ABSTRACT
Magnaporthe oryzae, causative agent of the rice blast disease, is a major concern for the loss in yield of rice crop across the globe. It is known for its characteristic melanised dome-shaped appressorium containing a dense melanin layer. The melanised layer is of considerable importance as it is required to generate turgor pressure for initiating peg formation, consequently rupturing the plant cuticle. Various virulence factors play an important role in the disease progression as well as pathogenesis of the fungus. Some of the proteins encoded by virulence genes are associated with signalling, secondary metabolism, protein deprivation, defence responses and conidiation. The purpose of this review is to describe various fungal virulence determinants and provide insights into the molecular mechanisms that are involved in progression of the disease. Besides, the recent molecular approaches being employed to combat the rice blast have also been elaborated.
KEYWORDS: Magnaporthe oryzae, rice blast, virulence genes, pesticides, mutant breeding
Introduction
Rice (Oryza sativa) is an essential food crop consumed by over 3.5 billion people across the world (Godfray et al. 2010). It is also an economically important crop as it is employed in textile, leather, cosmetics and food industry. However, pathogens such as fungi, bacteria, viruses and nematodes infect rice plants, causing a substantial loss in its yield (Asibi et al. 2019). Rice blast is one of the most significant and devastating disease occurring in the rice crop leading to almost 10% to 30% of crop loss every year (Sakulkoo et al. 2018).
Rice blast disease is caused by fungi Magnaporthe oryzae. It affects the foliar parts of the plant at the sprouting and mature stages. Rice cultivated in temperate and subtropical climates of Asia are highly vulnerable to this pathogen, while rice cultivation in tropical upland areas is prone to this pathogen only under irrigation (Nutsugah et al. 2008). The disease spreads frequently under moist conditions with relative humidity ranging from 80%-100% and temperatures ranging from 25 to 30°C (Talbot 2003). Significant harvest losses have been reported in many rice-growing countries of the South-east Asia (China, Sri Lanka, Indonesia, Bangladesh, India) (Wilson and Talbot 2009; Suprapta and Khalimi 2012; Kumar and Kalita 2017) and also in other regions like South America, Australia, Korea and Philippines (Greer and Webster 2001; Pena et al. 2007; Shahriar et al. 2020) (Figure 1). In an outbreak of rice blast disease in Malaysia, yield loss caused by panicle blast was as high as 50–70% (Ashkani et al. 2015; Zakaria and Misman 2018). Zhang et al. (2010) reported 40–50% rice yield loss due to blast infection in China. Yashaswini et al. (2017) reported blast disease percentage ranged from 50 to 74% in different districts of Telangana and Andhra Pradesh in India. Similar study has been reported from eastern India where rainfed rice yield loss was around 30% per annum (Jha et al. 2012).
Figure 1.
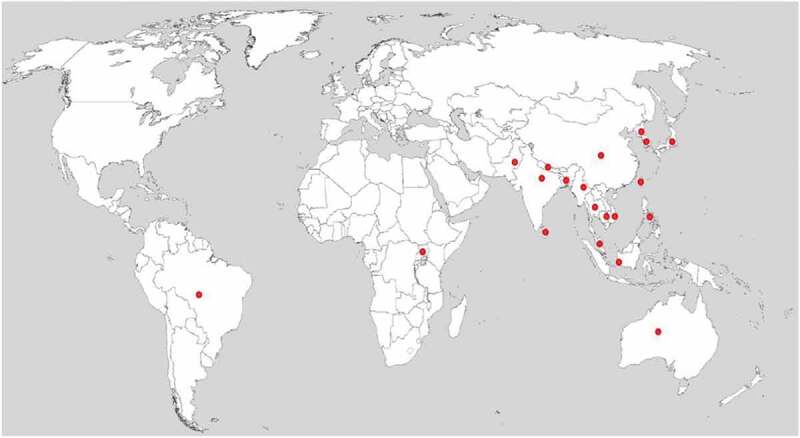
Global distribution of rice blast disease. Red spots indicate the countries with reports of rice blast
Pathogenesis and infection cycle of Magnaporthe oryzae
Rice blast symptoms rely on ecological conditions age and the level of resistance of the host plant. The pathogen predominantly infects foliage, causing blasting during vegetative growth phase, or in reproductive stage on the necks and panicles (Shahriar et al. 2020). These indications are extreme in case of blast of neck usually defined by the infection at panicle’s base and its rotting.
The disease is caused by a heterothallic ascomycete M. oryzae that produces asexual or sexual spores (ascospores) in structures called asci (Wilson and Talbot 2009). The pathogen initiates infection through a protracted biotrophic stage, where the fungus bounded by the invaginated plant plasma membrane, grows within host cells and proceeds to a necrotrophic stage leading to lesion development (Figure 2). The mycelium consists of branched, septate and uninucleate hyphae. Septate conidiophores with a dark base forming hyaline and pyriform conidia acrogenously. The pathogen produces lesions on leaves (leaf blast), leaf collars (collar blast), culms, culm nodes, panicle neck nodes (neck rot), and panicles (panicle blast). These lesions vary in colour and shape depending on environmental conditions and developmental stage of the plant (Law et al. 2017).
Figure 2.

Rice blast lesions on a leaf of rice plant
The infection begins when a three-celled conidium (Figures 3 (a,b)) comes in contact with the hydrophobic surface of the leaf and attaches itself by an adhesive called spore tip mucilage (Wilson and Talbot 2009). The germ tube formed after germination of conidia swells into appressorium (Figure 3 (c)), a specialised infection structure capable of penetrating the leaves and stems of the rice plant. As the appressorium undergoes maturation, a dense layer of melanin accumulates in the appressorium wall (Boddy 2016). Subsequently, hydrostatic turgor pressure of up to 8 MPa develops due to accumulation of glycerol in the appressorium, which provides sufficient mechanical force to perforate the leaf cuticle and enter the plant epidermal cells with the help of penetration peg (arising at the base of appressorium). Eventually, invasive hyphae prevent nutrients and water from reaching the kernels and also secrete effector molecules to suppress host immunity and aid infection (Giraldo et al. 2013). These effector molecules move into host cell cytoplasm by a biotrophic interfacial complex, a plant-derived membrane-rich structure (Giraldo et al. 2013; Mochizuki et al. 2015).
Figure 3.
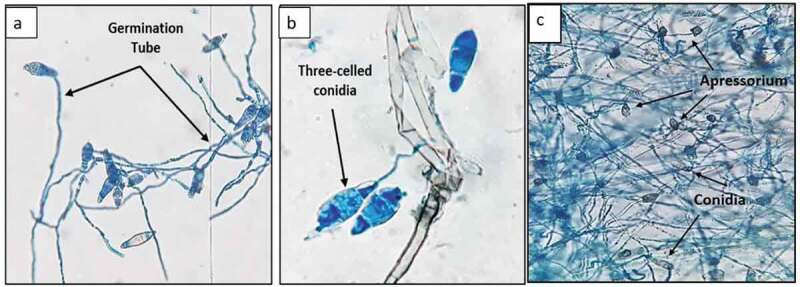
Microscopic view of M. oryzae (a) and (b) three-celled conidia with germination tube (40× magnification); and (c) appressorium formation (10× magnification)
The M. oryzae infection cycle has distinct developmental stages (Figure 4). The first stage is characterised by the attachment of the three-celled conidium on the hydrophobic surface of rice leaf and development of polarised germ tube. In the second stage, this tube further divides into a swelled dome-shaped appressorium for the initiation of infection. Third stage embodies the development of the penetration peg from appressorium that is dependent on various ecological aspects, such as hydrophobicity and firmness of the interacting surface with negligible availability of exogenous nutrients. Further, the biotrophic infection spreads to the neighbouring cells, and infected cells enter a necrotrophic phase in which thin filamentous secondary hyphae eventually kill the plant cells (Ebbole 2007).
Figure 4.
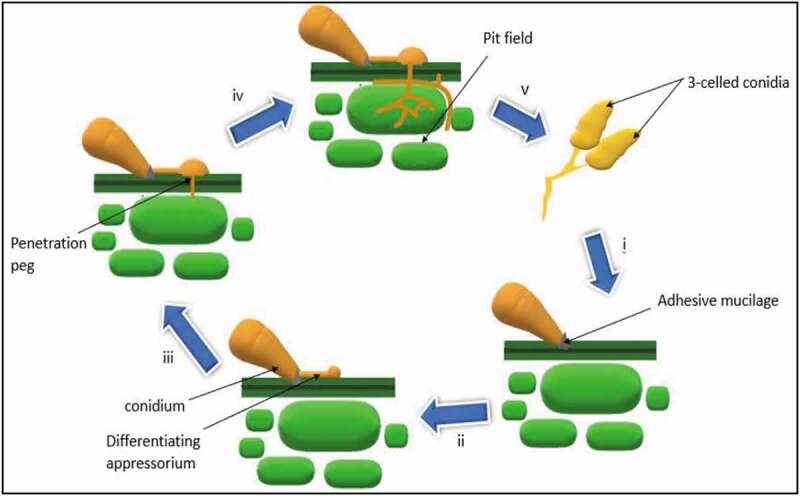
Infection cycle of M. oryzae: i) Attachment of conidia to the surface of host cell by adhesive mucilage ii) spore germination, development of germ tube and appressorium formation iii) emergence of penetration peg iv) invasive growth in the host plant. Modified from (Jia et al. 2016)
Molecular targets in appressorium cell wall, signal transduction pathways and host-pathogen interactions
Cell wall-associated factors
The pathogen M. oryzae penetrates into the plant cell wall through the melanin-rich dome-shaped appressorium. Melanin pigment plays an important role in imparting high turgor pressure to the appressorium, a major requirement for physical penetration and peg formation, a prerequisite to initiate infection (Galhano and Talbot 2011). Studies have shown that melanin-deficient fungal mutants with altered pigmentation (buffy, rosy, and albino) are non-pathogenic in nature (Langfelder et al. 2003). Polyketide synthase (PKS), encoded by the alb1 gene, is the first enzyme of the melanin synthesis pathway. It converts acetate units into 1,3,6,8-tetrahydroxynaphthalene (4THN), the first stable intermediate. 4THN is reduced to scytalone by 1,3,6,8-tetrahydroxynaphthalene reductase (4HNR), then dehydrated to 1,3,8-trihydroxynapthalene (3THN) by the scytalone dehydratase (SD). The 3THN is further reduced to vermelone by 1,3,8-trihydroxynapthalene reductase (3HNR) and dehydrated by SD to yield 1,8-dihydroxynaphthalene (DHN) (Langfelder et al. 2003). Blocking the enzymes involved in the pathway could restrict the production of melanin, thus restricting the appressorium penetration into the leaf surface. The reductase enzymes, especially 3HNR, are known targets of tricyclazole (Thompson et al. 2000).
Glycerol accumulation is additionally required to generate appressorium turgor pressure that facilitates hyphal penetration to rupture plant cuticle. Glycerol synthesis requires mobilisation of lipid bodies (storage polymers) to the maturing appressorium (Galhano and Talbot 2011). The amount of glycogen is determined not only by its synthesis, but also by rate of its degradation. The cytosolic glycogen degradation in M. oryzae, which requires glycogen phosphorylase Gph1p, plays an important role in the virulence of the fungus (Badaruddin et al. 2013). Cleavage of the α-1,4-glycosidic linkages of glycogen can be achieved by phosphorolysis, catalysed by Gph1p, or by hydrolysis. The function of phosphorylase is to decrease glycogen levels during the early stationary phase. Earlier findings have shown that lack of gph1 gene eliminated the transient drop in glycogen in the early stationary phase and resulted in an exaggerated re-synthesis phase (Badaruddin et al. 2013). The Δgph1 can prevent glycogen reserves from getting mobilised during development of appressorium and may significantly affect progression of disease.
Signal transduction pathways
The primary metabolic variation during appressorium maturation are controlled by a Tps facilitated genetic switch, which response to glucose-6-phosphate levels and NADPH/NADP balance in the cell (Badaruddin et al. 2013). Earlier findings have suggested that mutants with altered multi-functional fatty acid β-oxidation protein Mfp1 showed a considerable reduction in virulence (Wang et al. 2005). Moreover, mutants deficient in carnitine acetyltransferase enzyme meant for acetyl CoA transport across the mitochondrial or peroxisomal membrane, have been reported to be non-pathogenic (Wang et al. 2005).
The formation of appressorium on the artificial hydrophobic surfaces can be induced by exogenous cyclic Adenosine Monophosphate (cAMP). In absence of cAMP, conidia produce long germ tubes without tip differentiation on the hydrophilic surfaces. Molecular studies have confirmed the role of cAMP signalling in surface recognition and initiation of appressorium formation (Zhang et al. 2011). Besides surface hydrophobicity, other factors like surface hardness, cutin monomers and leaf waxes also affect appressorium formation in M. oryzae (Liu et al. 2007). Various physical and chemical signals have also been shown to affect appressorium formation in other plant pathogenic fungi, including Ustilago maydis and Colletotrichum species. It has also been reported that the Cap1 gene is associated with the actin cytoskeleton involved in cAMP pathway, Mac1 activation and transcription factors Mst1, Som1, Cdtf1 function downstream from the cAMP–PKA pathway. Of these, Som1 and Cdtf1 are two novel transcription factors vital for sporulation and appressorium development (Liu et al. 2011). Mst1 mutant has shown interrupted appressorium development due to delay in deployment of lipid bodies and transport of glycogens to appressoria, which is regulated by cAMP signalling (Soanes et al. 2012). There might be a cross-talk occurring with the cAMP pathway through the G-subunit protein MagB. The cAMP response pathway seems to be regulated by G proteins MagA and MagB, which potentially interacts with the Pth11 G protein-coupled receptor. Adenylate cyclase, Mac1 causes the accumulation of cAMP, which binds to the regulatory protein kinase A subunit Sum1, allowing detachment of the catalytic subunit CpkA (Figure 5).
Figure 5.
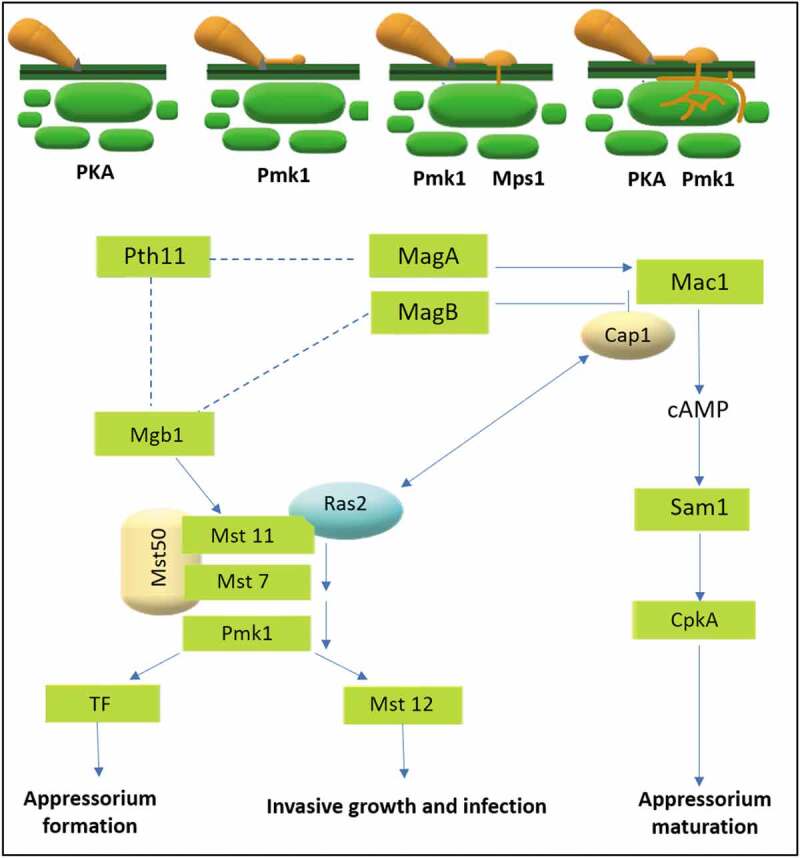
Schematic diagram of the Pmk1 mitogen-activated protein kinase (MAPK) pathway and the cyclic AMP (cAMP) response pathway in the rice blast fungus. Solid lines denote physical or genetic interactions that are supported by experimental evidence. Dotted lines denote tentative interactions Modified from (Wilson and Talbot 2009; Jiang et al. 2018)
Host-pathogen interaction
The rice-M.oryzae pathosystem has been extensively studied as a model for understanding plant-fungal interactions. Analysis of genetic and molecular interaction between avirulence effectors and their cognate resistance proteins have provided new insights into the resistance patterns in host plant as well as mode of pathogen infection. The formation as well as penetration of appressorium is regulated by Pmk1 mitogen-activated protein (MAP) kinase (Zhang et al. 2011). The Pmk1 MAP kinase gene plays a central role in appressorium formation and its growth beneath the plant cell surface. Its orthologs are also essential for numerous processes during plant infection stage in many other phytopathogenic fungi (Liu et al. 2011). A number of upstream genes involved in the activation of Pmk1 MAP kinase have been identified, including Mst50, Mst11, Mst7, Mgb1, and Ras2 genes (Zhao et al. 2005; Park et al. 2006).
Mst50 functions as an adaptor protein that binds with both the Mst7 and Mst11 kinases. The Mst7-Pmk1 interaction is a relatively transient weak interaction which may be stabilised or facilitated by additional components of the Pmk1 MAP kinase pathway during appressorium formation (Ding et al. 2009)). The null mutant of Pmk1 gene is defective in appressorium formation, infectious growth and has a lower phosphorylation level of Pmk1, but it still recognises hydrophobic surfaces and responds to exogenous cAMP (Figure 5). MAPK genes have also been reported to be essential for virulence, suggesting that components of MAPK signalling pathway involved in pathogenesis are widely conserved. The downstream transcription factor which regulates the Pmk1 gene is Mst12 involved in appressorial penetration and invasive growth (Li et al. 2011). Although key components of the cAMP signalling and Pmk1 pathways have been identified, fungal mechanisms for distinguishing physical and chemical signals of plant surfaces have not been well studied. The Pth11 gene, one putative receptor gene has been reported to be involved in surface sensing in M. oryzae (Kramer et al. 2009; Rispail et al. 2009). The Pth11 mutant demonstrated reduced virulence and appressorium formation on hydrophobic surfaces. The M. oryzae genome contains about 60 putative G-Protein Coupled Receptor (GPCR) genes, including several Pth11-like genes with the CEFM domain. Zhang et al. (2011) identified nine putative Pmk1-interacting genes, six and two of them being isolated from the appressorium and nitrogen starvation library, respectively. Only Pic1 gene was identified in both the libraries. The authors predicted that some of these gene products may be involved in stimulating or stabilising the Mst7-Pmk1 interaction during appressorium formation.
The firm attachment of spore and germ tube on the contact surface is one of the most important factors to induce appressorium differentiation. Another gene playing a significant role in surface recognition is Cbp1, which encodes a putative chitin-binding protein with signal peptide. The mutant study of Cbp1 revealed formation of abnormal appressoria on artificial surfaces but normal functional appressorium formed on leaf surface (Kamakura et al. 2002; Liu et al. 2011). It has been observed that loss of the Cbp1p caused reduction in cellular attachment, suggesting that it might be a component of adhesive materials for germ tube attachment n (Kamakura et al. 2002; Kuroki et al. 2017).
The Mpg1 gene of the fungal pathogen encodes a protein that modifies the leaf surface hydrophobicity before it adheres to it. Recombinant proteins under in vitro conditions can self-assemble on the surface of conidia (Xu et al. 2007), signifying that Mpg1p are capable of forming an amphipathic layer over surface of rice leaf, promoting adhesion of germ tube and acting as a signal for development of appressorium. Comparable to Mpg1, Mhp1 is over expressed during plant colonisation and conidiation (Soonok et al. 2005). The Mhp1 altered mutant had reduced conidiation, conidial propagation, appressorium development, and plant infection.
Therefore, various genes involved in the host–pathogen interaction can serve as key targets to stop the invasion and entry of pathogen in the rice plant if arrested at the point of initial contact.
Targets of rice immunity
Various factors can influence plant immunity in response to the pathogen invasion. Plants possess an efficient immune system to protect them by detecting the conserved pathogen-associated molecular patterns (PAMPs) of invading pathogen and elicits PAMP-triggered immune response (Zipfel and Felix 2005) or through eliciting an effector-triggered immunity activated by cognate intracellular immune receptors (Jones and Dangl 2006). PAMP-triggered and effector-triggered immunity plays a crucial role in plant preinvasive and post-invasive resistance, respectively, and inhibits the colonisation of invading pathogen (Li et al. 2014; Lee et al. 2009b; Chen and Ronald 2011; Liu et al. 2012)
The rice plant possesses two types of resistance genes, responsible for blast resistance: a) Major resistance (R) genes that confer race-specific resistance and b) quantitative trait loci (QTLs) that control partial, nonrace-specific resistance (Skamnioti and Gurr 2009). The Pik locus having cluster of six alleles (Pik, Pikm, Pikp, Piks, Pikh, and Pi1) is of prime importance due to its role in a number of blast R genes involved in rice breeding (Zhai et al. 2011a). Recently, it has been demonstrated that polymorphic residues in Pik-1 control the specificity of resistance (Carlos et al. 2018). Additional findings have recognised a new allele, Pikx, that is significantly associated with rice blast resistance at the Pik locus and is important for rice breeding against the M. oryzae (Shi et al. 2018). A total of 56 QTLs associated with blast resistance were identified in the rice genome (Hua et al. 2012; Liu et al. 2014). Only one QTL associated with resistance was present, and localised with the known R gene of Pik locus (Li et al. 2019).
Small interfering RNA (siRNA) and microRNA (miRNA) are expressed by plants under biotic stress conditions (Zhai et al. 2011b; Khraiwesh et al. 2012; Li et al. 2017a). The miRNAs present in rice have a vital role in providing immunity against associated pathogens. Innate immunity of rice is controlled by miR160a and miR398b against the blast fungus M. oryzae and their overexpression enhances resistance against blast disease (Li et al. 2014). Moreover, during the blast infection, dynamic balance of superoxide anion and hydrogen peroxide (H2O2) is maintained by superoxide dismutase2 targeted by miR398. MicroRNA miR169 acted as a negative regulator in rice immunity against M. oryzae by repressing the expression of nuclear factor Y-A genes (Li et al. 2017b). Likewise, jasmonic acid signalling pathway is a key regulator of plant defence responses against pathogens. M. oryzae infection blocks the conversion of α-linoleic acid to hydroxyoctadecadienoic acid, a critical step in jasmonic acid biosynthesis and facilitates its propagation and infection in the host plant. The miR319 is involved in the vital jasmonic acid biosynthetic step by genetic approaches (Zhang et al. 2018). OncemiR319 is induced in the host plant, specifically by the M. oryzae strain Guy11, which accomplishes the suppression of transcription factor gene OsTCP21, and its target genes OsLOX2 and OsLOX5 that encode key synthetic components of jasmonic acid (Zhang et al. 2018). Therefore, it has been suggested that miR319 may manipulate the plant innate response against M. oryzae by affecting jasmonic acid biosynthesis and signalling.
A recent study has also revealed an immune mechanism facilitated by phosphorylation of light-harvesting complex II (Liu et al. 2019). The light-harvesting complex II protein, LHCB5 in rice, has been reported to undergo light-induced phosphorylation at the time of blast infection. This step leads to accumulation of ROS in chloroplast, thereby enhancing broad-spectrum resistance of rice to M. oryzae.
Eradication of rice blast from agricultural fields using Pesticides/Fungicides
The fungicides are applied as spray or powder to protect the plant foliage. These fungicides can be either contact/surface acting or systemic. The mode of their action can range from inhibition of protein synthesis to targeting cell membrane and respiration (Yang et al. 2011). There are a variety of chemicals used as pesticides/fungicides to suppress the infection of rice blast in the agricultural fields.
One of the earliest, efficacious agricultural antibiotics developed in Japan was Blasticidin-S (Takeuchi et al. 1958). Blasticidin-S has shown better activity against blast disease of rice as compared to other synthetic chemicals used. The antibiotic obstructs the protein synthesis in M. oryzae and acts at peptidyl transfer site on the ribosomes (Svidritskiy et al. 2013). However, application of Blasticidin-S by spraying is known to cause conjunctivitis by accidental interaction, therefore, an enhanced formulation consisting of calcium acetate is used to eliminate this risk. Blasticidin-S gets easily degraded in presence of sunlight and microorganisms in soil (Matsunaka et al. 2013), and no deposit is usually spotted in the grains of rice.
Kasugamycin, an aminoglycoside isolated from Streptomyces kasugaensis is used against bacterial grain rot of rice, seedling rot and rice blast caused by Acidovorax avenae, Burkholderia glumae and M.oryzae, respectively (Vakulenko and Mobashery 2003). Cytotoxicity of Kasugamycin is observed to be low with no phytotoxicity in majority of the harvests. Kasugamycin exerts its effect by binding to the 30S subunit of bacterial ribosome at the mRNA binding region, which in turn blocks the interaction between codon and anticodon at initiation of translation, thereby inhibiting tRNA binding and translation (Schluenzen et al. 2006; Schuwirth et al. 2006).
Sodium hypochlorite has also been used to eliminate blast infection on seeds. Seeds soaked in low concentrations (0.8%-3.2%) of sodium hypochlorite were successful in reducing blast infection. Moreover, sodium hypochlorite concentrations and soak time required to eliminate P. grisea blast from infested seed resulted in stunted and chlorotic seedlings. The germination rates were reduced from >90% to <75% and seedling health was visibly reduced (Greer and Webster 2001).
Azoxystrobin is the most commonly used rice fungicide in the southern United States and Asian countries (Groth 2005). Information is limited on the optimum rates and timing of fungicide application for blast control as compared with sheath blight information (Prasanna et al. 2013). Timing of fungicide application is critical and is targeted towards protecting the panicle as it emerges from the flag leaf. Consequently, there has been a general trend to make a single application of azoxystrobin in attempt to control both blast and sheath blight (Groth 2005). It is an effective inhibitor of spore germination and is most effective when used as a protectant prior to infection (Bartett et al. 2001). Preliminary studies indicate the rate of azoxystrobin required to effectively control blast infections, as disease incidence varies with the susceptibility of the host (Uppala and Zhou 2018).
Pesticides targeting biosynthesis of phospholipids in rice blast
Iprobenphos (IBP) has been popularly used for controlling blast disease of rice along with Edifenphos (EDDP) after phosphorothiolate (PTL) compounds were discovered to exhibit fungicidal properties. IBP is a systemic fungicide, and EDDP is non-systemic fungicide showing effective fungicidal activity when used as a foliar applicant against the blast causing fungus (Gohel and Chauhan 2015). Isoprothiolane, malonate ester having two methylene hydrogens is also used as a fungicide to control a range of diseases including blast of rice. The chemical structure of PTL is apparently different from that of isoprothiolane, yet, cross-resistance between PTL fungicides and isoprothiolane proposes a parallelism in their mode of action (Fukuta et al. 2004). IBP, EDDP and isoprothiolane have shown to explicitly prevent the conversion of phosphatidylethanolamine to phosphatidylcholine (Upmanyu and Rana 2012). Inhibition of phosphatidylcholine synthesis leads to increased permeability in the cell membrane along with enzymatic activities that are membrane-associated, detrimental to fungal pathogen. Additionally, propiconazole inhibits sterol biosynthesis by inhibiting demethylation of ergosterol important for cellular growth of the fungus (Kumar and Veerabhadraswamy 2014; Uppala and Zhou 2018).
Pesticides for disrupting respiration and permeability of the membrane
Novel systemic fungicides, ferimzone and metominostrobin are also used to inhibit blast of rice. Fungistatic action of ferimzone in vitro involves efflux of acidic electrolytes from the mycelia of M.oryzae (Kawasaki 2004). Metominostrobin, belonging to the class of methoxyacrylate fungicides, prevents the electron chain during mitochondrial respiration by obstructing the flow of electron via the cytochrome bc1 segment. However, mycelial cells recover respiratory activity by inducing cyanide-resistant respiration to release the block by metominostrobin. Metominostrobin-dependent initiation mechanism is proposed to be dependent on the superoxide anion during cyanide-resistant respiration. When applied, the flavonoids have also been able to scavenge superoxide anion thus generated, by blocking the flow of electron through the cytochrome bc1 segment, thereby inhibiting metominostrobin-dependent induction of cyanide-resistant respiration (Yamaguchi 2004). Similarly, another chemical Kresoxim methyl, contact or local in nature, disrupts the respiration by blocking the electron transport by binding to the Qo site of the chain (Kumar and Veerabhadraswamy 2014).
Eprobenfos is another systemic fungicide that alters the structure of the membrane by blocking the phospholipid synthesis, thereby increasing the permeability and causing loss of key cellular components (Srivastava et al. 2017).
Melanin biosynthesis inhibition
Appressoria cells formed during the infection cycle of M.oryzae are matured by the development of a layer of melanin, which facilitates the generation of necessary turgor pressure required for its penetration into leaf. Thus, inhibitors of melanin biosynthesis have been reported to show a remarkable effect as blast controller (Kimura and Fukuchi 2018). Hydroxynaphthalene inhibitors and scytalone dehydratase inhibitors are the two categories of melanin biosynthesis inhibitors. The structure of inhibitors is shown in (Figure 6). Fthalide has an effective defensive action against rice blast for long duration (Yamaguchi 2004). It restricts melanin biosynthesis in appressoria crucial during invasion process of M. oryzae. Phytotoxicity as well as cytotoxicity to mammals from the main metabolites has been indicated to be negligible.
Figure 6.
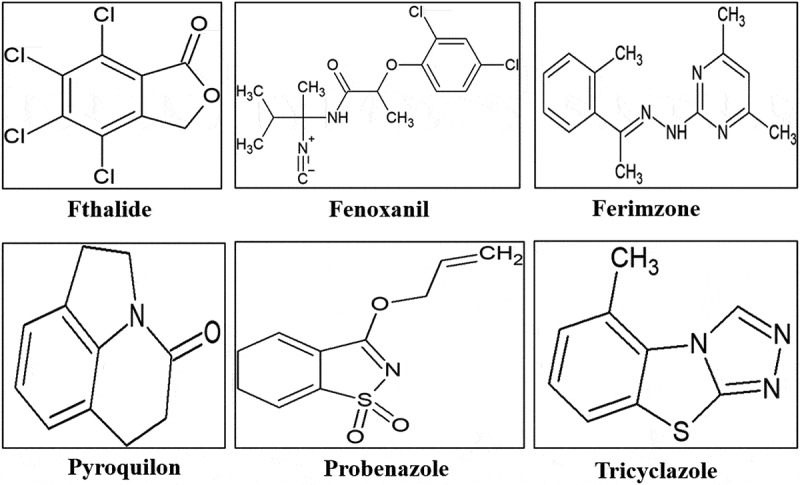
Structural representation of various pesticides used as inhibitors against rice blast infections
Tricyclazole, is a reductase inhibitor which results in the deposition of auto-oxidation intermediate products of the pathway (Kunova et al. 2013). It inhibits melanin biosynthesis not only in M. oryzae but also in C. lindemuthianum and C. lagenarium (Sisler 1986). Pyroquilon exerts similar modes of action i.e. scytalone and 2-hydroxyjuglone (2-HJ) accumulation in the cell. Increased concentrations of tricyclazole followed by pyroquilon induce flaviolin accumulation, signifying that alternative step of inhibition is present in the pathway. The chemical also obstructs secondary infection by decreasing the sporulation of M. oryzae under field settings. Field studies indicated long-lasting inhibitory potential of tricyclazole and pyroquilon (Kunova et al. 2014).
Carpropamid has been established as an effective regulatory root systemic agent against the rice blast. Various studies revealed that it is a competitive inhibitor that has tight-binding with an enzyme in the melanin biosynthesis pathway, scytalone dehydratase (STD). Other systemic fungicides that function as inhibitors to melanin biosynthesis are pyroquilone, diclocymet and fenoxanil (Nishimura and Hino 2002; Srivastava et al. 2017). Tebuconazole also acts as inhibitor of fungal cell wall development in addition to melanin inhibition (Ghazanfar et al. 2009).
New approaches to combat rice blast disease
Mutation breeding
The mutated genes with the help of tagging can be bought into a noble solitary breeding line making it easier to track consequent breeding programme (Shu 2009). The isolation and molecular characterisation of different blast resistance genes can lead to clarification of the actual allelic variants of these genes via various molecular breeding and transgenic approaches. The bioinformatics approach is helpful in understanding the evolution of new pathotypes of Magnaporthe isolates by studying major blast resistance (R) gene. This approach is utilised for designing better resistance breeding strategies. In this context, allele mining for resistance genes in all sequenced rice genomes shows the presence/absence of polymorphism and a large number of structural variations (Gowda et al. 2015; Mahesh et al. 2016). Approximately 100 quantitative blast R genes have been detected in rice, and 22 of these have been successfully cloned and characterised (Sharma et al. 2012; Ashkani et al. 2016). There are a number of varieties which have been formed by mutation breeding, like RD6, glutinous mutant of the prominent non-glutinous variety Khao Dawk Mali 105 (KDML105) induced by radiation. Chemo- mutagenesis with 0.1 and 0.2% ethylmethane sulphonate (EMS) has also been deployed to impart blast resistance in the Ratna (IR8/TKm6) variety (Kumar et al. 2017). However, the drawback of mutation breeding while protecting crop against rice blast infection is restricted in effecting the production of dominant alleles with less efficacy.
Resistance to blast through Marker-assisted selection (MAS)
MAS in comparison to the traditional breeding approaches, is convenient in breeding for acquiring resistance to blast, as only single or few genes are involved in coding of these resistant phenotypes (Srivastava et al. 2017). It is a suitable approach to control blast by manipulating the interaction among specific R gene and Avr gene of avirulence from host–pathogen interaction (Petit-Houdenot and Fudal 2017; Srivastava et al. 2017). MAS along with conventional breeding has facilitated R genes to be combined in elite rice varieties to improve their blast resistance and durability.
MAS also improves the success rate of traditional breeding by selecting those that could assist in achieving the required traits. A group of simple sequence repeats markers viz. RM168, RM8225, RM1233, RM6836, RM5961 and RM413, that have been reported to be linked to blast immunity trait, can be further utilised in MAS programs (Ashkani et al. 2012). Limitations of MAS programme are the high costs involved and less consistency, since they hamper the effective application for varietal development. Mapping and tagging of QTLs associated with blast resistance would be supportive in the cloning of major disease resistance genes as well as marker-assisted breeding program for development of resistant cultivars (Ashkani et al. 2012).
Blast disease management by miRNA
MicroRNAs (miRNAs) are one of the vital regulators for development and defence in eukaryotic species. miRNAs are implicated in providing immunity against spread of pathogenic organisms in plants, which could be used as biological control agents (Katiyar-Agarwal and Jin 2010). Expression of nine miRNAs has been identified upon infection by M. oryzae after inoculating with M. oryzae elicitors (Li et al. 2014). The involvement of miRNAs in providing resistance to rice plant against M. oryzae has been reported by (Katiyar-Agarwal and Jin 2010. Li et al. 2017b) also elucidated that the increased expression of miR398b or miR160a can improve immunity against the rice blast disease (Li et al. 2017b). Systematic silencing of M. oryzae has been directed and used for the improvement of rice plant resistance. The OsACDR1 gene is a resistant gene that encodes for MAPKKK, involved in producing defence-related pathways aimed at up-regulating the OsACDR1 transcript (Srivastava et al. 2017). Rice plants with increased expression of OsACDR1 illustrated impulsive hypersensitive response and heightened build-up of the phenolic compounds, required for the positive alteration leading to gain of resistance in the plant (Srivastava et al. 2017).
Transgenic approaches for blast resistance
Effective and stable integration of gene of interest into the rice genome has been confirmed through Agrobacterium-mediated transformation (Srivastava et al. 2017). Studies have shown aggravated resistance in rice cultivars against blast fungus by expressing chitinase gene in rice by the use of these approaches (Li et al. 2009). Insertion of RCC2 by the above-mentioned approach has shown that resistance levels increased in rice under in-vitro conditions (Asghar et al. 2007). Increasing the level of gene expression in transformed rice for OsCPK4 encoding for calcium-dependent protein kinase in transgenic rice gave resistance against the blast infection. Numerous significant genes have been effectively mutated in the selected varieties of rice for managing the blast disease in rice (Srivastava et al. 2017).
Perspective
Rice blast, caused by M. oryzae, is one of the most destructive rice diseases worldwide, causing substantial yield losses every year. Traditional strategies to manage this disease include the fertilisation of the different cultivars properly; usage of high quality and disease-free seeds etc. Besides, the other techniques used to manage blast infections are seed treatments (to prevent infection of the seedlings after germination) and application of fungicides in the fields (to prevent infection of leaves and panicles during the growing season). These techniques have been used to manage and reduce rice blast, but neither one is considered to be highly successful. Recent advances in molecular techniques have been used as an effective tool to understand the biological pathways as well as genes taking part in host-pathogen infection pattern, plant response and disease development.
The understanding of major molecular targets associated with cell wall, signal transduction pathways and host–pathogen interactions in M. oryzae could help to prevent the onset of blast disease, limiting the use of harmful chemicals as pesticides. Recent practices like mutation breeding, MAS for blast resistance in rice programme, miRNA in blast disease management, transgenic approaches for blast resistance can also play key role in monitoring blast disease.
Funding Statement
This work was supported by the Bayers’ AG [Grants4Targets- Novel targets for crop protection 2018-01-008].
Author contributions
Lovely Gupta: Manuscript writing, analysis and editing; Maansi Vermani: Manuscript analysis and editing; Simran Kaur Ahluwalia: Data search, manuscript writing; Pooja Vijayaraghavan: Conceptualization, review and editing.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Asghar A, Rashid H, Ashraf M, Khan MH, Chaudhry Z.. 2007. Improvement of basmati rice against fungal infection through gene transfer technology. Pakistan J Bot. 39:1277. [Google Scholar]
- Ashkani S, Rafii MY, Rusli I, Sariah M, Abdullah SNA, Rahim HA, Latif MA. 2012. SSRs for marker-assisted selection for blast resistance in rice (oryza sativa L.). Plant Mol Biol Report. 30(1):79–86. doi: 10.1007/s11105-011-0315-4. [DOI] [Google Scholar]
- Ashkani S, Rafii MY, Shabanimofrad M, Ghasemzadeh A, Ravanfar SA, Latif MA. 2016. Molecular progress on the mapping and cloning of functional genes for blast disease in rice (oryza sativaL.): current status and future considerations. Crit Rev Biotechnol. 36(2):353–367. doi: 10.3109/07388551.2014.961403. [DOI] [PubMed] [Google Scholar]
- Ashkani S, Yusop MR, Shabanimofrad M, Harun AR, Sahebi M, Latif MA. 2015. Genetic analysis of resistance to rice blast. : A study on the inheritance of resistance to the blast disease pathogen in an F3 F3 population of rice. J Phytopathol. 163(4):300–309. doi: 10.1111/jph.12323. [DOI] [Google Scholar]
- Asibi AE, Chai Q, Coulter JA. 2019. Rice blast: A disease with implications for global food security. Agronomy Agronomy. 9(8):451. doi: 10.3390/agronomy9080451. [DOI] [Google Scholar]
- Badaruddin M, Holcombe LJ, Wilson RA, Wang ZY, Kershaw MJ, Talbot NJ. 2013. Glycogen metabolic genes are involved in trehalose-6-phosphate synthase-mediated regulation of pathogenicity by the rice blast fungus magnaporthe oryzae. PLoS Pathog. 9(10):e1003604. doi: 10.1371/journal.ppat.1003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartett DW, Clough JM, Godfrey CRA, Godwin JR, Hall AA, Heaney SP, Maund SJ. 2001. Understanding the strobilurin fungicides. Pestic Outlook. 24:48. [Google Scholar]
- Boddy L. 2016. Pathogens of Autotrophs. Fungi. 3:245–292. [Google Scholar]
- Carlos DLCJ, Marina F, Abbas M, Hiromasa S, Ryohei T, Sophien K, Banfield MJ. 2018. Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nature Plants. 4(8):576–585. doi: 10.1038/s41477-018-0194-x [DOI] [PubMed] [Google Scholar]
- Chen X, Ronald PC. 2011. Innate immunity in rice. Trends Plant Sci. 16(8):451–459. doi: 10.1016/j.tplants.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Zhou X, Zhao X, Xu J-R. 2009. The PMK1 MAP kinase pathway and infection-related morphogenesis. In: Wang G-L, Valent B, editors. Advances in genetics, genomics and control of rice blast disease. Dordrecht: Springer Netherlands; p. 13–21. [Google Scholar]
- Ebbole DJ. 2007. magnaporthe as a model for understanding host-pathogen interactions. Annu Rev Phytopathol. 45(1):437–456. doi: 10.1146/annurev.phyto.45.062806.094346. [DOI] [PubMed] [Google Scholar]
- Fukuta Y, Yanoria MJ, Mercado-Escueta D, Ebron LA, Fujita Y, Araki E, Khush GS.. 2004. Rice blast: Interaction with rice and control. In Proceedings of the 3rd International Rice Blast Conference. Dordrecht: Springer. [Google Scholar]
- Galhano R, Talbot NJ. 2011. The biology of blast: Understanding how Magnaporthe oryzae invades rice plants. Fungal Biol Rev. 25(1):61–67. doi: 10.1016/j.fbr.2011.01.006. [DOI] [Google Scholar]
- Ghazanfar M, Wakil W, Sahi S, Kaku K. 2009. Influence of various fungicides on the management of rice blast disease. Mycopath. 7:29–34. [Google Scholar]
- Giraldo MC, Dagdas YF, Gupta YK, Mentlak TA, Yi M, Martinez-Rocha AL, Saitoh H, Terauchi R, Talbot NJ, Valent B. 2013. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat Commun. 4(1):1996. doi: 10.1038/ncomms2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C. 2010. Food security: The challenge of feeding 9 billion people. Science. 372(5967):812–818. doi: 10.1126/science.1185383. [DOI] [PubMed] [Google Scholar]
- Gohel NM, Chauhan HL. 2015. Integrated management of leaf and neck blast disease of rice caused by pyricularia oryzae. African J Agric Res. 10(19):2038–2040. doi: 10.5897/AJAR12.1960. [DOI] [Google Scholar]
- Gowda M, Shirke MD, Mahesh HB, Chandarana P, Rajamani A, Chattoo BB. 2015. Genome analysis of rice-blast fungus Magnaporthe oryzae field isolates from southern India. Genomics Data. 5:284–291. doi: 10.1016/j.gdata.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer CA, Webster RK. 2001. Occurrence, distribution, epidemiology, cultivar reaction, and management of rice blast disease in California. Plant Dis. 85(10):1096–1102. doi: 10.1094/PDIS.2001.85.10.1096. [DOI] [PubMed] [Google Scholar]
- Groth DE. 2005. Azoxystrobin rate and timing effects on rice sheath blight incidence and severity and rice grain and milling yields. Plant Dis. 89(11):1171–1174. doi: 10.1094/PD-89-1171. [DOI] [PubMed] [Google Scholar]
- Hua L, Wu J, Chen C, Wu W, He X, Lin F, Wang L, Ashikawa I, Matsumoto T, Wang L, et al. 2012. The isolation of pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor Appl Genet. 125(5):1047–1055. doi: 10.1007/s00122-012-1894-7. [DOI] [PubMed] [Google Scholar]
- Jha A, Singh K, Meena MS, Singh RKP. 2012. Constraints of rainfed rice production in eastern india: An overview. SSRN Electronic Journal com/abstract=2061953 or org/10.2139/ssrn.2061953 [Google Scholar]
- Jia Y, Zhou E, Lee S, Bianco T. 2016. Coevolutionary dyanamics of rice blast resistance gene pi-ta and Magnaporthe oryzae avirulence gene avr-pita 1. Phytopatho. 106(7):676–683. doi: 10.1094/PHYTO-02-16-0057-RVW. [DOI] [PubMed] [Google Scholar]
- Jiang C, Zhang X, Liu H, Xu JR. 2018. Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLoS Pathog. 14(3):1006875. doi: 10.1371/journal.ppat.1006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. 2006. The plant immune system. Nature. 444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kamakura T, Yamaguchi S, Saitoh KI, Teraoka T, Yamaguchi I. 2002. A novel gene, CBP1, encoding a putative extracellular chitin-binding protein, may play an important role in the hydrophobic surface sensing of magnaporthe grisea during appressorium differentiation. Mol Plant-Microbe Interact. 15(5):437–444. doi: 10.1094/MPMI.2002.15.5.437. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Jin H. 2010. Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol. 48(1):225–246. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S.2004. Rice blast: Interaction with rice and control. In Proceedings of the 3rd International Rice Blast Conference. Dordrecht: Springer. [Google Scholar]
- Khraiwesh B, Zhu JK, Zhu J. 2012. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta - Gene Regul Mech. 1819(2):137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N, Fukuchi A. 2018. Management of melanin biosynthesis dehydratase inhibitor (MBI-D)-resistance in pyricularia oryzae using a non-MBI-D fungicidal application program for nursery boxes and a diclocymet and ferimzone mixture for field foliar applications. J Pestic Sci. 43(4):287–292. doi: 10.1584/jpestics.D18-023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B, Thines E, Foster AJ. 2009. MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea. Fungal Genet Biol. 46(9):667–681. doi: 10.1016/j.fgb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Kumar D, Kalita P. 2017. Reducing postharvest losses during storage of grain crops to strengthen food security in developing countries. Foods. 6(1):8. doi: 10.3390/foods6010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MKP, Veerabhadraswamy LA. 2014. Appraise a combination of fungicides against blast and sheath blight diseases of paddy (oryza sativa L.). J Exp Biol Agric Sci. 2(1):49–57. [Google Scholar]
- Kumar V, Kumar A, Singh VP, Tomar A. 2017. Effectiveness measurement of bio-agents and botanicals against pyricularia oryzae. J Pure Appl Microbiol. 11(1):585–592. doi: 10.22207/JPAM.11.1.76. [DOI] [Google Scholar]
- Kunova A, Pizzatti C, Bonaldi M, Cortesi P. 2014. Sensitivity of nonexposed and exposed populations of magnaporthe oryzae from rice to tricyclazole and Azoxystrobin. Plant Dis. 98(4):512–518. doi: 10.1094/PDIS-04-13-0432-RE. [DOI] [PubMed] [Google Scholar]
- Kunova A, Pizzatti C, Cortesi P. 2013. Impact of tricyclazole and azoxystrobin on growth, sporulation and secondary infection of the rice blast fungus, Magnaporthe oryzae. Pest Manag Sci. 69(2):278–284. doi: 10.1002/ps.3386. [DOI] [PubMed] [Google Scholar]
- Kuroki M, Okauchi K, Yoshida S, Ohno Y, Murata S, Nakajima Y, Nozaka A, Tanaka N, Nakajima M, Taguchi H, et al. 2017. Chitin-deacetylase activity induces appressorium differentiation in the rice blast fungus Magnaporthe oryzae. Sci Reports. 7(1):9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder K, Streibel M, Jahn B, Haase G, Brakhage AA. 2003. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet Biol. 38(2):143–158. doi: 10.1016/S1087-1845(02)00526-1. [DOI] [PubMed] [Google Scholar]
- Law JWF, Ser H-LH-L, Khan TM, Chuah L-HL-H, Pusparajah P, Chan K-GK-G, Goh B-HB-H, Lee L-HL-H. 2017. The potential of streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (pyricularia oryzae). Front Microbiol. 8:3. doi: 10.3389/fmicb.2017.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-WS-W, Han S-WS-W, Sririyanum M, Park C-JC-J, Seo Y-SY-S, Ronald PC. 2009. A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science. 326(5954):850–853. doi: 10.1126/science.1173438. [DOI] [PubMed] [Google Scholar]
- Li C, Wang D, Peng S, Chen Y, Su P, Chen J, Zheng L, Tan X, Liu J, Xiao Y, et al. 2019. Genome-wide association mapping of resistance against rice blast strains in South China and identification of a new Pik allele. Rice. 12(1):47. doi: 10.1186/s12284-019-0309-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Zhou X, Kong L, Wang Y, Zhang H, Zhu H, Mitchell TK, Dean RA, Xu J-RJ-R, Idnurm A. 2011. Mosfl1 is important for virulence and heat tolerance in magnaporthe oryzae. PLoS One. 6(5):19951. doi: 10.1371/journal.pone.0019951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Pei Y, Sang X, Ling Y, Yang Z, He G. 2009. Transgenic indica rice expressing a bitter melon (momordica charantia) class I chitinase gene (McCHIT1) confers enhanced resistance to Magnaporthe grisea and rhizoctonia solani. Eur J Plant Pathol. 125(4):533. doi: 10.1007/s10658-009-9501-8. [DOI] [Google Scholar]
- Li S, Castillo-González C, Yu B, Zhang X. 2017a. The functions of plant small RNAs in development and in stress responses. Plant J. 90(4):654–670. doi: 10.1111/tpj.13444. [DOI] [PubMed] [Google Scholar]
- Li Y, Lu YG, Shi Y, Wu L, Xu YJ, Huang F, Guo XY, Zhang Y, Fan J, Zhao JQ, et al. 2014. Multiple rice MicroRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 164(2):1077–1092. doi: 10.1104/pp.113.230052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhao SL, Li JL, Hu XH, Wang H, Cao XL, Xu YJ, Zhao ZX, Xiao ZY, Yang N, et al. 2017b. Osa-miR169 negatively regulates rice immunity against the blast fungus magnaporthe oryzae. Front Plant Sci. 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li J-FJ-F, Ao Y, Qu J, Li Z, Su J, Zhang Y, Liu J, Feng D, Qi K, et al. 2012. Lysin motif-containing Motif–Containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell. 2416(8):3406–3419. doi: 10.1105/tpc.112.102475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Suresh A, Willard FS, Siderovski DP, Lu S, Naqvi NI. 2007. Rgs1 regulates multiple Gα subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. Embo J. 26(3):690–700. doi: 10.1038/sj.emboj.7601536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang S, Hu J, Sun W, Padilla J, He Y, Li Y, Yin Z, Liu X, Wang W, et al. 2019. Phosphorylation-guarded light-harvesting complex II contributes to broad-spectrum blast resistance in rice. Proc Natl Acad Sci U S A. 116(35):17572–17577. doi: 10.1073/pnas.1905123116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Zhou X, Li G, Li L, Kong L, Wang C, Zhang H, Xu JR. 2011. Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 7:1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WD, Liu JL, Leach JE, Wang GL. 2014. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol. 52:213–241. doi: 10.1146/annurev-phyto-102313-045926 [DOI] [PubMed] [Google Scholar]
- Mahesh HB, Shirke MD, Singh S, Rajamani A, Hittalmani S, Wang G-LG-L, Gowda M. 2016. Indica rice genome assembly, annotation and mining of blast disease resistance genes. BMC Genomics. 17(1):242. doi: 10.1186/s12864-016-2523-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaka S, Hutson DH, Murphy SD. 2013. Mode of action, metabolism and toxicology: Pesticide chemistry: Human welfare and the environment. Elsevier, Pergamon p. 3. [Google Scholar]
- Mochizuki S, Minami E, Nishizawa Y. 2015. Live-cell imaging of rice cytological changes reveals the importance of host vacuole maintenance for biotrophic invasion by blast fungus, Magnaporthe oryzae. Microbiologyopen. 425(6):952–966. doi: 10.1002/mbo3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura A, Hino I. 2002. Fenoxanil (ACHI-BUρ) - A new systemic fungicide for the control of rice blast. Agrochem Japan. 81:13–15. [Google Scholar]
- Nutsugah SK, Twumasi JK, Chipili J, Séré Y, Sreenivasaprasad S. 2008. Diversity of the rice blast pathogen populations in Ghana and strategies for resistance management. Plant Pathol J. 7106(1):109–113. doi: 10.3923/ppj.2008.109.113. [DOI] [Google Scholar]
- Park G, Xue C, Zhao X, Kim Y, Orbach M, Xu J-RJ-R. 2006. multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell. 18106(10):2822–2835. doi: 10.1105/tpc.105.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena FA, Perez LM, Cruz CMV, Ardales EY. 2007. Occurrence of rice blast fungus, pyricularia grisea in the Philippines. JIRCAS Working Report. 53: 65–70 [Google Scholar]
- Petit-Houdenot Y, Fudal I. 2017. Complex interactions between fungal avirulence genes and their corresponding plant resistance genes and consequences for disease resistance management. Front Plant Sci. 8:1072. doi: 10.3389/fpls.2017.01072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanna MK, Sidde DK, Moudgal R, Kiran N, Pandurange KT, Vishwanath K. 2013. Impact of fungicides on rice production in India, Fungicides - showcases integrated plant disease managment from around the world, Mizuho Nita, IntechOpen, DOI: 10.5772/51009. [DOI] [Google Scholar]
- Rispail N, Soanes DM, Ant C, Czajkowski R, Grünler A, Huguet R, Perez-Nadales E, Poli A, Sartorel E, Valiante V, et al. 2009. Comparative genomics of MAP kinase and calcium-calcineurin calcium–calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet Biol. 46(4):287–298. doi: 10.1016/j.fgb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Sakulkoo W, Osés-Ruiz M, Garcia EO, Soanes DM, Littlejohn GR, Hacker C, Correia A, Valent B, Talbot NJ. 2018. A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science. 359(6382):1399–1403. doi: 10.1126/science.aaq0892. [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Takemoto C, Wilson DN, Kaminishi T, Harms JM, Hanawa-Suetsugu K, Szaflarski W, Kawazoe M, ShirouzoShirouzu M, Nierhaus KH, et al. 2006. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat Struct Mol Biol. 13(10):871–878. doi: 10.1038/nsmb1145. [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Day JM, Hau CW, Janssen GR, Dahlberg AE, Cate JHD, Vila-Sanjurjo A. 2006. Structural analysis of kasugamycin inhibition of translation. Nat Struct Mol Biol. 13(10):879–886. doi: 10.1038/nsmb1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahriar SA, Imtiaz AA, Hossain MB, Husna A, Eaty MNK. 2020. Review: rice Blast Disease. Ann Res Review Biol. 35:50–64. doi: 10.9734/arrb/2020/v35i130180. [DOI] [Google Scholar]
- Sharma TR, Rai AK, Gupta SK, Vijayan J, Devanna BN, Ray S. 2012. Rice blast management through host-plant resistance: Retrospect and prospects. Agric Res. 1(1):37–52. doi: 10.1007/s40003-011-0003-5. [DOI] [Google Scholar]
- Shi X, Long Y, He F, Zhang C, Wang R, Zhang T, Wu W, Hao Z, Wang Y, Wang GL, et al. 2018. The fungal pathogen Magnaporthe oryzae suppresses innate immunity by modulating a host potassium channel. PLoS Pathog. 14(1):e1006878. doi: 10.1371/journal.ppat.1006878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu QY2009. Induced plant mutations in the genomics era. Food Agric Organ United Nations. Rome. [Google Scholar]
- Sisler HD. 1986. Control of fungal diseases by compounds acting as antipenetrants. Crop Prot. 5(5):306–313. doi: 10.1016/0261-2194(86)90108-0. [DOI] [Google Scholar]
- Skamnioti P, Gurr SJ. 2009. Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol. 27(3):141–150. doi: 10.1016/j.tibtech.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Soanes DM, Chakrabarti A, Paszkiewicz KH, Dawe AL, Talbot NJ. 2012. Genome-wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 8(2):1002514. doi: 10.1371/journal.ppat.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonok K, Il-Pyung A, Hee-Sool R, Yong-Hwan L. 2005. MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol Microbiol. 57(5):1224–1237. doi: 10.1111/j.1365-2958.2005.04750.x. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Shamim M, Kumar M, Mishra A, Pandey P, Kumar D, Yadav P, Siddiqui MH, Singh KN. 2017. Current Status of Conventional and Molecular Interventions for Blast Resistance in Rice. Rice Sci. 24(6):299–321. doi: 10.1016/j.rsci.2017.08.001. [DOI] [Google Scholar]
- Suprapta DN, Khalimi K. 2012. Potential of microbial antagonists as biocontrol agents against plant fungal pathogens. J Int Society for Southeast Asian Agril Sci. 18(2):1–8. [Google Scholar]
- Svidritskiy E, Ling C, Ermolenko DN, Korostelev AA. 2013. Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome. Proc Natl Acad Sci U S A. 110(30):12283–12288. doi: 10.1073/pnas.1304922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi S, Hirayama K, Ueda K, Sakai H, Yonehara H. 1958. Blasticidin S, a new antibiotic. J Antibiot (Tokyo). 11(1):1–5. [PubMed] [Google Scholar]
- Talbot NJ. 2003. On the Trail of a Cereal Killer: exploring the Biology of Magnaporthe grisea. Annu Rev Microbiol. 57(1):177–202. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- Thompson JE, Fahnestock S, Farrall L, Liao DI, Valent B, Jordan DB. 2000. The second naphthol reductase of fungal melanin biosynthesis in Magnaporthe grisea. J Biol Chem. 275(45):34867–34872. doi: 10.1074/jbc.M006659200. [DOI] [PubMed] [Google Scholar]
- Upmanyu S, Rana SK. 2012. Effect of fungicides on neck blast incidence and grain yield of rice in mid hills of himachal Pradesh. Pl Dis Res. 26(2): 196. [Google Scholar]
- Uppala S, Zhou XG. 2018. Field efficacy of fungicides for management of sheath blight and narrow brown leaf spot of rice. Crop Prot. 104:72–77. doi: 10.1016/j.cropro.2017.10.017. [DOI] [Google Scholar]
- Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev. 16(3):430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- . [Google Scholar]
- Wang ZY, Jenkinson JM, Holcombe LJ, Soanes DM, Veneault-Fourrey C, Bhambra GK, Talbot NJ. 2005. The molecular biology of appressorium turgor generation by the rice blast fungus Magnaporthe grisea. Biochem Soc Trans. 33(2):384–388. doi: 10.1042/BST0330384. [DOI] [PubMed] [Google Scholar]
- Wilson RA, Talbot NJ. 2009. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol. 7(3):185–195. doi: 10.1038/nrmicro2032. [DOI] [PubMed] [Google Scholar]
- Xu JR, Zhao X, Dean RA. 2007. From genes to genomes: A new paradigm for studying fungal pathogenesis in Magnaporthe oryzae. Adv Genet. 57:175–218. [DOI] [PubMed] [Google Scholar]
- Yamaguchi I. 2004. Overview on the chemical control of rice blast disease. In: Kawasaki S, editor. Rice blast: Interaction with rice and control. Dordrecht: Springer. 1–13. [Google Scholar]
- Yang C, Hamel C, Vujanovic V, Gan Y. 2011. Fungicide: Modes of action and possible impact on nontarget Microorganisms. ISRN Ecol. 2011:1–8. doi: 10.5402/2011/130289. [DOI] [Google Scholar]
- Yashaswini C, Reddy PN, Pushpavati B, Rao S, Madhav S. 2017. Prevalence of rice blast (Magnaporthe oryzae) incidence in South India ch. Bull Env Pharmacol Life Sci. 6:370–373. [Google Scholar]
- Zakaria L, Misman N. 2018. The pathogen and control management of rice blast disease. Malaysian J Microbiol. 14:705–714. [Google Scholar]
- Zhai C, Lin F, Dong Z, He X, Yuan B, Zeng X, Wang L, Pan Q. 2011a. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 189(1):321–334. doi: 10.1111/j.1469-8137.2010.03462.x. [DOI] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, de Paoli E, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA, et al. 2011b. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25(23):2540–2553. doi: 10.1101/gad.177527.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xue C, Kong L, Li G, Xu JR. 2011. A pmk1-interacting gene is involved in appressorium differentiation and plant infection in Magnaporthe oryzae. Eukaryot Cell. 10(8):1062–1070. doi: 10.1128/EC.00007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang Q, Cheng Z, Yang H, Zhou Y, Lu C. 2010. Resistance of four sets of rice varieties to 19 strains of Magnaporthe grisea in Jiangsu area. J Agril Sci. 26(5):948–953. [Google Scholar]
- Zhang X, Bao Y, Shan D, Wang Z, Song X, Wang Z, Wang J, He L, Wu L, Zhang Z, et al. 2018. Magnaporthe oryzae induces the expression of a MicroRNA to suppress the immune response in rice. Plant Physiol. 177(1):352–368. doi: 10.1104/pp.17.01665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Kim Y, Park G, Xu JR. 2005. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell. 17(4):1317–1329. doi: 10.1105/tpc.104.029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Felix G. 2005. Plants and animals: A different taste for microbes? Curr Opin Plant Biol. 8(4):353–360. doi: 10.1016/j.pbi.2005.05.004. [DOI] [PubMed] [Google Scholar]


