The circPrkcsh probes were synthesized by RiboBio Co., and the probe sequences are listed in Additional Table 1. A FISH Kit (RiboBio Co.) was used in accordance with the manufacturer’s instructions. 4′,6-Diamidino-2-phenylindole (blue) was used to label nuclei, while Cy-3 (red) was used to label circPrkcsh probes. Fluorescence detection was conducted using an Opera Phenix HCS system (PerkinElmer, Waltham, MA, USA) at Shandong University.
Additional Table 1.
The sequences of primers, plasmids and probes
| Sequence (5’-3’) | |
|---|---|
| Primers | |
| circPrkcs | Forward: CAG GTG AAC GAC GAC TAC T |
| h | |
| Reverse: TGT TAT GCG GCA GGA AGA | |
| miR-488 | Forward: TTG AAA GGC TGT TTC TTG GTC |
| Reverse: Universal Reverse Primer (Takara) | |
| Prkcsh | Forward: CTG GAC ACA GAT GGA GAT GGA |
| Reverse: GCC CAG ACA CGG TCA TAG AAG | |
| Ccl2 | Forward: AGC AGC AGG TGT CCC AAA GA |
| Reverse: GTG CTG AAG ACC TTA GGG CAG A | |
| Il-1β | Forward: TCC AGG ATG AGG ACA TGA GCA C |
| Reverse: GAA CGT CAC ACA CCA GCA GGT TA | |
| Il-10 | Forward: GCC AGA GCC ACA TGC TCC TA |
| Reverse: GAT AAG GCT TGG CAA CCC AAG TAA | |
| CD86 | Forward: ATA TGA CCG TTG TGT GTG TTC TGG A |
| Reverse: AGG GCC ACA GTA ACT GAA GCT GTA A | |
| Mrc-1 | Forward: AGA GCT GGC GAG CAT CAA GAG |
| Reverse: TTC CAT AGG TCA GTC CCA ACC AA | |
| Gapdh | Forward: AAA TGG TGA AGG TCG GTG TGA AC |
| Reverse: CAA CAA TCT CCA CTT TGC CAC TG | |
| U6 | Forward: GGA ACG ATA CAG AGA AGA TTA GC |
| Reverse:TGG AAC GCT TCA CGA ATT TGC G | |
| circRNA2 | Forward: ACC AGA GAT GGA CAG GAG AT |
| 947 | |
| Reverse: GAG AGG TCA GCA AAC TTG GA | |
| circRNA4 | Forward: CTC CCG GAG CAA TAA GCG AG |
| 72 | |
| Reverse: ATG GTC ATG GGC ACT GTC AAA | |
| circRNA7 | Forward: TGG TGG CTT CTC TAT CCT GC |
| 09 | |
| Reverse: TTT TGT CAT TGT CAC CGG AAG | |
| circPrkcsh | TTC AGA TGA GCC TGT GAG A |
| -si1 | |
| circPrkcsh | CAG ATG AGC CTG TGA GAG G |
| -si2 | |
| circPrkcsh | TGA GCC TGT GAG AGG CCA T |
| -si3 | |
| Plasmid | TGAGAGGCCATCGCGTATCTTCCTGCCGCATAACACACTTCCGGTCTCGGCCCGGACGG |
| of | GATGCTGCTGCTGCTGCTACTACTACTACCCCTCTGTTGGGCTGTAGAAGTTAAGAGACC |
| circPrkcsh | CCGGGGCGTTTCCCTCAGCAACCATCACTTCTATGAAGAATCTAAACCTTTCACCTGTTT |
| (pLenti-E | GGACGGCACAGCCACCATCCCATTCGATCAGGTGAACGACGACTACTGCGACTGTAAGG |
| F1a-EGFP | ACGGTTCAGATGAGCCTG |
| -F2A-Pur | |
| o-CMV-ci | |
| rcular | |
| sequence- | |
| circPrkcsh | |
| -circular) | |
| Overexpre | TGTTTGAATGAGGCTTCAGTACTTTACAGAATCGTTGCCTGCACATCTTGGAAACACTTG |
| ssion | CTGGGATTACTTCTTCAGGTTAACCCAACAGAAGGCTCGAGAAGGTATATTGCTGTTGA |
| vector for | CAGTGAGCAACTTGAAAGGCTGTTTCTTGGTCCTGTGAAGCCACAGATGAGGACCAAGA |
| miR-488 | AAGCCTATCATGTCGCCTACTGCCTCGGAATTCAAGGGGCTACTTTAGGAGCAATTATCT |
| (pRLenti- | TGTTTACTAAAACTGAATACCTTGCTATCTCTTTGATACATTTTTACAAAGCTGAATTAA |
| EF1a-EG | AATGGTATAAATTAAATCACT |
| FP-F2A-P | |
| uro-CMV- | |
| mmu-miR | |
| -488 | |
| precursor) | |
| Sponge | CGGGGAATTATTTTGACTGTAAACACAAAGATATTAGTACAAAATACGTGACGTAGAAA |
| vector for | GTAATAATTTCTTGGGTAGTTTGCAGTTTTAAAATTATGTTTTAAAATGGACTATCATAT |
| miR-488 | GCTTACCGTAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACG |
| (pLKD-C | AAACACCGGGACCAAGAAACAGCCTTTCAATTTTTTGAATTCGGATCCATTAGGCGGCC |
| MV-EGF | GCGTGGATAACCGTATTACCGCCATGCATTAGTTATTAATAGTAATCAATTACGGGGTC |
| P-2A-Pur | ATTAGTTCATAGCCCATATATGGAGTTCCGCGTTACATAACTTACGGTAAATGGCCCGCC |
| o-U6-shR | TGGCTGACCGCCCAACGACCCCCGCCCATTGACGTCAATAATGACGTATGTTCCCATAG |
| NA | TAACGCCAATAGGGACTTTCCATTGACGTCAATGGGTGGAGTATTTACGGTAAACTGCC |
| (mmu-mi | CACTTGGCAGTACATCAAGTGTATCATATGCCAAGTACGCCCCCTATTGACGTCAATGA |
| R-488 | CGGTAAATGGCCCGCCTGGCATTATGCCCAGTACATGACCTTATGGGACTTTCCTACTTG |
| sponge)) | GCAGTACATCTACGTATTAGTCATCGCTATTACCATGGTGATGCGGTTTTGGCAGTACAT CAATGGGCGTGGATAGCGGTTTGACTCACGGGGATTTCCAAGTCTCCACCCCATTGACG |
| TCAATGGGAGTTTGTTTTGGCACCAAAATCAACGGGACTTTCCAAAATGTCGTAACAAC | |
| TCCGCCCCATTGACGCAAATGGGCGGTAGGCGTGTACGGTGGGAGGTCTATATAAGCAG | |
| AGCTGGTTTAGTGAACCGTCAGATCCGCTAGCGCTACCGGACGCCACCATGGATGAGCA | |
| AGGGCGAGGAGCTGATTCCACCGGGGGTGGTGCCCAATCCTGGTCGAGCTGGACTGCGT | |
| ACGTAAACCGTCACAGTCAGCGTGTCAGCGAGGTCGAGGAGATGCCCACCTTACGGCTA | |
| GCC | |
| RNA | |
| pull-down | |
| circPrk | TGAGAGGCCATCGCGTATCTTCCTGCCGCATAACACACTTCCGGTCTCGGCCCGGACGG |
| csh-WT | GATGCTGCTGCTGCTGCTACTACTACTACCCCTCTGTTGGGCTGTAGAAGTTAAGAGACC |
| vector: | CCGGGGCGTTTCCCTCAGCAACCATCACTTCTATGAAGAATCTAAACCTTTCACCTGTTT |
| pcDNA3. | GGACGGCACAGCCACCATCCCATTCGATCAGGTGAACGACGACTACTGCGACTGTAAGG |
| 1(+) | ACGGTTCAGATGAGCCTG |
| circPrk | GTCCGAGTAGACTTGGCAGGAATGTCAGCGTCATCAGCAGCAAGTGGACTAGCTTACCC |
| csh-Mut- | TACCACCGACACGGCAGGTTTGTCCGTCCCTTAAATCTGGAGGGTATCTTCACTACCAAC |
| AS | GACTCCCTTTGCGGGGCCCCAGAGAATTGAAGATGTCGGGTTGTCTCCCCATCATCATC |
| vector: | ATCGTCGTCGTCGTCGTAGGGCAGGCCCGGCTCTGGCCTTCACACAATACGCCGTCCTTC |
| pcDNA3. | TATGCGCTACCGGAGAGT |
| 1(+) | |
Ccl2: Chemokine CC motif ligand 2; circRNA: circular RNA; Gapdh: glyceraldehyde-3-phosphate dehydrogenase;
Il: Interleukin; Mrc-1: monocyte chemoattractant protein 1; Prkcsh: protein kinase C substrate 80K-H.
Separation of nuclear and cytoplasmic fractions
Astrocytes were washed once with phosphate-buffered saline and collected by centrifugation at 72 × g for 3 minutes at 25°C. The PARIS™ Kit (Invitrogen, Shanghai, China) was used in accordance with the detailed protocols provided by the manufacturer. Nuclear and cytoplasmic RNA were extracted and collected separately for further reverse transcription-quantitative PCR (RT-qPCR) analysis.
RT-qPCR analysis
Total RNA from the cultured astrocytes and spinal cord tissue was extracted using TRIzol and stored at –80°C. The Bulge-Loop™ miRNA Reverse Transcription and RT-qPCR Starter Kits (RiboBio Co.) were then used to quantify miR-488 expression levels, using U6 as the endogenous control for normalization. To investigate messenger RNA (mRNA) expression, 1000 ng total RNA was used for reverse transcription, using the Evo M-MLV RT Kit with gDNA Clean for qPCR II (Cat# AG11711; Accurate Biology, Changsha, China). Next, RT-qPCR was performed with the SYBR® Green Premix Pro Taq HS qPCR Kit (Cat# AG11701; Accurate Biology) using the Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific, Wilmington, DE, USA). Amplifications were set up with 20 μL reaction volume containing 1.6 μL of primer (BioSune, Shanghai, China) and 2 μL of cDNA. The PCR conditions were: 30-second denaturation at 95°C, followed by 40 cycles of 5 seconds at 95°C and 31 seconds at 60°C, with a subsequent melt curve analysis. For the circRNA, the total RNA was incubated for 20 minutes in the presence or absence of 3 U/μg RNase R at 37°C, and was then purified with the total RNA isolation miRNeasy Mini Kit (Qiagen, Düesseldorf, Germany). The expression levels of mRNA or circRNA were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. Comparative quantitation was then performed using the 2–ΔΔCT method (Livak and Schmittgen, 2001). Each PCR reaction was conducted in triplicate. All primer sequences are provided in Additional Table 1.
Enzyme-linked immunosorbent assay
CCL2, interleukin (IL)-1β, and IL-10 concentrations in TNF-α-treated astrocytes were quantified using the corresponding CCL2, IL-1β, and IL-10 enzyme-linked immunosorbent assay (ELISA) kits (Invitrogen, Shanghai, China), following the manufacturer’s protocols. The colorimetric optical density was measured using a microplate reader (Multiskan Go, Shanghai, China).
Dual-luciferase reporter assay
HEK293T cells (Procell, Wuhan, China) were maintained in Dulbecco’s Modified Eagle Medium containing 100 IU/mL penicillin-streptomycin plus 10% fetal bovine serum supplementation. Twenty-four hours before transfection, the HEK293T cells were plated at 1 × 105 per well in 24-well plates. For circPrkcsh and miR-488, either wild-type (WT) or mutated (MUT) circPrkcsh fragments were inserted into a pGL3-firefly_luciferase vector (BioSune, Jinan, China). For Ccl2 and miR-488, either the WT or MUT Ccl2 fragment was inserted into a pGL3-firefly_luciferase vector (BioSune, Jinan, China). Next, the miR-488 mimic, plasmid, or negative control was transfected. The firefly and Renilla luciferase activities were detected after 48 hours of incubation using the dual-luciferase system (Promega, Madison, WI, USA). The cells were lysed in 1× passive lysis buffer on a gentle rocking platform at 25°C for 10 minutes before the cell lysates were collected. In a dedicated 96-well plate, 100 μL of LARII and 20 μL of cell lysate was added. A multi-function microplate reader (Berthold, Nanjing, China) was set to measure the firefly activity, while the Renilla luciferase activity was measured with the addition of 100 μL Stop & Glo® Reagent (Promega). The different luciferase activities between Renilla and firefly were calculated to determine the relative luciferase activity.
RNA pull-down assay
First, pcDNA3.1(+)-circPrkcsh, pcDNA3.1(+)-antisense-circPrkcsh, and pcDNA3.1(+) (Additional Table 1) were linearized with appropriate restriction enzymes for the template DNA preparation for transcription in vitro. The MEGAscript T7 Kit with biotin 16 UTP (Ambion, Shanghai, China) was used to produce biotin-labeled RNA transcripts. These products were then purified using MEGAclear Kits (Ambion) in vitro. Next, 3 µg biotinylated RNA was heated for 5 minutes at 90°C, and was then incubated at room temperature for 30 minutes. The RNA was then cooled to 4°C, mixed with 1 mg protein extracted from the astrocytes, and incubated for 2 hours at room temperature. Subsequently, 60 μL of streptavidin agarose beads (Invitrogen, Shanghai, China) was added to each individual binding reaction, incubated for 2 hours at room temperature, and washed five times to remove the beads. The pull-down RNA was then identified by RT-qPCR analysis.
RNA immunoprecipitation assay
The RNA immunoprecipitation (RIP) assay was conducted on astrocytes using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA). Approximately 1 × 107 cells were sedimented and then resuspended in RIP Lysis Buffer with an equal pellet volume containing protease and RNase inhibitors. Beads coated with Argonaute-2 (AGO2) antibody (5 μL; Millipore, Billerica, MA, USA) or rabbit IgG control were mixed into the cell lysates by overnight rotation at 4°C. Following treatment with proteinase K, the immunoprecipitated RNA was extracted using an RNeasy MinElute Cleanup Kit (Qiagen). The detection of RNA was conducted by RT qPCR analysis.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (version 8.0.1, GraphPad Software, La Jolla, CA, USA) and SPSS (v22.0; IBM, Chicago, IL, USA) software. Differences between two groups were analyzed using the Student’s t-test. Differences among more than two groups were analyzed using one-way analysis of variance followed by Bonferroni post hoc tests. Data are expressed as the mean ± standard deviation (SD). Statistically significant differences were determined if P < 0.05.
Results
circRNA and miRNA expression patterns in mouse injured and normal spinal cords
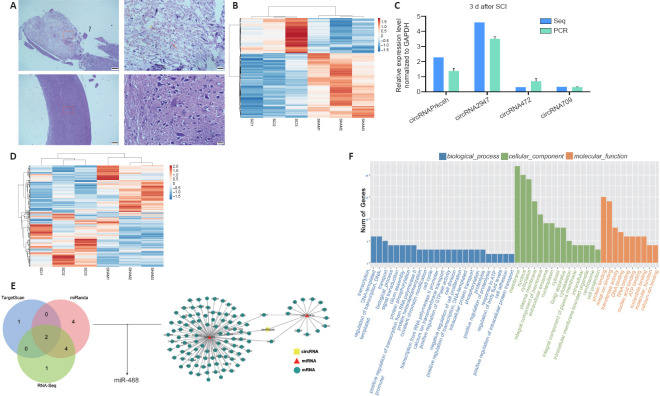
To generate circRNA and miRNA databases, RNA sequencing analyses of the total RNA from the SCI and sham groups were performed. When the spinal cord tissue of C57 mice was sectioned and HE staining was performed, it was revealed that the injury severely damaged the structural integrity of affected tissue and the blood-spinal cord barrier, inducing inflammatory cell infiltration, hemorrhage, and rupture (Figure 1A). An Illumina HiSeq 4000 system was used to sequence the samples, yielding at least 30 million reads. In addition, an Illumina HiSeq 2500 system was used to perform miRNA single-end sequencing. Using CIRCexplorer, we identified a total of 16,013 circRNAs, and the majority of the identified circRNAs in the study—8248 of 16 013 circRNAs (51.5%)—were supported by more than 10 reads. Moreover, 165 circRNAs were upregulated and 167 were downregulated (Figure 1B). To validate the RNA deep-sequencing results, RNA was extracted from tissue and four circRNAs were randomly selected to determine the differentially expressed circRNAs using RT-qPCR analysis (Figure 1C). For miRNA, 960 were identified, of which 241 were upregulated and 212 were downregulated (Figure 1D). According to our sequencing data, the expression of circPrkcsh and Ccl2 were significantly increased after SCI, while the expression of miR-488 was decreased. The circRNA and miRNA sequences were then analyzed using a combination of TargetScan and miRanda databases and sequencing data, and the possible targeted binding of circPrkcsh and miR-488 was identified (Figure 1E). The binding score for circPrkcsh and miR-488 was 97, which was the highest among the interactions between circRNA and miRNA. Based on these findings, the regulatory axis of circPrkcsh/miR-488/Ccl2 was chosen for further research. Furthermore, the GO analysis results are shown in Figure 1F.
Figure 1.
Differentially expressed circRNA and miRNA profiles between the SCI and sham groups.
(A) Hematoxylin and eosin staining on spinal cord tissue from the SCI and sham groups at 3 days post-surgery. Compared with the sham group, the SCI group showed hemorrhage (red arrow), infiltration of a large number of inflammatory cells (black arrow), and physalides (yellow arrow). The top row is the SCI group and the bottom row is the sham group. Scale bars: 200 µm (left) and 20 µm (right). (B) Heat map of differentially expressed circRNA in the SCI group compared with the sham group. (C) Four circRNAs were randomly selected to validate the results of sequencing through reverse transcription-quantitative polymerase chain reaction. Data are expressed as the mean ± SD based on triplicate independent experiments, and were analyzed using the Student’s t-test. (D) Heat map of differentially expressed miRNA in the SCI group compared with the sham group. (E) Targeting prediction performed by TargetScan and miRanda as well as sequencing results combined with the network analysis of the interactions among circRNA, miRNA, and mRNA. circPrkcsh was predicted to bind to miR-488. (F) Gene Ontology analysis of the differentially expressed circRNA. circRNA: Circular RNA; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; miRNA: microRNA; SCI: spinal cord injury.
circPrkcsh is different from Prkcsh mRNA
Three days after SCI, total RNA from the injured spinal cord tissue was extracted, and we observed the differential expression levels of circPrkcsh and Prkcsh (Figure 2A). To verify that circPrkcsh is indeed circular, we performed PCR (Figure 2B) and revealed that RNase R treatment significantly decreased the linear mRNA levels of Prkcsh, while the expression levels of circPrkcsh remained relatively unchanged. We then performed Sanger sequencing to confirm the head-to-tail splicing of the circPrkcsh PCR product by its predicted size (Figure 2C). Furthermore, PCR (Figure 2D) and FISH (Figure 2E) analyses revealed a high expression of cytoplasmic circPrkcsh.
Figure 2.
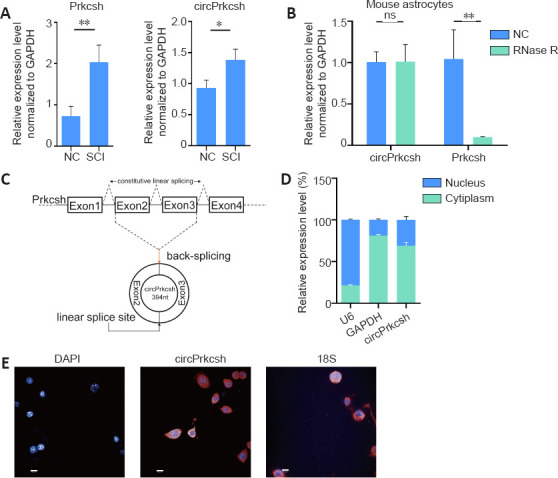
Circular RNA verification of circPrkcsh.
(A) The levels of Prkcsh and circPrkcsh expression were significantly different between tissue from the SCI and sham groups at 3 days post-surgery, based on RT-qPCR analysis. (B) The mRNA levels of circPrkcsh and Prkcsh in astrocytes with or without RNase R treatment were detected using RT-qPCR, and were then normalized to Gapdh. (C) Schematic illustration showing that circPrkcsh is formed by Prkcsh exon 2–3 circularization (black arrows). RT-qPCR was performed to confirm the presence of circPrkcsh, followed by Sanger sequencing. Head-to-tail circPrkcsh splicing sites are marked with a red arrow. (D) The circPrkcsh expression in different cell fractions was detected using RT-qPCR. U6 was used as the nuclear RNA internal control, and GAPDH was used as the cytoplasmic RNA control. Data are expressed as the mean ± SD based on triplicate independent experiments. *P < 0.05, **P < 0.01 (Student’s t-test). (E) RNA FISH revealed that circPrkcsh was predominantly located in the cytoplasm. DAPI was used to label the nuclei, while Cy-3 was used to label the circPrkcsh probes. The cellular localization of circPrkcsh was indicated using FISH with junction-specific probes. Scale bars: 50 µm. DAPI: 4′,6-Diamidino-2-phenylindole; FISH: fluorescence in situ hybridization; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; NC: normal control; ns: not significant; Prkcsh: protein kinase C substrate 80K-H; RT-qPCR: reverse transcription-quantitative polymerase chain reaction; SCI: spinal cord injury.
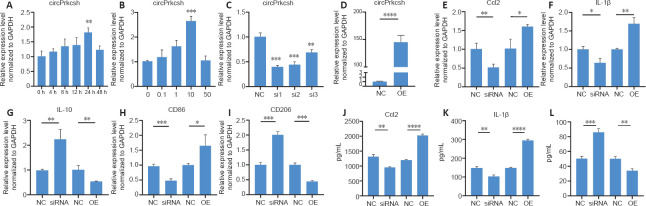
circPrkcsh reduces inflammatory cytokines secreted by TNF-α-treated astrocytes
To simulate the inflammatory microenvironment after SCI in vitro, we treated primary mouse astrocytes for 24 hours with 10 ng/mL TNF-α (Figure 3A and B). To investigate the role of circPrkcsh in inflammation after SCI, either siRNA or plasmids were transfected into mouse primary astrocytes to upregulate or downregulate circPrkcsh (Figure 3C and D). When circPrkcsh was upregulated, we observed that CCL2, IL-1β, and CD86 were increased compared with the NC group, while IL-10 and CD206 were decreased. In contrast, when circPrkcsh was downregulated, the mRNA levels of these inflammatory-related cytokines compared with the NC group showed an opposite trend (Figure 3E–I). Furthermore, ELISA was conducted to analyze the expression of the pro-inflammatory cytokines CCL2 and IL-1β (Figure 3J and K) and the anti-inflammatory cytokine IL-10 (Figure 3L) in the supernatant of each group after siRNA or plasmid transfection. The expression of CCL2 and IL-1β was increased in the OE group compared with the NC group, while it was decreased in the knockdown group compared with the NC group. The trends in IL-10 expression were the opposite of those in CCL2 and IL-1β expression.
Figure 3.
circPrkcsh expression influences the TNF-α-induced expression of inflammatory-related cytokines in astrocytes.
(A, B) Expression changes of circPrkcsh over time or with the concentration gradient of TNF-α in treated astrocytes were detected using RT-qPCR. (C, D) circPrkcsh expression in astrocytes with siRNA or circPrkcsh-overexpression plasmid transfection, as well as with the negative control (NC). (E–I) Expression of Ccl2, IL-1β, CD86, IL-10, and CD206 in astrocytes transfected with siRNA or plasmid, as detected by RT-qPCR. (J–L) Secretion of CCL2, IL-1β, and IL-10 from astrocytes treated with siRNA or plasmid, as measured using ELISA. In E–L, the first NC group is compared with the siRNA group, and was treated with siRNA-NC. The second NC group is compared with the OE group, with plasmid transfection, and was treated with plasmid-NC. Data (expressed as the fold change compared with the NC group) are expressed as the mean ± SD based on triplicate independent experiments (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Student’s t-test). Ccl2: Chemokine CC motif ligand 2; ELISA: enzyme-linked immunosorbent assay; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; IL: interleukin; OE: overexpression; RT-qPCR: reverse transcription-quantitative polymerase chain reaction; siRNA: small interfering RNA; TNF-α: tumor necrosis factor-α.
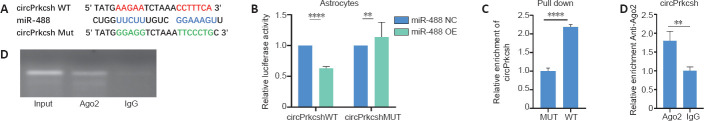
circPrkcsh binds to miR-488 as a miRNA sponge
In view of the abundant and stable content of circPrkcsh in the cytoplasm, we tested the binding ability of circPrkcsh to miRNA. As mentioned earlier, through sequence analysis, the expression of miR-488 was low after SCI, and it may have targeted binding to circPrkcsh (Figure 4A). We therefore selected miR-488 for further analysis. The binding between circPrkcsh and miR-488 was confirmed through the dual-luciferase reporter assay (Figure 4B). Because miRNAs take effect via the recruitment of the RNA-induced silencing complex (RISC), the AGO2 protein (serving as the heart of the RISC) may be combined with circPrkcsh (Bartel, 2018). Indeed, the binding of circPrkcsh to AGO2 proteins was further verified by RIP and RNA pull-down analyses (Figure 4C and D).
Figure 4.
Verification of specific binding between circPrkcsh and miR-488 in astrocytes treated with TNF-α.
(A) Prediction of putative binding sites between circPrkcsh and miR-488. (B) Relative luciferase activity, which was normalized to the NC. HEK293T cells were co-transfected with miR-488 mimics, pGL3-firefly_luciferase vector containing circPrkcsh WT/MUT fragments, or the NC. A dual-luciferase reporter assay was conducted to identify the binding sites between circPrkcsh and miR-488. (C) An RNA pull-down assay was conducted using lysates prepared from astrocytes, followed by reverse transcription-quantitative polymerase chain reaction analysis. The input levels were used to normalize the relative levels of circPrkcsh. (D) In astrocytes stably expressing AGO2, an AGO2 RIP assay was conducted to detect circPrkcsh levels (which were normalized to IgG). Data are expressed as the mean ± SD based on triplicate independent experiments. **P < 0.01, ****P < 0.0001 (Student’s t-test). AGO2: Argonaute-2; circPrkcsh: circular RNA protein kinase C substrate 80K-H; MUT: mutation; NC: negative control; RIP: RNA immunoprecipitation; WT: wild type.
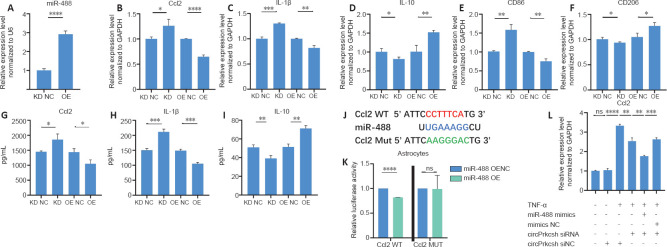
miR-488 regulates the secretion of inflammatory cytokines
To confirm that knocking down circPrkcsh induces the secretion of inflammatory cytokines in astrocytes after TNF-α treatment via the upregulation of miR-488, we transfected lentiviruses into astrocytes to overexpress or sponge miR-488 (Figure 5A). After TNF-α treatment, PCR revealed the decreased expression of CCL2, IL-1β, and CD86 and the increased expression of IL-10 and CD206 in cells overexpressing miR-488; in contrast, transfection with the miR-488 sponge lentivirus had the opposite effect (Figure 5B–F). ELISA was then performed to quantify the release of CCL2, IL-1β, and IL-10 after upregulating or downregulating miR-488 (Figure 5G–I). The binding sites between CCL2 and miR-488 were identified (Figure 5J), and these were further validated using the dual-luciferase reporter assay (Figure 5K). To confirm the functional role of circPrkcsh in regulating inflammatory cytokine secretion through miR-488/Ccl2, circPrkcsh siRNA was transfected into cells overexpressing miR-488. The expression of CCL2 was lower in the group with combined circPrkcsh siRNA and miR-488 mimics compared with the group with only knockdown of circPrkcsh (Figure 5L).
Figure 5.
miR-488 regulates inflammation via Ccl2 in astrocytes treated with TNF-α.
(A) miR-488 expression of astrocytes transfected with LV-mmu-miR-488 (miR-488 overexpression lentivirus; miR-488 OE) and its negative control (miR-488 overexpression lentivirus negative control; OE NC). (B–F) Expression of Ccl2, IL-1β, CD86, IL-10, and CD206 (which was normalized to the negative control [NC]) in astrocytes transfected with LV-mmu-miR-488-sponge (miR-488 sponge lentivirus; miR-488 KD) and its negative control (miR-488 sponge lentivirus negative control; KD NC), or miR-488 OE and OE NC, as detected by reverse transcription-quantitative polymerase chain reaction. (G–I) Quantification of CCL2, IL-1β, and IL-10 levels obtained from astrocytes after miR-488 LV transfection, as detected by enzyme-linked immunosorbent assay. (J) Prediction of putative binding sites between miR-488 and CCL2. (K) HEK293T cells were co-transfected with miR-488 mimics, pGL3-firefly_luciferase vector containing Ccl2 WT/MUT fragments, or the negative control. A dual-luciferase reporter assay was conducted to identify the binding sites between Ccl2 and miR-488. Data were normalized to the NC group. (L) miR-488 overexpression reversed circPrkcsh-induced inflammation in astrocytes. Data are expressed as the mean ± SD based on triplicate independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Student’s t-test). Ccl2: Chemokine CC motif ligand 2; circPrkcsh: circular RNA protein kinase C substrate 80K-H; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; IL: interleukin; LV: lentivirus; MUT: mutation; NC: normal control; ns: not significant; OE: overexpression; TNF-α: tumor necrosis factor-α; WT: wild type.
Discussion
There is currently no effective clinical therapy for SCI (O’Shea et al., 2017). To develop effective, feasible treatments to improve SCI, it is important to investigate the pathophysiological processes following SCI, including the promotion of recovery from neuronal apoptosis and limiting the inflammatory response (Vismara et al., 2017). circRNAs are endogenous non-coding RNAs that are conserved in mammals and exhibit a wide range of tissue specificity (Kristensen et al., 2019). There have been reports of associations between circRNAs and disease progression (Adams et al., 2017; Mehta et al., 2020). For example, in diabetic retinopathy, vascular dysfunction and endothelial proliferation can both be increased via the blocking of miR-30a function by circHIPK3 (Shan et al., 2017). Nevertheless, the function of circRNAs in SCI remains unknown. In the present research, we revealed that circPrkcsh was significantly upregulated after SCI, and that downregulating its expression in vitro significantly alleviated the inflammatory response.
circPrkcsh is produced by exons 2–3 back-splicing in the protein kinase C substrate 80K-H gene (Prkcsh, encoding the protein hepatocystin). Prkcsh functions as the non-catalytic β subunit of glucosidase II that normally resides in the endoplasmic reticulum lumen (Hu et al., 2009). It has also been reported that PRKCSH contributes to tumorigenesis by alternatively stimulating inositol-requiring enzyme 1 signaling (Shin et al., 2019). In addition, autosomal dominant polycystic liver disease may be related to the genetic loss of PRKCSH (Drenth et al., 2003; Li et al., 2003; Rauscher et al., 2018). In the current study, Prkcsh was similarly upregulated after SCI, but the expression of Prkcsh and circPrkcsh were different, and a change in the expression of circPrkcsh had no influence on the expression of Prkcsh. These results indicate that circPrkcsh has an independent role in inflammation following SCI. When it is taken into consideration that mRNA does not have the same function as circRNA, it is clear that Prkcsh cannot play the same regulatory role as circPrkcsh.
A growing number of reports have demonstrated that, during the pathophysiological process of many diseases (including SCI), competing endogenous RNA mechanisms play key roles (Yu et al., 2015; Xu et al., 2020; Xian et al., 2021). However, many studies are still in the stage of exploring the differential expression profiles of circRNA after SCI (Qin et al., 2018; Zhou et al., 2019). In the present study, we revealed that circPrkcsh contains an miR-488 binding site, and that CCL2, the only inflammatory cytokine that shows a binding target of miR-488, can be positively regulated by circPrkcsh. We therefore constructed a regulatory axis of circPrkcsh as an miR-488 sponge, regulating the inflammatory response in astrocytes after TNF-α treatment in vitro by altering the expression of CCL2. In view of the enrichment of circRNA in the CNS and the conservation of species, the regulation of circRNA may be more stable and reliable compared with that of miRNA.
Many studies have demonstrated that a range of differentially expressed miRNAs participate in the regulation of pathophysiological changes after SCI (Adams et al., 2017). For example, miR-21 regulates astrocyte proliferation and significantly promotes astrocyte apoptosis post-SCI through the transforming growth factor-β-mediated phosphoinositide 3-kinase/protein kinase B/mechanistic target of rapamycin pathway, thus promoting the recovery from SCI both in vitro and in vivo (Liu et al., 2018). Furthermore, miR-488 also participates in esophageal squamous cell carcinoma (Yang et al., 2019), non-small-cell lung carcinoma (Fang et al., 2017; Wang et al., 2019a), and colorectal cancer progression (Wang et al., 2019b). Additionally, miR-488-3p is sponged by circRNA cir-CCDC66 to modulate doublecortin expression in Hirschsprung’s disease (Wen et al., 2019). Nevertheless, the role of miR-488 in the inflammatory response is not completely understood. For the first time, our study demonstrated that miR-488 improves the inflammatory response after TNF-α treatment by targeting Ccl2. Therefore, the circPrkcsh/miR-488/Ccl2 axis has the potential to act as a treatment target for improving the inflammatory response in astrocytes after TNF-α treatment. However, we cannot rule out the existence of other potential targets of circPrkcsh related to the regulation of the inflammatory response after SCI.
This study has several limitations. The main objective of this study focused on the molecular mechanisms of the circPrkcsh/miR-488/Ccl2 axis during SCI pathology, and most of the data came from in vitro settings. However, treating astrocytes with TNF-α in vitro does not precisely imitate inflammation after SCI. Therefore, the inflammation-regulating function of circPrkcsh after SCI will be investigated in future in vivo studies. Furthermore, a detailed in vivo assessment of circPrkcsh and miR-488 co-localization in tissue will also be investigated in future studies. Moreover, previous studies reporting the role of astrocytes in inflammation (Liddelow and Barres, 2017) have aroused our interest. As a chemokine, CCL2 is mainly produced by astrocytes, and in the present study its expression was able to be regulated by the expression of circPrkcsh. The inflammation-related cells, such as microglia, that express circPrkcsh and are regulated by this circRNA will also be detected in future studies.
In short, regulating the circPrkcsh/miR-488/Ccl2 axis has been identified as a potential therapy to promote the recovery of inflammation in astrocytes after TNF-α treatment. circPrkcsh inhibition may alleviate inflammation. Additionally, for SCI treatment, miR-488 overexpression may be developed as a novel therapeutic strategy.
Additional files:
Additional Table 1: The sequences of primers, plasmids and probes.
Additional file 1: Open peer review reports 1 (88.9KB, pdf) and 2 (87.2KB, pdf) .
Additional file 2 (581.1KB, pdf) : Original data of the experiment.
Acknowledgments:
We are grateful to Dr. Rong-Han Liu, Dr. Lin Shen and Dr. Hui-Hui Wu from Jinan Central Hospital, Cheeloo College of Medicine, Shandong University, China for their valuable discussions and technical assistance. We thank Translational Medicine Core Facility of Shandong University for consultation and instrument availability that supported this work.
Footnotes
P-Reviewers: Ordaz J, Puelles E; C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Gardner B, Yu J, Song LP; T-Editor: Jia Y
Funding:This work was supported by the National Natural Science Foundation of China, Nos. 81771346, 82071383; the Taishan Scholar Program of Shandong Province of China, No. tsqn201812156; Spring City Leader Talent Support Plan, No. 201984; and Rongxiang Regenerative Medicine Foundation of Shandong University, No. 2019SDRX-23 (all to BN).
Conflicts of interest:The authors declare that there is no conflict of interests.
Financial support:This work was supported by the National Natural Science Foundation of China, Nos. 81771346, 82071383; the Taishan Scholar Program of Shandong Province of China, No. tsqn201812156; Spring City Leader Talent Support Plan, No. 201984; and Rongxiang Regenerative Medicine Foundation of Shandong University, No. 2019SDRX-23 (all to BN). The funders had no roles in the study design, conduction of experiment, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional review board statement:The study was approved by the Research Ethics Committee of Shandong University (Jinan, China) (approval No. KYLL-20170303) on March 3, 2017.
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:All raw sequencing data has been uploaded to GEO (accession number GSE158195). In addition, the datasets used in the project are available from the corresponding author.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open peer reviewers:Josue Ordaz, Indiana University School of Medicine, USA; Eduardo Puelles, Instituto de Neurociencias de Alicante, Universidad Miguel Hernandez, Consejo Superior de Investigaciones Científicas, Spain.
References
- 1.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127:761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahuja CS, Wilson JR, Nori S, Kotter MRN, Druschel C, Curt A, Fehlings MG. Traumatic spinal cord injury. Nat Rev Dis Primers. 2017;3:17018. doi: 10.1038/nrdp.2017.18. [DOI] [PubMed] [Google Scholar]
- 3.Alilain WJ, Horn KP, Hu H, Dick TE, Silver J. Functional regeneration of respiratory pathways after spinal cord injury. Nature. 2011;475:196–200. doi: 10.1038/nature10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambrosini E, Aloisi F. Chemokines and glial cells: a complex network in the central nervous system. Neurochem Res. 2004;29:1017–1038. doi: 10.1023/b:nere.0000021246.96864.89. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. Metazoan microRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding L, Fu WJ, Di HY, Zhang XM, Lei YT, Chen KZ, Wang T, Wu HF. Expression of long non-coding RNAs in complete transection spinal cord injury: a transcriptomic analysis. Neural Regen Res. 2020;15:1560–1567. doi: 10.4103/1673-5374.274348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drenth JP, te Morsche RH, Smink R, Bonifacino JS, Jansen JB. Germline mutations in PRKCSH are associated with autosomal dominant polycystic liver disease. Nat Genet. 2003;33:345–347. doi: 10.1038/ng1104. [DOI] [PubMed] [Google Scholar]
- 8.Fang C, Chen YX, Wu NY, Yin JY, Li XP, Huang HS, Zhang W, Zhou HH, Liu ZQ. MiR-488 inhibits proliferation and cisplatin sensibility in non-small-cell lung cancer (NSCLC) cells by activating the eIF3a-mediated NER signaling pathway. Sci Rep. 2017;7:40384. doi: 10.1038/srep40384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehlings MG. London: Future Medicine Ltd; 2013. Critical care in spinal cord injury. [Google Scholar]
- 10.Gene Ontology Consortium (2015) Gene Ontology Consortium: going forward. Nucleic Acids Res. 43:D1049–1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo MF, Zhang HY, Zhang PJ, Bai ZJ, Yu JW, Wang YY, Wei WY, Song LJ, Chai Z, Yu JZ, Ma CG. Fasudil inhibits lipopolysaccharide-induced astrocytic injury by regulating Nrf2/HO-1 signaling pathway. Zhongguo Zuzhi Gongcheng Yanjiu. 2021;25:5012–5017. [Google Scholar]
- 13.He J, Huang Z, He M, Liao J, Zhang Q, Wang S, Xie L, Ouyang L, Koeffler HP, Yin D, Liu A. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol Cancer. 2020;19:17. doi: 10.1186/s12943-019-1120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu D, Kamiya Y, Totani K, Kamiya D, Kawasaki N, Yamaguchi D, Matsuo I, Matsumoto N, Ito Y, Kato K, Yamamoto K. Sugar-binding activity of the MRH domain in the ER alpha-glucosidase II beta subunit is important for efficient glucose trimming. Glycobiology. 2009;19:1127–1135. doi: 10.1093/glycob/cwp104. [DOI] [PubMed] [Google Scholar]
- 15.Kechin A, Boyarskikh U, Kel A, Filipenko M. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol. 2017;24:1138–1143. doi: 10.1089/cmb.2017.0096. [DOI] [PubMed] [Google Scholar]
- 16.Kim RY, Hoffman AS, Itoh N, Ao Y, Spence R, Sofroniew MV, Voskuhl RR. Astrocyte CCL2 sustains immune cell infiltration in chronic experimental autoimmune encephalomyelitis. J Neuroimmunol. 2014;274:53–61. doi: 10.1016/j.jneuroim.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 18.Li A, Davila S, Furu L, Qian Q, Tian X, Kamath PS, King BF, Torres VE, Somlo S. Mutations in PRKCSH cause isolated autosomal dominant polycystic liver disease. Am J Hum Genet. 2003;72:691–703. doi: 10.1086/368295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liddelow SA, Barres BA. Reactive astrocytes: production, function, and therapeutic potential. Immunity. 2017;46:957–967. doi: 10.1016/j.immuni.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Liu R, Wang W, Wang S, Xie W, Li H, Ning B. microRNA-21 regulates astrocytic reaction post-acute phase of spinal cord injury through modulating TGF-β signaling. Aging (Albany NY) 2018;10:1474–1488. doi: 10.18632/aging.101484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayo L, Trauger SA, Blain M, Nadeau M, Patel B, Alvarez JI, Mascanfroni ID, Yeste A, Kivisäkk P, Kallas K, Ellezam B, Bakshi R, Prat A, Antel JP, Weiner HL, Quintana FJ. Regulation of astrocyte activation by glycolipids drives chronic CNS inflammation. Nat Med. 2014;20:1147–1156. doi: 10.1038/nm.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehta SL, Dempsey RJ, Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol. 2020;186:101746. doi: 10.1016/j.pneurobio.2020.101746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 26.O’Shea TM, Burda JE, Sofroniew MV. Cell biology of spinal cord injury and repair. J Clin Invest. 2017;127:3259–3270. doi: 10.1172/JCI90608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh JY, Hwang TY, Jang JH, Park JY, Ryu Y, Lee H, Park HJ. Muscovite nanoparticles mitigate neuropathic pain by modulating the inflammatory response and neuroglial activation in the spinal cord. Neural Regen Res. 2020;15:2162–2168. doi: 10.4103/1673-5374.282260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olmos G, Lladó J. Tumor necrosis factor alpha: a link between neuroinflammation and excitotoxicity. Mediators Inflamm. 2014;2014:861231. doi: 10.1155/2014/861231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park J, Wetzel I, Marriott I, Dréau D, D’Avanzo C, Kim DY, Tanzi RE, Cho H. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat Neurosci. 2018;21:941–951. doi: 10.1038/s41593-018-0175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paterniti I, Impellizzeri D, Di Paola R, Esposito E, Gladman S, Yip P, Priestley JV, Michael-Titus AT, Cuzzocrea S. Docosahexaenoic acid attenuates the early inflammatory response following spinal cord injury in mice: in-vivo and in-vitro studies. J Neuroinflammation. 2014;11:6. doi: 10.1186/1742-2094-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin C, Liu CB, Yang DG, Gao F, Zhang X, Zhang C, Du LJ, Yang ML, Li JJ. Circular RNA expression alteration and bioinformatics analysis in rats after traumatic spinal cord injury. Front Mol Neurosci. 2018;11:497. doi: 10.3389/fnmol.2018.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rauscher B, Heigwer F, Henkel L, Hielscher T, Voloshanenko O, Boutros M. Toward an integrated map of genetic interactions in cancer cells. Mol Syst Biol. 2018;14:e7656. doi: 10.15252/msb.20177656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothhammer V, Quintana FJ. Control of autoimmune CNS inflammation by astrocytes. Semin Immunopathol. 2015;37:625–638. doi: 10.1007/s00281-015-0515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shan K, Liu C, Liu BH, Chen X, Dong R, Liu X, Zhang YY, Liu B, Zhang SJ, Wang JJ, Zhang SH, Wu JH, Zhao C, Yan B. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136:1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 35.Shin GC, Moon SU, Kang HS, Choi HS, Han HD, Kim KH. PRKCSH contributes to tumorigenesis by selective boosting of IRE1 signaling pathway. Nat Commun. 2019;10:3185. doi: 10.1038/s41467-019-11019-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva NA, Sousa N, Reis RL, Salgado AJ. From basics to clinical: a comprehensive review on spinal cord injury. Prog Neurobiol. 2014;114:25–57. doi: 10.1016/j.pneurobio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vismara I, Papa S, Rossi F, Forloni G, Veglianese P. Current options for cell therapy in spinal cord injury. Trends Mol Med. 2017;23:831–849. doi: 10.1016/j.molmed.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Wang L, Zhang Y, Yan Z, Liu L, Chen G. PYCR1 promotes the progression of non-small-cell lung cancer under the negative regulation of miR-488. Biomed Pharmacother. 2019a;111:588–595. doi: 10.1016/j.biopha.2018.12.089. [DOI] [PubMed] [Google Scholar]
- 40.Wang G, Han B, Shen L, Wu S, Yang L, Liao J, Wu F, Li M, Leng S, Zang F, Zhang Y, Bai Y, Mao Y, Chen B, Yao H. Silencing of circular RNA HIPK2 in neural stem cells enhances functional recovery following ischaemic stroke. EBioMedicine. 2020;52:102660. doi: 10.1016/j.ebiom.2020.102660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W, Liu R, Su Y, Li H, Xie W, Ning B. MicroRNA-21-5p mediates TGF-β-regulated fibrogenic activation of spinal fibroblasts and the formation of fibrotic scars after spinal cord injury. Int J Biol Sci. 2018;14:178–188. doi: 10.7150/ijbs.24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang YB, Shi Q, Li G, Zheng JH, Lin J, Qiu W. MicroRNA-488 inhibits progression of colorectal cancer via inhibition of the mitogen-activated protein kinase pathway by targeting claudin-2. Am J Physiol Cell Physiol. 2019b;316:C33–47. doi: 10.1152/ajpcell.00047.2018. [DOI] [PubMed] [Google Scholar]
- 43.Wen Z, Shen Q, Zhang H, Su Y, Zhu Z, Chen G, Peng L, Li H, Du C, Xie H, Xu X, Tang W. Circular RNA CCDC66 targets DCX to regulate cell proliferation and migration by sponging miR-488-3p in Hirschsprung’s disease. J Cell Physiol. 2019;234:10576–10587. doi: 10.1002/jcp.27733. [DOI] [PubMed] [Google Scholar]
- 44.Xian S, Ding R, Li M, Chen F. LncRNA NEAT1/miR-128-3p/AQP4 axis regulating spinal cord injury-induced neuropathic pain progression. J Neuroimmunol. 2021;351:577457. doi: 10.1016/j.jneuroim.2020.577457. [DOI] [PubMed] [Google Scholar]
- 45.Xu S, Wang J, Jiang J, Song J, Zhu W, Zhang F, Shao M, Xu H, Ma X, Lyu F. TLR4 promotes microglial pyroptosis via lncRNA-F630028O10Rik by activating PI3K/AKT pathway after spinal cord injury. Cell Death Dis. 2020;11:693. doi: 10.1038/s41419-020-02824-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yadav A, Saini V, Arora S. MCP-1: chemoattractant with a role beyond immunity: a review. Clin Chim Acta. 2010;411:1570–1579. doi: 10.1016/j.cca.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 47.Yang Y, Li H, He Z, Xie D, Ni J, Lin X. MicroRNA-488-3p inhibits proliferation and induces apoptosis by targeting ZBTB2 in esophageal squamous cell carcinoma. J Cell Biochem. 2019;120:18702–18713. doi: 10.1002/jcb.29178. [DOI] [PubMed] [Google Scholar]
- 48.Yu B, Zhou S, Yi S, Gu X. The regulatory roles of non-coding RNAs in nerve injury and regeneration. Prog Neurobiol. 2015;134:122–139. doi: 10.1016/j.pneurobio.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Shi LL, Gao X, Jiang D, Zhong ZQ, Zeng X, Rao Y, Hu X, Li TZ, Li XJ, Li L, Chen JM, Xia Q, Wang TH. Lentivirus-mediated inhibition of tumour necrosis factor-α improves motor function associated with PRDX6 in spinal cord contusion rats. Sci Rep. 2015;5:8486. doi: 10.1038/srep08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou ZB, Du D, Chen KZ, Deng LF, Niu YL, Zhu L. Differential expression profiles and functional predication of circular ribonucleic acid in traumatic spinal cord injury of rats. J Neurotrauma. 2019;36:2287–2297. doi: 10.1089/neu.2018.6366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.