Abstract
Macrophomina phaseolina is a Catastrophic plant pathogen, which can cause serious reduction in crop production. In the current study, the mitochondrial genome of M. phaseolina is assembled and annotated. The mitogenome of M. phaseolina is a circular molecule of 101,198 bp. The overall nucleotide content is 34.95% A, 35.25% T, 13.30% C, 16.50% G, with a CG content of 29.80%. The mitogenome contains 42 genes, including 14 protein coding genes (PCGs), 2 ribosomal RNA (rRNA) genes and 26 transfer RNA (tRNA) genes. Phylogenetic analyses based on concatenated protein sequences from 15 taxa of five orders in Ascomycota indicated that M. phaseolina is clustered in the order Botryosphaeriales. This study would have a positive impact on the molecular biology research and biological control of Macrophomina fungi in the future.
Keywords: Macrophomina phaseolina, mitochondrial genome, phylogenetic analyses
Macrophomina phaseolina (Tassi) Goid., the type of the Macrophomina, belongs to order Botryosphaeriaceae (Botryosphaeriales, Ascomycota) (Crous et al. 2006; Phillips et al. 2013; Ghosh et al. 2018), is the most common species of the genus and synonymous with Tiarosporella phaseolina (Tassi) Vander Aa. In 1982, Goidanich named it Macrophomina phaseolina (Tassi) Goid, and this name is still in use today (Ghosh et al. 2018). M. phaseolina is an important soilborne plant pathogenic fungus that can cause charcoal rot disease on about 500 plant species of more than 100 families all over the world and can cause significant yield losses (Suriachandraselvan et al. 2005). The fungus can survive in soil for up to 15 years (Su et al. 2001; Babu et al. 2007; Ghosh et al. 2018). Soybean carbon rot caused by M. phaseolina is one the most serious diseases that can reduce the yield of soybean in the world. Under the conditions of heat and drought stress, soybean carbon rot caused up to 77% plant mortality, which seriously affected soybean yield and quality (Allen et al. 2017). Sun et al. (2019) first reported identification of charcoal rot caused by M. phaseolina on faba bean in China. This disease caused serious yield loss of faba bean, infected plants initially showed leaf chlorosis and wilting, and the plants eventually died with the leaves remaining attached. The identification of the phylogenetic position and the study of genetic diversity of M. phaseolina are conducive to the prevention and control of charcoal rot. However, our understanding of the genetic diversity and phylogenetic position based on mitochondrial genome (mitogenome) of M. phaseolina is lacking. The current study we first reported the complete mitogenome of M. phaseolina and determine its systematic position, which would have a positive impact on the molecular biology research and biological control of Macrophomina fungi in the future.
In this study, the M. phaseolina strain was isolated from the diseased root of Vicia faba L. collected in Fumin County, Yunnan, China (25°18′40″N, 103°07′25″E, alt. 1903 m). A specimen was deposited in the herbarium at Yunnan Academy of Agricultural Sciences (http://www.yaas.org.cn/, Haitian Yu, email: haitianlegume@outlook.com) under the voucher number YN23. Mycelia cultured on PDA at 20 °C for 2 weeks were used to extract total genomic DNA using MiniBEST Plant Genomic DNA Extraction Kit (TaKaRa, China). To validate the DNA integrity and quality, electrophoresis on 1.0% agarose gels was used. The purified DNA was sequenced by the Illumina sequencing platform (HiSeq PE150). Mitogenomic sequences of high quality reads were assembled by employing the programmes the SPAdes 3.9.0 with default parameterbe (Bankevich et al. 2012). The mitogenome annotation were conducted with MFannot tool and ARWEN web server, combined with manual corrections. The mitogenomic circular map of M. phaseolina was drawn by Organellar Genome DRAW tool (Lohse et al. 2007).
The complete mitogenome of M. phaseolina (GenBank accession: MT MW557546) is a circular molecule of 101,198 bp. The overall nucleotide content is 34.95% A, 35.25% T, 13.30% C, 16.50% G, with a CG content of 29.80%. The mitogenome contains 42 genes, including 14 protein coding genes (PCGs), 2 ribosomal RNA (rRNA) genes and 26 transfer RNA (tRNA) genes. Protein-encoding genes include two ATP synthase subunits (atp6 and atp9), three cytochrome oxidase subunits (cox1, cox2, and cox3), one apocytochrome b (cob), seven NADH dehydrogenase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6) and one ribosomal protein (rps3).
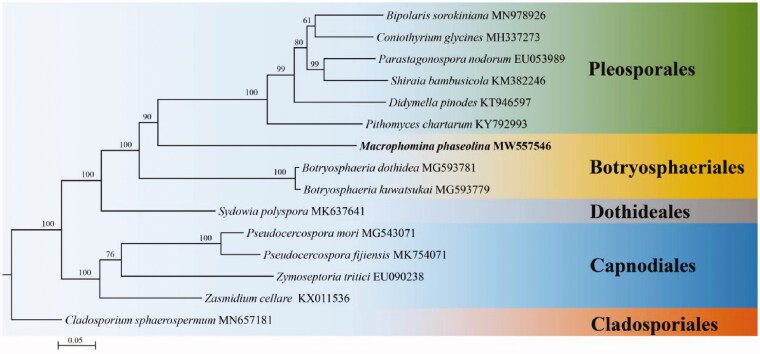
To verify the systematic position of M. phaseolina, mitogenomic sequences of 14 species in Ascomycota downloaded from NCBI were employed. Sequence alignment and phylogenetic analysis were conducted by complying with methods of Wang et al. (2020) and Wang et al.(2021). Fifteen concatenated mitochondrial PCGs were employed for phylogenetic analysis. Phylogenetic analysis was carried out using Maximum Likelihood (ML) method with RaxML 7.0.3 (Stamatakis 2006). The phylogenetic tree is composed of 5 orders (Figure 1), viz. Pleosporales, Botryosphaeriales, Dothideales, Cladosporiales and Mycosphaerellales. Phylogenetically, M. phaseolina is clustered in the order Botryosphaeriales with a high credible support by BI posterior probabilities (PP = 100%).
Figure 1.
Phylogenetic relationships among 15 species based on Maximum likelihood (ML) analysis from 15 concatenated mitochondrial protein-coding genes (PCGs). The 15 PCGs include subunits of the respiratory chain complexes (cob, cox1, cox2, cox3), ATPase subunits (atp6, atp8, atp9), NADH: quinone reductase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, nad6) and one ribosomal protein (rps3).
Funding Statement
This work was funded by Natural Science Foundation of Yunnan Province [202001AT070011], National Key Research Plan [2019YFD1001300/2019YFD1001303], and Chengxuzhen Expert Workstation Project of Qujing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
This mitogenome of Macrophomina phaseolina was submitted to GenBank under the accession number of MW557546 (https://www.ncbi.nlm.nih.gov/nuccore/MW557546). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA751633, SRR15329826, and SAMN20524426, respectively.
References
- Allen TW, Bradley CA, Sisson AJ, Byamukam E, Chilvers MI, Coker CM, Collins AA, Damicone JP, Dorrance AE, Dufault NS, et al. 2017. Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Prog. 18(1):19–27. [Google Scholar]
- Babu BK, Saxena AK, Srivastava AK, Arora DK.. 2007. Identification and detection of Macrophomina phaseolina by using species-specific oligonucleotide primers and probe. Mycologia. 99(6):797–803. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SL, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO, Philips AJL, Alves A, Burgess T, Barber P, Groenewald JZ.. 2006. Phylogenetic lineages in the Botryosphaeriaceae. Stud Mycol. 55:235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh T, Biswas MK, Guin C, Roy P.. 2018. A review on characterization, therapeutic approaches and pathogenesis of Macrophomina phaseolina. Plant Cell Biotechnol Mol Biol. 19(3&4):72–84. [Google Scholar]
- Lohse M, Drechsel O, Bock R.. 2007. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274. [DOI] [PubMed] [Google Scholar]
- Phillips AJL, Alves A, Abdollahzadeh J, Slippers B, Wingfield MJ, Groenewald JZ, Crous PW.. 2013. The Botryosphaeriaceae: genera and species known from culture. Stud Mycol. 76(1):51–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Su G, Suh SO, Schneider RW, Russin JS.. 2001. Host specialization in the charcoal rot fungus, Macrophomina phaseolina. Phytopathology. 91(2):120–126. [DOI] [PubMed] [Google Scholar]
- Suriachandraselvan M, Aiyyanathan KEA, Vimala R.. 2005. Host range and cross inoculation studies on Macrophomina phaseolina from sunflower. Madras Agric Res J. 92:238–240. [Google Scholar]
- Sun SL, Zhu ZD, Duan CX, Zhao P, Sun F, Deng D, He YH.. 2019. First report of charcoal rot caused by Macrophomina phaseolina on faba bean in China. Plant Dis. 103(6):1415. [Google Scholar]
- Wang X, Jia LH, Wang MD, Yang H, Chen MY, Li X, Liu HY, Li Q, Liu N.. 2020. The complete mitochondrial genome of medicinal fungus Taiwanofungus camphoratus reveals gene rearrangements and intron dynamics of Polyporales. Sci Rep. 10(1):16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Liang X, Hao X, Dang H, Hsiang T, Gleason ML, Zhang R, Sun G.. 2021. Comparison of mitochondrial genomes provides insights into intron dynamics and evolution in Botryosphaeria dothidea and B. kuwatsukai. Environ Microbiol. doi: 10.1111/1462-2920.15608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This mitogenome of Macrophomina phaseolina was submitted to GenBank under the accession number of MW557546 (https://www.ncbi.nlm.nih.gov/nuccore/MW557546). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA751633, SRR15329826, and SAMN20524426, respectively.